Significance
Understanding of how ClpA and other double-ring AAA+ enzymes perform mechanical work is limited. Using site-specific cross-linking and mutagenesis, we introduced ATPase-inactive AAA+ modules at alternating positions in individual ClpA rings, or in both rings, to investigate potential active-site coordination during ClpAP degradation. ClpA variants containing alternating active/inactive ATPase modules processively unfolded, translocated, and supported ClpP degradation of protein substrates with energetic efficiencies similar to, or higher than, completely active ClpA. These results impact current models describing the mechanisms of AAA+ family enzymes. The cross-linking/mutagenesis method we employed will also be useful for answering other structure-function questions about ClpA and related double-ring enzymes.
Keywords: AAA+ protease, ClpAP, double-ring ATPase, molecular machine, cysteine cross-linking
Abstract
ClpA is a hexameric double-ring AAA+ unfoldase/translocase that functions with the ClpP peptidase to degrade proteins that are damaged or unneeded. How the 12 ATPase active sites of ClpA, 6 in the D1 ring and 6 in the D2 ring, work together to fuel ATP-dependent degradation is not understood. We use site-specific cross-linking to engineer ClpA hexamers with alternating ATPase-active and ATPase-inactive modules in the D1 ring, the D2 ring, or both rings to determine if these active sites function together. Our results demonstrate that D2 modules coordinate with D1 modules and ClpP during mechanical work. However, there is no requirement for adjacent modules in either ring to be active for efficient enzyme function. Notably, ClpAP variants with just three alternating active D2 modules are robust protein translocases and function with double the energetic efficiency of ClpAP variants with completely active D2 rings. Although D2 is the more powerful motor, three or six active D1 modules are important for high enzyme processivity, which depends on D1 and D2 acting coordinately. These results challenge sequential models of ATP hydrolysis and coupled mechanical work by ClpAP and provide an engineering strategy that will be useful in testing other aspects of ClpAP mechanism.
Enzymes of the AAA+ (ATPases associated with various cellular activities) superfamily harness the chemical energy from ATP hydrolysis to remodel macromolecules in all kingdoms of life (1, 2). Simple AAA+ proteases consist of a hexameric AAA+ unfoldase and a self-compartmentalized peptidase, encoded either in a single polypeptide chain (e.g., Lon or FtsH) or as distinct proteins (e.g., ClpAP, ClpXP, or HslUV). These proteases recognize protein substrates via specific peptide degradation tags (degrons) and then, in energy-dependent reactions, unfold and processively translocate substrates through the central channel of the unfoldase into the degradation chamber of the peptidase (Fig. 1A) (3, 4). ATP-dependent unfoldases and related protein-remodeling machines differ in having either one or two AAA+ modules per subunit. Hexamers with one module per subunit form a single ring with 6 ATPase active sites, whereas those with two modules form a double-ring enzyme with 12 active sites.
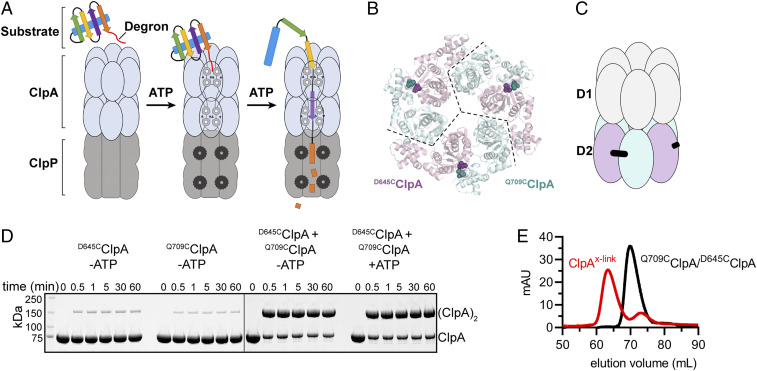
Fig. 1.
Engineered cross-linked ClpA hexamers allow sequence changes in individual subunits. (A) ClpAP recognizes a degron in a protein substrate and then uses cycles of ATP hydrolysis to unfold the substrate and translocate it into the inner chamber of ClpP for degradation. (B) Homology model of the ClpA D2 hexameric ring (cartoon representation) with potential sites for subunit cross-linking across the dimer interface shown in CPK. Pink D2 modules contain D645C and teal D2 modules contain Q709C mutations for cross-linking. (C) Diagram of the cross-linked ClpA pseudohexamer with D2 cross-linking modules (teal and pink) and cross-link (black) between engineered cysteines. (D) Bismaleimide cross-linking time course assayed by SDS/PAGE in the presence or absence of ATP. (E) Separation of cross-linked dimers from uncross-linked species by size-exclusion chromatography. After cross-linking of D645CClpA and Q709CClpA (red trace), ClpAx dimers eluted in a peak centered near ∼63 mL, whereas uncross-linked monomers eluted in a peak near ∼73 mL. Uncross-linked D645CClpA or Q709CClpA (black trace) elutes as a mixture of multimers with a peak near ∼71 mL.
Escherichia coli ClpAP, a double-ring AAA+ protease, consists of the ClpA6 unfoldase and the ClpP14 peptidase (5–7). Each ClpA monomer has an N-domain and two AAA+ modules, termed D1 and D2, which belong to evolutionarily distinct clades (1, 8, 9). Contributions of the D1 and D2 rings toward overall ClpA function have been investigated by eliminating ATP hydrolysis in one ring or the other via mutation of the catalytic residues in each AAA+ module. These studies suggest that the D1 and D2 rings hydrolyze ATP independently, with D2 catalyzing 80–90% of the total ATP hydrolysis (10–12). In addition to being the major ATPase component of ClpA, the D2 ring is also the principal unfoldase and translocase: a variant with an ATPase-inactive D2 ring (but an ATPase-active D1 ring) fails to unfold and translocate many protein substrates (12, 13). In contrast, the D1 ring functions as a regulatory/auxiliary motor that assists ATP hydrolysis-coupled mechanical work performed by D2 (14). Despite these insights, we do not know how ATP hydrolysis-coupled work in individual AAA+ modules is coordinated to promote ClpA unfolding and translocation.
Here, we use site-specific cysteine cross-linking and mutagenesis to engineer ClpA hexamers with alternating ATPase-active and ATPase-inactive subunits in the D1 ring, the D2 ring, or both rings to investigate coordination of activities between individual subunits and the two rings. We find that AAA+ modules in both rings operate as units where the number of active subunits, but not their positions or relative orientations, affects ring activity. Moreover, degradation and ATPase experiments provide evidence for cooperative action within and between the D1 and D2 rings during the processing of protein substrates. These results challenge sequential models of ATP hydrolysis and coupled mechanical work by ClpA and demonstrate how cross-linking and mutagenesis can be used to interrogate AAA+ enzyme mechanism.
Results
Design and Formation of Cross-Linked ClpA Dimers.
Covalent linkage of AAA+ subunits, either through genetic fusion or cysteine cross-linking, has been used to probe the contributions of individual subunits toward AAA+ enzyme function in the E. coli ClpXP and HslUV proteases, the PspF transcriptional regulator, and the Thermus thermophilus ClpB disaggregase (15–18). Genetic fusion is most successful when the C terminus of one subunit is close in space to the N terminus of a neighboring subunit, which is not the case for ClpA (15). Hence, we developed a cysteine-cross-linking strategy to investigate the potential coordination among the 12 ATPase active sites of ClpA.
