Significance
Memory CD8 T lymphocytes are a long-lived population of immune cells that provide formidable protection against infections and malignancy and are thus often central to vaccine and cancer immunotherapy efficacy. Here we refine the identity, developmental relationships, and functional roles of memory CD8 T cells in mice through delineation of a discrete population that confers robust protection against reinfection and exhibits an unexpected molecular program bearing typically divergent characteristics of short-lived effector cells and long-lived memory T cells. Mass cytometry and single-cell RNA-sequencing analyses revealed an analogous population of cells in humans. These findings hold broad implications for both informing immunotherapy treatments and predicting their efficacy.
Keywords: infection, T cells, memory T cells, immunology
Abstract
Memory CD8 T cells provide durable protection against diverse intracellular pathogens and can be broadly segregated into distinct circulating and tissue-resident populations. Paradigmatic studies have demonstrated that circulating memory cells can be further divided into effector memory (Tem) and central memory (Tcm) populations based on discrete functional characteristics. Following resolution of infection, we identified a persisting antigen-specific CD8 T cell population that was terminally fated with potent effector function but maintained memory T cell qualities and conferred robust protection against reinfection. Notably, this terminally differentiated effector memory CD8 T cell population (terminal-Tem) was conflated within the conventional Tem population, prompting redefinition of the classical characteristics of Tem cells. Murine terminal-Tem were transcriptionally, functionally, and developmentally unique compared to Tem cells. Through mass cytometry and single-cell RNA sequencing (RNA-seq) analyses of human peripheral blood from healthy individuals, we also identified an analogous terminal-Tem population of CD8 T cells that was transcriptionally distinct from Tem and Tcm. Key findings from this study show that parsing of terminal-Tem from conventionally defined Tem challenge the reported characteristics of Tem biology, including enhanced presence in lymphoid tissues, robust IL-2 production, and recall potential, greater than expected homeostatic fitness, refined transcription factor dependencies, and a distinct molecular phenotype. Classification of terminal-Tem and clarification of Tem biology hold broad implications for understanding the molecular regulation of memory cell states and harnessing immunological memory to improve immunotherapies.
Memory CD8 T cells are critical mediators of long-lived immunity and provide dynamic protection against intracellular pathogens and malignancies. Given the unique attributes of memory T cells, including specificity, durability, and rapid effector functions, leveraging this population of cells is a key objective of diverse immunotherapies and vaccines. Memory CD8 T cells can be broadly segregated into recirculating Tcm and Tem subsets predominantly found in the blood and lymphoid tissues as well as tissue-resident memory T cells (Trm), which are primarily localized to nonlymphoid sites (1). Numerous studies have established that Tcm and Tem populations are phenotypically and functionally distinct. Tcm are generally considered to exhibit greater homeostatic proliferation and longevity, multipotency, recall potential, and lymph node homing capacity due to characteristically elevated expression of CD62L and CCR7 (1–6). Conversely, Tem are thought to be relatively shorter-lived and terminally fated, exhibit an effector phenotype marked by granzyme production, and primarily localized in the vasculature relative to lymphoid tissues and, in certain contexts, to recirculate through or populate nonlymphoid sites (2, 7–9). This partitioning of functional attributes and localization properties within the circulating memory compartment provides a division of labor as well as flexibility in the mounting of robust defenses to diverse pathogens and neoplasms.
Memory T cells are primarily derived from a multipotent memory precursor (MP) population distinguished by elevated expression of the IL-7 receptor (CD127) and low expression levels of KLRG1 observed in the effector phase of the response to acute infection, whereas CD127loKLRG1hi terminally differentiated effector (TE) cells are generally considered to be short-lived and terminally fated (10–12), although ex-KLRG1-expressing cells have been shown to form multiple memory populations (13). As our understanding of T cell states and fates has expanded, it has become evident that diverse intracellular and extracellular cues dictate CD8 T cell differentiation and homeostasis, ultimately through the dynamic activity of fate-specifying transcription factors (3, 14). Canonical proeffector transcription factors linked to the formation of TE cells and Tem include T-bet (11), Blimp1 (15, 16), Zeb2 (17, 18), Stat4 (19), and Id2 (20–23), whereas promemory transcription factors required for optimal MP and Tcm differentiation include Eomes (24), Bcl6 (25, 26), Foxo1 (27–29), Stat3 (26), and Id3 (22, 30). However, our ability to probe the molecular underpinnings of Tcm and Tem fates is limited by our capacity to accurately define and delineate these distinct memory populations.
In humans, Tem and Tcm are frequently defined as CD45ROhiCCR7lo and CD45ROhiCCR7hi, respectively (1, 4, 5, 31). However, our understanding of the function, differentiation, and molecular regulation of memory T cells is primarily derived from mouse models, wherein a common strategy for discriminating Tem and Tcm is often based solely on CD62L expression (4, 8, 32–40). A number of studies have reported heterogeneity within the Tem compartment, including a persisting “effector-like” population of KLRG1hi cells (41), CD43loCD27lo cells (42), or CX3CR1int peripheral memory cells (43), and some reports suggest alternative or complementary (in addition to CD62L for example) approaches for demarcation of memory populations based on differing levels of CX3CR1 (43, 44), CXCR3 (43, 45), CD27 (4, 46), CD28 (46), KLRG1 (13, 17), and CD127 (10, 47, 48). Here, we report that the conventional CD62Llo Tem population is conflated with a transcriptionally and functionally distinct population of CD127loCD62Llo terminally differentiated effector memory cells (terminal-Tem). This finding has a number of implications, but the principal relevance is that the general understanding of the phenotype, function, and transcriptional regulation of Tem cells has been confounded by a contaminating terminal-Tem population, requiring reexamination of Tem biology. Further, we show terminal-Tem exhibit potent cytotoxic activity but have limited multipotency and recall potential. Thus, we here support a definition of Tem as CD127hiCD62Llo and provide a framework for ascribing novel (or previously muddled) characteristics of Tem including an enhanced presence in lymphoid tissues, robust IL-2 production and recall potential, greater than expected homeostatic fitness, and a distinct molecular phenotype. Additionally, we refine roles for the key lineage-specifying transcription factors T-bet, Blimp1, Bcl6, and Foxo1. Last, we identify an analogous terminal-Tem subset within the human circulating memory CD8 T cell compartment with discrete transcriptional and phenotypic qualities compared to Tem, Tcm, and effector cells. Given that memory T cells are linked to prevention and progression of diverse diseases, an in-depth understanding and elucidation of circulating memory states hold widespread implications from molecular analyses to clinical studies and allow greater context for pinpointing which memory state might be most therapeutically relevant for a given disease setting.
Results
Terminal-Tem Are a Distinct Subset of CD8 T Cells Contained within the Conventional CD62Llo Tem Population.
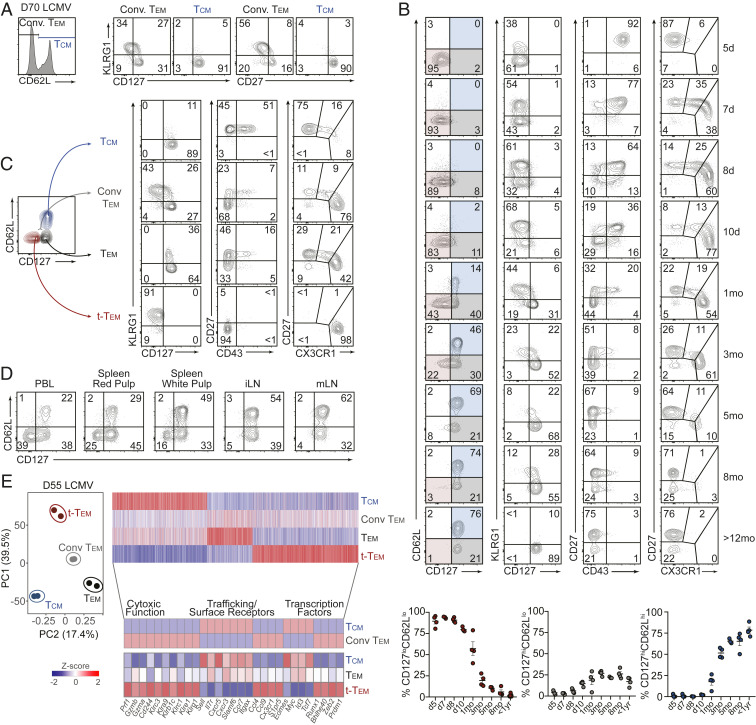
Tem are most often classified as CD62LloCCR7lo and Tcm as CD62LhiCCR7hi (1, 3, 31). However, CCR7 staining is not typically incorporated in murine memory CD8 T cell subsetting as the staining process (43) is not as straightforward as for CD62L, and both molecules are generally coexpressed (4, 5). Therefore, a majority of the studies investigating circulating memory T cell populations utilize differential CD62L expression as the defining molecule to distinguish Tcm and Tem (4, 8, 32–40). Expression of CD127 has also been widely used to define antigen-experienced CD8 T cell populations with memory qualities (10, 16, 47–49). Utilizing the lymphocytic choriomeningitis virus (LCMV) infection system, we found that LCMV GP33–41-specific tetramer+ memory CD8 T cells (SI Appendix, Fig. S1A) or TCR transgenic P14 cells with low expression levels of CD62L (i.e., conventional Tem) were heterogeneous for CD127, memory-associated costimulatory molecule CD27, and the TE- or short-lived effector-associated molecule KLRG1 (Fig. 1A). Additionally, circulating antigen-specific CD62Llo memory cells generated following Listeria monocytogenes infection also displayed this heterogeneity (SI Appendix, Fig. S1B). More than 2 mo after LCMV infection, >30% of CD62Llo cells remained CD127lo and CD27lo (Fig. 1A) prompting the questions: Are CD127lo cells at a “memory timepoint” considered memory cells? Should the CD127lo cells be grouped into the conventional “Tem” population? How do these CD127lo cells fit with other strategies for distinguishing memory T cell subsets (e.g., CX3CR1, CD43, and CD27)?
