Abstract
Background
Carbapenemase-resistant Enterobacteriaceae (CRE) cause many serious infections resulting in increasing treatment cost, prolonged hospitalization, and mortality rate. Reduced expression and/or mutations of porins and the presence of carbapenemase promote Enterobacteriaceae survival under carbapenem treatments. Development of accurate methods for the detection of antimicrobial resistance is required not only for therapy but also to monitor the spread of resistant bacteria or resistance genes throughout the hospital and community. In this study, we aimed to evaluate the phenotypic methods, Modified Hodge test (MHT), modified carbapenem inactivation method (mCIM), and EDTA-CIM (eCIM) for the detection of carbapenemase-producing Enterobacteriaceae (CPE).
Results
The results showed that mCIM had a sensitivity of 100% and a specificity of 100%, whereas the MHT had a sensitivity of 84.8% and a specificity of 97.8% for the 195 CRE isolates tested (105 CPE and 90 non-CPE isolates). The sensitivity of the mCIM/eCIM to detect metallo-carbapenemases in this study was 89.3% and the specificity was 98.7% as compared to the genotypic PCR detection.
Conclusions
These findings indicate that the mCIM combined with eCIM is useful for detecting and distinguishing different types of carbapenemase in Enterobacteriaceae.
Keywords: Carbapenemase, Enterobacteriaceae, eCIM, mCIM, Phenotypic detection
Background
Enterobacteriaceae are Gram-negative, facultatively anaerobic, non-spore-forming rods and one of the most prevalent causes of invasive infections and nosocomial infections [1]. Infections caused by Enterobacteriaceae are often difficult to treat due to the high antimicrobial resistance among clinical isolates [2]. Carbapenems (imipenem, ertapenem, meropenem, and doripenem) are considered as “antibiotics of last-resort” in the treatment of critically ill patients with a variety of bacterial infections due to their broad spectrum among β-lactam antibiotics and relative resistance to hydrolysis by most β-lactamases [3, 4]. However, carbapenem-resistant Enterobacteriaceae (CRE) have emerged and become a public health threat worldwide in the past decade [5].
While carbapenems enter bacterial cells via specific porins, the reduced expression and/or mutation of porins plays a critical role in the resistance to carbapenems [6, 7]. Moreover, the presence of carbapenemase genes on conjugative plasmids for the hydrolysis of carbapenems to promote Enterobacteriaceae survival under antibiotic treatments is associated with the rapid emergence of CRE [8]. β-lactamases are categorized according to sequence homology into four molecular classes: A, B, C and D [9], whereas carbapenemases are the members of class A, B, and D β-lactamases [10]. Moreover, based on the participation of divalent cations in enzyme activation, carbapenemases are segregated into non-metallo-β-lactamases (zinc-independent serine carbapenemases, classes A and D) and metallo-β-lactamases (MBLs, zinc-dependent, class B) [11]. Therefore, EDTA or dipicolinic acid can serve as chelators to block class B carbapenemases activity by binding zinc [12]. MBL genes such as blaNDM, blaIMP, blaVIM, and non-metallo-carbapenemase genes, blaOXA (class D) and blaKPC (class A), have been frequently reported in CRE, called carbapenemase-producing Enterobacteriaceae (CPE) [8, 13–15].
The characterization of underlying mechanisms leading to carbapenem resistance of clinical isolates is not undertaken by most clinical microbiology laboratories for therapeutic decision-making; however, understanding if an isolate is CPE has significant epidemiological implications for monitoring local epidemiology and also lead to more effective treatment of infections caused by CPE (e.g. ceftazidime-avibactam or meropenem-vaborbactam, which have activity against KPC-producer) [16].
The Modified Hodge test (MHT) is the first Clinical & Laboratory Standards Institute (CLSI) recommended growth-based carbapenemase detection test in 2009 with high level of sensitivity and specificity in detecting carbapenemases [17, 18]. Currently, the modified carbapenem inactivation method (mCIM) has been reported to accurately identify carbapenemases but cannot distinguish between serine and metallo-carbapenemases [19, 20]. Therefore, a further modification to mCIM with the addition of EDTA (eCIM) has been endorsed in the CLSI M100-S28 supplement in 2018 to specifically identify metallo-carbapenemases [19, 20]. In this study, we aimed to evaluate the phenotypic detection methods MHT, mCIM, and eCIM for detecting CPE.