When this work was initiated, there were no reported high-resolution structures of ClpA hexamers. Thus, we used homology modeling to identify potential sites to cross-link neighboring ClpA subunits. We generated several homology models using SWISS-MODEL based on structural alignment of the ClpA D1 or D2 modules from a monomeric ClpA structure (Protein Data Bank [PDB] ID code 1R6B) to structures of hexameric AAA+ unfoldases (19, 20). The ClpA D2 modules could be aligned reasonably well with a hexameric HslU structure (PDB ID code 1G41). Using this model and the Disulfide-by-Design algorithm, we identified Asp645 and Gln709 as potential sites for cysteine substitutions predicted to allow cross-linking between D2 dimer interfaces within a ClpA hexamer (21). Fig. 1B shows a model of a ClpA hexamer in which three subunits harbor the D645C mutation (pink) and three subunits contain the Q709C mutation (teal). Because D1 and D2 are in the same polypeptide chain, successful cysteine cross-linking at these two positions would generate a pseudohexamer consisting of a trimer of cross-linked dimers (Fig. 1C). In two recent cryo-electron microscopy (EM) structures of substrate-engaged ClpAP (6W1Z and 6W21) (22), the distance between the Cβ atoms of residues 645 and 709 in adjacent subunits of ClpA was too long (6–8.6 Å) to form a disulfide bond (∼5.5 Å) but close enough for cross-linking using a 10.9-Å bismaleimide cross-linker.
ClpA purified in the absence of ATP exists in an equilibrium of different multimeric states (23, 24). To ensure a single cross-link forms between neighboring subunits, the three native cysteines in WT ClpA (ClpAwt) were changed to serines (C47S, C203S, C243S) to generate a cysteine-free variant (ClpAcf) before introducing the D645C or Q709C mutations to produce D645CClpA or Q709CClpA, respectively. D645CClpA and Q709CClpA were combined, incubated to allow subunit mixing, and then treated with 1,4-bismaleimidobutane. Analysis by reducing SDS/PAGE indicated that cross-linking of the D645CClpA/Q709CClpA mixture was highly specific and ∼80% efficient (with or without ATP), whereas only 5–8% cross-linking was observed with either D645CClpA or Q709CClpA alone (Fig. 1D). Thus, most cross-linking occurred between the two cysteines in neighboring subunits in a hexamer, as predicted from the design. The cross-linked D645CClpA–Q709CClpA dimer was purified from uncross-linked species by size-exclusion chromatography under nucleotide-free conditions that do not support hexamer formation (Fig. 1E). The pseudohexamer (ClpAx) was then assembed from cross-linked dimers by ATP addition for all subsequent activity assays.
Construction of ClpA Complexes to Probe ATPase-Module Function.
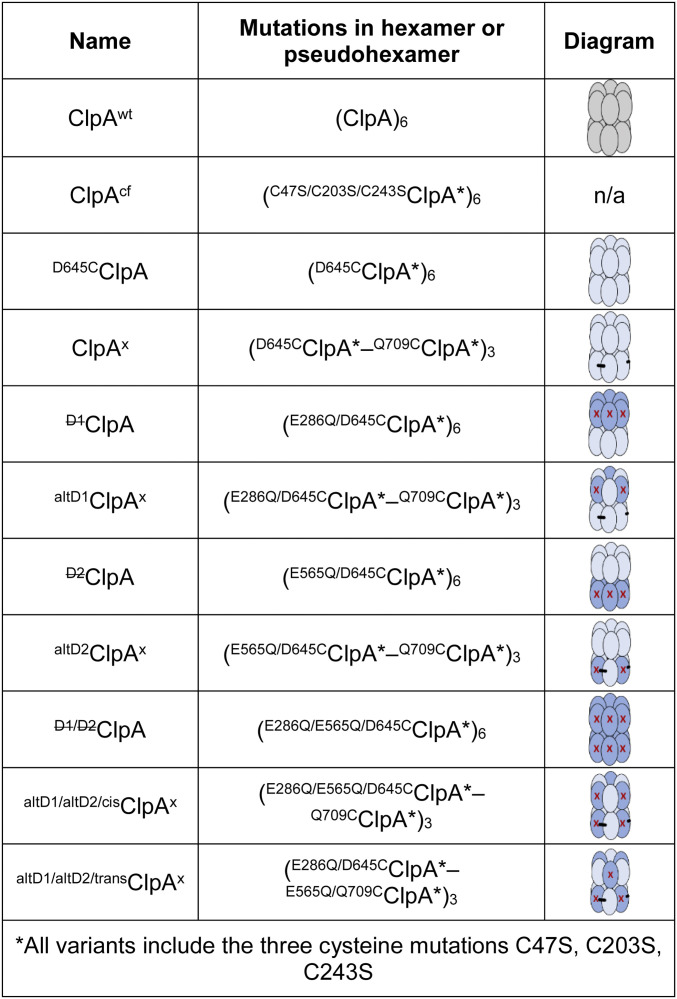
Mutation of the catalytic Walker-B glutamate in the active sites of the D1 (E286Q) or the D2 (E565Q) modules allows ATP to bind but dramatically slows hydrolysis (12–16, 25). As shown in Fig. 2, we constructed pseudohexamers with multiple configurations of ATPase modules that were either active or inactive. These variants included enzymes with completely inactive D1 rings (D1ClpA), D2 rings (D2ClpA), or both rings (D1/D2ClpA), and cross-linked variants with alternating active/inactive subunits in D1 (altD1ClpAx), alternating active/inactive subunits in D2 (altD2ClpAx), and two variants with alternating active/inactive modules in both rings (altD1/altD2/cisClpAx and altD1/altD2/transClpAx). This set of enzymes was then used to interrogate the contributions of the number and/or configuration of AAA+ modules to the ATPase and protease functions of ClpAP.
Fig. 2.
ClpA variants used in this study. Names, relevant mutations, and cartoons are shown for each ClpA variant. In the diagrams of hexamers, light blue modules have WT ATPase active sites, whereas darker blue modules with red X’s contain Walker-B ATPase mutations (E286Q or E565Q). Black bars represent the presence of subunit-subunit cross-links between the engineered cysteines (D645C and Q709C).
Cross-Linked ClpA Variants Are Functional Enzymes.
