Significance
Digital PCR (dPCR) has long been recognized as a powerful tool in nucleic acid quantitative analysis, but there still remain problems that hinder its wide popularity. Sample loss is one major challenge that compromises the certainty of dPCR quantification and handicaps its ability to detect and quantify nucleic samples of low copy number. Another main difficulty is to prevent this single-copy-sensitivity experiment from possible contaminations of PCR product exposure. The CLEAR-dPCR introduced herein solves these two problems by integrating optically cleared monodisperse droplet generation and light-sheet fluorescence microscopy and fulfills a genuinely lossless and contamination-free digital PCR approach. Such an emulsion clearing method pioneers a way of high-throughput in situ signal capture in mass droplets.
Keywords: digital PCR, light-sheet microscopy, clear droplet, contamination-free clinical test
Abstract
The realization of the vast potential of digital PCR (dPCR) to provide extremely accurate and sensitive measurements in the clinical setting has thus far been hindered by challenges such as assay robustness and high costs. Here we introduce a lossless and contamination-free dPCR technology, termed CLEAR-dPCR, which addresses these challenges by completing the dPCR sample preparation, PCR, and readout all in one tube. Optical clearing of the droplet dPCR emulsion was combined with emerging light-sheet fluorescence microscopy, to acquire a three-dimensional (3D) image of a half million droplets sealed in a tube in seconds. CLEAR-dPCR provides ultrahigh-throughput readout results in situ and fundamentally eliminates the possibility of either sample loss or contamination. This approach exhibits improved accuracy over existing dPCR platforms and enables a greatly increased dynamic range to be comparable to that of real-time quantitative PCR.
The desire for better quantitative assessment of genetic information is always increasing. Techniques for sequence-specific nucleic acid quantification are key tools in biology and medicine, especially in scenarios involving rare genetic variant detection where both high sensitivity and high specificity are warranted (1–3). Unlike real-time quantitative PCR (qPCR), where relative quantification is achieved using a standard curve, digital PCR (dPCR) (4) enables absolute quantification with up to single-molecule resolution. Soon after the first demonstration of dPCR through microtiter plate compartmentalization (4, 5), various improvements using microfluidics-based methods were developed (6–10). Nanoliter or subnanoliter (11) microfluidic compartmentalization reduces reagent cost, increases reaction efficiency, facilitates large dynamic-range Poisson distribution, and simplifies the experimental process (12–16).
A typical dPCR process comprises three major steps: partitioning, amplification, and counting. Partitioning can be implemented via droplets (9, 17) or microchambers (12). Counting is also done via two major approaches: serial reading and planar imaging. Serial reading naturally fits the transferability of the fluidic emulsion and has been widely coupled with droplet-based compartmentalization. Meanwhile, planar imaging is more compatible with array-based compartmentalization (13).
Currently, droplet dPCR is gaining in popularity (14), partly because the emulsion can be easily transferred into conventional microcentrifuge tubes and the amplification can be conducted in conventional PCR machines. However, the prevailing flow-focusing-based microfluidic droplet generation strategy often used in this scenario suffers from sample loss during the flow stabilization period. When serial counting is applied, further sample loss and contamination are inevitable during sample transfer from the PCR tube to the reading device. Recently we have demonstrated the use of microcapillary array (MiCA) for lossless emulsion generation (SI Appendix, Fig. S1) and subsequent dPCR (15). However, the counting step employed a serial counting strategy that still suffered from the same sample loss and contamination issues as other dPCR strategies. Here, we demonstrate a dPCR strategy, characterized by centrifugation-based droplet generation, optical clearing of PCR emulsion, and in situ counting based on light-sheet fluorescence microscopy (LSFM) (Fig. 1). With this approach named CLEAR-dPCR, we are able to achieve lossless and contamination-free dPCR.
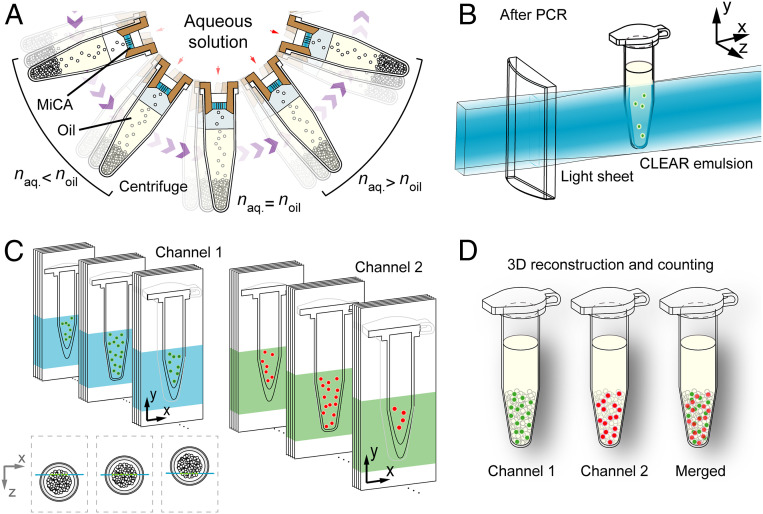
Fig. 1.
