Significance
Over 600 proteases work together to maintain the normal functions and homeostasis of cells in the human body. Determining protease specificity and their natural substrates is critical to understanding their biology. We present a facile, inexpensive, general, and global means to profile the natural specificity of human proteases at unprecedented depth. Using two genetically encoded substrate phage libraries, we deeply profile the substrate specificities of two important protease families, caspases involved in cell death and ADAMs family proteases that shed membrane proteins. We validated our results by recapitulating and expanding on known consensus substrates with up to 1,000-fold greater coverage.
Keywords: protease specificity, human proteome, substrate phage library, NGS
Abstract
Proteolysis is a major posttranslational regulator of biology inside and outside of cells. Broad identification of optimal cleavage sites and natural substrates of proteases is critical for drug discovery and to understand protease biology. Here, we present a method that employs two genetically encoded substrate phage display libraries coupled with next generation sequencing (SPD-NGS) that allows up to 10,000-fold deeper sequence coverage of the typical six- to eight-residue protease cleavage sites compared to state-of-the-art synthetic peptide libraries or proteomics. We applied SPD-NGS to two classes of proteases, the intracellular caspases, and the ectodomains of the sheddases, ADAMs 10 and 17. The first library (Lib 10AA) allowed us to identify 104 to 105 unique cleavage sites over a 1,000-fold dynamic range of NGS counts and produced consensus and optimal cleavage motifs based position-specific scoring matrices. A second SPD-NGS library (Lib hP), which displayed virtually the entire human proteome tiled in contiguous 49 amino acid sequences with 25 amino acid overlaps, enabled us to identify candidate human proteome sequences. We identified up to 104 natural linear cut sites, depending on the protease, and captured most of the examples previously identified by proteomics and predicted 10- to 100-fold more. Structural bioinformatics was used to facilitate the identification of candidate natural protein substrates. SPD-NGS is rapid, reproducible, simple to perform and analyze, inexpensive, and renewable, with unprecedented depth of coverage for substrate sequences, and is an important tool for protease biologists interested in protease specificity for specific assays and inhibitors and to facilitate identification of natural protein substrates.
Proteolysis is one of the most common post translational modifications (PTMs) and plays essential roles in diverse aspects of cellular functions, from protein degradation to specific protein activation (1, 2). The roughly 600 human proteases—around 2% of the genome—work together to maintain the normal functions and homeostasis of cells and tissues in the body. Aberrant protease activities propagate cancer (3), inflammation (4), and infectious diseases (5). Understanding the substrate specificities of proteases and their substrates helps define protease functions in cellular processes and provides insights into inhibitor design for both research and therapeutic purposes.
In the past two decades, synthetic peptide libraries (6, 7) have been used to characterize the linear recognition sequence specificities for proteases. While these are very useful for evaluating 100s to 1,000s of possible synthetic substrates, they do not deeply sample the possible sequences over the six- to eight-residue stretch that proteases typically recognize, nor do these random sequences cover exact human sequences. In the past decade, mass spectrometry methods have been developed for identifying intact human protein substrates (8–12). While proteomics approaches have enabled a broader understanding of protease substrates on intact proteins, they require significant amounts of lysate and miss low abundance proteins and those simply not expressed in cell lines tested that typically express only half their genomes (13).
To potentially screen a larger and more diverse sequence space, investigators have developed genetically encoded substrate phage (14, 15) or yeast display libraries (16, 17). Degenerate DNA sequences (up to 107) encoding random peptides were fused to a phage or yeast coat protein gene for a catch-and-release strategy or with the assistance of cell sorting, respectively. In the case of the substrate phage, the library is bound to an affinity support, exposed to a protease of interest to release, propagated, and enriched for sensitive and resistant clones that are individually sequenced. However, it is difficult to determine the exact proteolytic site by gene sequencing, and, until now, only short five- to six-residue random linear peptide libraries (up to 107 members) have been individually screened, not ones specifically covering the human proteome.
To allow deep substrate profiling, we present a next generation of genetically encoded phage display libraries containing either random 10-mers (up to 109 sequence diversity) or human proteome-wide tiled sequences (up to 106 members). We validate these libraries and the method on members of the caspases and ADAMs family proteases. Coupling substrate phage display with next-generation sequencing (SPD-NGS) allowed profiling of protease specificity at 103- to 104-fold greater depth than classical synthetic peptide libraries and identified specific human sequences capable of being cleaved much beyond what has been reported by proteomics. We deployed state-of-the-art position-specific scoring matrix (PSSM) methods to precisely identify cleavage sites within each selected clone and produce high confidence consensus sequence motifs for the proteases validated by literature. Structural bioinformatics was used to triage the candidate linear substrates for the identification of potential natural protein substrates. We believe these genetically encoded libraries, screening, and computational methods provide a simpler, less expensive, and more comprehensive companion to synthetic peptide libraries and proteomics.
Results
Substrate Phage Library Strategy, Design, Assembly, and Quality Control.
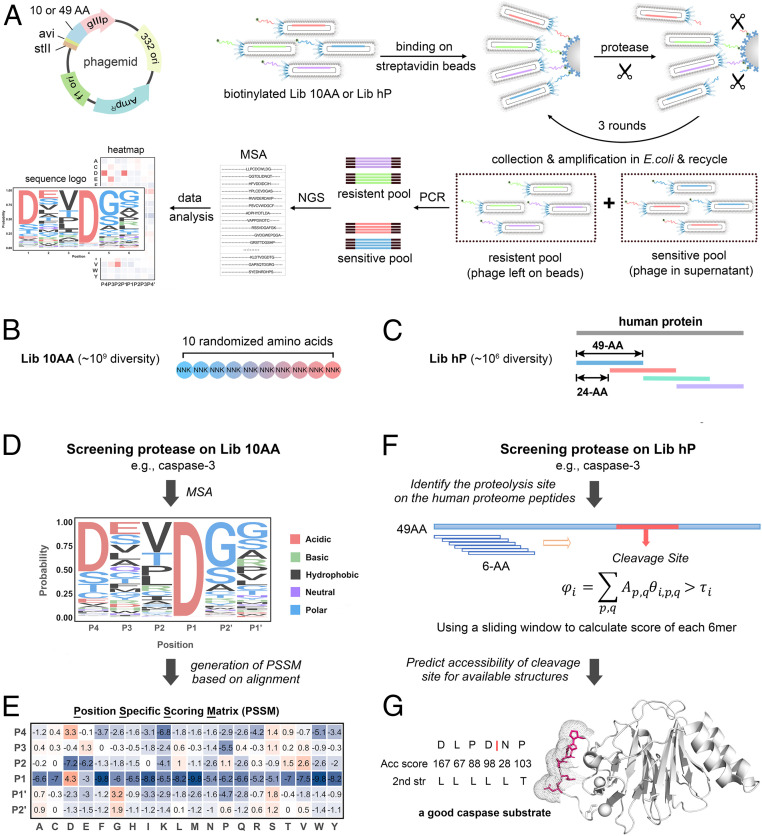
The overall strategy for SPD-NGS is diagramed in Fig. 1A. Peptides are displayed as a fusion protein on the surface of M13 bacteriophage. We utilized a monovalent phage display system to avoid avidity and ensure that a single cleavage event per phage is sufficient to release the avidin bound phage. We constructed two SPD libraries, Lib 10AA and Lib hP (human proteome), for complementary and mutually reinforcing purposes (Fig. 1 B and C and SI Appendix, Figs. S1–S3). The two libraries were produced using synthetic DNA, and the quality and diversity were validated by NGS (SI Appendix, Figs. S1 and S2). Lib 10AA contains highly diverse (∼109 unique sequences) and fully randomized 10-amino acid substrate segments (Fig. 1B and SI Appendix, Figs. S1 and S3). The fact that the codon frequency at each position and throughout the 10 amino acid window was uniform and matched the expected input synthetic DNA suggests that there is little cloning or expression bias for the displayed peptides. The strength of the Lib 10AA is to identify highly preferred substrates based on NGS counts from the protease-sensitive pool of substrate phage. Consensus cleavage motifs are generated through multiple sequence alignments (MSAs) (Fig. 1D), based on which a PSSM (Fig. 1E) is derived for high confidence prediction of precise cleavage sites within each selected sequence.
Fig. 1.
