Abstract
Type 1 diabetes (T1D) is a common autoimmune disease that is characterized by insufficient insulin production. The onset of T1D is the result of gene-environment interactions. Sociodemographic and behavioural factors may contribute to T1D, and the gut microbiota is proposed to be a driving factor of T1D. An integrated preventive strategy for T1D is not available at present. This case–control study attempted to estimate the exposure linked to T1D to identify significant risk factors for healthy children. Forty children with T1D and 56 healthy controls were included in this study. Anthropometric, socio-economic, nutritional, behavioural, and clinical data were collected. Faecal bacteria were investigated by molecular methods. The findings showed, in multivariable model, that the risk factors for T1D include higher Firmicutes levels (OR 7.30; IC 2.26–23.54) and higher carbohydrate intake (OR 1.03; IC 1.01–1.05), whereas having a greater amount of Bifidobacterium in the gut (OR 0.13; IC 0.05 – 0.34) was a protective factor for T1D. These findings may facilitate the development of preventive strategies for T1D, such as performing genetic screening, characterizing the gut microbiota, and managing nutritional and social factors.
Subject terms: Microbiology, Molecular biology, Biomarkers, Diseases, Risk factors
Introduction
Type 1 diabetes (T1D) is a multifactor disease caused by β-cell destruction (which is mostly immune-mediated) and absolute insulin deficiency. At present, the management of T1D has been improved, but the disease remains incurable. T1D onset is most common in childhood. T1D represents approximately 5–10% of all diabetes diagnoses1. Between 70 and 90% of T1D patients at diagnosis exhibit evidence of an immune-mediated process with β-cell autoantibodies. T1D onset is preceded by a preclinical period that lasts approximately 3 years, in which autoantibodies appear in the circulatory system2. Immune destruction of the β-cells can be detected by the evaluation of some haematic markers3. The disease has strong HLA associations, which explain nearly half of the genetic disease predisposition, while the remainder is due to other genetic polymorphisms3,4.
Analysis of genetic disease susceptibility suggests that there is a greater risk of T1D development when the father is affected by the disease than when the mother is affected5. On the other hand, there is evidence that a critical role is played by non-genetic factors, including both environmental and host-related factors, which are considered to play decisive roles in the disease process, leading to the manifestation of clinical T1D6.
The worldwide incidence of T1D in the age group of 0–15 years varies considerably by region (from 0.5 to 60 per 100,000 children), and the yearly increase ranges from 0.6% to 9.3%. In Europe, the percentage of cases in the age group of 0–15 years will rise by 70%7. In the Piedmont region, up to 2013, there were approximately 8,000 cases in this age group with an incidence of 27 new diagnoses per 100,0008. Migrant populations tend to show an incidence of diabetes similar to that of most host populations; therefore, a higher T1D incidence in migrant children was observed in Europe6,9,10. Such a pronounced increase in incidence cannot be attributable to genetic factors alone. Other major risk factors may include the environment, Western lifestyle and nutrition10. Other diseases with immune involvement, such as allergies, exhibit a similar trend, suggesting an inductor role for exogenous factors regarding the increased predisposition to autoimmunity11. Preventive measures to reduce the incidence of T1D have not been defined to date. Various factors seem to be involved in modulating the incidence of T1D, including birth delivery mode, feeding, birth weight, infections (especially viral), dietary behaviour, and pharmaceutical use (especially antibiotics). Such factors may contribute to T1D development during the early disease stage12; however, compared with genetic factors, environmental factors are less well characterized13. β- Cell vulnerability to stress factors has been discussed as the basis of the overload hypothesis14. Associations among the microbiome, metabolome, and T1D were shown, highlighting a host-microbiota role in the onset of the disease12,15. The origin of the disease process was suspected to be gut microbiota dysbiosis (imbalances in the composition and function of intestinal microbes) associated with altered gut permeability and a major vulnerability of the immune system6. Accordingly, evidence obtained from both animal models and human studies suggests that the gut microbiota and the immune system interact closely, emphasizing the role of the intestinal microbiota in the maturation and development of immune functions16. Recently, mycobiome-bacteriome interactions, as well as intestinal virome and islet autoimmunity, were hypothesized to be drivers of dysbiosis17. Several studies have specifically investigated microbiota composition in children with T1D18–20, but the results have not been consistent. Interestingly, most studies are in agreement regarding the reduced microbial diversity observed in subjects with T1D compared with controls; moreover, the microbiota structure in T1D subjects was found to be different from that of control subjects21,22. To date, a typical T1D-associated microbiota has not been identified23–26. The research also determined that T1D clinical management could be improved by in-depth analysis of the partial remission phase27; however, preventive measures are limited and generally focus only on genetic susceptibility28 and general population screening for islet autoimmunity29. The development of an integrated prediction strategy could be useful for increasing early diagnosis while avoiding onset complications by identifying children at risk of T1D to place under observation and, in the future, to treat with preventive methods10.
The aim of this study is to identify environmental, behavioural, and microbial risk factors of T1D onset to develop an integrated T1D preventive management strategy that is suitable for paediatricians in the Piedmont region.
Results
Subject description and origin factor analysis
To analyse the origin factor, the study population was subdivided by the children's origins (Italian and migrant, 69 and 27 children, respectively). An analysis of the socio-demographic and behavioural factors examined in the study showed many differences between Italian and migrant children, while other variables appear to be quite homogeneous (Table 1). In the studied cohort, migrant status did not produce a significant increase in T1D onset.
Table 1.
Summary of the population anthropometric characteristics, comparing cases and controls: number of children involved, sex, age and anthropometrics as the mean and standard deviation.
| Type 1 diabetes patients | Healthy controls | ||
|---|---|---|---|
| Subjects (number) | 40 | 56 | |
| Gender | Male (%) | 28 (70.0%) | 40 (71.4%) |
| Female (%) | 12 (30.0%) | 16 (28.6%) | |
| Age (years) | 8.23 ± 1.42 | 7.87 ± 1.72 | |
| Height (m) | 1.33 ± .11 | 1.30 ± .12 | |
| Weight (kg) | 29.73 ± 8.06 | 29.25 ± 9.83 | |
| BMI (kg/m2) | 16.51 ± 2.77 | 17.01 ± 2.79 | |
Approximately 79% of the children in the cohort had siblings; approximately 40% of the included children lived with a pet in the house, and more than 65% of the children took antibiotics during the first two years of life. The residency zone was notably different between Italians and migrants: the percentage of migrant children living in urban sites was higher but not significant following the adjusted model. Regular sports activities seem to be practised more by Italian children than by migrant children (73.5% vs 51.8%, p = 0.054). A total of 77.9% of Italian children and 55.6% of migrant children were subjected to regular health check-ups (p = 0.017). A significant difference was confirmed for the ages of the migrant mother and father (Table 1), meanly 6 years and 4 years younger respectively at recruitment, respect the Italians (p = 0.017 and p = 0.0425). The analysis of eating habits and nutritional intake revealed that the majority of the children were breastfed. Moreover, the weaning age was 6 months, as recommended. Migrant children showed higher total carbohydrate intake (+ 12%, p = 0.044) and simple carbohydrate intake (+ 24%, p = 0.0045). Moreover, among migrants, the children tended to access food by themselves and to consume meals alone. The percentage of migrant children who ate meals while watching TV was higher but not significant. Finally, the one-course meal was more frequent in migrant families (ratio 1:3, p = 0.006).
The analysis of microbiota and bioindicator species displayed no significant differences between Italian and migrant children: the qRT-PCR measurements showed a trend of greater value for the total bacteria (both for the experimental design with and without probe), Bacteroides and M. smithii (both using 16S rDNA and nifH) in migrant children. The DGGE profile and dendrogram analysis did not show a different clustering pattern based on the origin, and the migrant group showed a trend towards greater α-diversity of the faecal microbiota profiles (Shannon index + 5%). Additionally, the α-diversity analyses in next generation sequencing (NGS) showed a difference in taxonomic units (OTUs), i.e., there were more OTUs in migrants than in Italians, but the difference was not significant, though it was close to the limit of significance (p = 0.057). Furthermore, the phylogenetic diversity index (Faith PD) suggested that the origin of the subjects could influence the structure of the microbial community. Although the overall number of OTUs did not change significantly, the phylogenetic distance of the individual OTUs was greater in the migrant group than in the Italian group, as the OTUs occupied a broader ecological niche in the migrant group.
