Significance
Ig CSR occurs through the nonhomologous end-joining pathway. In NHEJ-deficient cells, substantial CSR can be achieved through the Alt-EJ pathway. High-throughput analyses of IgH CSR junctions in NHEJ-deficient cells show that in contrast to endonuclease-generated breaks, Alt-EJ-mediated CSR can occur without CtIP-mediated DNA resection. The first temporal analyses of CSR junctions in NHEJ-proficient cells further showed that early Sμ internal deletion junctions are enriched for MH. Taken together, the results identified repetitive core IgH switch regions as favorable substrates for MH-mediated recombination independent of resection.
Keywords: alternative end-joining, CtIP, class switch recombination
Abstract
To generate antibodies with different effector functions, B cells undergo Immunoglobulin Heavy Chain (IgH) class switch recombination (CSR). The ligation step of CSR is usually mediated by the classical nonhomologous end-joining (cNHEJ) pathway. In cNHEJ-deficient cells, a remarkable ∼25% of CSR can be achieved by the alternative end-joining (Alt-EJ) pathway that preferentially uses microhomology (MH) at the junctions. While A-EJ-mediated repair of endonuclease-generated breaks requires DNA end resection, we show that CtIP-mediated DNA end resection is dispensable for A-EJ-mediated CSR using cNHEJ-deficient B cells. High-throughput sequencing analyses revealed that loss of ATM/ATR phosphorylation of CtIP at T855 or ATM kinase inhibition suppresses resection without altering the MH pattern of the A-EJ-mediated switch junctions. Moreover, we found that ATM kinase promotes Alt-EJ-mediated CSR by suppressing interchromosomal translocations independent of end resection. Finally, temporal analyses reveal that MHs are enriched in early internal deletions even in cNHEJ-proficient B cells. Thus, we propose that repetitive IgH switch regions represent favored substrates for MH-mediated end-joining contributing to the robustness and resection independence of A-EJ-mediated CSR.
In peripheral lymphoid organs, B lymphocytes undergo Ig CSR to generate antibodies with different effector functions. At the DNA level, CSR replaces the Cμ exons that encode the Ig heavy chain constant region of IgM antibodies with exons encoding other isotypes (e.g., Cγ1 for IgG1) to generate antibodies with different effector functions (1). CSR is initiated by the B cell-specific activation-induced cytidine deaminase (AID), which converts cytosine to uracil in the transcribed switch (S) regions preceding each set of constant region exons (2). The mismatches generated by AID are eventually converted to DNA double-strand breaks (DSBs) in proliferating B cells. The IgH isotype switch is then achieved by joining a DSB in the initially expressed IgM switch region (Sμ) with a DSB in a downstream switch region (e.g., Sγ1 for IgG1). Two DSBs within the Sμ region can also be joined to form an internal deletion that removes intervening Sμ sequences without achieving isotype switch. The ligation of these AID-initiated DSBs is primarily mediated by classical cNHEJ that can directly ligate two unprocessed or minimally processed DSB ends. B cells deficient for core cNHEJ factors, such as LIG4 or its obligatory binding partner XRCC4, can still achieve a remarkable 25% of normal CSR, using the mechanistically distinct A-EJ pathway (3, 4). In addition to the specific repair pathways, ATM kinase and its chromatin-bounded substrates (e.g.,53BP1, H2AX, etc.) promote efficient CSR (5). However, whether ATM specifically promotes cNHEJ-mediated CSR or both cNHEJ- and A-EJ-mediated CSR remains elusive.
Studies using endonuclease-generated DSBs have shown that the A-EJ and the related MH-mediated end-joining (MMEJ) pathways both depend on DNA end resection, a nucleolytic process that converts the two ends of a DSB into 3′ single-strand DNA overhangs (6, 7). End resection is important for A-EJ by exposing the flanking MH (8, 9) short stretches of MH within the two opposing overhangs can, then, anneal to each other and, after further nucleolytic processing (e.g., flap removal by FEN1 nuclease), converting a DSB to two single-stranded DNA gaps or nicks, which can, then, be filled in and joined by LIG1 or LIG3, both of which have been implicated in A-EJ-mediated CSR (10, 11). Thus, the most widely recognized feature of A-EJ- or MMEJ-mediated repair is the prevalence of MH at the junctions. In this context, the high-throughput genome-wide translocation sequencing (HTGTS) (12), a linear-amplification-based method to recover thousands of junctions involving a single bait DSB has greatly improved the efficiency for isolating CSR junctions (13, 14). When the bait break site is placed at the 5′ Sμ region, HTGTS can efficiently recover thousands of CSR junctions and internal-deletion junctions. Using HTGTS, we and others have shown that the A-EJ events arising in cNHEJ-deficient (e.g., Xrcc4−/− or DNA-PKcs−/−) B cells are markedly enriched for junctions bearing both short (1–3-nucleotides [nt]) and long (≥4-nt) MHs (13–15). In addition, A-EJ-mediated CSR correlated with extensive use of junctions beyond the 3′ (distal) boundaries of the core Sμ and Sγ1 regions, suggesting DNA end resection (13–15). Nevertheless, why A-EJ-mediated repair is particularly efficient during CSR and whether resection is required for A-EJ-mediated CSR (and, if so, the pathway of resection), remains poorly understood.
In addition to A-EJ, DNA resection is also essential for homologous recombination (HR) that repairs DSBs using homology sequencing. In eukaryotes, DNA resection can be initiated by two pathways, DNA2 together with BLM or WRN helicases, or the nucleolytic activities of the MRE11-RAD50-NBS1 (MRN) complex in association with the CtIP protein (6). While both the DNA2 and the MRN-CtIP pathways have been implicated in A-EJ-mediated repair of endonuclease-generated breaks (7, 16), the study of their yeast orthologs suggests that CtIP promotes both DNA2- and MRN-mediated end resections (17). Specifically, the CtIP-mediated end resection is most active in the S and G2 phases of the cell cycle due to high CtIP protein expression (18) and CDK-mediated phosphorylation of CtIP at residue T847 (19). Moreover, upon DNA damage, the ATM and ATR kinases further promote end resection by phosphorylating CtIP at T859 (T855 in mouse) and other sites (20–23). CtIP is specifically implicated in A-EJ. Bennardo et al., (2008) showed that CtIP facilitates A-EJ-mediated repair of an MMEJ reporter in murine embryonic stem (ES) cells, presumably by promoting end resection and thereby exposing the flanking MHs in the resulting single-strand DNA overhangs (7). In this regard, several studies have shown that CtIP depletion significantly reduces CSR in B cells activated in vitro (22, 24–26). In addition to a direct role in A-EJ (24), other possible mechanisms have also been proposed to explain how CtIP loss impairs CSR, including its ability to modulate AID function (24) and CDK2 regulation (25). However, since the loss of CtIP abrogates the proliferation of activated B cells (22, 26), and proliferation is essential for successful CSR, whether the impact of CtIP loss on CSR is due to impaired end resection or through other indirect mechanisms, including impairing proliferation, remains difficult to sort out. Using the HTGTS assay, we recently compared the CSR and Sμ internal deletion junctions that arise in CtIP+/+ and CtIP∆/∆ B cells. To our surprise, the CSR junctions recovered from these cells were largely indistinguishable for their MH usage and resection patterns (22). Likewise, the T855A phosphorylation site mutation, which partially blocks ATR/ATM-dependent induction of the CtIP-mediated resection, also has no impact on the efficiency, end resection, or MH usage of CSR (22). These findings suggest that CtIP, and by extension, CtIP-mediated end resection may be dispensable for end ligation required for CSR, at least, when measured within cNHEJ-proficient cells. However, whether CtIP contributes to A-EJ-mediated CSR specifically remains unknown.
