Abstract
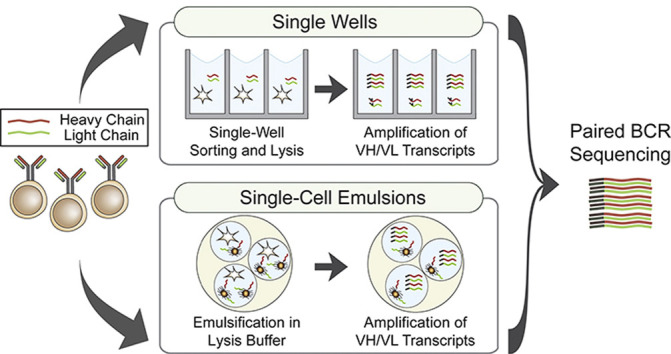
Recent advancements in paired B-cell receptor sequencing technologies have accelerated the development of simpler, high-throughput pipelines for generating native antibody heavy and light chain pairs used to elucidate novel antibodies and provide insights into antibody response against pathogenic targets. These technologies involve single-cell isolation, using either single wells or emulsified droplets to maintain physical separation of individual cells, followed by sequencing. The development of novel single wells and emulsion-based workflows addresses key challenges by improving throughput of single-cell analyses, reducing method complexity, and integrating functional assays into existing workflows. Enabled by paired B-cell receptor sequencing, functional characterization of pathogen-specific antibodies reveals immunological insights beyond bulk sequencing.
Keywords: BCR sequencing, Antibody repertoire, Antibody sequencing, VH:VL pair
Graphical abstract
Introduction
Adaptive immunity is dependent on both B-cell receptors (BCRs) and T-cell receptors that mediate various immune responses and confer protection against diverse pathogens; the unique ensemble of these receptors is known as the adaptive immune receptor repertoire. A principal component of humoral immune response is the repertoire of antibodies in both membrane-bound receptor form on the B-cell surface and in soluble form as immunoglobulin (Ig) molecules. Antibodies consist of two identical heavy (H) chains and two identical light (L) chains connected by disulfide bonds, with both H and L chains containing variable regions (called VH and VL, respectively) and constant domains. As B cells develop, the recombination of gene segments, known as V(D)J recombination, and somatic hypermutation, triggered by BCR–antigen engagement, result in highly diverse antibody repertoires [1]. This diversity in the antibody repertoire allows for protection against a plethora of targets [2], and it is constantly reshaped by continuous exposures to pathogens and vaccines across one's lifetime.
Recent and rapid advancements in antibody sequencing technologies have enabled unprecedented opportunities in studying B-cell immunobiology, informing both antibody engineering research [3] and the design of novel immunogens [4,5]. As the assembly of full-length antibodies involves two separate mRNA transcripts which encode the H and L chains, lysing and sequencing bulk B cells prevents elucidation of cognate H and L chain matches [6]. While the development of paired BCR sequencing [7, 8, 9] enabled determination of VH:VL pairs, it was limited by low cell throughput and a lack of pipelines to profile antibody–antigen interactions. Recent technological advances with application for paired BCR sequencing, summarized in Table 1 , have bolstered our ability to understand humoral immunity and have proven essential particularly in the rapid response to the global outbreak of coronavirus disease 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), facilitating discovery of monoclonal antibodies (mAbs) with therapeutic potential and elucidating the pathogenesis of coronavirus disease 2019. In this review, we highlight paired BCR sequencing advancements, categorized by methods in single-well and emulsified single-cell isolation, with a focus on publications from the past two years.
Table 1.
Summary of highlighted technologies applicable for paired BCR sequencing.
| Platform | Approach | Cell throughput | Strengths | Reference |
|---|---|---|---|---|
| Primer Well Barcoding | Single-well | 102–103 | Allows use of NGS for higher sequencing throughput and lower costs | [22, 23, 24, 25, 26] |
| Fluidigm C1 | Single-well | 103–104 | Minimal manual handling and preparation, options for automated staining or microscopy of cells | [29,30] |
| Seq-Wella | Single-well | ∼105 | High cell processing throughput at low cost | [31] |
| LIBRA-Seq | Single-cell emulsions | 104 | Built into the 10x Chromium system, allows interrogation of BCR binding to multiple antigens | [35] |
| CelliGo | Single-cell emulsions | 104 | Identifies secreted antibody–antigen binding from antibody-secreting cells to either secreted or membrane-bound antigens | [36] |
| RTX-NPS | Single-cell emulsions | 103–105 | No specialized equipment, simple workflow, generates physically linked VH:VL sequences | [39] |
| ER Microsomes | Single-cell emulsions | 103–106 | No specialized equipment, high cell processing throughput, generates physically linked VH:VL sequences | [40] |
BCR, B-cell receptor; TCR, T-cell receptor; LIBRA-seq, linking BCR to antigen specificity through sequencing; RTX, reverse transcription xenopolymerase; RTX-NPS, reverse transcription xenopolymerase natively paired sequencing; OE, overlap extension.
