Abstract
Progress in understanding molecular mechanisms contributing to chlamydial pathogenesis has been greatly facilitated by recent advances in genetic manipulation of C. trachomatis. Valuable approaches such as random, chemical-induce mutagenesis or targeted, insertion-based gene disruption have led to significant discoveries. We describe herein a technique for generating definitive null strains via complete deletion of chromosomal genes in C. trachomatis. Fluorescence-reported allelic exchange mutagenesis (FRAEM), using the suicide vector pSUmC, enables targeted deletion of desired chromosomal DNA. The protocol provided here describes steps required to produce transformation competent chlamydiae, generate of a specific allelic exchange plasmid construct, carry out mutagenesis, and isolate clonal populations of resulting mutant strains.
Keywords: Allelic exchange, FRAEM, mutagenesis
1. Introduction
Allelic exchange by homologous recombination is a commonly used approach for generating targeted deletion mutants in genetically tractable bacteria [1]. One of the methods typically employed in producing these null mutations requires transformation of bacteria with a replicative-incompetent plasmid termed a suicide vector. This vector contains i) chromosomal up- and down-stream sequences flanking a target gene to provide elements for homologous recombination within the bacterial chromosome and ii) an antibiotic cassette necessary for appropriate selection. The antibiotic cassette is generally placed between the upstream and downstream homologous sequences. Bacteria transformed with suicide vector constructs undergo two sequential rounds of recombination between the homologous plasmid-encoded and chromosomal sequences. The first recombination event (occurring within one of the homology arms) results in integration of plasmid DNA into the genome to yield a heterozygous merodiploid. To accomplish allelic exchange, the second recombination event must involve the opposite homology arm to result in resolution of the suicide vector backbone and replacement of the targeted gene with the antibiotic resistance cassette. Cultivation under conditions conducive for plasmid curing coupled with antibiotic selection provides a mechanism to recover successfully mutagenized strains. Although such genetic manipulation of numerous bacterial species has been feasible for decades, genetic manipulation of obligate intracellular bacteria has remained challenging. Here we demonstrate a novel approach for generation of chromosomal deletion mutants in Chlamydia trachomatis, serovar L2.
Chlamydiae are gram-negative, obligate intracellular bacteria that undergo a biphasic developmental cycle [2]. Eukaryotic host cells are infected with the elementary bodies (EBs), which are the chlamydial developmental forms specialized for transmission of the microorganism. Upon invasion, the EB differentiates into the intracellular, vegetative form termed a reticulate body (RB). Reticulate bodies grow and multiply in the host cytoplasm within a vacuole termed the chlamydial inclusion. After multiple rounds of bacterial cell division, the RBs asynchronously convert back to the infectious EBs, which are released upon exit from the infected eukaryotic cell. The entire developmental cycle during infection with C. trachomatis, L2 takes about 40-48 hours. Overall, this biology poses significant barriers for successful genetic manipulation of chlamydiae—including generation of targeted deletion mutations in the chlamydial genome.
Many, but not all, chlamydial species contain a 7.5 kb cryptic plasmid that expresses at least 8 genes [3]. C. trachomatis plasmid (pL2)-encoded pgp6 is essential for maintenance of the plasmid by chlamydiae [4]. Deletion of pgp6 leads to loss of the cryptic plasmid after a few rounds of chlamydial infection. We leveraged these findings to engineer a Chlamydia-specific conditionally replicative vector, pSUmC [5]. Initially, the pL2 plasmid isolated from C. trachomatis, serovar L2/434Bu was utilized to produce the chlamydial cloning plasmid pBOMB4-Tet-mCherry [6]. We used this expression plasmid as a backbone to generate pSUmC, where expression of pgp6 was placed under control of a tetracycline (Tet) inducible promoter. The presence of anhydrotetracycline (ATc) inducer in the culture medium permits expression of pgp6, and confers the ability of C. trachomatis to maintain pSUmC. When ATc is removed, the Tet repressor binds to the operator positioned upstream of pgp6, and expression of this gene is inhibited, resulting in loss of pSUmC [5]. A constitutively expressed bla coding region is located downstream of pgp6, followed by elements encoding green-fluorescent protein (GFP), a pMB1 origin of replication, and red-fluorescent protein (mCherry). Although previous versions of pSUmC required iPCR-based techniques to insert chlamydial DNA representing homology arms [5, 7], the bla-gfp cassette in pSUmC described herein is flanked by unique SalI and SbfI sites. This feature enables convenient application of Gibson Assembly technology [8] to clone up- and down-stream homology arms into the 5’ and 3’ flanks of bla-gfp, respectively.