Results
Detection of carbapenemase genes by PCR
To detect the presence of carbapenemase genes among our 419 CRE isolates, we performed PCR on 7 Citrobacter koseri, 8 Klebsiella aerogenes, 21 Enterobacter cloacae, 73 E. coli, and 310 Klebsiella pneumoniae isolates. The PCR results showed that 105 (25.1%, 105/419) CRE isolates were genetically characterized to carry carbapenemase genes: blaOXA-48 was detected in 41 (39.0%) isolates, blaKPC was detected in 34 (32.4%) isolates, blaNDM in 15 (14.3%) isolates, blaIMP in 7 (6.7%) isolates, and blaVIM in 3 (2.9%) isolates. Five isolates carried two types of carbapenemase genes (blaNDM and blaOXA-48 in three E. coli isolates, and blaKPC and blaOXA-48 in 2 K. pneumoniae isolates).
Phenotypic detection of carbapenemase-producing Enterobacteriaceae
The ability of CRE isolates to produce carbapenemases was tested by MHT first. Ninety non-carbapenemase-producing CRE isolates were randomly selected as negative controls. Out of the 195 isolates (105 CPE and 90 non-CPE), 91 were MHT-positive for carbapenemase production (2 false-positive isolates) (Table 1). Moreover, 16 isolates showed false-negative MHT results (Table 1). MHT had excellent sensitivity for the detection of blaOXA-48 (95.1%) and blaKPC (100%), but not blaNDM (46.7%), blaIMP (57.1%), and blaVIM (33.3%) (Table 1). The overall sensitivity and specificity of MHT in this study for the detection of CPE was 84.8 and 97.8%, respectively (Table 1).
Table 1.
Sensitivity and specificity of Modified Hodge test for the detection of carbapenemase-producing Enterobacteriaceae
| OXA-48 | KPC | NDM | IMP | VIM | NDM/OXA-48 | KPC/OXA-48 | carbapenemase (−) | total | |
|---|---|---|---|---|---|---|---|---|---|
| TP (n) | 39 | 34 | 7 | 4 | 1 | 2 | 2 | – | 89 |
| FN (n) | 2 | 0 | 8 | 3 | 2 | 1 | 0 | – | 16 |
| TN (n) | – | – | – | – | – | – | – | 88 | 88 |
| FP (n) | – | – | – | – | – | – | – | 2 | 2 |
| Sensitivity (%)a | 95.1 | 100 | 46.7 | 57.1 | 33.3 | 66.7 | 100 | – | 84.8 |
| Specificity (%)b | – | – | – | – | – | – | – | 97.8 | 97.8 |
aSensitivity = TP/TP + FN
bSpecificity = TN/TN + FP
TP true positive, FN false negative, TN true negative, FP false positive
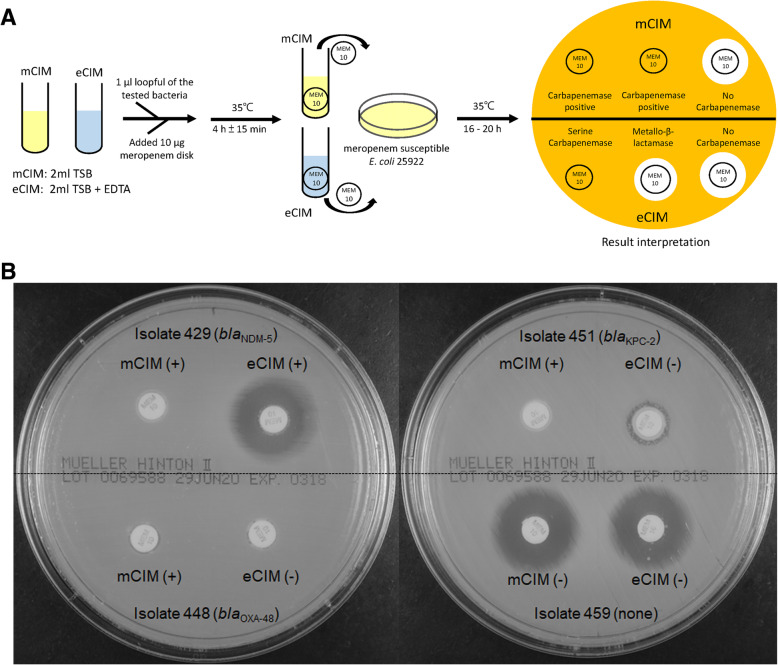
The mCIM and eCIM procedures and interpretation are shown in Fig. 1a. All CPE isolates and 90 non-CPE isolates were used to evaluate the mCIM and mCIM combined with eCIM (mCIM/eCIM) to detect carbapenemase producers. The interpretation of mCIM and eCIM results of clinical isolates were shown in Fig. 1b. Isolate 459 was a non-carbapenemase producer and isolates 448 (blaOXA-48 positive) and 451 (blaKPC-2 positive) were serine-carbapenemase producers (Fig. 1b). In contrast, isolate 429 expressed a metallo-carbapenemase (blaNDM-5 positive) and thus served as mCIM/eCIM positive control (Fig. 1b).