This method has been used to sequence TCRs but can be analogously applied to study BCRs.
Facile and versatile paired BCR sequencing using single-well cell isolation
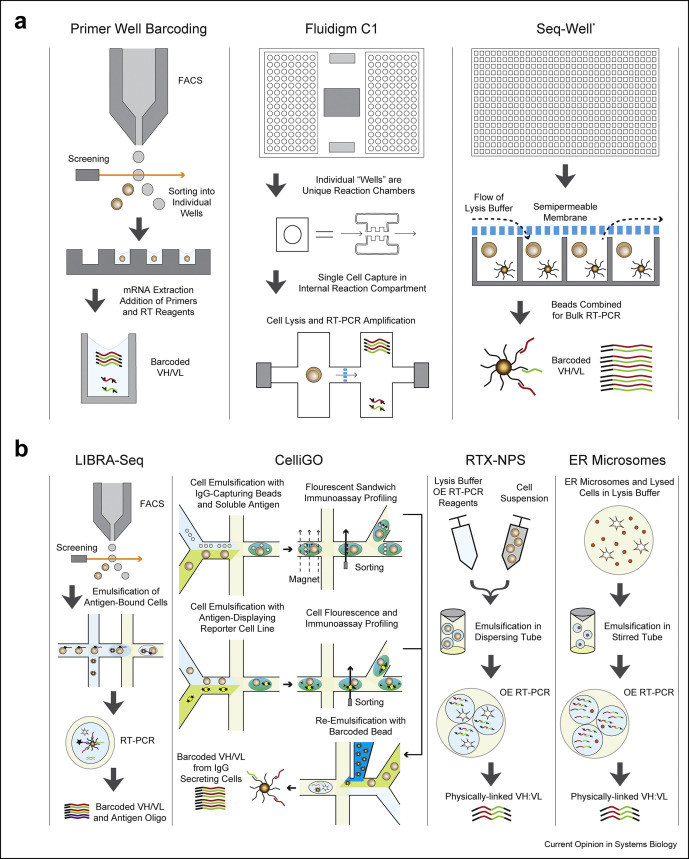
Interrogation of natively paired VH:VL sequences can rely on a simple method of isolating single B cells into individual wells using flow cytometry. Fluorescently labeled antigens and B-cell surface marker probes enable index sorting of specific subsets of B cells with desired antigen specificity [10], while having their phenotypic signatures recorded. Once single cells have been isolated into separate wells, they can be lysed, keeping individual cell contents constrained in the well, including the H and L chain transcripts [11] (Figure 1 a). Antibodies from sorted B cells can be subsequently cloned and recombinantly expressed for further characterization, as exemplified by recent articles rapidly isolating mAbs that target SARS-CoV-2 [12,13].
Figure 1.