Fig. 1.
Terminal-Tem are a distinct subset of CD8 T cells contained within the conventional Tem population. P14 CD8 T cells were adoptively transferred into congenically distinct recipient mice that were subsequently infected with LCMV. (A) Expression of KLRG1, CD127 (Left), and CD27 (Right) by Tcm (CD62Lhi) and conventional (Conv.) Tem (CD62Llo) within peripheral blood lymphocytes (PBLs) on day 70 of infection. (B) Expression of indicated molecules on P14 PBLs in response to LCMV infection. Frequency of CD127loCD62Llo (highlighted red), CD127hiCD62Llo (highlighted gray), and CD127hiCD62Lhi (highlighted blue) in PBLs following LCMV infection (Bottom). (C) Representative expression patterns of indicated molecules by terminal-Tem, Tem, Conv. Tem, and Tcm. (D) Representative flow cytometry plots demonstrating the frequency of terminal-Tem, Tem, and Tcm in indicated tissues (>30 d postinfection). Splenic red and white pulp localized P14 cells were discriminated by intravascular staining of CD8α. (E) Principal component analysis of the RNA-seq transcriptional profile of splenic terminal-Tem, Tem, Conv. Tem, and Tcm P14 cells on day 55 of infection (Left), and heatmap illustrating differentially expressed genes (≥1.5-fold) ordered through k-means clustering (Top Right) among terminal-Tem, Tem, Conv. Tem, and Tcm, or highlighted key genes (Bottom). Numbers in plots are the frequency of cells in the indicated gate (A–D). All data are from two independent experiments with n = 3 to 5 per timepoint, and RNA-seq samples consist of two biological replicates wherein each replicate is comprised of cells pooled from two mice. Graphs indicate mean ± SEM, and symbols represent an individual mouse (B).
In an attempt to develop a comprehensive and unifying understanding of the dynamics of well-studied molecules often used to distinguish circulating memory CD8 T cell populations, we measured expression patterns of CD62L, CD127, KLRG1, CD43, CD27, and CX3CR1 over the course of >1 y following LCMV infection. Although CD43 and CD27 have previously been reported to distinguish circulating memory cells with differing levels of effector function (41, 42) and recent investigations have established varying CX3CR1 levels can delineate distinct populations of circulating memory cells (43, 44), CD62Lhi Tcm and conventional CD62Llo Tem classification is widely used to subset circulating memory populations. Thus, an in-depth characterization of how so-called “memory markers” fully describe memory CD8 T cell states is needed. As expected, the proportion of CD127hi circulating memory T cells increased over time after LCMV infection, and 1 y after infection nearly all P14 cells were CD127hi in the blood (Fig. 1B). We noted a prominent CD127loCD62Llo population of cells persisting up to 5 mo after infection, and therefore sought to investigate this apparent heterogeneity within the CD62Llo population of memory cells as well as to clarify the molecular identity, ontogeny, and function of Tem in relation to the CD127loCD62Llo population.
CD127loCD62Llo cells at memory timepoints (>30 d after infection) exhibited expression patterns consistent with a terminal effector phenotype, including elevated KLRG1 expression, low expression of CD43 and CD27, and robust expression of CX3CR1; furthermore, while longer lived than TE cells, the persisting CD127loCD62Llo population ultimate wanes after 3 to 5 mo and thus have limited durability compared to CD127hiCD62Llo and Tcm populations (Fig. 1B). Therefore, we refer to CD127loCD62Llo cells as terminally differentiated effector memory T cells (terminal-Tem, Fig. 1C). We also redefine Tem as CD127hiCD62Llo cells and Tcm as CD127hiCD62Lhi, although CD62L staining alone is sufficient to distinguish Tcm, as nearly all CD62Lhi cells are CD127hi. We next utilized this revised memory T cell nomenclature to investigate the phenotypic characteristics and unique qualities of circulating memory T cells with an emphasis on contrasting terminal-Tem with Tem.
Following LCMV infection, terminal-Tem were primarily localized in the vasculature and less abundant in lymph nodes (Fig. 1D), consistent with reports of CX3CR1hi cells predominating in the blood (43). However, we find a considerable proportion of CD127hiCD62Llo Tem localized not only in the blood but also in splenic white pulp and lymph nodes as well. Thus, clarifying conventional Tem into discrete CD127hiCD62Llo Tem and CD127loCD62Llo terminal-Tem subsets reveals relevant biology about memory T cells; classically, conventional Tem abundance in lymphoid tissues was thought to be relatively lower than blood (2, 3), but here we demonstrate that this was chiefly due to a contaminating terminal-Tem population and that Tem in steady-state conditions are abundant in the spleen and draining lymph nodes.
We next profiled the transcriptome of the newly defined terminal-Tem and CD127hiCD62Llo Tem populations compared to Tcm and conventional CD62Llo Tem to understand their transcriptional relationship as well as to elucidate differential gene-expression programs contributing to the fate and homeostasis of these populations. Principal component analysis (PCA) illustrated that, indeed, terminal-Tem are transcriptionally distinct from Tcm and Tem, but importantly also demonstrated that redefined Tem and Tcm are distinct populations and not simply a uniform population of cells with varying expression levels of CD62L. We detected >5,000 genes differentially expressed among the three memory subsets, and closer evaluation of mRNA expression levels of key functional and transcriptional molecules further confirmed that terminal-Tem displayed elevated expression of key cytotoxic, effector, and migratory molecules (Fig. 1E). Inclusion of the transcriptional profile of conventionally defined Tem (CD62Llo) in the transcriptomic analyses highlighted how redefining Tem provides clarification into regulatory gene-expression programs and the lineage relationship of memory T cells. For example, contrary to current paradigms, Tem express relatively low levels of Prdm1 and Zeb2 but elevated levels of Tcf7 and Id3 after parsing of CD127loCD62Llo terminal-Tem and CD127hiCD62Llo Tem populations (Fig. 1E). Last, as Trm are a distinct memory T cell population with robust effector features (50, 51), we compared the transcriptional relationship of terminal-Tem, Tem, Tcm, and intestinal Trm, which also revealed unique gene-expression patterns between each population and supported the underlying notion that terminal-Tem are a discrete CD8 T cell population (SI Appendix, Fig. S2 A and B).
Distinct Fate, Homeostasis, and Effector Phenotype of CD127loCD62Llo Terminal-Tem and CD127hiCD62Llo Tem.