Fig. 1.
The procedure and interpretation of mCIM and eCIM. a. A 1-μL loopful of test CRE isolate is resuspended in two tubes containing 2 mL of TSB. One tube is devoid of EDTA (mCIM), while the other is supplemented with EDTA (eCIM). A meropenem (MEM) disk is submerged in each tube, and the tubes are incubated at 35 °C for 4 h ± 15 min. The disks are then removed from the tubes and placed on MH agar plates upon which a carbapenem-susceptible reporter E. coli ATCC 25922 has been freshly applied. The plates are incubated at 35 °C for 16 to 20 h before the zone sizes are recorded. b. Interpretation of mCIM and eCIM tests of 4 clinical K. pneumoniae isolates. Isolate 459 was carbapenemase negative; 429 had metallo-β-lactamase (blaNDM-5); 448 had serine carbapenemase (blaOXA-48); 451 had serine carbapenemase (blaKPC-2)
Sensitivity and specificity of mCIM and eCIM for the detection of CPE
The distribution of carbapenemase genes and results of phenotypic detection of carbapenemases in our CRE isolates were shown in Table 2. The MHT, mCIM, and eCIM tests were replicated by two independent investigators to ensure reproducibility, and the results showed full reproducibility of these tests. Importantly, two isolates (1 K. pneumoniae and 1 K. aerogenes) with a false-positive result on MHT showed a negative result on mCIM. Moreover, 16 isolates with a false-negative result on MHT were mCIM positive (Table 2). The sensitivity and specificity for the mCIM to detect carbapenemase were both 100% in this study.
Table 2.
Carbapenemase genes, MHT, mCIM, eCIM, and antibiotic susceptibility of 195 CRE isolates
| Species | Carbapenemase-encoding genesa | No. of strains | MHT (n) | mCIM (n) | eCIM (n) | Carbapenem resistance (n)b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neg | Pos | Neg | Pos | Neg | Pos | IPM | ETP | MEM | DOP | |||
| Citrobacter koseri | blaOXA-48 | 4 | 0 | 4 | 0 | 4 | 4 | 0 | 4 | 4 | 4 | 4 |
| None | 3 | 3 | 0 | 3 | 0 | ND | ND | 2 | 3 | 2 | 2 | |
| Enterobacter cloacae | blaIMP | 2 | 1 | 1 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 |
| blaOXA-48 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | |
| None | 9 | 9 | 0 | 9 | 0 | ND | ND | 4 | 9 | 2 | 3 | |
| Escherichia coli | blaKPC | 5 | 0 | 5 | 0 | 5 | 5 | 0 | 5 | 5 | 5 | 5 |
| blaNDM | 11 | 6 | 5 | 0 | 11 | 0 | 11 | 11 | 11 | 11 | 11 | |
| blaOXA-48 | 13 | 1 | 12 | 0 | 13 | 12 | 1 | 10 | 13 | 10 | 11 | |
| blaNDM/blaOXA-48 | 3 | 1 | 2 | 0 | 3 | 2 | 1 | 3 | 3 | 3 | 3 | |
| None | 18 | 18 | 0 | 18 | 0 | ND | ND | 10 | 16 | 6 | 5 | |
| Klebsiella aerogenes | blaVIM | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| blaNDM | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | |
| None | 6 | 5 | 1 | 6 | 0 | ND | ND | 5 | 6 | 3 | 2 | |
| Klebsiella pneumoniae | blaKPC | 29 | 0 | 29 | 0 | 29 | 29 | 0 | 29 | 29 | 29 | 29 |
| blaIMP | 5 | 2 | 3 | 0 | 5 | 1 | 4 | 5 | 5 | 4 | 4 | |
| blaVIM | 2 | 1 | 1 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | |
| blaNDM | 3 | 2 | 1 | 0 | 3 | 0 | 3 | 3 | 3 | 3 | 3 | |