Schematic illustration of the CLEAR-dPCR process. (A) The generation of droplet emulsion using MiCA. Droplet PCR samples are centrifuged in a swing-bucket rotor and jet through the through holes on MiCA plates, yielding massive droplets monodispersed in silicone oil. CLEAR emulsion is formed by matching aqueous and oil phases’ refractive indices. (B) The high-throughput readout of bulk PCR droplets, optically transparent and densely packed, in the PCR tube using light-sheet illumination. A laser light sheet scans the droplets in the PCR tube in situ, and the plane fluorescence images are sequentially acquired using a high-speed sCMOS camera. (C) The dual-channel light-sheet fluorescence image sequences, in which all of the fluorescently positive droplets are recorded (red: HEX/VIC, green: FAM). (D) The volumetric reconstruction of dual-channel images and accurate droplet counting can be implemented based on the 3D data.
Results
CLEAR Digital PCR.
In order to achieve lossless counting via optical readout in situ, we generate an optically transparent dPCR emulsion termed CLEAR emulsion, by dispersing an aqueous dPCR mix with a refractive index close to that of the oil phase using MiCA centrifugation (Fig. 1A). After PCR, in situ noninvasive and high-throughput readout of whole-tube emulsion is accomplished by rapid three-dimensional (3D) light-sheet fluorescence imaging (Fig. 1B) of the same tube. By recording a series of planar images of the CLEAR emulsion (Fig. 1C), we are able to rapidly reconstruct the 3D structure of the whole PCR emulsion and achieve digital counting of the positive droplets (Fig. 1D). This high-throughput approach streamlines dPCR in a convenient operation, thoroughly eliminating sample loss and contamination, and greatly reducing the cost of dPCR assays.
Transparent Emulsion via Refractive Index Matching.
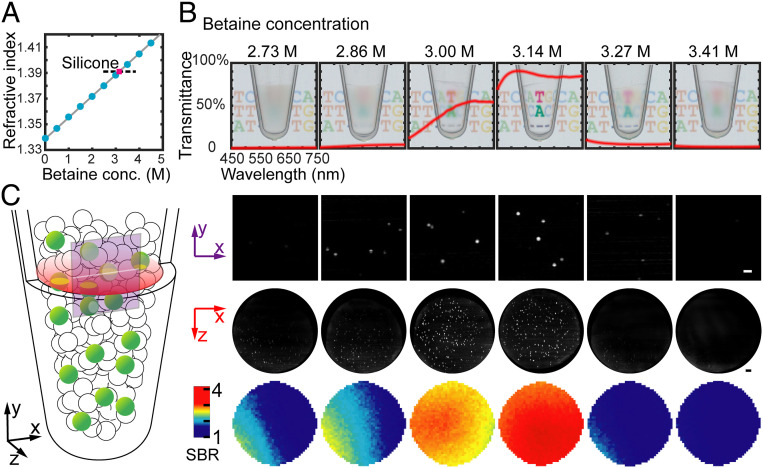
In typical water-in-oil (w/o) emulsions, the massive number of droplets densely packed in the PCR tube creates an opaque emulsion due to the light scattering at the water–oil interfaces. We serendipitously discovered that a common PCR additive, betaine (16), effectively increases the refractive index of the aqueous solution to be similar to that of the carrier oil, thereby minimizing the light scattering in the PCR emulsion. We carefully titrated the betaine addition from 0 to 5 mol/L (Fig. 2A and SI Appendix, Fig. S2) and found that an optimized concentration of 3.14 mol/L generates a PCR mix with a refractive index similar to that of the oil-surfactant mix (low-viscosity silicone, n = 1.3903). At this betaine concentration, the whole emulsion is completely transparent (Fig. 2B). Brightfield photographs show the highest light transmission rate (∼85%) at 3.14 mol/L. As a result of the greatly improved clarity, this optimized CLEAR-dPCR emulsion exhibited the highest image quality and signal-to-background ratio in light-sheet fluorescence imaging (Fig. 2C). We further verified that PCR reactions inside these CLEAR-ed picoliter droplets are not affected by the addition of betaine (SI Appendix, Fig. S3). We carefully weighed the mass of the MiCA tube before and after centrifugation using high-precision balance. The results have verified that no observable liquid loss was induced during the MiCA emulsification process.
