Abstract
The primary concentration and molecular process are critical to implement wastewater-based epidemiology for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the previously developed methods were optimized for nonenveloped viruses. Few studies evaluated if the methods are applicable to the efficient recovery of enveloped viruses from various types of raw sewage. This study aims (1) to compare the whole process recovery of Pseudomonas phage φ6, a surrogate for enveloped viruses, among combinations of primary concentration [ultrafiltration (UF), electronegative membrane vortex (EMV), and polyethylene glycol precipitation (PEG)] and RNA extraction methods (spin column-based method using QIAamp Viral RNA Mini Kit and acid guanidinium thiocyanate–phenol–chloroform extraction using TRIzol reagent) for three types of raw sewage and (2) to test the applicability of the method providing the highest φ6 recovery to the detection of SARS-CoV-2 RNA. Among the tested combinations, PEG+TRIzol provided the highest φ6 recovery ratio of 29.8% to 49.8% (geometric mean). UF + QIAamp Viral RNA Mini Kit provided the second highest φ6 recovery of 6.4% to 35.8%. The comparable φ6 recovery was observed for UF + TRIzol (13.8–30.0%). PEG + QIAamp Viral RNA Mini Kit provided only 1.4% to 3.0% of φ6 recovery, while coliphage MS2, a surrogate for nonenveloped viruses, was recovered comparably with PEG + TRIzol. This indicated that the nonenveloped surrogate (MS2) did not necessarily validate the efficient recovery for enveloped viruses. EMV + QIAamp Viral RNA Mini Kit provided significantly different φ6 recovery (1.6–21%) among the types of raw sewage. Then, the applicability of modified PEG + TRIzol was examined for the raw sewage collected in Tokyo, Japan. Of the 12 grab samples, 4 were positive for SARS-CoV-2 CDC N1 and N3 assay. Consequently, PEG + TRIzol provided the highest φ6 recovery and allowed for the detection of SARS-CoV-2 RNA from raw sewage.
Keywords: SARS-CoV-2, Wastewater-based epidemiology, Virus concentration, Pseudomonas phage φ6, Polyethylene glycol precipitation
Graphical abstract
Highlights
-
•
The combinations of primary concentration and RNA extraction were compared.
-
•
PEG + TRIzol provided the highest φ6 recovery ranging from 29.8–49.8%.
-
•
Nonenveloped surrogates did not necessarily validate the efficient recovery of φ6.
-
•
SARS-CoV-2 RNA was detected by modified PEG+TRIzol from raw sewage in Japan.
1. Introduction
The ongoing disease outbreaks caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have tremendous damage to global health and world economics. The main transmission of the respiratory viruses is via close contact with respiratory secretions expelled by an infected person (World Health Organization, 2020). Although the SARS-CoV-2 predominantly caused respiratory diseases, its genome was detected not only from the respiratory tract but also from stools (Wölfel et al., 2020). Therefore, the detection of human pathogenic viruses from wastewater suggests the presence of infected people in the catchment area. This approach has been expected to be feasible to evaluate the eradication of poliovirus in the community (Lodder et al., 2012) and the prevalence of norovirus (Kazama et al., 2016), known as wastewater-based epidemiology (WBE). Currently, the monitoring of the SARS-CoV-2 concentration is expected to be a powerful tool for the early detection of outbreaks and the prediction of prevalence in the catchment (Kitajima et al., 2020; Murakami et al., 2020; Rusiñol et al., 2020).
Recent studies have detected SARS-CoV-2 RNA in the raw sewage or secondary effluent by applying primary concentration methods followed by RNA extraction and reverse transcription (RT)-quantitative polymerase chain reaction (qPCR) (Ahmed et al., 2020a; Haramoto et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). The adopted primary concentration methods included polyethylene glycol precipitation (PEG) (Hata et al., 2020; Wu et al., 2020), ultrafiltration (UF) (Medema et al., 2020; Sherchan et al., 2020), electronegative membrane vortex (EMV) (Haramoto et al., 2020), and adsorption–extraction methods (Ahmed et al., 2020a). Considering that the constraints on financial capacity, laboratory equipment, and labor resources differ among the implementers of WBE, it is preferable to validate a variety of primary concentration methods.
The conventional primary concentration has been optimized to detect waterborne enteric viruses, which are mostly nonenveloped. Enveloped viruses possess a lipid membrane at the outermost layer, thus exhibiting different physical properties (i.e., more hydrophobic) from nonenveloped viruses (Lytle and Routson, 1995). Few studies have evaluated the applicability to the recovery of enveloped viruses from water. A previous study adopted the filter-lysis method and struggled with efficient recovery of enveloped viruses: poliovirus (nonenveloped) for 45% versus koi herpes virus (enveloped) for 3.6% (Haramoto et al., 2009). Thus, the conventional process should be optimized for the efficient recovery of enveloped viruses.
A recent study compared a variety of concentration methods for the recovery of murine hepatitis virus (MHV), a surrogate of SARS-CoV-2, from raw sewage (Ahmed et al., 2020b). They reported the highest MHV recovery rate for the adsorption–extraction methods with MgCl2 pretreatment (65.7%). They also reported a 56.0% recovery for the UF using Amicon Ultra-15 centrifugal filter device. Although these methods achieved a high recovery efficiency enough for detecting enveloped viruses in the raw sewage, expensive equipment (e.g., bead-beating system) and consumable supplies (e.g., centrifugal device) are needed for sample processing. Moreover, previous studies did not investigate how the water quality and the type of the molecular process affect the whole recovery efficiency of enveloped viruses. Thus, the recovery efficiency of enveloped viruses from the raw sewage should be further validated.
Bacteriophage φ6 is an enveloped double-stranded RNA virus, with a diameter of 85 nm, belonging to Cystoviridae, infecting gram-negative Pseudomonas syringae. To evaluate the recovery efficiency during primary concentration and the persistence in the aqueous phase, this virus has been frequently used as a surrogate for pathogenic enveloped viruses (i.e., human coronavirus and influenza virus) (Casanova and Weaver, 2015a; Ye et al., 2016). A recent study adopted it as a molecular process control (MPC) (Sherchan et al., 2020). Moreover, φ6 and its host, Pseudomonas syringae, are not pathogenic to humans and require minimal containment for laboratory facilities (i.e., BSL1) (Casanova and Weaver, 2015b). Finally, φ6 can be propagated within 1 day with high titers (up to 1010 PFU/mL); thus, it is practically adoptable as a surrogate for the human pathogenic enveloped viruses.
This study aims (1) to compare the combination of primary concentration (UF, EMV, and PEG) and RNA extraction (QIAamp Viral RNA Mini Kit and TRIzol) for the whole process recovery of nonenveloped and enveloped virus surrogates and (2) to test the applicability of the method providing the highest φ6 recovery to detect SARS-CoV-2 RNA from the raw sewage. In this study, F-specific coliphage MS2, as a surrogate of nonenveloped viruses, and Pseudomonas phage φ6, as a surrogate of enveloped viruses, were adopted as a whole process control (WPC) (Haramoto et al., 2018). Furthermore, the murine norovirus (MNV) was used as an MPC to evaluate the inhibitory effects or recovery loss during the RNA extraction and RT-qPCR process.