| blaOXA-48 | 23 | 1 | 22 | 0 | 23c | 23 | 0 | 22 | 23 | 22 | 21 | |
| blaKPC/blaOXA-48 | 2 | 0 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | |
| None | 54 | 53 | 1 | 54 | 0 | ND | ND | 28 | 54 | 25 | 23 | |
| Total | 195 | 10 | 91 | 90 | 105 | 79 | 26 | 149 | 193 | 138 | 134 | |
aClass A carbapenemase, KPC; Class B carbapenemase, IMP, NDM, and VIM; Class D carbapenemase, OXA-48
bAntibiotic susceptibility to carbapenems was determined by disc diffusion. IPM, imipenem; ETP, ertapenem; MEM, meropenem; DOP, doripenem
cThe results of 2 mCIM tests were interpreted as “indeterminate” (pinpoint colonies were present within a 16- to 18-mm zone)
Pos positive, Neg negative
The sensitivity for the mCIM combined with eCIM to detect different classes of carbapenemases was shown in Table 3. Twenty-five metallo-carbapenemase producers showed positive results by eCIM. However, a total of 3 isolates escaped the detection of metallo-carbapenemase, including 2 E. coli isolates having both blaNDM and blaOXA-48, and 1 K. pneumoniae having blaIMP. One out of 77 isolates characterized to carry the serine carbapenemase genes showed positive results by eCIM (Table 3). The sensitivity of the eCIM observed in this study was 89.3% (25/28) and the specificity was 98.7% (76/77) as compared to the genotype (Table 3).
Table 3.
Sensitivity and specificity for the mCIM combined with eCIM to detect different types of carbapenemase
| mCIM + eCIM | blaOXA-48 | blaKPC | blaNDM | blaIMP | blaVIM | blaNDM/blaOXA-48d | blaKPC/blaOXA-48 | total |
|---|---|---|---|---|---|---|---|---|
| TP (n) | – | – | 15 | 6 | 3 | 1 | – | 25 |
| FN (n) | – | – | 0 | 1 | 0 | 2 | – | 3 |
| TN (n) | 40c | 34 | – | – | – | – | 2 | 76 |
| FP (n) | 1 | 0 | – | – | – | – | 0 | 1 |
| Sensitivity (%)a | – | – | 100 | 85.7 | 100 | 33.3 | – | 89.3 |
| Specificity (%)b | 97.6 | 100 | – | – | – | – | 100 | 98.7 |
aSensitivity = TP/TP + FN
bSpecificity = TN/TN + FP
cThe results of 2 mCIM tests were interpreted as “indeterminate” (pinpoint colonies were present within a 16- to 18-mm zone)
dIsolates with blaNDM/blaOXA-48 were defined as metallo-carbapenemase positive
TP true positive, FN false negative
mCIM/eCIM assay is designed to simultaneously detect and distinguish the types of carbapenemase. Sfeir et al. showed that the sensitivity and specificity of eCIM was both 100%, in the presence of 5 mM EDTA [21]. Our results showed that the sensitivity and specificity for the mCIM/eCIM to detect MBLs is 89.3 and 98.7%, respectively (Table 3). K. pneumoniae isolate 456, an IMP-8 producer, showed a false-negative result by mCIM/eCIM (Table 4). Our sequencing data identified a wild-type blaIMP-8 in isolate 456. However, isolate 456 showed low resistance to carbapenems (Table 4). Minimum inhibitory concentrations (MICs) of isolate 456 to imipenem, ertapenem, and meropenem were 0.5, 2, and ≤ 0.25, respectively (Table 4). E. coli isolates 488 and 492 containing blaOXA-48/blaNDM-5 also showed false-negative results by mCIM/eCIM (Table 4).