T1D risk factors
Previous results indicated that being a migrant child in the Piedmont region is not a significant risk factor for T1D onset30. Table 2 shows single logistic regressions performed to estimate the impact of the different variables on the outcome. Notably, the analysis of socio-demographic, behavioural, and nutritional determinants revealed that having parents with at least a high school certificate seems to be a protective factor for T1D onset, even if not significant after adjusted comparisons.
Table 2.
Oligonucleotide primers, probes and genomic standards used in biomolecular analyses.
| Microbial Target | Sequences | Standard Genomic DNA | Ref | |
|---|---|---|---|---|
|
Total Bacteria 16 s rDNA |
F R |
5′ACTCCTACGGGAGGCAGCAG3' 5′ATTACCGCGGCTGCTGG3' |
Desulfovibrio vulgaris ATCC 29579D-5 |
36 |
|
Total Bacteria 16 s rDNA |
F R Probe |
5′AGAGTTTGATCMTGGCTCAG3’ 5′TTACCGCGGCKGCTGGCAC3’ 5′CCAKACTCCTACGGGAGGCAGCAG3’ |
Desulfovibrio vulgaris ATCC 29579D-5 |
36 |
|
Bacteroidetes 16 s rDNA |
F R |
5′CATGTGGTTTAATTCGATGAT3' 5′AGCTGACGACAACCATGCAG3' |
Bacteroides fragilis ATCC 25285D-5 |
19,38 |
|
Bacteroides 16 s rDNA |
F R |
5′GAGAGGAAGGTCCCCCAC3' 5′CGCTACTTGGCTGGTTCAG3' |
Bacteroides fragilis ATCC 25285D-5 |
19,38 |
|
Firmicutes 16 s rDNA |
F R |
5′ATGTGGTTTAATTCGAAGCA3' 5′AGCTGACGACAACCATGCAC3' |
Clostridium acetobutylicum ATCC 824D-5 |
40 |
|
Bifidobacteria 16 s rDNA |
F R |
5′CTCCTGGAAACGGGTGG3' 5′GGTGTTCTTCCCGATATCTACA3' |
Bifidobacterium longum infantis ATCC 15697D-5 |
39 |
|
Akkermansia muciniphila 16 s rDNA |
F R |
5′CAGCACGTGAAGGTGGGGAC3' 5′CCTTGCGGTTGGCTTCAGAT3' |
Akkermansia municiphila ATCC-BAA835D-5 |
37 |
|
M. smithii 16 s rDNA |
Smit.16S-740 F Smit.16S-862 R Smit.16S FAM |
5′CCGGGTATCTAATCCGGTTC-3’ 5′CTCCCAGGGTAGAGGTGAAA3’ 5′CCGTCAGAATCGTTCCAGTCAG3’ |
M. smithii DSM 861 |
36 |
|
M. smithii nifH |
Mnif 202 F Mnif 353 R Mnif Probe |
5′GAAAGCGGAGGTCCTGAA3' 5′ACTGAAAAACCTCCGCAAAC3' 5′CCGGACGTGGTGTAACAGTAGCTA3' |
M. smithii DSM 861 |
21 |
|
Bacterial 16 s rRNA |
357 F-GC 518 R |
5′GCclampCTCCTACGGGAGGCAGCAG3' 5′GTATTACCGCGGCTGCTGG3' |
34 | |
| 16 s rDNA V3-V4 |
Pro 341 F Pro 805 R |
5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNBGCASCAG3' 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGCAGGACTACNVGGGTATCTAATCC3' |
41 | |
High total caloric intake, as well as high protein intake and consumption of total carbohydrates, are associated with only a slightly increased risk of T1D onset.
The DGGE gel and the results of the cluster analysis are shown in Fig. 1. The Pearson similarity clustering showed macro beta-diversity differences between the T1D patients and healthy children, with the main division being in two different clusters.
Figure 1.
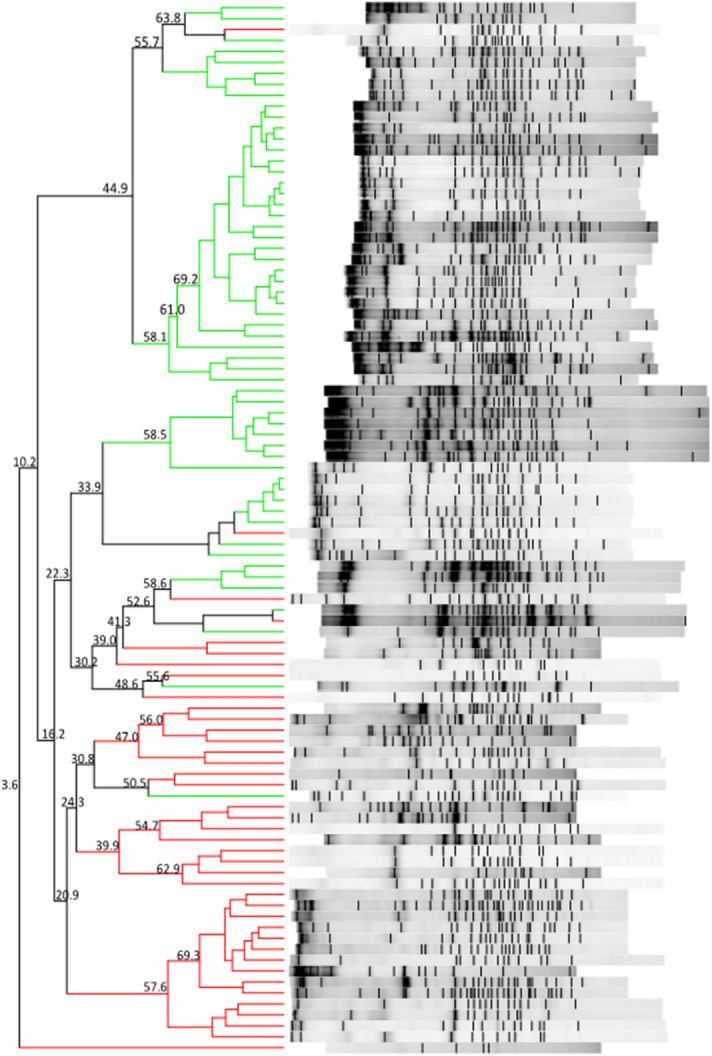
DGGE banding patterns and the results of the analysis in which the Pearson coefficient (numbers reported near the nodes) was used for measuring similarity in banding patterns. The cluster identifies T1D patients (red lines) and healthy children (green lines).
Firmicutes and Bacteroidetes followed by Proteobacteria and Actinobacteria (Table 3) predominantly composed the gut microbiota of all children. In the children with diabetes, an increase in the levels of three members of Bacteroidetes (Alistipes senegalensis, Bacteroides timonensis, and Barnesiella intestinihominis) and three members of Firmicutes (Christensenella timonensis, Ruminococcus bromii, and Urmitella timonensis) was observed by sequencing.
Table 3.