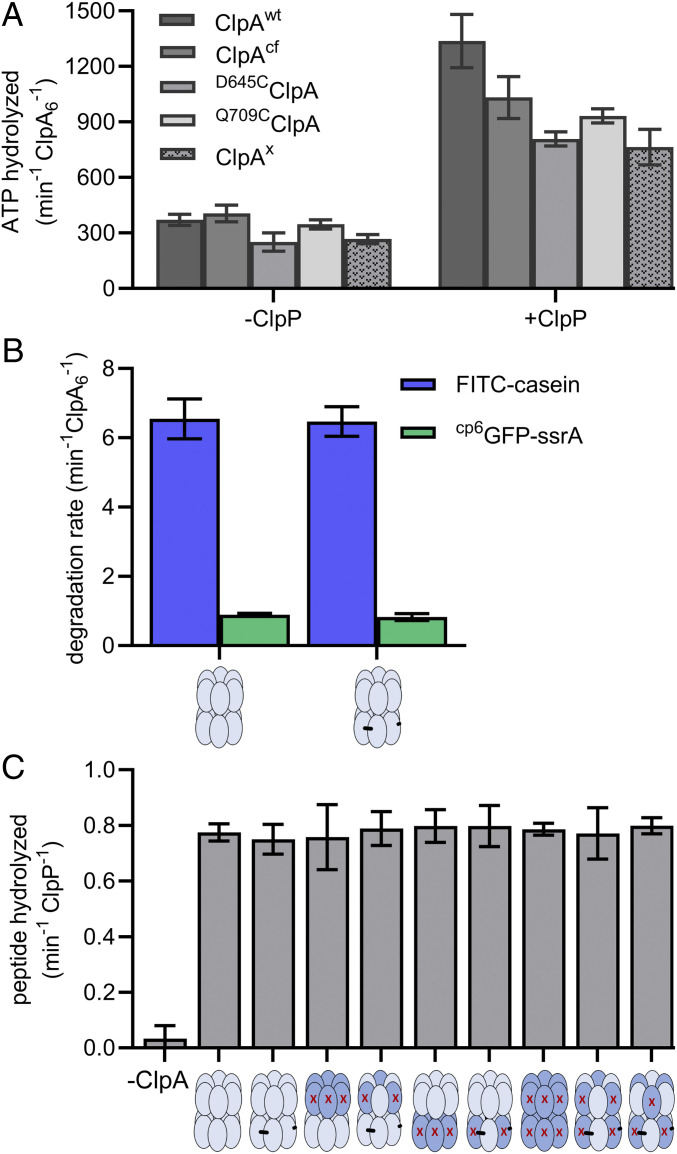
By cross-linking ClpA subunits to construct our experimental pseudohexamers, we considered that ClpA may be rendered more rigid, thus potentially perturbing motions that occur with the ATPase cycle, or harming docking between ClpA and ClpP. Therefore, to assess if cross-linking is detrimental to ClpA activity, we carried out a set of assays to compare the ATPase rates, degradation activities, and ClpP interactions of uncross-linked to cross-linked ClpA variants. Assays with ClpAx revealed that it hydrolyzed ATP with or without ClpP at similar rates to the uncross-linked D645CClpA and Q709CClpA enzymes (Fig. 3A). Although D645CClpA, Q709CClpA, and ClpAx all had modestly reduced ClpP-stimulated ATPase activities compared to ClpAwt (∼57–70%), their activities were comparable to ClpAcf, suggesting that removing the three native cysteines (but not cross-linking) was responsible for most of the reduction in ClpP-stimulated ATPase activity (Fig. 3A). Further, cross-linked ClpAx/ClpP degraded two protein substrates—molten globule-like FITC-casein (26) and a circularly permuted variant of green fluorescent protein, cp6GFP-ssrA (27)—at nearly identical rates to the variant we chose as our uncross-linked control, D645CClpA/ClpP (Fig. 3B). These results demonstrate that cross-linking does not impede ClpAP movements associated with unfolding (cp6GFP-ssrA) and translocation (FITC-casein, cp6GFP-ssrA) of protein substrates; further, they suggest that a functional ClpA-ClpP interface was maintained in ClpAx/ClpP, thereby allowing ClpAx/ClpP to display normal degradation activity compared to D645CClpA/ClpP. Degradation and ATPase activities of our full panel of ClpA variants will be discussed in the sections below, but these initial assays comparing ClpAx to D645CClpA gave us confidence that constructing ClpA pseudohexamers from covalently cross-linked dimers did not inhibit interaction of ClpA with ATP, protein substrates, or ClpP.
Fig. 3.
Cross-linking does not inhibit ClpA interactions with ClpP and/or substrates. (A) Hydrolysis of ATP (5 mM) by ClpA variants (0.25 µM) without inactivating ATPase mutations in the absence or presence of ClpP (0.75 µM). Values are averages (n ≥ 3) ± 1 SD. (B) Rates of degradation of cp6GFP-ssrA (20 µM, green) or FITC-casein (50 µM, blue) by uncross-linked D645CClpA or cross-linked ClpAx (0.25 µM each) with ClpP (0.75 µM). Values are averages (n ≥ 6) ± 1 SD. (C) ClpA variants interact with ClpP as assayed by a pore-opening assay. ClpP cleavage of a fluorogenic decapeptide of RseA (15 µM) was assayed in the presence of ATPγS (2 mM), different ClpA variants (0.50 µM), and ClpP (0.25 µM). Values are averages (n ≥ 3) ± 1 SD.
Although our ClpA variants with different combinations of ATPase mutations are expected to change ATPase and degradation rates (10–12), differences in these variants’ abilities to bind and activate ClpP could affect the interpretation of experimental results. To assess the integrity and function of the ClpA-ClpP interface, we used a “pore opening assay.” This assay specifically measures ClpA’s ability to bind and open the ClpP pore, allowing degradation of long peptides (for example, the 10-residue peptide used here) that ClpP cannot efficiently degrade on its own (28). Importantly, ATP hydrolysis-dependent unfolding and/or translocation of the peptide substrate by ClpA are not required for the observed ClpP cleavage, as the ATP analogs ATPγS and AMP-PNP (both not hydrolyzed at a detectable level) work as effectively as ATP in stimulating ClpP cleavage of long peptides (25, 28). In our assay, all ClpAP variants degraded a fluorescent decapeptide with similar rates (Fig. 3C). Therefore, regardless of the number or orientation of inactive ATPase modules, all of our ClpA variants form active complexes with ClpP, which requires proper ClpA to ClpP docking and opening of the ClpP pore. Thus, we conclude that the major differences observed in our degradation and ATPase assays with the mutated ClpA variants in the following sections are principally due to the inactivating ATPase mutations and not caused by deformation of the ClpA hexamer or poor interactions between ClpA and ClpP.
Only Three Active Modules in the D2 Ring Are Needed for Degradation.
Four protein substrates were used for degradation assays: FITC-casein, cp6GFP-ssrA, ssrA-(V15Ptitin)4-Halo, and Halo-(V15Ptitin)4-ssrA. ssrA-(V15Ptitin)4-Halo and Halo-(V15Ptitin)4-ssrA are multidomain substrates, each with a N- or C-terminal ssrA degron (thus initiating degradation from opposite termini), four repeated native titinI27 domains containing the destabilizing V15P mutation (V15Ptitin) (29), and a C-terminal or N-terminal HaloTag domain (Halo) covalently linked to a fluorescent Halo-TMR ligand (30). These multidomain substrates both require unfolding and translocation for degradation by ClpAP, although translocation is rate limiting for the N-to-C direction substrate (ssrA-(V15Ptitin)4-Halo), whereas unfolding is rate limiting for the C-to-N direction substrate (Halo-(V15Ptitin)4-ssrA) (14, 30). The rates of FITC-casein and cp6GFP-ssrA degradation were monitored by changes in fluorescence, whereas degradation of the multidomain substrates was monitored by SDS/PAGE and fluorimetry of the TMR group present on the intact substrate and/or degradation products at different time points.
D645CClpA/ClpP (all active modules) degraded FITC-casein and cp6GFP-ssrA with comparable rates to ClpAx/ClpP (all active modules) (Fig. 3B and datasets 1 and 2 in Fig. 4A). These enzymes also degraded the V15Ptitin domains of ssrA-(V15Ptitin)4-Halo at nearly identical rates, but, as anticipated from studies using WT ClpAP (14, 30), did not efficiently degrade Halo (datasets 1 and 2 in Fig. 4 C and D). Although D2ClpA/ClpP (six active D1 modules and no active D2 modules) and D1/D2ClpA/ClpP (no active modules) were highly defective in ATP hydrolysis, we observed higher than background (no enzyme) degradation of FITC-casein for both variants (datasets 5 and 7 in Fig. 4A). However, when we measured the FITC-casein degradation rates in the presence of ATPγS, D2ClpA/ClpP, D1/D2ClpA/ClpP, and D645CClpA/ClpP all gave the same result: no degradation above background was observed (Fig. 4B). Thus, for this molten globule-like substrate, the small degradation activity observed (10–15% of D645CClpA/ClpP’s activity; Fig. 4A) appears largely due to slow, residual ATP hydrolysis by D2ClpA and D1/D2ClpA (SI Appendix, Fig. S1).
Fig. 4.