For mutagenesis, C. trachomatis L2 is transformed with a pSUmC-based allelic exchange construct using the CaCl2 method [9], and cultures are maintained with penicillin G and ATc. Once transformants are detected, as determined by the appearance of red and green chlamydial inclusions, cultures are shifted to media lacking ATc. Loss of red fluorescence confirms plasmid curing whereas the presence of penicillin G selects for transformed chlamydiae that have undergone spontaneous homologous recombination, integrating the bla-gfp cassette into the chromosome in exchange for the chlamydial target gene. Transformants are typically bright red and green indicating the presence of multiple copies of pSUmC. Chlamydial mutants however will have lost the plasmid-encoded mCherry and only retain a single copy of the bla-gfp cassette in the genome, and therefore exhibit no red fluorescence and comparably dim green fluorescence. An isogenic population of mutant chlamydiae is obtained by the limiting dilutions method.
2. Materials
All solutions are prepared and handled in a class II biological safety cabinet. McCoy cells are routinely maintained according to the supplier’s instructions. Chlamydia trachomatis serovar L2/434Bu is a Biosafety Level 2 pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms. Use tissue culture grade water for all solutions prepared for cell culture experiments. We typically do not add antibiotics during routine maintenance of eukaryotic host cells.
2.1. Generation of chlamydial inocula for transformation
RPMI-1640 tissue culture medium supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (complete RPMI).
Hanks’ Balanced Salt Solution (HBSS).
Chlamydia trachomatis, serovar L2/434Bu of known titer. Chlamydiae can be obtained from ATCC. The routine propagation and purification of C. trachomatis has been previously described in detail [10]. For this procedure high purity of the inocula is not critical.
6-well plastic sterile tissue culture plates.
Cell scrapers (1.8 cm blade)
SPG (sucrose-phosphate-glutamic acid) buffer: Dissolve 75 g of sucrose, 2.47g of Na2HPO4 and, 0.36 g of NaH2PO4 and 0.72 g of L-glutamic acid in 900 ml of water, then adjust pH to 7.4, and adjust volume to 1 l. Filter sterilize and store SPG at 4°C.
McCoy cells (mouse fibroblasts) are obtained from ATCC.
Temperature controlled clinical centrifuge, sealed, with swinging bucket rotor and microplate buckets.
Temperature controlled bench top centrifuge for microcentrifuge tubes
Cycloheximide: Prepare 1000 x stock solution by dissolving 10 mg of cycloheximide in 9 ml of water then adjust volume to 10 ml, filter sterilize, aliquot and store at −20°C.
Methanol
PBS (phosphate-buffered saline): Dissolve 7.89 g of NaCl, 0.201 g of KCl, 1.42 g of Na2HPO4 and 0.313 g of KH2PO4 in 900 ml of water, then adjust pH to 7.2 and adjust volume to 1 l. Autoclave to sterilize and store PBS at room temperature.
Primary monoclonal or polyclonal antibodies specific to Chlamydia
Inverted fluorescence microscope
15-ml sterile conical tube
CaCl2 buffer: prepare 10 mM Tris pH 7.4 and 50 mM CaCl2 in water, filter sterilize and store at 4°C.
2-ml Safe-Lock microcentrifuge tubes
1.5 ml sterile microcentrifuge tubes with silicone O-rings
Tissue culture incubator
Class II biological safety cabinet
2.2. Generation of chlamydial suicide vector
C. trachomatis genomic DNA
Chlamydial suicide vector, pSUmC with SalI and SbfI restriction sites flanking the bla-gfp cassette. pSUmC-sal-sbf is available upon request from the corresponding author.
Q5® High-Fidelity DNA Polymerase
Engineered primers to amplify Chlamydia-specific DNA and insert into vector via Gibson Assembly reaction.
SalI and SbfI restriction enzymes
A gel DNA Recovery Kit for purification of PCR amplicons
Agarose, molecular biology grade
Gibson assembly cloning kit
Electro-competent E. coli NEB10β
dam−/dcm− Chemically competent E. coli
Plastic Petri dishes for bacteria
Carbenicillin: Prepare 1000 x stock solution by dissolving 500 mg of carbenicillin in 9 ml of water then adjust volume to 10 ml, filter sterilize, aliquot and store at −20°C.