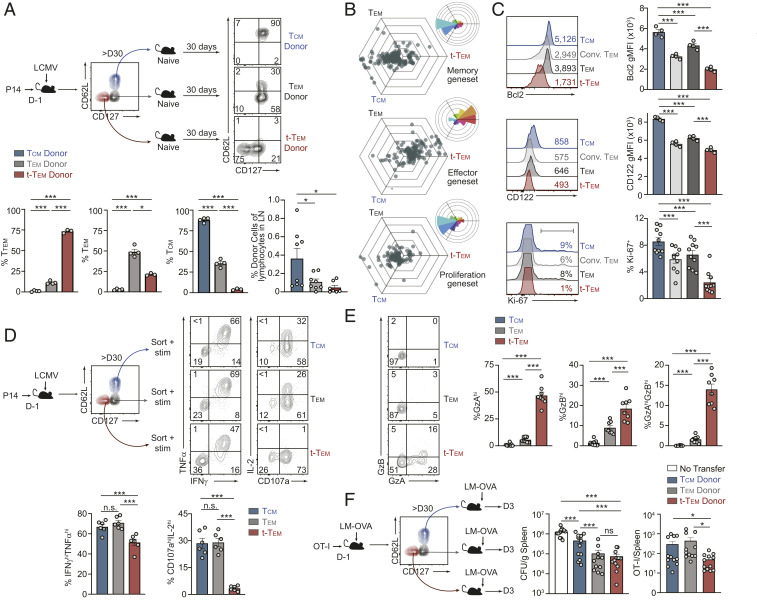
Profiling of population dynamics in the blood revealed that the terminal-Tem population declines over time while Tcm become the predominant memory population, as expected (Fig. 1B). It was unclear if the loss of terminal-Tem was due to limited homeostatic proliferation, impaired survival, or transdifferentiation. Although we hypothesized limited flexibility in terminal-Tem fate given their resemblance to TE, we first tested if terminal-Tem were able to transdifferentiate into other memory cell types, potentially contributing to population waning. Terminal-Tem, Tem, and Tcm were sorted and transferred into naive recipient mice, and ∼30 d after adoptive transfer, the donor cells were phenotyped (Fig. 2A). We found that terminal-Tem primarily retained a CD127loCD62Llo phenotype with minimal conversion to other memory subsets, consistent with their designation as a terminally differentiated population. In contrast, ∼50% of Tem were able to up-regulate CD62L (ostensibly converting to Tcm), and transferred Tcm remained phenotypically stable, consistent with previous reports (4). The relative fixed fate of terminal-Tem was also emphasized by their limited presence in lymph nodes upon retransfer into naive recipient mice (Fig. 2A).
Fig. 2.
Distinct fate, homeostasis, and effector phenotype of CD127loCD62Llo terminal-Tem and CD127hiCD62Llo Tem. (A) More than 30 d after LCMV infection, terminal-Tem (CD127loCD62Llo), Tem (CD127hiCD62Llo), and Tcm (CD127hiCD62Lhi) P14 memory subsets were sorted and transferred into naive, congenically distinct mice. After >30 d posttransfer, the frequency of each donor population in the spleen was analyzed by flow cytometry. Representative plots (Top) and quantification of the phenotype (Bottom) are shown as well as frequency of donor cells recovered from lymph nodes (Bottom Right). (B) Three-way comparisons of differentially expressed genes (≥1.5-fold) within the indicated gene-expression signature were plotted in a hexagonal diagram in which the magnitude of differential expression is reflected by distance from the origin. Rose plots (Upper Right corner of each hexagonal plot) indicate the percentages of genes in each orientation. Memory- (Top), effector- (Middle), and proliferation-associated (Bottom) genesets are displayed. (C) Flow cytometry analysis of terminal-Tem, Tem, Conv. Tem, and Tcm on day >D35 of infection. Representative plots (Left) and quantification of geometric mean fluorescence intensity (gMFI) or % positive (Right) are shown. (D) Memory subsets were sorted and cultured ex vivo with GP33–41 peptide and congenically distinct splenocytes for assessment of TNFα, IFNγ, IL-2 production, and degranulation (CD107a). Representative flow cytometry plots (Top) and quantification of the frequency of cytokine production by each subset (Bottom) are shown. (E) GzA and GzB expression by terminal-Tem, Tem, and Tcm was analyzed by flow cytometry (Left) and quantified (Right). (F) OT-I CD8 T cells were transferred into congenically distinct mice that were infected with LM-OVA the following day. More than 30 d after infection, terminal-Tem (CD127loCD62Llo), Tem (CD127hiCD62Llo), and Tcm (CD127hiCD62Lhi) subsets were sorted and transferred to naive recipients that were challenged with LM-OVA. On day 3 of challenge, bacterial load in spleens was assessed (CFU/gram of tissue) and abundance of donor cells in recipient spleens were enumerated by flow cytometry. Numbers in plots represent gMFI or the frequency of cells in the indicated gate (A, C, and E). All data are from one representative experiment of three independent experiments with n = 3 to 4 per group (A and C), or data are combined from three independent experiments (Ki-67 staining in C and F), or data are combined from two independent experiments (E), or combined from three independent experiments where each experiment consisted of two biological replicates and each replicate was sorted from one or two mice combined (D); n.s., not significant, *P < 0.05, ***P < 0.005. Graphs indicate mean ± SEM, and symbols represent an individual mouse or an individual replicate.
As terminal-Tem exhibit relatively minimal conversion into other memory T cell populations, we next assessed their capacity for homeostatic proliferation and long-term survival. Through three-way comparisons, we found that terminal-Tem expressed lower levels of signature T cell proliferation genes (52) as well as memory signature genes, but elevated levels of characteristic terminal effector genes relative to Tem and Tcm (Fig. 2B). Flow cytometry analyses confirmed the distinct transcriptional differences observed, further demonstrating terminal-Tem express relatively low levels of prosurvival molecules CD122 and Bcl2 and undergo lower levels of homeostatic proliferation indicated by diminished Ki-67 staining (Fig. 2C). Taken together, the loss of the terminal-Tem population over time (Fig. 1B) is likely due to diminished homeostatic fitness (i.e., the capacity for homeostatic proliferation and long-term survival).
We next sought to determine if terminal-Tem and the redefined Tem population are functionally distinct from each other as well as Tcm. To assay cytokine production, terminal-Tem, Tem, and Tcm populations were sorted (due to diminished surface abundance of CD127 and CD62L upon ex vivo stimulation) (53) and incubated with cognate peptide. Terminal-Tem exhibited reduced polyfunctionality compared to Tem and Tcm, with a lower frequency of IFNγhiTNFαhi cells (Fig. 2D). Consistent with an impaired homeostasis phenotype, the percentage of IL-2 producing terminal-Tem was approximately eightfold lower compared to Tcm and Tem, while in contrast to current paradigms (2) an equivalent frequency of Tem produced IL-2 compared to Tcm (Fig. 2D). Next, given the robust expression of effector molecules in terminal-Tem (Fig. 1E), we evaluated granzyme abundance and the effector capacity of the distinct memory populations. Consistent with gene-expression levels, terminal-Tem had elevated levels of both GzA and GzB molecules compared to Tem and Tcm, while Tem had elevated expression of GzA and GzB compared to Tcm (Fig. 2E). To determine how the diverse functional, phenotypic, and migratory/localization qualities of terminal-Tem are integrated to mediate pathogen clearance, we transferred sorted populations of congenically distinct terminal-Tem, Tem, and Tcm OT-I cells into naive recipient mice subsequently challenged with Listeria monocytogenes expressing OVA. We found that terminal-Tem conferred equivalent protection compared to Tem despite limited recall expansion (Fig. 2F). Therefore, terminal-Tem have the most potent effector activity on a per cell basis following Listeria challenge, likely due to constitutively elevated expression of granzymes, perforin, and key migratory molecules (Figs. 1E and 2E). These data also demonstrate that the redefined population of Tem are functionally distinct from Tcm, which is notable as our reclassification suggests greater than previously appreciated similarity between Tem and Tcm, especially regarding homeostasis and recall potential. The functional relevance of the three circulating memory populations was also investigated in a tumor vaccination model, in which cognate peptide and polyIC were administered following adoptive transfer of sorted terminal-Tem, Tem, and Tcm P14 cells into B16-GP33–41 tumor-bearing mice (SI Appendix, Fig. S3). Consistent with results from similar experiments (13, 54), Tcm conferred the greatest protection, likely due to their elevated lymph node homing capacity, recall proliferation, and enhanced survival. However, terminal-Tem provided minimal protection despite elevated expression levels of cytotoxic and migratory molecules, thus highlighting that the activity and protective role of each memory subset is likely dependent on the disease or therapeutic context.
Terminal-Tem Possess Key Characteristics of Memory T Cells with Features of Terminal Effector CD8 T Cells.
