Abstract
Background
Dysregulated circular RNAs (circRNAs) are involved in the development of glioma. This paper aims to analyze the role and mechanism of circRNA tau tubulin kinase 2 (circ-TTBK2) in glioma progression.
Methods
The glioma samples and normal brain tissues were collected. The levels of circ-TTBK2, microRNA-1283 (miR-1283) and chromodomain helicase DNA-binding protein 1 (CHD1) were examined via quantitative reverse transcription polymerase chain reaction or Western blot. Cell proliferation, migration, invasion and glycolysis were determined via 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide, transwell assay, Western blot, glucose and lactate assay kits. The target relationship was analyzed via dual-luciferase reporter assay. The xenograft model was established using U251 cells.
Results
circ-TTBK2 expression was increased in glioma tissues and cells. circ-TTBK2 knockdown suppressed glioma cell proliferation, migration, invasion and glycolysis. circ-TTBK2 was a sponge for miR-1283, and knockdown of miR-1283 reversed the effect of circ-TTBK2 silence on glioma progression. CHD1 was targeted via miR-1283, and miR-1283 repressed glioma cell proliferation, migration, invasion and glycolysis via decreasing CHD1. Knockdown of circ-TTBK2-reduced CHD1 expression by mediating miR-1283. Silence of circ-TTBK2 reduced xenograft tumor growth.
Conclusion
Down-regulation of circ-TTBK2 suppressed glioma development by regulating miR-1283 and CHD1, providing a new mechanism for understanding glioma pathogenesis.
Keywords: glioma, circ-TTBK2, miR-1283, CHD1, migration, glycolysis
Introduction
Glioma is the most common brain tumor with low 5-year overall survival.1 Great advances have gained on understanding the biology of glioma because of the development of experimental models of glioma in vivo and in vitro.2 However, the outcomes of patients remain poor, and the effective strategies are limited. The proliferation, migration, invasion and glucose metabolism are the main malignant processes in glioma.3–5 Hence, exploring the mechanism of these processes may help find a new target for the treatment of glioma.
Circular RNAs (circRNAs) are a group of noncoding RNAs with a closed continuous loop, which have important roles in brain disorders.6 Many circRNAs are dysregulated in glioma, and they can act as competitive endogenous RNAs (ceRNAs) for microRNAs (miRNAs).7 Tau tubulin kinase 2 (TTBK2) participates in the neurotoxicity and neurodegeneration in brain diseases.8,9 The circ-TTBK2 (hsa_circ_0000594) is derived from TTBK2, which can facilitate glioma cell proliferation, migration and invasion by regulating miR-217 and hepatocyte nuclear factor-1beta (HNF1β).10 However, the mechanism underlying circ-TTBK2 in glioma is complex, and more explorations are needed.
miRNAs play important roles in the angiogenesis, invasion, metabolism and therapy of glioma.11 miR-1283 has been reported to be associated with vascular injury, melanoma malignancy and endometriosis.12–14 More importantly, the previous study suggests that miR-1283 can suppress glioma cell proliferation and invasion via regulating activating transcription factor 4 (ATF4).15 The Circular RNA Interactome online predicts that miR-1283 may bind with circ-TTBK2. Nevertheless, whether miR-1283 is implicated in circ-TTBK2-mediated regulatory mechanism in glioma is unknown. Moreover, chromodomain helicase DNA-binding protein 1 (CHD1) is a pivotal biomarker involved in DNA-damage signaling.16 Emerging evidence indicates that CHD1 exhibits a pro-tumor role in glioma development.17 TargetScan online predicts that CHD1 is a target of miR-1283, and the complementary sequence of CHD1 is same to circ-TTBK2. Therefore, we assume that circ-TTBK2 could regulate glioma progression by miR-1283/CHD1 axis.
In this research, we examined the abundance of circ-TTBK2 in glioma samples and cells. Furthermore, we assessed the function of circ-TTBK2 knockdown on glioma cell proliferation, migration, invasion, glycolysis and xenograft tumor growth. Besides, we explored the ceRNA network of circ-TTBK2/miR-1283/CHD1 in glioma cells.