2. Materials and methods
2.1. Preparation of coliphage MS2, Pseudomonas phage φ6, and murine norovirus
Bacteriophage MS2 (NBRC 102619, National Institute of Technology and Evaluation (NITE), Tokyo, Japan) was propagated as described elsewhere (Torii et al., 2019). Pseudomonas phage φ6 (NBRC 105899, NITE) was propagated using Pseudomonas syringae (NBRC14084, NITE) as a host strain. Briefly, 3 mL of phosphate buffer (PB) (10 mM, pH 7.0) was added on the soft LB plates (0.7% agar) semiconfluent with φ6 plaques (approximately 1000 plaques/plates) and incubated at room temperature for 5 h. The suspension and soft agar were then removed from the plate and transferred to a centrifuge tube. Next, 10 mL of recovered viruses were clarified by centrifugation at 3500g for 15 min and filtered through a cellulose acetate filter (0.2 μm, DISMIC-25CS, Advantec, Tokyo, Japan). MNV S7-PP3 strain was propagated using RAW.264.7 cell as a host (Kitajima et al., 2008). The concentrations of propagated stocks were approximately 1011 PFU/mL, 5 × 109 PFU/mL, and 1011 copies/mL for MS2, φ6, and MNV, respectively. The propagated stocks were stored at 4 °C. Before the primary concentration, the MS2 and φ6 stocks were diluted by 1000-fold and 100-fold, respectively, to prepare the diluted stocks.
2.2. Raw sewage
Three raw sewages were used for the comparison: raw sewage A was a grab sample collected from the influent of a wastewater treatment plant (WWTP) located in Niigata Prefecture, raw sewage B was a composite sample collected from the influent of a WWTP located in Kanagawa Prefecture, and raw sewage C was a composite sample collected from the influent of WWTP located in Tokyo metropolis. The physicochemical water qualities of each raw sewage is shown in Table 1 . The composite samples (raw sewages B and C) were generated as described in the Supporting Information (SI). All the samples were stored at −20 °C until the primary concentration.
Table 1.
Physicochemical water quality parameters of each raw sewage.
| Raw sewage | pH | Total suspended solids (TSS) (mg/L)a | UV260 |
|---|---|---|---|
| A | 7.56 | 225 ± 27 | 0.231 |
| B | 8.19 | 344 ± 41 | 0.583 |
| C | 8.45 | 334 ± 49 | 0.529 |
TSS was measured three times. Average and standard deviation are described.
2.3. Primary concentration
Three primary concentration methods were tested: ultrafiltration after pre-centrifugation (UF), electronegative membrane vortex (EMV), and polyethylene glycol precipitation after pre-centrifugation (PEG).
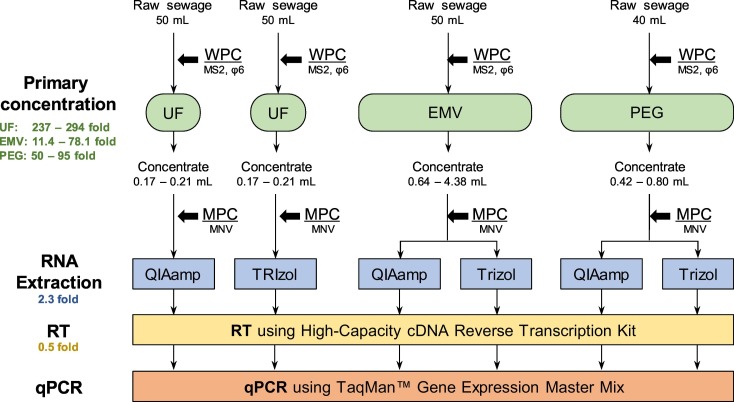
A 50-mL (for UF and EMV) or 40-mL (for PEG) of raw sewage was inoculated with 1/1000 amount of diluted stocks of MS2 and φ6 (i.e., 50 μL for UF and EMV, 40 μL for PEG) and incubated at 4 °C for 1.5 h – 2.5 h. The incubation of raw sewage for 1.5–2.5 h is expected to reach the liquid–solid partitioning of spiked viruses at equilibrium, which allows for mimicking the actual partitioning conditions of viruses in raw sewage (Ye et al., 2016). For a single day, ranging from 4 to 14 of aliquots of spiked raw sewage were prepared. As a control, 50 mL of MilliQ water spiked with 50 μL of diluted stocks of MS2 and φ6 were prepared in duplicate every single day. The procedure of each primary concentration method was conducted four times per type of raw sewage (i.e., raw sewages A, B, and C).
UF: A 50-mL aliquot of raw sewage was centrifuged at 3500g for 15 min to remove large particles. The supernatant was further centrifuged at 3500g for 20 min to filter through the Centricon Plus-70 centrifugal device with a molecular weight cutoff of 30 kDa (Merck Millipore, Billerica, MA, USA). The final volume of the concentrates ranged from 0.17 to 0.21 mL. Thus, the concentration factor in UF ranged from 237- to 294-fold.
EMV: EMV was performed based on a previous study (Haramoto et al., 2020). A 50 mL aliquot of raw sewage inoculated with 500 μL of 2.5 M MgCl2 was filtered through a mixed cellulose-ester membrane (HAWP04700, pore size, 0.45 μm; diameter, 47 mm; Merck Millipore) by vacuum aspiration. Subsequently, 10 mL of elution buffer, containing 0.2 g/L of sodium polyphosphate, 0.3 g/L of ethylenediaminetetraacetic acid trisodium salt trihydrate (C10H13N2O8Na3 3H2O), and 0.1 mL/L of Tween 80, was added to the filtered membrane in a 50-mL centrifuge tube. Elution was performed by vigorous vortexing using a football-shaped stirring bar. This step was repeated using a 5 mL of elution buffer after transferring a 10-mL of eluent to a different centrifuge tube. The resultant eluent (approximately 15 mL) was centrifuged at 2000g for 10 min at 4 °C. The supernatant was filtered through a cellulose acetate filter (0.2 μm, DISMIC-25CS, Advantec, Tokyo, Japan). The filtrate was centrifuged at 1500g for 10 min twice followed by 5 min in a Centriprep YM-50 UF device (Merck Millipore) for concentration. The final volume of the concentrate ranged from 0.64 to 4.38 mL. Thus, the concentration factor in EMV ranged from 11.4- to 78.1-fold.
PEG: A 40-mL aliquot of raw sewage was centrifuged at 3500g for 5 min to remove large particles. The supernatant was supplemented with 4 g of PEG8000 (Sigma-Aldrich, MO, USA) and 2.35 g of NaCl (Wako, Osaka, Japan) to the final concentrations of 10% (w/v) and 1.0 M, respectively, and incubated at 4 °C overnight in a shaker (Hata et al., 2020). Thereafter, the mixture was centrifuged at 10,000g for 30 min. The pellet was resuspended with 10 mM PB. The final volume of the concentrate ranged from 0.42 to 0.8 mL. Thus, the concentration factor in PEG ranged from 50- to 95-fold.