Studies of A-EJ-mediated CSR are complicated by the fact that junctional MHs, a characteristic feature of A-EJ, can also be generated by cNHEJ. Although the MHs of cNHEJ junctions are thought to be shorter than those generated by A-EJ, the cutoff is not absolute, and junctions with short MHs (1–3 nt) cannot be definitively attributed to either DSB repair pathway. To circumvent this ambiguity, we examined CSR in B cells that are genetically deficient for cNHEJ owing to loss of the core cNHEJ factor XRCC4, an obligatory partner of LIG4. The role of CtIP in A-EJ-mediated CSR in Xrcc4−/− murine B cells was then evaluated by either conditional inactivation of CtIP or nonphosphorylatable CtIP mutation (T855A). Unexpectedly, we found that isotype switching in Xrcc4−/− B cells was unaffected by either deletion or mutation (T855A) of CtIP. Moreover, despite a significant reduction in end resection, the CSR junctions recovered from Xrcc4-deficient alone and Xrcc4-deficient and CtIP-T855A mutant cells displayed similar MH usage, suggesting that MH-mediated end ligation can occur without extensive end resection during CSR. Similarly, ATM kinase inhibitor (ATMi) also attenuated end resection without affecting MH usage during A-EJ-mediated CSR. The analysis of ATMi-treated Xrcc4−/− cells also identified an end resection independent role for ATM in regulating the orientation of CSR by both cNHEJ and A-EJ. Finally, when CSR junctions were analyzed by cell division, MHs (4–15 bps) are significantly enriched in early internal deletion junctions even in cNHEJ-proficient B cells. Taken together, our findings show that A-EJ-mediated CSR can occur independent of CtIP-mediated end resection, suggesting that the repetitive IgH switch regions are uniquely prone to MH-mediated repair.
Results
CtIP and ATR/ATM-Mediated Phosphorylation of CtIP(T855) Are Dispensable for A-EJ-Mediated Class Switching.
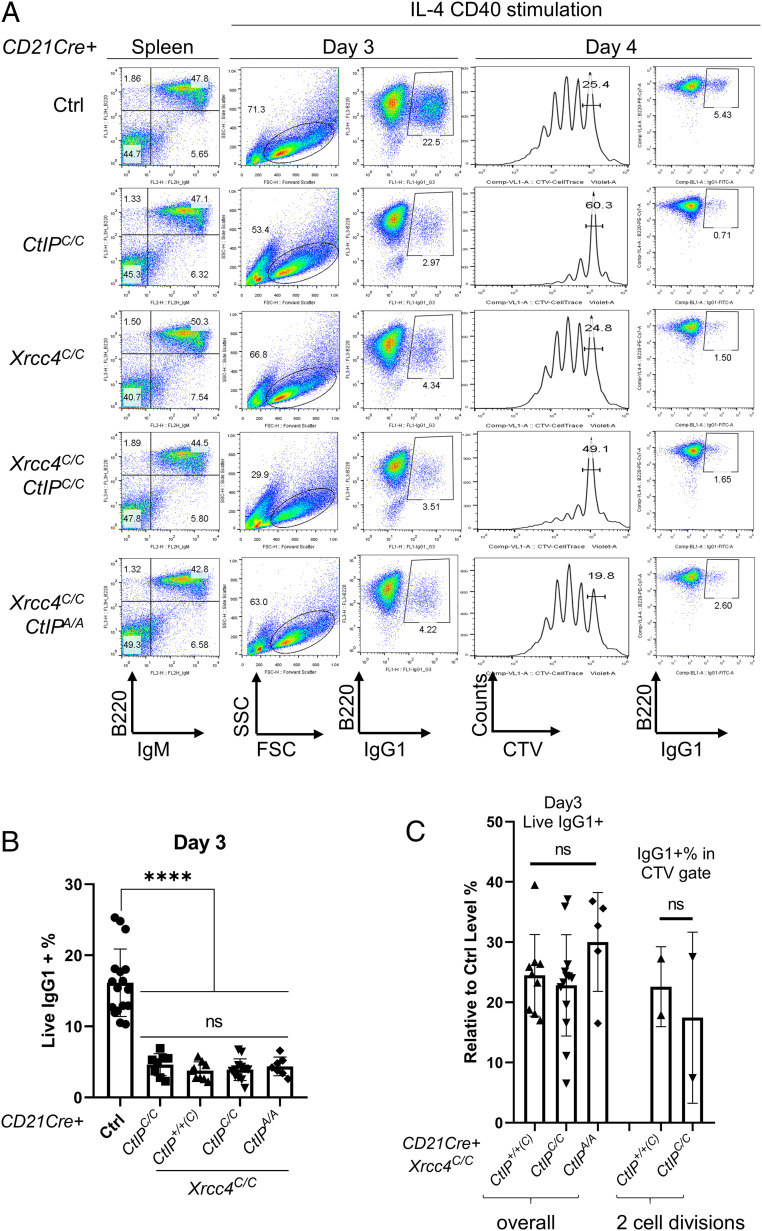
Xrcc4 and CtIP are both essential for early B cell development (22, 27). To investigate the role of CtIP-dependent resection in A-EJ-mediated CSR, we used a CD21-driven Cre recombinase to conditionally inactivate the Xrcc4 and/or CtIP genes in naive B cells (3, 22, 28). The efficiency of CtIP and Xrcc4 inactivation in splenic B cells was validated by genomic PCR and Western blotting (SI Appendix, Fig. S1A and B). As noted previously (22), successful Cre-mediated recombination of the CtIPC allele generates CtIPΔ, which encodes a truncated CtIP protein that lacks most of the oligomerization domain (SI Appendix, Fig. S2 A–D). The oligomerization domain of CtIP is essential for the end-resection function of CtIP (29). Accordingly, CtIPΔ/Δ mice died during embryonic development (30), consistent with the CtIP-null mice described before (31). Somatic inactivation of CtIPC/C (converted to CtIPΔ/Δ) in progenitor B cells abrogates proliferation and normal B cell development (22, 26), suggesting that CtIPΔ acts as a CtIP null allele in vivo. Inactivation of Xrcc4 alone (CD21CreXrcc4C/CCtIP+/+(C)), CtIP alone (CD21CreXrcc4+/+(C) CtIPC/C) or both together (CD21CreXrcc4C/C CtIPC/C) did not affect the frequency of the IgM+B220+ B cells in the spleen (Fig. 1A and SI Appendix, Fig. S3A). In the presence of anti-CD40 and IL-4, purified splenic B cells undergo CSR to express IgG1 or IgE (32). Xrcc4-deletion reduced IgG1 switching to ∼25% of control levels at both days 3 and 4 (at day 4, IgG1+% = 29.02 ± 6.83% in control vs. 8.25 ± 1.23% in CD21CreXrcc4C/CCtIP+/+(C)) (Fig. 1 A and B and SI Appendix, Fig. S3B) (3, 4). Although CtIP inactivation compromised B cell proliferation (Fig. 1A) (22, 26), CtIP inactivation did not further reduce the frequency of IgG1+live cells% from Xrcc4-deficient B cells (CD21CreXrcc4C/C CtIPC/C) (Fig. 1 A and B and SI Appendix, Fig. S3B), suggesting that CtIP is not required for A-EJ-mediated CSR.