Materials and Methods
Patient Tissues
Thirty glioma tissues and normal brain samples from traumatic patients were collected from the Sixth Medical Center of PLA General Hospital. The patients did not receive chemotherapy and radiotherapy before surgery. All samples were pathologically diagnosed by pathologists and classified according to the 2007 WHO classification. The patients rank all grades of glioma. The tissue samples were stored at −80°C. The written informed consents were obtained, and this study was approved via the Ethics Committee of the Sixth Medical Center of PLA General Hospital.
Cell Culture and Treatment
Normal human astrocytes (NHA) and glioma cell lines (T98G and U251) were purchased from Bena Culture Collection (Beijing, China). Cells were grown in DMEM (Procell, Wuhan, China) containing 10% fetal bovine serum (Thermo Fisher, Wilmington, DE, USA), and 1% penicillin/streptomycin (Thermo Fisher) at 37°C in 5% CO2. To detect the stability of circ-TTBK2, cells were incubated with 1 μg/mL of Actinomycin D (Abcam, Cambridge, MA, USA) for different time points.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The RNA was isolated through TRIzol reagent (Takara, Tokyo, Japan). To obtain high quality of circRNAs, the isolated RNA was challenged via RNase R (Geneseed, Guangzhou, China) for 30 min. 500 ng RNA was reversely transcribed to cDNA using the specific reverse transcription kit (Thermo Fisher). The cDNA was mixed with SYBR Green (Takara) and specific primers, and qRT-PCR was conducted on an ABI 7900 system (ABI, Foster City, CA, USA). The primers (Sangon, Shanghai, China) were listed as: circ-TTBK2 (sense, 5ʹ-GCTTGGCTCGACAATTTACC-3ʹ; antisense, 5ʹ-AGTGCAACATTTTCCCTGGT-3ʹ), TTBK2 (sense, 5ʹ-GGGCTTTGGAGAAATTTACG-3ʹ; antisense, 5ʹ-GGTCTTTCCCTTGCAGCTTT-3ʹ), CHD1 (sense, 5ʹ-TCAGGAACAGAACGAACAGG-3ʹ; antisense, 5ʹ-GGAATGGATTTGTGCAAAGG-3ʹ), miR-1283 (sense, 5ʹ-CGCGCTCTACAAAGGAAAGCG-3ʹ; antisense, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ), U6 (sense, 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ; antisense, 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ), GAPDH (sense, 5ʹ-TCCTGCACCACCAACTGTTT-3ʹ; antisense, 5ʹ-GGATGATGTTCTGGTGGGCA-3ʹ). GAPDH and U6 acted as internal references, and the relative gene expression was calculated according to the 2-ΔΔCt method.18
Cell Transfection
The overexpression vector of CHD1 (pcDNA-CHD1) was generated via using pcDNA3.1 (Thermo Fisher), with pcDNA3.1 empty vector as negative control (pcDNA). The siRNA against circ-TTBK2 (si-circ-TTBK2#1, 5ʹ-GUCAGACCAUUGAGAAAGAUU-3ʹ, si-circ-TTBK2#2, 5ʹ-ACGUCAGACCAUUGAGAAAGA-3ʹ, si-circ-TTBK2#3, 5ʹ-AGACCAUUGAGAAAGAUUGGG-3ʹ), siRNA negative control (si-NC, 5ʹ-CACUGACGGUGACCAGAACAAAGAU-3ʹ), miR-1283 mimic (5ʹ-UCUACAAAGGAAAGCGCUUUCU-3ʹ), mimic negative control (miR-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-1283 inhibitor (anti-miR-1283, 5ʹ-AGAUGUUUCCUUUCGCGAAAGA-3ʹ), and corresponding negative controls (anti- NC, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) were formed via Ribobio (Guangzhou, China). T98G and U251 cells with 70% confluence were transfected with these oligonucleotides (40 nM) or vector (600 ng) using Lipofectamine 3000 reagent (Thermo Fisher) for 24 hrs.
3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT)
The proliferation of T98G and U251 cells was determined via MTT assay. Cells (5 × 103 cells/well) were added into 96-well plates and maintained at 37°C. After 24, 48 or 72 hrs, each well was added with 10 μL MTT reagent (Beyotime, Shanghai, China), and cells were cultured for another 4 hrs. Next, 100 μL DMSO (Beyotime) was added. The optical density (OD) value was examined at 490 nm with a microplate reader (Bio-Gene Technology, Guangzhou, China).