Fig. 2.
The optical clearing of the dPCR emulsion, which substantially improves the transparency and enables large-depth, whole-tube scanning. (A) The change of the PCR mix’s refractive index versus the concentration of betaine. (B) Brightfield transmission images of the emulsions generated with different concentrations of betaine in PCR buffer. The red lines plot the emulsions’ transmission spectra (light path: 1 cm). A total of 3.14 mol/L betaine provides the best refractive index matching. (C) The fluorescence images of emulsions at different betaine concentrations. (Top) Light-sheet fluorescence images taken by camera. (Middle) Reconstructed x-z plane images of the emulsions. (Bottom) Signal-to-background ratio distribution in x-z plane. (Scale bars: 200 µm.)
Light-Sheet Sectioning the Transparent Emulsion.
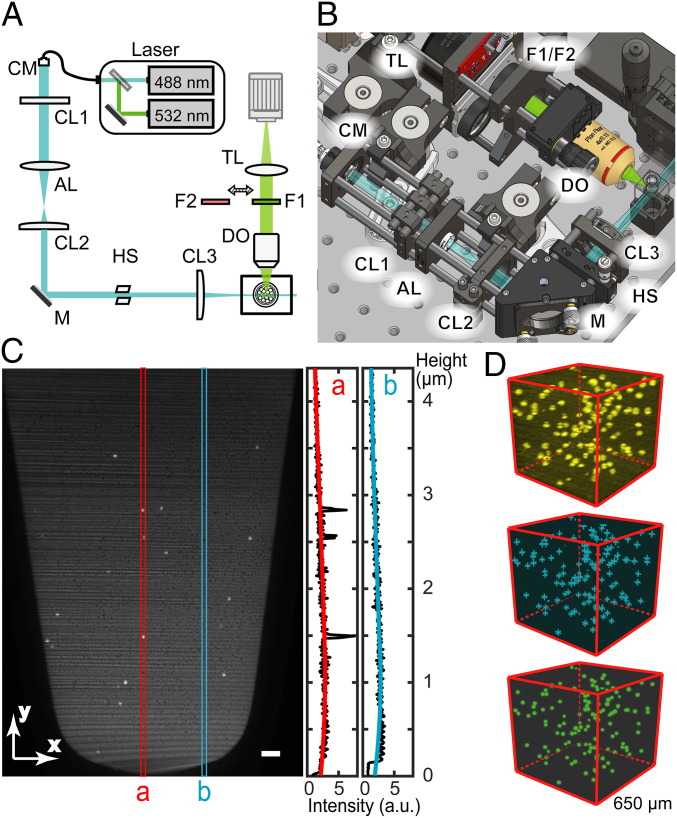
We built a compact light-sheet illumination microscope (Fig. 3 A and B) to read the fluorescence signals in the bulk CLEAR-dPCR emulsion layer by layer. Our light-sheet readout geometry was customized for PCR tube scanning (SI Appendix, Fig. S4). To make our design easily replicable, all parts except the sample holder are commercially available (SI Appendix, Table S1) Dual-wavelength (488/532 nm) 3D imaging was implemented for duplex dPCR with FAM and HEX/VIC labeling. The combination of plane illumination and a scientific complementary metal–oxide–semiconductor (sCMOS) camera provided a large imaging field of view of ∼4.6 × 3.6 mm, covering the entire emulsion (Fig. 3C). Together with a rapid acquisition rate of 100 frames/s, high-speed scanning of the whole tube can be completed within 6 s. The spatial resolution of our light-sheet imaging is 6.5 µm (lateral) and 20 µm (axial), sufficient to identify the 41-µm droplets in all three dimensions (SI Appendix, Fig. S5). By stacking the raw light-sheet images into a 3D volume, we further identified and counted the positive droplets using a customized algorithm (Fig. 3D and SI Appendix, Fig. S6).
Fig. 3.