Fig. 1.
CtIP deficiency or mutation did not reduce A-EJ-mediated CSR in Xrcc4-deficient B cells. (A) Representative flow cytometry analysis of purified splenic B cells, stimulated with IL-4 and anti-CD40, at day 3 or day 4, from control (WT or CD21CreXrcc4C/+CtIP+/+(C)), CD21CreXrcc4C/CCtIP+/+(C), CD21CreXrcc4C/C CtIPC/C, and CD21CreXrcc4C/C CtIPT855A/T855A mice with representative CTV labeling for cell proliferation upon stimulation. Cells that underwent two cell divisions were gated based on CTV dilution and plotted for IgG1+ frequency. (B) The percentage of live and IgG1+ cells at day 3 after stimulation. Two-tailed Mann–Whitney U test, ****P < 0.0001. (C) The relative (to control analyzed in parallel) percentage of live and IgG1+ cells at day 3 after stimulation in cells with two divisions, measured by CTV.
Given the important role of CtIP in proliferation, we used two additional approaches to ascertain whether CtIP has a proliferation-independent role in A-EJ-mediated CSR. First, we labeled activated B cells with a surface dye (CTV) to monitor cell division and then compared CSR efficiency between cells with the same number of divisions. As shown in Fig. 1A and quantified in Fig. 1C, CtIP loss has no impact on A-EJ-mediated IgG1 switching in Xrcc4-deficient B cells even after controlling for cell division. Second, we took advantage of the CtIPT855A allele, which harbors a phosphorylation site mutation that partially blocks ATR/ATM-induced CtIP-mediated end resection without affecting B cell proliferation (Fig. 1A) (22). IgG1 switching is comparable in CD21CreXrcc4C/C or CD21CreXrcc4C/CCtIPT855A/T855A B cells, even after controlling for cell division status (Fig. 1 A and B). DNA2 is essential for murine embryonic development (33) and a conditional allele for DNA2 is not yet available. We found that a selective inhibitor for DNA2—C5 (34)—compromised the viability, but not the A-EJ-mediated CSR to IgG1 in CD21CreXrcc4C/C B cells (SI Appendix, Fig. S3C). Together, these data indicate that CtIP, CtIP phosphorylation at T855, and DNA2 activity are dispensable for A-EJ-mediated CSR.
CtIP and Its Resection Activity Are Necessary for Extensive S-Region Resection in cNHEJ-Deficient B Cells.
Next, we performed HTGTS analyses of CSR junctions. Briefly, a linear amplification-based method was used to identify DNA junctions involving AID-generated breaks at the 5′ Sμ region (called the “bait”) and another DSB (called the “prey”) (12, 14, 15). In switching B cells, most preys reside in the switch regions, representing internal deletion (to another Sμ break) or CSR (to breaks in Sγ1 or Sε) (Fig. 2A). Notably, HTGTS recovers both types of junctions and the orientation of the joining products (Fig. 2A). For this study, we analyzed >5,000 junctions from three independent mice of each genotype (SI Appendix, Fig. S3D). While the majority of the prey resided in the IgH locus (∼6% of the prey junctions from wild type (WT) B cells mapped outside the IgH locus (Fig. 2B). The frequency of non-IgH prey increased significantly in Xrcc4-deficient (14% in CD21CreXrcc4C/CCtIP+/+(C)) and double-deficient (14% in CD21CreXrcc4C/CCtIPC/C and 13.5% in CD21CreXrcc4C/C CtIPT855A/T855A) B cells (Fig. 2B), consistent with increased chromosomal translocations. Fig. 2C illustrates the relative distribution of prey junctions within the IgH locus. In WT B cells, roughly 20% of all IgH preys reside in Sμ (majorities are the products of Sμ-Sμ internal deletions), 40% in Sγ1, and another 40% in Sε (Fig. 2C). Xrcc4 deficiency preferentially affected the Sμ-Sε junction frequency (to 15% and P = 0.016) without affecting Sμ-Sγ1 junctions (∼40% and P > 0.05) (Fig. 2C). These results are consistent with sequential switching, thus, two end-joining events are often needed to achieve Sε switching in adult B cells (35). Correspondingly, selective depletion of Sμ-Sε junctions has also been noted for ATM or DNA-PKcs-deficient B cells (13, 15). CtIPT855A mutation (CD21CreXrcc4C/C CtIPT855A/T855A) did not affect the IgH prey distribution in Xrcc4-deficient B cells (Fig. 2C).
Fig. 2.
CtIP deficiency reduces end resection in Xrcc4-deficient B cells. (A) Diagram of the HTGTS assay with a single bait site at the 5′ Sμ region. The IgH locus is drawn in its native orientation on mouse chromosome 12. The solid and dashed lines represent junctions made in the (−) (normal internal deletion and CSR) and (+) (inversion) orientations, respectively. The different switch regions are color coded. The bait site is marked with a red arrow. The bait starts from 114,664,910 near the telomere of Chr. 12. (B) The distribution of prey sites inside (IgH all) and outside (non-IgH) the IgH locus. (C) The distribution of IgH prey sites by switch region—Sμ, Sγ1, Sε, and others. (D and E) Relative frequency of distal preys among the prey in each respective switch region. Two-tailed Mann–Whitney U test, *P < 0.05; ns is not marked. (D) for Sμ, distal = downstream/centromeric to Chr12: 114,662,100. (E) for Sγ1, distal = downstream/centromeric to Chr12:114,568,500. For (B) to (E), the data represent greater than or equal to three independent repeats of each genotype. Error bar = SD.