Transwell Assay
The migrated and invasive abilities of T98G and U251 cells were detected using 24-well transwell chambers (Costar, Corning, NY, USA). To detect migrated ability, cells (1 × 104 cells/well) in medium without serum were seeded into the upper chambers. The lower chamber was filled with medium containing 10% serum. After culturing for 24 hrs, the cells passed the membranes were fixed and stained with 0.5% crystal violet (Beyotime). Subsequently, the number of migrated cells was counted under a ×100 magnification microscope (Olympus, Tokyo, Japan). Three random fields were selected. For invasion assay, cells (5 × 104 cells/well) were placed into the upper chambers coated with Matrigel (BD Bioscience, San Jose, CA, USA), and the other procedures were same to those in the migration assay.
Glucose Consumption and Lactate Production Assays
The glycolysis was analyzed via glucose consumption and lactate production. T98G and U251 cells (1 × 105 cells/well) were added into 12-well plates and cultured for 72 h. Next, the medium was harvested, and the amounts of glucose and lactate were detected via a glucose or lactate assay kit (Sigma, St. Louis, MO, USA) following the instructions. The relative glucose consumption and lactate production were normalized to the control group.
Western Blot
The protein was extracted using RIPA reagent (Solarbio, Beijing, China) with 1 mM PMSF (Solarbio), quantified via a BCA assay kit (Amyjet, Wuhan, China) and denatured at 100°C for 10 min. Next, 20 μg protein samples were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes (Solarbio). After the transfer, the membranes were interacted in 5% non-fat milk and incubated with specific primary antibodies (Abcam) as well as secondary antibody. The antibodies used in this research were listed as: anti-CDK4 (ab137675, 1:2000 dilution), anti-Cyclin D1 (ab226977, 1:3000 dilution), anti-Snail (ab216347, 1:500 dilution), anti-Vimentin (ab137321, 1:2000 dilution), anti-E-cadherin (ab15148, 1:500 dilution), anti-HK2 (ab227198, 1:5000 dilution), anti-CHD1 (ab242098, 1:1000 dilution), and anti-β-actin (ab227387, 1:5000 dilution), and horseradish peroxidase-conjugated IgG (ab97051, 1:5000 dilution). The protein bands were visualized via ECL reagent (Abcam). The relative protein expression was normalized to β-actin level and the control group.
Dual-Luciferase Reporter Assay
The binding site of circ-TTBK2 and miR-1283 was searched via Circular RNA Interactome online (https://circinteractome.nia.nih.gov/). The complementary site of miR-1283 and CHD1 3ʹUTR was predicted via TargetScan online (http://www.targetscan.org/vert_72/). The wild-type sequence of circ-TTBK2 or CHD1 3ʹUTR containing miR-1283 seed site was inserted into the psiCHECK-2 vector (Promega, Madison, WI, USA) to generate corresponding constructs, named as circ-TTBK2-WT or CHD1 3ʹUTR-WT. The mutant-type constructs were generated via mutating the seed sites of miR-1283, named as circ-TTBK2-MUT or CHD1 3ʹUTR-MUT, respectively. T98G and U251 cells were co-transfected with the wild-type or mutant-type constructs and miR-1283 mimic or miR-NC using Lipofectamine 3000 reagent. After 24 h post-transfection, the luciferase activity was measured via a dual-luciferase assay kit (Thermo Fisher).
Xenograft Model
The BALB/c nude mice (male, 4-week-old) were purchased from Charles River (Beijing, China) and acclimatized for 1 week before experiments. The shRNA for circ-TTBK2 (sh-circ-TTBK2) and negative control (sh-NC) were synthesized via Ribobio and transfected into U251 cells. The cells (2 × 106 cells/mouse) stably transfected with sh-circ-TTBK2 or sh-NC were subcutaneously inoculated into mice (n=5 per group). Tumor volume was measured every 7 days and calculated (Volume=length × width2/2). Until 35 days after cell injection, animals were killed via cervical dislocation under anesthesia. The tumor tissues were collected and the weight was detected. Moreover, the levels of circ-TTBK2, miR-1283 and CHD1 were examined in tumor tissues. The animal experiments were carried out under the approval of the Institutional Animal Care and Use Committee of the Sixth Medical Center of PLA General Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
The experiments were carried out with 3 biological replicates and 3 technical replicates. The data were expressed as mean ± SD. The boxplots in Figures 1A, 3D and 5G were made and analyzed via GraphPad Prism 6 (GraphPad, La Jolla, CA, USA). The linear correlation in Figures 3F, 5I and 7C was analyzed via Pearson correlation analysis. Student’s t-test was performed in Figures 1–3, 5 and 8; ANOVA followed via Tukey’s test was conducted in Figures 4, 6 and 7. The difference was significant when P<0.05.