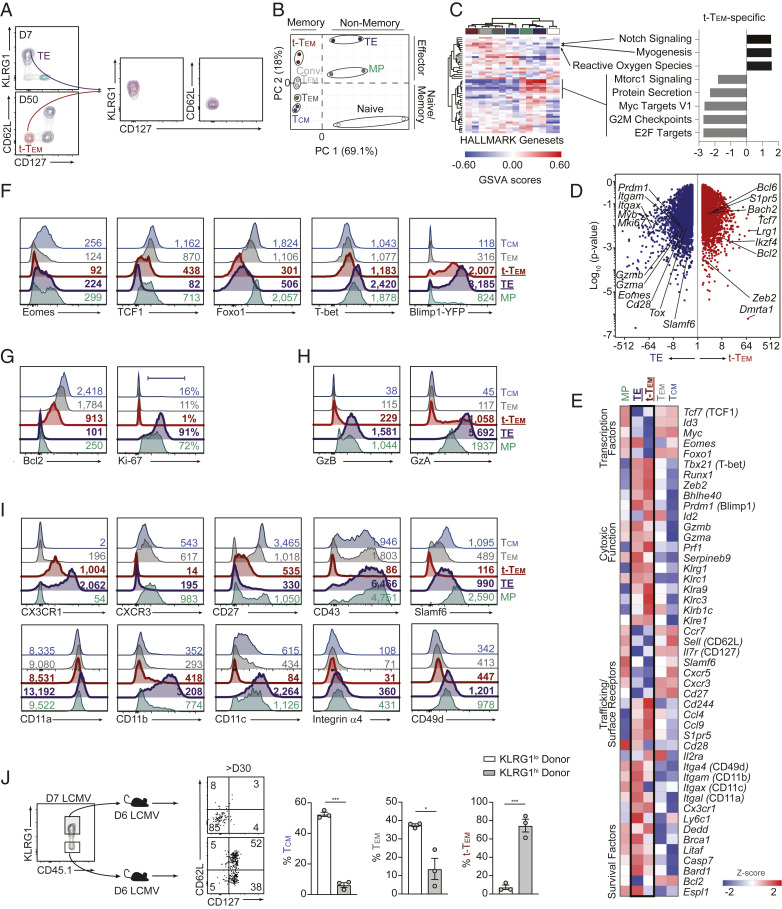
The relatively low expression levels of CD62L and CD127 and elevated levels of KLRG1 on terminal-Tem resembled the surface phenotype of TE cells (Fig. 3A), prompting the questions: 1) At late infection time points, are terminal-Tem true memory cells; and 2) How are terminal-Tem, which phenotypically resemble short-lived TE cells, able to persist for >200 d following infection? Principal component analysis (Fig. 3B) and a gene-expression similarity matrix (SI Appendix, Fig. S4A) revealed that while terminal-Tem were transcriptionally distinct from D7 TE cells and more transcriptionally related to memory populations, terminal-Tem do exhibit some transcriptional similarly to effector cells relative to Tcm and Tem (Fig. 3B and SI Appendix, Fig. S4A). Further, we utilized HALLMARK genesets of key molecular processes for an unbiased evaluation of the biological relationship of terminal-Tem to memory, effector, and naive cells. Notably, the terminal-Tem subset clustered more closely to memory populations, further supporting the idea that terminal-Tem are memory T cells (Fig. 3C). Additionally, a key defining characteristic distinguishing terminal-Tem from all other CD8 T cell populations was an apparent deficiency of certain biological processes related to proliferation such as “Myc Targets,” “G2M checkpoint,” and “E2F targets” (Fig. 3C), while oxidative stress and notch signaling may be distinctive features of terminal-Tem.
Fig. 3.
Terminal-Tem are memory T cells with features of terminal effector CD8 T cells. Memory cells were generated as in Fig. 1. (A) Flow cytometric analyses of splenic P14 CD8 T cells on days 7 or 50 of infection. (B) Principal component analysis of RNA-seq data. (C) GSVA of HALLMARK genesets, wherein rows and columns are ordered by hierarchical clustering (Left) with key molecular processes unique to terminal-Tem compared with all other populations highlighted (Right). Heatmap populations are denoted by color as in B. (D) Volcano plot illustrating differentially expressed genes. (E) Heatmap of select genes. (F–I) Representative expression of key molecules on day 7 (TE/MP) or 55 post-LCMV infection. (J) KLRG1hi or KLRG1lo P14 CD8 T cells were sorted from spleens on day 7 of LCMV infection and transferred into infection-matched, congenically distinct mice (Left). Representative phenotype of donor cells and quantification of frequency of each memory population (Right) >30 d after transfer. Numbers in plots represent the frequency of cells in the indicated gate or gMFI (F–J). All data are from one representative experiment out of three independent experiments (A) or two independent experiments (F–J) with n = 3 to 4 per group; *P < 0.05, ***P < 0.005. Graphs indicate mean ± SEM, symbols represent an individual mouse (J).
Next, we sought to further understand how CD127loCD62LloKLRG1hi (i.e., seemingly terminal or short-lived cells) CD8 T cells are able to persist for extended periods of time into the memory phase of infection. We compared the transcriptome and molecular profile of TE, MP, terminal-Tem, Tem, and Tcm to begin parsing molecular characteristics distinct to terminal-Tem, with an emphasis on the relationship of TE and terminal-Tem. Focused analyses of TE and terminal-Tem populations revealed a number of unique factors that may facilitate the surprising long-lived nature of terminal-Tem, including elevated levels of Tcf7/TCF1 and Bcl2/Bcl2 as well as lower levels of Prdm1/Blimp1 compared to TE (Fig. 3 D–F). However, terminal-Tem also shared a number of characteristics with TE cells relative to Tcm or Tem, including expression of elevated levels of TE-signature molecules such as members of the killer cell lectin-like receptor family (e.g., Klrg1, Klrc1, and Klra9), Prdm1/Blimp1, and effector molecules such as granzymes and perforin (Fig. 3 D–H). Further, consistent with the biological pathway analysis (Fig. 3C) and Fig. 2C, terminal-Tem exhibited minimal homeostatic proliferation evidenced by diminished Ki-67 staining, whereas nearly all TE cells expressed Ki-67 (Fig. 3G). Taken together, terminal-Tem are more “effector-like” than Tem and Tcm but also acquire key memory characteristics compared to TE, which may allow them to persist to memory timepoints.
To understand the ontogeny of terminal-Tem, we adoptively transferred sorted KLRG1hi and KLRG1lo populations from day 7 infected mice into infection-matched recipient mice; ∼30 d postinfection, we evaluated memory T cell populations (Fig. 3J). Terminal-Tem were primarily derived from KLRG1hi cells, whereas Tem and Tcm were primarily derived from KLRG1lo cells. Thus, terminal-Tem and Tem have unexpectedly distinct developmental pathways. These studies further clarify the ontogeny of memory T cells, demonstrating that KLRG1lo effector cells are indeed the precursors of longer-lived memory cells (i.e., Tem and Tcm) but are not necessarily the originating population of all memory T cells, consistent with the finding that certain ex-KLRG1 cells can give rise to memory populations (13). Without parsing terminal-Tem from the Tem population, it would potentially be inferred that Tem are derived from both KLRG1hi and KLRG1lo effector cells.
Terminal-Tem and Tem Exhibit Differential Dependencies on Key Lineage-Specifying Transcription Factors.
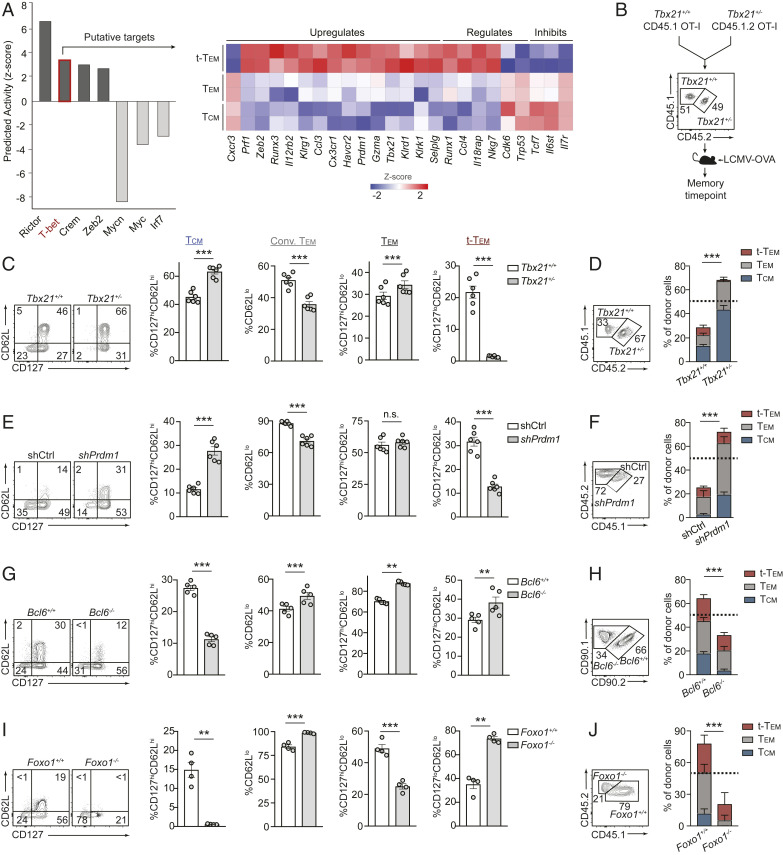
We next utilized Ingenuity Pathway Analysis software to predict transcription factors putatively supporting terminal-Tem relative to Tem and Tcm lineages (Fig. 4A). Rictor, T-bet, and Zeb2 were among the top predicted candidates promoting the terminal-Tem fate; all three factors have previously established roles in supporting TE formation (11, 17, 18, 55). T-bet is also widely considered to be essential for conventional Tem formation (3, 56); however, our computational approach predicted T-bet to be more influential in supporting terminal-Tem compared to both Tem and Tcm. To evaluate the fate-specifying role of T-bet on Tem and terminal-Tem differentiation, we cotransferred Tbx21+/+ or Tbx21+/− OT-I cells at 1:1 ratio into congenically distinct recipient mice subsequently infected with LCMV-OVA (Fig. 4B). This mixed transfer approach permitted investigation of a cell-intrinsic role for T-bet in regulating memory subset formation. Consistent with current paradigms, Tbx21 heterozygosity resulted in a reduced frequency of conventional Tem (3, 56) (Fig. 4C); however, the frequency of the redefined CD127hiCD62Llo Tem population was unexpectedly unchanged or even slightly enhanced by diminished T-bet levels (Fig. 4C). Notably, there was a dramatic loss in the frequency of terminal-Tem (Fig. 4C), concurrent with the computational predictions (Fig. 4A). On closer examination, the population frequencies did not necessarily translate to absolute abundance, and a key advantage of the cotransfer system is that it provides a sensitive method for directly comparing the relative abundance of Tbx21+/+ or Tbx21+/− memory populations given the input ratio of transferred cells was 1:1. With this analysis, we found that Tbx21 heterozygosity resulted in enhanced accumulation of total splenic OT-I cells at a memory timepoint compared to Tbx21+/+ OT-I cells (Fig. 4D), and notably, Tbx21 heterozygosity resulted in an approximately threefold increase in Tem (gray bars) but nearly a complete loss of terminal-Tem (red bars, Fig. 4D and SI Appendix, Fig. S4B). In summary, clarification of memory T cell nomenclature provided a framework for predicting critical roles for key regulatory molecules (e.g., T-bet, Zeb2, and Rictor) and allowed fine-tuning of the lineage-specifying role of T-bet, ultimately revealing that T-bet is essential for terminal-Tem but actually suppresses the formation of Tem and Tcm.