Two-pronged strategy used to identify protease substrates using SPD-NGS. (A) The schematic illustration of a high-throughput platform for SPD-NGS to profile the protease substrate specificity. stII: signaling peptide, avi: avi-Tag, gIIIp: M13 bacteriophage coat protein gIIIp, ori: origin of replication, AmpR: ampicillin-resistant gene, f1 ori: phage-derived ori. (B) Lib 10AA contains a highly diverse (∼109 unique sequences) and fully randomized 10 amino acid substrate segment encoded by NNK degenerate codons. (C) Lib hP displays tiled peptides covering the human proteome in 49 amino acid blocks and overlapping in a 25 amino acid sliding window. (D–G) Workflow to determine the substrate specificity of a protease in vitro, such as caspase-3. A fully randomized 10 amino acid peptide phage library (Lib 10AA) is screened to identify optimal linear peptide sequences/motifs from which a scoring function can be generated (D and E). A 49 amino acid human proteome tiled library (Lib hP) is screened to identify specific human sequences and sites that can be cut (F and G). (D) A representative six-residue sequence logo based on SPD-NGS from Lib 10AA for a protease of interest, such as caspase-3. The alignment of the top ∼20,000 peptides enriched from selection affords exhaustive generation of a sequence consensus. The data allow us to calculate the probability of each amino acid at each of six positions (P4-P2′ was determined based on existing knowledge). (E) The PSSM of caspase-3 substrates is generated according to the alignment. (F) Determining the precise cut site(s) within each of the positively selected 49 amino acid clones based only on 6 amino acid segments can be informed by the scoring matrix derived from the Lib 10AA. (G) Solvent accessibility (Acc score) and secondary structure (2nd str) of the potential cleavage site, calculated by DSSP, are used to enhance the prediction of whether the folded protein is a potential protease substrate (adapted from PDB: 4N7I).
The Lib hP library contains nearly complete coverage of human proteome sequences in 49 amino acid blocks, tiled with 25 amino acid overlaps for duplicate coverage, containing ∼730,000 individual sequences (Fig. 1C and SI Appendix, Figs. S2 and S3). The Lib hP has the advantage that all substrate sequences are from the human proteome, and it provides direct information about what linear sequences can be cut in the human proteome. The Lib hP also provides broader coverage than one can typically achieve by proteomics or RNA sequencing (RNA-seq) because no single cell line expresses the entire genome (SI Appendix, Fig. S2 E and F). When coupled with sequence and structural bioinformatics, the Lib hP data identify candidate substrates for more detailed analysis at the protein level (Fig. 1G).
Each substrate phage displays an avi-Tag on its N terminus to permit quantitative biotinylation and immobilization on streptavidin magnetic beads (Fig. 1A). For the input library, we biotinylated the displayed peptide in vitro. We confirmed the quantitative presence of biotin attached to the peptide displayed on phage using a phage enzyme-linked immunosorbent assay (ELISA) (SI Appendix, Fig. S4 A–C). In subsequent rounds of selection to facilitate biotinylation, we propagated enriched substrate phage pools using Escherichia coli XL-1 Blue cells that we engineered to express intracellular biotin ligase BirA with pBirAcm so biotinylation can be simultaneously done during phage amplification (SI Appendix, Fig. S4 C and D).
Using SPD-NGS to Profile the Linear Specificity of Caspases.
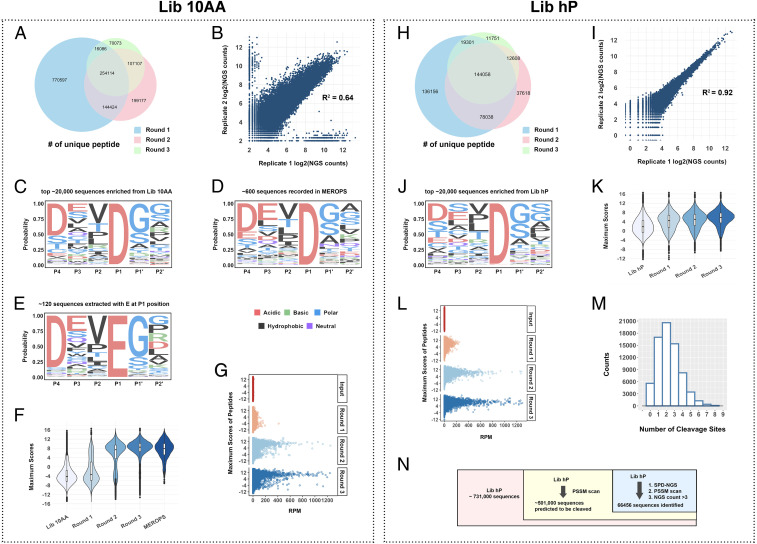
Caspases are of high biological interest because of their powerful roles in cell death and differentiation (18). These are excellent proteases to validate the SPD-NGS approach because they have been extensively studied using traditional synthetic peptide libraries and proteomics. We began by profiling caspase-3 (Fig. 2). It is challenging to know a priori the ideal concentration of enzyme, incubation time, and number of rounds of selection for optimal profiling. Thus, we determined these empirically by covering a 1,000-fold range of enzyme concentrations and monitoring enrichment as a function of multiple rounds of selection. Briefly, avidin magnetic beads were incubated with the Lib 10AA phage and washed extensively to remove nonbound phage, and freshly expressed caspase-3 was added for 30 min at room temperature at enzyme concentrations varying from 1 to 1,000 nM. We measured the number of released phage by counting infectious units as a function of round of selection (SI Appendix, Fig. S5). The number of released phage over the untreated substrate phage beads increased by 10- to 1,000-fold as a function of round of selection and enzyme concentration, suggesting strong positive selection.
Fig. 2.
Validation of SPD-NGS libraries to identify protease specificity for caspase-3. (A) The Venn diagram of unique peptides identified from Lib 10AA as a function of rounds of selection. The numbers decrease due to increasing enrichment of better substrates with rounds of the selection. (B) The relatively strong correlation between two biological replicates (R2 = 0.64) indicates the reproducibility of the selection strategy. (C) The sequence logo for caspase-3 substrates generated by aligning the top 20,000 sequences based on counts identified from screening Lib 10AA (round 3, P4-P2′ was determined based on existing knowledge). (D) The sequence logo for caspase-3 substrates generated from substrates compiled in the MEROPS database. (E) A similar motif is obtained by selecting only proteolytic sites with a glutamate (E) at the P1 position as seen previously by N-terminomics (23). (F) Violin plot for distribution of the maximum scores of 10 amino acid sequences for the input library, round 1, 2, and 3 outputs, and the scores of peptides in the MEROPS database. The peptides with higher scores get progressively enriched in round 2 and 3, and round 3 match scores are seen in MEROPS. (G) The plot of maximum score vs. RPM for peptides in the input library and round 1, 2, and 3 outputs. The peptides with higher scores get enriched faster. (H) The Venn diagram of unique peptides identified from Lib hP decreases with each of three rounds of the selection as enrichment increases as was seen for Lib 10AA. (I) The correlation for two biological replicates (round 3) for the Lib hP. A stronger correlation (R2 = 0.9) between two biological replicates indicates the reliability of the selection strategy and the reduced starting library size for Lib hP (∼106) compared to Lib 10AA (∼109). (J) The sequence logo of caspase-3 substrate consensus generated by the top 20,000 cleavage events identified from Lib hP. (K) Violin plot of the maximum scores of the Lib hP input library and progressive enrichment of substrates as one progresses from outputs of round 1, 2, and 3, which then aligns with MEROPS seen in Fig. 3D. (L) The plot of maximum score vs. NGS RPM for peptides in input library and round 1, 2, and 3 outputs. The peptides with higher scores get enriched faster. (M) Distribution of frequency observed as a function of number of cuts in the 49 amino acid peptides. Most have one cut, but some have multiple cuts that decay monotonically. (N) Venn diagram of the library (Lib hP), the peptides passing PSSM scan, and the peptides identified from SPD-NGS.
After each round of selection, NGS was applied to sequence the protease sensitive pools. With increasing rounds of selection, we would expect the number of unique reads to decrease and NGS counts per sequence to increase as good substrates are selected over poor ones. Indeed, from an average of 500,000 NGS reads per round, we identified roughly 377,000, 272,000, and 187,000 unique sequence reads from rounds 1, 2, and 3, respectively (Fig. 2A). The selection for better substrates was further supported by the Venn diagram (Fig. 2A) showing that generally more than 60% of the substrates identified in a later round were also present in the set from the previous round. Two biological replicates were performed for each of the three rounds of positive selections for caspase-3 at 1 µM; these showed a strong correlation (R2 = 0.64), indicative of the good reproducibility of the selections (Fig. 2B). This is not a perfect correlation, which is likely the result of NGS sampling only a small portion (∼105 to 106) of the possible unique sequences due to high diversity of the starting Lib 10AA (∼109). The reads per million (RPM) for each unique sequence ranged from a few to over 1,000, indicative of the wide dynamic range and variability in rates of hydrolysis for individual substrates.