LB broth, Miller (Luria-Bertani)
Agar, bacteriological, powder
Plasmid miniprep kit
Plasmid maxiprep kit
Sterile Erlenmeyer flasks for bacterial cultures
2.3. Transformation of Chlamydia trachomatis
RPMI-1640 tissue culture medium supplemented with 10% (vol/vol) heat-inactivated FBS (complete RPMI).
Hanks’ Balanced Salt Solution (HBSS).
Chlamydia trachomatis, crude inoculum of known titer.
6-well plastic sterile tissue culture plates.
Sterile 1.5 ml microcentrifuge tubes
McCoy cells.
Temperature controlled centrifuge, sealed, with swinging bucket rotor with microplate buckets.
Cycloheximide: 1000 x stock solution (1 mg/ml)
Penicillin G: Prepare 10,000 x stock solution (6mg/ml) by dissolving 60 mg of the antibiotic in 9 ml of water then adjust volume to 10 ml, filter sterilize, aliquot and store at −20°C.
Anhydrotetracycline (ATc): Prepare 10, 000 x stock solution by dissolving 500 μg of ATc in 1 ml of DMSO (cell culture grade). Aliquot (20 μl) and freeze the stock solution at −20°C. Do not thaw and re-freeze ATc for more than 3 times.
CaCl2 buffer: prepare 10 mM Tris pH 7.4 and 50 mM CaCl2 in water, filter sterilize and store at 4°C.
Cell scrapers (1.8 cm blade)
2-ml Safe-Lock Microcentrifuge Tubes.
Unmethylated plasmid DNA for deletion construct at 1.0-1.5 μg/μl.
Class II biological safety cabinet
2.4. Isolation of deletion mutant by limiting dilution
RPMI-1640 tissue culture medium supplemented with 10% (vol/vol) heat-inactivated FBS (complete RPMI).
Hanks’ Balanced Salt Solution (HBSS).
Chlamydia trachomatis, deletion mutant of known titer.
McCoy cells.
Temperature controlled clinical centrifuge, sealed, with swinging bucket rotor with microplate buckets.
Temperature controlled bench top centrifuge for microcentrifuge tubes.
Cycloheximide: 1000 x stock solution (1 mg/ml)
Penicillin G: 10,000 x stock solution (6 mg/ml)
Crude stock of C. trachomatis deletion mutant of known titer
384-Well Tissue Culture Plates
Bio-Pure reagent reservoirs
Multichannel pipette
Sterile 50-ml conical tube
Class II biological safety cabinet
3. Methods
Except for specified centrifugation steps, all experiments are carried out at room temperature. All experiments with McCoy cells and viable chlamydiae are performed in a Class II biological safety cabinet under aseptic conditions.
3.1. Preparation of C. trachomatis crude stock for transformation
Seed four 6-well plates with McCoy cells at 1-1.5 x 106 of cells per one well in complete RPMI medium. Incubate for 24 hrs at 37°C in an atmosphere of 5% CO2/95% humidified air
Remove medium from confluent McCoy cultures and add C. trachomatis at the MOI of 1 (2-3 x 106 of chlamydiae per well) diluted in 2 ml of HBSS per well. Infect McCoy cells by centrifugation at 900 x g for 1 hour at 20°C in a centrifuge equipped with swinging bucket rotor and microplate buckets.
After centrifugation, remove inoculum, add 2 ml of complete RPMI medium supplemented with 1 μg/ml of cycloheximide per well, and incubate at 37°C in an atmosphere of 5% CO2/95% humidified air, for 40 hours.
Harvest chlamydiae-infected McCoy cells by gentle scraping of cell cultures with a cell scraper and collect the contents of each well into 2-ml Safe-Lock sterile microcentrifuge tubes.
Pellet harvested cell material (24 tubes) at 20,000 x g for 30 minutes at 4°C.
Remove supernatant and resuspend all 24 pellets in a total of 4 ml of SPG. Vortex well and centrifuge again at 200 x g for 5 minutes at 4°C.
Finally, collect supernatant into a sterile 15-ml conical tube and disperse obtained inoculum into sterile microcentrifuge tubes with silicone O-rings in 50 μl aliquots. This crude chlamydial preparation is stored (in SPG) at −80°C.