Main characteristics of type 1 diabetes patients and healthy children enrolled by subject origin.
| Italians (n. 69) | Migrants (n. 27) | p-value | Adj p-value | ||
|---|---|---|---|---|---|
| Socio-demographic factors | |||||
| Age at onset | 7.85 (± 1.75) | 8.46 (± 1.04) | 0.093 | 0.1581 | |
| Gender | Female | 24 (34.8%) | 4 (14.8%) | 0.079 | 0.149 |
| Male | 45 (65.2%) | 23 (85.2%) | |||
| Percentile BMI | 51.38 (± 32.35) | 66.46 (± 36.27) | 0.053 | 0.121 | |
| BMI categories | Underweight | 1 (1.5%) | 2 (7.7%) | 0.016 | 0.091 |
| Normal weight | 50 (73.5%) | 11 (42.3%) | |||
| Overweight | 11 (16.2%) | 7 (26.9%) | |||
| Obese | 6 (8.8%) | 6 (23.1%) | |||
| Residency (urban) | 43 (63.2%) | 23 (85.2%) | 0.048 | 0.121 | |
| Sport activity | 50 (73.5%) | 14 (51.8%) | 0.054 | 0.121 | |
| The child has siblings | 52 (76.5%) | 22 (81.5%) | 0.785 | 0.861 | |
| The child does regular health check-up | 53 (77.9%) | 15 (55.6%) | 0.043 | 0.121 | |
| The child used antibiotics in the first two years of life | 46 (67.6%) | 17 (63.0%) | 0.810 | 0.861 | |
| Breast feeding | 57 (83.8%) | 24 (88.9%) | 0.750 | 0.861 | |
| Weaning age (months) | 5.91 (± 2.47) | 5.76 (± 1.62) | 0.760 | 0.861 | |
| Presence of pets in the house | No pets | 14 (24.6%) | 7 (28.0%) | 0.057 | 0.121 |
| Dogs or cats | 24 (42.1%) | 4 (16.0%) | |||
| Other pets | 19 (33.3%) | 14 (56.0%) | |||
| Mother age at recruitment | 40.61 (± 4.77) | 34.18 (± 5.41) | < 0.001 | 0.017 | |
| Mother education (at least high school) | 50(73.5%) | 15 (55.6%) | 0.141 | 0.218 | |
| Father age at recruitment | 44.19 (± 5.99) | 40.29 (± 5.93) | 0.005 | 0.0425 | |
| Father education (at least high school) | 38 (55.9%) | 17 (63.0%) | 0.646 | 0.861 | |
| Important changes in the family contest in the last year | 7 (10.3%) | 3 (11.1%) | 1.000 | 1.000 | |
| Nutritional anamnesis | |||||
| Total caloric intake (Kcal/die) | 1760.23 (± 349.43) | 1891.48 (± 372.29) | 0.108 | 0.262 | |
| Delta Kcal | -116.15 (± 346.65) | -11.11 (± 366.43) | 0.192 | 0.408 | |
| Delta Kcal % | -5.20 (± 17.54) | -0.33 (± 19.06) | 0.236 | 0.446 | |
| Total supply of proteins (g) | 60.53 (± 13.61) | 60.56 (± 13.91) | 0.991 | 1.000 | |
| Total supply of lipids (g) | 65.40 (± 12.92) | 66.52 (± 16.08) | 0.724 | 0.879 | |
| Total supply of carbohydrates (g) | 232.51 (± 58.51) | 259.70 (± 59.09) | 0.044 | 0.1496 | |
| Total supply of CHO RA (g) | 71.02 (± 26.39) | 88.85 (± 28.45) | 0.0045 | 0.0255 | |
| The child has access to food by himself when he/she is at home | 39 (57.4%) | 23 (85.2%) | 0.016 | 0.0544 | |
| The child consumes meals alone | Always alone | 3 (4.4%) | 12 (44.4%) | < 0.001 | 0.006 |
| Always with an adult | 61 (89.7%) | 16 (37.1%) | |||
| Both | 4 (5.9%) | 5 (18.5%) | |||
| Number of extra meals a day | 0 | 1 (1.5%) | 0 (0%) | 0.330 | 0.4675 |
| 1 | 2 (2.9%) | 0 (0%) | |||
| 2 | 36 (52.9%) | 10 (37.1%) | |||
| 3 | 21 (30.9%) | 10 (37.1%) | |||
| 4 | 8 (11.8%) | 7 (25.8%) | |||
| The child consumes meals while watching TV | 43 (63.2%) | 22 (81.5%) | 0.094 | 0.262 | |
| The child consumes sweets more than three times a week | 39 (57.3%) | 16 (59.3%) | 1.000 | 1.000 | |
| Child family consumes meals all together | 62 (91.2%) | 22 (81.5%) | 0.284 | 0.4675 | |
| Family talks during the meal | 61 (89.7%) | 22 (81.5%) | 0.312 | 0.4675 | |
| The child often asks for supplementary portions of food | 35 (51.5%) | 16 (59.2%) | 0.649 | 0.849 | |
| The main meal of child | Lunch | 14 (20.9%) | 7 (25.9%) | 0.892 | 1.000 |
| Dinner | 51 (76.1%) | 20 (74.1%) | |||
| Both | 2 (2.99%) | 0 (0%) | |||
| Meals | One course meals | 11 (16.2%) | 14 (51.9%) | 0.001 | 0.006 |
| Not one course meals | 54 (79.4%) | 12 (44.4%) | |||
| Both | 3(4.4%) | 1 (3.7%) | |||
| Microbiota | |||||
|
Akkermansia muciniphila (Log gene copies/g stool) |
6.21 (± 1.29) | 6.66 (± 1.44) | 0.1475 | 0.228 | |
|
Bacteroides spp. (Log gene copies/g stool) |
8.56 (± 0.91) | 9.08 (± 0.74) | 0.0092 | 0.060 | |
|
Bacteroidetes (Log gene copies/g stool) |
8.38 (± 1.31) | 8.81 (± 0.83) | 0.124 | 0.228 | |
|
Total bacteria Probe (Log gene copies/g stool) |
9.48 (± 0.96) | 9.96 (± 0.74) | 0.019 | 0.062 | |
|
Total bacteria SYBR (Log gene copies/g stool) |
9.95 (± 0.63) | 10.27 (± 0.63) | 0.025 | 0.065 | |
|
Firmicutes (Log gene copies/g stool) |
10.38 (± 0.77) | 10.59 (± 0.87) | 0.259 | 0.306 | |
|
Bifidobacterium spp. (Log gene copies/g stool) |
6.89 (± 1.16) | 7.02 (± 0.97) | 0.597 | 0.597 | |
|
Methanobrevibacter smithii 16S (Log gene copies/g stool) |
5.24 (± 1.22) | 6.15 (± 1.74) | 0.004 | 0.052 | |
|
Methanobrevibacter smithii Nihf (Log gene copies/g stool) |
5.16 (± 1.05) | 5.83 (± 1.53) | 0.0151 | 0.062 | |
| Simpson index | 0.11 (± 0.04) | 0.10 (± 0.05) | 0.1580 | 0.228 | |
| Shannon index | 2.45 (± 0.29) | 2.58 (± 0.31) | 0.0402 | 0.087 | |
| Firmicutes/Bacteroidetes ratio | 1.29 (± 0.37) | 1.22 (± 0.17) | 0.3224 | 0.349 | |
| Margalef index | 2.53 (± 0.68) | 2.72 (± 0.65) | 0.2145 | 0.279 | |
The continuous variables are expressed as means and standard deviations; the categorical variables are expressed as absolute numbers and percentages. Adj p-value: adjusted for multiple comparisons.
Furthermore, other notable results were obtained by NGS analyses. The taxonomic analysis revealed that the gut microbiota of the study participants was composed of nine relevant phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, Euryarchaeota, Tenericutes, Cyanobacteria, and an unclassified phylum.
Moreover, beta-diversity analyses were carried out to highlight the differences among the samples based on the structures of their microbial communities. The weighted UniFrac metric showed that the samples were not subdivided into clusters. The intragroup and intergroup distances were comparable, and there was no separation between the clusters. These findings were confirmed by the Permanova test. Finally, analyses of the differential abundance were performed to compare the increase or decrease in the abundance of one or more bacteria in the case and control groups. DeSeq2 showed 48 significantly abundant OTUs (p < 0.001). The most abundant OTU was Rikenellaceae followed by Prevotellaceae (Prevotella copri), Barnesiellaceae, Lachnospiraceae, and Ruminococcaceae (Ruminococcus bromii), which were significantly more abundant in children with diabetes.