We next analyzed the enrichment based on Z-scores over the starting library for each of the 20 amino acids as a function of round of selection and enzyme concentration used over the 10 amino acid substrate window (SI Appendix, Fig. S6). We saw obvious enrichments for particular amino acids (Asp, Glu, and Gly) and deenrichment for others (Leu and Arg), especially in the middle of the 10 amino acid window in round 3 (SI Appendix, Fig. S6 C, F, and I). Also, simple analysis of residue preferences across the 10 amino acid substrate window showed a strong selected amino acid preference in the middle six to seven residues favoring Asp, Glu, Val, and Gly and disfavoring Phe, His, Lys, Arg, and Trp (SI Appendix, Figs. S6 and S7A). These preferences increased with increasing round of selection and enzyme concentration, further suggesting positive selective pressure for some residues and not others. These data are consistent with structural data showing caspase-3 binds a six-residue linear stretch of peptide (SI Appendix, Fig. S7B) with known subsite preferences, such as P4 (acidic), P3 (acidic), P2 (hydrophobic), P1 (acidic), and P1′ (small). We generated a sequence logo based on a simple MSA of the top 20,000 unique peptides cleaved by caspase-3 using the DECIPHER package (SI Appendix, Fig. S7C). A clear DEVDGG motif showed up in the sequence logo. We then extracted the motif and defined the P4-P2′ position based on existing knowledge of caspase-3 substrates. Based on the motif, we generated the Z-score enrichment values over the input library for a six-residue window (SI Appendix, Fig. S7D). The top scoring residues reflected the classic caspase-3 motif, DEVD|GG. We can also represent these data in a familiar sequence logo that expresses the probability of each amino acid at each of the six positions (Fig. 2C). This logo based on ∼20,000 sequences is remarkably close to what has been seen from synthetic peptide libraries and proteomics compiled in the MEROPS database of about ∼600 sequences (19–22) (Fig. 2D and SI Appendix, Fig. S7E). It is also known that caspase-3 can cleave with Glu at P1 (23). Indeed, a sequence logo derived from positively selected peptides containing a Glu at P1 position after MSA (Fig. 2E) was virtually identical to the motif for Asp (Fig. 2C) (DEVE|GG versus DEVD|GG), but not surprisingly a much smaller percentage (0.56%).
Generating an Unbiased PSSM for Caspase-3 Using Consensus Data from Lib 10AA.
To more rigorously compare the quality of substrates and to predict the precise cleavage sites on the 49 amino acid sequences identified from Lib hP, we developed a traditional PSSM. A peptide is predicted to be cleaved by caspase-3 if:
where is a substrate score, A is an indicator of peptide sequence ( if the amino acid at position p of the peptide is q, and otherwise), θi,p,q is a number in PSSM reflecting the preference of a protease for an amino acid (q) at a certain position (p), and is a scoring threshold, specific to the protease (Fig. 1C and SI Appendix, Fig. S8) (24). θi,p,q is positive if a certain amino acid is preferred and otherwise negative. An example showing the calculation of substrate score is shown in SI Appendix, Fig. S8E. In most cases, is 0 although one can set a higher threshold for only good substrates. We arbitrarily assumed that the contribution of each peptide position to selective recognition is additive. For caspase-3, our model takes into account a 6 amino acid peptide: positions P4-P2′ defined based on existing knowledge, according to alignment and caspase-3 crystal structure (SI Appendix, Fig. S7 B and C).
The PSSM model, described further in the Materials and Methods, generated a quantitative score for 6-mer substrate peptides scanned over the selected sequences. Applying the PSSM to the caspase-3 NGS dataset from each round at the highest concentration (1 µM, 30-min treatment), we generated violin plots showing the distribution of the number of peptides versus the maximum PSSM score a 6 amino acid peptide within the 18 amino acid window (4 amino acid [flanking region] on each side + 10 amino acid) (Fig. 2F and SI Appendix, Fig. S8F). The majority of sequences in the input library had relatively low scores, and there was little change at round 1. After round 2, the majority were high scoring sequences, and, by round 3, almost all sequences were of high scores. The violin plots were generated based on more than 4× 500,000 sequences. If we set for caspase-3 to be a zero threshold, the PSSM model derived from our data produced a true-positive rate >95% of the MEROPS database. Another way to view the enrichment as a function of round of selection was simply by plotting the NGS counts (represented by RPM) for each peptide versus their corresponding maximum PSSM score (Fig. 2G). There is a clear increase in the number of highly scoring peptides as the selection proceeds through the three rounds relative to the unselected input.
Selection of Caspase-3 Substrates from Lib hP.
We next applied the SPD-NGS protocols that were optimized for caspase-3 on Lib 10AA to the human focused Lib hP. As for Lib 10AA, we conducted three rounds of selection and similarly saw the number of unique selected sequences decreases significantly (Fig. 2H). Again, about 80 ∼ 90% of the sequences found in the round 2 pool were present in round 1 and, similarly for round 3, captured in round 2, suggesting the better sequences were winning over poorer sequences. We also analyzed the data by filtering out low abundance peptides having fewer than three reads and obtained similar results, albeit with an almost twofold lower number of candidate substrates (SI Appendix, Fig. S9A). However, the overlap between rounds was more suggesting the unfiltered set would have more false positives so we retained a threshold of greater than three reads. We also tested the reproducibility of the selections by conducting two biological replicates for three rounds on Lib hP. A plot of the number of NGS counts per identical peptide sequence for replicate 1 versus 2 (Fig. 2I) showed a linear and remarkably high correlation (R2 = 0.92). We believe the higher correlation coefficient seen among these replicates from Lib hP is a result of the 1,000-fold lower diversity of this library compared to Lib 10AA, allowing more complete capture of positive clones.
Subjecting the top selected 49 amino acid sequences to a simple MSA to get a reliable consensus is much more challenging given the many possible alignments one could generate in a 49 amino acid window from Lib hP versus the 10 amino acid window in Lib 10AA. However, it is reasonable to assume the sequence preferences for caspase-3 are the same whether cut in a 10 amino acid window or a 49 amino acid window. Thus, we applied the six-residue PSSM derived from Lib 10AA to Lib hP for identifying potential cleavage site(s) and derived a sequence logo for the top 20,000 enriched sequences as we did for Lib 10AA (Fig. 2J). All potential cleavage sites (with a score greater than 0) were included in the sequence logo. To do this, we calculated the score of each possible 6-mer on every cleaved sequence in the 49 amino acid NGS dataset after three rounds of selection and considered it a cleavage site if . Indeed, the sequence logo derived from these data (Fig. 2J) is remarkably close to that seen in the consensus from Lib 10AA and the MEROPS database (Fig. 2 C and D). Violin plots were generated from these and revealed a consistent increase in the average NGS counts as the selection proceeded from round 1 to 3 (Fig. 2K). The change in distribution as a function of rounds is also evident from the increase in counts for specific sequences as selections proceeded (Fig. 2L) as was seen in the Lib 10AA data (Fig. 2G). The increases in NGS counts per round for Lib hP were less dramatic than seen for Lib 10AA and likely reflect the fact that the average number of cuts within the 49 amino acid window was 1.5 and ranged from 0 to 6 (Fig. 2M and SI Appendix, Fig. S9B). We also tested if we could simply apply the PSSM score derived from the Lib 10AA to identify potential cleavage sites in the human proteome. Applying the PSSM on the Lib hP sequences identified ∼500,000 peptides that could potentially be cleaved by caspase-3, which was about 10 times more than the actual ∼60,000 peptides (NGS counts >3) enriched from SPD-NGS screening of the Lib hP (Fig. 2N). Although the MSA and PSSM are useful for identifying the most probably cut site within a selected clone, they are not a substitute for the experimental selections.