The light-sheet fluorescence reader and droplet counting. (A) The schematic light path and major components. A wide laser light sheet which can illuminate the entire emulsion in the PCR tube is generated by the line focusing of a collimated elliptical beam. CM: collimator, CL: cylindrical lens, AL: aspherical lens, HS: horizontal slit, DO: detection objective, F: filter, TL: tube lens, M: mirror. (B) The compact (30 × 30 cm) layout of the setup. (C) Signal and background measurement in a raw image of CLEAR-dPCR. The red line cut a includes three positive droplets while the blue line cut b includes only the background. Uneven light field will be further corrected and the background signal will be subtracted. (Scale bar: 200 µm.) (D) The 3D reconstruction of dPCR images. Raw images are first reconstructed into a 3D intensity matrix (Top, yellow). Then in silico scanning detects the spatial local intensity maxima (Middle, cyan crosses) as candidate positive signals. Finally, the fluorescently positive droplets (Bottom, green dots) are located and counted based on the identified candidates.
Absolute Quantification.
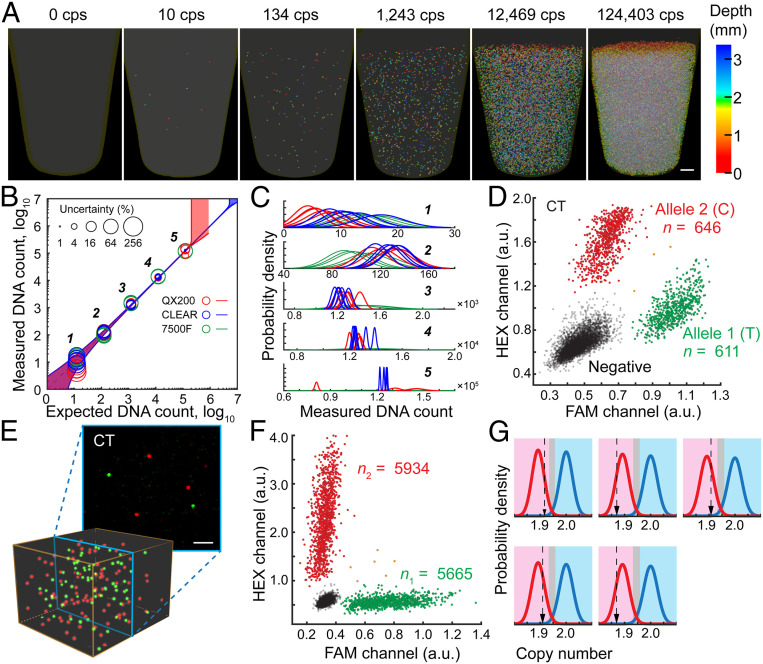
To evaluate the performance of CLEAR-dPCR, we used three assays to benchmark it against dPCR industry leaders: Bio-Rad droplet dPCR (QX200-ddPCR) and ABI 7500 Fast Real-Time PCR (7500F-qPCR). For the first benchmark assay, we performed absolute quantification of a 280-bp amplicon DNA from the Listeria monocytogenes genome (SI Appendix, Table S2). We serially diluted the template DNA to different concentrations spanning five orders of magnitude (101 to 105 copies/aliquot). The results show that CLEAR-dPCR and QX200-ddPCR both had excellent linearity (R = 0.9999) within the range of 10–1.3 × 105 copies/aliquot, while 7500F-qPCR demonstrated lower linearity (R = 0.9975) (Fig. 4 A and B and SI Appendix, Tables S3–S5). The 7500F-qPCR quantitation relies on a standard curve and some of the measurements apparently deviated from those obtained from the digital methods with much lower precision (Fig. 4C and SI Appendix, Table S5), which makes 7500F-qPCR unsuitable for absolute counting. The relatively higher measurement uncertainty of low copy number samples was mainly due to sampling error, which is independent of specific methods.
Fig. 4.
Performance of CLEAR-dPCR with different assays. (A) Absolute in situ quantification of positive droplets in a PCR tube, with 105 dynamic range. (Scale bar: 500 µm.) (B) Comparison of quantification performance by CLEAR-dPCR, QX200-ddPCR, and 7500F-qPCR, with their uncertainty levels (95% confidence) shown as the circle sizes. The shaded areas along the diagonal represent the 95% confidence boundaries. (C) Probability density distribution of each measurement. (D) SNP detection and allele counting. Scatterplot results of a heterozygous sample at SNP rs10092491. (E) The heterozygote’s merged images (Top) and 3D reconstructions (Bottom) of both channels. (Scale bar: 200 µm.) (F) Clinical copy number variation detection. Copy numbers of Chr 1 uc13 (red) and the TSC2 gene (green) were detected in a mock sample with 5% deletion of TSC2 exons. (G) Results of five TSC tests of mock-affected samples using a likelihood ratio classifier. Relative copy number in the magenta region is classified as affected, in light blue as healthy, and in gray undetermined.