Fig. 4.
Terminal-Tem and Tem exhibit differential dependencies on key lineage-specifying transcription factors. (A) Regulatory transcription factors were predicted by Ingenuity Pathway Analysis based on gene-expression profiles of terminal-Tem, Tem, and Tcm (Left), and relative expression of key predicted targets of T-bet (Right). (B) Summary of experimental model. (C) Frequency of splenic memory CD8 T cell subsets on day 72 of infection. (D) Ratio of recovered splenic OT-I cells (Left), and the relative abundance of terminal-Tem, Tem, and Tcm. (E) Congenically distinct P14 cells were transduced with Prdm1 or Cd19 (control) shRNA encoding retroviruses, mixed 1:1, and transferred into recipient mice subsequently infected with LCMV. Frequency of splenic memory CD8 T cell subsets on day 25 of infection. (F) Ratio of recovered P14 cells (Left), and the relative abundance of each subset (Right). (G) Congenically distinct CD4-Cre-Bcl6fl/fl P14 and Bcl6fl/fl P14 cells were mixed 1:1 and transferred to recipient mice subsequently infected with LCMV. Quantification of the frequency of splenic memory CD8 T cell subsets on day 35 of infection (Right). (H) Ratio of recovered P14 cells (Left), and the relative abundance of each subset (Right). (I) Congenically distinct dLck-Cre-Foxo1fl/fl P14 and Foxo1fl/fl P14 cells were mixed 1:1 and transferred to recipient mice subsequently infected with LCMV. Frequency of splenic memory CD8 T cell subsets on day 30 of infection (Right). (J) Ratio of recovered P14 cells (Left), and the relative abundance of each subset (Right). All data are from one representative experiment of two independent experiments with n = 4 to 6 per group (C–I); n.s. not significant, **P < 0.01, ***P < 0.005. Graphs indicate mean ± SEM, and symbols represent an individual mouse.
A distinguishing characteristic of terminal-Tem included elevated expression levels of Blimp1 compared to Tem and Tcm (Fig. 3F). Similar to T-bet, Blimp1 is widely considered to be critical for Tem formation (3, 56), and given our relatively unexpected finding that T-bet was not essential for Tem formation, we next evaluated a role for Blimp1 in differentially supporting terminal-Tem and Tem populations. Prdm1 RNAi resulted in a reduced percentage of CD62Llo conventional Tem (Fig. 4E), consistent with current paradigms (3, 56). However, we found that Blimp1 deficiency did not alter the frequency of the redefined Tem population (Fig. 4E). Analogous to diminished T-bet levels, knockdown of Prdm1 resulted in a greater accumulation of total P14 cells in the spleen compared to control P14 cells at a memory timepoint (Fig. 4F), and consistent with previous reports (15, 16), Blimp1 deficiency resulted in a greater abundance of Tcm (blue bars, Fig. 4F and SI Appendix, Fig. S4B). However, despite robust Prdm1 expression in the terminal-Tem population, Prdm1 knockdown did not impair the overall formation of terminal-Tem (red bars) and actually enhanced the accumulation of Tem (gray bars) in the spleen (Fig. 4F). Therefore, we also clarify the regulatory role of Blimp1 as a suppressor of Tem and Tcm formation.
We next examined the memory lineage-specifying roles of Bcl6 and Foxo1, transcription factors canonically recognized to support Tcm formation relative to Tem (3, 25–29). As expected, loss of Bcl6 or Foxo1 resulted in a reduced frequency of Tcm (3, 25–28) (Fig. 4 G and I). However, Bcl6 deficiency resulted in the enhanced frequency of Tem and terminal-Tem, whereas Foxo1 deficiency resulted in a lower frequency of Tem but increased frequency of terminal-Tem. Converse to the effects of diminished expression of T-bet or Blimp1, the relative abundance of donor cells revealed conditional deletion of Bcl6 or Foxo1 resulted in a loss of total memory P14 cells, including fewer Tcm (blue bars) compared to control P14 cells (Fig. 4 H and J). Unexpectedly, we also found that loss of Foxo1 also resulted in a reduced abundance of CD127hiCD62Llo Tem (Fig. 4 H and J and SI Appendix, Fig. S4B). Therefore, clarification of memory T cell nomenclature has implications for how the roles of critical fate-specifying transcription factors are understood. In summary, we established T-bet as a central regulator of terminal-Tem, demonstrated that Foxo1 was unexpectedly essential for optimal Tem formation, and found that T-bet and Blimp1 may actually suppress development of Tem.
Molecular Profile of Human Terminal-Tem Revealed by Single-Cell RNA-Seq.
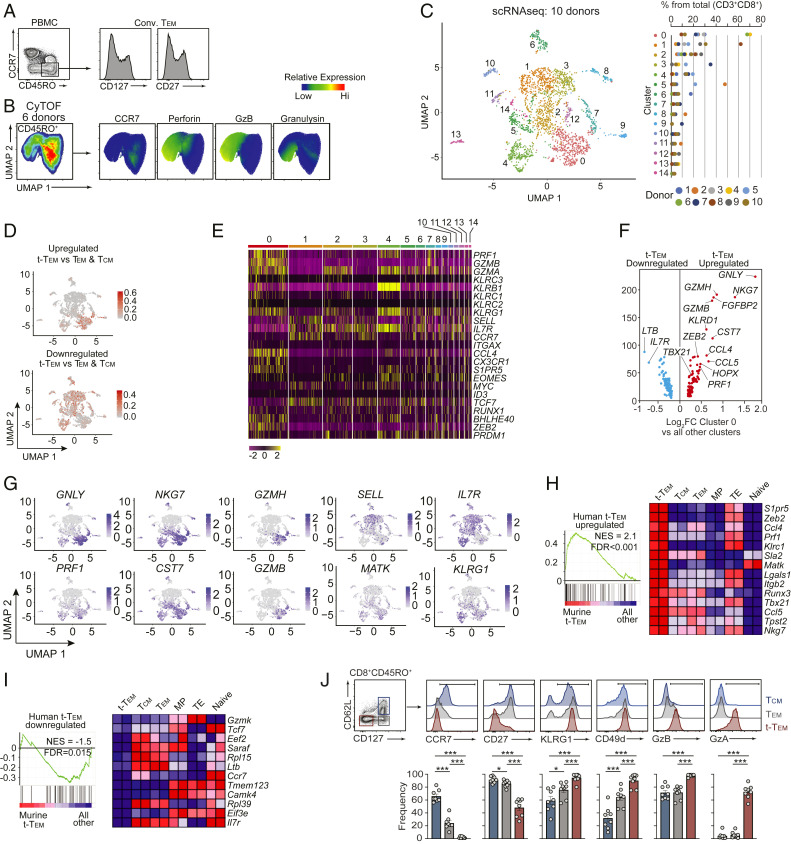
Although molecules such as CD28, CD27, and CD43 have been used to subset human CD8 T cells (46), circulating memory T cell populations in humans are most often distinguished based on expression of CD45RO (or CD45RAlo) and CCR7 expression, wherein CD45ROhiCCR7hi cells are considered Tcm and CD45ROhiCCR7lo are Tem (3, 5, 31). Utilizing this paradigm for subsetting human memory T cells, we found that conventionally defined CD45ROhiCCR7lo Tem exhibit heterogeneous expression levels of CD127 and CD27 (Fig. 5A), analogous to conventionally defined mouse Tem (Fig. 1A). Further, multiparameter mass cytometry (CyTOF) analysis of human peripheral blood mononuclear cells (PBMCs) from six healthy donors also demonstrated heterogeneous expression of effector molecules perforin, granzyme B, and granulysin within the CCR7lo memory CD8 T cell population (Fig. 5B and SI Appendix, Fig. S5 A and B), supporting the presence of a functionally distinct human terminal-Tem population that would otherwise be grouped into conventionally defined Tem.