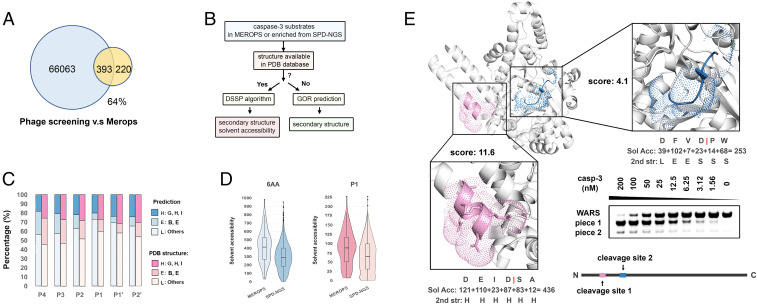
We next compared the unique human sequences identified by SPD-NGS (∼60,000) to those found by mass spectrometry (∼600). As seen in the Venn diagram in Fig. 3A, nearly 64% of the caspase-3 substrates recorded in MEROPS database is found in the Lib hP dataset (Fig. 3A). There are ∼700,000 Asp in the human proteome, and only 4% are cut in the Lib hP, indicating much more is required than simply a P1 Asp for efficient cutting. We also find about 100-fold more substrates than have been identified by mass spectrometry. This could be for a number of reasons. Mass spectrometry of lysates would typically miss the ∼4,000 membrane and extracellular proteins that would not be cleaved by intracellular caspases. In addition, cells only express about half of their genomes, and low abundance proteins would be missed by mass spectrometry methods (SI Appendix, Fig. S2 E and F).
Fig. 3.
Triaging the candidate linear substrates by structural bioinformatics. (A) SPD-NGS identifies 64% of caspase-3 substrate sequences recorded in the MEROPS database. (B) The workflow showing to use the secondary structure and solvent accessibility of the cleaved peptides enriched in SPD-NGS in a protein context (if PDB structures are available) as supplement to determine whether the protein where the peptide comes from is a potential substrate. The GOR algorithm was applied to calculate the secondary structure of those peptides enriched from Lib hP whose structures are unavailable in the PDB database. (C) Caspase-3 has a known structural preference for cutting loop>helix>sheets, and this matches structural bioinformatics for sites identified from Lib hP. The DSSP (structure available: structure could be found in PDB database) or GOR method was used for secondary structure prediction. We have mapped results “H,” “G,” and “I’ to α-helix (H); “B” and “E” to β-sheet (E); and “S,” “T,” and “L” to loop (L). (D) The violin plots of solvent accessibility for the residue at P1 or sum of the solvent accessibility of the 6-mer (structure available) identified from Lib hP and recorded in the MEROPS database. (E) Validation of an unreported protein substrate of caspase-3, tryptophanyl-tRNA synthetase (WARS) identified by SPD-NGS. The ribbon diagram shows the two sites (1 and 2) with preference score of 11.6 and 4.1 and solvent accessibility of 436 and 253, respectively. SDS gel shows the most accessible site 1 is cleaved at lower caspase-3 concentrations compared to less accessible site 2. (PDB structure 5UJJ).
Using Lib hP Data Plus Solvent Accessibility to Predict New Protein Substrates.
Although the Lib hP can broadly identify what can be cut when exposed, we anticipate the majority of these sequences may not be accessible to the protease in the native folded protein substrate. Thus, to help triage the candidate linear substrates, we found it useful to apply a global modeling and surface accessibility score, Dictionary of Secondary Structure of Proteins (DSSP) (25), to rank cleavage sites based upon surface accessibilities. The application of DSSP requires a three-dimensional (3D) structure which only applies to a subset. For those substrates where Protein Data Bank (PDB) structures are not available, we estimated their secondary structure using the Garnier–Osguthorpe–Robson (GOR) algorithm (26) (Fig. 3B). Previous proteomics studies have shown that caspases have a preference for cleaving loops over helices over sheets in native proteins (19). When we apply our candidate sequences from the Lib hP to secondary structure prediction algorithms or look at the structures in a protein context, we find a similar preference for loop, followed by helix and then sheet (Fig. 3C and SI Appendix, Fig. S9C). A plot of the solvent accessibility values for those substrates (6-mer) or residues at the P1 position of known structure shows a broad Gaussian distribution (Fig. 3D). We made the same calculation for those substrates found in the MEROPS dataset for the caspase-3 (Dataset S1) and found a higher mean score (Fig. 3D), which emphasizes the role of accessibility.
Realizing that site accessibility is critical for cleavage by caspase-3 (27), we scanned our dataset to identify potential substrates with a known structure. For caspase-3, we filtered the sequence list based on the intracellular location expected for substrates of caspase-3. For a solved structure or a comparative model, we used the DSSP program to assess secondary structure (mapping results “H,” “G,” and “I” to helix; “B” and “E” to sheet; and “S,” “T,” and “L” to loop) and solvent accessibility (25). When a structure or model was not available, we used sequence-based GOR algorithms to predict secondary structure (Datasets S2–S6) (26).
We chose to test a protein that had not been reported before, tryptophanyl-tRNA synthetase (WARS). We identified two candidate sites in the WARS protein with solvent accessibility of 436 (DEID|SA, cleavage site 1) and 253 (DFVD|PW, cleavage site 2) (Fig. 3E and SI Appendix, Fig. S10 A and B). The cleavage site 1 was located in a helix and had a PSSM score of 11.6 whereas the cleavage site 2 was located in a sheet and had lower PSSM score of 4.1. DSSP is a tool that can be used to calculate the solvent accessibility (Sol Acc) and secondary structure (2nd str) of individual amino acids in a protein using the structure information (25). We summed up the accessibility of each amino acid of the 6-mers as the parameter (solvent accessibility). After expressing the WARS protein in E. coli, we treated it with different concentrations of caspase-3 and used sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) to probe for these two cleavage events. Indeed, we observed two cleavage sites and confirmed the molecular weight of each piece with liquid chromatography–mass spectrometry (LC–MS) (SI Appendix, Fig. S10). Moreover, the DEID|SA sequence was cleaved much more rapidly than the DFVD|PW site, as predicted by both higher solvent accessibility and PSSM score. In addition to WARS, we observed many more caspase-3 substrates that have been reported previously (22, 28). For example, the apoptotic protease-activating factor1 (APAF1) can be cleaved by caspase-3 at the SVTD|SV site, which locates between a helix and loop structure with a PSSM score of 10.3 and solvent accessibility of 232. All these parameters indicate that the SVTDSV sequence on APAF1 is highly preferred by capase-3 in the protein context. We list some of the more prominent examples in SI Appendix, Table S1.
Generalization of SPD-NGS to Other Caspases.
Having built and validated the two complementary substrate phage libraries and established a workflow to collect and analyze the SPD-NGS data for caspase-3, we expanded the approach to other caspases involved in cell death for which extensive proteomics work has been applied, including caspases-2, -6, -7, and -8. We conducted the two-step substrate phage selections with Lib 10AA and Lib hP as performed on caspase-3, using freshly expressed recombinant active caspases-2, -6, -7, and -8 (SI Appendix, Figs. S11–S15). The caspase-2 used in this study contained no N-terminal caspase recruitment domain (known as ΔCARD-caspase-2), and caspase-8 had the death-effector domain removed (known as ΔDED-caspase-8). The caspase activities were validated on fluorogenic substrate Ac-DEVD-R110 (SI Appendix, Fig. S15). As for caspase-3, each of the selections using Lib 10AA showed increasing enrichments as the three rounds proceeded. This permitted us to generate a PSSM for each caspase and to construct sequence logos (SI Appendix, Figs. S11–S14). Similarly, the selections with the Lib hP proceeded with enrichment through the three rounds, and application of the Lib 10AA PSSM allowed generation of sequence logos from both Lib 10AA and Lib hP data.
