Significance
Physiological signals that modulate glutamate receptor trafficking and function remain enigmatic. Here, we identify the auxiliary glutamate receptor subunit dSol-1 to be necessary for the homeostatic control of presynaptic neurotransmitter release at the Drosophila neuromuscular junction. dSol-1 promotes the accumulation of kainate receptors at presynaptic terminals to enable the synapse-specific expression of homeostatic plasticity.
Keywords: glutamate receptor, auxiliary subunit, Drosophila, homeostatic plasticity, synapse
Abstract
Presynaptic glutamate receptors (GluRs) modulate neurotransmitter release and are physiological targets for regulation during various forms of plasticity. Although much is known about the auxiliary subunits associated with postsynaptic GluRs, far less is understood about presynaptic auxiliary GluR subunits and their functions. At the Drosophila neuromuscular junction, a presynaptic GluR, DKaiR1D, localizes near active zones and operates as an autoreceptor to tune baseline transmission and enhance presynaptic neurotransmitter release in response to diminished postsynaptic GluR functionality, a process referred to as presynaptic homeostatic potentiation (PHP). Here, we identify an auxiliary subunit that collaborates with DKaiR1D to promote these synaptic functions. This subunit, dSol-1, is the homolog of the Caenorhabditis elegans CUB (Complement C1r/C1s, Uegf, Bmp1) domain protein Sol-1. We find that dSol-1 functions in neurons to facilitate baseline neurotransmission and to enable PHP expression, properties shared with DKaiR1D. Intriguingly, presynaptic overexpression of dSol-1 is sufficient to enhance neurotransmitter release through a DKaiR1D-dependent mechanism. Furthermore, dSol-1 is necessary to rapidly increase the abundance of DKaiR1D receptors near active zones during homeostatic signaling. Together with recent work showing the CUB domain protein Neto2 is necessary for the homeostatic modulation of postsynaptic GluRs in mammals, our data demonstrate that dSol-1 is required for the homeostatic regulation of presynaptic GluRs. Thus, we propose that CUB domain proteins are fundamental homeostatic modulators of GluRs on both sides of the synapse.
Synaptic strength is dynamically tuned during both Hebbian and homeostatic forms of plasticity. One major mechanism that achieves this modulation targets the abundance, localization, and/or functionality of ionotropic glutamate receptors (GluRs). For instance, the expression of long-term potentiation and depression requires bidirectional changes in the abundance of postsynaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors to adjust synaptic strength (1–3). Furthermore, some forms of homeostatic plasticity also tune the abundance of N-methyl-d-aspartate (NMDA), AMPA, and kainate receptors at postsynaptic densities to stabilize neuronal activity (4–6). Auxiliary subunits associated with GluRs are key factors that control GluR trafficking and dynamics during plasticity, where transmembrane AMPA receptor regulatory protein (TARP), Cornichon, and Neto subunits orchestrate AMPA and kainate receptor function and plasticity (7–9). Although much is known about the GluR subtypes and associated auxiliary subunits that regulate GluR trafficking, abundance, and functionality at postsynaptic densities during both Hebbian and homeostatic plasticity, far less is understood about these mechanisms at presynaptic release sites.
Presynaptic autoreceptors have emerged as important regulators of neurotransmitter release at glutamatergic synapses. For example, presynaptic kainate receptors are present in hippocampal mossy fibers, where autocrine feedback facilitates neurotransmission during trains of activity (10–12). In addition, presynaptic NMDA receptors in the hippocampus mediate presynaptic inhibition in response to excess glutamate release as well as presynaptic facilitation following the induction of long-term potentiation (13). Furthermore, presynaptic metabotropic receptors play critical roles in various forms of plasticity and can bidirectionally tune presynaptic neurotransmitter release (14–16). Finally, ionotropic neurotransmitter receptors at presynaptic terminals can modulate release at neuromuscular junctions (NMJs) in Caenorhabditis elegans and Drosophila (17, 18). While it is now clear that presynaptic autoreceptors are important bidirectional modulators of neurotransmitter release, how the levels, activity, and localization of these receptors are controlled to establish baseline function, and to what extent they are further modified during plasticity, remains unclear.
A kainate-type ionotropic GluR, DKaiR1D, was previously shown to be necessary at the Drosophila NMJ for the expression of presynaptic homeostatic potentiation (PHP) (17). PHP is a fundamental form of synaptic plasticity in which pharmacological and genetic challenges that diminish postsynaptic neurotransmitter receptor functionality trigger a transsynaptic retrograde signal that enhances presynaptic neurotransmitter release to precisely compensate for reduced postsynaptic excitability (19, 20). PHP has been observed at NMJs of Drosophila (21), rodents (22–24), and humans (25, 26) and was recently demonstrated to be rapidly expressed in the mammalian central nervous system (27). DKaiR1D was identified in a forward genetic screen to be required for the rapid expression of PHP at the fly NMJ (17). DKaiR1D receptors form homomers that are permeable to both sodium and calcium (28), localized near presynaptic release sites, and proposed to homeostatically regulate presynaptic voltage following autocrine activation by glutamate (17). This DKaiR1D-dependent enhancement in neurotransmitter output implies a rapid modulation in the abundance, functionality, and/or localization of these receptors must occur in the course of PHP induction. This regulation could, in principle, be achieved through interactions with auxiliary GluR subunits (29, 30). However, the precise mechanisms that control DKaiR1D and enable robust and stable neurotransmission at baseline and during plasticity, and whether auxiliary factors are involved, are unknown.
We have performed a candidate screen of Drosophila GluR modulators and auxiliary subunits to identify potential functions in PHP expression. This effort has discovered an uncharacterized auxiliary GluR subunit that functions in neurons to promote neurotransmitter release and enable homeostatic potentiation. This factor, a homolog of the C. elegans auxiliary GluR subunit Sol-1 (31), contains multiple CUB domains and is structurally similar to the Neto/Sol-2 family of auxiliary GluR subunits (6, 32–34). dSol-1 mutants essentially phenocopy DKaiR1D mutants in neurotransmission and PHP. Further experiments demonstrate that dSol-1 functions to homeostatically modulate presynaptic glutamate release by promoting the rapid accumulation of DKaiR1D receptors near active zones. Together, these data indicate that the interactions between CUB domain auxiliary subunits and their associated GluRs are fundamental physiological targets of homeostatic signaling.
Results
A Screen of Drosophila GluR Modulators and Putative Auxiliary Subunits Identifies dSol-1 to Be Necessary for PHP Expression.
The kainate receptor DKaiR1D functions at presynaptic terminals of the Drosophila NMJ to homeostatically increase presynaptic neurotransmitter release following pharmacological attenuation of postsynaptic GluRs using philanthotoxin-433 (PhTx; schematized in Fig. 1 A and B) (17). Auxiliary subunits that associate with AMPA and kainate GluR subtypes in rodents and worms have been identified (7, 35, 36). In Drosophila, the homolog of neto/sol-2 is highly expressed in muscle and functions with postsynaptic GluRs at the NMJ (34, 37). We established a candidate list of Drosophila genes that are homologs of auxiliary GluR subunits in other systems or that modulate GluR levels in flies to screen for roles in PHP at the NMJ.
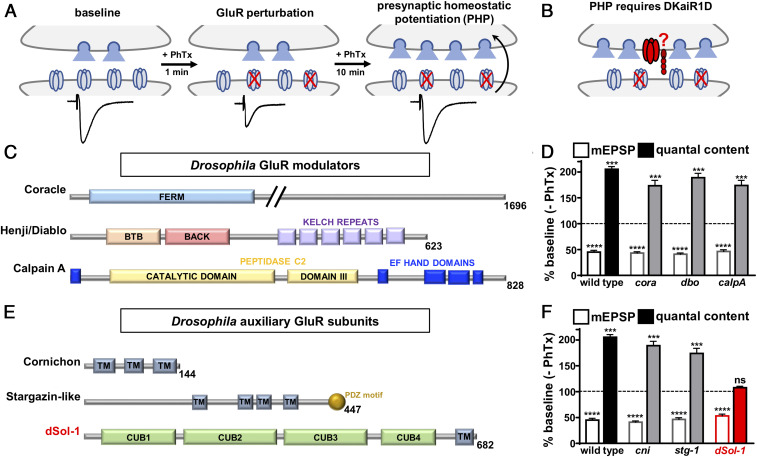
Fig. 1.