The difference in the results observed between methods is an interesting discussion point. The methods are characterized by different sensitivities; they represent different molecular perspectives regarding the faecal microbiota. When a method with a higher sensibility is used (NGS), a flattening effect is possible. On the other hand, the major abundance of such genera as Ruminococcus was confirmed by different microbiota study methods, which is in keeping with the qRT-PCR results. A group of 23 samples showed different clusterization compared to the others (Fig. 2, left). This small group was not different from the main group regarding any characteristics. The only significant difference was observed for the M. smithii presence and the A. muciniphila levels, both of which were higher in the separated group (Fig. 2, right). A. muciniphila was proposed as a probiotic31, while M. smithii has been characterized as the most abundant methanogen in the gut32.
Figure 2.
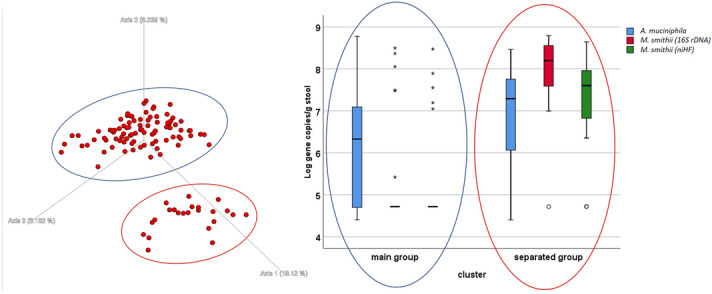
Left-Unweighted UniFrac graph of the NGS results. There are two identifiable groups: the blue circle (main group) and the red circle (separated group). No experimental hypothesis was confirmed for the cluster definition. On the Right: box plot of the qRT-PCR results for some microbiological targets (Akkermansia muciniphila and Methanobrevibacter smithii), the difference between the groups is significant (t-test p < 0.05).
The qRT-PCR gut microbiota analysis indicated significant differences among T1D patients and healthy children (Table 2). The logistic regression analysis showed that the increase in the Margalef index was associated with a decrease in the likelihood of disease onset (OR 0.20; 95% CI 0.09–0.46, p = 0.000). Increased Firmicutes levels and decreased Bacteroidetes levels were significant risk factors for T1D (OR 7.49; 95% CI 3.25–17.28, p = 0.0001; OR 0.28; 95% CI 0.15–0.51 p = 0.0001, respectively). Moreover, Bifidobacterium spp. was a protective factor for T1D onset (OR 0.20; 95% CI 0.10–0.38, p = 0.0001).
The multivariable analysis produced a R2 = 0.6259 (p < 0.001). After adjusting for confounding factors, the likelihood of having diabetes is significantly higher in those with higher amount of Firmicutes, lower amount of Bifidobacterium spp and a higher amount of total carbohydrate intake (Table 4).
Table 4.
Logistic regressions: likelihood of having diabetes.
| Likelihood of having diabetes | |||||
|---|---|---|---|---|---|
| OR | 95% IC | p-value | Adj p-value | ||
| Socio-demographic factors | |||||
| Age at recruitment | 1.15 | 0.89—1.49 | 0.290 | 0.6195 | |
| Gender (female) | 1.07 | 0.43—2.61 | 0.879 | 0.925 | |
| Percentile BMI | 0.99 | 0.98—1.003 | 0.179 | 0.507 | |
| BMI categories | Underweight | 0.39 | 0.034—4.61 | 0.461 | 0.7785 |
| Overweight | 0.25 | 0.02—3.34 | 0.295 | 0.6195 | |
| Obese | 0.36 | 0.02—5.11 | 0.448 | 0.779 | |
| Residency (rural) | 0.96 | 0.39—2.32 | 0.924 | 0.925 | |
| The child consumes meals at school | 0.51 | 0.19—1.40 | 0.193 | 0.507 | |
| The child consumes meals at home more than two times a week | 0.42 | 0.17—1.07 | 0.070 | 0.2555 | |
| Sport activity | 1.23 | 0.51—2.95 | 0.641 | 0.785 | |
| Having siblings | 0.75 | 0.28—1.99 | 0.563 | 0.785 | |
| Having done regular health check-up | 0.37 | 0.15—0.93 | 0.036 | 0.252 | |
| Use of antibiotics in the first two years of life | 0.74 | 0.31—1.75 | 0.503 | 0.7785 | |
| Breastfeeding | 0.68 | 0.22—2.14 | 0.519 | 0.7785 | |
| Weaning age (months) | 0.68 | 0.46—1.03 | 0.068 | 0.2555 | |
| Presence of dogs and/or cats in the house | 0.95 | 0.37—2.44 | 0.925 | 0.925 | |
| Mother age at recruitment | 0.98 | 0.91—1.06 | 0.673 | 0.785 | |
| Mother education (at least high school) | 0.34 | 0.14—0.83 | 0.018 | 0.189 | |
| Father age at recruitment | 1.01 | 0.95—1.08 | 0.671 | 0.785 | |
| Father education (at least high school) | 0.33 | 0.14—0.77 | 0.011 | 0.189 | |
| Important changes in the family contest in the last year | 3.68 | 0.88—15.22 | 0.073 | 0.2555 | |
| Nutritional anamnesis | |||||
| Total caloric intake (Kcal/die) | 1.0023 | 1.0009—1.0036 | 0.001 | 0.005 | |
| Total supply of proteins (g) | 1.06 | 1.02—1.10 | 0.002 | 0.007 | |
| Total supply of lipids (g) | 1.03 | 1.002—1.069 | 0.036 | 0.072 | |
| Total supply of carbohydrates (g) | 1.01 | 1.005—1.022 | 0.001 | 0.005 | |
| Total supply of CHO RA (g) | 1.03 | 1.007—1.045 | 0.006 | 0.015 | |
| The child consumes more than two extra meals | 1.67 | 0.73—3.82 | 0.224 | 0.329 | |
| The child consumes meals while watching TV | 0.58 | 0.24—1.40 | 0.230 | 0.329 | |
| The child consumes sweets more than three times a week | 1.08 | 0.47—2.47 | 0.859 | 0.859 | |
| Child family consumes meals all together | 0.54 | 0.15—1.91 | 0.339 | 0.4238 | |
| The child often asks for supplementary portions of food | 1.44 | 0.62—3.28 | 0.389 | 0.432 | |
| Microbiota | |||||
| Akkermansia muciniphila (Log gene copies/g stool) | 0.84 | 0.62—1.14 | 0.260 | 0.423 | |
| Bacteroides spp. (Log gene copies/g stool) | 0.79 | 0.50—1.25 | 0.317 | 0.443 | |
| Bacteroidetes (Log gene copies/g stool) | 0.28 | 0.15—0.51 | 0.000 | 0.000 | |
| Total bacteria Probe (Log gene copies/g stool) | 0.72 | 0.46—1.12 | 0.147 | 0.318 | |
| Total bacteria SYBR (Log gene copies/g stool) | 0.69 | 0.37—1.31 | 0.259 | 0.423 | |
| Firmicutes (Log gene copies/g stool) | 7.49 | 3.25—17.28 | 0.000 | 0.000 | |
| Bifidobacterium spp. (Log gene copies/g stool) | 0.20 | 0.10—0.38 | 0.000 | 0.000 | |
| Methanobrevibacter smithii 16S (Log gene copies/g stool) | 0.97 | 0.73—1.30 | 0.858 | 0.930 | |
| Methanobrevibacter smithii Nihf (Log gene copies/g stool) | 1.04 | 0.75—1.44 | 0.824 | 0.930 | |
| Simpson index | 1.14 | 0.00005—23,610.2 | 0.980 | 0.980 | |
| Shannon index | 0.51 | 0.13—2.03 | 0.341 | 0.443 | |
| Firmicutes/Bacteroidetes ratio | 34,288.2 | 637.20—1,845,077 | 0.000 | 0.000 | |
| Margalef index | 0.20 | 0.09—0.46 | 0.000 | 0.000 | |
The continuous variables are shown on a light grey background; the categorical variables are shown on a white background. Adj p-value: adjusted for multiple comparisons. Significant p-values are bolded.