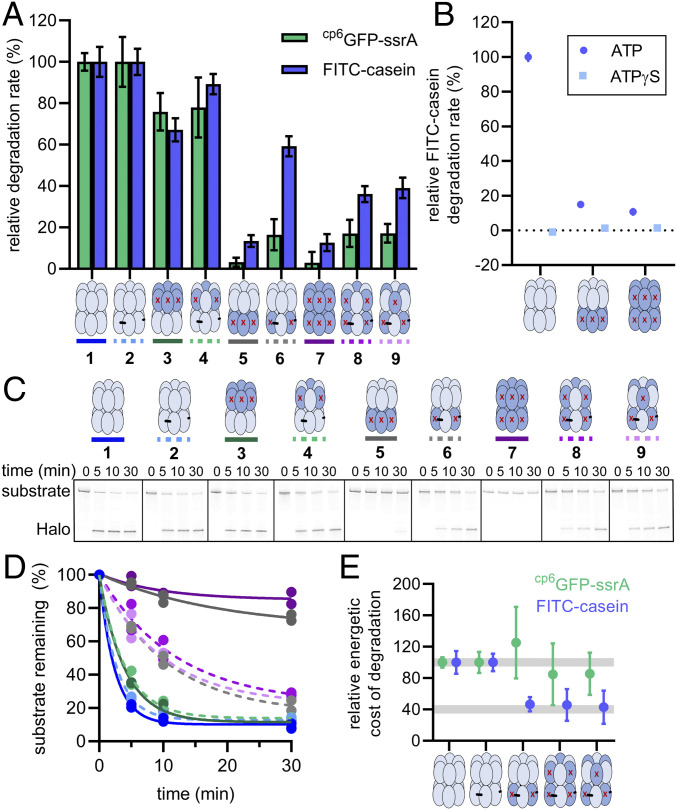
ClpA variants have different protein degradation profiles. (A) Relative rates of degradation of cp6GFP-ssrA (20 µM, green) or FITC-casein (50 µM, blue) by ClpAP variants. All rates were normalized to the 100% activity of each variant’s “parental” enzyme, either D645CClpA/ClpP or ClpAx/ClpP. In each case, the background rate (observed with no enzyme) was subtracted from the data before normalization. Values are averages (n ≥ 5) ± 1 SD. (B) FITC-casein degradation in the presence of 4 mM ATP or ATPγS. The concentrations of D645CClpA, D2ClpA, and D1/D2ClpA were 0.25 µM, and the concentration of ClpP was 0.75 µM. Data were normalized to the D645CClpA + ATP rate. Values are averages (n ≥ 3) ± 1 SD. (C) Degradation of ssrA-(V15Ptitin)4-Halo (0.5 µM) by ClpAP variants (0.5 µM ClpA, 1 µM ClpP) monitored by SDS/PAGE and imaged for TMR fluorescence. (D) Quantification of disappearance of the intact substrate over time (from experiments like that in C); degradation curves for each variant (n = 2 independent experiments) were independently fit to a single exponential. The correspondence between the line color and the identity of each variant is shown over the gel in C). (E) Energetic cost of degradation of FITC-casein or cp6GFP-ssrA by ClpAP variants. Energetic costs were calculated by dividing the ATPase rate of each variant in the presence of substrate (SI Appendix, Fig. S1) by the degradation rate, with the ratios for D645CClpA and ClpAx normalized to 100. Data points represent averages (n ≥ 5) ± propagated error.
The altD2ClpAx/ClpP variant, with six active D1 modules and three active D2 modules, degraded FITC-casein, cp6GFP-ssrA, and ssrA-(V15Ptitin)4-Halo faster than D2ClpA/ClpP, with six active D1 modules and no active D2 modules, but slower than the parental enzymes with fully active D2 rings (datasets 1, 2, 5, and 6 in Fig. 4 A and C; Fig. 4D). Further, the altD1/altD2/cisClpAx/ClpP and altD1/altD2/transClpAx/ClpP variants, which have three active D1 modules in addition to three active D2 modules, functioned at similar rates to altD2ClpAx/ClpP (datasets 8 and 9 in Fig. 4 A and C; Fig. 4D). The fact that we observed substantially increased degradation rates for three substrates with altD2ClpAx/ClpP, altD1/altD2/cisClpAx/ClpP, and altD1/altD2/transClpAx/ClpP compared to D2ClpA/ClpP and D1/D2ClpA/ClpP (no active modules) reveals that D2 rings with only three ATPase-active modules are functional protein unfoldases and translocases, albeit working at lower rates than D2 rings with six active modules. These results indicate that a reduced complement of active D2 modules slows translocation, the rate-limiting step for degradation of FITC-casein and ssrA-(V15Ptitin)4-Halo. Fewer active D2 modules also slows unfolding, the slowest step in degradation of cp6GFP-ssrA (27, 31), with unfolding being more sensitive than translocation to fewer active D2 modules.
ClpA Is “Overpowered” for Degradation of Some Substrates.
ClpAP variants with three D2 modules degraded substrates more slowly than variants with six active D2 modules (Fig. 4 A, C, and D) and had lower ATPase activity during substrate processing (SI Appendix, Fig. S1). To determine if the reduced degradation rates were a direct consequence of slower ATP hydrolysis, we calculated the energetic cost of degrading cp6GFP-ssrA or FITC-casein by dividing the working ATPase rates, measured with saturating substrate and ClpP, by the corresponding degradation rates. Interestingly, compared to ClpA variants with six active D2 modules, those with three active D2 modules used ∼60% less ATP to degrade each molecule of FITC-casein (Fig. 4E). In contrast, the energetic costs of cp6GFP-ssrA degradation were about the same for enzymes with three versus six D2 ATPase modules. Thus, the ∼2.5-fold lower energetic cost of FITC-casein degradation by altD2ClpAx/ClpP and the altD1/altD2/cisClpAx/ClpP and altD1/altD2/transClpAx/ClpP variants shows that ATP hydrolysis by three alternating active D2 modules is used more efficiently to promote degradation of this substrate. This observation, in turn, suggests that variants with six active D2 modules are “overpowered” for degradation of FITC-casein, with many ATP hydrolysis events apparently uncoupled from mechanical work. Because major energetic differences were not as clear for cp6GFP-ssrA degradation by three versus six active D2 modules, the increased efficiency of ClpAP with alternating active D2 modules may chiefly occur with substrates that only need to be translocated for degradation, as opposed to ones that must be unfolded and translocated.
D1 Modules Boost Unfolding/Translocation Powered by the Complete D2 Ring.
The D2 ring is the more powerful unfolding and translocation motor in ClpA (10, 12). However, the D1 and D2 rings appear to bind and translocate a single polypeptide simultaneously (22), so we looked for evidence of coupled ring activity. Notably, we found that the ability of active D1 modules to enhance unfolding and translocation rates of ClpAP was dependent on the number of active D2 modules. For example, the degradation rate of all substrates tested by D645CClpA/ClpP (six active D1 and six active D2 modules) was ∼30% higher than that of D1ClpA/ClpP (no active D1 and six active D2 modules) (datasets 1 and 3 in Fig. 4 A and C; Fig. 4D). However, when the D2 ring was completely inactive, the degradation rates for all three substrates by D2ClpA/ClpP (six active D1 and no active D2 modules) were comparable to D1/D2ClpA/ClpP (no active D1 or D2 modules) (datasets 5 and 7 in Fig. 4 A and C; Fig. 4D). Together, these data show that D1 modules only work to help promote degradation when active D2 modules are also engaged in unfolding and translocation, demonstrating coordination between the mechanical activities of the two rings for ClpA function. This coordination appears especially critical for the unfolding and degradation of more stable substrates (12).
D1 and D2 Communicate to Enhance Processivity During Substrate Processing.
We next investigated how D1 modules might coordinate with alternating active/inactive D2 rings. Degradation rates of FITC-casein, cp6GFP-ssrA, and ssrA-(V15Ptitin)4-Halo by the altD2ClpAx (six active D1 and three active D2 modules), altD1/altD2/cisClpAx/ClpP, and altD1/altD2/transClpAx/ClpP (each with three active D1 and D2 modules) variants were similar, especially for cp6GFP-ssrA and ssrA-(V15Ptitin)4-Halo (datasets 6, 8, and 9 in Fig. 4 A and C; Fig. 4D). These results initially suggested that active D1 modules only function well in coordination with fully active D2 rings.