Table 4.
Characteristics of isolates with false mCIM/eCIM results
| Isolatea | Carbapenemase | MIC (μg/ml) | Disc zone (mm) | Phenotypic detection | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | ETP | MEM | IPM | ETP | MEM | DOP | mCIM | eCIM | MHT | mCIM/eCIMa | ||
| False-positive | ||||||||||||
| E. coli 514 | blaOXA-48 | 8 | ≥ 8 | ≥ 16 | 16 | 12 | 15 | 16 | 6 | 23 | – | + |
| False-negative | ||||||||||||
| K. pneumoniae 456 | blaIMP-8 | 0.5 | 2 | ≤0.25 | 22 | 18 | 25 | 25 | 19 | 20 | – | – |
| E. coli 488 | blaOXA-48/blaNDM-5 | ≥ 16 | ≥ 8 | 8 | 6 | 6 | 6 | 6 | 6 | 6 | + | – |
| E. coli 492 | blaOXA-48/blaNDM-5 | ≥ 16 | ≥ 8 | ≥ 16 | 6 | 6 | 6 | 6 | 6 | 6 | + | – |
aAn isolate is positive for metallo-carbapenemase production when the eCIM zone size increases by ≥5 mm compared to the zone size observed for the mCIM and is considered negative for a metallo-carbapenemase if the increase in zone size is < 4 mm
IPM imipenem, ETP ertapenem, MEM meropenem
Discussion
The first CLSI recommended growth-based carbapenemase detection test was the MHT in 2009. Here, we showed that MHT had excellent sensitivity for the detection of blaOXA-48 (95.1%) and blaKPC (100%) (non-metallo-carbapenemase genes) (Table 1). However, the MHT has some limitations, notably insensitivity for the detection of MBL enzymes [22]. Our results (Table 1) are consistent with previous studies which showed a high rate of false-negative MHT results with MBL-producers [19, 23, 24]. Moreover, MHT results are often difficult to interpret, and false-positive results are observed for isolates producing ESBL or AmpC β-lactamase with porin loss [25].
The overall sensitivity and specificity of MHT in this study for the detection of CPE was 84.8 and 97.8%, respectively (Table 1). However, 16 isolates with a false-negative result on MHT were mCIM positive (Table 2). Our results showed that the sensitivity of mCIM to detect MBLs is 100% and the mCIM is more accurate compared to MHT. mCIM/eCIM assay is designed to simultaneously detect and distinguish the different types of carbapenemase. Previous studies showed that the sensitivity and specificity of eCIM was both 100%, in the presence of 5 mM EDTA [21]. However, the number of E. coli and K. pneumoniae isolates used to evaluate mCIM/eCIM assay for identifying CPE in Sfeir’s report was limited [21]. In this study, 50 E. coli (32 carbapenemase producers) and 118 K. pneumoniae (64 carbapenemase producers) were enrolled to evaluate mCIM/eCIM for detecting CPE. Our results showed that the sensitivity and specificity for the mCIM/eCIM to detect MBLs is 89.3 and 98.7%, respectively (Table 3). Therefore, the inconsistency of sensitivity and specificity for the mCIM/eCIM assay to detect MBLs between our results and Sfeir’s findings might be caused by the number of tested isolates. In this study, we used PCR targeting carbapenemase genes as the gold standard to evaluate the performance of phenotypic tests for identifying carbapenemase producers. This might be a limitation as new or truncated carbapenemase genes might not be identified consistently with the phenotype.