It is worth noting that, while both dPCR systems presented equivalent linearity, QX200-ddPCR measurements had higher uncertainty for the more concentrated samples (Fig. 4 B and C, 104 and 105 copies/aliquot). We attributed this to the combination of low droplet number and high sample loss of QX200-ddPCR. As shown in SI Appendix, Fig. S7, target molecule distribution in QX200-ddPCR becomes oversaturated when the expected copy number approaches 105; in this regime, the standard Poisson correction is insufficient and leads to error. With nearly half a million droplets and complete elimination of sample loss, CLEAR-dPCR maintains reasonable uncertainty with over 106 target molecules (Fig. 4B). This dynamic range is comparable to that of qPCR (Ct value 15 to 33) and typically not feasible in dPCR; our CLEAR-dPCR method is distinctly advantageous in maintaining absolute quantification while greatly expanding the dynamic range of dPCR.
Precise measurement of low copy number targets further relies on low false-positive rates. In the second benchmark assay, we investigate possible false-positive counting against a high background, by using human genomic DNA as template instead of Listeria DNA in the same way as the first assay. When ∼60 fg of DNA (equivalent to 20 copies of the human genome) per droplet was used as the input, we found no false-positive signals in the CLEAR-dPCR results.
Single Nucleotide Resolution.
Digital PCR has been intensively applied to clinical diagnostics for its unrivaled accuracy, which is an essential requirement for identifying single nucleotide variation in clinical samples, such as in circulating DNA-based liquid biopsy (1, 2, 18, 19). For the third benchmark, we designed a TaqMan genotyping assay with two probes, each targeting an allele of SNP rs10092491 with FAM or HEX labeling (SI Appendix, Table S6 and Fig. S8). Genomic DNA from eight human peripheral blood samples was quantified by two-color CLEAR-dPCR and QX200-ddPCR, yielding three types of positive droplet patterns for the three different genotypes (Fig. 4D and SI Appendix, Fig. S9 and Table S7). For heterozygous samples, both channels yielded almost equal numbers of positive counts. CLEAR-dPCR demonstrated higher accuracy with a mean count ratio of 99.6%, compared with the ratio of 104.9% for QX200-ddPCR. By checking the two-color 3D images taken from the heterozygous samples, we discovered that most positive signals in the two channels did not overlap (Fig. 4E and SI Appendix, Fig. S10). A few droplets were positive in both channels, which is consistent with the Poisson statistical expectation (Fig. 4D). For homozygotes, both CLEAR-dPCR and QX200-ddPCR detected zero false-positive signals (SI Appendix, Table S7).
Application in Copy Number Quantification.
We further evaluated CLEAR-dPCR on copy number variation (CNV) measurements. The detection relies on measurement of the relative abundances of a target region and a reference region. We first tested the performance of CLEAR-dPCR duplex digital counting on sex determination by Y chromosome (Chr) copy number quantification. Two TaqMan assays targeting the SRY gene on Chr Y (FAM) or ultraconserved region (uc) 13 on Chr 1 (HEX) were used (SI Appendix, Table S8). Both CLEAR-dPCR and QX200-ddPCR confirmed the presence of Chr Y in all male samples, with count ratio between Chr Y and Chr 1 approaching 1:2 (SI Appendix, Fig. S11 and Table S9). CLEAR-dPCR demonstrated comparable accuracy to QX200-ddPCR. In female samples, CLEAR-dPCR detected zero Chr Y signals among 3,074 Chr 1 signals, while QX200-ddPCR gave one false-positive Chr Y count among 1,341 Chr 1 signals (SI Appendix, Table S7).
Prenatal Fetal DNA Exonal Deletion Identification.
Prenatal genetic diagnosis using cell-free DNA in maternal blood is an area where dPCR can play an indispensable role. Existing noninvasive prenatal test methods only focus on large CNV or point mutations. Exon deletion has rarely been covered due to technical difficulties (20), despite its essential role in many diseases (21, 22). With CLEAR-dPCR, we demonstrated an exon copy number counting assay for tuberous sclerosis complex (TSC), an autosomal dominant Mendelian disorder caused by mutations in the TSC1 or TSC2 gene. Owing to the relatively large sizes of the two genes, exonal deletions are present in as many as 10% of clinical cases (22). A previous multiplex ligation-dependent probe amplification (MLPA) test in a TSC patient revealed a heterozygous deletion spanning exons 1 to 16 of TSC2. We hence prepared mock samples by mixing genomic DNA from this patient and the patient’s mother at a 1:9 ratio. The exact patient DNA fraction (10.8%) could be measured by multiplex genotyping dPCR (19). We then designed a dPCR assay targeting the deleted region in TSC2 and another assay targeting Chr 1 as reference (SI Appendix, Table S10). We conducted five independent measurements on the mock samples as well as negative controls, and all of the results agreed with the corresponding genotypes using a likelihood ratio classifier (Fig. 4 F and G, Methods, and SI Appendix, Table S11). These findings demonstrate the readiness of CLEAR-dPCR for further clinical assay development.