Highlight of methods applicable for Paired BCR sequencing. (a) Single-well methods. (1) Primer well barcoding is accomplished using FACS. Cells are sorted and isolated into single wells where they are lysed. RNA is extracted from each well, unique well-barcoded primers are added, and BCR sequences are amplified by RT-PCR. (2) The C1 chip isolates cells into single nodes (represented by circles) where the cell is captured. These cells are lysed, RNA is passed into a secondary containment chamber, and the flow of RT-PCR reagents into this chamber allows amplification of barcoded sequences. (3) Seq-Well captures a single uniquely barcoded bead and an individual cell into a subnanoliter volume well. A semipermeable seal is applied, and lysis buffer is flowed over the plate, lysing the cell and hybridizing RNA to the barcoded beads. The seal is removed, beads are combined, and RT-PCR generates barcoded sequences. This method has been used to sequence TCRs but can be analogously applied to study BCRs. (b) Single-cell emulsion methods. (1) LIBRA-seq uses FACS to sort barcoded-antigen and B-cell mixtures, selecting for antigen-bound B cells. Cells are emulsified with uniquely barcoded primer beads and lysis buffer using microfluidics. Cell-barcoded BCR transcript and antigen oligos are amplified by RT-PCR, and barcodes are used to bioinformatically match antigen specificity with BCR sequences. (2) CelliGo sorts antibody-secreting cells for antibody binding to either secreted antigens (top) or membrane-bound antigens (middle). For secreted antigen sorting, cells are profiled by antibody–antigen capture on magnetic beads where the antigen's fluorescent signature can be read. For membrane-bound antigen sorting, cells are emulsified with a reporter cell line that is fluorescent and displays membrane-bound antigens. Fluorescently labeled Fc-specific F(ab’)2 anti-IgG antibodies bind to secreted IgG, and the fluorescent signal from both the reporter cell and the anti-IgG antibody is read. Target cells are emulsified with barcoded beads and lysis buffer (bottom), the beads are isolated and aggregated, and RT-PCR generates barcoded amplicon. (3) In the RTX-NPS approach, cells, lysis buffer, overlap extension primers, and a RT-PCR mixture containing RTX are joined at a Y junction and emulsified in oil. Physically linked VH:VL amplicon is amplified in emulsion by OE RT-PCR for use in sequencing. (4) In the ER microsome method, cycloheximide-treated cells are incubated in a sucrose and digitonin buffer, forming ER microsomes and lysing the cell. After purification by low-speed centrifugation, microsomes are emulsified with RT-PCR reagents and overlap extension primers. Physically linked VH:VL amplicon is generated by OE RT-PCR. BCR, B-cell receptor; TCR, T-cell receptor; LIBRA-seq, linking BCR to antigen specificity through sequencing; RTX, reverse transcription xenopolymerase; RTX-NPS, reverse transcription xenopolymerase natively paired sequencing; OE, overlap extension; FACS: fluorescence-activated cell sorting; ER: endoplasmic reticulum.
With the precise control provided by single-well sorting, single B-cell cultures enable functional characterization of secreted antibodies [14, 15, 16, 17]. This approach has elucidated unique cross-reactive antibodies to influenza hemagglutinin [18] and provided insights into a novel antigen epitope [19]. Alternatively, a nanofluidic optoelectronic single B lymphocyte antibody screening technique processes single antibody-secreting cells by sequestering them into small nanopens, where a fluorescent binding assay is used to distinguish cells that secrete antigen-specific antibodies and export them for plate-based sequencing [20]. Although functional profiling has proven a useful addition, paired BCR sequencing has additionally been hindered by the high cost and low throughput of Sanger sequencing.
The desire to sequence a greater number of BCRs parallelly, while decreasing cost, has led to the utilization of next-generation sequencing (NGS) platforms [21], although Sanger sequencing may be preferred in certain applications for its simplicity, high accuracy, and absence of sequencing biases. Three predominant approaches use well-specific primers to barcode amplicon for NGS (primer well barcoding). Linnarsson's group [22] created single-cell tagged reverse transcription sequencing to append well-specific six-base nucleotide barcodes to transcript. Sandberg and colleagues developed a switching mechanism at the 5′ end of RNA template sequencing (Smart-seq) technology [23], which uses well-specific oligos for extension and amplification of cDNA from transcripts. This has been further refined in the Smart-seq2 [24] and Smart-seq3 [25] technologies, minimizing cDNA purification steps, changing reverse transcriptase, and optimizing reverse transcription polymerase chain reaction (RT-PCR) conditions. The Wardemann lab introduced barcoded primer matrices [26] using 5′ and 3′ wells and antibody-specific primers to allow for parallel sequencing from thousands of individual wells. Applying NGS with the simplicity of single-well technologies is perhaps best demonstrated by the identification of SARS-CoV-2–neutralizing antibodies. Sorting B cells for spike protein and receptor-binding domain binding in infected patients and hamsters, well-barcoded BCR amplicons from thousands of cells were parallelly sequenced using NGS, leading to the identification and functional characterization of thousands of antigen-binding mAbs, some of which demonstrate potent neutralization activities against SARS-CoV-2, in as short as ten days [27,28].
To address the need for higher cell throughput to study the diverse BCR repertoire, advances have also focused on increasing the number of B cells profiled per experiment, using the sequencing throughput of NGS. The Fluidigm C1 system allows facile preparation of barcoded transcripts from up to 104 cells and provides a low-labor approach using a unique chip which isolates cells into individual reaction chambers, lyses cells, and generates a cDNA library for each cell automatically. This system has been used to study BCR class switching [29] and has recently facilitated profiling the autoreactive BCR repertoire in patients with rheumatoid arthritis [30]. An alternative workflow developed by Shalek and colleagues [31], which has massively expanded the cell processing throughput of single-well approaches, is the Seq-Well platform, using ∼86,000 subnanoliter wells in a custom plate to confine single cells and reagents for cDNA amplification and sequencing. Recently, applied in the study of T-cell receptors by sequencing peanut allergen–specific T cells and identifying unique T-cell clonotype expression patterns [32], the authors acknowledge application of this approach to analogously study BCRs.