Candidate screen of GluR modulators and auxiliary subunits identifies dSol-1 to be necessary for PHP. (A) Schematic illustrating presynaptic homeostatic potentiation at the Drosophila NMJ. Application of the postsynaptic glutamate receptor antagonist philanthotoxin-433 to the larval NMJ initially causes an ∼50% reduction in EPSC amplitude. After 10 min, EPSC amplitudes return to baseline values due to a homeostatic enhancement in presynaptic neurotransmitter release (quantal content). (B) Schematic showing the presynaptic kainate receptor DKaiR1D (noted in red) functions as an autoreceptor near release sites to enhance neurotransmitter release following PhTx application. The putative auxiliary subunit (also in red) that may function with DKaiR1D is unknown. (C) The domain structures of known proteins in Drosophila that modulate GluR levels and trafficking are shown. (D) Quantification of average mEPSP amplitude and quantal content values in mutations of coracle (cora: w;coraMI00820), diablo/henji (dbo: w;dbo8), and calpain A (calpA: w;calpAKG13868) after PhTx treatment normalized to baseline values (−PhTx). A reduction in mEPSP amplitude but concomitant increase in quantal content demonstrates PHP is expressed in these genotypes. (E) The domain structure of putative auxiliary GluR subunits in Drosophila. TM, transmembrane domain. (F) Quantification of average mEPSP amplitude and quantal content values in putative mutations or RNA interference lines of cornichon (cni: OK6-Gal4/+;UAS-cornichon-RNAi/+), stargazin-like (stg-1: stg-1EY06948), and dSol-1 (dSol-1: w;dSol-1MI14035) after PhTx application normalized to baseline values (−PhTx). dSol-1 mutants fail to increase presynaptic release (quantal content) following PhTx application. Error bars indicate ±SEM. ***P < 0.001, ****P < 0.0001; ns, not significant. Details of the mutations and RNAi lines screened, their source, and absolute values of electrophysiology data have been summarized in SI Appendix, Table S1.
We first obtained mutations in three genes that regulate postsynaptic GluR levels at the Drosophila NMJ: coracle (38), diablo/henji (39), and calpain A (40) (Fig. 1C and SI Appendix, Table S1). In addition, we identified three homologs of auxiliary GluR subunits that have not been characterized in Drosophila: cornichon, stargazin-like, and CG34402/dSol-1 (Fig. 1E and SI Appendix, Table S1). We obtained mutations or RNA interference (RNAi) lines targeting each of these genes (SI Appendix, Table S1) and assessed PHP expression following PhTx application. Baseline transmission was consistent with previously published data for mutations in these genes (SI Appendix, Table S1). Following application of PhTx to the NMJ of these mutants, miniature excitatory postsynaptic potential (mEPSP) amplitude was reduced by ∼50%, as expected (Fig. 1 D and F). A homeostatic increase in presynaptic neurotransmitter release (quantal content) was robustly expressed in mutations targeting coracle, diablo, calpain A, cornichon, and stargazin-like (Fig. 1 D and E and SI Appendix, Table S1). However, while mEPSP amplitude was reduced in CG34402 mutants after PhTx application, no increase in quantal content was observed, demonstrating a block in PHP expression (Fig. 1F). The allele of CG34402 we screened (MI14035) contains a MiMIC (Minos-mediated integration cassette) transposon insertion into the second intronic region, predicted to generate a premature stop codon before the first CUB domain. PHP was blocked in this allele when homozygous and in trans with a deficiency (Fig. 1F and SI Appendix, Table S2). Thus, of all candidate mutations screened, PHP failed to be expressed in a single mutation targeting CG34402.
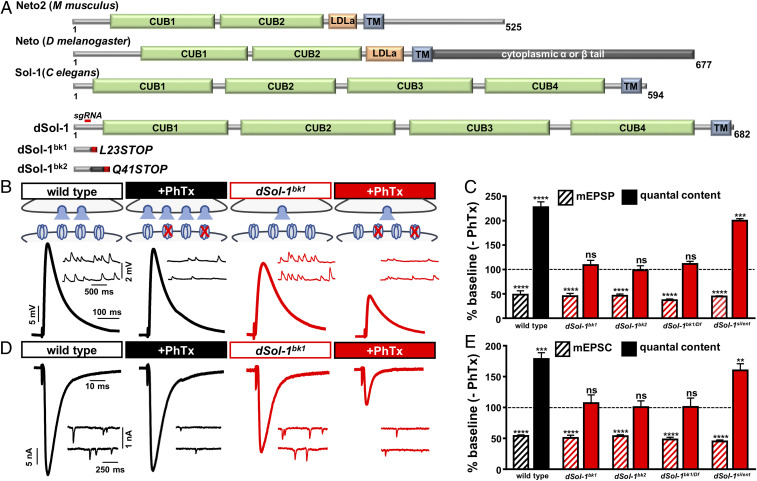
CG34402 encodes a protein of orthologous homology to Sol-1, an auxiliary GluR subunit previously identified and characterized in C. elegans (31). We therefore named this gene dSol-1. dSol-1 encodes a protein with a high degree of structural similarity to C. elegans Sol-1, each containing four extracellular CUB domains and a single transmembrane domain (Fig. 2A) (31, 41, 42). Sol-1/dSol-1 shares structural homology with the Sol-2/Neto family of auxiliary GluR subunits, which contains two extracellular CUB domains, a single transmembrane domain, and an additional LDL (low-density lipoprotein) domain (Fig. 2A). In Drosophila, two isoforms of neto are expressed, neto-α and neto-β, which differ in the intracellular C-terminal region (Fig. 2A). Sol-1 and dSol-1 have both been characterized in heterologous cells, where each can regulate the desensitization kinetics of the C. elegans AMPA-type GluR GLR1 (42). dSol-1 has not been previously studied in Drosophila, so we generated independent null mutations in dSol-1 using CRISPR-Cas9 gene editing. A single-guide RNA (sgRNA) was targeted to the first exon of the dSol-1 open reading frame (ORF), which generated several independent indels (Experimental Procedures and Fig. 2A). We chose two alleles that were predicted to lead to early stop codons, truncating the protein before the first CUB domain, which we named dSol-1bk1 and dSol-1bk2 (Fig. 2A). We characterized baseline synaptic function in these dSol-1 mutant alleles. Although we found no significant change in miniature excitatory postsynaptic current (mEPSC) amplitude, baseline EPSC amplitudes were significantly reduced in dSol-1 mutants (Fig. 2 B and D and SI Appendix, Table S2). This suggests a role for dSol-1 in promoting basal presynaptic glutamate release, without any obvious roles in regulating postsynaptic sensitivity to glutamate.
Fig. 2.
Auxiliary glutamate receptor subunit dSol-1 is required for rapid PHP expression. (A) Protein structure of Neto2, Neto, Sol-1, and dSol-1 (from the indicated organisms), with the region targeted by the single-guide RNA to generate the dSol-1bk1 and dSol-1bk2 mutant alleles and their predicted protein products indicated below. (B) Representative mEPSP and EPSP traces in wild type (w1118), dSol-1bk1 (w;dSol-1bk1), dSol-1bk2 (w;dSol-1bk2), dSol-1bk1/Df [w;dSol-1bk1/Df(3R)Exel7315], and dSol-1silent (w;dSol-1silent) at baseline and after PhTx application. While PhTx application reduces mEPSP amplitudes in all genotypes, as expected, EPSP amplitude fails to return to baseline levels in all dSol-1 mutant alleles and combinations following PhTx application due to a failure to enhance presynaptic neurotransmitter release (quantal content). (C) Quantification of mEPSP amplitude and quantal content values in the indicated genotypes following PhTx application relative to baseline (−PhTx). (D) Representative mEPSC and EPSC traces in two-electrode voltage-clamp configuration in the indicated genotypes at baseline and after PhTx application. (E) Quantification of mEPSC amplitude and quantal content TEVC values in the indicated genotypes and conditions. Error bars indicate ±SEM. **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant. Absolute values for normalized data are summarized in SI Appendix, Table S2.