Discussion
Based on comparisons with existing dPCR methods, CLEAR-dPCR has three major advantages. First, CLEAR-dPCR is a lossless approach. Microfluidic droplet dPCR methods typically lose >30% of the sample during the compartmentalization and detection steps. In CLEAR-dPCR, centrifugal force guarantees that the aqueous phase is completely transformed into droplets through MiCA centrifugation (SI Appendix, Fig. S1). Optical clearing and in situ 3D imaging through light-sheet illumination ensures thorough interrogation of the entire volume of the emulsion. Using CLEAR-dPCR, true absolute counting is intrinsically achievable with no need for extrapolation to compensate for material loss, and lossless interrogation can further reduce measurement uncertainty (SI Appendix, Fig. S7). These advantages are most crucial when the nucleic acids being interrogated are precious and low in abundance. While insufficient sensitivity by sample loss has so far prevented the more widespread applications of dPCR, especially when extremely high accuracy and low error rates are required (such as the SARS-CoV-2 viral nucleic acids detection during the COVID-19 pandemic), our lossless CLEAR-dPCR pipeline can solve these problems and achieves improved throughput in the meantime (quantitative analysis in SI Appendix, Supporting Methods and Fig. S12).
Besides overcoming the sample loss issue, which we believe is one of the most challenging problems in the dPCR field, CLEAR-dPCR’s second advantage is that it is contamination-free. For most clinical applications that repeatedly amplify specific regions, exposure of the postamplification products to the laboratory environment poses a great risk for contamination of subsequent operations. In CLEAR-dPCR, once the sample is added into the PCR tube on top of the MiCA, the lid can be closed for good. All subsequent steps including compartmentalization, thermal cycling, and signal detection are performed with the lid closed and the emulsion fully sealed by oil. Third, CLEAR-dPCR is robust and easy to operate. The use of conventional equipment, such as a centrifuge and standard PCR machine, renders CLEAR-dPCR a simple experiment. Optical clearing can be achieved with a premixed reaction buffer and the total hands-on time is less than 1 min per sample, with the total time being less than an hour for dPCR assays. The samples are well sealed, allowing easy operation of the reading instrument, and omitting fluidic manipulation. We have validated the biocompatibility of a silicon-based surfactant through multiple times of PCR reactions and light-sheet readout experiments, and obtained results consistent with established digital PCR methods. Meanwhile, our CLEAR-dPCR emulsion is also highly stable, remaining intact for days and allowing hundreds of times of repeated scanning (SI Appendix, Fig. S13).
In summary, our CLEAR-dPCR approach based on the combination of MiCA-facilitated emulsion generation, betaine-mediated emulsion clearing, and light-sheet illumination imaging, has greatly simplified the dPCR experimental process while further achieving higher sensitivity and accuracy. Through our demonstrations, CLEAR-dPCR has shown several key advantages over existing dPCR techniques, including complete compartmentalization from bulk solution to monodispersed droplet, comparable dynamic range with qPCR, elimination of false positives through fully sealed operation, and fast digital counting at single-copy-resolution level without sample loss (SI Appendix, Table S12). We envision that this simple and inexpensive method will lower the technical and economic barriers of dPCR and have broad application in biology and medicine.
Methods
Clinical Samples.
Peripheral blood DNA samples of a pediatric TSC patient and a healthy family member were obtained from a previous TSC study. Both samples were deidentified prior to use in our study.
Refractive Index Matching via Addition of Certain Reagents.
Transferring postamplification PCR products is time and labor consuming. Furthermore, it raises the risk of contaminating subsequent assays. However, due to the light scattering at the interface of droplet and carrier fluid, it is very difficult to extract fluorescence signals directly from the emulsion, containing massive densely packed droplets. In order to read the intact PCR emulsion at a high throughput, we developed a refractive index matching strategy to produce optically clear PCR emulsion for parallel light-sheet excitation. On the one hand, considering usually a big refractive index (RI) difference between water and oil, we sought a suitable oil with a relatively low RI close to the water-phase reagent. Among a few available candidates, silicone was selected for its lower RI value (from 1.37 to 1.41), thus quite applicable in this case compared to fatty esters (1.43 to 1.46), aromatic hydrocarbon (higher than 1.50), and liquid fatty hydrocarbons (higher than 1.40). In our demonstration, we chose silicone DMS-T01.5 (Gelest) with a low refractive index (1.3880) and viscosity (1.5 cSt), as the base oil. Silicon-based surfactant ES5612 (Evonik) (5% wt/wt) was added to the base silicone to produce the final emulsion oil, with refractive index being 1.3903.