Discussion
T1D is an important disease that affects health with onset primarily occurring in childhood. At present, there is no cure for this disease, and only disease management is possible. The disease burden of T1D is immense, especially considering the number of years of life lost due to disability but also the years of life lost due to premature death. The life expectancy for T1D patients is approximately 16 years shorter than that of the comparable healthy population33. Even if relevant risk factors are known, to date, such scientific determinants do not include a screening programme for preventive purposes. Of course, preventive action must be considered as a systematic process that focuses on the main risk factors to identify children at higher risk of T1D and to suggest efficacious preventive treatments. In the study, the main T1D onset risk factors seem to be identifiable in the composition of the microbiota and, in particular, the microbiota α-diversity, Firmicutes and Bacteroidetes levels and their ratio, as well as the Bifidobacterium level. Similar evidence was obtained by other studies, which observed both higher Bacteroidetes in T1D patients34,35 and less abundant anti-inflammatory genera in children with multiple islet autoantibodies36. Reduced microbial diversity appears to become significant between seroconversion and overt T1D15. A significant difference in the Bifidobacterium level was observed in different studies, including both a small cohort of autoimmune children37,38 and a larger population associated with such protective factors as breastfeeding21. At the genus level, a significant difference in, for example, Blautia (increased in patients), was observed39; however, in other studies, different single species (Bacteroides ovatus) seem to be more abundant in patients than in the controls18. However, prior studies suggest the presence of duodenal mucosa abnormalities in the inflammatory profile for T1D patients22,40 and on the T1D-related changes in the gut microbiota, even if proving the causality of these factors has remained challenging21.
The characterization of the microbiota is rapidly evolving. Traditional methods that are not as sensitive as PCR-DGGE are still suitable, while NGS methods are expanding. Sophisticated whole-genome sequencing methods integrated with metabolomics and proteomics have been proposed. However, the large amount of data, being affected by multiple confounding factors, has not had a clear impact on T1D prevention strategies. The development of a simple method to describe microbiota modulation using validated biomarkers, which could serve as a rapid screening test, may be warranted.
Another risk factor is the occurrence of stress due to a traumatic or emotional experience. This stress seems to be able to affect the autoimmunity process. Therefore, particular attention could be paid to such risk factors for T1D risk in children.
A high education level of one or both parents could be also protective, suggesting that socioeconomic factors affect the T1D risk. Other factors, identified as significant risk modulators among behavioural and nutritional factors, had minor effects.
The study has some potential limitations, including susceptibility to bias in recollection about exposure and reverse causality. The exposure recollection could be biased, but this issue can be less influential at the onset, as in this study. Moreover, recruitment at the onset guarantees a temporal coherence of the exposure with respect to the disease onset.
T1D is one of the most frequently diagnosed diseases in children; however, it is not a high-incidence disease. The prospective inclusion of a large number of healthy children, which is needed for the observation of enough cases, requires a very long time of observation. Moreover, a restricted age range was necessary in children for the rapid changes in behaviour and microbiota. This requirement resulted in an additional included subject restriction. On the other hand, the study of multifactorial diseases with poorly understood pathogenic pathways is imperative, even if it is at risk for obtaining less conclusive evidence. Of course, such a study alone could not elucidate the causation process, but the evidence obtained could be important for the selection of higher-risk subpopulations, planning of future research, and improving prevention.
Identification of a higher-risk subpopulation is strictly relevant for the subsequent validation of an efficient preventive screening to be produced with a prospective method. Of course, the pathogenesis of type 1 diabetes has not been fully elucidated to date; however, in this study, various factors (associated with both the disease and the microbiota composition) were included, such as the origin of the children, the age of the mother, the age of breastfeeding and the age of weaning. Other possible confounding factors not included in our analysis are viral infections, particularly enteroviruses, and preterm birth; however, there was no clear consensus regarding these novel factors at the beginning of the study.
Concerning the microbiota, the knowledge is still incomplete, and various factors can interact to produce a T1D risk modulation that is not explainable at present. Moreover, the results obtained using different techniques were also dissimilar (for example, clusterization due to β-diversity analysis). This finding is likely due to the different sensitivities of the applied methods41. Furthermore, even if the time between the symptom comparison and the diagnosis is very short, there is a danger of biased estimates due to reverse causality.
In conclusion, this study confirmed that T1D onset risk is modulated by compositional changes in the gut microbiota and that such evidence must be employed to devise preventive measure. The results showed that the gut microbial indicators found in children with T1D differ from those found in healthy children. These findings also pave the way for new research attempting to develop strategies to control T1D development by modifying the gut microbiota. However, a better knowledge of gut microbial composition associated with the development of T1D must be obtained to choose the best treatment10,42–45.
In brief, direct or indirect manipulations of the intestinal microbiome may provide effective measures for preventing or delaying the disease process leading to the manifestation of clinical T1D. At present, a preventive strategy could be developed that includes the main genetic and microbiome risk factors. Then, this strategy could be applied to healthy children to reduce the burden of T1D.
Methods
Study design and participants
The case–control study began in January 201646 and ended in September 2018 (case–control phase of clinicaltrial.gov Protocol ID: G12114000080001). The work was conducted following the STROBE Statement for a case–control study. The activity is bicentric and includes the two main paediatric hospitals in the Piedmont region (located in Torino and Novara), which cover the clinical management for cases of T1D in the region. The ethics committees of the two hospitals approved the research activities during 2015 (“Comitato etico interaziendale A.O.U. Ordine Mauriziano di Torino ASLTO1” with record number 0117120 and “Comitato etico Interaziendale A.O.U. “Maggiore della Carità” ASL BI, NO, VCO” record number 631/CE).
The recruitment included 40 paediatric patients with T1D (cases) and 56 healthy children (controls), who were comparable in terms of age, gender, and ethnicity to avoid bias. The included subjects represent the most convenient sample possible. The inclusion criteria were age (5–10 years), normal weight, and residence in Piedmont. Exclusion criteria were celiac disease, chronic disease diagnosis, eating disorders, active infections, use of antibiotics and/or probiotics and/or any other medical treatment that influences intestinal microbiota during the 3 months before recruitment and children with parents of mixed origins (Italian and migrant) for the exclusion of important confounding factors due to genetic and cultural mixed backgrounds19.
The T1D children were integrated into the study at disease onset, with hyperglycaemia, with or without ketoacidosis, polyuria symptoms, a high value of glycated haemoglobin (HbA1c > 42 mmol/mol) and T1D-specific autoantibody positivity. Healthy children were contacted by paediatricians in the territory of the acute care system. The guardians of the enlisting children read, understood, and then signed informed consent forms following the declaration of Helsinki. A module is prepared for parents, children, and mature children47. All the following methods were carried out following relevant guidelines and regulations when available. A questionnaire was given to the parents containing items and questions to retrieve data on the family contest with particular regards to emotive stressors, such as mourning or separation, anthropometrics, and socio-demographic, nutritional, and behavioural information.
Anthropometric and nutritional data included weight, height, body mass index (BMI), food frequency based on 24-h recall and a food frequency questionnaire (FFQ), neonatal feeding, and age of weaning. The anthropometric parameters (weight and height) were measured according to standard recommendations. The BMI values were interpreted according to the WHO criterion. The 24-h recall technique reconstructed the meals and food intake on a recent "typical" day, estimating the bromatological inputs according to a food composition database for epidemiological studies in Italy (BDA). The FFQ, developed for the study, focused on the consumption of certain food categories (those containing sugars, fibre, omega-3, calcium, vitamin D, condiments, and cereals) and eating habits (e.g., alone or with adults, in front of the TV).