HTGTS analyses of WT B cells confirm that most CSR junctions fall within the core S regions (13, 14, 15) where AID hotspots and (R, purine; G, guanine; Y, pyrimidine; W, A, or T) (RGYW) motifs are slightly enriched (Fig. 3 for Sμ, Fig. 4 for Sγ1, and SI Appendix, Fig. S4 for Sε). In contrast, in Xrcc4-deficient cells, a significant fraction of CSR junctions fall beyond the 3′ boundaries of the core Sμ (∼22% vs. 2.7% in control and P = 0.016) (Figs. 2D and 3), and Sγ1 (3.5% vs. 0.6% in control and P = 0.016) (Figs. 2E and 4) regions (13, 14, 15), which was defined as extensive resection. We, thus, categorize junctions beyond the core switch region (114,662,100< in Sμ, 114,568,500 < in Sγ1, and 114,510,500 < in Sε) as extensive resection. The exact location of the boundary and their base pair numbers are included in the figures. Distal junctions are less prevalent in Sε (1.9–3.5% and P = 0.56) (SI Appendix, Fig. S4), potentially due to the low number of Sμ-Sε junctions. These distal junctions could arise by extensive resection of unrepaired switch region breaks due to cNHEJ deficiency. Alternatively, they could reflect the joining with AID-initiated breaks outside the core switch region generated due to the lack of productive CSR. CtIP deletion (CD21CreXrcc4C/CCtIPC/C) largely eliminated the formation of distal CSR junctions at all three switch regions in Xrcc4-deficient B cells (Fig. 3 for Sμ, Fig. 4 for Sγ1, and SI Appendix, Fig. S4 for Sε) and CtIPT855A mutation markedly reduced distal junctions %, especially in Sμ, and to a much lesser extent in Sγ1 and Sε (Figs. 2D, 3, and 4 and SI Appendix, Fig. S4). Together, these observations indicate that CtIP-mediated end resection is essential for the formation of distal CSR junctions outside the core switch region.
Fig. 3.
Diagram and relative distribution of Sμ preys in Xrcc4-deficient B cells. The diagram of ∼10 kb (chr12: 114,658,100–114,668,100, mm10) Sµ region and the counts of RGYW motifs per 50 bp by orientation ([+], blue and [−], red). The percentage of preys that fall into the distal region (<Chr 12: 114,662,100, mm10, indicating extensive resection, 2.810 kb from the bait) are marked on the left and the frequency of (+) and (–) oriented preys are marked on the right. The Sµ preys were divided into three regions: proximal, middle, and distal as marked on the graph. The distal region is the same as the extensively resected region. The data represent the pool of, at least, three independent libraries per genotype per treatment.
Fig. 4.
Diagram and relative distribution of Sγ1 preys in Xrcc4-deficient B cells. The diagram of ∼30 kb (chr12: 114,558,500–114,588,500, mm10) Sγ1 region and the counts of RGYW motifs per 150 bp by orientation ([+], blue and [−], red). The percentage of preys that fall into the distal region (<Chr 12: 114,568,500, mm10, indicating extensively resection region, 96.410 kb from the bait) are marked on the left and the frequency of (+) and (–) oriented preys are marked on the right. The Sγ1 preys were divided into three regions: proximal, middle, and distal as marked on the graph. The distal region is the same as the extensively resected region. The data represent the pool of, at least, three independent libraries/mice per genotype per treatment.
MH-Mediated CSR in cNHEJ-Deficient Cells Requires CtIP but Not T855 Phosphorylation.
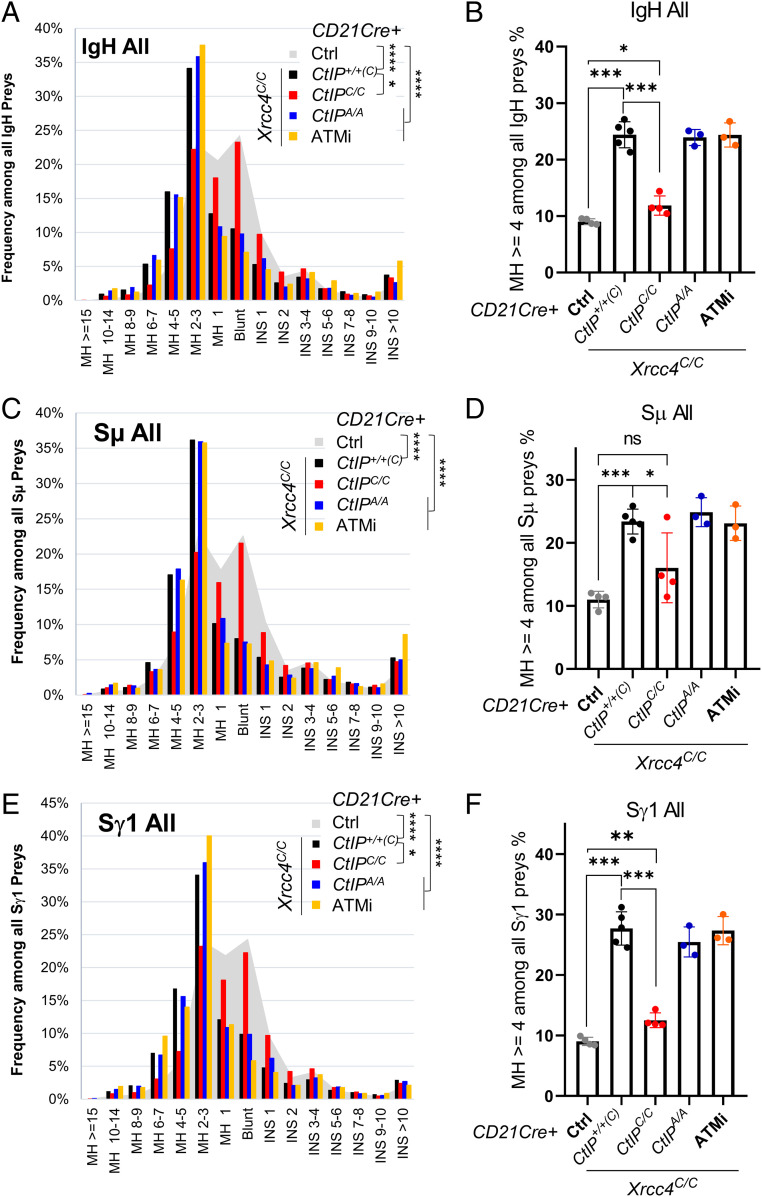
MH is the characteristic feature of A-EJ-mediated DSB repair. We, then, asked whether reduced resection associated with CtIP deficiency affects MH usage during A-EJ-mediated CSR. To do so, we binned the CSR junctions according to the extent of MH (0 = blunt, 1, 2 to 3, 4 to 5, etc.), and the size of insertions (INS, 1–10 nt) (Fig. 5). Since MH usage cannot be determined at junctions bearing insertions, the INS and MH junctions are mutually exclusive. In WT cells, roughly 25% of all IgH junctions are blunt, 25% have 1-nt MH, and 25% have 2- to 3-nt MH (Fig. 5A). In WT cells, MH distribution is similar in Sμ-Sμ (Fig. 5C) and Sμ-Sγ1 (Fig. 5E) junctions. As expected, Xrcc4 deficiency (CD21CreXrcc4C/CCtIP+/+(C)) increased the proportion of junctions with 2- to 3-nt (from 25 to 35%) and ≥4-nt MH (from ∼9% total in WT to ∼25%) with a concurrent drop in blunt (from 25 to 8%) and 1-nt MH (from 25 to 10%) junctions (P < 0.0001, Kolmogorov–Smirnov test) (Fig. 5 A and B) (3). Codeletion of both Xrcc4 and CtIP (CD21CreXrcc4C/CCtIPC/C) largely