K. pneumoniae isolate 456 (blaIMP-8 positive) showed a false-negative result by mCIM/eCIM (Table 4). Although wild-type blaIMP-8 was identified in isolate 456, the expression level of blaIMP-8 in isolate 456 remains to be determined. Isolate 456 showed low resistance to carbapenems (Table 4). In addition, no additional carbapenemase genes (blaGES, blaIMI, blaSME, blaSPM, blaSIM, blaDIM, and blaGIM) were detected in isolate 456. Therefore, it is worth investigating whether the carbapenem resistance level of bacteria is associated with the accuracy of mCIM/eCIM to detect carbapenemase. E. coli isolates 488 and 492 having blaOXA-48/blaNDM-5 showed false-negative results for MBL detection by mCIM/eCIM (Table 4). These results suggest the expression of both carbapenemase genes can cause the misidentification of MBLs by mCIM/eCIM. E. coli isolate 514, which carried only the serine carbapenemase genes (blaOXA-48), showed false-positive results by eCIM based on the PCR targeting carbapenemase genes as the gold standard in this study (Table 4). However, we could not rule out the possibility of a novel carbapenemase gene in isolate 514, and therefore, an entire genome analysis of isolate 514 is worth investigating in the future.
Conclusion
Understanding the mechanism(s) causing carbapenem resistance of Enterobacteriaceae has important clinical implications and results in different prevention measurements and individualized antibiotic therapy. In this study, our results indicate that the phenotypic detection, mCIM combined with eCIM, showed high sensitivity and specificity to detect carbapenemase-producing Enterobacteriaceae, compared with MHT.
Methods
Identification of carbapenem-resistant Enterobacteriaceae isolates
Enterobacteriaceae isolates were recovered in En Chu Kong hospital, 2011 to 2019. These isolates were identified in the clinical laboratory by colony morphology, Gram stain, biochemical tests, and the Vitek 2 system (bioMérieux, Marcy l′Etoile, France) according to the manufacturer’s recommendations. Susceptibility to third-generation cephalosporins (ceftazidime or ceftriaxone, 30 μg/disc, BD BBL™ Sensi-Disc™, Sparks, MD, USA) for Enterobacteriaceae isolates were determined by the disk diffusion method on Mueller-Hinton (MH) agar plates (Bio-Rad, Marne la Coquette, France) based on the CLSI guidelines (M100-S30) [26]. Third-generation cephalosporin -resistant isolates were further tested for their susceptibility to carbapenems, including imipenem, ertapenem, meropenem, and doripenem (10 μg/disc, BD BBL™, USA). MICs to imipenem, ertapenem, and meropenem were further determined by Vitek 2 using the AST-N322 card according to the manufacturer’s instructions. A total of 419 CRE isolates were identified and stored at − 80 °C in tryptic soy broth (TSB) containing 20% glycerol (v/v) until used.
Carbapenemase gene detection
Bacterial genomic DNA was isolated from bacteria grown overnight at 37 °C in 3 mL LB broth. Bacterial culture was centrifuged for 1 min at 12,000 rpm, and the supernatant was removed. Crude DNA extracts were obtained by suspending the pellet in 300 μL distilled water and boiling at 95 °C for 10 min, followed by centrifugation at 12,000 rpm for 5 min. The supernatant containing DNA was transferred to a new eppendorf tube, and the DNA samples were stored at 4 °C until testing.
PCR targeting carbapenemase genes was used as the standard to assess the performance of phenotypic tests. Therefore, PCR amplification for the detection of β-lactamase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaOXA-48, blaGES, blaIMI, blaSME, blaSPM, blaSIM, blaDIM, and blaGIM) was carried out on a iCycler iQ5 real time PCR system (Bio-Rad, USA) with the HotStar PCR SuperMix (GeneDireX, USA). Primers and PCR procedures used in β-lactamase genes detection were described in previous studies [15, 27–29]. The PCR products were analyzed by electrophoresis with 2% agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer. The gels were stained with Novel Juice (GeneDireX, USA), and the PCR products were visualized with UV light. Clinical K. pneumoniae isolates harboring blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48 were used as PCR positive controls. In accessibility to have strains with blaGES, blaIMI, blaSME, blaSPM, blaSIM, blaDIM, or blaGIM as our PCR controls. The PCR products with relevant expected size were purified and verified by sequencing.