Twenty-eight percent of the involved population is migrants (both parents not Italian). Such data are consistent with the percentage of newborns from non-Italian mothers, which is approximately 30% in northern Italy48. The migrant group included children coming mainly from northern Africa and Eastern Europe. The migration involved the parents and sometimes the children; on average, the included children as migrants were residents in Italy for less than 5 years. At the end of recruitment, no significant differences were observed between the case and control groups for age, sex composition, and origins (criteria for pairing) or for height, weight, and BMI (T-test, p > 0.05) (Table 5).
Table 5.
Bacterial species identified by sequencing of the most representative DGGE bands amplified from fecal DNA of type 1 diabetes (cases) and healthy children (controls).
| Closet Relative | Identity | Phylum | Class | Order | Family | Genus | Cases | Controls |
|---|---|---|---|---|---|---|---|---|
| Alistipes putredinis | 100% | Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | 10% (4) | 35.7% (20) |
| Alistipes senegalensis | 96% | Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | 75% (30) | 50% (28) |
| Bacteroides coprocola | 98% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 5% (2) | 0% (0) |
| Bacteroides dorei | 100% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 67.5%(27) | 80.3% (45) |
| Bacteroides faecis | 99% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 15% (6) | 16.1% (9) |
| Bacteroides finegoldii | 94% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 17.5% (7) | 46.4% (26) |
| Bacteroides intestinalis | 97% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 90% (36) | 92.8% (52) |
| Bacteroides timonensis | 100% | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 20% (8) | 10.7% (6) |
| Barnesiella intestinihominis | 99% | Bacteroidetes | Bacteroidia | Bacteroidales | Barnesiellaceae | Barnesiella | 72.5% (29) | 66.1% (37) |
| Bifidobacterium faecale | 100% | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | 0% (0) | 28.6% (16) | |
| Christensenella timonensis | 93% | Firmicutes | Clostridia | Clostridiales | Christensenellaceae | Christensenella | 40% (16) | 16.1% (9) |
| Clostridium dakarense | 97% | Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | Romboutsia | 5% (2) | 5.3% (3) |
| Colidextribacter massiliensis | 93% | Firmicutes | Clostridia | Clostridiales | Colidextribacter | 42.5% (17) | 39.3% (22) | |
| Coprobacter fastidiosus | 82% | Bacteroidetes | Bacteroidia | Bacteroidales | Barnesiellaceae | Coprobacter | 0% (0) | 5.3% (3) |
| Dialister propionicifaciens | 89% | Firmicutes | Negativicutes | Veillonellales | Veillonellaceae | Dialister | 10% (4) | 25% (14) |
| Dialister succinatiphilus | 100% | Firmicutes | Negativicutes | Veillonellales | Veillonellaceae | Dialister | 5% (2) | 30.3% (17) |
| Escherichia coli | 99% | Proteobacteria | Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | Escherichia | 65% (26) | 76.8% (43) |
| Eubacterium rectale | 100% | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 3 5% (14) | 33.9% (19) | |
| Fusicatenibacter saccharivorans | 100% | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Fusicatenibacter | 87.5% (35) | 100% (56) |
| Megasphaera massiliensis | 99% | Firmicutes | Negativicutes | Veillonellales | Veillonellaceae | Megasphaera | 37.5% (15) | 41.1% (23) |
| Negativibacillus massiliensis | 91% | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Negativibacillus | 20% (8) | 26.8% (15) |
| Parabacteroides johnsonii | 95% | Bacteroidetes | Bacteroidia | Bacteroidales | Tannerellaceae | Parabacteroides | 10% (4) | 23.2% (13) |
| Prevotella copri | 99% | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 57.5% (23) | 76.8% (43) |
| Pseudoflavonifractor phocaeensis | 94% | Firmicutes | Clostridia | Clostridiales | Pseudoflavonifractor | 20% (8) | 51.8% (29) | |
| Romboutsia timonensis | 100% | Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | Romboutsia | 82.5% (33) | 87.5% (49) |
| Roseburia faecis | 99% | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia | 60% (24) | 83.9% (47) |
| Ruminococcus bromii | 93% | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus | 45% (18) | 41.1% (23) |
| Subdoligranulum variabile | 97% | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | 57.5% (23) | 78.6% (44) |
| Urmitella timonensis | 88% | Firmicutes | Tissierellia | Tissierellales | Tissierellaceae | Urmitella | 40% (16) | 39.3% (22) |
Sample collection and DNA extraction
A kit for stool collection was delivered to each study participant following a validated procedure49,50 and using a Fecotainer device (Tag Hemi VOF, Netherlands). Faecal samples were homogenized within 24 h in the laboratory, and five 2 g aliquots were stored at − 80 °C until DNA isolation was performed. Total DNA extractions from the stool samples were performed using the QiaAmp PowerFecal DNA Kit (QIAGEN, Hilden, Germany). The nucleic acids were quantified using a NanoQuant Plate (TECAN Trading AG, Switzerland), which allows quantification using a spectrophotometer read at 260 nm. The spectrophotometer used was the TECAN Infinite 200 PRO, and the software was i-Control (version 1.11.10). The extracted DNA concentrations ranged from 1.1–155.5 ng/μl (mean 41.35 ± 38.70 ng/μL). Samples were stored at –20 °C until molecular analysis was performed.
PCR-DGGE
The PCR products for denaturing gradient gel electrophoresis (DGGE) were obtained by amplifying the bacterial 16S rRNA genes following a marker gene analysis approach51. The primer pairs were 357F-GC and 518R (Table 6)52. All PCRs were performed with the T100 Bio-Rad Thermocycler in a 25-μl reaction volume containing 1X Master Mix (166–5009, Bio-Rad, Berkeley, CA, USA), 0.02 bovine serum albumin (BSA), 0.4 μM of each primer, and 2 μl of DNA diluted 1:10 in sterile DNase-treated water. DGGE was carried out using a DCode System (Bio-Rad) with a 30–50% denaturing gradient of formamide and urea53. Electrophoresis ran at 200 V for 5 h at 60 °C in 1X TAE buffer. Gels were stained for 30 min with SYBR Green I nucleic acid gel stain (10.000X in DMSO, S9430, Sigma-Aldrich, USA) and were visualized using the D-Code XR apparatus from Bio-Rad. Then, DGGE bands were excised, incubated overnight at − 20 °C, washed, and crushed in 20 μl of molecular-grade water. The supernatant (2 μl) was used as a template and reamplified, as previously described, without BSA and using modified linker-PCR bacterial primers (357F-GC; 518R-AT-M13) (Table 6) 19,52,54–60. The obtained PCR products were sequenced with Sanger sequencing (Genechron-Ylichron S.r.l.). The sequence similarities were obtained by the National Centre for Biotechnology Information (NCBI) database using nucleotide Basic Local Alignment Search Tool (BLASTn) analysis.
Table 6.
Multivariable logistic regression model assessing potential risk factors of T1D.
| OR* | 95% CI** | |
|---|---|---|
| Total charbohydrate intake (g) | 1.03 | 1.01—1.05 |
| Firmicutes (Log gene copies/g stool) | 7.30 | 2.26—23.54 |
| Bifidobacterium spp (Log gene copies/g stool) | 0.13 | 0.05—0.34 |
*Odds Ratio, adjusted also for age and gender.