Fig. 5.
Molecular phenotype of human terminal-Tem. (A) Representative expression levels of CD127 and CD27 in conventional Tem from 8 healthy donors. (B) PBMCs isolated from 6 healthy donors were analyzed by mass cytometry. Data were combined from all 6 donors, down-sampled to 3 × 104 events, and UMAP highlighting heterogeneity of pregated CD45+CD3+CD8β+CD45RO+ T cells was constructed (Left). (C–I) Single-cell RNA-seq analysis was performed on PBMCs from 10 healthy donors. (C) CD8 T cells were prefiltered based on expression of CD3D, CD3E, CD8A, and CD8B, and unbiased UMAP analysis revealed 15 distinct clusters (Left). Frequency of each cluster per donor. (D) Relative enrichment of the murine-derived terminal-Tem gene-expression signature (compared to both Tem and Tcm, see SI Appendix, Fig. S6B). (E) Relative expression of key molecules between the 15 distinct clusters from C. (F) Differential expression of highlighted genes uniquely up-regulated or down-regulated in cluster 0. (G) Relative expression of indicated genes from C. (H) GSEA of the human terminal-Tem gene-expression signature in murine populations, and (Right) relative expression in murine populations of leading edge genes up-regulated in both murine and human terminal-Tem. (I) GSEA of the human nonterminal-Tem gene-expression signature in murine populations. (J) Representative gating of human terminal-Tem, Tem, and Tcm populations. All data are from one representative experiment of 8 donors (A and J), 6 donors (B), and 10 donors (C–I). *P < 0.05, ***P < 0.005. Graphs indicate mean ± SEM, and symbols represent an individual donor. NES, normalized enrichment score; FDR, false discovery rate.
As there is often a disconnect between “markers” used to distinguish murine memory populations and human populations, we utilized single-cell RNA-seq analysis for unbiased identification of human terminal-Tem. We performed single-cell RNA-seq analysis on PBMCs from 10 healthy individuals (Fig. 5C and SI Appendix, Fig. S6A) as well as utilized a publicly available human PBMC dataset (SI Appendix, Fig. S7 A–D). Through these analyses, we detected extensive heterogeneity and diverse CD8 T cell states ranging from naive to memory populations yielding a total of 15 discrete subpopulations (Fig. 5C). We identified cluster “0” as enriched with the mouse-generated terminal-Tem gene signature as well as partial enrichment in clusters “2,” “7,” and “4” (Fig. 5D and SI Appendix, Fig. S6B). Gene-expression analysis of key regulatory molecules also supported the finding that cluster 0 likely represents a terminal-Tem population as this cluster exhibited up-regulation of cytotoxic molecules (GZMA, GZMB, and PRF1), members of the killer cell lectin-like receptor family (KLRC3, KLRB1, KLRC1, KLRC2, and KLRG1), CX3CR1, CCL4, S1PR5, BLHE40, ZEB2, and PRDM1 as well as relatively low levels of SELL, IL7R, CCR7, MYC, ID3, and TCF7 (Fig. 5 E–G). Further, through gene-set enrichment analyses we confirmed that the cluster 0 gene-expression signature (i.e., genes uniquely up-regulated in cluster 0 compared to all other clusters) was enriched in murine terminal-Tem (compared to MP, TE, Tcm, Tem, and naive cells) (Fig. 5H) and a gene-expression signature of transcripts uniquely down-regulated in human terminal-Tem was depleted from murine terminal-Tem compared to other memory, effector, and naive populations (Fig. 5I). Last, in analyses of PBMCs from eight healthy donors, we found that subsetting human memory (CD45RO+) CD8 T cells based on CD127 and CD62L expression distinguishes a human terminal-Tem population, and this approach more clearly delineates human memory populations compared to CCR7 expression levels alone (Fig. 5J). These data are consistent with profiling of murine terminal-Tem in that our analyses of human CD8 T cells revealed terminal-Tem to be transcriptionally and phenotypically distinct from Tem and Tcm populations.
Discussion
Defining functional and phenotypic memory CD8 T cell states is valuable for understanding the secondary immune response and the molecular regulation of memory T cell differentiation. For example, the detection of a given subset can be informative for predicting memory T cell protection in the context of a given pathogen or cancer (4, 42, 48, 54, 56–58). As previously recognized by others (16, 41, 47, 48), we noted heterogeneity within CD62Llo memory T cells, including a population of CD127loCD62Llo cells able to persist for several months after infection; this finding prompted the investigation and clarification of the CD62Llo memory T cell compartment. Here, we refine the definition of conventional CD62Llo Tem to include expression of CD127 (16, 44, 47, 48, 59), ultimately allowing parsing and characterization of a CD127loCD62Llo terminally differentiated Tem population (i.e., terminal-Tem) separate from Tem. A key finding from this study, distinct from prior investigations of long-lived or persisting CD8 T cells (41–44), was that our revised framework for examining circulating memory cells permitted an improved understanding of Tem biology as well as clarification of the roles of widely studied lineage-specifying transcription factors.
Human memory T cells with discrete lymphoid homing properties were first described by Sallusto et al. (5), yielding the definition of canonical memory T cell subsets Tcm and Tem. While the ontogeny and defining attributes of Tcm and Tem have been somewhat contentious (2–4, 60), general features of Tcm include heightened presence in secondary lymphoid organs, greater recall potential, enhanced IL-2 production, and greater homeostatic fitness (proliferation and survival) (2, 56, 61). Our studies confirm previous findings describing Tcm but redefine the attributes of the Tem population. Conventional Tem are characteristically considered to be more cytolytic, have a greater capacity to traffic to infected sites, have limited recall proliferation potential, generally accepted to be more terminally fated, and do not persist to the same extent as Tcm (2, 4, 56, 61). Through our refining of Tem identity, we reveal that Tem exhibit an enhanced presence in lymphoid tissues, more robust IL-2 production and recall potential, greater than expected homeostatic fitness (i.e., elevated homeostatic proliferation and expression of survival molecules such as Bcl2), heightened multipotency with the ability to give rise to Tcm, and a distinct molecular phenotype compared to prior paradigms (2, 3). These discrete functional attributes make certain memory populations better suited for handling specific infections, wherein previous studies have demonstrated that Tcm are superiorly equipped to provide protection against systemic LCMV Cl13 infection (4, 33) and malignancy (54), whereas conventional Tem confer enhanced protection to vaccinia virus infection (48) and in some cases Listeria infection (53). We found that terminal-Tem and redefined CD127hiCD62Llo Tem conferred equivalent protection to Listeria infection, but terminal-Tem conferred the most robust protection on a per cell basis. Although terminal-Tem displayed higher expression of cytolytic genes and up-regulation of granzymes, they also exhibited limited recall proliferation, decreased lymphoid tissue presence, and impaired cytokine production compared to Tem. Conversely, Tcm were most protective in a tumor vaccine model compared to both Tem and terminal-Tem, likely due to an enhanced lymph node homing capacity (resulting in increased access to vaccine antigen) (54) and heightened recall proliferation. Taken together, the relative protection afforded by each memory T cell subset is dependent on the context of the disease or therapy, and the contribution of each subset is a culmination of unique molecular features impacting their location, expansion, survival, and functional activity. Resolving the terminal-Tem population provides clarity to this notion and may impact future immunotherapy approaches. In connection, we have also identified putative markers of human terminal-Tem that could inform vaccination studies or provide insight for pinpointing therapeutic or pathogenic roles for each memory state in a given therapy or disease setting.