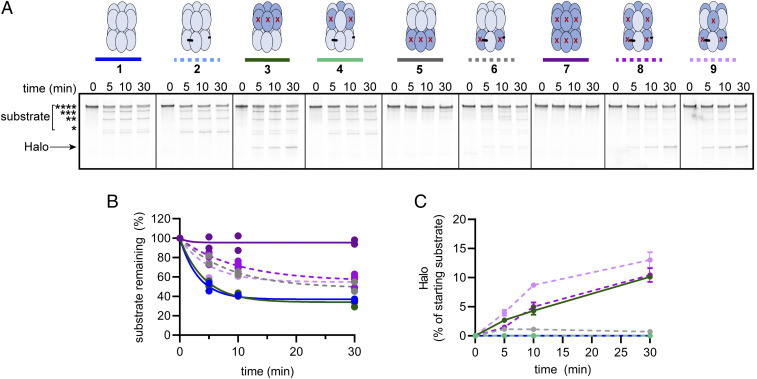
The D1 motor antagonizes stalling and is important for processive substrate degradation following committed substrate engagement (14); processivity of degradation can be best monitored using the multidomain substrates and observing degradation products by SDS/PAGE. During Halo-(V15Ptitin)4-ssrA degradation by ClpAP, for example, some of the enzymes dissociate after reaching different domain-domain junctions in the multidomain substrate, leading to an accumulation of incompletely degraded products with different numbers of titin domains and/or the Halo domain remaining intact (32, 33). The results in Fig. 5 A–C suggest that degradation of Halo-(V15Ptitin)4-ssrA by altD2ClpAx/ClpP (six active D1 modules and three active D2 modules) is more processive than that performed by altD1/altD2/cisClpAx/ClpP or altD1/altD2/transClpAx/ClpP (each with three active D1 and D2 modules). For example, although degradation by all three variants resulted in loss of the full-length substrate at similar rates (Fig. 5B), there were clear differences in the accumulation of incompletely degraded products between the ClpA variants. Most notably, a fragment of the approximate size of the released Halo domain appeared during the time courses with specific variants. We observed Halo accumulate to 11 ± 1 and 13 ± 1% of the starting substrate over 30 min by altD1/altD2/cisClpAx/ClpP and altD1/altD2/transClpAx/ClpP, respectively, whereas only ∼1% of the substrate accumulated in the Halo band during degradation by altD2ClpAx/ClpP (datasets 6, 8, and 9 in Fig. 5A; Fig. 5C). This greater ability of altD2ClpAx/ClpP to degrade the multidomain substrate to completion demonstrates that six active D1 modules paired with three active D2 modules enhances the processivity of degradation and also reveals that D1 modules can assist even partially active D2 rings. The Halo domain also accumulated during degradation by D1ClpA/ClpP (no active D1 modules and six active D2 modules) but not by altD1ClpAx/ClpP (three active D1 modules and six active D2 modules) (datasets 3 and 4 in Fig. 5A; Fig. 5C). In this case, the change in enzyme processivity indicates that three active D1 modules are sufficient to boost the processivity of fully active D2 rings. Thus, this analysis reveals that several different architectural arrangements of active D1 and D2 molecules are sufficient to empower ClpAP to processively degrade a long, multidomain substrate, including the well-folded terminal Halo domain.
Fig. 5.
Degradation of a multidomain substrate reveals contributions of D1 and D2 to enzyme processivity. (A) Kinetics of degradation of Halo-(titinV15P)4-ssrA assayed by SDS/PAGE and TMR imaging. The uppermost band is full-length substrate. Lower bands correspond to fragments containing fewer domains. The number of titinV15P domains in each species is denoted by the number of asterisks. (B) Quantification of the Halo-(titinV15P)4-ssrA remaining, with the zero time point set to 100%. The time course for each variant (n ≥ 2 independent experiments) was fit to a single exponential. The correspondence between the line color and the identity of each variant is shown over the gel in A). (C) Kinetic quantification of the Halo fragment during degradation by selected ClpAP variants. Each point represents the Halo-fragment-TMR intensity divided by the total TMR intensity in the time 0 lane. Values for D1ClpA, altD1/altD2/cisClpAx/ClpP, and altD1/altD2/transClpAx/ClpP are averages (n = 3) ± 1 SD.
Functional Communication Between the D1 and D2 ATPase Sites.
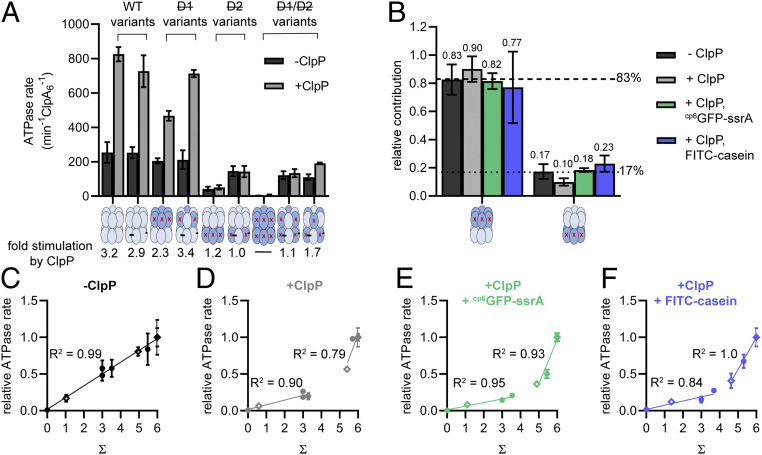
Previous work demonstrated that ClpP only stimulates ATP hydrolysis by the D2 ring of ClpA (12). As shown in Fig. 6A, ClpP stimulated ATP hydrolysis by ClpA variants with six active D2 modules ∼threefold, but did not stimulate variants with zero or three active D2 modules. In principle, lack of stimulation might result from poor ClpP binding. However, our cross-linked and uncross-linked ClpA variants were all equally active in stimulating ClpP cleavage of a fluorogenic peptide by the pore opening assay presented above (Fig. 3C), which requires ClpA–ClpP complex formation, but not mechanical activity by ClpA. Thus, ClpP stimulation of ClpA is markedly reduced when only three D2 modules are active ATPases.
Fig. 6.
ATP hydrolysis by ClpA variants differs without and with ClpP. (A) Rates of hydrolysis of ATP (5 mM) by ClpA variants (0.25 µM) in the absence (light gray bars) or presence of ClpP (0.75 µM; dark gray bars). Values are averages (n ≥ 5) ± 1 SD. Fold stimulation is the rate in the presence of ClpP divided by the rate in the absence of ClpP. (B) Relative contributions of the D2 and D1 rings to ATP hydrolysis under different conditions. Contributions were calculated by dividing the ATPase rate of D1ClpA or D2ClpA by the sum of these rates (A and SI Appendix, Fig. S1). Calculated fractional contributions are listed above each bar that represent averages (n ≥ 3) ± propagated error. On average, the D2 ring contributed ∼83% and the D1 ring contributed ∼17% toward the overall ATPase rate. (C–F) Normalized observed ATPase rates under different conditions plotted against the value predicted for each variant’s ATPase rate if each module contributed independently (Σ) (see text and SI Appendix, Table S1) for cross-linked (filled circles) and uncross-linked (open diamonds) variants. Each value is an average (n ≥ 3) ± 1 SD. Linear regressions are shown for all values of Σ in C, or for Σ = 0–3 and Σ = >3–6 in D–F.
Weber-Ban and coworkers posited that the D1 and D2 rings contribute unequally and independently to overall ATP hydrolysis (12). In agreement with this model, we found that the sum of the high ATP hydrolysis rate of D1ClpA (no active D1 modules, six active D2 modules) and the low rate of D2ClpA (six active D1 modules, no active D2 modules) was roughly equal to the rate of the parental enzyme (D645CClpA) (SI Appendix, Fig. S2). Based on experiments performed in the presence of ClpP and protein substrates, however, the two rings appear to contribute to ATP hydrolysis in a more complex fashion. For example, with ClpP or ClpP/substrate present, adding the ATP-hydrolysis rates of D1ClpA and D2ClpA resulted in a net activity that was 45–60% of the rate of the parental enzyme (SI Appendix, Fig. S2). Nevertheless, in each case, the D2 ring contributed the majority (∼80% on average) of the observed activity (Fig. 6B and SI Appendix, Fig. S2). Thus, there is modest synergy in ATP hydrolysis between the D1 and D2 rings when ClpA is functioning with ClpP.