**Confidence Interval.
NGS
High-throughput DNA sequencing and analysis were conducted by BMR Genomics s s.r.l. The V3-V4 region of 16S rDNA was amplified using the MiSeq 300PEPro341F and Pro805R primer pair6. The sample reads were above 12*106. The reaction mixture (25 μl) contained 3–10 ng/μl genomic DNA, Taq Platinum HiFi (Invitrogen, Carlsbad, CA), and 10 μM of each primer. The PCR conditions for amplification of DNA were as follows: 94 °C for 1 min (1X), 94 °C for 30 s, 55 °C for 30 s, 68 °C for 45 s (25X), and 68 °C for 7 min (1X). PCR products were purified through Agencourt XP 0.8X Magnetic Beads and amplified shortly with the Index Nextera XT. The amplicons were normalized with SequalPrep (Thermo Fisher) and multiplexed. The pool was purified with Agencourt XP 1X Magnetic Beads, loaded onto MiSeq, and sequenced with the V3 chemistry-300PE strategy.
qRT-PCR
Starting from the extracted DNA, the following microbial targets were quantified by qRT-PCR using a CFX Touch Real-Time PCR Detection System (Bio-Rad-Hercules, CA) and CFX Manager (3.1 Software): total Bacteria, Bacteroidetes, Bacteroides spp., Firmicutes, Bifidobacterium spp., Akkermansia muciniphila, and Methanobrevibacter smithii. Total bacteria and M. smithii were detected following two reaction designs. For M. smithii, the analysis was performed using as target both the 16S rDNA and then a specific functional gene (nifH). For total bacteria, quantification was carried out using a protocol with or without a probe. For the determination of total bacteria (method without probe), Bacteroidetes, Bacteroides spp., Firmicutes, Bifidobacterium spp. and Akkermansia muciniphila, 2 µl of 1:10 extracted DNA was added to a reaction mixture consisting of 10 µl Sso Advance SYBR Green Supermix (172–5261, Bio-Rad), 0.5 µl each of the forward and reverse primers (10 µM final concentration) and 7 µl of ultrapure water in a 20 µl final reaction volume. The reaction conditions were set as follows: 95 °C for 3 min (1X), 95 °C for 10 s, and 59 °C for 15 s (57 °C for Bacteroidetes spp. and 60 °C for Firmicutes), 72 °C for 10 s (39X), 65 °C for 31 s, 65 °C for 5 s + 0.5 °C/cycle, ramp 0.5 °C/s (60X). Moreover, for the determinations of M. smithii and total bacteria (method with probe), the reaction was as follows. Two microlitres of 1:10 extracted DNA was added to a reaction mixture consisting of 10 µl IQ Multiplex PowerMix (Bio-Rad-Hercules, CA), 0.2 µl of the molecular probe (10 µM), 0.5 µl each of the forward and reverse primers (10 µM final concentration) and 6.8 µl of ultrapure water in a 20 µl final reaction volume. The reaction conditions were 95 °C for 3 min (1X), 95 °C for 10 s, 59 °C for 15 s, 72 °C for 15 s (39X), and 72 °C for 5 min. Standard curves were produced with serial six-fold dilutions of genomic DNA from the microorganism target, provided by ATCC (Manassas, Virginia, USA) or DSMZ (Braunschweig, Germany). All PCR tests were carried out in triplicate. Table 6 provides detailed information regarding oligonucleotide sequences and genomic standards19,54–60. The PCR efficiencies were always between 90 and 110%. To confirm the amplification of each target, gel electrophoresis was performed on 2% agarose gels.
Data elaboration and statistical analyses
The statistical analysis was performed using STATA version 11.0. Moreover, the data on the included T1D patients and healthy controls were elaborated to highlight the likelihood of having diabetes. A descriptive analysis of the variables was conducted. The data were reported as absolute numbers and percentages for categorical variables and as means and standard deviations for continuous variables. Moreover, the subjects were divided by individual origins into two groups: Italian and migrant, considering the origin of the children and their families, to show differences in the distribution of disease determinants and to assess whether being a migrant could be associated with T1D onset. Differences between Italian and migrant children were assessed using the χ2 test with Fisher’s correction for categorical variables and Student’s t-test for continuous variables. Univariable logistic regression was then performed to estimate the impact of sociodemographic, nutritional, and microbiota-related variables on the outcome. These associations were expressed as odds ratios (OR) at a 95% confidence interval (CI). Moreover, the adjusted p-value for multiple comparisons was calculated using the Benjamini and Hochberg false discovery rate method. We conducted multivariable analyses including various variables (age, gender, Firmicutes, Bifidobacterium spp., and total carbohydrate intake) and the risk of type 1 diabetes using logistic regression models. The Spearman rank-order correlation coefficient was also determined to assess the relationships between variables. A p-value p < 0.05 was considered significant for all analyses.
The DGGE gel analysis was performed with Bionumerics 7.2. The hierarchical classification was performed with a UPGMA system (1% tolerance and optimization level) and Pearson correlation. Simpson's diversity index, Shannon’s index, and Margalef index were calculated for each DGGE profile to evaluate alpha diversity.
NGS bioinformatics analysis was performed with the software pipeline Qiime2. The reads were cleaned up by the primers using the software Cutadapt (version 2018.8.0) and processed with the software DADA2. The sequences were trimmed at the 3′ end (forward: 270 bp; reverse 260 bp), filtered by quality, and merged with default values. Subsequently, the sequences were elaborated to obtain unique sequences. In this phase, the chimaeras (denoised-paired) are also eliminated. The sequences were clustered against unique sequences at 99% similarity. The taxonomies of both GreenGenes (version 13–8) and Silva (version 132) were assigned to the OTU sequences. Alpha-diversity analyses were performed on all samples using the observed OTUs, Shannon, Pielou's evenness, and Faith PD indices, and for each index, the Kruskal–Wallis test was used to verify the significance of the comparisons between samples. Beta-diversity analyses were performed on all samples using the Bray–Curtis, Jaccard, and UniFrac metrics (weighted and unweighted). Multivariable statistical analyses were performed using the PERMANOVA, Adonis, and ANOSIM tests; instead, the analysis of the differential abundance was based on the packages of R (MetagenomeSeq, DeSeq2, and ANCOM).
Acknowledgements
The authors are grateful to the Italian Ministry of Health (RF-2011-02350617), the University of the Study of Torino and the Città della salute e e della scienza di Torino and the Hospital “Maggiore della Carità" di Novara for co-funding this project. Moreover, the authors wish to thank dr. Barbara Di Stefano (Sanitary Direction AOU Novara) and Mrs Rim Maatoug, Mrs Shpresa Xheka, and Mrs Daniela Elena Zelinschi (cultural intermediaries) at Novara Hospital for the translation of the questionnaire for migrant people. Finally, the authors make a special acknowledgement to the participant children and their families.
Author contributions
F.C. and R.S. coordinate the work. F.C., I.R., R.S., D.T.: design the work. F.C., I.R., S.S., and F.C.: patient inclusion and questionnaire administration. C.V., D.C.: clinical data collection, Torino and Novara, respectively. I.R.: patient sample collection and transport, questionnaire elaboration. D.T., G.C.: sample processing and extraction, molecular analysis. G.S., U.A., D.T. : statistical analysis and bioinformatics. M.D., A.C., A.F.: nutritional data elaboration. G.C., G.S.: drafted the work. F.C., I.R., R.S., M.D.: revised the work. D.T.: substantively revised the work.
Data availability
The database includes human data that are available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.National Center for Chronic Disease Prevention and Health Promotion. National Diabetes Statistics Report, 2017. Estimates of Diabetes and Its Burden in the United States. CDC (2017).
- 2.Mikael Knip, Md, P. et al. Prediction of Type 1 Diabetes in the General Population. Diabetes Care33, 1206–1212 (2010). [DOI] [PMC free article] [PubMed]
- 3.American Diabetes Association Standard medical care in diabetes - 2018. Diabetes Care. 2018;41:1–159. doi: 10.2337/dci18-0007. [DOI] [PubMed] [Google Scholar]
- 4.Pociot F, Lernmark Å. Genetic risk factors for type 1 diabetes. Lancet. 2016;387:2331–2339. doi: 10.1016/S0140-6736(16)30582-7. [DOI] [PubMed] [Google Scholar]
- 5.Turtinen, M. et al. Characteristics of familial type 1 diabetes : effects of the relationship to the affected family member on phenotype and genotype at diagnosis. (2019). [DOI] [PMC free article] [PubMed]
- 6.Knip M, Luopajärvi K, Härkönen T. Early life origin of type 1 diabetes. Semin. Immunopathol. 2017;39:653–667. doi: 10.1007/s00281-017-0665-6. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Global Report on Diabetes. (2016).