Prepare one 6-well plate with confluent McCoy cells as described above.
Perform a sham transformation reaction in order to determine the most accurate titer of the crude inoculum. To accomplish this set out 6 sterile microcentrifuge tubes with 50 μl of CaCl2 buffer in each.
Thaw out an aliquot of the crude chlamydial inocula and add 0.5 μl into the first microcentrifuge tube, 1 μl into the second, 2 μl into the third tube and so on. Once 5 μl is added into the sixth tube, mix chlamydiae in the buffer by gently flicking each tube and incubate the bacteria in CaCl2 buffer for 30 minutes.
Meanwhile, remove medium from one 6-well plate with McCoy cells and add 1 ml of HBSS per well.
The incubation of C. trachomatis in calcium buffer is stopped by adding 1 ml of HBSS into each reaction.
Pipette the contents of one microcentrifuge tube into one well in the 6-well plate and infect McCoy cells by centrifugation as described above. Incubate infected cells in complete RPMI supplemented with 1 μg/ml of cycloheximide as described above.
At ~24 hours post infection, fix infected cell cultures with methanol for 5 min., wash wells 3 times with PBS and stain chlamydial inclusions with anti-C. trachomatis specific primary antibody followed by staining with a fluorophore conjugated secondary antibody according to the manufacturers’ instructions.
The number of inclusion forming units (IFUs) per ml is obtained by enumeration of chlamydial inclusions using a fluorescent microscope as described by [10].
3.2. Generation of chlamydial suicide vector
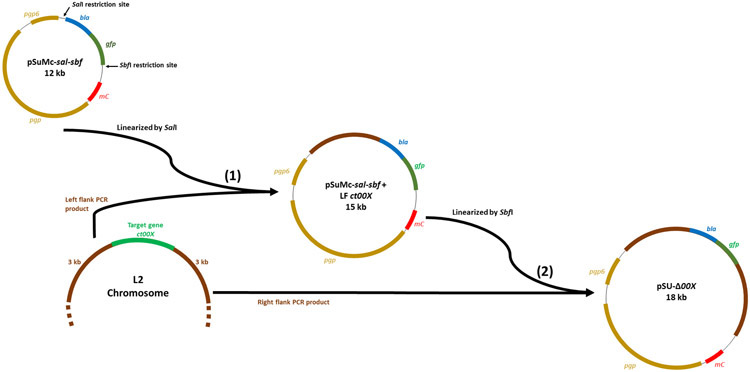
Our previously described pSUmC required multiple steps of iPCR-based cloning to generate vectors for allelic exchange mutagenesis [5, 7]. However, Gibson Assembly has gained traction as an effective molecular biology technique—particularly in the construction of allelic exchange constructs [11]. pSUmC-sal-sbf contains unique SalI and SbfI restriction sites immediately flanking the 5’ and 3’ regions of the bla-gfp cassette, respectively. Two, sequential Gibson Assembly steps can be employed to insert (±) 3-kb left flank (upstream) and (±) 3-kb right flank (downstream) of the targeted locus (gene 00X) to the SalI and SbfI sites of the backbone plasmid, respectively (Fig. 1).
Figure 1-.
Schematic of overall cloning strategy used for constructing allelic exchange plasmid. (1) The 3 kb left flank (upstream) of gene of interest (ct00X) is PCR-amplified from C. trachomatis L2 genomic DNA and cloned into linearized (SalI) pSuMc-sal-sbf by Gibson Assembly to yield pSUmC-sal-sbf + LF. (2) The 3 kb right flank (downstream) of gene of interest (ctl00X) is then mobilized into linearized (SalI) pSuMc-sal-sbf + LF ct00X to yield the final construct pSU-Δ00X.
-
Design primers for amplifying (±) 3-kb left flank (upstream) of the targeted gene (gene 00X) that include specific overlap sequence (underlined) for the SalI digested site of the backbone plasmid (pSUmC-sal-sbf).
Forward and reverse Chlamydia-specific sequences should then be added to the 3’ end of X-LF-F and X-LF-R, respectively, to produce the complete primer sequences:
00X-LF-F: CGTCACTGCAGGTACCGGTCGA- Specific sequence to the left flank of targeted gene.