On the other hand, the refractive index of the original PCR mix (∼1.34) is only slightly higher than that of pure water (1.33). Thus, we needed to tune the ingredients of the PCR mix, and hence to increase its refractive index to a value close to 1.39. In practice, we added betaine into the PCR mix to reduce this RI difference. Compared to other commonly used PCR additives, such as glycerol, dimethyl sulfoxide (DMSO), tetramethylammonium chloride (TMAC), formamide, and bovine serum albumin (BSA) that usually work at low concentration, betaine has a higher applied concentration up to several molar, which can induce a large tunable range of RI. Furthermore, as a zwitterion quaternary amino acid, betaine is neutral in electrical charge and thereby will not destabilize the emulsion. We verified that through the addition of betaine with 3 to 3.2 M concentration (depending on the concentration of other solutes), a highly clear PCR emulsion with well-matched RI can be successfully prepared for high-speed light-sheet readout (Fig. 2). We also note that bulk PCR reactions with such a high concentration of betaine in regular microcentrifuge tubes barely take place, but if the reaction mix is partitioned in a digital format, the amplification works (SI Appendix, Supporting Methods). We hypothesize that as the amplicons are more concentrated in the picoliter droplets, they can trigger the chain reaction of subsequent circles more effectively.
Emulsification.
In our previous work (15), we used microarray channels to generate monodisperse droplets for digital PCR. Here we increased the number of the through-hole channels from 7 to 37, to compensate for the loss of droplet generation rate due to higher viscosity. The reaction mixes were added into the well on the MiCA and centrifuged for 4 min at 15,000 relative centrifuge force (rcf). Around 443,000 droplets were generated in 200-μL regular PCR microcentrifuge tubes with an average diameter of 41 μm. The transparency of the generated emulsion varied with the concentration of betaine addition (Fig. 2).
In Situ 3D Imaging of Emulsion Using LSFM.
LSFM has recently emerged as a technique of choice that can image large samples with relatively low phototoxicity and at high speed. Here we built a small-format, macroview light-sheet fluorescence microscope to noninvasively read the fluorescent signals of massive droplets that stack inside the sealed PCR tube. In our experiment, 488- and 532-nm light sheets with thickness of around 22 μm (1/e2 value) were generated to optically section the CLEAR emulsion with FAM and HEX/VIC fluorescence, respectively. The 200-μL centrifuge tube (Axygen) containing dPCR emulsion inside was submerged into a rectangular glass cuvette filled with silicone oil (mixed by two kinds of raw silicone oil, one with high viscosity and RI, the other low, final RI = 1.390), to minimize the light deflection by the conical plastic tube. Then the droplets stacked in the tube were quickly scanned across the stationary laser light sheet by a motorized actuator (KSTM25E/M, Thorlabs). A 2× wide-field detection path consecutively collected the fluorescence signals from the light-sheet illuminated planes of the emulsion. An sCMOS camera (ORCA-Flash 4.0 V2) sequentially recorded the fluorescence images with a fine step size of 6.5 μm. For each fluorescence channel, 560 plane images (z depth 3.6 mm) of 550 × 700 pixels were captured within ∼6 s. Then by simply stacking the 560 planes together, the fluorescence distribution of the entire clear emulsion was reconstructed at ∼6.5-μm lateral and ∼22-μm axial resolution that were sufficient to resolve single droplets in three dimensions (SI Appendix, Fig. S5). Finally, the two-color visualization of the emulsion was readily created by merging the two-channel results together.
Image-Based Droplets Counting.
We developed a computation procedure to process the raw LSFM image stacks, counting the positive droplets at high accuracy. We first corrected the vertical background variance induced by Gaussian laser illumination in the vertical direction and removed the stripe noise caused by the laser diffraction in the Fourier space. Then we searched for intensity local maxima in the 3D image matrices and put down their coordinates in three dimensions (Fig. 3C and SI Appendix, Fig. S6). Then the candidate coordinates were filtered based on intensity, template correlation, contrast, and distance to remove noise and artifact signals. Finally the nucleic acid copy number and concentration was obtained with the number of identified positive droplets by Poisson distribution (see SI Appendix, Supporting Methods for details). Our custom codes implemented in current study are available either at https://github.com/MengchengJ/CLEAR_code or from the corresponding authors upon request.