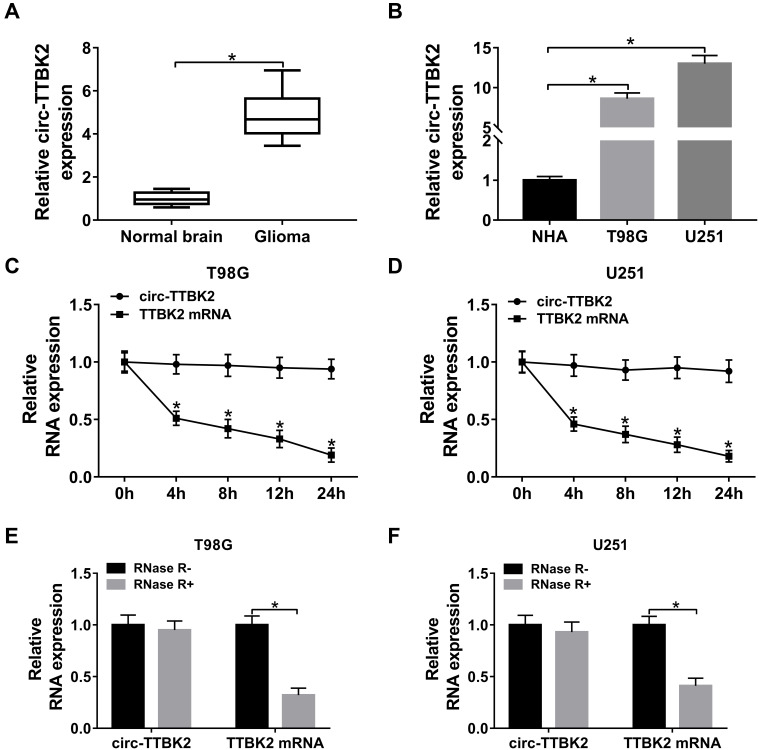
Figure 1.
The level of circ-TTBK2 in glioma tissues and cells. (A) The level of circ-TTBK2 was detected in glioma samples and normal brain samples via qRT-PCR. n=30. (B) The expression of circ-TTBK2 was examined in glioma cell lines (T98G and U251) and NHA cells via qRT-PCR. (C and D) The levels of circ-TTBK2 and TTBK2 mRNA were measured in T98G and U251 cells treated by Actinomycin D via qRT-PCR. (E and F) The levels of circ-TTBK2 and TTBK2 mRNA were detected after stimulation of RNase R via qRT-PCR. *P<0.05.
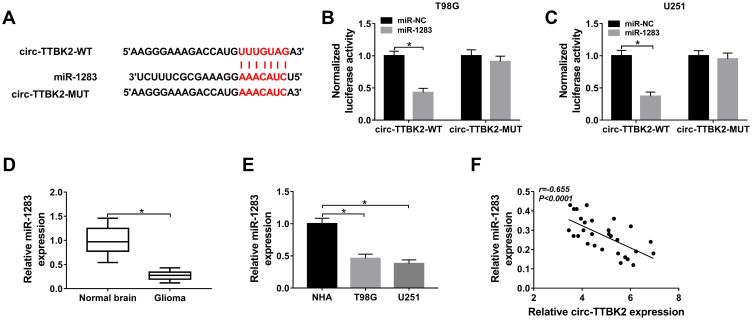
Figure 3.
The association between circ-TTBK2 and miR-1283. (A) The complementary sequence of circ-TTBK2 and miR-1283 was predicted via Circular RNA Interactome. (B and C) Luciferase activity was measured in T98G and U251 cells with transfection of circ-TTBK2-WT or circ-TTBK2-MUT. (D and E) The expression of miR-1283 was detected in glioma tissues and cells as well as corresponding controls. (F) The linear correlation of circ-TTBK2 and miR-1283 was analyzed. *P<0.05.