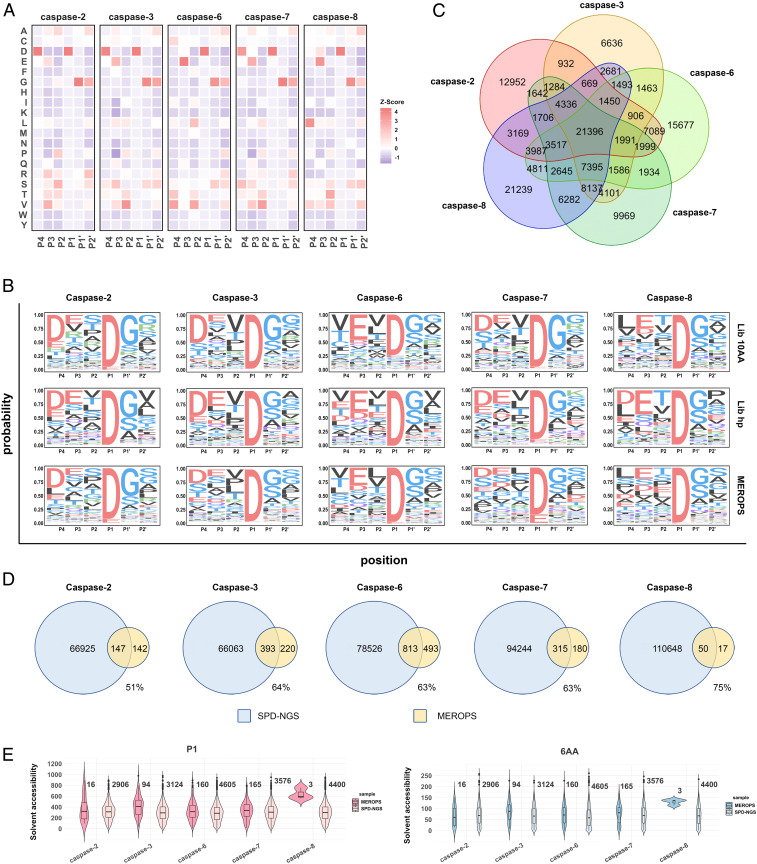
These experiments generated a massive amount of data which was compiled to allow comparison among the apoptotic caspases both from the SPD-NGS and existing proteomics data (Fig. 4). We first compared the Z-score enrichment heat maps for all five caspases generated from the Lib 10AA data to obtain general comparative features (Fig. 4A and SI Appendix, Figs. S11–S14 and S16). Using hierarchical clustering, which incorporates all of the data, it is apparent that caspases-3 and -7 are most similar; this is not surprising, given their central role as executioner caspases. Both have the dominant DEVD|GG motif and are almost indistinguishable in other general subsite details. Caspase-2 is more closely related to these two executioners, except for subtle differences at P2 showing a dominant DESD|GG motif. The biggest difference is the preference for polar residues (S, T, R) at P2 compared to the V at P2 for caspases-3 and -7. This raises the possibility that caspase-2 is more executioner-like in its function compared to caspase-6 and -8 (29). In contrast, caspase-6 and -8 are more similar to each other, having a dominant VEVD|GG and LETD|GG motif, respectively. These two differ most at P4 from the executioners. Although caspase-6 has long been thought to be more executioner-like because of higher sequence homology, more recent studies suggest it plays a more independent role and may be more of an initiator, like caspase-8 (30, 31). Interestingly, the heat maps also show that, although Asp is clearly dominant for all of the caspases at P1, the next most enriched residue is Glu, suggesting it should not be ignored at possible cleavage sites, assuming other subsites are strongly satisfied.
Fig. 4.
SPD-NGS for profiling substrates of other caspases. (A) Z-score heat map showing positional enrichment or deenrichment of each amino acid at P4-P2′ compared to the input libraries (Lib 10AA). (B) Sequence logos of substrates for purified caspase-2, -3, -6, -7, and -8 generated from the top 20,000 sequences selected by SPD-NGS from Lib 10AA, Lib hP, and for 300 to 1,300 sequences reported in the MEROPS database. (C) Venn diagrams for the overlap of substrates found by SPD-NGS for each caspase. (D) Venn diagrams of the sequences enriched from Lib hP (ranging from 43,000 to 127,000) and those reported in the MEROPS database (ranging from 300 to 1,300). The SPD-NGS from the Lib hP captured 50 to 75% of those reported in MEROPS and generally identified 100x more cut sites. (E) The violin plots of solvent accessibility for the residue at P1 or sum of the solvent accessibility of the 6-mer (structure available) identified from Lib hP and recorded in the MEROPS database. The proteolyses recorded in the MEROPS database are real cleavages, most of which are exposed.
We generated three sequence logos for each of the caspases from the Lib 10AA, Lib hP, and MEROPS database for comparison (Fig. 4B). There is remarkable agreement in the patterns seen when comparing the data obtained by either SPD-NGS method to proteomics for each caspase. When we compare across caspases, we come to the same interpretation we did from the heat map data in Fig. 4A.
We next analyzed specific human substrates found in the SPD-NGS Lib hP dataset for all five caspases, caspase-2, -3, -6, -7, and -8, uploaded into Datasets S2–S6, respectively. These resource datasets present gene name of each substrate, the 49 amino acid sequence, cleavage site predicted, PSSM score, subcellular location, PDB structures if available, solvent accessibility, and secondary structure of the predicted cleavage site. For caspase-2, -3, -6, -7, and -8, we identified ∼60,000 unique sequences having greater than three NGS reads from ∼10,000 proteins, respectively. The two- to three-fold range in numbers of unique substrates identified is narrow and generally tracks with the relative specific activity of each caspase. We also compared the overlap of substrates identified in Lib hP among the five caspases (Fig. 4C). Interestingly, there was an overlap of ∼20,000 common substrates, suggesting some redundancy, but a larger set of 6,000 to 20,000 that were unique to each caspase. We do not believe the unique sets are due to sampling issues since we found high reproducibility in the biological replicates (Fig. 2I).
In Fig. 4D, we compare the overlap of substrates found by SPD-NGS from the Lib hP and corresponding MEROPS datasets. Remarkably, on average, more than 50% of the substrates identified by proteomics are contained in the respective SPD-NGS datasets. Eukaryotic translation initiation factor 5A-1 (EIF5A), a known caspase-2 substrate with reported cleavage sites of ETGD|AG and DDLD|FE, was identified in SPD-NGS. Both sites are very solvent accessible (Sol Acc >600) and on a loop, with PSSM scores 6.7 and 1.7, respectively. Annexin A5 (ANXA5), reported to be cleaved by caspase-6 at VVGD|TS and LEDD|VV sites, showed up as well in SPD-NGS datasets. The former one has a PSSM score of 4.3 and solvent accessibility of 270, and the latter one has a PSSM score of 2.6 and solvent accessibility of 243; both sites are on helix structure. Transitional endoplasmic reticulum ATPase (VCP or p97) is a very good caspase-6 substrate, known to be cleaved at multiple sites (e.g., VTMDDF, VAPDTV, DELDSI, VGYDDI, DDVDLE, and TEMDGM), most of which were detected in SPD-NGS. Caspase-7 also cleaves EIF5A and VCP at multiple sites like caspase-2 and -7 do. The SPD-NGS data show caspase-8 cleaves known BH3-interacting domain death agonist (BID) and caspase-8 precursor (CASP8) at LQTD|GN for BID and VETD|SE and LEMD|LS for CASP8. More examples can be found in Datasets S2–S6.
The number of human substrates identified from the caspase-3 Lib hP dataset was 66,000 substrates compared to about 600 so far mostly identified by proteomics, a difference of 400-times more in the SPD-NGS dataset. We believe greater accessibility of the linear peptides in the Lib hP versus accessibility in the folded proteins in the proteome is the major factor that accounts for much of this difference. To compare these datasets based on accessibility, we filtered the SPD-NGS dataset for those substrates where a structure is available in the PDB to allow calculation of accessibility by DSSP (values shown in Datasets S2–S6). A plot of the solvent accessibility value for those substrates of known structure shows a broad Gaussian distribution (Fig. 4E). We made the same calculation for those substrates found in the MEROPS dataset for the five caspases (Dataset S1) and found a higher mean score (Fig. 4E). We would expect that the true protein substrates would have a distribution more like the proteomics data. Since the sample sizes of true protein substrates for caspases are small, finding an accessibility threshold for the linear peptides in their native folded protein is rather challenging. Substrates identified in SPD-NGS from the Lib hP with a P1 accessibility >25 and 6 amino acid stretch accessibility >100 would be top candidates for further detailed validation in vitro.