Through profiling of terminal-Tem gene-expression patterns, we demonstrated that terminal-Tem were transcriptionally distinct from Tem, Tcm, and Trm. Further, despite a phenotypic resemblance to TE cells, terminal-Tem were more transcriptionally related to memory cells and sort-retransfer experiments demonstrated terminal-Tem are able to persist for extended periods of time (although not to the same extent as Tem and Tcm). Therefore, we conclude that terminal-Tem are a genuine memory T cell population with robust features of TE cells. We also found that compared to TE, terminal-Tem exhibited up-regulation of prosurvival molecule Bcl2 as well as promemory transcription factors TCF1 and Bcl6, providing a possible explanation of their prolonged survival and potentially facilitating a dual terminal effector/memory phenotype. Although our data suggest terminal-Tem are also distinct from TE, this conclusion comes with the caveat that differences in TE and terminal-Tem may partially be driven by differing levels of antigen and inflammation. In connection, we refine paradigms of transcription factor-mediated regulation of memory T cell fate. To date, T-bet (11), Blimp1 (15, 16), Zeb2 (17, 18), Stat4 (19), and Id2 (20–23) have been considered essential for the formation of TE or conventional Tem cells, whereas Eomes (24), Bcl6 (25, 26), Foxo1 (27–29), Stat3 (26), and Id3 (22, 30) have been linked to the differentiation of MP or Tcm (3). Here, we demonstrated that optimal formation of the redefined Tem population was not impaired by diminished T-bet or Blimp1 expression. Further, we also highlighted that Tem actually require the promemory transcription factors Bcl6 and Foxo1. Notably, we found that loss of T-bet and Bcl6 resulted in a reduced abundance of terminal-Tem.
The physiological role of memory T cells has been linked to both the resolution as well as the instigation of disease (1, 3, 14, 62, 63). The overall protective or pathogenic impact of memory T cells for a given disease setting is determined by both the quantity and quality of responses, which is likely dependent on the overall makeup and phenotype of the memory T cell compartment. While it is well established that the circulating memory CD8 T cell population displays meaningful heterogeneity, it is also clear that T cells can exist in a broad spectrum of cell states rather than discrete subsets (1), complicating categorization of memory T cells into distinct lineages. Indeed, compartmentalization of terminal-Tem, Tem, and Tcm does not fully capture circulating memory CD8 T cell heterogeneity. Despite their terminal nature, we find that ∼20% of terminal-Tem are capable of giving rise to CD127hi memory cells approximately 1 mo following retransfer (compared to >85% CD127hi for Tcm and Tem donors). This finding reflects that our approach does not completely enompass all aspects of circulating T cell heterogeneity, as it is likely a rare population of KLRG1lo cells within the terminal-TEM pool is capable of up-regulation of CD127. Nonetheless, defining distinct states and delineating characteristics of terminal-Tem, Tem, and Tcm emphasize the division of labor within memory T cell populations and highlight how the respective attributes of each subset may be complimentary both temporally and anatomically. Clarification of memory T cell nomenclature and lineage-specific characteristics provides insight for understanding the complex activity and molecular regulation of memory cell states and holds implications for the timing and delivery of immunotherapies.
Materials and Methods
Mice.
All mouse strains were bred and housed in pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California San Diego (UCSD). P14 mice (with transgenic expression of H-2Db-restricted TCR specific for LCMV glycoprotein33–41), OT-I mice (with transgenic expression of H-2Kb-restricted TCR specific for ovalbumin257–264), Tbx21+/− mice, Bcl6fl/flCd4Cre, Bcl6fl/flErt2Cre mice, Foxo1fl/fldLckCre mice, Blimp1-YFP reporter mice, CD45.1 mice, and Thy1.1 mice were purchased from The Jackson Laboratory and bred at UCSD.
Cell Transfers and Infections.
To generate memory T cells, naive P14 or OT-I CD8 T cells (5 × 104) were adoptively transferred into congenically distinct recipient mice, which were infected the next day with 2 × 105 plaque-forming unit (PFU) LCMV-Armstrong i.p. or with 8 × 104 CFU L. monocytogenes-OVA (LM-OVA) i.v. For KLRG1hi/KLRG1lo transfers, P14 effector cells were sorted from spleens based on KLRG1 expression on day 7 of infection and 1 × 106 cells were transferred into congenically distinct, infection-matched mice. For fate studies, memory P14 cells were sorted >30 d postinfection from spleens based on CD62L and CD127 expression, and 1 × 105 cells were transferred into naive recipients.
For rechallenge assays, memory OT-I subsets were sorted from recipient spleen and lymph nodes at >30 d after LCMV-OVA infection and 1 × 105 cells were transferred i.v. into congenically distinct naive recipients that were subsequently infected with 8 × 104 CFU LM-OVA i.v. On day 3 of infection, spleens from recipient mice were homogenized in 0.2% igepal (Sigma-Aldrich) and serial dilutions in brain heart infusion (BHI) media were plated onto BHI plates in the presence of antibiotic selection. Colonies were counted the following day and normalized to splenic weight. For tamoxifen-induced deletion, 1 mg of tamoxifen (Cayman Chemical Company) was emulsified in 100 μL of sunflower seed oil (Sigma-Aldrich) and administered i.p. from days 2 to 7 after infection.
To assess the activity of each memory subset in a tumor vaccine model, 5 × 105 B16-GP33–41 cells were transplanted s.c. into naive recipient mice and allowed to progress for 5 d. Next, 7.5 × 104 congenically distinct cells of each sorted memory population were adoptively transferred and tumor-bearing mice were subsequently immunized with 10 μg of GP33–41 and 2 μg of poly(I:C) (GE Healthcare) s.c. into the ipsilateral flank. Tumors were monitored daily and mice with ulcerated tumors or tumors exceeding 1,500 mm3 were euthanized.
Antibodies, Flow Cytometry, and Cell Sorting.
CD8 T cell subsets were first gated on congenically distinct P14 cells or CD44+Tetramer+ cells (as indicated), and then defined as terminal-Tem: CD127loCD62Llo; Tem: CD127hiCD62Llo; Tcm: CD127hiCD62Lhi; conventional Tem: CD62Lhi; TE: D7 KLRG1hiCD127lo; and MP: D7 KLRG1loCD127hi. Detailed information on antibodies, flow cytometry, and cell sorting can be found in SI Appendix, Materials and Methods.
RNAi Studies.
shRNAmirs targeting mouse Prdm1 or Cd19 (control) cloned into a pLMPd-Amt vector were utilized, and retroviral supernatants were generated as previously described (32). For transductions, naive P14 cells from spleen and lymph nodes were enriched via negative selection using MACS magnetic beads according to the manufacturer’s protocol (Miltenyi Biotec). Enriched P14 cells (2 × 106) were activated in six-well plates coated with 100 μg/mL goat anti-hamster IgG (H+L, Thermoscientific), 1 μg/mL anti-CD3 (145–2C11), and 1 μg/mL anti-CD28 (37.51) (eBioscience) for 18 h. Culture media was replaced with retroviral supernatant supplemented with of 50 μM beta-mercaptoethanol and 8 μg/mL polybrene (Millipore), and cells were centrifuged for 60 min at 2,000 rpm, 37 °C. After 24 h, P14 cells transduced with Prdm1 shRNAmir encoding- or Cd19 shRNAmir encoding-retroviruses were mixed at 1:1 ratio and a total of 5 × 105 cells were transferred to congenically distinct recipient mice that were infected with LCMV-Armstrong strain.
RNA-Seq Analysis.
On day 55 of LCMV-Armstrong infection, 1 × 103 splenic Tem (CD127hiCD62Llo) P14 cells were sorted into TCL buffer (Qiagen) with 1% 2-mercaptoethanol. RNA was isolated and RNA-seq library preparation was carried out as per Immgen Protocols (https://www.immgen.org/Protocols/11Cells.pdf). D7 MP and TE as well as naive RNA-seq datasets were previously published (32), and terminal-Tem (CD127loCD62Llo) and conventional Tem (CD62Llo), Tcm (CD127hiCD62Lhi) and small intestine epithelial Trm (CD103hiCD62Llo) datasets at D55 are from GSE147080 and were sorted in the same experiment as Tem (CD127hiCD62Llo). Gene expression analysis was performed in the Multiplot Studio module within Genepattern (with >1.5 fold differential expression cutoff and expression threshold >10). Heatmaps were generated using Morpheus software (https://software.broadinstitute.org/morpheus). K-means clustering was set according to three groups, maximum number of iterations = 1,000. Hexagonal plots as well as rose plots were created with log2 transformed data using the Triwise R package and following https://zouter.github.io/triwise/rd.html. PCA was performed in R studio using Deseq2 normalized expression values. GSVA scores for each sample were calculated using gene set variation analysis (GSVA) R module (64) and the Hallmarks v7 geneset collection (MSigDb, Broad Institute). Unsupervised clustering was performed with one minus Pearson correlation metric. Gene set enrichment analysis (GSEA) was run with a signal-to-noise ratio for metric ranking, 1,000 permutations based on geneset and the Mouse Gene Symbol Remapping MSigDB.v7.0 chip. “Memory” and “effector” genesets were generated from analysis of populations from the Immgen database (www.immgen.org); specifically, D8 OT-I and D100 OT-I cells from LM-OVA mice were compared and memory genes were defined as up-regulated in the D100 population relative to the D8 population and effector genes were down-regulated in the D100 population relative to the D8 population. The “proliferation” geneset was from module 2 of Best et al. (52). For computational prediction of putative regulators of terminal-Tem (Fig. 4A), the Inguenity Pathway Analysis (Qiagen, https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/) was used. Naive P14, LCMV D7 TE, LCMV D7 MP RNA-seq datasets are from Milner et al. (32).