eliminated the skew toward longer MHs (Fig. 5A, P < 0.0001, Kolmogorov–Smirnov test, relative to CD21CreXrcc4C/CCtIP+/+(C)) in all IgH junctions (Fig. 5 A and B), suggesting that, although dispensable for A-EJ CSR efficiency, CtIP promotes MH-end-joining during CSR. In contrast, although the CtIPT855A mutant (CD21CreXrcc4C/C CtIPT855A/T855A cells) reduced resection, it did not alter the MH pattern of Xrcc4-deficient (CD21CreXrcc4C/CCtIP+/+(C)) cells (Fig. 5 A–F, P = 0.67, Kolmogorov–Smirnov test). Correspondingly, ATM inhibition also reduced resection without affecting MH usage (Fig. 5 A–F). Thus, in contrast to endonuclease-generated breaks, end resection, and MH usage can be uncoupled during A-EJ-mediated CSR.
Fig. 5.
The MH usage of IgH junctions recovered from Xrcc4-deficient B cells. (A) The graphic distribution of all IgH preys by MH, blunt, and INS. (B) The percentage of IgH junctions with ≥4-nt MH by genotype. (C) The graphic distribution of all Sμ preys by MH, blunt, and INS. (D) The percentage of Sμ junctions with ≥4-nt MH by genotype. (E) The graphic distribution of all Sγ1 preys by MH, blunt, and INS. (F) The percentage of Sγ1 junctions with ≥4-nt MH by genotype. (A), (C) and (E) represent the pool of, at least, three independent libraries per genotype. For A, C, and E, Kolmogorov–Smirnov test, *P < 0.05, and ****P < 0.0001. In B, D, and F, the bar and error bars represent the average and the standard derivations. The P values in B, D, and F were calculated via unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.0005, and ****P < 0.0001.
At endonuclease-generated breaks, end resection facilitates A-EJ by revealing flanking MH in the single-strand DNA overhangs (8). If it is also true during A-EJ-mediated CSR, we expect some correlation between the junction location and the MH usage. To test this, we divided the Sμ and Sγ1 into three mutually exclusive regions, proximal, middle, and distal (Figs. 3 and 4) and analyzed MH usage in each region separately (SI Appendix, Fig. S5). In WT cells where the CSR junctions are largely mediated by cNHEJ, there is a clear preference to MH in the middle and distal Sμ regions (SI Appendix, Fig. S5A), consistent with a correlation between resection (distal junctions) vs. MH usage. In contrast, in Xrcc4−/− cells where all junctions were mediated by the A-EJ pathway, there is no skew of the MH usage (SI Appendix, Fig. S5B), suggesting that end resection and MH usage are uncoupled in A-EJ-mediated CSR. There were insufficient junctions that fell into the Sγ1 distal region in WT cells, so we only compared the proximal and middle regions (SI Appendix, Fig. S5C). In both WT and Xrcc4−/− cells, MH usage did not correlate with prey location (SI Appendix, Fig. S5C). Together, these findings confirm that end resection and MH usage can be uncoupled during A-EJ-mediated CSR.
ATM Signaling Promotes A-EJ-Mediated CSR through Both CtIP-T855 Phosphorylation-Dependent and CtIP-Independent Mechanisms.
ATM/ATR phosphorylates several other substrates in addition to CtIP (T855) to promote end resection. Next, we tested the impact of ATM inhibition on end resection and A-EJ-mediated CSR. In contrast to the CtIP-T855A mutation, ATM kinase inhibition (ATMi, KU55933, 7.5 μM) significantly reduced IgG1 switching of Xrcc4-deficient splenic B cells (Fig. 6 A and B), indicating a CtIP-independent role of ATM in A-EJ-mediated CSR. HTGTS data show that ATM inhibition reduced the extensive resection at Sμ (Figs. 2D and 3) and Sγ1 (Figs. 2E and 4) in Xrcc4−/− B cells without significantly affecting the MH usage in Xrcc4−/− B cells (CD21CreXrcc4C/CCtIP+/+(C)) (Fig. 5). Thus, like in CtIP-mutant cells, end resection can be uncoupled from MH usage during A-EJ-mediated CSR by ATM inhibition.
Fig. 6.
The impact of ATM inhibition on A-EJ-mediated CSR. (A) Representative flow cytometry of Xrcc4−/− B cells with or without ATMi (7.5 µM, KU55933) at day 4 of stimulation. (B) Quantification of live IgG1+% at day 4 after stimulation, with and without 7.5-μM ATMi (KU55933). Two-tailed Mann–Whitney U test, **P < 0.01, and *P < 0.05. (C) The percentage of IgH preys that fall into each switch region by genotype. (D) The quantification of the fraction of Sε preys among all IgH preys. (E) The ratio of Sμ preys that fall into (−) vs. (+) orientations. For B–E, the bars and error bars represent average and SDs. Two-tailed Mann–Whitney U test, **P < 0.01, and *P < 0.05.