We generated a metric to determine if the ATP hydrolysis is independent between ATPase modules in the two rings. We defined a variable, Σ, as the calculated ATPase rate expected for any variant assuming that each module contributes independently of any other module. We calculated Σ for each ClpA variant in the absence of ClpP, the presence of ClpP, or the presence of ClpP and substrate, by multiplying the number of active D1 sites by their fractional ATPase contributions (range 0.10–0.23; Fig. 6B) and adding this value to the number of active D2 sites multiplied by their fractional contributions (range 0.77–0.90; Fig. 6B) (SI Appendix, Table S1). A strong positive linear correlation between Σ and the observed ATPase rates for our ClpA variants (R2 ≥ 0.8) would support an independent model of ATP hydrolysis.
In Fig. 6 C–F, we plotted the ATPase rates of our ClpA variants (normalized to their parent enzymes, see Fig. 6A) against (Σ). In the absence of ClpP, the linearity of Fig. 6C (R2 = 0.99) provides strong evidence that the active sites in the D1 and D2 rings contribute independently to ATPase activity; importantly, there is no requirement for two active modules to be adjacent either within the D1 or the D2 rings, or within the individual subunits that form these stacked rings, to observe each module’s individual contribution to enzyme activity.
In the presence of ClpP or ClpP/substrate, by contrast, these observed vs. calculated ATPase plots were biphasic, with a shallow linear phase at lower Σ values and a steeper linear phase at higher Σ values (Fig. 6 D–F); these biphasic plots clearly did not support an independent model of ATP hydrolysis by each of the modules. Rather, the steeper slope of the high Σ region of the graph is consistent with ClpP and active D1 modules stimulating ATP hydrolysis by complete D2 rings, as observed in Fig. 6A. The linear nature (R2 ≥ 0.8) between ClpA variants in each of the two phases, however, indicates that ATP hydrolysis increases proportionally to the number of active modules under two conditions: 1) when an incomplete D2 ring is not stimulated by ClpP or D1 modules (low Σ), and 2) when a complete D2 ring is activated by ClpP and is also increasingly activated by the presence of more D1 modules (e.g., zero vs. three vs. six) (high Σ). Overall, we conclude that interaction of ClpA with ClpP is the key feature that “breaks” the independent activities of the D1 and D2 ATPases and, thereby, promotes the more coordinated ATP hydrolysis and mechanical activity needed by the fully assembled protease.
Discussion
Cross-Linked, Mutated Variants Reveal ClpA D1 and D2 Ring Functions.
Our experiments provide insights into the functional contributions of the 12 ATPase modules in the D1 and D2 rings of ClpA. We developed a cross-linking/mutagenesis strategy to introduce active-site mutations that severely diminished ATP hydrolysis in every other module of the D1 ring, the D2 ring, or both rings. These variants retained the ability to hydrolyze ATP and power ClpAP degradation of protein substrates with different rates and efficiencies depending on the number of active modules in each variant. Previous work established that the D2 ring provides the major motor activity required for mechanical work, whereas the D1 ring antagonizes enzyme stalling and promotes processive translocation and unfolding by the D2 ring (10–14). However, these studies were unable to elucidate how individual ATPase modules in the D1 and D2 rings contribute to ClpAP function.
We find that ClpA variants with alternating active and inactive modules in the D2 ring combine with ClpP to promote ATP-dependent unfolding, translocation, and degradation of folded and unfolded protein substrates. Thus, the D2 motor need not be fully active or even have two adjacent active subunits to carry out mechanical work. Surprisingly, variants with three active D2 modules degrade FITC-casein using half the number of ATPs as variants with six active D2 modules, suggesting that fully active ClpAP is energetically inefficient at degrading this unfolded substrate. Consistent with previous findings (12), we observe that more D2 modules are needed for robust unfolding and translocation of increasingly stably folded substrates such as cp6GFP-ssrA; however, our results show that variants with three active D2 modules degrade cp6GFP-ssrA with similar energetic costs as variants with six active D2 modules, reinforcing the notion that incomplete D2 motors are slower than complete D2 motors, but effective.
Our results also reinforce the idea that D1 functions as an important “booster motor” to increase ClpAP’s efficiency and processivity during unfolding and translocation. When the D2 ring is fully active, we find that three alternating active D1 modules are sufficient to perform these functions. Although the number of active D1 and D2 modules affects rates of ClpAP unfolding, translocation, stalling, and degradation, our combined results demonstrate that there is no requirement for hydrolytically active modules to be adjacent either within the D1 or the D2 rings, or within the same subunit, for function.
Implication of Mixed ATPase Motors for Models of Work.
The D1 and D2 rings of ClpA are members of different subfamilies of AAA+ unfoldases. The D1 ring is part of the classic clade, which includes the single-ring Rpt1–6, PAN, and FtsH enzymes, double-ring NSF, p97, and Vps4 enzymes, and the D1 rings of ClpB, ClpC, and Hsp104. The D2 ring is a member of the HCLR clade, which includes the single-ring ClpX, HslU, and Lon enzymes and the D2 rings of ClpB, ClpC, and Hsp104 (8, 9). Cryo-EM structures of classic and HCLR clade enzymes have been used to support a mechanism in which each module in the AAA+ ring cycles sequentially through six distinct ring positions and, thus, each subunit takes a turn hydrolyzing ATP to drive the next power stroke required for mechanical work (34–41). However, if the D1 or D2 rings of ClpA were to operate by this strictly sequential mechanism, then rings with alternating hydrolytically active and inactive subunits should have been mechanically inert rather than merely slower.
A competing model posits that probabilistic ATP hydrolysis at many positions within a AAA+ ring can drive a power stroke, thereby eliminating the requirement for adjacent subunits to fire in a strictly sequential pattern (15, 16). This model is also consistent with observed states in recent cryo-EM structures (42–44), mixed-ring experiments showing that ClpX and HslU can function with only a subset of ATPase active AAA+ modules (15, 16), and single-molecule experiments that demonstrate ClpXP translocation occurs with different step sizes in a stochastic pattern (45, 46). Our ClpA experiments lend strong support that ClpA and ClpAP function via a nonstrictly sequential model for both ATP hydrolysis and work by the D1 and D2 rings.
Ring-Ring Coordination.
In the absence of ClpP and substrate, the ATP-hydrolysis activities of the D1 and D2 rings of ClpA appear to be independent, as originally shown by Kress et al. (12) and confirmed here (Fig. 6 B and C and SI Appendix, Fig. S2). When ClpP or ClpP/substrate are present, however, our results indicate that the D1 and D2 rings communicate both in terms of mechanical function and with respect to ATP hydrolysis. Although ClpP binds all of our ClpA variants well enough to promote degradation of model substrates, we find that ClpP does not stimulate ATP hydrolysis by an incomplete D2 ring. When the D2 ring is completely active, three or six D1 modules and ClpP stimulate ATP hydrolysis by D2, leading to enhanced unfolding, translocation, and processivity of ClpAP. This coordination makes sense for efficient machine function, as two unsynchronized motor rings would often be expected to oppose each other. For example, one ring in a substrate-gripping mode might antagonize the other ring in a pulling mode. Recent cryo-EM structures of ClpAP–substrate complexes show that conserved D1 and D2 pore loops in the axial channel of the ring contact the substrate polypeptide (22). Thus, it is unlikely that one ring disengages while the other ring pulls. Our observations that altD1/altD2/cisClpAx/ClpP and altD1/altD2/transClpAx/ClpP display similar ATPase and degradation rates provide evidence that there is no strict requirement for active modules to be oriented in the same subunit for coordinated function; instead, our data support that it is the number of active D1 and D2 modules (zero, three, or six) that has the major effect on the activity of ClpAP variants rather than the arrangement of active modules. We were, however, limited in the types of arrangements we could probe, due to the strategy of building our enzymes out of cross-linked dimers.