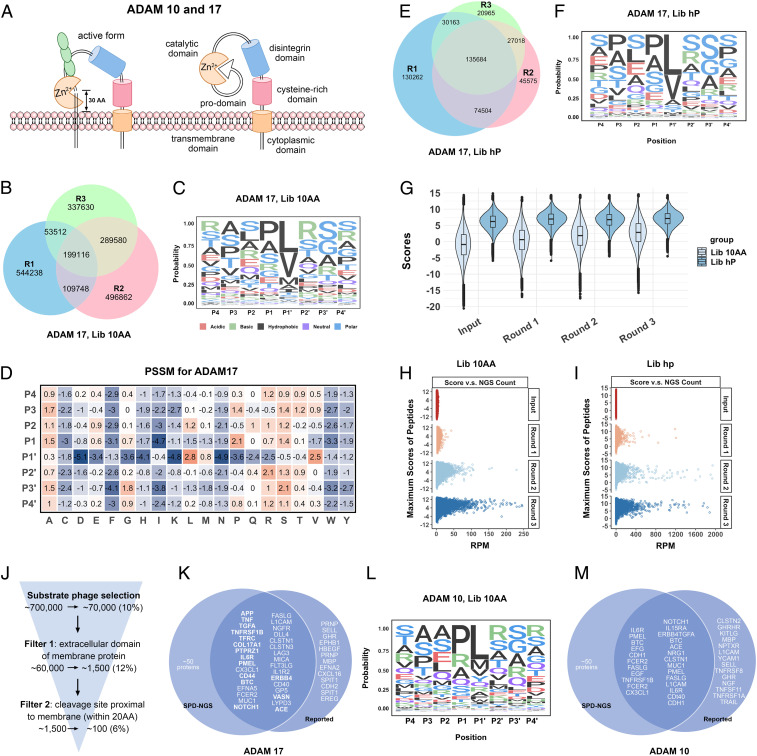
Generalizing the Protocol to the ADAMs Family Sheddases—ADAM10 and ADAM17.
ADAM10 and -17 are membrane-bound proteases involved in shedding of extracellular domains in signaling (Fig. 5A). They are produced as inactive zymogens that become activated. They are more highly activated in the tumor microenvironment (32), and identifying substrates is of high interest in cancer. Although only 50 substrates are known for ADAM17, many are of high biological interest, such as TNF, NOTCH1, APP, and EGF. The specificity of ADAM10 and -17 has been studied using a library of 200 synthetic 10-mer peptides (33).
Fig. 5.
Mapping ADAM10 and -17 proteases specificity. (A) Schematic illustration of ADAM10 or -17 and the cleavage of their substrates proximal to the membrane (within 30 amino acids range). (B) The Venn diagram of individual peptides identified from Lib 10AA in each of the three rounds of the selection with ADAM17. (C) The sequence logo of ADAM17 substrate consensus generated by aligning the top 10,000 most frequently observed sequences from screening Lib 10AA. (D) The PSSM of ADAM17 substrates generated according to the alignment. A score of every 8 amino acid peptide window was calculated based on the PSSM. (E) The Venn diagram of unique peptides identified from Lib hP in each of the three rounds of the selection with ADAM17. (F) The sequence logo of ADAM17 substrates generated from the top 50,000 cleavage events identified from Lib hP. (G) Violin plot of the maximum scores of either Lib 10AA or Lib hP peptides in the input library, round 1, 2, and 3 outputs. The peptides with higher scores get enriched with more rounds of selection. (H and I) The plot of maximum score vs. NGS counts RPM for Lib 10AA or Lib hP peptides, respectively, for each input library and round 1, 2, and 3 outputs. (J) The filters applied to identify protein substrates of ADAMs. (K) Venn diagram of ADAM17 substrates identified in our screening and ones that have been reported in the literature (the substrates with a known cleavage site are in bold, and details are in Table 1). (L) The sequence logo of ADAM10 substrate consensus generated by 10,000 events identified from Lib 10AA. (M) Venn diagram of ADAM10 substrates identified in our screening and the ones reported as a substrate (some of the substrates without a known cleavage site).
We applied our SPD-NGS libraries for deeper sequence coverage and to identify optimal sequences and potentially identify new protein targets. To identify the cleavage sites of ADAMs, we first expressed extracellular domains (ECD) of ADAM10 and -17 in human Expi293 cells. Briefly, after purifying them with nickel-nitrilotriacetic acid (Ni-NTA) resin, we confirmed their activities by monitoring the cleavage of the known fluorogenic peptide substrate, Mca-KPLGL-Dpa-AR-NH2 (SI Appendix, Fig. S17). After three rounds of phage selection against Lib 10AA using ADAM17 (Fig. 5B and SI Appendix, Fig. S18A), we generated a sequence consensus from the NGS dataset using MSA of the top ∼10,000 enriched sequences from round 3 and extracted the 8-mer motif by fixing the large hydrophobic residue at P1′ (Fig. 5C). It is known that ADAM10 and -17 have a preference for large hydrophobic residue at P1′. (34) Remarkably, the sequence logo was similar to that derived from ∼20 substrates from MEROPS sequence data (SI Appendix, Fig. S18B). Based on the alignment, we then generated a PSSM (Fig. 5D).
We next used the Lib hP to select for human-derived sequences for ADAM17. After three rounds of selection (Fig. 5E), we generated a sequence logo (Fig. 5F), largely matching the sequence logo derived from Lib 10AA (Fig. 5C). We constructed violin plots of the maximum scores of the 10 amino acid or 49 amino acid sequences to monitor enrichment as a function of round of selection (Fig. 5G). We saw the change in the violin plots was more subtle than seen for the caspase selections (Fig. 2 F and K), which likely reflects lower specificity requirements for the ADAM17 substrates. The lower specificity of substrates for ADAM17 relative to caspases likely explains the larger number of substrates identified (Fig. 5 B and E). This is further supported by the lower raw NGS counts seen for top selectants as a function of round of selection for both Lib 10AA and Lib hP for ADAM17 (Fig. 5 H and I) compared to caspase-3 (Fig. 2 G and L) of >250 RPM versus >1,200 RPM for ADAM17 and caspase-3, respectively.
The 10AA Lib results revealed a broad proteolytic specificity of ADAM17 for linear peptides. We next sought to identify human protein sequences. From Lib hP, we found >100,000 substrates with greater than three NGS counts; this is clearly a gross overestimate for what to expect for the roughly 50,000 protein isoforms in the human genome. To triage the candidate list of the ∼100,000 cleavable linear human sequences, we applied two filters. First, we know that ADAM17 works on extracellular domains of membrane proteins; applying this criterion reduced the number of candidate sequences to roughly 1,000 (Fig. 5J). Previous studies of the identified substrates show that cleavage occurs within a 30 amino acid window beyond their transmembrane ectodomains (Fig. 5A and Table 1) (35, 36). This is reinforced by a recent X-ray structure of ADAM10, the closest relative of ADAM17, that has been solved (33). When we apply this structural filter for extracellular sequences within 30 amino acid juxtamembrane extracellular regions for type I or II membrane proteins, we are left with about ∼100 putative substrates from the SPD-NGS dataset (Dataset S7). Remarkably, this set captures virtually all of the annotated substrates for ADAM17 that have been validated in the past two decades (Fig. 5K and Table 1). The exact proteolytic sites for lots of the known protein substrates have not been validated yet for the reasons that 1) ectodomain shedding occurs in the immediate extracellular juxtamembrane region, which is also where O-glycosylation is often found; 2) multiple cleavages occur on the same stalk due to the low specificity; and 3) the involvement of other proteases, like other ADAMs family proteases and peptidases. According to our PSSM, most 49-mers enriched from SPD-NGS have multiple cleavage sites if we set threshold τi as 0, indicating the low specificity of ADAM17. In addition to type I and II membrane proteins, we also identified cleavage events in proximal-membrane regions of the extracellular domain of multipass transmembrane proteins (SI Appendix, Fig. S18C) and glycosylphosphatidylinositol (GPI)-anchored proteins. We are not aware of others reporting cleavage of a multipass transmembrane protein by ADAM17 substrates because the fragments are not shed. We provide here a list of all predicted cleavage sites proximal to the cell membrane (Dataset S7).
Table 1.
ADAM17 cleavage sites in protein substrates reported and identified by SPD-NGS
| Gene | Cleavage site | PSSM score | Distance to membrane (no. of amino acids) |
| TNF | LAQAVRSS | 11.56 | 11 |
| TGFA | DLLAVVAA | 8.6 | 9 |
| TNFR | PAEGSTGD | 1.73 | 4 |
| IL1R2 | EASSTFSW | 1.3 | −3 |
| COL17A1 | EKDRLQGM | 2.26 | 35 |
| TFRC | ECERLAGT | 5.5 | 18 |
| IL6R | TSLPVQDS | 5 | 7 |
| NOTCH1 | KIEAVQSE | 4.2 | 15 |
| PTPRZ1 | LAEGLESE | 10.4 | 6 |
| APP | HHQKLVFF | <0 | 14 |
| PMEL | VSTQLIMP | 1.5 | 12 |
| VASN | TPPAVHSN | 2.21 | 17 |
| CD44 | SHGSQEGG | 0.6 | 19 |
| BTC | DLFYLQQD | 1.8 | 7 |
| ERBB4 | STLPQHAR | 5 | 6 |
| ACE | NSARSEGP | 3.2 | 24 |
| TNFRSF1B | FALPVGLI | 1.02 | 0 |
Reported cleavage sites are shown for proteins established as ADAM17 substrates through genetic, knockdown, or proteomics approaches. The same cleavage events were identified in SPD-NGS with PSSM scores, and their distances to membrane are listed. The distance to membrane was obtained from UniProtKB topological domain information.