What is the CtIP-independent function of ATM in A-EJ-mediated CSR? ATMi-treated cells display several features that are not found in CtIP-deficient cells. First, ATM inhibition but not CtIP deletion further reduced the relative frequency of Sμ-Sε junctions in Xrcc4−/− cells (P = 0.036) (Fig. 6 C and D), suggesting ATM promotes A-EJ-mediated end ligation. Second, ATM inhibition altered the orientation distribution of the Sμ-Sμ junctions (Fig. 6E). As noted in Fig. 2A, the bait break at the 3′Sμ can either be joined in the centromere-to-telomere orientation (defined as positive, +) or in the telomere-to-centromere orientation (defined as negative, −). Since the IgH locus resides in a telomere- (5′-IgH) to-centromere (3′-IgH) orientation on murine chromosome 12 (Fig. 2A), the productive CSR junctions and the Sμ-Sμ internal deletions both fall in the negative orientation (telomere-to-centromere) (Figs. 3 and 4, and SI Appendix, Fig. S4). Indeed, in control cells, the vast majority of IgH preys (76.1% Sμ, 91.8% Sγ1, and 94.9% Sε) are in the negative orientation (Figs. 3 and 4, and SI Appendix, Fig. S4), consistent with bona fide CSR and internal deletion (36). Meanwhile, prey breaks on other chromosomes (non-Chr12) are evenly distributed between (+) and (–) orientations (SI Appendix, Fig. S6A). IgH preys that fall in the positive orientation could arise by true inversional joining (as shown in the Fig. 2A diagram) or through interhomolog or intersister chromatid translocation (13, 15). Loss of Xrcc4 (CD21CreXrcc4C/CCtIP+/+(C)) increased chromosomal translocation and moderately increased the frequencies of positively oriented preys (24–30% in Sμ-Sμ, 8.2–16.5% in Sμ-Sγ1, P = 0.016, and 5.1–7.1% in Sμ-Sε) (Fig. 6E). Further loss of the CtIP or CtIP-T855A mutation did not affect the +/− ratio of Sμ junctions recovered from Xrcc4-deficient B cells (Fig. 6E). In contrast, ATM inhibition significantly increased the +/− ratio of Sμ preys in both WT cells and Xrcc4-deficient cells (P = 0.029 and P = 0.036, respectively) (Figs. 3 and 6E), indicating an important role of ATM in promoting orientation-specific joining mediated by either cNHEJ or A-EJ. Taken together, these observations confirm a role of ATM in end resection and identify a CtIP-independent role of ATM in suppressing interchromosomal/intersister recombination to promote CSR.
Earlier Sμ Internal Deletions Tend to Use MH-Mediated End-Joining Even in cNHEJ Proficient Cells.
A-EJ-mediated CSR is thought to be the backup for cNHEJ and occurs slower, potentially due to the time needed for resection (37). Consistent with the dispensible role of end resection in A-EJ-mediated CSR, CSR junctions recovered from Xrcc4−/− cells at day-3 or day-4 poststimulation show no difference in MH usage (SI Appendix, Fig. S6B). Next, we asked whether MH usage changes in a temporal manner in cNHEJ-proficient cells. To our surprise, WT CSR junctions harvested at day 3 display moderately higher MH usage than those obtained at day 4 (SI Appendix, Fig. S6B). In particular, early Sμ-Sμ events preferentially use MHs (2–15 bp) at day 3, followed by an accumulation of blunt joints at day 4 (Fig. 7A). Meanwhile, MH usage at Sμ-Sγ1 junctions does not differ significantly between days 3 and 4 (Fig. 7A).
Fig. 7.
CSR junction analyses by cell division. (A) The MH, blunt, and INS distributions of WT Sμ and Sγ1 junctions at day 3 or 4 by switch region (Sμ or Sγ1). Kolmogorov–Smirnov test, **P < 0.01. (B) Representative CTV labeling analyses of B cells from WT BL6 mice. The IgG1+% of cells with the defined number of cell divisions plotted below. The underlines marked the division groups used for sequence analyses. Cells with the least divisions were in group I and the most divisions in group IV. (C) The relative distribution of IgH preys among different switch regions by cell divisions. The bars and error bars represent average and SDs. (D) The MH, blunt, and INS usages among Sμ preys by cell division. Kolmogorov–Smirnov test, *P < 0.05, and **P < 0.01.
As revealed by the cell trace violet (CTV) analyses (Fig. 1A), the cells harvested at day 4 contain a mixture of B cells with different poststimulation proliferation histories (from two to seven cell divisions each). To better understand the kinetics of MH-mediated joining, we sorted the cells by cell division according to CTV staining (Fig. 7B): group I with one or two divisions and moderate CSR (9.33% ± 2.90% IgG1+%), group II with three divisions (13.9% ± 2.43% IgG1+%), group III with four divisions (28.4% ± 4.56% IgG1+%), and group IV with five or more divisions (41.8% ± 3.77% IgG1+%). As shown in Fig. 7B, productive isotype switching steadily increased with the number of cell divisions. Correspondingly, the fraction of Sε preys increases from 24% in group I to 42% in group IV (Fig. 7C), consistent with sequential CSR from Sμ-Sγ1 first, then, to Sε (35). Most notably, MHs larger than 4 nt was significantly more common in groups I + II (22%) than either group III (15%) or group IV (10%) cells, while 1-nt MHs were rare in groups I + II compared to groups III and IV (10%, 17%, and 17%, respectively) cells (P = 0.039 and P = 0.0053) (Fig. 7D and SI Appendix, Fig. S6C). Again, this difference is only observed within Sμ-Sμ but not Sμ-Sγ1 junctions. Collectively, these findings support a model in which MH, at the repetitive Sμ regions, promotes cNHEJ-mediated internal deletion in the earlier phases of CSR.
Discussion
Ig CSR is mediated primarily by cNHEJ. However, in cNHEJ-deficient cells, a remarkable 25% of CSR can be achieved by the A-EJ repair pathway characterized by extensive usage of MHs in the junctions (3). While subsequent studies established that, in addition to its lymphocyte-specific role in CSR, the A-EJ pathway can also contribute to DSB repair during the DNA damage response in a range of cell types, why CSR is particularly prone to robust A-EJ remains elusive. In this context, Bennardo et al., (2008) showed that efficient A-EJ of an endonuclease-generated DSB in ES cells requires CtIP, presumably reflecting a role for DNA resection in unmasking the flanking MH sequences necessary for A-EJ. Using HTGTS, we examined the role of CtIP and end resection in A-EJ-mediated CSR. In contrast to endonuclease-mediated breaks (7), CtIP is dispensable for A-EJ-mediated CSR. Indeed, the ability of cNHEJ-deficient naive splenic B cells to achieve IgG1 expression was unaffected by either CtIP deletion or a CtIP mutation (T855A).
What accounts for the different dependence on CtIP by A-EJ-mediated repair at endonuclease-initiated breaks vs. during CSR? Perhaps the difference lies in the unique structural and functional features of the switch regions. As these regions are composed largely of repetitive sequences enriched for somatic hypermutation hotspot motifs, AID can generate multiple DSBs within a given S region (2, 38), mitigating the need for extensive CtIP-mediated end resection to expose complementary MHs. Moreover, the closely spaced nicks generated by the base excision repair pathway can lead to staged breaks directly or crease the entry point for Exo1 without MRN-CtIP or DNA2 (38). This notion is supported by the HTGTS analyses, which revealed that A-EJ-mediated CSR occurs efficiently with CtIPT855A and ATM inhibition and maintains a strong skew toward longer MH stretches. Accordingly, the role of CtIP in A-EJ may be context dependent: required for general DSB repair by A-EJ in most cell types for a single DSB (7) but dispensable for CSR by A-EJ in B lymphocytes.