Next, we assessed PHP expression in the dSol-1 mutant alleles. Following acute inhibition of postsynaptic GluRs by PhTx application, mEPSP values were reduced, as expected, while EPSP values were similarly reduced compared with baseline (Fig. 2 B and C and SI Appendix, Table S2), demonstrating a failure to homeostatically increase presynaptic release (Fig. 2 B and C). Similar results were observed in both dSol-1 alleles when homozygous and in trans to a deficiency that removes the entire dSol-1 locus (Fig. 2 B and C). We further assayed PHP expression in these genotypes using a two-electrode voltage-clamp configuration (TEVC), and found similar results to current clamp (Fig. 2 D and E and SI Appendix, Table S2). Thus, dSol-1 promotes baseline neurotransmitter release and is essential for the rapid homeostatic potentiation of presynaptic release following GluR perturbation at the NMJ.
dSol-1 Functions in Neurons to Promote Baseline Neurotransmission and PHP.
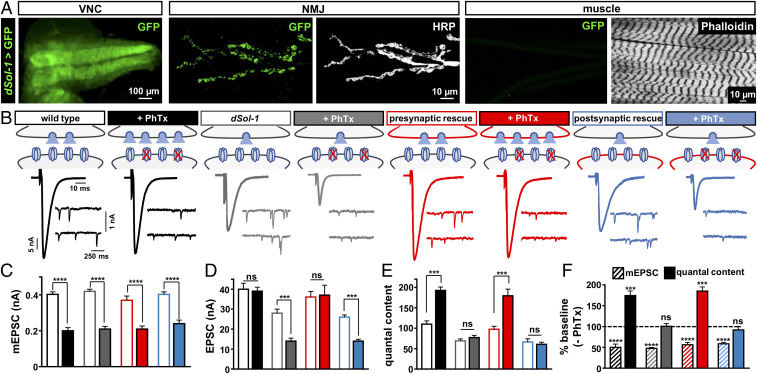
Auxiliary GluR subunits in mammals have distinct expression patterns and functions in the central nervous system (7, 29). To determine the expression of dSol-1, we cloned a 4-kb region upstream of the dSol-1 locus and generated a promotor fusion with the Gal4 transcriptional activator. In combination with a green fluorescent protein (GFP) reporter, we observed dSol-1 expression throughout the larval brain as well as in motor neurons, while no expression was observed in the postsynaptic muscle (Fig. 3A). It is important to note that while this promotor fusion is capable of rescuing PHP in dSol-1 mutants (see below), it is possible that not all dSol-1 regulatory information was captured in this promotor and the GFP reporter itself does not provide information about the subcellular localization of dSol-1. This neuronal expression of dSol-1 is distinct from that of Drosophila neto-β, which is highly expressed in the larval muscle (34). Together, these data indicate that dSol-1 is exclusively expressed in the nervous system and is absent from the postsynaptic muscle at the Drosophila larval NMJ.
Fig. 3.
Presynaptic dSol-1 expression is necessary to promote basal neurotransmitter release and homeostatic potentiation. (A) Representative larval ventral nerve cord (VNC) and NMJ images of GFP expression driven by the dSol-1 promoter and amplified with a Gal4-inducible tubulin-Gal4 cassette (w;dSol-1-Gal4/Tub-FRT-STOP-FRT-Gal4,UAS-FLP,UAS-CD8-GFP). Anti-HRP (neuronal membrane marker) and anti-phalloidin (muscle actin marker) are shown. dSol-1 is exclusively expressed in the nervous system with no detectible signal in the muscle. (B) Rapid expression of PHP requires dSol-1. Representative EPSC and mEPSC traces for wild type, dSol-1bk1 mutants, presynaptic rescue (neuronal expression of dSol-1 in dSol-1 mutants; w;OK6-Gal4/UAS-dSol-1;dSol-1bk1), or postsynaptic rescue (muscle expression of dSol-1 in dSol-1 mutants; w;G14-Gal4/UAS-dSol-1;dSol-1bk1) at baseline and after PhTx application. Presynaptic expression of dSol-1 fully restores PHP expression, while PHP fails in the postsynaptic rescue condition. (C–E) Quantification of mEPSC amplitude (C), EPSC amplitude (D), and quantal content (E) values at baseline and after PhTx treatment. (F) Quantification of mEPSC amplitude and quantal content values following PhTx application relative to baseline (−PhTx) in the indicated genotypes. Error bars indicate ±SEM. ***P < 0.001, ****P < 0.0001; ns, not significant. Absolute values for normalized data are summarized in SI Appendix, Table S2.
Next, we assessed baseline synaptic transmission in dSol-1 mutants and determined the synaptic compartment in which dSol-1 was required to enable PHP expression. Consistent with dSol-1 not being expressed in muscle, we found no significant change in mEPSC amplitude nor in the levels of postsynaptic GluRs in dSol-1 mutants (Fig. 3 B and C and SI Appendix, Fig. S1 A and B). However, baseline EPSC amplitude was significantly decreased in dSol-1 mutants, with a concomitant reduction in quantal content (Fig. 3 B, D, and E). In principle, alterations in synaptic growth or number could contribute to this reduced quantal content, but we did not find any significant changes in synaptic growth or active zone density in dSol-1 mutants (SI Appendix, Fig. S1 C–F). Importantly, expression of dSol-1 using either dSol-1>Gal4 or a motor neuron-specific driver (OK6-Gal4) restored synaptic strength to wild-type levels when expressed in dSol-1 mutant backgrounds (Fig. 3 B–E and SI Appendix, Table S2), indicating that dSol-1 functions in motor neurons to promote basal neurotransmitter release. Finally, expression of dSol-1 in motor neurons, but not in muscle, rescued the block in PHP expression in dSol-1 mutants (Fig. 3 B–F). These results demonstrate that dSol-1 is expressed in the nervous system, where it functions in motor neurons to promote baseline neurotransmitter release and PHP at the NMJ.
dSol-1 and DKaiR1D Act in the Same Genetic Pathway to Promote Presynaptic Neurotransmitter Release.
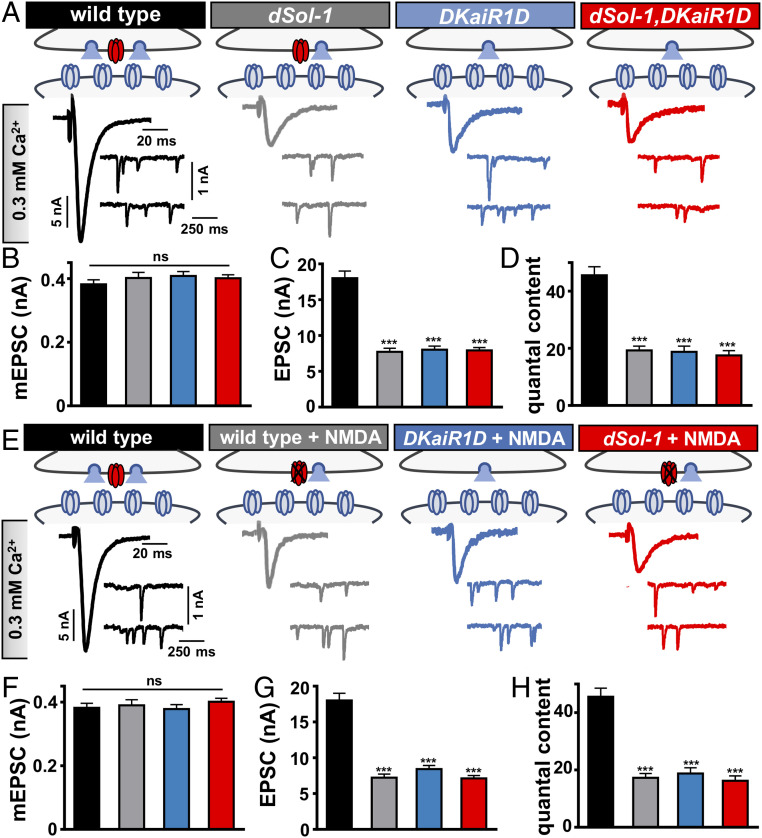
It has previously been shown that the presynaptic kainate receptor DKaiR1D is also required in motor neurons to promote neurotransmitter release (17). In particular, recording in lowered extracellular Ca2+ revealed a reduction in transmission in DKaiR1D mutants compared with controls (17). We therefore tested whether dSol-1 may function in the same genetic pathway as DKaiR1D by probing synaptic transmission in DKaiR1D or dSol-1 mutant synapses, as well as in dSol-1,DKaiR1D double mutants. Synaptic strength was reduced in dSol-1 mutants in 0.3 mM Ca2+ to levels similar in magnitude to that observed in DKaiR1D mutants, and did not change further in dSol-1,DKaiR1D double mutants (Fig. 4 A–D). These data are consistent with dSol-1 and DKaiR1D functioning together in the same genetic pathway to promote baseline neurotransmission.