Single-well technologies remain one of the most widely used approaches for paired BCR sequencing. With unparalleled control, pipelines for application-specific uses can be easily customized, and procedure simplicity obviates the need for specialized equipment or staff. Recent technological developments have expanded the utility of this approach by profiling antibody–antigen interactions, providing both Sanger and NGS workflows, while expanding the BCR sequencing throughput with customized equipment.
High-throughput paired BCR sequencing using single-cell emulsions
Single-cell emulsions generated by aqueous and organic phase separation provide a high-throughput alternative to single-well methods (Figure 1b). Two earlier technologies are highlighted as representative workflows for emulsion-based paired BCR sequencing. Drop-seq, an approach which utilizes microfluidics, merges a stream of B-cell suspension with uniquely barcoded beads, emulsifying them in an oil channel to create nanoliter droplets containing a single cell and a barcoded bead which is used to amplify transcript while retaining single-cell information [33]. In contrast, an ultrahigh-throughput approach requires a custom flow focusing device that joins two streams of poly-dT magnetic beads and B cells, emulsifying them in oil to create nanoliter droplets. Magnetic beads with hybridized mRNA are separated and re-emulsified in an RT-PCR solution where they undergo overlap extension (OE) RT-PCR to generate physically linked VH:VL amplicons [34]. Single-cell emulsion methods typically have a much larger throughput (104-106 cells per experiment) than single-well methods but are highly complex and rigid, preventing facile integration of antibody analysis pipelines.
Enabling simultaneous BCR profiling against a diverse panel of antigens, Georgiev and colleagues developed the linking BCR to antigen specificity through sequencing technology [35], which uses a panel of DNA-barcoded fluorescent antigens to sort and sequence antigen-binding B cells along with antigen barcodes. This method can interrogate an array of BCR–antigen interactions at once, deconvoluting antibodies with specificity to multiple antigens while also generating paired BCR sequences. CelliGO [36], on the other hand, develops on a previous antibody profiling method, DropMap [37], by identifying and sorting cells with (B cells and plasmablasts) and without (plasma cells) BCRs based on antigen specificity for secreted Ig, while also providing paired sequencing information. This broadens the utility of single-emulsion approaches, as fluorescence-based Ig analysis is now available in a high-throughput single-cell manner.
Recent technical developments have also provided solutions for reducing the high complexity and requirement for specialized training in emulsion-based methods. Engineering of reverse transcription xenopolymerase (RTX) [38], a cell lysate–tolerant xenopolymerase that acts as both a DNA polymerase and reverse transcriptase, has led to RTX natively paired sequencing [39], which has eliminated the need for microfluidics and other specialized equipment by directly emulsifying cells in lysis buffer with OE RT-PCR mixtures using two syringes, a Y junction, and a dispersing tube to generate paired VH:VL amplicons. An alternative approach uses naturally occurring rough endoplasmic reticulum microsomes to encapsulate H and L chain transcripts. By lysing the cell and using a series of density-based isolation steps through centrifugation, transcripts are contained in rough endoplasmic reticulum microsomes which can be emulsified with RT-PCR reagents to amplify paired transcripts [40], although this method has yet to be widely used. Physically linked VH:VL amplicon generated from OE RT-PCR, however, is too large to be fully read on many short-length NGS platforms and often requires additional sequencing. Developed as an alternative, repertoire and gene expression by sequencing (RAGE-seq) [41] combines the depth of short-length NGS with longer, full amplicon NGS reads, and a 3′ shearing method, where the 3′ end of 5' barcoded amplicons is sheared to generate short fragments [42], allows the entire physically linked cDNA to be read.