The D1 and D2 rings of the ClpB and Hsp104 protein-remodeling machines share strong sequence and structural homology with those of ClpA (8, 22, 47). However, based on the findings in this study, coordination of ATPase modules within and between the D1 and D2 rings appears to be different in ClpA (18, 48–51). For example, doping just one or two ATPase-inactive subunits into a ClpB hexamer abrogates ATPase activity (52). Further, allosteric networks in Hsp104/ClpB, including the interaction of these enzymes’ “middle domains” (absent in ClpA family enzymes) and folding chaperones with both rings, may make ATP hydrolysis in one module much more highly dependent on the nucleotide states of other modules in either ring (18, 51, 53). However, for ClpA, our alternating active-inactive D2 variants performed ATP-dependent work at similar or greater energetic efficiencies than fully active ClpA variants, and altD1/altD2/cisClpAx/ClpP and altD1/altD2/transClpAx/ClpP (each with three active D1 and three active D2 modules) had similar ATPase and degradation activities, demonstrating that ClpA’s two ATPase modules within one subunit do not exhibit high allosteric interdependence. These differences in ring-ring coordination may also reflect the fact that Hsp104/ClpB act primarily as protein disaggregases in collaboration with several cofactors (54–57), whereas ClpA functions in concert with ClpP as a proteolytic machine (22, 38, 47).
Going forward, important questions about ClpA function include how coordination between the D1 and D2 rings is accomplished structurally and whether D1 or D2 rings with fewer than three hydrolytically active modules are also functional. The methods developed here will also enable selective mutation of other important ClpA residues in the pore-1, pore-2, and IGL-motifs, and at the D1-D2 ring interfaces for functional analysis.
Materials and Methods
Protein Purification.
Standard PCR techniques were used to introduce mutations into a pET23b-plasmid-borne gene encoding E. coli ClpAΔC9 with an N-terminal SUMO solubility tag (the ∆C9 deletion prevents autodegradation) (58). All variants described here contain the ∆C9 deletion. Plasmids encoding each ClpA variant were transformed into E. coli strain BL21(DE3). For purification, a 4-L culture was grown at 25 °C to an OD600 of ∼0.5, ClpA expression was induced with 0.5 mM isopropyl 1-thio-d-galactopyranoside, and the culture was grown for 3–4 h at 22 °C before harvesting. Cell paste was resuspended in 3 mL of lysis buffer (50 mM Tris-Cl, pH 7.5, 2 mM ethylenediaminetetraacetic acid [EDTA], 10% [vol/vol] glycerol, 2 mM dithiothreitol [DTT]) per gram of paste and stored at –80 °C. ClpA was purified as described (59) with several modifications. After lysis by French press, ∼2,000 units of benzonase and 10 µL of Calbiochem Protease Inhibitor III mixture (EMD Millipore) were added, and the lysate was incubated at 4 °C for 30 min, before clearing the lysate by centrifugation. The supernatant was dialyzed overnight against S-Sepharose Buffer (25 mM Hepes-KOH, pH 7.5, 0.1 mM EDTA, 10% (vol/vol) glycerol, 2 mM DTT) with the addition of 200 mM KCl and 0.2 µM Ulp1 protease, which removes the N-terminal SUMO domain, before cation exchange chromatography on an S-Sepharose column (GE Healthcare). Peak fractions were then chromatographed on a HiLoad 16/10 Phenyl Sepharose HP column, and final fractions containing ClpA were dialyzed in HO activity buffer (50 mM Hepes-KOH, 20 mM MgCl2, 0.3 M NaCl, 10% [vol/vol] glycerol, 2 mM tris(2-carboxyethyl)phosphine) for protein storage. ClpP, cp6GFP-ssrA, Halo-(V15Ptitin)4-ssrA substrate, and the cys-(V15Ptitin)4-Halo protein were purified as described (27, 30, 46). An ssrA peptide containing an N-terminal maleimide was attached to cys-(V15Ptitin)4-Halo to generate ssrA-(V15Ptitin)4-Halo for biochemical experiments (30). FITC-casein was bovine milk type III labeled with fluorescein isothiocyanate (Sigma-Aldrich) dissolved in HO buffer for biochemical experiments.
Cross-Linking.
Purified D645CClpA and Q709CClpA variants containing Walker-B mutations (E286Q and/or E565Q) or no Walker-B mutations were exchanged into cross-linking buffer (50 mM Hepes-KOH, pH 7, 300 mM NaCl, 20 mM MgCl2, 10% glycerol, 5 mM EDTA) using Zeba Spin Desalting Columns (ThermoFisher Scientific). Cross-linking reactions containing ClpA variants (30–50 µM total monomer equivalents at a 1:1 ratio of D645CClpA and Q709CClpA) and 100 µM 1,4-bismaleimidobutane in cross-linking buffer were incubated for 20–60 min at room temperature and then quenched by the addition of 50 mM DTT. Cross-linked dimers were separated from monomers using a Superdex 200 16/600 size-exclusion column (GE Healthcare) equilibrated in HO buffer, which does not support hexamer assembly (no ATP). Fractions containing cross-linked dimers were stored at –80 °C.
Biochemical Assays.
ATPase assays were performed at 30 °C in HO buffer. Hydrolysis of 5 mM ATP was measured using an NADH-coupled assay (60) with an ATP-regeneration system (20 U/mL pyruvate kinase, 20 U/mL lactate dehydrogenase, 7.5 mM phosphoenolpyruvate, and 0.2 mM NADH) by monitoring loss of absorbance at 340 nm using a SpectraMax Plus 384 Microplate Reader (Molecular Devices).
All activity assays were performed in HO buffer at 30 °C. To monitor pore opening of ClpP by ClpA, reactions contained 0.50 µM ClpA6, 0.25 µM ClpP14, 2 mM ATPγS, and 15 µM RseA (Abz-KASPVSLGYNO2D) decapeptide (where Abz is the fluorophore 2-aminobenzoic acid and YNO2 is the quencher 3-nitrotyrosine). Fluorescence (excitation 320 nm; emission 420 nm) was monitored using a SpectraMax M5 Microplate Reader (Molecular Devices). To monitor ClpAP degradation of cp6GFP-ssrA or FITC-casein, reactions contained 0.25 µM ClpA6, 0.75 µM ClpP14, 4 mM ATP, an ATP regeneration system (50 μg/mL creatine kinase [Millipore-Sigma], 5 mM creatine phosphate [Millipore-Sigma]), and either 20 µM cp6GFP-ssrA or 50 µM FITC-casein (determined as the concentration of casein due to variability of FITC labeling among casein molecules [ε = 11,460 M−1·cm−1]). Loss of cp6GFP-ssrA fluorescence (excitation 467 nm; emission 511 nm) or increase in FITC-casein fluorescence (excitation 340 nm; emission 460 nm) was monitored using a SpectraMax M5 Microplate Reader.
To assay degradation of ssrA-(V15Ptitin)4-Halo or Halo-(V15Ptitin)4-ssrA by ClpAP variants, the substrate was initially incubated with an equimolar concentration of HaloTag TMR Ligand (Promega) in HO buffer at 30 °C for 30 min. ClpA6 (0.5 µM), ClpP14 (1 µM), ATP (5 mM), and the ATP regeneration system described above were preincubated at 30 °C for 2 min, and either ssrA-(V15Ptitin)4-TMR-Halo or TMR-Halo-(V15Ptitin)4-ssrA (0.5 µM) was added to initiate degradation. Samples were taken at different time points, quenched by addition of SDS-sample buffer and rapid freezing, and later thawed and electrophoresed on a Mini-PROTEAN TGX 4–15% (wt/vol) precast gel (Bio-Rad). TMR fluorescence in the gel was imaged using a Typhoon FLA 9500 scanner (GE Healthcare) and quantified with ImageQuant 8.1 (GE Healthcare).