Fig. 4.
dSol-1 and DKaiR1D act in the same genetic pathway to promote baseline neurotransmitter release. (A) Schematics and representative electrophysiological recordings at 0.3 mM extracellular Ca2+ in wild type, dSol-1 mutants (w;dSol-1bk1), DKaiR1D mutants (w;DKaiR1D2), and dSol-1,DKaiR1D double mutants (w;dSol-1bk1,DKaiR1D2). Note that the EPSC amplitude is not further reduced in dSol-1,DKaiR1D double mutants compared with either mutant alone. (B–D) Quantification of mEPSC amplitude (B), EPSC amplitude (C), and quantal content (D) values in the indicated genotypes. (E) Schematics and representative traces recorded in the presence of the DKaiR1D antagonist NMDA in 0.3 mM extracellular Ca2+ in the indicated genotypes. Application of NMDA to wild-type NMJs reduces baseline transmission but has no effect on DKaiR1D and dSol-1 mutants. (F–H) Quantification of mEPSC amplitude (F), EPSC amplitude (G), and quantal content (H) values in the indicated genotypes and conditions. Error bars indicate ±SEM. ***P < 0.001; ns, not significant. Absolute values for normalized data are summarized in SI Appendix, Table S2.
The NMDA receptor agonist NMDA was shown to function as an antagonist of DKaiR1D receptors in vitro and in vivo (17, 28). To further test the relationship between dSol-1 and DKaiR1D in basal neurotransmission, we applied NMDA to wild type and DKaiR1D and dSol-1 mutants. First, we confirmed that acute application of NMDA reduces baseline release in wild type at lowered extracellular Ca2+ to the same level observed in DKaiR1D mutants, with no effect on mEPSCs (Fig. 4 E–G). In addition, application of NMDA to DKaiR1D mutants had no impact, as expected (Fig. 4 E–G). Finally, NMDA application to dSol-1 mutant synapses also had no impact on EPSC amplitude or quantal content (Fig. 4 E–H). However, in elevated extracellular Ca2+, DKaiR1D has no apparent functions in basal neurotransmission nor in PHP expression (17), properties we confirmed were also shared by dSol-1 (SI Appendix, Fig. S2). Together, these results are consistent with a model in which dSol-1 and DKaiR1D function together to promote baseline neurotransmitter release.
DKaiR1D Is Required for dSol-1 to Potentiate Neurotransmitter Release.
There is precedent for enhanced expression of auxiliary GluR subunits to alter the synaptic delivery, abundance, stabilization, and/or activity of associated receptors. For example, increased TARP expression enhances AMPA receptor number, distribution, and gating in the hippocampus (43, 44), while muscle overexpression of Drosophila neto-β reduces postsynaptic GluR levels at the NMJ (34). Similarly, overexpression of Sol-1 increases glutamate-gated currents through functional modulation of associated GluRs in C. elegans (41). Given that loss of dSol-1 diminishes presynaptic glutamate release, we next asked whether elevated expression of dSol-1 in motor neurons is sufficient to promote neurotransmitter release. Overexpression of dSol-1 (dSol-1-OE) using a motor neuron-specific driver in an otherwise wild-type background potentiated baseline synaptic strength through an increase in EPSC amplitude, without significantly changing mEPSC amplitude (Fig. 5 A–C and SI Appendix, Table S2). Correspondingly, quantal content was increased by over 50% compared with baseline values (Fig. 5D), demonstrating that enhanced neuronal expression of dSol-1 is sufficient to potentiate glutamate release from motor neurons.
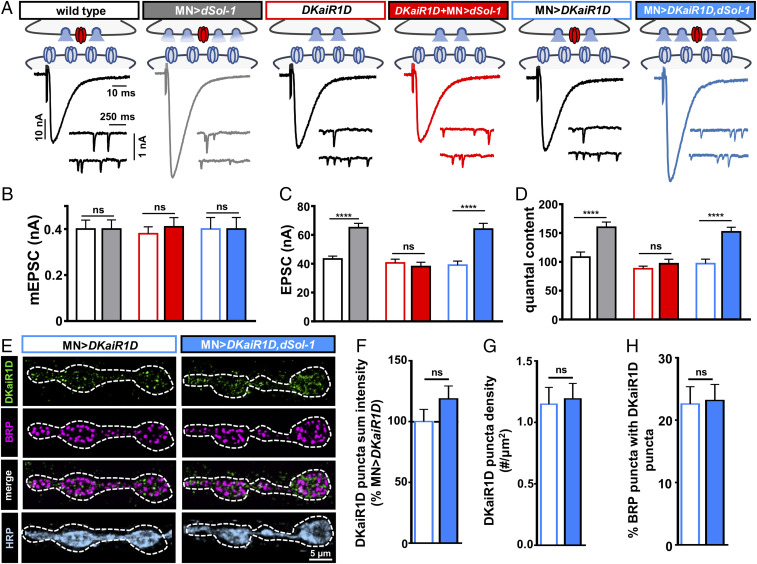
Fig. 5.
DKaiR1D is required for dSol-1 to potentiate baseline neurotransmitter release. (A) Schematics and representative EPSC and mEPSC traces showing dSol-1 overexpression in motor neurons enhances presynaptic neurotransmitter release in wild type (MN>dSol-1: w;OK6-Gal4/UAS-dSol-1). However, dSol-1 overexpression in motor neurons fails to increase presynaptic release in the absence of DKaiR1D (DKaiR1D+MN>dSol-1: w;OK6-Gal4/UAS-dSol-1;DKaiR1D2). Overexpression of DKaiR1D at wild-type NMJs (MN>DKaiR1D: w;OK6-Gal4/UAS-DKaiR1D) does not impact neurotransmission and does not enhance release further in conjunction with dSol-1 overexpression (MN>DKaiR1D,dSol-1: w;UAS-dSol-1/UAS-DKaiR1D;D42-Gal4/+). (B–D) Quantification of mEPSC amplitude (B), EPSC amplitude (C), and quantal content (D) values in the indicated genotypes. (E) Representative images of NMJs with DKaiR1D overexpressed in wild type (MN>DKaiR1D) or in combination with dSol-1 (MN>DKaiR1D,dSol-1) immunostained with anti-DKaiR1D, -BRP, and -HRP. Note that no change in the DKaiR1D signal is observed in MN>DKaiR1D,dSol-1 compared with MN>DKaiR1D. (F–H) Quantification of DKaiR1D punctum sum intensity (F), DKaiR1D punctum density (G) and percentage of BRP-positive active zones colocalized with DKaiR1D (H) reveals no significant change in MN>DKaiR1D,dSol-1 compared with MN>DKaiR1D. Error bars indicate ±SEM. ****P < 0.0001; ns, not significant. Absolute values for normalized data are summarized in SI Appendix, Table S2.
Since dSol-1 and DKaiR1D function in the same genetic pathway to promote release, we considered whether dSol-1-OE–mediated enhancement in release requires DKaiR1D. In moderate extracellular Ca2+ conditions, loss or overexpression of DKaiR1D has no impact on baseline synaptic transmission (Fig. 5 A–D) (17). However, overexpression of dSol-1 in DKaiR1D mutant backgrounds failed to potentiate release (Fig. 5 A–D). Thus, increased dSol-1 expression in neurons can potentiate transmitter release, but this ability requires DKaiR1D.