Duplex CLEAR-dPCR for prenatal test of TSC.
DNA concentrations were first quantitated by fluorometer (Qubit, Thermo Fisher) as a rough reference. We then set the threshold of presumption of an affected fetus to be , whereas that of a healthy fetus is . Values between the two thresholds were thought to be invalid. Pure healthy and affected samples were first tested by CLEAR-dPCR for validation of the assay as well as to further quantify the DNA concentration. A healthy donor DNA sample mixed with the affected patient DNA was used as mock maternal cell-free DNA of a mother carrying an affected fetus (fetal fraction = 10.8%). Each independent assay consisted of six tubes for CLEAR-dPCR so that the DNA counts were enough to give a solid result (18). We carried five parallel assays and the results came as expected, all being positive (Fig. 4 F and G and SI Appendix, Tables S10 and S11).
Supplementary Material
Acknowledgments
We thank Dr. Wenxiong Zhou for fruitful discussions. This work was supported by the National Natural Science Foundation of China (21927802 to Y.H., P.F., and J.W.; 21525521 to Y.H.; 21874052 to P.F.; and 21675098 to J.W.), the National Key R&D Program of China (2017YFA0700500 to P.F.; 2018YFA0108100 to Y.H.; and 2018YFC1002300 to J.W.), the Innovation Fund of the Wuhan National Laboratory for Optoelectronics, and the Beijing Advanced Innovation Center for Genomics.
Footnotes
Competing interest statement: Peking University and Huazhong University of Science and Technology have applied for patents related to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002448117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Wong Y. K. et al., Applications of digital PCR in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2, 177–186 (2017). [Google Scholar]
- 2.Olmedillas-López S., García-Arranz M., García-Olmo D., Current and emerging applications of droplet digital PCR in oncology. Mol. Diagn. Ther. 21, 493–510 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Robin J. D., Ludlow A. T., LaRanger R., Wright W. E., Shay J. W., Comparison of DNA quantification methods for next generation sequencing. Sci. Rep. 6, 24067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B., Kinzler K. W., Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 96, 9236–9241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sykes P. J. et al., Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13, 444–449 (1992). [PubMed] [Google Scholar]
- 6.Ottesen E. A., Hong J. W., Quake S. R., Leadbetter J. R., Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Heyries K. A. et al., Megapixel digital PCR. Nat. Methods 8, 649–651 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Shen F., Du W., Kreutz J. E., Fok A., Ismagilov R. F., Digital PCR on a SlipChip. Lab Chip 10, 2666–2672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatch A. C. et al., 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 11, 3838–3845 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Lo Y. M. D. et al., Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. U.S.A. 104, 13116–13121 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dangla R., Kayi S. C., Baroud C. N., Droplet microfluidics driven by gradients of confinement. Proc. Natl. Acad. Sci. U.S.A. 110, 853–858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreutz J. E. et al., Theoretical design and analysis of multivolume digital assays with wide dynamic range validated experimentally with microfluidic digital PCR. Anal. Chem. 83, 8158–8168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao P., Huang Y., Digital PCR: Endless frontier of “divide and conquer”. Micromachines 8, 231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker M., Digital PCR hits its stride. Nat. Methods 9, 541–544 (2012). [Google Scholar]
- 15.Chen Z. et al., Centrifugal micro-channel array droplet generation for highly parallel digital PCR. Lab Chip 17, 235–240 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Kang D. K. et al., Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 5, 5427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henke W., Herdel K., Jung K., Schnorr D., Loening S. A., Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25, 3957–3958 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camunas-Soler J. et al., Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR. Clin. Chem. 64, 336–345 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Kuliev A., Rechitsky S., Preimplantation genetic testing: Current challenges and future prospects. Expert Rev. Mol. Diagn. 17, 1071–1088 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Willis A. S., van den Veyver I., Eng C. M., Multiplex ligation-dependent probe amplification (MLPA) and prenatal diagnosis. Prenat. Diagn. 32, 315–320 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Jones A. C. et al., Comprehensive mutation analysis of TSC1 and TSC2-and phenotypic correlations in 150 families with tuberous sclerosis. Am. J. Hum. Genet. 64, 1305–1315 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin K. R. et al., The genomic landscape of tuberous sclerosis complex. Nat. Commun. 8, 15816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.