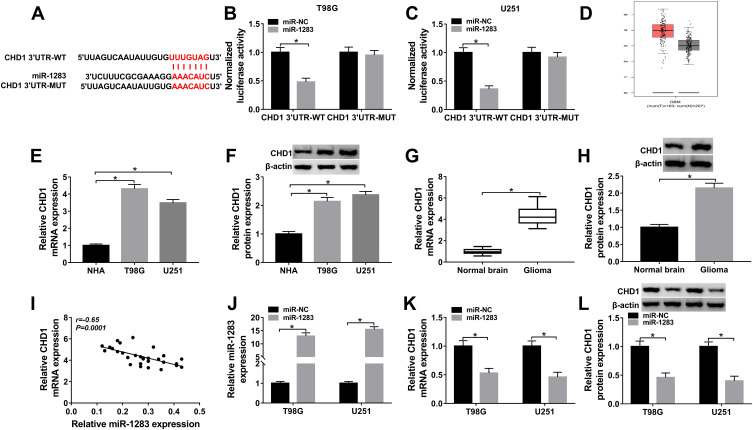
Figure 5.
The association between miR-1283 and CHD1. (A) The target sites of miR-1283 and CHD1 were searched via TargetScan. (B and C) Luciferase activity was measured in T98G and U251 cells transfected with CHD1 3ʹUTR-WT or CHD1 3ʹUTR-MUT. (D) CHD1 expression in glioma tissues was predicted via GEPIA. (E–H) The mRNA and protein levels of CHD1 were measured in glioma cells and tissues. (I) The linear correlation between the levels of CHD1 and miR-1283 were analyzed. (J) The expression of miR-1283 was measured in T98G and U251 cells transfected with miR-NC or miR-1283. (K and L) The mRNA and protein levels of CHD1 were detected in T98G and U251 cells transfected with miR-NC or miR-1283. *P<0.05.
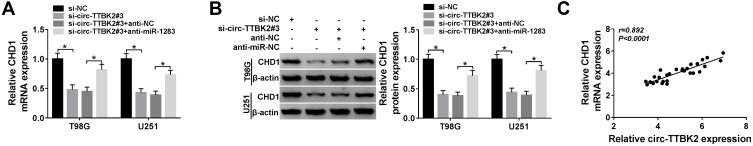
Figure 7.
The effect of circ-TTBK2 on CHD1 expression. (A and B) The mRNA and protein levels of CHD1 were measured in T98G and U251 cells transfected with si-NC, si-circ-TTBK2#3, si-circ-TTBK2#3 + anti-NC or anti-miR-1283. (C) The linear relationship between CHD1 and circ-TTBK2 was analyzed. *P<0.05.
Figure 8.
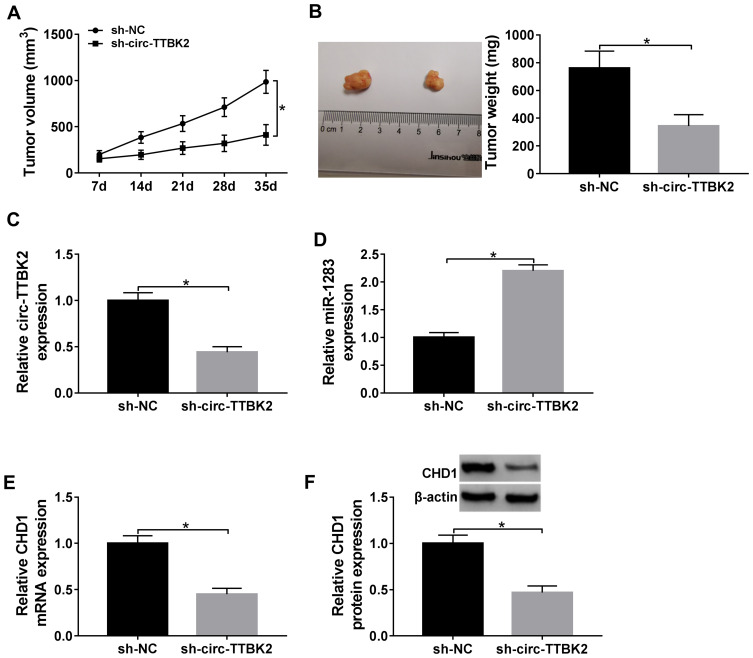
The effect of circ-TTBK2 on xenograft tumor growth. (A and B) Tumor volume and weight were detected in sh-circ-TTBK2 and sh-NC group. (C–F) The levels of circ-TTBK2, miR-1283, CHD1 were detected in sh-circ-TTBK2 and sh-NC group. *P<0.05.