In principle, overexpression of dSol-1 could increase the abundance and/or gating properties of DKaiR1D receptors to promote neurotransmitter release. However, in C. elegans, Sol-1 functions to modify the gating properties of GLR1, slowing desensitization and promoting open states, without having any apparent roles in modifying the abundance of surface GLR1 receptors at synapses or in heterologous cells (31, 41, 42). To gain insight into how overexpression of dSol-1 potentiates release in a DKaiR1D-dependent way, we immunostained synapses with an antibody against DKaiR1D at wild-type and dSol-1-OE NMJs. Overexpression of DKaiR1D in motor neurons (MN>DKaiR1D) is necessary for anti-DKaiR1D antibodies to reliably detect these receptors at NMJ terminals (17). MN>DKaiR1D does not significantly impact neurotransmission compared with wild type (Fig. 5 A–D), and we observed anti-DKaiR1D puncta that colocalized with the active zone marker bruchpilot (BRP), as has been previously shown (Experimental Procedures and Fig. 5E) (17). Interestingly, when both DKaiR1D and dSol-1 were overexpressed together, release was potentiated to levels similar to dSol-1-OE alone (Fig. 5 A–D), but no increase in DKaiR1D punctum intensity, density, or localization relative to active zones was observed (Fig. 5 E–H). Although the mechanism through which dSol-1-OE potentiates neurotransmitter release is uncertain, these findings are consistent with overexpression of dSol-1 potentiating presynaptic release through functional changes to existing DKaiR1D receptors, without apparent alterations to DKaiR1D receptor abundance or localization at individual release sites. Such a mechanism would parallel the means through which C. elegans Sol-1 modulates synaptic strength (41, 42).
dSol-1 Promotes the Rapid Accumulation of DKaiR1D Receptors near Release Sites during PHP Signaling.
Neuronal overexpression of dSol-1 potentiates neurotransmitter release to levels similar in magnitude to what is observed during PHP. One possibility is that dSol-1-OE promotes presynaptic release through a common mechanism shared with the adaptations that occur during PHP. Alternatively, dSol-1-OE may enhance release through a novel mechanism, distinct from the signaling that happens during PHP. In this case, increased quantal content induced by PHP expression would be expected to be additive with dSol-1-OE. We sought to distinguish between these possibilities by assessing PHP expression at dSol-1-OE NMJs. PhTx application to wild-type and dSol-1-OE NMJs reduced mEPSC amplitudes, as expected (Fig. 6 A and B and SI Appendix, Table S2). However, while wild-type EPSCs recovered to baseline levels after PhTx, EPSC values were correspondingly reduced in dSol-1-OE, failing to return to baseline levels (Fig. 6 A and B and SI Appendix, Table S2). This demonstrates that dSol-1-OE occludes additional PHP signaling, perhaps because PHP has effectively already been triggered.
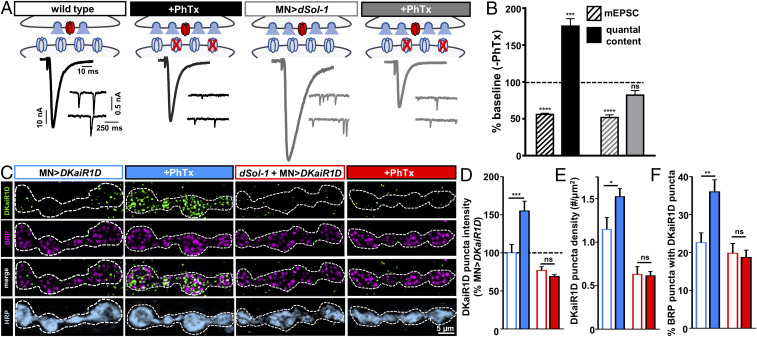
Fig. 6.
PHP signaling requires dSol-1 to promote rapid accumulation of DKaiR1D receptors near release sites. (A) Schematics and representative traces in the indicated genotypes showing presynaptic neurotransmitter release is not further enhanced following PhTx application at NMJs overexpressing dSol-1 in motor neurons. (B) Quantification of mEPSC and quantal content values following PhTx application normalized to baseline (−PhTx) demonstrates quantal content is not further increased following PhTx application to MN>dSol-1 NMJs. (C) Representative images of boutons immunostained with anti-DKaiR1D, -BRP, and -HRP at NMJs with DKaiR1D overexpressed in wild type (MN>DKaiR1D: w;OK6-Gal4/UAS-DKaiR1D) or dSol-1 mutants (dSol-1+MN>DKaiR1D: w;OK6-Gal4,dSol-1bk1/UAS-DKaiR1D,dSol-1bk1) at baseline and following PhTx application. Note the increase in DKaiR1D signal after PhTx observed in wild type is not found in dSol-1 mutants. (D–F) Quantification of DKaiR1D punctum sum intensity (D) and density (E) and percentage of BRP-positive active zones colocalized with DKaiR1D (F) reveals increases in MN>DKaiR1D following PhTx application, while no enhancement is observed in dSol-1 mutants. Error bars indicate ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Absolute values for normalized data are summarized in SI Appendix, Table S2.
Auxiliary subunits can function to increase the surface expression and synaptic localization of their respective GluRs (7, 9, 35, 45). In addition, auxiliary subunits can also affect the kinetics and gating properties of GluRs to modulate their activity (42, 43, 46). These mechanisms are not mutually exclusive. Neuronal overexpression of dSol-1 potentiates neurotransmitter release through DKaiR1D without measurably changing receptor levels at presynaptic terminals (Fig. 5). In our final set of experiments, we sought to test whether PHP signaling modulates DKaiR1D receptor levels, and how dSol-1 functions in this process.
We quantified DKaiR1D receptor puncta by immunostaining transgenically overexpressed DKaiR1D in controls and in dSol-1 mutant backgrounds before and after PhTx application. In baseline conditions, relatively low levels of DKaiR1D puncta colocalized with BRP puncta, as observed previously (Fig. 5E) (17). Remarkably, after a 10-min incubation in PhTx, we found a substantial increase in DKaiR1D signals, including in the intensity and density of individual DKaiR1D puncta (Fig. 6 C–E), with a significant increase in the number of DKaiR1D puncta colocalized with BRP puncta at active zones (Fig. 6F). Importantly, no increase in DKaiR1D levels was observed following PhTx application in dSol-1 mutants (Fig. 6 C–F), indicating that dSol-1 is required to rapidly enhance DKaiR1D receptors at presynaptic terminals during PHP signaling. Thus, dSol-1 has two separable functions in promoting neurotransmitter release through DKaiR1D receptors. First, increased dSol-1 expression enhances baseline neurotransmitter release without impacting DKaiR1D levels. However, dSol-1 has an additional function during PHP signaling to promote the rapid accumulation of DKaiR1D receptors at presynaptic release sites.
Discussion
We have identified an auxiliary GluR subunit that functions in motor neurons to promote glutamate release during both baseline activity and homeostatic plasticity at the Drosophila NMJ. Genetic, pharmacological, and cell biological evidence suggests dSol-1 functions with DKaiR1D to enhance neurotransmitter release. In the context of baseline neurotransmission, dSol-1 promotes release without measurably changing the abundance or localization of DKaiR1D receptors, indicating a functional role in modulating DKaiR1D activity. However, dSol-1 is necessary during PHP signaling to drive a rapid increase in DKaiR1D receptor abundance at presynaptic terminals. These findings define a CUB domain auxiliary GluR subunit as a central target for the presynaptic modulation of synaptic efficacy and homeostatic plasticity.
Several lines of evidence suggest that dSol-1 enhances baseline neurotransmitter release by targeting DKaiR1D receptor functionality. First, dSol-1 promotes baseline neurotransmission in low extracellular Ca2+, a function shared with DKaiR1D (17). In addition, neurotransmitter release is reduced in dSol-1 mutants and enhanced by neuronal overexpression of dSol-1, indicating a capacity for dSol-1 expression levels to bidirectionally tune release. However, this potentiation in baseline transmission occurs without a significant increase in DKaiR1D receptor abundance, at least when both dSol-1 and DKaiR1D are overexpressed in motor neurons (Fig. 5), suggesting a change in DKaiR1D functionality. Interestingly, in C. elegans, Sol-1 regulates GLR1 functionality by modulating channel gating, promoting the open state, and slowing sensitization, without an apparent change in glr1 expression (31, 41, 42). In heterologous cells, both Sol-1 and dSol-1 promote GLR1 function without altering expression levels (42). This indicates a potentially conserved function between sol-1 and dSol-1 to confer similar modulations to GluR functionality. In mammals, Neto auxiliary subunits selectively associate with kainate-subtype GluRs, while TARPs such as Stargazin associate with AMPA-type receptors (9, 29, 30). In Drosophila, Neto is an important auxiliary subunit for the postsynaptic GluRs at the NMJ, which, like DKaiR1D, are generally characterized as non-NMDA, kainate-type GluRs (28, 47, 48). However, in C. elegans, both Sol-1 and Sol-2/Neto form a complex together with the AMPA receptor subtype GLR1 (33), suggesting some level of promiscuity, at least in invertebrates, between AMPA and kainate GluRs and their auxiliary subunits. One possibility is that at baseline states, a substantial proportion of DKaiR1D receptors do not interact with dSol-1. By increasing levels of dSol-1, more DKaiR1D receptors may become associated with dSol-1, perhaps leading to changes in gating properties that enhance DKaiR1D receptor function. However, we cannot rule out the possibility that dSol-1 somehow regulates DKaiR1D through a more indirect mechanism. While DKaiR1D receptors can form homomers and traffic to the cell surface when expressed alone in heterologous cells (28), future in vitro studies will be needed to determine the precise role dSol-1 has in DKaiR1D receptor trafficking and/or functionality.