After each primary concentration, the resultant concentrate and control samples were stored at −20 °C until the molecular process. Freeze-thawing of primary concentrates was limited up to once.
2.4. RNA extraction
For the Section 3.1, two types of viral RNA extraction methods were tested: spin column-based nucleic acid purification using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) and acid guanidinium thiocyanate–phenol–chloroform extraction (Chomczynski and Sacchi, 1987) using TRIzol reagent (Thermo Fisher Scientific, MA, USA). First, a 140-μL aliquot of the virus concentrate was seeded with 5 μL of nonenveloped MNV (corresponding to 4.32 ± 0.19 copies) as an MPC. As a control, a 140-μL aliquot of MilliQ spiked with the same amount of MNV was prepared in duplicate. The comparison of MNV concentrations between concentrated samples and control samples allows evaluating the molecular process recovery ratio as described in 2.6. The spiked concentrates were processed following the manufacturer's instruction to obtain an RNA extract with a final volume of 60 μL. Thus, the concentration factor in the RNA extraction process was 2.3-fold. The extracted RNA was subjected to the RT process within the same day.
2.5. RT-qPCR
Before the RT-qPCR step, the RNA was incubated at 95 °C for 5 min followed by 4 °C for 1 min (Mijatovic-Rustempasic et al., 2013) to denature dsRNA of φ6. A preliminary investigation indicates that the heating step did not significantly affect the quantification results of MS2 suspended in MilliQ water (data not indicated).
First, 20 μL of the RNA extract was subjected to the RT step. A High-Capacity cDNA RT Kit (Thermo Fisher Scientific) was used to obtain cDNA with a final volume of 40 μL. The concentration factor in RT process was 0.5-fold. The obtained cDNA was stored at 4 °C and subjected to qPCR within 2 days.
Then, TaqMan™ Gene Expression Master Mix was used to perform qPCR following the manufacturer's protocol. A 5 μL of the cDNA was mixed with 15 μL of the reaction mixture containing 10 μL of the master mix, virus-specific forward primers and reverse primers, and TaqMan probe. The sequence of primers and the probe (Gendron et al., 2010; Kitajima et al., 2008; Wolf et al., 2010) were described in Table S1. The conditions of thermal cycling were as follows: 50 °C for 2 min and 95 °C for 10 min, followed by 45 cycles of 94 °C for 15 s and 60 °C for 60 s (for quantification of MS2 and φ6), or followed by 94 °C for 15 s and 56 °C for 60 s (for MNV quantification).
A standard curve generated from tenfold serial dilution of standard DNA (plasmid DNA or oligo DNA) containing the target sequence (105–101 or 5 × 105 to 5 × 100 copies/reaction) was used to determine the number of viral genome copies per qPCR reaction. A negative control was included in all qPCR assays. The amplification efficiencies for MS2, φ6, and MNV averaged 93.5%, 97.2%, and 99.8%, respectively. The coefficient of determination (R 2) for MS2, φ6, and MNV assay averaged 0.998, 0.999, and 0.999.
To test the impact of RT-qPCR kit on the whole or molecular process recovery (see Section 3.3), the RT-qPCR steps described earlier (referred to Method 1) was compared with one-step RT-qPCR using QuantiTect Probe RT-PCR kit (QIAGEN) (Method 2). Method 2 was performed following the manufacturer's instruction. A 5 μL of the RNA extract was mixed with 15 μL of the reaction mixture, containing 10 μL of master mix (QIAGEN), 0.2 μL of QuantiTect RT mix, virus-specific forward primers and reverse primers, and TaqMan probe. The conditions of thermal cycling were as follows: 50 °C for 30 min and 95 °C for 15 min, followed by 45 cycles of 94 °C for 15 s and 60 °C for 60 s (for quantification of MS2 and φ6), or followed by 94 °C for 15 s and 56 °C for 60 s (for MNV quantification).
2.6. Calculation of whole process recovery and molecular process recovery
The whole process recovery ratio (W), molecular process recovery ratio (M), and sample limit of detection (SLOD) (copies/mL) were presented as Eqs. (1), (2), (3), respectively. The cDNA concentration of MS2 and φ6 in control, measured by QIAamp Viral RNA Mini Kit, followed by RT-qPCR, were 5.55 ± 0.31 log and 5.61 ± 0.27 log copies/mL, respectively.
| (1) |
| (2) |
| (3) |
where C obs_WPC indicates the cDNA concentration of WPC in concentrated samples (copies/mL), C ini_WPC indicates the cDNA concentration of WPC in the control sample (copies/mL), x indicates the concentration factor during the whole process (primary concentration, RNA extraction, and RT), C obs_MPC indicates the cDNA concentration of MPC in concentrated samples (copies/mL), C ini_MPC indicates the cDNA concentration of MPC in the control sample (copies/mL) and ALOD indicates the assay limit of detection defined as the minimum copy number with a 95% probability detection (copies/mL).
The concentrated samples and corresponding control samples were always subjected to RT-qPCR simultaneously to avoid the potential bias on the whole and molecular process recovery ratio. Furthermore, the number of freeze-thawing (i.e., none or once) was made consistent between concentrated and control samples.
2.7. Detection of SARS-CoV-2 RNA from raw sewage
2.7.1. Refinement of PEG+TRIzol by RNeasy PowerMicrobiome Kit
The PEG + TRIzol method was slightly modified by combining TRIzol LS reagent (Thermo Fisher Scientific) with RNeasy PowerMicrobiome Kit (QIAGEN) (TRIzol + RNeasy PowerMicrobiome Kit). This method contains two steps: sample lysis by TRIzol LS reagent and RNA precipitation, purification and elution using RNeasy PowerMicrobiome Kit. The theoretical advantages of this method are to increase the input volume for RNA extraction, which contribute to minimize the SLOD, and to reduce the sample processing time compared to TRIzol.
Specifically, a 250 μL aliquot of PEG concentrate was mixed with 750 μL of TRIzol LS reagent and vigorously vortexed for 30 s. After a 5 min incubation, 200 μL of chloroform was added to the mixture and vigorously vortexed for 15 s. The mixture was then centrifuged at 12,000g for 15 min at 4 °C. A colorless upper aqueous phase was mixed with 150 μL of solution IRS, which is designed to effectively remove PCR inhibitors (Ahmed et al., 2020b). Thereafter, the RNA was precipitated and filtered through a silica-based column followed by washing and elution, as described in the manufacturer's instructions, without DNase treatment. The final volume of eluted RNA was 60 μL. Thus, the concentration factor of this process was 4.2-fold.