Expansion of emulsion-based method' accessibility from recent technical advancements and commercial solutions such as 10x Genomics provide expertise-free alternatives, assisting in a deeper profiling of B-cell–mediated immunity and integrated into a rapid response pipeline for antiviral discovery and therapeutic potency verification [43]. The promise of using single-cell emulsions is best demonstrated by a study from Cao et al. [44], where SARS-CoV-2–binding BCRs were sequenced and characterized in eight days, leading to the identification of hundreds of antigen-binding mAbs, 14 of which showed viral neutralization in a murine model. Challenges still remain, however, in emulsion-based paired BCR sequencing, namely dealing with errors and biases in NGS (i.e., high base pair substitution error rates in sequencing and primer bias in multiplex PCR) [45] and inaccurate barcoding in emulsions [46]. These highlight the need for new advancements to increase the accuracy of these analyses.
Beyond BCR sequencing
While paired BCR sequencing has driven antibody discovery and provided unique insights into B-cell development, there is a need to relate BCRs with functionally relevant (i.e., abundantly present) Ig molecules in circulation, which directly provide protection against pathogens. This is particularly important as not all B cells secrete Ig molecules. Using donor-specific BCR sequences as a database for proteomic analysis through high-resolution liquid chromatography tandem mass spectrometry and protein-level analyses enables the identification, quantification, and characterization of the secreted antibody repertoire, where antigen-specific antibody molecules can be profiled from affinity purification [47]. A recent study used the technology to longitudinally track influenza hemagglutinin–specific Ig molecules in the serum, which showed a small number of antibody clonotypes persisting over 5 years and dominating the serum response [48]. In addition, this proteomic approach assisted in the discovery of broadly neutralizing antibodies in both norovirus and HIV studies [49,50]. In addition, proteomic analysis can elucidate H and L chain pairs by analyzing intact mAb, Fab, F(ab')2, and light chain masses [51]. Proteomic analysis, in combination with BCR sequencing, provides quantitative details about the circulating antibody repertoire which is essential for distinguishing highly abundant protective antibodies from sequencing information.
Conclusions
Interrogating the BCR repertoire using paired BCR sequencing is imperative for both discovery and characterization of novel mAbs and understanding B-cell immunity. New technological developments have expanded throughput, decreased complexity, and provided integrated pipelines for profiling antibody–antigen engagement which have been critical in studying the humoral immune response to pathogens. Ultimately, a comprehensive understanding of the complexity of our immune system relies on both immune receptor sequencing and relevant proteomic information; the close interplay between these two fields will transform our ability to react to novel viral threats, understand and treat autoimmunity and cancer, and develop therapeutics and vaccines. Despite improvements in paired BCR sequencing, new innovations in both sequence preparation and sequencing methods are still needed to address outstanding challenges that exist in generating accurate and bias-free sequences.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge Dr. Seungmin Shin and Meredith Bailey for their assistance in editing this manuscript. This work was supported by the National Institutes of Health [P20GM113132], the Cystic Fibrosis Foundation [STANTO19R0] (JL), and the National Science Foundation [1840344] (NCC).
This review comes from a themed issue on Systems immunology & host-pathogen interaction (2020)
Edited by Ramit Mehr
References
- 1.Rees A.R. Understanding the human antibody repertoire. mAbs. 2020;12:1729683. doi: 10.1080/19420862.2020.1729683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoehn K.B., Fowler A., Lunter G., Pybus O.G. The diversity and molecular evolution of B-cell receptors during infection. Mol Biol Evol. 2016;33:1147–1157. doi: 10.1093/molbev/msw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grobben M., Stuart R.A., van Gils M.J. The potential of engineered antibodies for HIV-1 therapy and cure. Curr Opin Virol. 2019;38:70–80. doi: 10.1016/j.coviro.2019.07.007. [DOI] [PubMed] [Google Scholar]
- Steichen J.M., Lin Y.-C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J.R., Toy L., Sok D., Liguori A., Kratochvil S. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science. 2019;366 doi: 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Steichen et al. utilized antibody sequencing data to express naïve bnAb-like antibodies targeting a highly conserved HIV envelope protein epitope. They then used directed evolution with these antibodies to produce N332-GT2, a HIV envelope protein-like antigen. This antigen was able to mature germline murine antibodies into bnAbs and was also shown to select for rare bnAb precursors in naïve human B cells, enabling maturation of bnAb-like precursors into broadly neutralizing HIV antibodies.