00X-LF-R: GCGGATACATATTGAATGGTCGA- Specific sequence to the left flank of targeted gene.
PCR-amplify left flank of 00X gene from chlamydial genomic DNA using X-LF-F and X-LF-R primers with Q5 High-Fidelity DNA Polymerase according to manufacturer’s instructions. Separate the PCR reaction on a 0.8% agarose gel and extract the PCR product with a Gel DNA Recovery Kit.
Linearize backbone plasmid by digestion with SalI. Separate the linearized plasmid on a 0.8% agarose gel and extract the correct product with a Gel DNA Recovery Kit.
Assemble linearized backbone plasmid and left flank of 00X gene by a Gibson Assembly according to the manufacturer’s instructions.
Transform NEB® 10-beta Electro-competent E. coli according to manufacturer’s instructions. Spread the transformation on LB agar plates containing 50 μg/ml carbenicillin and incubate overnight at 37°C.
Choose colonies that display both green and red florescence for screening and incubate positive colonies in 3 ml LB liquid medium containing 50 μg/ml carbenicillin overnight at 37°C. Extract the plasmid DNA with a plasmid miniprep Kit and confirm the insertion by restriction digestion using appropriate restriction sites within the left flank DNA. Check the correct sizes by separating digested plasmids on a 0.8% agarose gel. PCR-based screening can also be employed depending on investigator preferences.
-
Design primers for amplifying (±) 3-kb right flank (downstream) of 00X that include specific overlap sequence (underlined) for the SbfI site of the backbone plasmid (pSUmC-sal-sbf +LF).
Forward and reverse Chlamydia-specific sequences should then be added to the 3’ end of X-RF-F and X-RF-R, respectively, to produce the complete primer sequences:
X-RF-F: CTATACAAGTAACCTGCA- Specific sequence to the downstream of targeted gene.
X-RF-R: GGTCTGACGCTCCCTGCA- Specific sequence to the downstream of targeted gene.
PCR-amplify right flank of 00X gene from chlamydial genomic DNA using X-RF-F and X-RF-R primers with Q5 High-Fidelity DNA Polymerase according to manufacturer’s instructions. Separate the PCR reaction on a 0.8% agarose gel and extract the PCR product with a Gel DNA Recovery Kit.
Linearize backbone plasmid by digestion with SbfI. Separate the linearized plasmid on a 0.8% agarose gel and extract the correct product with a Gel DNA Recovery Kit.
Assemble linearized backbone plasmid and right flank of 00X gene by a Gibson Assembly as described in step 4.
Transform E. coli and screen resulting colonies as described in steps 5 and 6. DNA sequence the engineered regions of final plasmids to confirm correct insertion of chlamydial DNA.
3.3. Transformation of Chlamydia trachomatis, L2
Prepare two 6-well plates with confluent McCoy cells as described above. Based on chlamydial titer determined in the section 3.1, calculate the number of chlamydiae required to infect 12 wells at the MOI of 2.
Thaw out an aliquot(s) of crude C. trachomatis preparation, transfer required amount of chlamydial inocula into a 1.5 ml microcentrifuge tube and pellet bacteria by centrifugation at 20,000 x g for 30 minutes at 4°C.
Meanwhile, thaw out the unmethylated deletion-construct plasmid DNA generated in 3.2, and set out 5 sterile 1.5 ml microcentrifuge tubes.
Next, resuspend chlamydial pellet in 600 μl of CaCl2 buffer and pipette 100 μl of the chlamydial inocula in each microcentrifuge tube (6 tubes total). Then quickly add 5 μg of the deletion construct DNA into each tube and mix contents by flicking each microcentrifuge tube every 10 minutes. Incubate a total of 30 min at room temperature.
Supplement each transformation reaction with 1 ml of HBSS.
Remove complete RPMI from the two 6-well plates and replace with 1.5 ml of fresh HBSS and 550 μl of the transformation reaction into each well.
Centrifuge plates at 900 x g for 1 hour at 20°C, remove chlamydial inoculum, and add 2 ml of complete RPMI per well supplemented with 1 μg/ml of cycloheximide. Incubate at 37°C in an atmosphere of 5% CO2 and 95% humidified air for 7 hours.
Replace the medium with 2 ml of complete RPMI supplemented with 1 μg/ml cycloheximide, 0.6 μg/ml of penicillin G, and 50 ng/ml of anhydrotetracycline (ATc), and return plates back to the incubator.