The applicability of PEG + TRIzol + RNeasy PowerMicrobiome Kit was confirmed by raw sewages A, B, and C. The whole process recovery of φ6 was evaluated in duplicate for each raw sewage. The RT-qPCR process follows method 1 (two-step RT-qPCR using High-Capacity cDNA Reverse Transcription Kit and TaqMan™ Gene Expression Master Mix).
2.7.2. Processing of raw sewage from COVID-19 epidemic areas
A 400–500 mL of grab sample of raw sewage was collected on June 30, July 7, 16, 22, and 29; and August 5, 2020 at municipal WWTPs D and E, located in Tokyo Metropolis, Japan. The samples were stored at −20 °C until processing. A total of 12 samples were further processed as below.
A 40-mL aliquot of each raw sewage was concentrated by PEG. Then, a 250 μL of the concentrate was processed by TRIzol + RNeasy PowerMicrobiome Kit to obtain a 60 μL of RNA extract.
Then, 35 μL of RNA extract was subjected to RT to obtain 70 μL of cDNA as a final volume. Four published assays (NIID_2019-nCOV_n, CDC N1, CDC N2, and CDC N3 assays (Centers for Disease Control and Prevention, 2020; Shirato et al., 2020)) were performed to detect the SARS-CoV-2 RNA from raw sewage. We also tested the whole process recovery ratio of φ6.
A standard curve generated from tenfold serial dilution of standard DNA [plasmid DNA (Integrated DNA Technologies) for CDC assays and gBlocks for NIID assay] containing the target sequence (5 × 104 to 5 × 100 copies/reaction) was used to determine the number of viral genome copies per qPCR reaction. Negative control was included in duplicate in all qPCR assays. The amplification efficiencies for NIID_2019-nCOV_n, CDC N1, CDC N2, and CDC N3 averaged 97.1%, 94.5%, 92.2%, and 95.2%, respectively. The coefficient of determination (R 2) for NIID_2019-nCOV_n, CDC N1, CDC N2, and CDC N3 averaged 0.995, 0.995, 0.999, and 0.998, respectively. The assay limit of quantification (ALOQ) (i.e., lowest copy number detected at 100%) was 1 copies/μL.
2.8. Statistical analysis
Paired t-test and analysis of variance (ANOVA) were conducted using R (R Core Team, 2017).
3. Results and discussion
3.1. Whole process recovery ratio of MS2 and φ6
The comparisons among the combination of primary concentration and RNA extraction methods were performed as shown in Fig. 1 . For each type of raw sewage, three primary concentration methods were performed four times. Each primary concentrate was then subjected to two types of RNA extraction methods. Note that UF concentration was performed separately to conduct both RNA extraction methods; The limited volume of single UF concentrate (i.e., 170–210 μL) did not allow for conducting both methods, which requires a total of 280 μL of concentrate. UF + TRIzol was not performed for raw sewage A due to the limited amount of the sample.
Fig. 1.
Flow diagram of sample processing for the comparison of whole process recovery ratio. The concentration factor in each process was described as **-fold, below the name of the processing.
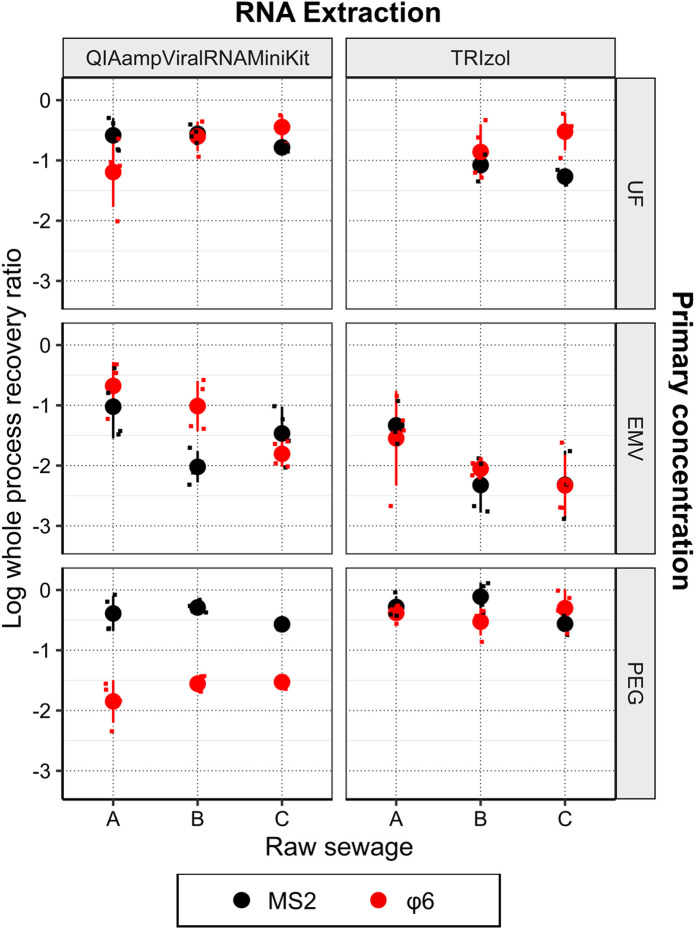
The whole process recovery ratio of MS2 and φ6 by each method (primary concentration + RNA extraction) as a function of the type of raw sewage is shown in Fig. 2 . The mean log whole process recovery ratio along with its corresponding standard deviation (SD) is shown in Table 2 .
Fig. 2.
Whole process recovery ratio of MS2 and φ6 by each method (primary concentration + RNA extraction) as a function of the type of raw sewage. Each panel shows the achievable sample limit of detection (SLOD) of MS2 and φ6 with respect to the type of raw sewage. Black color indicates MS2 recovery, whereas red color indicates φ6 recovery. Circles with error bars indicate mean values and standard deviation. Smaller square is jittered plots, indicating each data of SLOD. Note that the SLOD by UF + TRIzol (raw sewage A) was not available.
Table 2.
MS2 and φ6 recovery ratio of each method (primary concentration + RNA extraction).