- 5.Andreano E., Seubert A., Rappuoli R. Human monoclonal antibodies for discovery, therapy, and vaccine acceleration. Curr Opin Immunol. 2019;59:130–134. doi: 10.1016/j.coi.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Robinson W.H. Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery. Nat Rev Rheumatol. 2015;11:171–182. doi: 10.1038/nrrheum.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagerkvist A., Furebring C., Borrebaeck C. Single, antigen-specific B cells used to generate Fab fragments using CD40-mediated amplification or direct PCR cloning. Biotechniques. 1995;18:862–869. [PubMed] [Google Scholar]
- 8.Babcook J.S., Leslie K.B., Olsen O.A., Salmon R.A., Schrader J.W. A novel strategy for generating monoclonal antibodies from single, isolated lymphocytes producing antibodies of defined specificities. Proc Natl Acad Sci Unit States Am. 1996;93:7843–7848. doi: 10.1073/pnas.93.15.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meijer P.-J., Andersen P.S., Haahr Hansen M., Steinaa L., Jensen A., Lantto J., Oleksiewicz M.B., Tengbjerg K., Poulsen T.R., Coljee V.W. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J Mol Biol. 2006;358:764–772. doi: 10.1016/j.jmb.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 10.Scheid J.F., Mouquet H., Feldhahn N., Walker B.D., Pereyra F., Cutrell E., Seaman M.S., Mascola J.R., Wyatt R.T., Wardemann H. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kantor A.B., Merrill C.E., Mackenzie J.D., Herzenberg L.A., Hillson J.L. Development of the antibody repertoire as revealed by single-cell PCR of FACS-sorted B-cell subsets. Ann N Y Acad Sci. 1995;764:224–227. doi: 10.1111/j.1749-6632.1995.tb55831.x. [DOI] [PubMed] [Google Scholar]
- 12.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that bnds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 15.Nojima T., Haniuda K., Moutai T., Matsudaira M., Mizokawa S., Shiratori I., Azuma T., Kitamura D. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 16.Kuraoka M., Schmidt A.G., Nojima T., Feng F., Watanabe A., Kitamura D., Harrison S.C., Kepler T.B., Kelsoe G. Complex antigens drive permissive clonal selection in germinal centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 18.McCarthy K.R., Watanabe A., Kuraoka M., Do K.T., McGee C.E., Sempowski G.D., Kepler T.B., Schmidt A.G., Kelsoe G., Harrison S.C. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity. 2018;48:174–184. doi: 10.1016/j.immuni.2017.12.009. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., McCarthy K.R., Kuraoka M., Schmidt A.G., Adachi Y., Onodera T., Tonouchi K., Caradonna T.M., Bajic G., Song S. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell. 2019;177:1124–1135. doi: 10.1016/j.cell.2019.03.048. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]; The Kelsoe and Harrison groups profiled thousands of memory B cells from recently immunized donors. By expanding B cells in single-cell cultures and sequencing using single-well technologies, they profiled broadly reactive and protective antibodies which guided the elucidation of a highly conserved immunogenic epitope of Influenza hemagglutinin
- Winters A., McFadden K., Bergen J., Landas J., Berry K.A., Gonzalez A., Salimi-Moosavi H., Murawsky C.M., Tagari P., King C.T. Rapid single B cell antibody discovery using nanopens and structured light. mAbs. 2019;11:1025–1035. doi: 10.1080/19420862.2019.1624126. [DOI] [PMC free article] [PubMed] [Google Scholar]; Winters et al. utilized a novel assay platform controlled by nanopens to isolate antibody-secreting cells. Individual cells were screened for IgG secretion and secreted mAbs were profiled for antigen specificity using an in-channel fluorescent assay. IgG + antigen-specific B cells are then exported from the nanopens using OptoElectro Positioning for single-well BCR sequencing.