In cNHEJ-deficient cells but not WT cells, a proportion of CSR junctions (e.g., 1–5%) falls beyond the 3′ boundaries of the core Sμ and Sγ1 regions (13, 14, 15). Using HTGTS, our data provide the unequivocal proof that the formation of distal CSR junctions in Xrcc4-deficient cells is, indeed, achieved by CtIP-mediated end resection and significantly reduced by CtIP deletion, the CtIP-T855A mutation, or ATM kinase inhibition. Moreover, the observation that CtIP is required for the generation of CSR junctions lying distal to the switch regions but not for junctions in the core switch regions further supports the argument that core switch sequences are compatible for A-EJ even without additional resections. In addition to the repetitive nature of the switch region, the AID-generated breaks might contain natural small overhangs. Notably, A-EJ-mediated CSR junctions recovered from KU-deficient B cells have less long MHs (4), suggesting that while long MHs promote A-EJ, they are not required for A-EJ-mediated CSR.
By phosphorylating multiple distinct substrates, ATM can influence different aspects of CSR beyond end resection. Using HTGTS analysis together with ATM inhibitor treatment, our results indicate that ATM kinase activity regulates the orientation of CSR to promote intrachromosomal deletions, including distal Sµ-Sε joins while suppressing nonproductive interchromosomal or intersister translocations. Interestingly, the ATM-enforced preference for intrachromosomal deletions affects CSR events mediated by either cNHEJ or A-EJ—in both cNHEJ-proficient and cNHEJ-deficient cells (Fig. 6C). While the exact mechanisms remain unclear, ATM is known to phosphorylate several chromatin-bound factors, including H2AX and 53BP1, that have been implicated in establishing and maintaining topologically associated domains during CSR (14, 36). Moreover, ATM-phosphorylated H2AX, and the MDC1 protein that binds phosphorylated H2AX, facilitate the lateral spread of DNA damage signals in cis along the chromosome (39). Since CSR requires germ line transcription (40) and ATM facilitates DNA damage-induced transcriptional suppression, ATM may also influence CSR patterns by limiting breaks on sister chromatids or homologs (41).
Using a surface dye to monitor cell division after B cell activation, we were able to monitor the formation of CSR junctions over time. Interestingly, in both cNHEJ-proficient and cNHEJ-deficient cells, we observed that Sμ-Sε switching occurs later than Sμ- Sγ1 switching and is preferentially suppressed by either cNHEJ deficiency or ATM inhibition. These findings can be potentially explained by the sequential CSRs—first to Sγ1, then, to Sε, which would predict that two or more DSB repair events are needed to achieve IgE switching (35). Moreover, Sε is relatively small (4 kb in the BL6 strain used here) and distal from the Sμ. Moreover, we found that short MH (2–1- nt) -mediated joining occurs earlier in Sμ-Sμ even in cNHEJ-proficient cells, suggesting annealing mediated by short MHs might promote cNHEJ as well. This finding agrees with the previous in vitro characterization of cNHEJ using a plasmid substrate, which showed that cNHEJ can occur efficiently with 1 to 2 nt and up to 4 nt of MH (42).
Together, the combination of the CtIP and XRCC4 conditional alleles and HTGTS allowed us to uncover several unique features of A-EJ in switch regions and uncovered the temporal regulation of CSR at nucleotide resolution. The results explain the robust CSR in both cNHEJ- and CtIP-deficient cells and why CSR is a unique biological system to study A-EJ in vivo.
Materials and Methods
Mice.
CD21Cre (43), CtIPC (44), CtIPT855A (22), and Xrcc4C (3) mice were previously described. All animal works were conducted in a specific pathogen-free facility, and all procedures were approved by the Institutional Animal Care and Use Committee at Columbia University Medical Center.
CSR with CTV.
CSR experiments were performed with 6–10-wk-old mice as previously described (22, 15). Splenic B cells were purified with antimurine CD43 magnetic beads (MACS; Miltenyi Biotech) and cultured at 1 × 106 cells mL−1 in Roswell Park Memorial Institute medium supplemented with 15% fetal bovine serum, anti-CD40 (1 ng/mL; BD Pharmingen), and IL-4 (20 ng/mL; R&D). Purified B cells were analyzed by flow cytometry, stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD43 (BD Pharmigen), phycoerythrin-conjugated anti-IgM (Southern Biotech), Cy5-conjugated anti-B220 (eBioscience), and allophycocyanin-conjugated anti-Ter119 to confirm over 90% purity. Each CSR sample was collected on day 3 and day 4 after activation for flow cytometry with FITC-conjugated anti-IgG1 (BD PharMingen) and Cy5-conjugated antimurine B220 (eBioscience). For the CTV experiment, purified B cells were stained at 6 × 106 cells mL−1 in phosphate-buffered saline with 1 μL of 5-mM CTV dye dissolved in dimethyl sulfoxide (Thermo Scientific), following the standard protocol with two additional washes with the medium. Stained B cells were stimulated the same way and harvested on day 4 for flow cytometry analysis, stained with FITC-conjugated anti-IgG1 (BD PharMingen) and Cy7-conjugated antimurine B220 (eBioscience). For the WT CTV sorting experiment, splenic B cells from three WT 6-wk-old C57BL/6J mice purchased from the Jackson Laboratory were pooled together and stained following the same protocol, analyzed, and sorted on day 4 after stimulation. Flow cytometry was performed on a fluorescence-activated cell sorting calibur flow cytometer (BD Bioscience), LSR II (BD Bioscience), or an Attune NxT flow cytometer (Thermo Fisher Scientific). Sorting was performed with an Aria II cell sorter (BD Bioscience).
HTGTS.