| Raw sewage | Primary concentration | RNA extraction kit | MS2 |
φ6 |
||||
|---|---|---|---|---|---|---|---|---|
| Meana | SDb | Meanc (%) | Meana | SDb | Meanc (%) | |||
| A | UF | QIAamp Viral RNA Mini Kit | −0.58 | 0.53 | 26.1 | −1.19 | 0.58 | 6.4 |
| EMV | QIAamp Viral RNA Mini Kit | −1.02 | 0.30 | 9.5 | −0.68 | 0.40 | 21.0 | |
| EMV | TRIzol | −1.33 | 0.30 | 4.6 | −1.55 | 0.79 | 2.8 | |
| PEG | QIAamp Viral RNA Mini Kit | −0.39 | 0.18 | 40.8 | −1.85 | 0.35 | 1.4 | |
| PEG | TRIzol | −0.28 | 0.28 | 52.4 | −0.38 | 0.13 | 41.6 | |
| B | UF | QIAamp Viral RNA Mini Kit | −0.56 | 0.26 | 27.6 | −0.60 | 0.25 | 25.0 |
| UF | TRIzol | −1.08 | 0.19 | 8.4 | −0.86 | 0.46 | 13.8 | |
| EMV | QIAamp Viral RNA Mini Kit | −2.02 | 0.46 | 1.0 | −1.01 | 0.42 | 9.7 | |
| EMV | TRIzol | −2.32 | 0.10 | 0.5 | −2.05 | 0.15 | 0.9 | |
| PEG | QIAamp Viral RNA Mini Kit | −0.29 | 0.24 | 51.4 | −1.56 | 0.14 | 2.8 | |
| PEG | TRIzol | −0.11 | 0.13 | 77.6 | −0.53 | 0.23 | 29.8 | |
| C | UF | QIAamp Viral RNA Mini Kit | −0.78 | 0.44 | 16.5 | −0.45 | 0.20 | 35.8 |
| UF | TRIzol | −1.26 | 0.10 | 5.2 | −0.52 | 0.31 | 30.0 | |
| EMV | QIAamp Viral RNA Mini Kit | −1.47 | 0.70 | 3.4 | −1.80 | 0.22 | 1.6 | |
| EMV | TRIzol | −2.58 | 0.07 | 0.3 | −2.33 | 0.51 | 0.5 | |
| PEG | QIAamp Viral RNA Mini Kit | −0.57 | 0.13 | 27.0 | −1.53 | 0.09 | 3.0 | |
| PEG | TRIzol | −0.56 | 0.05 | 27.5 | −0.30 | 0.31 | 49.8 | |
Mean values of log whole process recovery ratio.
Standard deviation of log whole process recovery ratio.
Anti-logarithm of mean values of log whole process recovery ratio.
PEG + TRIzol provided the highest mean φ6 recovery ratio of −0.38 ± 0.13 log (41.6%), −0.53 ± 0.23 log (29.8%), and −0.30 ± 0.31 log (49.8%) for raw sewages A, B, and C, respectively. The second highest recovery was achieved by UF + QIAamp Viral RNA Mini Kit [−0.60 ± 0.25 log (25.0%) for B, −0.45 ± 0.20 log (35.8%) for C] or EMV + QIAamp Viral RNA Mini Kit [−0.68 ± 0.40 log (21.0%)] for A. Two methods (PEG + QIAamp Viral RNA Mini Kit and EMV + TRIzol) did not provide >−1.0 log (>10%) recovery for φ6. These results suggested that PEG + TRIzol can be an efficient recovery method of enveloped viruses from a variety of raw sewages.
PEG + TRIzol showed the highest φ6 recovery among the five combinations. A similar approach was performed for the detection of SARS-CoV-2 RNA from the raw sewage (Wu et al., 2020). A previous study reported a comparable whole process recovery ratio of MHV [−0.39 log (44%)] with our results (Ahmed et al., 2020b). These studies further confirmed the applicability of PEG + TRIzol for the efficient recovery of enveloped viruses.
PEG + QIAamp Viral RNA Mini Kit provided significantly lower φ6 recoveries than PEG +TRIzol by averaged 1.2 log (paired t-test, P < 0.001), whereas the MS2 recoveries were comparable (paired t-test, P > 0.05). This might be because the particles in the PEG concentrate might hamper the extraction of φ6 by QIAamp Viral RNA Mini Kit. A previous study also reported the lower RNA extraction efficiency for enveloped influenza virus than QIAamp Viral RNA Mini Kit under the presence of particles (Fabian et al., 2009). TRIzol was originally designed to extract RNA from cell and tissue samples, whereas QIAamp Viral RNA Mini Kit was designed to extract from relatively turbid-free samples (i.e., plasma, serum, and other cell-free body fluids) according to the manufacturer. These results may contribute to the superior performance of TRIzol over QIAamp Viral RNA Mini Kit.
UF was chosen as a primary concentration method in several studies (Medema et al., 2020; Sherchan et al., 2020) for the detection of SARS-CoV-2 RNA from raw sewage. Our results indicated that UF concentration provided ranging from −1.19 to −0.45 log (6.4–35.8%) and from −0.86 to −0.52 log (13.8–30.0%) when combined with QIAamp Viral RNA Mini Kit and TRIzol, respectively. These results were comparable with Ahmed et al. (2020b), which have reported MHV recovery of −0.55 log (28%) using UF [i.e., Centricon Plus-70 (10 kDa)]. A slightly lower recovery of UF+ QIAamp Viral RNA Mini Kit in raw sewage A was due to the outlier (φ6 recovery of −2.06 log) (see Fig. 2). Interestingly, the use of TRIzol did not significantly improved the whole process φ6 recovery of UF, contrary to the results observed in PEG. This was possibly because the positive effect of using TRIzol for φ6 extraction was offset by the inhibition during molecular process. The MNV recovery was lower in UF + TRIzol than UF + QIAamp Viral RNA Mini Kit by 0.37 to 1.09 log (see Fig. S1 and Table S2). Accordingly, the MS2 recovery was lower in TRIzol than QIAamp Viral RNA Mini Kit, as shown in Fig. 2. Thus, the both effects, higher extraction efficiency but stronger inhibition, leads to the comparable whole φ6 recovery ratio of UF + TRIzol to UF + QIAamp Viral RNA Mini Kit.
EMV + QIAamp Viral RNA Mini Kit has been originally developed to concentrate viruses and protozoa simultaneously (Haramoto et al., 2012). This method provided favorable φ6 recovery ratio for raw sewage A. However, the φ6 recovery was decreased to −1.01 ± 0.42 (9.7%) for B and −1.80 ± 0.22 (1.6%) for C. Interestingly, the result of ANOVA suggested that the water matrix significantly affected the recovery ratio of MS2 (P < 0.05) and φ6 (P < 0.01). The lower recovery in raw sewages B and C might be due to the lower elution efficiency, caused by the higher turbidity and organic concentration in wastewater matrix (higher TSS and UV260) (see Table 1). The turbid water matrix promoted membrane fouling, which prevented the elution buffer from contacting with the surface of the membrane, where viruses attached, during the vigorous vortex. In the filtration of raw sewage supplemented with Mg2+ ions through negatively charged membranes, not only viruses but also various components of feed water (e.g., humic acids, silica, and clays) are co-deposited on the membrane surface (Hata et al., 2011). Previous research also reported the negative impact of membrane fouling on the elution efficiency (Shi et al., 2016). In fact, the original work developing EMV + QIAamp Viral RNA Mini Kit validated the whole process recovery ratio of nonenveloped coliphage Qβ and poliovirus for river water and tap water, but did not test for raw sewage (Haramoto et al., 2012). A recent work detected SARS-CoV-2 RNA from the secondary effluent and did not detect from the raw sewage (Haramoto et al., 2020). These results suggest that EMV + QIAamp Viral RNA Mini Kit can be applied only to concentrate enveloped viruses for relatively clean water samples (e.g., secondary effluent or raw sewage containing low TSS and UV260, such as A) and not universally recommended for raw sewage.