dSol-1 enables PHP expression through a mechanism that is distinct from its role in baseline transmission, although both functions converge on DKaiR1D. Our data suggest that PHP signaling leads to a rapid accumulation of DKaiR1D receptors near presynaptic release sites that requires dSol-1. This dSol-1–dependent increase in DKaiR1D levels may be a unique feature of homeostatic signaling in Drosophila, as there is no evidence for worm sol-1 to promote surface levels or changes in GLR1 localization in vivo or in vitro (31, 41, 42). Although the rapid increase in DKaiR1D receptor levels at synaptic terminals during PHP signaling is surprising, it is not unprecedented. DKaiR1D receptors are present near presynaptic release sites (17), and many other active zone components rapidly accumulate and/or remodel following PhTx application (49–54). An attractive possibility is that DKaiR1D receptors participate in this process of rapid active zone remodeling during PHP signaling. Mechanistically, new protein synthesis of DKaiR1D is unlikely to be involved, as PHP expression (51, 55) and active zone remodeling (49) can occur without new translation. Recently, the lysosomal kinesin adaptor arl-8 and other axonal transport factors were identified to be necessary for the rapid increase in active zone components during PHP signaling (49, 50). Thus, it is tempting to speculate that DKaiR1D receptors might be cotransported during PHP as part of this pathway. In vertebrates, auxiliary subunits traffic GluRs during synaptic plasticity (1, 45, 56), so dSol-1 may function similarly in delivering DKaiR1D receptors to the plasma membrane and/or to release sites during PHP signaling. Finally, it is possible that DKaiR1D receptors are constitutively degraded under baseline conditions, and that PHP signaling through dSol-1 inhibits this degradation. The role of protein degradation in PHP signaling has been recently studied in both pre- and postsynaptic compartments at the Drosophila NMJ (57, 58). Interestingly, inhibition of proteasomal degradation in presynaptic compartments is capable of rapidly enhancing neurotransmission (58) to levels similar to what is observed after overexpression of dSol-1. In both cases, no further increase in neurotransmitter release is observed after PhTx application. While there are apparently distinct roles for dSol-1 in baseline function and homeostatic plasticity, a common point of convergence is DKaiR1D.
CUB domains define a structural motif in a large family of extracellular and plasma membrane-associated proteins present in invertebrates to humans (59). While the specific four extracellular CUB domains that define sol-1/dSol-1 are unique to invertebrates, genes containing multiple CUB domains (between two and eight) are present throughout vertebrate species and function in diverse processes including intercellular signaling, developmental patterning, inflammation, and tumor suppression (60, 61). CUB domains mediate dimerization and binding to collagen-like regions; this interaction may be relevant to its role in promoting PHP, as the Drosophila collagen member Multiplexin is present in the extracellular matrix and has been proposed to be part of the homeostatic retrograde signaling system (57, 62). Our characterization of dSol-1 contrasts with what is known about another CUB domain auxiliary glutamate receptor in Drosophila, Neto-β. neto-β is highly expressed in the larval muscle, where it is clearly involved in the trafficking and/or stabilization of postsynaptic GluRs at the NMJ (34, 47). In contrast, dSol-1 is exclusively expressed in the nervous system. Another interesting distinction is that while both dSol-1 and Neto contain multiple extracellular CUB domains and a single transmembrane domain, dSol-1 lacks any intracellular domain while two isoforms of Neto are expressed with one of two alternative intracellular C-terminal cytosolic domains, Neto-α or Neto-β (37). Neto-β is clearly the major isoform and performs the key functions in controlling postsynaptic GluR levels and composition (37), while neto-α was recently proposed to function in motor neurons with DKaiR1D and to be necessary for PHP (63). Interestingly, the C. elegans receptor GLR1 requires the auxiliary subunits Sol-1, Stargazin, and Neto/Sol-2 for functionality in vivo and in vitro (33, 42). It is therefore possible that Drosophila Neto-α and/or Stargazin-like interact with both dSol-1 and DKaiR1D. In mammals, there is a large body of evidence demonstrating that presynaptic GluRs, including kainate receptors, modulate presynaptic function (29). While postsynaptic kainate receptors in mammals associate with the CUB domain auxiliary GluR subunit Neto2 to regulate synaptic function and homeostatic plasticity (6, 33), to what extent Neto2 or other auxiliary subunits function with presynaptic kainate receptors remains enigmatic (29, 64). Our study indicates that CUB domain proteins may be fundamental modulators of GluRs in synaptic function and plasticity on both sides of the synapse.
Experimental Procedures
Fly Stocks.
All Drosophila stocks were raised at 25 °C on standard molasses food. The w1118 strain was used as the wild-type control unless otherwise noted because this is the genetic background in which all genotypes are bred. Details of all stocks and their sources are listed in SI Appendix, Tables S1 and S3.
Molecular Biology.
dSol-1bk1 and dSol-1bk2 mutants were generated using a CRISPR-Cas9 genome-editing strategy as described (65). Briefly, a target Cas9 cleavage site was selected early in the first coding exon of the dSol-1 ORF without containing obvious off-target sequences in the Drosophila genome (sgRNA sequence: 5′-CTTGGCTCTGGGATTAACCGTGG-3′; protospacer-adjacent motif (PAM) sequence: underlined; off targets: 0; strand: +). This construct was sent to BestGene, Inc. for targeted insertion into the VK18 attP site on the second chromosome (66). Flies with the corresponding sgRNA sequences were crossed to a vas-Cas9 line on the second chromosome to induce active germline CRISPR mutagenesis and 10 lines were screened for mutations. We found 4 lines that exhibited no mutations in the dSol-1 locus (including dSol-1silent) and identified 6 independent indels in the dSol-1 gene. The two lines that were predicted to induce the earliest stop codons (L23STOP and Q41STOP) were chosen for further analysis and named dSol-1bk1 and dSol-1bk2, respectively. To control for possible off-target lesions due to CRISPR-Cas9 mutagenesis, we also characterized a dSol-1silent allele, which was derived from a chromosome that underwent the same Cas9 and sgRNA crosses that were used to generate the dSol-1 mutant alleles but did not mutate the dSol-1 locus.
To generate a UAS-dSol-1 transgene, we obtained an expressed sequence tag (EST) (IP10972) encoding the entire dSol-1 ORF from the Berkeley Drosophila Genome Project (https://www.fruitfly.org). We inserted the dSol-1 complementary DNA into the pACU2 vector (67) using standard approaches. To generate the dSol-1>Gal4 promotor fusion, we PCR-amplified a 4-kb region upstream of the dSol-1 start codon and cloned this product into the pDEST-APIGH Gal4 expression vector using the Gateway expression system (Invitrogen). All constructs were sequenced to confirm fidelity and orientation and were sent to BestGene, Inc. for targeted insertion into the VK18 attP site on the second chromosome.
Immunochemistry.
Third-instar larvae were dissected in ice-cold 0 Ca2+ HL-3 solution and immunostained as described (68, 69). Briefly, larvae were fixed in either Bouin’s fixative (Sigma; HT10132-1L) or 4% paraformaldehyde in phosphate-buffered saline (PBS) (Sigma; F8775). Larvae were washed with PBS containing 0.1% Triton X-100 (PBST) for 30 min and blocked for 1 h in 5% normal donkey serum followed by overnight incubation in primary antibodies at 4 °C, washing in PBST, incubation in secondary antibodies for 2 h, washing again in PBST, and equilibration in 70% glycerol in PBST. Samples were mounted in VectaShield (Vector Laboratories). Details of all antibodies, their source, dilution, and references are listed in SI Appendix, Table S3.