HTGTS experiments and analyses were performed as previously described (12, 22, 15). We harvested B cells after 3 or 4 d of cytokine stimulation to obtain genomic DNA. From each sample, about 20 µg of genomic DNA was sonicated (Diagenode Bioruptor) to 300-bp–1-kb fragments and amplified with an Sµ-specific biotinylated primer (5′/5BiosG/CAGACCTGGGAATGTATGGT3′) and nested (5′CACACAAAGACTCTGGACCTC3′) primers. Restriction enzyme (NEB) AflII was used to remove the germ line sequence. When mice with mixed background were used, the IgH locus was genotyped to distinguish the 129 vs. BL6 alleles (primers: IgH_F: TGTGGACCTCCTTGTGAGACTG, and IgH_R: GGGACAGCAGAGGGATTAGG, the 129 allele shows an ∼300-bp band and the BL6 allele shows an ∼360-bp band). Only mice with both IgH loci in the C57/BL6 background were used for the HTGTS analyses. All sequencing reads were aligned to the mm9 genome. Downstream analyses were performed as described before (12, 14) with the pipeline deposited on GitHub (https://github.com/robinmeyers/transloc_pipeline). In brief, we took the bowtie2-ranked top alignments with a score bigger than 50 and ran through the best-path searching algorithm (45) to identify optimal sequences. Several filters were applied additionally, including but not limited to mispriming PCR products, multiple joinings, and duplications. To be specific, if the bait and prey sequences from two individual reads were aligned within 2 nt from each other, respectively, only one unique read will be kept after applying the duplication filter. The overlapping homologous sequences between the bait and the prey sites are characterized as MHs. The additionally unaligned nucleotides in between bait and prey sites were defined as insertions. Reads contain no MHs or insertions were counted as blunt junctions. To compare the MHs use in distal vs. proximal areas within each switch region, the Sµ was divided at Chr 12: 114,662,500 and the Sγ1 at Chr 12: 114,570,500. We deposited all of the sequencing raw data used in this paper to the Gene Expression Omnibus database with the accession no. GSE156392.
Western Blot.
Cells were lysed in radioimmunoprecipitation assay buffer (50-mM 2-Amino-2-(hydroxymethyl)propane-1,3-diol hydrochloride, pH 8.0, 150-mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% Nonidet P-40, and fresh proteinase inhibitor mixture) following standard Western blotting protocols and blotted with antibodies anti-CtIP (1:1,000, ref. 18), and anti-XRCC4 (1:1,000; Santa Cruz, sc-8285) antibodies. Horseradish peroxidase-conjugated anti-mouse secondary antibody (GE Healthcare) and anti-goat secondary antibody (Santa Cruz, sz2020) were used, and an ECL Western blotting detection system (ThermoScientific) was used for detection.
Supplementary Material
Acknowledgments
We thank Dr. Lorraine Symington at Columbia University for helpful discussions and Dr. Binghui Shen at the City of Hope National Cancer Center for sharing the specific inhibitor for DNA2. We thank the members of the Alt Lab, especially Dr. Pei-Chi Wei for their technical assistance and advice for the HTGTS analyses. This work was supported, in part, by grants NIH R01CA158073, R01CA215067, and R01CA226852 to S.Z.; 1P01CA174653 to R.B., S.Z., R.R., and J.G.; NIH R01CA172272 to R.B., and NIH R35 CA197606 to J.G. X.S.W. was, in part, supported by CTSA/NIH TL1 TR000082. S.Z. was a Leukemia Lymphoma Society Scholar. This research was funded, in part, through the NIH/NCI Cancer Center Support Grant P30CA013696 to the Herbert Irving Comprehensive Cancer Center of Columbia University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010972117/-/DCSupplemental.
Data Availability.
Sequencing data have been deposited in the Gene Expression Omnibus database (GSE) (accession no. GSE156392).
References
- 1.Chaudhuri J. et al., Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94, 157–214 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Hwang J. K., Alt F. W., Yeap L. S., Related mechanisms of antibody somatic hypermutation and class switch recombination. Microbiol. Spectr. 3, MDNA3-0037-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan C. T. et al., IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Boboila C. et al., Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 207, 417–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco S., Alt F. W., Manis J. P., Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst.) 5, 1030–1041 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Symington L. S., Gautier J., Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Bennardo N., Cheng A., Huang N., Stark J. M., Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sfeir A., Symington L. S., Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McVey M., Lee S. E., MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 24, 529–538 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masani S., Han L., Meek K., Yu K., Redundant function of DNA ligase 1 and 3 in alternative end-joining during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 113, 1261–1266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boboila C. et al., Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc. Natl. Acad. Sci. U.S.A. 109, 2473–2478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J. et al., Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 11, 853–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panchakshari R. A. et al., DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc. Natl. Acad. Sci. U.S.A. 115, 762–767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J. et al., Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature 525, 134–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crowe J. L. et al., Kinase-dependent structural role of DNA-PKcs during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 115, 8615–8620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard S. M., Yanez D. A., Stark J. M., DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 11, e1004943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T. Y., Kimble M. T., Symington L. S., Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection. Proc. Natl. Acad. Sci. U.S.A. 115, E11961–E11969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X., Baer R., Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275, 18541–18549 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Sartori A. A. et al., Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson S. E. et al., Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell 49, 657–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makharashvili N. et al., Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X. et al., CtIP is essential for early B cell proliferation and development in mice. J. Exp. Med. 216, 1648–1663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H. et al., The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9, e1003277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Theilen M., Matthews A. J., Kelly D., Zheng S., Chaudhuri J., CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18, 75–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buis J., Stoneham T., Spehalski E., Ferguson D. O., Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 19, 246–252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polato F. et al., CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 211, 1027–1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y. et al., A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95, 891–902 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Pelanda R. et al., Receptor editing in a transgenic mouse model: Site, efficiency, and role in B cell tolerance and antibody diversification. Immunity 7, 765–775 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Davies O. R. et al., CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bothmer A. et al., Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J. Exp. Med. 210, 115–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P. L. et al., Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25, 3535–3542 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wesemann D. R. et al., Immature B cells preferentially switch to IgE with increased direct Sμ to Sε recombination. J. Exp. Med. 208, 2733–2746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin W. et al., Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 32, 1425–1439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W. et al., A selective small molecule DNA2 inhibitor for sensitization of human cancer cells to chemotherapy. EBioMedicine 6, 73–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong P., Wesemann D. R., Molecular mechanisms of IgE class switch recombination. Curr. Top. Microbiol. Immunol. 388, 21–37 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Zhang X. et al., Fundamental roles of chromatin loop extrusion in antibody class switching. Nature 575, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han L., Yu K., Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J. Exp. Med. 205, 2745–2753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu K., Lieber M. R., Current insights into the mechanism of mammalian immunoglobulin class switch recombination. Crit. Rev. Biochem. Mol. Biol. 54, 333–351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savic V. et al., Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell 34, 298–310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews A. J., Zheng S., DiMenna L. J., Chaudhuri J., Regulation of immunoglobulin class-switch recombination: Choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv. Immunol. 122, 1–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A., ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieber M. R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraus M. et al., Interference with immunoglobulin (Ig)alpha immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation modulates or blocks B cell development, depending on the availability of an Igbeta cytoplasmic tail. J. Exp. Med. 194, 455–469 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reczek C. R. et al., The DNA resection protein CtIP promotes mammary tumorigenesis. Oncotarget 7, 32172–32183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faust G. G., Hall I. M., YAHA: Fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics 28, 2417–2424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Gene Expression Omnibus database (GSE) (accession no. GSE156392).