EMV + TRIzol provided a lower recovery of both MS2 and φ6 (<5%). The comparison between the RNA extraction kits in EMV was discussed in SI.
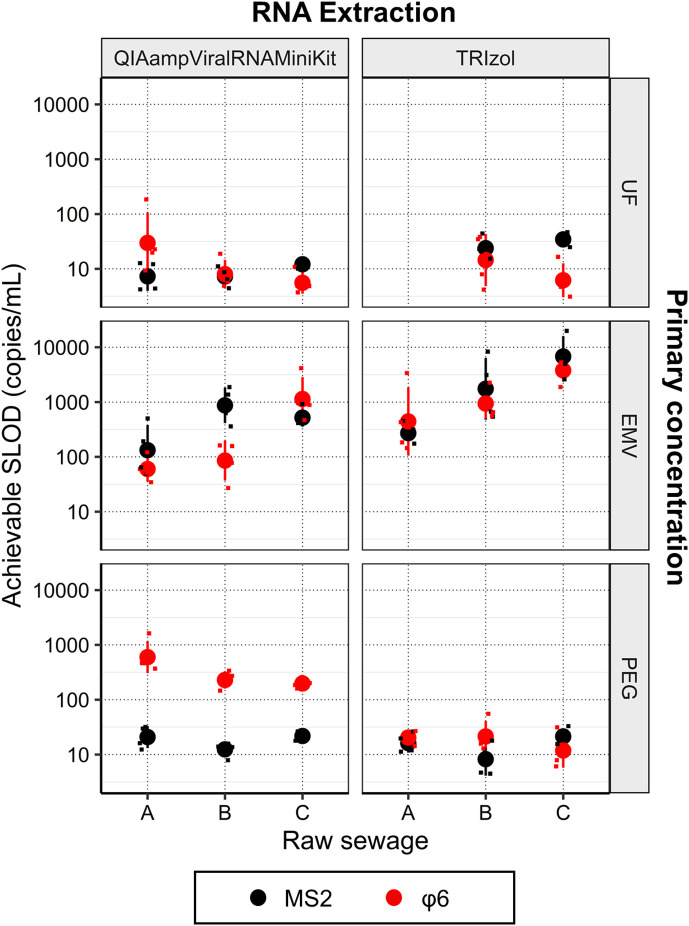
3.2. Achievable sample limit of detection (SLOD)
Achievable SLOD in each combination of primary concentration and RNA extraction method was shown in Fig. 3 . In this study, we adopted the observed values of concentration factor and whole process recovery ratio and assumed ALOD to be the most sensitive value theoretically possible, namely, 3 copies/5 μL (Bustin et al., 2009).
Fig. 3.
Sample limit of detection (SLOD) by each method (primary concentration + RNA extraction) as a function of the type of raw sewage. Each panel shows the achievable sample limit of detection (SLOD) of MS2 and φ6 with respect to the type of raw sewage. Black color indicates MS2 recovery, whereas red color indicates φ6 recovery. Circles with error bars indicate mean values and standard deviation. Smaller square is jittered plots, indicating each data of SLOD. Note that the SLOD by UF + TRIzol (raw sewage A) was not available.
PEG + TRIzol achieved the lowest SLOD of φ6 for A [1.31 ± 0.12 log copies/mL (20 copies/mL)] and the third lowest SLOD of φ6 for B and C [1.33 ± 0.28 log copies/mL (21 copies/mL) and 1.07 ± 0.31 copies/mL (12 copies/mL), respectively]. UF + QIAamp Viral RNA Mini Kit achieved the second lowest SLOD of φ6 for A [1.47 ± 0.56 log copies/mL (30 copies/mL)] and the lowest SLOD of φ6 for B and C [0.90 ± 0.26 log (8.0 copies/mL) and 0.75 ± 0.20 (5.6 copies/mL), respectively]. UF + TRIzol achieved the second lowest SLOD of φ6 for B and C [1.16 ± 0.48 log copies/mL (14 copies/mL) and 0.79 ± 0.31 copies/mL (6.2 copies/mL), respectively]. Hence, the PEG + TRIzol, UF + QIAamp Viral RNA Mini Kit, and UF + TRIzol provided the most sensitive SLOD among the tested combination of primary concentration and RNA extraction.
Achievable SLOD depends on the concentration factor, whole process recovery ratio, and ALOD. Thus, each method still can be potentially improved by higher concentration factors (i.e., increased initial sample volume or reduced concentrate volume). For example, previous studies adopted higher initial volume for primary concentration: up to 200 mL for UF concentration (Medema et al., 2020), up to 200 mL for EMV (Haramoto et al., 2020), and up to 250 mL for PEG (La Rosa et al., 2020). It should be noted that the concentration factor and whole/molecular process recovery are generally trade-offs; highly concentrated samples contain a higher amount of inhibitor, reducing the efficiency of RT and qPCR process. Hence, the removal of inhibitory substances during RNA extraction, optimized dilution of primary concentrate, and selection of reverse transcriptase qPCR master mix resistant to inhibition will be required if the primary concentration is performed with a higher concentration factor.
3.3. Impact of RT-qPCR kit on the whole process recovery ratio
The impact of the RT-qPCR kit selection on the whole process recovery was evaluated for three primary concentration methods using raw sewage A as a test sample and QIAamp Viral RNA Mini Kit as an RNA extraction kit (Fig. S2). The stored RNA extract of QIAamp Viral RNA Mini Kit was subjected to RT-qPCR process of method 2. The RT-qPCR recovery was compared between the two types of methods.
The whole process MS2 and φ6 recovery is presented in Fig. S3. Method 2 provided MS2 recovery of −2.53 ± 0.32 log, −3.74 ± 0.72 log, and −1.22 ± 0.11 log and φ6 recovery of −3.38 ± 0.11 log, −2.67 ± 0.49 log, and −2.26 ± 0.16 log for UF, EMV, and PEG, respectively. For all the primary concentration methods, the whole process MS2 and φ6 recovery by method 2 were significantly lower than method 1 (paired t-test, P < 0.05). Moreover, the molecular process MNV recovery was −2.17 ± 0.54 log, −2.42 ± 0.36 log, and −0.59 ± 0.22 log for UF, EMV, and PEG, respectively (Fig. S4). They were significantly lower in method 2 than method 1 (paired t-test, P < 0.05). These results indicate that method 2 was not effective for the quantification of both MS2 and φ6 from raw sewage processed by three types of primary concentration methods.
The observed difference was due to the inhibition during the RT-qPCR process, as revealed by the molecular process recovery of MNV. Both methods are different in terms of master mix composition, the type of primer (method 1, random primer; method 2, specific primer), and the inclusion of RNase inhibitor (method 1, included; method 2, not included). Moreover, in method 1, 0.5-fold RNA extract was subjected to qPCR [due to the dilution in the RT process (Fig. 1)], potentially contributing to the mitigation of PCR inhibition. A previous study reported that QIAGEN QuantiTect Probe RT-PCR Kit was susceptible to organic PCR inhibitor, such as lactoferrin, than other RT kits (Stephens et al., 2010). These results indicate that the RT-qPCR kit affects the quantification of the virus concentrate for all the methods in raw sewage. Thus, the RT-qPCR steps should also be optimized for efficient recovery.