We applied the same workflow used for ADAM17 to ADAM10, its closest sequence relative (SI Appendix, Fig. S19 and Dataset S8). We conducted three rounds of selection with Lib 10AA, generated a sequence logo (Fig. 5L) and PSSM from the MSA, and then applied these to Lib hP round 3 selectants. We then triaged the ADAM10 substrate list from the Lib hP selectants based on sequences within 30 amino acid of the juxtamembrane extracellular regions for type I or II membrane proteins and identified ∼100 candidate sequences (Fig. 5M). Remarkably, this analysis captured more than two-thirds of substrates reported in the literature or in MEROPS for ADAM10.
Discussion
Substrate phage methods were developed more than two decades ago (14) but only recently have started to emerge as a useful tool for analysis of protease specificity (16, 17). More recent advancements in the fields of synthetic DNA, NGS, and bioinformatics now enable far deeper profiling of protease substrate specificity to identify consensus linear sequences and even candidate protein substrates at unprecedented and proteome-wide scales. SPD-NGS allows for deep profiling of protease specificity of linear peptides sequences at >1,000-fold depth over traditional natural peptide libraries. The sequence logos identified by SPD-NGS are in close agreement with literature-curated proteomic datasets from the two classes we evaluated. In addition to the expanded sequence depth of coverage, there are several other important advantages. These libraries are genetically encoded so provide a simple, cheap, and renewable source accessible by simple molecular biology techniques. They are easily modified, allowing synthesis of more focused libraries. These studies validate a broad framework for comprehensive protease profiling that complements and expands upon existing synthetic peptide and proteomics technologies.
We found the randomized Lib 10AA to be useful to first calibrate cleavage conditions and to develop consensus logos over the typical six- to eight-residue stretch that proteases engage. The depth of coverage allows one to analyze potential subsite cooperativity, a property that would be difficult to analyze with sparse substrate libraries. We analyzed this in the case of the caspase-3. The sequence logo shown in Fig. 2C for caspase-3 substrates was generated with the MSA of the top ∼20,000 sequences enriched from SPD-NGS with Lib 10AA. Extracting all of the sequences from the ∼20,000 sequences with a fixed most abundant residue at each position from P4-P2′ allows the generation of position-specific sequence logos as shown in SI Appendix, Fig. S20. For example, the preferred amino acid distribution at P2 didn’t significantly change with different residues at P4, P3, P2, P1′, or P2′. This was true at other positions, indicating little cooperative effect. It was also possible to develop simple PSSMs based on MSAs for both caspases and the less specific ADAMs proteases. This provided high confidence predictions for cleavage sites that matched literature expectations. After three rounds of selection, we found substrates that range over 1,000-fold in NGS counts (ranging from 3 to 4,000 NGS counts), demonstrating a broad dynamic range of substrate quality. Moreover, each of the five caspases and two ADAMs proteases that we tested had their own unique preferred sequence motif, which highlights the sensitivity of the selections and the ability to generate highly specific protease fingerprints.
We show the Lib hP to be useful for identifying candidate linear substrates in the human proteome. The PSSMs developed from the Lib 10AA were applied to the larger 49-mer peptides in Lib hP to produce high confidence predictions for cleavage sites even when multiple cut sites were identified within the 49-mer. The logos generated from both the Lib 10AA and Lib hP were in close agreement, and selections were highly reproducible. The Lib hP selections routinely captured 10 to 100 times more candidate substrates than those previously identified from a decade of proteomics studies; most of the substrates found from proteomics were contained within the Lib hP selected sets. The SPD-NGS from the Lib hP captured 50 to 75% of those reported in the MEROPS database, but not 100% (Fig. 4D). There are several possible reasons for incomplete sampling. Some sequences may be lost in library construction, phage propagation, PCR amplification, and workup. (SI Appendix, Fig. S2B). Poor substrates with low read numbers could be filtered out. Finally to reduce background, we did three rounds of selections and overselection could miss some of the poor substrates.
Although the Lib hP can broadly identify what can be cut, not all will be accessible to the protease in the context of the folded protein. Thus, to help triage the candidate linear substrates, we found it useful to filter cleavage sites based upon surface accessibilities. The WARS protein served as one example. Secondly, it is useful to triage substrates based upon the cellular location of the protease. In the case of the intracellular caspases, one can exclude targets that are extracellular or within vesicles. This was especially useful in the case of the ADAMs proteases for triaging 60,000 candidate 49-mer substrates that were cleaved by the ectodomain of the two ADAMs proteases. These candidate substrates triaged to 1,000, when considering only domains that were extracellular, and to 100, when considering the structural restrictions that ADAMs cleave substrates within 30 amino acid of the nearest transmembrane ectodomain. Indeed, this triage approach captured virtually all of the known substrates and for both of the ADAMs.
Although SPD-NGS from Lib hP identifies far more candidate human substrates than are likely present in folded proteins, the data can be informatically triaged by compartment and structural considerations as described above. This work will offer tremendous opportunities to deeply profile protease substrate specificities in pure or complex mixtures of proteases, on cells, and in conditioned media and tissue lysate. The dataset generated in this work, much like our Degrabase for caspase cleaved products (37), provides insights for protease inhibitor design and for the determination of the biological function of proteolysis. No single cell line expresses the entire human proteome so the unprecedented sequence coverage of Lib hP can greatly supplement proteomics approaches regarding the identification of unexpressed or low-abundant protein substrates. Moreover, we believe the data generated by SPD-NGS could be a very useful companion for targeted mass spectrometry experiments by providing lookup lists for parallel reaction monitoring (11, 19).
SPD-NGS has limitations. To be precise, the technique does not explicitly define the P1-P1′ site of cleavage for each peptide identified but rather a motif for protease recognition. In the case of caspases and ADAMs, the definition of cleavage site (P1-P1′) is supported by extensive proteomic investigation over many decades. For novel proteases, synthesizing several peptides (∼20) identified in selection and using mass spectrometry to determine the fragment size are required to define a P1-P1′ position. Although Lib hP provides unprecedented protein sequence coverage and allows profiling the entire human proteome against a target protease, the results may not truly reflect the physiological substrate profile because a number of parameters may play a role in regulating proteolysis signature. In addition to the parameters we used as filters to triage datasets, such as spatiotemporal localization, solvent accessibility, and secondary structure at the putative site of cleavage, the relative abundance and concentration of substrate could also affect the substrate profile in cells. Another limitation is that the system is currently restricted to screening purified endo-proteases, not exo-proteases, because the N terminus and C terminus are blocked on the phage. Moreover, this approach based on linear peptides does not take into account the possibility of distal exo-sites that have been seen in rare cases for caspase-7 (38). Specifically, caspase-7 is better at cleaving poly(ADP ribose) polymerase 1 (PARP), despite a lower intrinsic activity than caspase-3. This is because the key lysine residues (K38KKK) within the N-terminal domain of caspase-7 bind PARP and improve its cleavage rate. One way to expand the system to native proteins would be to display whole proteome libraries of folded proteins that may be in reach, given the low cost of synthetic DNA and orfeome complementary DNA (cDNA) libraries. Despite these current limitations, we believe SPD-NGS is now a useful first pass companion to more traditional peptide library and proteomics methods, given the simplicity of the selections and analysis, renewability, reproducibility, depth of coverage, speed, and low cost.
Materials and Methods
Protein Expression and Purification.
ΔCARD-caspase-2, caspase-3, -6, and -7, ΔDED-caspase-8, and WARS were cloned into a His6-affinity tag containing vector pET23b. The active enzymes were expressed in E. coli BL21(DE3) pLysS cells (Promega), and WARS was expressed in E. coli strain BL21(DE3). Details are available in SI Appendix, Table S2. ADAM10 and -17 were expressed in a custom pFUSE (InvivoGen) vector using Expi293 cells and an ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific).
Construction and Characterization of Lib 10AA and Lib hP.