- 21.Slatko B.E., Gardner A.F., Ausubel F.M. Overview of next-generation sequencing technologies. Curr Prot Mol Biol. 2018;122 doi: 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.-B., Lönnerberg P., Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsköld D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picelli S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 25.Hagemann-Jensen M., Ziegenhain C., Chen P., Ramsköld D., Hendriks G.-J., Larsson A.J.M., Faridani O.R., Sandberg R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38:708–714. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- 26.Busse C.E., Czogiel I., Braun P., Arndt P.F., Wardemann H. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur J Immunol. 2014;44:597–603. doi: 10.1002/eji.201343917. [DOI] [PubMed] [Google Scholar]
- 27.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollmers C., Penland L., Kanbar J.N., Quake S.R. Novel exons and splice variants in the human antibody heavy chain identified by single cell and single molecule sequencing. PloS One. 2015;10 doi: 10.1371/journal.pone.0117050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titcombe P.J., Wigerblad G., Sippl N., Zhang N., Shmagel A.K., Sahlström P., Zhang Y., Barsness L.O., Ghodke-Puranik Y., Baharpoor A. Pathogenic citrulline-multispecific B cell receptor clades in rheumatoid arthritis. Arthr Rheumatol. 2018;70:1933–1945. doi: 10.1002/art.40590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gierahn T.M., Wadsworth M.H., Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Love J.C., Shalek A.K. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu A.A., Gierahn T.M., Monian B., Morgan D.M., Mehta N.K., Ruiter B., Shreffler W.G., Shalek A.K., Love J.C. TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures. Nat Immunol. 2019;20:1692–1699. doi: 10.1038/s41590-019-0544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeKosky B.J., Kojima T., Rodin A., Charab W., Ippolito G.C., Ellington A.D., Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L. High-throughput mapping of B cell receptor sequences to antigen specificity. Cell. 2019;179:1636–1646. doi: 10.1016/j.cell.2019.11.003. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Setliff et al. introduce LIBRA-seq, a high-throughput methodology which combines BCR sequencing data with antigen specificity. Using DNA barcoded antigens, cells are sorted for antigen binding and emulsified using droplet microfluidics. Single emulsions include bead-delivered oligos which, when reverse transcribed, add information from the target antigen to the BCR transcript, allowing for a sequencing readout that includes antigen binding information.
- Gérard A., Woolfe A., Mottet G., Reichen M., Castrillon C., Menrath V., Ellouze S., Poitou A., Doineau R., Briseno-Roa L. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0466-7. [DOI] [PubMed] [Google Scholar]; The Brenan, Bruhns, and Griffiths groups introduce CelliGO, a combined antibody profiling and sequencing platform. CelliGO profiles secreted antibody binding to either membrane-bound or secreted antigen, allowing functional profiling of antibody-secreting cells without surface antibodies. Using a microfluidic circuit to emulsify either cells, bioassay components, and secreted antigen, or cells, bioassay components, and reporter cells with membrane-bound antigen, single cells are encapsulated, profiled, and sequenced in single emulsions.
- 37.Eyer K., Doineau R.C.L., Castrillon C.E., Briseño-Roa L., Menrath V., Mottet G., England P., Godina A., Brient-Litzler E., Nizak C. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat Biotechnol. 2017;35:977–982. doi: 10.1038/nbt.3964. [DOI] [PubMed] [Google Scholar]
- 38.Ellefson J.W., Gollihar J., Shroff R., Shivram H., Iyer V.R., Ellington A.D. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science. 2016;352:1590–1593. doi: 10.1126/science.aaf5409. [DOI] [PubMed] [Google Scholar]
- Tanno H., McDaniel J.R., Stevens C.A., Voss W.N., Li J., Durrett R., Lee J., Gollihar J., Tanno Y., Delidakis G. A facile technology for the high-throughput sequencing of the paired VH:VL and TCRβ:TCRα repertoires. Sci Adv. 2020;6 doi: 10.1126/sciadv.aay9093. [DOI] [PMC free article] [PubMed] [Google Scholar]; Georgiou and colleagues utilize the cell-lysate resistant properties of RTX to enable OE RT-PCR of BCR transcript in emulsions containing debris from lysed cells and lysis buffer. This unique quality facilitates a simple pipeline where cells, RT-PCR reagents, and cell lysis buffer are combined and emulsified into single-cell emulsions which are used to generate physically linked VH:VL amplicons.
- 40.Devulapally P.R., Bürger J., Mielke T., Konthur Z., Lehrach H., Yaspo M.-L., Glökler J., Warnatz H.-J. Simple paired heavy- and light-chain antibody repertoire sequencing using endoplasmic reticulum microsomes. Genome Med. 2018;10 doi: 10.1186/s13073-018-0542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; The Warnatz lab uses naturally occurring rough endoplasmic reticulum microsomes, which contains antibody transcript, to physically encapsulate and isolate transcript from single cells. Cells are lysed in a lysis buffer which induces ER microsome formation, microsomes are purified through a series of centrifugation steps and are emulsified with overlap-extension primers and RT-PCR reagents to generate cognate-paired VH:VL amplicons.
- Singh M., Al-Eryani G., Carswell S., Ferguson J.M., Blackburn J., Barton K., Roden D., Luciani F., Giang Phan T., Junankar S. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun. 2019;10:3120. doi: 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Swarbrick, Smith, Goodnow and colleagues develop RAGE-seq, which uses long-read sequencing to elucidate BCR sequences while additionally profiling the cell transcriptome and multiplexing BCR sequence reads using short-read sequencing. Applied to study breast cancer, they utilize transcriptome profiles in classifying B cell subsets and BCR sequences to identify 3′ alternative splicing of antibody transcript which produces predominantly secreted IgA antibodies.