3.4. Detection of SARS-CoV-2 RNA from raw sewage
The applicability of PEG + TRIzol + RNeasy PowerMicrobiome Kit was investigated for the detection of SARS-CoV-2 RNA from raw sewage. First, the PEG + TRIzol + RNeasy PowerMicrobiome Kit provided the φ6 recovery ratio of −0.10 ± 0.10 (79%), −0.56 ± 0.10 (27.3%), and −0.75 ± 0.03 (17.8%) for A, B, and C, respectively, which were comparable with the PEG + TRIzol method. The SLOD was 0.72 ± 0.12 log copies/mL (5.3 copies/mL), 1.18 ± 0.09 log copies/mL (15.1 copies/mL), and 1.34 ± 0.03 log copies/mL (11.8 copies/mL). Thus, the SLOD can be slightly lowered than PEG + TRIzol.
The PEG + TRIzol + RNeasy PowerMicrobiome Kit was then applied to the detection of SARS-CoV-2 RNA from the raw sewage collected from Tokyo Metropolis. The results of four SARS-CoV-2 qPCR assays along with corresponding values of whole process recovery of φ6 were reported (Table 3 ). The whole process recovery of φ6 averaged −0.49 ± 0.32 log (33%). The sample limit of quantification (SLOQ), given by ALOQ (1 copies/μL) divided by the concentration factor and by φ6 whole process recovery ratio, averaged 4.29 ± 0.32 log copies/L (2.0 × 104 copies/L). Of the 12 grab raw sewage samples, SARS-CoV-2 RNA was detected in 4 samples, collected on July 7 (CDC N1:<4.1 × 104 copies/L), 16 (CDC N3:<6.0 × 103 copies/L), and 29 (CDC N1, <1.4 × 104 copies/L) in Plant D and on July 29 in Plant E (CDC N1, <1.1 × 104 copies/L). The statistical information of confirmed cases in the Tokyo Metropolis is presented in Table S3. The discussion between confirmed cases and the SARS-CoV-2 RNA is described in SI. Overall, the PEG + TRIzol was successfully applied to the detection of SARS-CoV-2 RNA from wastewater with a favorable whole process recovery ratio of φ6.
Table 3.
Detection of SARS-CoV-2 RNA from raw sewage.
| Date | Plant | Concentration factor | log10 Wa | SLOQ (copies/mL) |
SARS-CoV-2 |
|||
|---|---|---|---|---|---|---|---|---|
| CDC N1 |
CDC N2 |
CDC N3 |
NIID | |||||
| June 30 | D | 160 | −0.41 | 16.1 | Neg | Neg | Neg | Neg |
| E | 149 | −0.76 | 38.5 | Neg | Neg | Neg | Neg | |
| July 7 | D | 152 | −0.79 | 40.6 | Posb | Neg | Neg | Neg |
| E | 163 | −0.65 | 27.5 | Neg | Neg | Neg | Neg | |
| July 16 | D | 152 | 0.04 | 6.0 | Neg | Neg | Posc | Neg |
| E | 146 | −0.29 | 13.2 | Neg | Neg | Neg | Neg | |
| July 22 | D | 159 | −1.14 | 86.6 | Neg | Neg | Neg | Neg |
| E | 160 | −0.34 | 13.5 | Neg | Neg | Neg | Neg | |
| July 29 | D | 159 | −0.36 | 14.3 | Posd | Neg | Neg | Neg |
| E | 160 | −0.26 | 11.3 | Pose | Neg | Neg | Neg | |
| August 5 | D | 154 | −0.2 | 10.4 | Neg | Neg | Neg | Neg |
| E | 154 | −0.68 | 31.3 | Neg | Neg | Neg | Neg | |
Neg: negative, Pos: positive.
Whole process recovery ratio (W) of φ6 was determined at n = 1 for each sample.
SARS-CoV-2 CDC N1 gene was positive; the Ct was 38.7 (in one of two PCR reactions).
SARS-CoV-2 CDC N3 gene was positive; Ct was 38.9 (in one of two PCR reactions).
SARS-CoV-2 CDC N1 gene was positive; the Ct was 40.9 (in one of two PCR reactions).
SARS-CoV-2 CDC N1 gene was positive; the Ct was 37.7 (in one of two PCR reactions).
3.5. Implications for the wastewater-based epidemiology
The results suggest three implications for the successful application of WBE. First, to validate the quantification results of enveloped viruses, it is preferable to include enveloped surrogates. Earlier studies detecting the SARS-CoV-2 RNA used nonenveloped surrogates, such as indigenous F-RNA phages (Medema et al., 2020) and coliphage MS2 (Kumar et al., 2020), for the validation partially due to the resource limitation at that time. However, nonenveloped and enveloped surrogates can be recovered with different efficiency as highlighted in the case of PEG + QIAamp Viral RNA Mini Kit (see Fig. 2). This indicates the danger of reliance on nonenveloped surrogates.
In addition, not only primary concentration but also the molecular process (RNA extraction and RT-qPCR) should be optimized. Our study suggested that effective primary concentration is just the first step. An appropriate molecular process (i.e., RNA extraction and RT-qPCR) is required, as was proven by the comparison between PEG + QIAamp Viral RNA Mini Kit and PEG + TRIzol (see Fig. 2) and by the comparison between RT-qPCR methods (Fig. S3).
Finally, the universal applicability of primary concentration methods cannot be judged from a single type of wastewater matrix. The results of the whole process recovery by EMV indicate that the efficiency of primary concentration differs depending on the wastewater matrix (see Fig. 2). It may be necessary to validate the quantification process in house by adopting the appropriate WPC because the RNA extraction kits, RT-qPCR kits, and the water quality differ in every laboratory.
A further consideration is required for adopting φ6 as a process control to validate the quantification results of enveloped viruses. The possible rationales and limitations of adopting φ6 are presented in Table 4 . Under the limited BSL facility (e.g., WWTP), the choice of φ6 will be practically best considering the broad and commercial availability compared with other enveloped bacteriophages. However, φ6 has double-stranded RNA and an envelope derived from Pseudomonas syringae, which might not fully reflect the properties of viruses in interest (e.g., SARS-CoV-2 and influenza). Thus, further research should confirm the comparability of the fate of φ6 during primary concentration and molecular process with the coronavirus surrogates (MHV, porcine epidemic diarrhea virus (Randazzo et al., 2020), bovine attenuated coronavirus (Gonzalez et al., 2020)) and ideally indigenous SARS-CoV-2 in wastewater. To the best of our knowledge, no studies have evaluated the comparability of whole process recovery of surrogate viruses. De facto standardization of these surrogate viruses should be avoided before the comparison conducted in the future.