Collection of Human PBMCs and Mass Cytometry (CyTOF).
The Human Research Protection Programs at UCSD and the San Diego VA Healthcare System approved the study. Collection of human PBMCs and details on mass cytometry experiments are further detailed in SI Appendix, Materials and Methods.
Single-Cell RNA-Seq Analysis.
Cells were washed and resuspended in phosphate buffered saline + 0.04% bovine serum albumin. Single-cell libraries were prepared according to the protocol for 10× Genomics Chromium Single Cell Gene Expression. Approximately 2 × 104 sorted CD45+ cells were loaded and partitioned into Gel Bead In-Emulsions (GEMs). scRNA libraries were sequenced on a HiSeq4000 (Illumina). scRNAseq analysis was performed using cellranger software and the developmental version of Seurat 3.1 (65, 66) in R Studio. Cellranger was used with default parameters with the exception of cellranger aggr mapped = none. Seurat analysis of 10× count matrices was done by following these steps: low-quality cells, identified by percent mitochondria <15, nFeature_RNA < 200 or >2,000, were removed, counts were normalized with SCTransform, dimensionality reduction and cluster identification were done with uMAP (dims = 1:40), FindNeighbors (dims = 1:40), and FindClusters (resolution = 1). The 10K PBMC scRNAseq dataset was downloaded from https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/pbmc_10k_protein_v3. CD8 T cells were subsetted based on the expression of CD3D, CD3E, CD8A, and CD8B expression. Differential expression of genes per cluster was done with FindAllMarkers function with default parameters and min.pct = 0.1 and logfc.threshold = 0.2.
Statistics.
Two-tailed, paired and unpaired Student’s t test was used for comparisons between two groups. Statistical analysis was performed using GraphPad Prism software. All statistical tests were performed with GraphPad Prism software. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This study was supported by NIH P01AI13212 (A.W.G. and J.T.C.), NIH U19AI109976 (A.W.G.), the Kenneth Rainin Foundation (J.T.C.), NIH K99/R00 CA234430 (J.J.M.), T32DK007202 (M.T.), CRI Irvington Postdoctoral Fellowship (M.R.-C.), and S10OD018499 (CyTOF La Jolla Institute of Immunology).
Footnotes
Competing interest statement: A.W.G. serves on the scientific advisory boards of Pandion Therapeutics and Arsenal Bio.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008571117/-/DCSupplemental.
Data Availability.
All data discussed in this paper are included within the article and SI Appendix, and are deposited and accessible in Gene Expression Omnibus (GEO) (accession number GSE157072).
References
- 1.Jameson S. C., Masopust D., Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang J. T., Wherry E. J., Goldrath A. W., Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wherry E. J., et al. , Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4, 225–234 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A., Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Mueller S. N., Gebhardt T., Carbone F. R., Heath W. R., Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31, 137–161 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Masopust D., Vezys V., Marzo A. L., Lefrançois L., Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Steinert E. M., et al. , Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slütter B., et al. , Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2, eaag2031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaech S. M., et al. , Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Joshi N. S., et al. , Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar S., et al. , Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herndler-Brandstetter D., et al. , KLRG1+ effector CD8+ T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity 48, 716–729.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milner J. J., Goldrath A. W., Transcriptional programming of tissue-resident memory CD8+ T cells. Curr. Opin. Immunol. 51, 162–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutishauser R. L., et al. , Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallies A., Xin A., Belz G. T., Nutt S. L., Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 31, 283–295 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Dominguez C. X., et al. , The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omilusik K. D., et al. , Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212, 2027–2039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mollo S. B., Ingram J. T., Kress R. L., Zajac A. J., Harrington L. E., Virus-specific CD4 and CD8 T cell responses in the absence of Th1-associated transcription factors. J. Leukoc. Biol. 95, 705–713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omilusik K. D., et al. , Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8+ T cells. J. Exp. Med. 215, 773–783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannarile M. A., et al. , Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7, 1317–1325 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Yang C. Y., et al. , The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knell J., et al. , Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J. Immunol. 190, 1501–1509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee A., et al. , Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185, 4988–4992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z., et al. , Cutting edge: Transcription factor BCL6 is required for the generation, but not maintenance, of memory CD8+ T cells in acute viral infection. J. Immunol. 203, 323–327 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Cui W., Liu Y., Weinstein J. S., Craft J., Kaech S. M., An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35, 792–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess Michelini R., Doedens A. L., Goldrath A. W., Hedrick S. M., Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med. 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M. V., Ouyang W., Liao W., Zhang M. Q., Li M. O., The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39, 286–297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utzschneider D. T., et al. , Active maintenance of T cell memory in acute and chronic viral infection depends on continuous expression of FOXO1. Cell Rep. 22, 3454–3467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Y., et al. , Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12, 1230–1237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farber D. L., Yudanin N. A., Restifo N. P., Human memory T cells: Generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 14, 24–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milner J. J., et al. , Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolz J. C., Harty J. T., Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity 34, 781–793 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klonowski K. D., et al. , CD8 T cell recall responses are regulated by the tissue tropism of the memory cell and pathogen. J. Immunol. 177, 6738–6746 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obar J. J., Lefrançois L., Early signals during CD8 T cell priming regulate the generation of central memory cells. J. Immunol. 185, 263–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z., et al. , miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep. 20, 2584–2597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., et al. , The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skon C. N., et al. , Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngblood B., et al. , Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552, 404–409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakaradov B., et al. , Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renkema K. R., et al. , KLRG1+ memory CD8 T cells combine properties of short-lived effectors and long-lived memory. J. Immunol. 205, 1059–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson J. A., McDonald-Hyman C., Jameson S. C., Hamilton S. E., Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38, 1250–1260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerlach C., et al. , The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Böttcher J. P., et al. , Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat. Commun. 6, 8306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slütter B., Pewe L. L., Kaech S. M., Harty J. T., Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity 39, 939–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamann D., et al. , Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186, 1407–1418 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huster K. M., et al. , Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. U.S.A. 101, 5610–5615 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann M. F., Wolint P., Schwarz K., Jäger P., Oxenius A., Functional properties and lineage relationship of CD8+ T cell subsets identified by expression of IL-7 receptor alpha and CD62L. J. Immunol. 175, 4686–4696 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Yu B., et al. , Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 705–705 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Schenkel J. M., Masopust D., Tissue-resident memory T cells. Immunity 41, 886–897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casey K. A., et al. , Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Best J. A. et al.; Immunological Genome Project Consortium , Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huster K. M., et al. , Unidirectional development of CD8+ central memory T cells into protective Listeria-specific effector memory T cells. Eur. J. Immunol. 36, 1453–1464 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Klebanoff C. A., et al. , Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc. Natl. Acad. Sci. U.S.A. 102, 9571–9576 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L., et al. , Mammalian target of rapamycin complex 2 controls CD8 T cell memory differentiation in a foxo1-dependent manner. Cell Rep. 14, 1206–1217 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Martin M. D., Badovinac V. P., Defining memory CD8 T cell. Front. Immunol. 9, 2692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klebanoff C. A., Gattinoni L., Restifo N. P., Sorting through subsets: Which T-cell populations mediate highly effective adoptive immunotherapy? J. Immunother. 35, 651–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang X., et al. , Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 483, 227–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackay L. K., et al. , The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Manjunath N., et al. , Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J. Clin. Invest. 108, 871–878 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin M. D., et al. , Phenotypic and functional alterations in circulating memory CD8 T cells with time after primary infection. PLoS Pathog. 11, e1005219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park C. O., Kupper T. S., The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 21, 688–697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devarajan P., Chen Z., Autoimmune effector memory T cells: The bad and the good. Immunol. Res. 57, 12–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this paper are included within the article and SI Appendix, and are deposited and accessible in Gene Expression Omnibus (GEO) (accession number GSE157072).