For details, see SI Appendix. The fully randomized 10 amino acid sequences encoded by synthetic DNA [(NNK)10; IDT] were incorporated into the phagemid template by Kunkel mutagenesis (SI Appendix, Fig. S1) (39, 40). The Lib hP was generated by subcloning from a human proteome tiled T7 phage library in 49 amino acid blocks with 25 amino acid overlaps as previously described (41) (SI Appendix, Fig. S2). There is a vestige Flag tag from the subcloning of the Lib hP from the T7 library. This sequence does not affect the selected sequences based on the logos obtained and redundant overlapping tiles, which do not show a Flag sequence effect.
General Phage Selection Protocol.
One milliliter of biotinylated substrate phage library was incubated with 100 µL of magnetic StreptAvidin (SA) beads for 30 min, and, after that, the SA beads were then stringently washed with phosphate-buffered saline (PBS) buffer supplemented with 0.05% Tween 20 and 0.2% bovine serum albumin (BSA). After the SA beads were treated with protease of interest, released phage in supernatant were collected and propagated as a protease-sensitive pool. After three rounds of selection, PCR was conducted, followed by NGS to identify sensitive substrate sequences in each pool. After sequencing, we tallied the NGS read for each peptide and processed the data by a custom R script: https://github.com/crystaljie/NGS_data_process_sample_script_for_substrate_phage_paper_JZHOU. Details are available in SI Appendix.
NGS Sample Preparation.
We used overnight cultures (42) for NGS sample preparation after boiling the phage to release single-stranded DNA (ssDNA) as templates for PCR. For samples from both Lib 10AA and Lib hP, a barcoding PCR process was performed using primers shown in SI Appendix, Fig. S21. The thermal PCR profile for Lib 10AA was as follows: 98 °C (20 s), 63 °C (15 s), 72 °C (30 s), 15 cycles. The thermal profile for Lib hP was as follows: 98 °C (30 s), 72 °C (30 s), 15 cycles. PCR products were gel purified on 1.2% agarose. Illumina library quality was assessed by the Agilent DNA 1000 Bioanalyzer kit according to the manufacturer’s instructions. Libraries were sequenced on a NextSeq or HighSeq 4000 (Illumina) (50 or 150 single read). Custom sequencing primers were of Tm = 67 °C (primer melting temperature), GC% = 50 to 52 (primer sequence GC concent).
Supplementary Material
Acknowledgments
We thank the members of the J.A.W. laboratory and Antibiome Center for inspiring and helpful discussions. We thank M. Hornsby, X. Zhou, and M. Zhong for sharing tips and tricks to construct phage libraries with Kunkel mutagenesis; M. Raghavan and C. Mandel-Brehm for suggestions on constructing the human proteome library; X. Zhou for sharing the pKM0128 phagemid vector; and Y. Yang for coding in bash. J.A.W. thanks the Chan Zuckerberg Initiative and Biohub Investigator Program, as well as National Cancer Institute (NCI) Grant P41CA196276 for financial support of this work. J.Z. is supported by NCI Postdoctoral Fellow Grant F32CA236151-02. J.Y.Z. is supported by NSF Computing and Communication Foundations Grant 1763191, NIH Grant R21 MD012867-01, NIH Grant P30AG059307, and grants from the Silicon Valley Foundation and the Chan Zuckerberg Initiative.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009279117/-/DCSupplemental.
Data Availability.
NGS sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE154923) (43). All study data are included in the article, SI Appendix, and Datasets S1–S8.
References
- 1.Goldberg A. L., Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P. A., Henkel T., Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 12, 141–179 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Mason S. D., Joyce J. A., Proteolytic networks in cancer. Trends Cell Biol. 21, 228–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed C. E., Kita H., The role of protease activation of inflammation in allergic respiratory diseases. J. Allergy Clin. Immunol. 114, 997–1008 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Urban S., Making the cut: Central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7, 411–423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backes B. J., Harris J. L., Leonetti F., Craik C. S., Ellman J. A., Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 18, 187–193 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Wood W. J. L., Patterson A. W., Tsuruoka H., Jain R. K., Ellman J. A., Substrate activity screening: A fragment-based method for the rapid identification of nonpeptidic protease inhibitors. J. Am. Chem. Soc. 127, 15521–15527 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Bowyer P. W., Simon G. M., Cravatt B. F., Bogyo M., Global profiling of proteolysis during rupture of Plasmodium falciparum from the host erythrocyte. Mol. Cell. Proteomics 10, 001636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taoka M. et al., Global PROTOMAP profiling to search for biomarkers of early-recurrent hepatocellular carcinoma. J. Proteome Res. 13, 4847–4858 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Hartmann E. M., Armengaud J., N-terminomics and proteogenomics, getting off to a good start. Proteomics 14, 2637–2646 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Wiita A. P., Seaman J. E., Wells J. A., “Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini” in Regulated Cell Death Pt A: Apoptotic Mechanisms, Ashkenazi A., Yuan J., Wells J. A., Eds. (Methods in Enzymology, 2014), pp. 327–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wildes D., Wells J. A., Sampling the N-terminal proteome of human blood. Proc. Natl. Acad. Sci. U.S.A. 107, 4561–4566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djebali S. et al., Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews D. J., Wells J. A., Substrate phage: Selection of protease substrates by monovalent phage display. Science 260, 1113–1117 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Scholle M. D. et al., Mapping protease substrates by using a biotinylated phage substrate library. ChemBioChem 7, 834–838 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Yi L. et al., Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc. Natl. Acad. Sci. U.S.A. 110, 7229–7234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q. et al., Profiling protease specificity: Combining yeast ER sequestration screening (YESS) with next generation sequencing. ACS Chem. Biol. 12, 510–518 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Li J., Yuan J., Caspases in apoptosis and beyond. Oncogene 27, 6194–6206 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Mahrus S. et al., Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien O. et al., Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles. Proc. Natl. Acad. Sci. U.S.A. 113, E2001–E2010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimbo K. et al., Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc. Natl. Acad. Sci. U.S.A. 109, 12432–12437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings N. D., Barrett A. J., Bateman A., MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seaman J. E. et al., Cacidases: Caspases can cleave after aspartate, glutamate and phosphoserine residues. Cell Death Differ. 23, 1717–1726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiffler M. A. et al., PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 26.Sen T. Z., Jernigan R. L., Garnier J., Kloczkowski A., GOR V server for protein secondary structure prediction. Bioinformatics 21, 2787–2788 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkan D. T. et al., Prediction of protease substrates using sequence and structure features. Bioinformatics 26, 1714–1722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings N. D., Barrett A. J., MEROPS: The peptidase database. Nucleic Acids Res. 28, 323–325 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouchier-Hayes L., The role of caspase-2 in stress-induced apoptosis. J. Cell. Mol. Med. 14, 1212–1224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray D. C., Mahrus S., Wells J. A., Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 142, 637–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravalin M. et al., Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases. Nat. Chem. Biol. 15, 786–794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy G., The ADAMs: Signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 8, 929–941 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Seegar T. C. M. et al., Structural basis for regulated proteolysis by the alpha-secretase ADAM10. Cell 171, 1638–1648.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caescu C. I., Jeschke G. R., Turk B. E., Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem. J. 424, 79–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer A., Bhakdi S., Reiss K., How membrane asymmetry regulates ADAM17 sheddase function. Cell Cycle 15, 2995–2996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommer A. et al., Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat. Commun. 7, 11523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford E. D. et al., The DegraBase: A database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher D., Blais V., Denault J. B., Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc. Natl. Acad. Sci. U.S.A. 109, 5669–5674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tonikian R., Zhang Y., Boone C., Sidhu S. S., Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2, 1368–1386 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Kunkel T. A., Roberts J. D., Zakour R. A., Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367–382 (1987). [DOI] [PubMed] [Google Scholar]
- 41.O’Donovan B., et al. , Exploration of Anti-Yo and Anti-Hu paraneoplastic neurological disorders by PhIP-Seq reveals a highly restricted pattern of antibody epitopes. bioRxiv: 10.1101/502187 (20 December 2018). [DOI] [Google Scholar]
- 42.Pollock S. B. et al., Highly multiplexed and quantitative cell-surface protein profiling using genetically barcoded antibodies. Proc. Natl. Acad. Sci. U.S.A. 115, 2836–2841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., et al. , Peptide gene sequences of substrate phage for different proteases. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154923. Deposited 22 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE154923) (43). All study data are included in the article, SI Appendix, and Datasets S1–S8.