Confocal Imaging and Analysis.
Samples were imaged using a Nikon A1R resonant scanning confocal microscope equipped with NIS Elements software and a 100× APO 1.4 numerical aperture (NA) oil-immersion objective using separate channels with laser lines 488, 561, and 637 nm as described (70). Z stacks were obtained using identical settings for all genotypes, with z-axis spacing between 0.15 and 0.5 µm within an experiment and optimized for detection without saturation of the signal. Fluorescence-intensity measurements were taken on muscles 6/7 and muscle 4 of segment A3 for at least 10 NMJs acquired from at least 6 different animals. For fluorescence quantifications of the GluR subunits, the general analysis toolkit in the NIS Elements software was used as the binary for GluRIIA to measure values in GluRIIB and GluRIID channels. To measure synaptic growth, both type Ib and Is boutons were counted using vGlut-, horseradish peroxidase (HRP)-, and Dlg-stained NMJ terminals on muscles 6/7 and muscle 4 of segment A3, considering each vGlut punctum to be a bouton. The general analysis toolkit in the NIS Elements software was used to quantify BRP punctum number per NMJ and density was quantified by dividing the BRP punctum number by the neuronal membrane area masked by the HRP channel. For analysis of DKaiR1D puncta, BRP and DKaiR1D puncta were counted within an area also labeled by HRP and controls were performed to establish and subtract background levels of nonspecific signals due to the anti-rat secondary antibody used. To determine colocalization of DKaiR1D and BRP puncta, a binary mask was created for BRP puncta and only the DKaiR1D signal within that mask was quantified. BRP and DKaiR1D intensities were normalized to the HRP signal intensity and then normalized to wild-type values. Punctum density and intensity measurements based on confocal images were taken from at least 12 synapses acquired from at least 6 different animals.
Electrophysiology.
Electrophysiological recordings were performed from muscle 6 in abdominal segments 2 and 3 from wandering third-instar larvae at room temperature as described (71, 72). Recordings were made in modified hemolymph-like saline (HL-3) containing 70 mM NaCl, 5 mM KCl, 10 mM MgCl2, 10 mM NaHCO3, 115 mM sucrose, 5 mM trehalose, 5 Hepes, and 0.5 CaCl2 (unless specified) (pH 7.2) from cells with an initial Vm between −60 and −75 mV and input resistances >6 MΩ. Sharp intracellular electrodes with resistances of 12 to 25 MΩ filled with 3 M KCl were used. Recordings were performed with an Olympus BX61WI microscope using a 40×/0.80 NA water-dipping objective and acquired using an Axoclamp 900A amplifier, Digidata 1440A acquisition system, and pClamp 10.5 software (Molecular Devices). Electrophysiological sweeps were digitized at 10 kHz with a 1-kHz filter. For all TEVC recordings, muscles were clamped at −70 mV with a leak current below 10 nA and a settling time of the voltage clamp of 22.3 ms. For recordings at or above 1 mM Ca2+, evoked EPSCs were recorded with a combination of HS-9Ax10 and HS-9Ax1 head stages (Axon CNS; Molecular Devices). For recordings at <1 mM Ca2+, two HS-9Ax1 head stages were used. From each muscle cell, 100 mEPSCs were recorded in the absence of stimulation. Twenty EPSCs were acquired for each cell under stimulation at 0.5 Hz and stimulus duration of 0.5 ms, and with stimulus intensity adjusted with an ISO-Flex Stimulus Isolator (A.M.P.I.). Quantal content was calculated by dividing the average EPSC amplitude by the average mEPSC amplitude for each cell. To acutely block postsynaptic receptors, larvae were incubated with or without philanthotoxin-433 (20 µM; Sigma) in HL-3 for 10 min as described (55). For the acute blockade of DKaiR1D by NMDA, larvae were dissected and, following a 10-min incubation with PhTx, the central nervous system was removed and the larvae were incubated with 1 mM NMDA (Abcam; ab120052; resuspended in deionized H2O) for 5 min, with recordings performed in the continued presence of NMDA as described (17). Data were analyzed using Clampfit 10.7 (Molecular Devices), MiniAnalysis (Synaptosoft), Excel (Microsoft), and Prism (GraphPad Software).
Statistical Analysis.
Data were analyzed using GraphPad Prism (version 7.0) or Microsoft Excel software (version 16.22). Datasets were tested for normality using the D’Agostino and Pearson omnibus test to ensure that the assumption of normality was not violated. Values were then compared using either a one-way ANOVA, followed by Tukey’s multiple-comparison test, or an unpaired two-tailed Student’s t test with Welch’s correction. All data are presented as mean ± SEM; P denotes the level of significance assessed (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant). Statistics of all experiments are summarized in SI Appendix, Table S2.
Acknowledgments
We acknowledge the Developmental Studies Hybridoma Bank (Iowa) for antibodies and the Bloomington Drosophila Stock Center for fly stocks (NIH Grant P40OD018537). This work was supported by a grant from the NIH (NS091546) (to D.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915464117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Diering G. H., Huganir R. L., The AMPA receptor code of synaptic plasticity. Neuron 100, 314–329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herring B. E., Nicoll R. A., Long-term potentiation: From CaMKII to AMPA receptor trafficking. Annu. Rev. Physiol. 78, 351–365 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Derkach V. A., Oh M. C., Guire E. S., Soderling T. R., Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat. Rev. Neurosci. 8, 101–113 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Otaño I., Ehlers M. D., Homeostatic plasticity and NMDA receptor trafficking. Trends Neurosci. 28, 229–238 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury D., Hell J. W., Homeostatic synaptic scaling: Molecular regulators of synaptic AMPA-type glutamate receptors. F1000 Res. 7, 234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W. et al., A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron 61, 385–396 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissen D., Foss F., Acker-Palmer A., AMPA receptors and their minions: Auxiliary proteins in AMPA receptor trafficking. Cell. Mol. Life Sci. 76, 2133–2169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita S., Regulation of ionotropic glutamate receptors by their auxiliary subunits. Physiology 25, 41–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson A. C., Nicoll R. A., The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro P. S. et al., GluR7 is an essential subunit of presynaptic kainate autoreceptors at hippocampal mossy fiber synapses. Proc. Natl. Acad. Sci. U.S.A. 104, 12181–12186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz D., Mellor J., Frerking M., Nicoll R. A., Presynaptic kainate receptors at hippocampal mossy fiber synapses. Proc. Natl. Acad. Sci. U.S.A. 98, 11003–11008 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott R., Lalic T., Kullmann D. M., Capogna M., Rusakov D. A., Target-cell specificity of kainate autoreceptor and Ca2+-store-dependent short-term plasticity at hippocampal mossy fiber synapses. J. Neurosci. 28, 13139–13149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee A., Larsen R. S., Philpot B. D., Paulsen O., Roles of presynaptic NMDA receptors in neurotransmission and plasticity. Trends Neurosci. 39, 26–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanik L. et al., The Drosophila metabotropic glutamate receptor DmGluRA regulates activity-dependent synaptic facilitation and fine synaptic morphology. J. Neurosci. 24, 9105–9116 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittaluga A., Presynaptic release-regulating mGlu1 receptors in central nervous system. Front. Pharmacol. 7, 295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upreti C., Zhang X. L., Alford S., Stanton P. K., Role of presynaptic metabotropic glutamate receptors in the induction of long-term synaptic plasticity of vesicular release. Neuropharmacology 66, 31–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiragasi B., Wondolowski J., Li Y., Dickman D. K., A presynaptic glutamate receptor subunit confers robustness to neurotransmission and homeostatic potentiation. Cell Rep. 19, 2694–2706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayanagi-Kiya S., Zhou K., Jin Y., Release-dependent feedback inhibition by a presynaptically localized ligand-gated anion channel. eLife 5, e21734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis G. W., Müller M., Homeostatic control of presynaptic neurotransmitter release. Annu. Rev. Physiol. 77, 251–270 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Frank C. A., James T. D., Müller M., Homeostatic control of Drosophila neuromuscular junction function. Synapse 74, e22133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank C. A., Homeostatic plasticity at the Drosophila neuromuscular junction. Neuropharmacology 78, 63–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., McIntosh J. M., Rich M. M., Muscle nicotinic acetylcholine receptors may mediate trans-synaptic signaling at the mouse neuromuscular junction. J. Neurosci. 38, 1725–1736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Pinter M. J., Rich M. M., Reversible recruitment of a homeostatic reserve pool of synaptic vesicles underlies rapid homeostatic plasticity of quantal content. J. Neurosci. 36, 828–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr B. O. et al., Presynaptic homeostasis opposes disease progression in mouse models of ALS-like degeneration: Evidence for homeostatic neuroprotection. Neuron 107, 95–111.