Next day prepare two 6-well plates with fresh McCoy monolayer.
At 48 hours post infection re-passage infected cells by harvesting all cultures, including media, using a cell scraper. Collect cultures into 2-ml safe-lock microcentrifuge tubes and centrifuge tubes at 20,000 x g for 30 minutes at 4°C.
Pipette off supernatants and resuspend each pellet in 1 ml of HBSS, then vortex well and spin the harvested material down at 200 x g for 5 minutes at 4°C to remove crude debris from eukaryotic host cells.
Meanwhile, remove complete RPMI from two plates containing new confluent McCoy cells and add 1 ml of fresh HBSS per well.
Next, add supernatants into this new cell culture, supernatant of one tube per well, and centrifuge 6-well plates at 900 x g for 1 hour as described above.
Finally, remove HBSS and add complete RPMI supplemented with 1 μg/ml cycloheximide, 0.6 μg/ml of penicillin G, and 50 ng/ml of ATc. Incubate cells in the incubator for 48 hours and then perform another re-passage as described above.
Keep re-passaging cells every 48 hrs until inclusions develop which are both bright red- and green-fluorescence, as observed by microscopy. However, if after 5 passages, no fluorescent inclusions develop, restart the transformation protocol (beginning at step 1 above).
Once fluorescent inclusions are detected, keep re-passing chlamydiae as described above but the infected cells are now cultured in complete RPMI lacking ATc. After several following passages, chlamydial inclusion will become apparent that are dim green and not red, as observed by microscopy, indicating the generation of the deletion mutant and loss of the suicide vector. Maintain an apparent MOI of 0.1 during these steps.
Sufficiently expand the number of dim green inclusions and prepare a small amount of a crude chlamydial stock and accurately determine the C. trachomatis titer as described in the section 3.1.
3.4. Isolation of deletion mutant by limiting dilution
Prepare confluent monolayer of McCoy cells in a 384-well plate in complete RPMI.
Pipette 25 ml of HBSS into a 50-ml conical tube and add 50 chlamydiae. Vortex and transfer the diluted chlamydial mutant into a bio-pure reagent reservoir. Remove media from McCoy cells and apply 50 μl of chlamydial inoculum into each well employing multichannel pipette.
Centrifuge plate at 900 x g for 1 hour at 20°C.
Remove HBSS from the plate and add complete RPMI supplemented with 1 μg/ml cycloheximide, 0.6 μg/ml of penicillin G. Incubate the plate at 37°C in an atmosphere of 5% CO2/95% humidified air.
About five to ten days post infection individual wells should contain McCoy cell monolayers containing multiple chlamydial inclusions. Identify wells containing C. trachomatis by fluorescence microscopy.
Depending on the number of positive wells, prepare confluent McCoy cells in an appropriate number of wells in a 6-well plate.
Then harvest these positive samples by scraping the wells with pipette tips and transferring the contents to 2-ml safe-lock microcentrifuge tubes containing 2 ml HBSS. Vortex obtained samples and apply each to one well in a 6-well plate.
Centrifuge plate(s) at 900 x g for 1 hour at 20°C. Incubate plate in complete RPMI supplemented with 1 μg/ml cycloheximide as described above.
A second round of isolation from a 384 well plate is typically employed to ensure clonal populations are obtained.
Once deletion mutants are isolated and purified, no selective pressure is required during chlamydial growth.
Acknowledgements
This work was supported by Public Health Service grants from the National Institutes of Health, NIAID (AI065530 and AI124649) to K.A. Fields.
Footnotes
. Although C. trachomatis is routinely propagated in human HeLa cells, murine McCoy cells are employed here because, in our hands, this cell line can handle the entire process of chlamydial transformation much better with regard to the cell viability and overall fitness of the host cell culture.
. The number of 6-well plates used either in preparation of chlamydial crude stock or in chlamydial transformation can be adjusted.
. Primers specific for Left or Right flank sequences (18-30 bp) should be designed having a melting temperature between 58°C to 68°C and lacking hairpin at temperatures not higher than 50°C. For complete gene deletion, cloned 5’, left-flanking DNA should include up to the initiation codon for 00X, whereas 3’, right-flanking DNA should include up to the stop codon for 00X.