Table 4.
Rationales and limitations of φ6 as a process control for SARS-CoV-2.
| Rationales | Limitations |
|---|---|
| ・Similar morphologies Having an envelope, thus showing relatively similar adsorptive characteristics with murine hepatitis virus (MHV) compared with MS2 (Ye et al., 2016). ・Minimal containment for laboratory Not required for BSL2 facility (Handling of MHV and other enveloped virus surrogates required BSL2). This feature allows for on-site usages, such as a wastewater treatment plant, as a whole process control for enveloped viruses. ・Easy handling Pseudomonas syringae and φ6 can be easily propagated with high concentration. The φ6 titers of 1010 PFU/mL can be achieved. |
・Additional steps required for quantification Having a double-stranded RNA. This may require an additional step (e.g., heat denaturation of dsRNA before RT step) for quantification. ・No proof of comparability to SARS-CoV-2 The structural difference between φ6 and SARS-CoV-2 and between freshly prepared enveloped viruses and indigenous SARS-CoV-2 might affect the recovery efficiency. |
4. Conclusions
-
▪
Polyethylene glycol precipitation (PEG) followed by acid guanidinium thiocyanate-phenol-chloroform extraction (PEG + TRIzol) provided the highest whole process recovery ratio of Pseudomonas phage φ6 ranged from −0.53 to −0.30 log (29.8–49.8%) in three types of raw sewage.
-
▪
Ultrafiltration (UF) followed by QIAamp Viral RNA Mini Kit provided a comparable whole process recovery of φ6, ranging from −1.19 to −0.45 log (6.4–35.8%), with MS2.
-
▪
Electronegative membrane vortex (EMV) provided significantly different whole process recovery of MS2 and φ6 depending on the water quality of raw sewage; the recovery was reduced in raw sewage containing higher TSS and UV260.
-
▪
The successful recovery of the enveloped virus by PEG precipitation might need an appropriate RNA extraction method (e.g., acid guanidinium thiocyanate-phenol-chloroform RNA extraction). Not only primary concentration but also the following molecular process should be optimized for the efficient recovery of enveloped viruses.
-
▪
Non-enveloped surrogate (MS2 and MNV) did not necessarily validate the success of the primary concentration and molecular process of φ6 (e.g., PEG+QIAamp Viral RNA Mini Kit). This indicates enveloped viruses should be spiked to primary concentrate as whole process control and molecular process control to validate the quantification of enveloped viruses from raw sewage.
-
▪
The modified PEG + TRIzol method was successfully applied to detect SARS-CoV-2 RNA by CDC N1 and N3 assay from raw sewage collected on 7th, 16th, and 29th in July 2020 in Tokyo Metropolis.
CRediT authorship contribution statement
Shotaro Torii: Investigation, Resources, Funding acquisition, Writing - original draft. Hiroaki Furumai: Project administration, Writing - review & editing. Hiroyuki Katayama: Resources, Project administration, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by Grant-in-Aid for JSPS Fellows (20J10268), Grant-in-Aid for Scientific Research (A) 20H00259, Japan Society for the Promotion of Science (JSPS), Japan Institute of Wastewater Engineering and Technology, and the “Startup Research Program for Post-Corona Society” of Academic Strategy Office, School of Engineering, the University of Tokyo. Graphical abstract was created with Biorender.com.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.143067.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J., Simpson S., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. doi: 10.1021/acs.estlett.5b00029. [DOI] [Google Scholar]
- Casanova L.M., Weaver S.R. Evaluation of eluents for the recovery of an enveloped virus from hands by whole-hand sampling. J. Appl. Microbiol. 2015;118:1210–1216. doi: 10.1111/jam.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Fabian P., McDevitt J.J., Lee W.-M., Houseman E.A., Milton D.K. An optimized method to detect influenza virus and human rhinovirus from exhaled breath and the airborne environment. J. Environ. Monit. 2009;11:314–317. doi: 10.1039/B813520G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron L., Verreault D., Veillette M., Moineau S., Duchaine C. Evaluation of filters for the sampling and quantification of RNA phage aerosols. Aerosol Sci. Technol. 2010;44:893–901. doi: 10.1080/02786826.2010.501351. [DOI] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 2009;32:297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Asami M., Akiba M. Development of a novel method for simultaneous concentration of viruses and protozoa from a single water sample. J. Virol. Methods. 2012;182:62–69. doi: 10.1016/j.jviromet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kitajima M., Visvanathan C., Nol C., Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl. Environ. Microbiol. 2011;77:4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Honda R., Hara-Yamamura H., Meuchi Y. 2020. Identification of SARS-CoV-2 in Wastewater in Japan by Multiple Molecular Assays-implication for Wastewater-based Epidemiology (WBE). medRxiv 2020.06.09.20126417. [DOI] [Google Scholar]
- Kazama S., Masago Y., Tohma K., Souma N., Imagawa T., Suzuki A., Liu X., Saito M., Oshitani H., Omura T. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res. 2016;92:244–253. doi: 10.1016/j.watres.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Tohya Y., Matsubara K., Haramoto E., Utagawa E., Katayama H., Ohgaki S. Use of murine norovirus as a novel surrogate to evaluate resistance of human norovirus to free chlorine disinfection in drinking water supply system. Environ. Eng. Res. 2008;45:361–370. doi: 10.11532/proes1992.45.361. [DOI] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Buisman A.M., Rutjes S.A., Heijne J.C., Teunis P.F., de Roda Husman A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012;78:3800 LP–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle C.D., Routson L.B. Minimized virus binding for tests of barrier materials. Appl. Environ. Microbiol. 1995;61:643–649. doi: 10.1128/aem.61.2.643-649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S., Tam K.I., Kerin T.K., Lewis J.M., Gautam R., Quaye O., Gentsch J.R., Bowen M.D. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J. Clin. Microbiol. 2013;51:3047–3054. doi: 10.1128/JCM.01192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020;54:5311. doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Heal. 2020 doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Xagoraraki I., Parent K.N., Bruening M.L., Tarabara V.V. Elution is a critical step for recovering human adenovirus 40 from tap water and surface water by cross-flow ultrafiltration. Appl. Environ. Microbiol. 2016;82:4982 LP–4993. doi: 10.1128/AEM.00870-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. advpub. 2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Stephens K.W., Hutchins R.J., Dauphin L.A. Cross-platform evaluation of commercial real-time reverse transcription PCR master mix kits using a quantitative 5′nuclease assay for Ebola virus. Mol. Cell. Probes. 2010;24:370–375. doi: 10.1016/j.mcp.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Torii S., Hashimoto T., Do A.T., Furumai H., Katayama H. Impact of repeated pressurization on virus removal by reverse osmosis membranes for household water treatment. Environ. Sci. Water Res. Technol. 2019;5:910–919. doi: 10.1039/C8EW00944A. [DOI] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Greening G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010;76:1388–1394. doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions [WWW Document] 2020. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material