Figure 4.
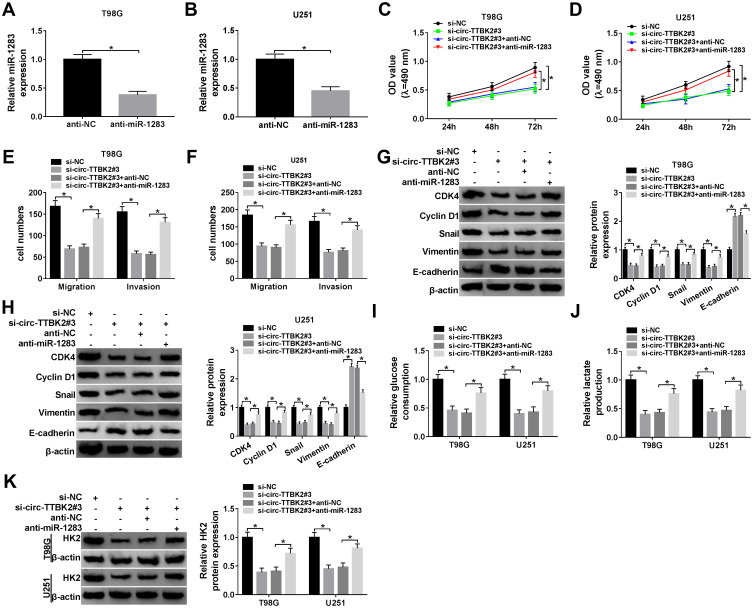
The effects of circ-TTBK2 and miR-1283 on glioma cell proliferation, migration, invasion and glycolysis. (A and B) The level of miR-1283 was examined in T98G and U251 cells transfected with anti-miR-1283 or anti-NC via qRT-PCR. Cell proliferation (C and D), migration and invasion (E and F), protein levels of CDK4, Cyclin D1, Snail, Vimentin and E-cadherin (G and H), glucose consumption (I), lactate production (J) and HK2 protein level (K) were examined in T98G and U251 cells transfected with si-NC, si-circ-TTBK2#3, si-circ-TTBK2#3 + anti-NC or anti-miR-1283. *P<0.05.
Figure 6.
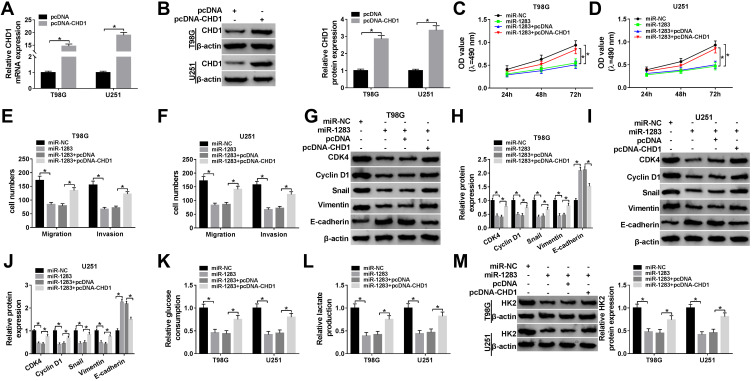
The effects of miR-1283 and CHD1 on glioma cell proliferation, migration, invasion and glycolysis. (A and B) The mRNA and protein levels of CHD1 were measured in T98G and U251 cells transfected with pcDNA-CHD1 or pcDNA. Cell proliferation (C and D), migration and invasion (E and F), protein levels of CDK4, Cyclin D1, Snail, Vimentin and E-cadherin (G–J), glucose consumption (K), lactate production (L) and HK2 protein level (M) were measured in T98G and U251 cells transfected with miR-NC, miR-1283, miR-1283 + pcDNA or pcDNA-CHD1. *P<0.05.