Phenotypic detection of carbapenemase-producing Enterobacteriaceae
MHT, mCIM, and eCIM were performed on CRE isolates according to the CLSI guidelines to detect the presence of carbapenemase [20]. The mCIM and eCIM procedures are illustrated in Fig. 1a. In brief, a 1-μL loopful of bacteria was resuspended in a 2-mL tube of TSB. Another 1-μL loopful of bacteria was resuspended in a 2-mL tube of TSB supplemented with EDTA (Thermo Fisher Scientific, Carlsbad, CA, USA) at a final concentration of 5 mM (add 20 μL of 0.5 M EDTA to 2 mL of TSB). A meropenem disk was placed in each tube, and the tubes were incubated at 35 °C for 4 h ± 15 min. Subsequently, the disks were removed and applied to MH agar plates freshly plated with a 0.5 McFarland suspension of a carbapenem-susceptible E. coli ATCC 25922 strain (Fig. 1a). The plates were incubated at 35 °C for 16 to 20 h and the mCIM and eCIM results were interpreted as previously described [21, 30]. The mCIM is considered negative if the zone size is ≥19 mm, positive if the zone size is 6 to 15 mm, or intermediate (defined as positive) if pinpoint colonies are present within a 16- to 18-mm zone [21, 30]. An isolate is positive for metallo-carbapenemase production when the eCIM zone size increases by ≥5 mm compared to the zone size observed for the mCIM and is considered negative for a metallo-carbapenemase if the increase in zone size is < 4 mm [21, 30]. According to CLSI guidelines [20], K. pneumoniae ATCC BAA-1706 (carbapenemase negative), K. pneumoniae ATCC BAA-1705 (blaKPC positive), and K. pneumoniae ATCC BAA-2146 (blaNDM positive) were used as internal controls for mCIM and eCIM tests. The MHT, mCIM and eCIM tests were replicated by two independent investigators to ensure reproducibility.
Acknowledgements
The authors thank Adam Schaenzer of McMaster University for his thoughtful comments and efforts towards improving our manuscript.
Abbreviations
- CLSI
Clinical & Laboratory Standards Institute
- CPE
Carbapenemase-producing Enterobacteriaceae
- CRE
Carbapenemase-resistant Enterobacteriaceae
- DOP
Doripenem
- eCIM
EDTA carbapenem inactivation method
- ETP
Ertapenem
- FN
False negative
- FP
False positive
- IPM
Imipenem
- MBL
Metallo-β-lactamases
- MEM
Meropenem
- mCIM
Modified carbapenem inactivation method
- MHT
Modified Hodge test
- MIC
Minimum inhibitory concentration
- TN
True negative
- TP
True positive
Authors’ contributions
YMT, SW, and HCC were responsible for the study design and the interpretation of results of bacterial identification, antibiotic susceptibility tests, and phenotypic detection of carbapenemases. CYK and LLW were responsible for manuscript preparation. All authors read and approved the final manuscript. YMT and SW contributed equally to this paper (co-first authors). CYK and LLW contributed equally to this paper (co-corresponding authors).
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (grant numbers MOST 108–2911-I-010-508 and 109–2320-B-010-036-MY3); and the En Chu Kong hospital, Taiwan (grant number ECKH_W10901).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ya-Min Tsai and Shining Wang contributed equally to this article.
Li-Li Wen and Cheng-Yen Kao contributed equally to this article.
Contributor Information
Cheng-Yen Kao, Email: kaocy@ym.edu.tw.
Li-Li Wen, Email: eckwen@yahoo.com.