- Goldstein L.D., Chen Y.-J.J., Wu J., Chaudhuri S., Hsiao Y.-C., Schneider K., Hoi K.H., Lin Z., Guerrero S., Jaiswal B.S. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun Biol. 2019;2:1–10. doi: 10.1038/s42003-019-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Goldstein et al. developed a paired VH:VL sequencing pipeline utilizing 3′ shearing to profile over 250,000 B cells. The 3′ end of 5′ barcoded DNA from the 10x Chromium pipeline was sheared, allowing short-read NGS sequencing to sequence the entire physically linked VH:VL transcript after in silico reconstruction.
- 43.Gilchuk P., Bombardi R.G., Erasmus J.H., Tan Q., Nargi R., Soto C., Abbink P., Suscovich T.J., Durnell L.A., Khandhar A. Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nature Biomed Eng. 2020 doi: 10.1038/s41551-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedensohn S., Khan T.A., Reddy S.T. Advanced methodologies in high-throughput sequencing of immune repertoires. Trends Biotechnol. 2017;35:203–214. doi: 10.1016/j.tibtech.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Lareau C.A., Ma S., Duarte F.M., Buenrostro J.D. Inference and effects of barcode multiplets in droplet-based single-cell assays. Nat Commun. 2020;11:866. doi: 10.1038/s41467-020-14667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buenrostro and others demonstrate that the emulsion-based 10x Chromium pipeline may provide inaccurate single-cell sequencing as multiple barcodes were present in over 10% of obtained sequences. Among the barcode multiplets, 4% likely contain multiple beads and 5% may contain multiple oligonucleotide barcodes. They also develop a computational workflow to identify barcode multiplets in single-cell data to deconvolute potentially misidentified sequences.
- 47.Lavinder J.J., Wine Y., Giesecke C., Ippolito G.C., Horton A.P., Lungu O.I., Hoi K.H., DeKosky B.J., Murrin E.M., Wirth M.M. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. 2014;vol. 111:2259. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Paparoditis P., Horton A.P., Frühwirth A., McDaniel J.R., Jung J., Boutz D.R., Hussein D.A., Tanno Y., Pappas L. Persistent antibody clonotypes dominate the serum response to influenza over multiple years and repeated vaccinations. Cell Host Microbe. 2019;25:367–376. doi: 10.1016/j.chom.2019.01.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee et al. utilize a flow-focusing device for paired BCR sequencing of B cells from an influenza patient over the span of five years in addition to Ig-Seq, a proteomics analysis pipeline which quantitatively analyses Ig abundance in serum. Utilizing the BCR sequences as a reference for their Ig-seq proteomic database, they demonstrate persistence in certain antibody clonotypes that dominate over repeated vaccinations. The longitudinal quantitative profiling of antibodies using Ig-Seq was essential in demonstrating high-abundance persisting antibody clonotypes, characterizing and quantifying immunity from circulating, protective molecules.
- 49.Lindesmith L.C., McDaniel J.R., Changela A., Verardi R., Kerr S.A., Costantini V., Brewer-Jensen P.D., Mallory M.L., Voss W.N., Boutz D.R. Sera antibody repertoire analyses reveal mechanisms of broad and pandemic strain neutralizing responses after human norovirus vaccination. Immunity. 2019;50:1530–1541. doi: 10.1016/j.immuni.2019.05.007. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sajadi M.M., Dashti A., Rikhtegaran Tehrani Z., Tolbert W.D., Seaman M.S., Ouyang X., Gohain N., Pazgier M., Kim D., Cavet G. Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell. 2018;173:1783–1795. doi: 10.1016/j.cell.2018.03.061. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.B., Liu W., Vasil′ev Y.V., Bracken C.C., Malhan N., Guthals A., Beckman J.S., Voinov V.G. Direct determination of antibody chain pairing by top-down and middle-down mass spectrometry using electron capture dissociation and ultraviolet photodissociation. Anal Chem. 2020;92:766–773. doi: 10.1021/acs.analchem.9b03129. [DOI] [PMC free article] [PubMed] [Google Scholar]; Voinov's laboratory develops an electron capture dissociation and ultraviolet photodissociation method to cleave heavy and light chain-linking disulfide bonds. Using a set of both intact and cleaved mAbs, they demonstrate the ability to determine heavy and light chain pairing from mass spectrometry analysis.