. 3 kb flanking arms are routinely employed, but it is anticipated that shorter lengths are possible. Regardless, 3 kb arms are an efficient starting length unless cloning necessitates shorter DNA lengths.
. When infecting McCoy cells with C. trachomatis, always check cell cultures for the desired level of infection and the condition of eukaryotic hosts by microscopy. If McCoy cells appear to be detaching and dying at 24 hours post infection, there is no point in going on with the planned procedure. It is better to start the experiments over with a fresh monolayer of McCoy cells and re-adjusted number of chlamydial MOI.
. During infection of McCoy cells with C. trachomatis by centrifugations in 6-well plates, 2 ml of HBSS per well is required, as lower volume often results in shifting of the cell monolayer onto one site and thus leads to an uneven chlamydial infection.
. The 7 hour recovery of chlamydiae after transformation has been determined to be the most optimal for transformation efficiency but could vary from lab to lab. In our hands, shorter recovery will result in low efficiency of bacterial transformation. Longer than 7 hour recovery leads to the host cell lysis next day due to the inability of the selective antibiotic to sufficiently inhibit chlamydial propagation.
. All antibiotics are aliquoted and stored at −20°C however, once an aliquot is thawed out it is kept at 4°C: Penicillin G for one week, whilst cycloheximide and carbenicillin for one month.
. If the chlamydial transformation is unsuccessful, there are several steps that can be augmented. Instead of employing two 6-well plates, use ≥four 6-well plates with an MOI of 2 as described above. Next, check the pH of CaCl2 buffer and make sure there are no precipitates in the buffer. The condition of the unmethylated suicide construct is also important. Run 0.8% agarose gel to confirm that the plasmid isn’t nicked or otherwise damaged. Importantly, it is advisable to include a positive control, empty pSUmC or GFP::SW2 [9], in transformation experiments to ensure an adequate level of bacterial competence.
. In some cases, merodiploids may not resolve to yield mutants, even after 10-20 passages. In this instance, DNA can be extracted from cultures and PCR used to confirm that cross-over events are occurring in both homology arms. Design primer sets where one primer anneals within the bla-gfp cassette and the Chlamydia-specific primer anneals outside the engineered homology arm. Perform PCR and assess recombination as described [11]. If there is evidence for recombination within only one homology arm, verify that the allelic exchange construct is correct. If evidence indicates that recombination is occurring in both homology arms, it is likely that the targeted gene is essential and cannot be deleted via this protocol.
. It is important to maintain a low MOI during passage of transformants, especially if the deletion results in significant attenuation of chlamydial fitness. In some cases, it may be necessary to isolate inclusions corresponding to mutant strains using micromanipulator or cloning cylinder approaches.
5. References
- [1].Nakashima N, Miyazaki K. Bacterial Cellular Engineering by Genome Editing and Gene Silencing. International Journal of Molecular Sciences 2014;15:2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].AbdelRahman Y, Belland R. The chlamydial developmental cycle. FEMS Microbiol Rev 2005;29:949 – 59. [DOI] [PubMed] [Google Scholar]
- [3].Zhong G Chlamydial Plasmid-Dependent Pathogenicity. Trends in Microbiology 25:141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, et al. Chlamydia trachomatis Plasmid-Encoded Pgp4 Is a Transcriptional Regulator of Virulence-Associated Genes. Infection and Immunity 2013;81:636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mueller KE, Wolf K, Fields KA. Gene Deletion by Fluorescence-Reported Allelic Exchange Mutagenesis (FRAEM) in Chlamydia trachomatis. mBio 2016;7:e01817–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 2014;196:1325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mueller KE, Wolf K, Fields KA. Chlamydia trachomatis transformation and allelic exchange mutagenesis. . Curr Protocol Microbiol 2017;45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison Iii CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 2009;6:343. [DOI] [PubMed] [Google Scholar]
- [9].Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a Transformation System for Chlamydia trachomatis: Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector. PLoS Pathog 2012;7:e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Scidmore MA. Cultivation and Laboratory Maintenance of Chlamydia trachomatis Current Protocols in Microbiology, John Wiley & Sons, Inc, 2005. [DOI] [PubMed] [Google Scholar]
- [11].Silayeva O, Barnes AC. Gibson Assembly facilitates bacterial allelic exchange mutagenesis. Journal of Microbiological Methods 2018;144:157–63. [DOI] [PubMed] [Google Scholar]