References
- 1.Wang Q, Zhang Y, Yao X, Xian H, Liu Y, Li H, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. 2016;35(10):1679–1689. doi: 10.1007/s10096-016-2710-0. [DOI] [PubMed] [Google Scholar]
- 2.Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ. 2016;352:h6420. doi: 10.1136/bmj.h6420. [DOI] [PubMed] [Google Scholar]
- 3.Barry AL, Jones RN, Thornsberry C, Ayers LW, Kundargi R. Imipenem (N-formimidoyl thienamycin): in vitro antimicrobial activity and beta-lactamase stability. Diagn Microbiol Infect Dis. 1985;3(2):93–104. doi: 10.1016/0732-8893(85)90017-3. [DOI] [PubMed] [Google Scholar]
- 4.Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by Carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10:80. doi: 10.3389/fmicb.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suay-Garcia B, Perez-Gracia MT. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics (Basel). 2019;8(3):122. [DOI] [PMC free article] [PubMed]
- 6.Sugawara E, Kojima S, Nikaido H. Klebsiella pneumoniae Major Porins OmpK35 and OmpK36 allow more efficient diffusion of beta-lactams than their Escherichia coli homologs OmpF and OmpC. J Bacteriol. 2016;198(23):3200–3208. doi: 10.1128/JB.00590-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Y, Xu L, Han Y, Chen Z, Liu C, Ming L. Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp Ther Med. 2018;15(1):1143–1149. doi: 10.3892/etm.2017.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann N Y Acad Sci. 2019;1457(1):61–91. doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 9.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond Ser B Biol Sci. 1980;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 10.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen Stuart J, Leverstein-Van Hall MA. Dutch working party on the detection of highly resistant M, guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents. 2010;36(3):205–210. doi: 10.1016/j.ijantimicag.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Martinez L, Gonzalez-Lopez JJ. Carbapenemases in Enterobacteriaceae: types and molecular epidemiology. Enferm Infecc Microbiol Clin. 2014;32(Suppl 4):4–9. doi: 10.1016/S0213-005X(14)70168-5. [DOI] [PubMed] [Google Scholar]
- 15.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noval M, Banoub M, Claeys KC, Heil E. The Battle is on: new Beta-lactams for the treatment of multidrug-resistant gram-negative organisms. Curr Infect Dis Rep. 2020;22(1):1. doi: 10.1007/s11908-020-0710-9. [DOI] [PubMed] [Google Scholar]
- 17.Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother. 2016;28(1):1–19. doi: 10.1179/1973947815Y.0000000063. [DOI] [PubMed] [Google Scholar]
- 18.Clinical & Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 19 2009. [Google Scholar]
- 19.Tamma PD, Simner PJ. Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J Clin Microbiol. 2018;56(11):e01140–18. [DOI] [PMC free article] [PubMed]
- 20.Clinical & Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 28 2018. [Google Scholar]
- 21.Sfeir MM, Hayden JA, Fauntleroy KA, Mazur C, Johnson JK, Simner PJ, et al. EDTA-Modified Carbapenem Inactivation Method: a Phenotypic Method for Detecting Metallo-beta-Lactamase-Producing Enterobacteriaceae. J Clin Microbiol. 2019;57(5):e01757–18. [DOI] [PMC free article] [PubMed]
- 22.Hoang CQ, Nguyen HD, Vu HQ, Nguyen AT, Pham BT, Tran TL, et al. Emergence of New Delhi Metallo-Beta-lactamase (NDM) and Klebsiella pneumoniae Carbapenemase (KPC) production by Escherichia coli and Klebsiella pneumoniae in southern Vietnam and appropriate methods of detection: a cross-sectional study. Biomed Res Int. 2019;2019:9757625. doi: 10.1155/2019/9757625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(2):477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamma PD, Opene BN, Gluck A, Chambers KK, Carroll KC, Simner PJ. Comparison of 11 phenotypic assays for accurate detection of Carbapenemase-producing Enterobacteriaceae. J Clin Microbiol. 2017;55(4):1046–1055. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasteran F, Mendez T, Rapoport M, Guerriero L, Corso A. Controlling false-positive results obtained with the Hodge and Masuda assays for detection of class a carbapenemase in species of enterobacteriaceae by incorporating boronic acid. J Clin Microbiol. 2010;48(4):1323–1332. doi: 10.1128/JCM.01771-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical & Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 30 2020. [Google Scholar]
- 27.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50(12):3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 29.Mlynarcik P, Roderova M, Kolar M. Primer evaluation for PCR and its application for detection of Carbapenemases in Enterobacteriaceae. Jundishapur J Microbiol. 2016;9(1):e29314. doi: 10.5812/jjm.29314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, et al. Modified Carbapenem inactivation method for phenotypic detection of Carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55(8):2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.