Results
The Expression of circ-TTBK2 is Increased in Glioma
To measure the abundance of circ-TTBK2 in glioma, 30 glioma tissues and normal samples were harvested. qRT-PCR assay showed that circ-TTBK2 level was clearly higher in glioma tissues than normal brain samples (Figure 1A). Moreover, the abundance of circ-TTBK2 was markedly increased in T98G and U251 cells in comparison to NHA cells (Figure 1B). Besides, circ-TTBK2 exhibited higher stability than linear TTBK2 mRNA after treatment of Actinomycin D (Figure 1C and D) or RNase R (Figure 1E and F). These data indicated that high expression of circ-TTBK2 might play an important role in glioma development.
Knockdown of circ-TTBK2 Suppresses Proliferation, Migration, Invasion and Glycolysis in Glioma Cells
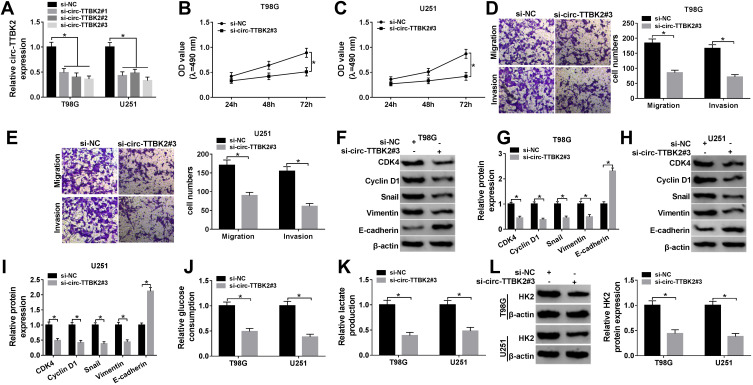
To assess the function of circ-TTBK2 in glioma progression, circ-TTBK2 was knocked down in T98G and U251 cells. circ-TTBK2 abundance was effectively decreased via transfection of si-circ-TTBK2 #1-#3 (Figure 2A), and si-circ-TTBK2 #3 with relative highest efficacy was used for further experiments. Moreover, knockdown of circ-TTBK2 significantly reduced the proliferation of T98G and U251 cells (Figure 2B and C). In addition, the abilities of migration and invasion were obviously decreased via the down-regulation of circ-TTBK2 in the two cell lines (Figure 2D and E). Cyclin D1 is a key biomarker for cell proliferation, which could activate CDK4 to regulate the G1-to-S-phase transition.19,20 E-cadherin is a representative epithelial surface marker and Vimentin is an important mesenchymal marker; the loss of E-cadherin during epithelial-to-mesenchymal transition is mediated via the transcription factor Snail.21 The epithelial-to-mesenchymal transition process contributes to the migrated and invasive capacities in glioma.4 Hence, these biomarkers were detected in our study. As shown in Figure 2F–I, silence of circ-TTBK2 led to an elevation of E-cadherin and reduction of CDK4, Cyclin D1, Snail and Vimentin. Besides, HK2 is an important regulator of glycolysis in cancers.22 The results displayed that knockdown of circ-TTBK2 suppressed glioma cell glycolysis by decreasing glucose consumption, lactate production and HK2 protein expression (Figure 2J–L). These results indicated that knockdown of circ-TTBK2 inhibited glioma progression in vitro.
Figure 2.
The effect of circ-TTBK2 on glioma cell proliferation, migration, invasion and glycolysis. (A) The abundance of circ-TTBK2 was detected in T98G and U251 cells transfected with si-circ-TTBK2#1, #2, #3 or si-NC via qRT-PCR. (B and C) The proliferation of T98G and U251 cells transfected with si-circ-TTBK2#3 or si-NC was detected at 24, 48 and 72 h by MTT. (D and E) Cell migration and invasion were analyzed in T98G and U251 cells transfected with si-circ-TTBK2#3 or si-NC by trans-well assy. (F–I) The protein levels of CDK4, Cyclin D1, Snail, Vimentin and E-cadherin were measured in T98G and U251 cells transfected with si-circ-TTBK2#3 or si-NC by Western blot. (J–L) The levels of glucose consumption, lactate production and HK2 protein were measured in T98G and U251 cells transfected with si-circ-TTBK2#3 or si-NC. *P<0.05.