- 8.Bruno, G. Il registro diabete Piemonte. Ital. Heal. Policy Br. 1–8 (2016).
- 9.Regnell SE, Lernmark Å. Early prediction of autoimmune (type 1) diabetes. Diabetologia. 2017;60:1370–1381. doi: 10.1007/s00125-017-4308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knip M, Honkanen J. Modulation of type 1 diabetes risk by the intestinal microbiome. Curr. Diab. Rep. 2017;17:4–11. doi: 10.1007/s11892-017-0933-9. [DOI] [PubMed] [Google Scholar]
- 11.Bach J-F, Chatenoud L. The hygiene hypothesis : an explanation for the increased frequency of insulin. Cold Sping Harb. Perpect. Med. 2012;2:a007799. doi: 10.1101/cshperspect.a007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zununi Vahed S, Moghaddas Sani H, Rahbar Saadat Y, Barzegari A, Omidi Y. Type 1 diabetes: through the lens of human genome and metagenome interplay. Biomed. Pharmacother. 2018;104:332–342. doi: 10.1016/j.biopha.2018.05.052. [DOI] [PubMed] [Google Scholar]
- 13.Butalia S, Kaplan GG, Khokhar B, Rabi DM. Environmental risk factors and type 1 diabetes: past, present, and future. Can. J. Diabetes. 2016;40:586–593. doi: 10.1016/j.jcjd.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2019;15:635–650. doi: 10.1038/s41574-019-0254-y. [DOI] [PubMed] [Google Scholar]
- 15.Siljander H, Honkanen J, Knip M. Microbiome and type 1 diabetes. EBioMedicine. 2019;46:512–521. doi: 10.1016/j.ebiom.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper LV, Littman DR, Macpherson AJ, Program MP. Interactions between the microbiota and the immune system. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis-richardson, A. G. & Triplett, E. W. On the role of gut bacteria and infant diet in the development of autoimmunity for type 1 diabetes. Reply to Hänninen ALM and Toivonen RK [ letter ]. 2197–2198 (2015). doi:10.1007/s00125-015-3701-x [DOI] [PubMed]
- 18.Giongo A, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murri, M. et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children : a case-control study. 1–12 (2013). [DOI] [PMC free article] [PubMed]
- 20.Mejìa-Leòn, M. E., Petrosino, J. F., Ajami, N. J., Domìnguez-Bello, M. G. & Calderòn de la Barca, M. Fecal microbiota imbalance in Mexican children with type 1 diabetes. 4, 1–5 (2013). [DOI] [PMC free article] [PubMed]
- 21.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vatanen T, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Goffau MC, et al. Fecal Microbiota Composition Differs Between Children With Beta-Cell Autoimmunity and Those Without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis-Richardson AG, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front. Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostic AD, et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemppainen KM, et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38:329–332. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong T, et al. The remission phase in type 1 diabetes: changing epidemiology, definitions and emerging immuno-metabolic mechanisms. Diabetes Metab. Res. Rev. 2019 doi: 10.1002/dmrr.3207. [DOI] [PubMed] [Google Scholar]
- 28.Winkler C, et al. Identification of infants with increased type 1 diabetes genetic risk for enrollment into Primary Prevention Trials—GPPAD-02 study design and first results. Pediatr. Diabetes. 2019 doi: 10.1111/pedi.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziegler, A.-G. et al. Screening for asymptomatic β-cell autoimmunity in young children No Title. Lancet Child Adolesc. Heal.May, 288–290 (2019). [DOI] [PubMed]
- 30.Rabbone I, Traversi D, Scaioli G, &,, et al. Microbiota, epidemiological and nutritional factors related to ketoacidosis at the onset of type 1 diabetes. Acta Diabetol. 2020 doi: 10.1007/s00592-020-01555-z. [DOI] [PubMed] [Google Scholar]
- 31.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dridi B, Raoult D, Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe. 2011;17:56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Rawshani A, et al. Excess mortality and cardiovascular disease in young adults with type 1 diabetes in relation to age at onset: a nationwide, register-based cohort study. Lancet. 2018;392:477–486. doi: 10.1016/S0140-6736(18)31506-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mejía-León ME, Petrosino JF, Ajami NJ, Domínguez-Bello MG, De La Barca AMC. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci. Rep. 2014;4:1–5. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alkanani AK, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64:3510–3520. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harbison JE, et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: a prospective cohort study. Pediatr. Diabetes. 2019;20:574–583. doi: 10.1111/pedi.12865. [DOI] [PubMed] [Google Scholar]
- 37.Maffeis, C. et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. 700–709 (2016). doi:10.1002/dmrr [DOI] [PubMed]
- 38.Murri, M. et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. 1–12 (2013). doi:10.1002/dmrr [DOI] [PubMed]
- 39.Qi, C. J. et al. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese Children. 129, 1298–1304 (2016). [DOI] [PMC free article] [PubMed]
- 40.Pellegrini S, et al. Duodenal mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J. Clin. Endocrinol. Metab. 2017;102:1468–1477. doi: 10.1210/jc.2016-3222. [DOI] [PubMed] [Google Scholar]
- 41.Putignani L, Del Chierico F, Petrucca A, Vernocchi P, Dallapiccola B. The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood. Pediatr. Res. 2014;76:2–10. doi: 10.1038/pr.2014.49. [DOI] [PubMed] [Google Scholar]
- 42.Regueiro L, et al. Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol. Res. 2012;167:581–589. doi: 10.1016/j.micres.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Uusitalo U, et al. Association of Early Exposure of Probiotics and Islet Autoimmunity in the TEDDY Study. JAMA Pediatr. 2015;33612:1–9. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panigrahi P. Probiotics and prebiotics in neonatal necrotizing enterocolitis: New opportunities for translational research. Pathophysiology. 2014;21:35–46. doi: 10.1016/j.pathophys.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Brüssow H. Biome engineering-2020. Microb. Biotechnol. 2016;9:553–563. doi: 10.1111/1751-7915.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Traversi D, et al. Gut microbiota diversity and T1DM onset: Preliminary data of a case-control study. Hum. Microbiome J. 2017;5–6:11–13. doi: 10.1016/j.humic.2017.11.002. [DOI] [Google Scholar]
- 47.World Health Organization. ICF Parental Consent-clinicalstudies. (2018).
- 48.Ministero della Salute. Certificato di assistenza al parto (CeDAP). Analisi dell’evento nascita - Anno 2015. (2018).
- 49.Franzosa EA, et al. Relating the metatranscriptome and metagenome of the human gut. PNAS. 2014;111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.IHMS Consortium. IHMS-SOP 02 V2: Standard Operating Procedure for Fecal Samples Self ‐ Collection Laboratory Analysis Handled Within 4 To 24 Hours. (2015).
- 51.Knight, R. et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol.16, (2018). [DOI] [PubMed]
- 52.Muyzer G, Waal ECDE, Uitierlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/AEM.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webster NS, Negri AP. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 2006;8:1177–1190. doi: 10.1111/j.1462-2920.2006.01007.x. [DOI] [PubMed] [Google Scholar]
- 54.Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE. 2009;4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dao MC, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity : relationship with gut microbiome richness and ecology. Gut Microbiota. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 56.Guo X, et al. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008;47:367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- 57.Johnston C, Ufnar JA, Griffith JF, Gooch JA, Stewart JR. A real-time qPCR assay for the detection of the nifH gene of Methanobrevibacter smithii, a potential indicator of sewage pollution. J. Appl. Microbiol. 2010;109:1946–1956. doi: 10.1111/j.1365-2672.2010.04824.x. [DOI] [PubMed] [Google Scholar]
- 58.Matsuki T, et al. Quantitative PCR with 16S primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 2004;70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakayama T, Oishi K. Influence of coffee (Coffea arabica) and galacto-oligosaccharide consumption on intestinal microbiota and the host responses. FEMS Microbiol. Lett. 2013;343:161–168. doi: 10.1111/1574-6968.12142. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The database includes human data that are available upon reasonable request.