Supplementary Material
Acknowledgments
We thank T. Bell and I. Levchenko (Massachusetts Institute of Technology) for materials, and members of our laboratories for advice and helpful discussions. This work was supported by NIH Grants GM-101988 (to R.T.S.) and AI-016892 (to R.T.S. and T.A.B.), the Howard Hughes Medical Institute, and the NSF Graduate Research Fellowship under Grant 174530.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014407117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Snider J., Thibault G., Houry W. A., The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9, 216.1-216.8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T. A., Sauer R. T., ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer R. T., Baker T. A., AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Olivares A. O., Baker T. A., Sauer R. T., Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama-Fujimura Y., Gottesman S., Maurizi M. R., A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262, 4477–4485 (1987). [PubMed] [Google Scholar]
- 6.Hwang B. J., Woo K. M., Goldberg A. L., Chung C. H., Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J. Biol. Chem. 263, 8727–8734 (1988). [PubMed] [Google Scholar]
- 7.Maurizi M. R., ATP-promoted interaction between Clp A and Clp P in activation of Clp protease from Escherichia coli. Biochem. Soc. Trans. 19, 719–723 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Erzberger J. P., Berger J. M., Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Iyer L. M., Leipe D. D., Koonin E. V., Aravind L., Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Singh S. K., Maurizi M. R., Mutational analysis demonstrates different functional roles for the two ATP-binding sites in ClpAP protease from Escherichia coli. J. Biol. Chem. 269, 29537–29545 (1994). [PubMed] [Google Scholar]
- 11.Seol J. H., Baek S. H., Kang M. S., Ha D. B., Chung C. H., Distinctive roles of the two ATP-binding sites in ClpA, the ATPase component of protease Ti in Escherichia coli. J. Biol. Chem. 270, 8087–8092 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Kress W., Mutschler H., Weber-Ban E., Both ATPase domains of ClpA are critical for processing of stable protein structures. J. Biol. Chem. 284, 31441–31452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baytshtok V., Baker T. A., Sauer R. T., Assaying the kinetics of protein denaturation catalyzed by AAA+ unfolding machines and proteases. Proc. Natl. Acad. Sci. U.S.A. 112, 5377–5382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotamarthi H. C., Sauer R. T., Baker T. A., The Non-dominant AAA + ring in the ClpAP protease functions as an anti-stalling motor to accelerate protein unfolding and translocation. Cell Rep. 30, 2644–2654.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin A., Baker T. A., Sauer R. T., Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Baytshtok V. et al., Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation. J. Biol. Chem. 292, 5695–5704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joly N., Buck M., Single chain forms of the enhancer binding protein PspF provide insights into geometric requirements for gene activation. J. Biol. Chem. 286, 12734–12742 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamasaki T., Oohata Y., Nakamura T., Watanabe Y. H., Analysis of the cooperative ATPase cycle of the AAA+ chaperone ClpB from Thermus thermophilus by using ordered heterohexamers with an alternating subunit arrangement. J. Biol. Chem. 290, 9789–9800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biasini M. et al., SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T., Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7, 10480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig D. B., Dombkowski A. A., Disulfide by design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinformatics 14, 346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez K. E. et al., Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis. Nat. Struct. Mol. Biol. 27, 406–416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veronese P. K., Stafford R. P., Lucius A. L., The Escherichia coli ClpA molecular chaperone self-assembles into tetramers. Biochemistry 48, 9221–9233 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Maurizi M. R., Singh S. K., Thompson M. W., Kessel M., Ginsburg A., Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry 37, 7778–7786 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Thompson M. W., Maurizi M. R., Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem. 269, 18201–18208 (1994). [PubMed] [Google Scholar]
- 26.Thompson M. W., Singh S. K., Maurizi M. R., Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 269, 18209–18215 (1994). [PubMed] [Google Scholar]
- 27.Nager A. R., Baker T. A., Sauer R. T., Stepwise unfolding of a β barrel protein by the AAA+ ClpXP protease. J. Mol. Biol. 413, 4–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M. E., Baker T. A., Sauer R. T., Control of substrate gating and translocation into ClpP by channel residues and ClpX binding. J. Mol. Biol. 399, 707–718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenniston J. A., Baker T. A., Fernandez J. M., Sauer R. T., Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell 114, 511–520 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Olivares A. O., Kotamarthi H. C., Stein B. J., Sauer R. T., Baker T. A., Effect of directional pulling on mechanical protein degradation by ATP-dependent proteolytic machines. Proc. Natl. Acad. Sci. U.S.A. 114, E6306–E6313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin A., Baker T. A., Sauer R. T., Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol. 15, 139–145 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Kenniston J. A., Baker T. A., Sauer R. T., Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc. Natl. Acad. Sci. U.S.A. 102, 1390–1395 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A., ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7, 627–637 (2001). [DOI] [PubMed] [Google Scholar]
- 34.de la Peña A. H., Goodall E. A., Gates S. N., Lander G. C., Martin A., Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gates S. N. et al., Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ripstein Z. A., Vahidi S., Houry W. A., Rubinstein J. L., Kay L. E., A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. eLife 9, e52158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deville C., Franke K., Mogk A., Bukau B., Saibil H. R., Two-Step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor. Cell Rep. 27, 3433–3446.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizo A. N. et al., Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase. Nat. Commun. 10, 2393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumder P. et al., Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc. Natl. Acad. Sci. U.S.A. 116, 534–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puchades C. et al., Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358, eaao0464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han H., Monroe N., Sundquist W. I., Shen P. S., Hill C. P., The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife 6, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fei X. et al., Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. eLife 9, 1–52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin M. et al., Structural basis for distinct operational modes and protease activation in AAA+ protease Lon. Sci. Adv. 6, eaba8404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Y. et al., Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen M. et al., The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell 155, 636–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordova J. C. et al., Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158, 647–658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duran E. C., Weaver C. L., Lucius A. L., Comparative analysis of the structure and function of AAA+ Motors ClpA, ClpB, and Hsp104: Common threads and disparate functions. Front. Mol. Biosci. 4, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werbeck N. D., Zeymer C., Kellner J. N., Reinstein J., Coupling of oligomerization and nucleotide binding in the AAA+ chaperone ClpB. Biochemistry 50, 899–909 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Mogk A. et al., Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278, 17615–17624 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Zeymer C., Fischer S., Reinstein J., trans-Acting arginine residues in the AAA+ chaperone ClpB allosterically regulate the activity through inter- and intradomain communication. J. Biol. Chem. 289, 32965–32976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franzmann T. M., Czekalla A., Walter S. G., Regulatory circuits of the AAA+ disaggregase Hsp104. J. Biol. Chem. 286, 17992–18001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werbeck N. D., Schlee S., Reinstein J., Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 378, 178–190 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Zeymer C., Barends T. R. M., Werbeck N. D., Schlichting I., Reinstein J., Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: A structure-based mechanistic dissection of a molecular motor. Acta Crystallogr. D Biol. Crystallogr. 70, 582–595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glover J. R., Lindquist S., Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B., Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zolkiewski M., ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 274, 28083–28086 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Desantis M. E. et al., Conserved distal loop residues in the Hsp104 and ClpB middle domain contact nucleotide-binding domain 2 and enable Hsp70-dependent protein disaggregation. J. Biol. Chem. 289, 848–867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maglica Z., Striebel F., Weber-Ban E., An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J. Mol. Biol. 384, 503–511 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Hou J. Y., Sauer R. T., Baker T. A., Distinct structural elements of the adaptor ClpS are required for regulating degradation by ClpAP. Nat. Struct. Mol. Biol. 15, 288–294 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Kim Y. I. et al., Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8, 230–233 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.