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cull-Candy S. G., Miledi R., Trautmann A., Uchitel O. D., On the release of transmitter at normal, myasthenia gravis and myasthenic syndrome affected human end-plates. J. Physiol. 299, 621–638 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Rich M. M., Homeostatic synaptic plasticity at the neuromuscular junction in myasthenia gravis. Ann. N. Y. Acad. Sci. 1412, 170–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delvendahl I., Kita K., Müller M., Rapid and sustained homeostatic control of presynaptic exocytosis at a central synapse. Proc. Natl. Acad. Sci. U.S.A. 116, 23783–23789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y. et al., Novel functional properties of Drosophila CNS glutamate receptors. Neuron 92, 1036–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lerma J., Marques J. M., Kainate receptors in health and disease. Neuron 80, 292–311 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Nicoll R. A., Tomita S., Bredt D. S., Auxiliary subunits assist AMPA-type glutamate receptors. Science 311, 1253–1256 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y., Mellem J. E., Brockie P. J., Madsen D. M., Maricq A. V., SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature 427, 451–457 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Ng D. et al., Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol. 7, e41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R. et al., The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron 75, 838–850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y. J., Bao H., Bonanno L., Zhang B., Serpe M., Drosophila Neto is essential for clustering glutamate receptors at the neuromuscular junction. Genes Dev. 26, 974–987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copits B. A., Swanson G. T., Dancing partners at the synapse: Auxiliary subunits that shape kainate receptor function. Nat. Rev. Neurosci. 13, 675–686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straub C., Tomita S., The regulation of glutamate receptor trafficking and function by TARPs and other transmembrane auxiliary subunits. Curr. Opin. Neurobiol. 22, 488–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos C. I., Igiesuorobo O., Wang Q., Serpe M., Neto-mediated intracellular interactions shape postsynaptic composition at the Drosophila neuromuscular junction. PLoS Genet. 11, e1005191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen K., Merino C., Sigrist S. J., Featherstone D. E., The 4.1 protein coracle mediates subunit-selective anchoring of Drosophila glutamate receptors to the postsynaptic actin cytoskeleton. J. Neurosci. 25, 6667–6675 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M. et al., Dbo/Henji modulates synaptic dPAK to gate glutamate receptor abundance and postsynaptic response. PLoS Genet. 12, e1006362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metwally E., Zhao G., Li W., Wang Q., Zhang Y. Q., Calcium-activated calpain specifically cleaves glutamate receptor IIA but not IIB at the Drosophila neuromuscular junction. J. Neurosci. 39, 2776–2791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y. et al., SOL-1 is an auxiliary subunit that modulates the gating of GLR-1 glutamate receptors in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 103, 1100–1105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker C. S. et al., Conserved SOL-1 proteins regulate ionotropic glutamate receptor desensitization. Proc. Natl. Acad. Sci. U.S.A. 103, 10787–10792 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milstein A. D., Zhou W., Karimzadegan S., Bredt D. S., Nicoll R. A., TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron 55, 905–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouach N. et al., TARP gamma-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat. Neurosci. 8, 1525–1533 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Huganir R. L., Nicoll R. A., AMPARs and synaptic plasticity: The last 25 years. Neuron 80, 704–717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheng N., Shi Y. S., Lomash R. M., Roche K. W., Nicoll R. A., Neto auxiliary proteins control both the trafficking and biophysical properties of the kainate receptor GluK1. eLife 4, e11682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han T. H., Dharkar P., Mayer M. L., Serpe M., Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 112, 6182–6187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid A. et al., Non-NMDA-type glutamate receptors are essential for maturation but not for initial assembly of synapses at Drosophila neuromuscular junctions. J. Neurosci. 26, 11267–11277 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Böhme M. A. et al., Rapid active zone remodeling consolidates presynaptic potentiation. Nat. Commun. 10, 1085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goel P. et al., Homeostatic scaling of active zone scaffolds maintains global synaptic strength. J. Cell Biol. 218, 1706–1724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goel P., Li X., Dickman D., Disparate postsynaptic induction mechanisms ultimately converge to drive the retrograde enhancement of presynaptic efficacy. Cell Rep. 21, 2339–2347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gratz S. J. et al., Endogenous tagging reveals differential regulation of Ca2+ channels at single active zones during presynaptic homeostatic potentiation and depression. J. Neurosci. 39, 2416–2429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weyhersmüller A., Hallermann S., Wagner N., Eilers J., Rapid active zone remodeling during synaptic plasticity. J. Neurosci. 31, 6041–6052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mrestani A., et al. , Active zone compaction in presynaptic homeostatic potentiation. bioRxiv:10.1101/802843 (22 May 2020). [Google Scholar]
- 55.Frank C. A., Kennedy M. J., Goold C. P., Marek K. W., Davis G. W., Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron 52, 663–677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson A. C., Nicoll R. A., Stargazin (TARP γ-2) is required for compartment-specific AMPA receptor trafficking and synaptic plasticity in cerebellar stellate cells. J. Neurosci. 31, 3939–3952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kikuma K. et al., Cul3 and insomniac are required for rapid ubiquitination of postsynaptic targets and retrograde homeostatic signaling. Nat. Commun. 10, 2998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wentzel C., Delvendahl I., Sydlik S., Georgiev O., Müller M., Dysbindin links presynaptic proteasome function to homeostatic recruitment of low release probability vesicles. Nat. Commun. 9, 267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bork P., Beckmann G., The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 231, 539–545 (1993). [DOI] [PubMed] [Google Scholar]
- 60.Blanc G. et al., Insights into how CUB domains can exert specific functions while sharing a common fold: Conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J. Biol. Chem. 282, 16924–16933 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Gaboriaud C. et al., Structure and properties of the Ca(2+)-binding CUB domain, a widespread ligand-recognition unit involved in major biological functions. Biochem. J. 439, 185–193 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Wang T., Hauswirth A. G., Tong A., Dickman D. K., Davis G. W., Endostatin is a trans-synaptic signal for homeostatic synaptic plasticity. Neuron 83, 616–629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han T. H. et al., Neto-α controls synapse organization and homeostasis at the Drosophila neuromuscular junction. Cell Rep. 32, 107866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyeth M. S. et al., Neto auxiliary subunits regulate interneuron somatodendritic and presynaptic kainate receptors to control network inhibition. Cell Rep. 20, 2156–2168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kikuma K., Li X., Kim D., Sutter D., Dickman D. K., Extended synaptotagmin localizes to presynaptic ER and promotes neurotransmission and synaptic growth in Drosophila. Genetics 207, 993–1006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venken K. J., He Y., Hoskins R. A., Bellen H. J., P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Han C., Jan L. Y., Jan Y. N., Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 108, 9673–9678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perry S., Han Y., Das A., Dickman D., Homeostatic plasticity can be induced and expressed to restore synaptic strength at neuromuscular junctions undergoing ALS-related degeneration. Hum. Mol. Genet. 26, 4153–4167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry S. et al., Developmental arrest of Drosophila larvae elicits presynaptic depression and enables prolonged studies of neurodegeneration. Development 147, dev186312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goel P. et al., A screen for synaptic growth mutants reveals mechanisms that stabilize synaptic strength. J. Neurosci. 39, 4051–4065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goel P., Li X., Dickman D., Estimation of the readily releasable synaptic vesicle pool at the Drosophila larval neuromuscular junction. Bio. Protoc. 9, e3127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X. et al., The BLOC-1 subunit pallidin facilitates activity-dependent synaptic vesicle recycling. eNeuro 4, 1–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and SI Appendix.