TTBK2 is a Sponge for miR-1283
To probe the mechanism mediated via circ-TTBK2, Circular RNA Interactome online predicted the potential targets of circ-TTBK2. The binding site of circ-TTBK2 and miR-1283 was exhibited in Figure 3A. The dual-luciferase reporter assay with circ-TTBK2-WT and circ-TTBk2-MUT was performed in T98G and U251 cells to analyze this prediction. As shown in Figure 3B and C, addition of miR-1283 induced approximately 60% reduction of luciferase activity in the circ-TTBK2-WT group, but this effect was abrogated in the circ-TTBK2-MUT group. Furthermore, the level of miR-1283 was remarkably decreased in glioma tissues and cells (T98G and U251) compared with that in normal brain samples or NHA cells (Figure 3D and E). Additionally, the level of miR-1283 in glioma tissues was inversely correlated with circ-TTBK2 (Figure 3F). These findings suggested that circ-TTBK2 could act as a sponge for miR-1283 in glioma cells.
Down-Regulation of miR-1283 Reverses the Inhibitive Effect of circ-TTBK2 Knockdown on Proliferation, Migration, Invasion and Glycolysis in Glioma
To explore whether circ-TTBK2-mediated glioma development required miR-1283, T98G and U251 cells were transfected with si-NC, si-circ-TTBK2#3, si-circ-TTBK2#3 + anti-NC or anti-miR-1283. The transfection of anti-miR-1283 effectively decreased the abundance of miR-1283 in T98G and U251 cells (Figure 4A and B). Moreover, down-regulation of miR-1283 attenuated knockdown of circ-TTBK2-induced proliferation inhibition in T98G and U251 cells (Figure 4C and D). In addition, inhibition of miR-1283 rescued the abilities of migration and invasion that were decreased via silence of circ-TTBK2 (Figure 4E and F). Furthermore, knockdown of miR-1283 alleviated the regulatory effect of circ-TTBK2 silence on the protein levels of CDK4, Cyclin D1, Snail, Vimentin and E-cadherin (Figure 4G and H). Besides, down-regulation of miR-1283 protected against the silence of circ-TTBK2-mediated suppression of glucose consumption, lactate production and HK2 expression (Figure 4I–K). These data uncovered that silence of circ-TTBK2 suppressed glioma progression by regulating miR-1283.
CHD1 is a Target of miR-1283
To explore the mechanism mediated via miR-1283 in glioma, the target of miR-1283 was searched via TargetScan. CHD1 was one of the potential targets and the binding site is shown in Figure 5A. In order to confirm the target association between miR-1283 and CHD1, the CHD1 3ʹUTR-WT and CHD1 3ʹUTR-MUT were constructed. The results of dual-luciferase reporter analysis displayed that luciferase activity in the CHD1 3ʹUTR-WT group was obviously reduced by miR-1283 overexpression, whereas it was not changed in the CHD1 3ʹUTR-MUT group (Figure 5B and C). High expression of CHD1 was predicted via GEPIA (Figure 5D). Furthermore, the mRNA and protein levels of CHD1 were obviously up-regulated in glioma cells (T98G and U251) and tissues (Figure 5E–H). Meanwhile, the expression of CHD1 in glioma tissues was negatively associated with miR-1283 level (Figure 5I). Additionally, the effect of miR-1283 on CHD1 expression was investigated in T98G and U251 cells. The efficacy of miR-1283 mimic is identified in Figure 5J, and miR-1283 overexpression markedly decreased the abundances of CHD1 at mRNA and protein levels (Figure 5K and L). These findings uncovered that CHD1 was targeted via miR-1283 in glioma cells.
MiR-1283 Inhibits Proliferation, Migration, Invasion and Glycolysis by Targeting CHD1 in Glioma Cells
To explore the role of miR-1283 in glioma progression and whether it was associated with CHD1, T98G and U251 cells were transfected with miR-NC, miR-1283 mimic, miR-1283 mimic + pcDNA or pcDNA-CHD1. As displayed in Figure 6A and B, the mRNA and protein levels of CHD1 were effectively increased via the introduction of pcDNA-CHD1. Moreover, the functional assays showed that overexpression of miR-1283 significantly repressed cell proliferation, migration, invasion and glycolysis in the two cell lines (Figure 6C–M). Besides, these events were attenuated via up-regulation of CHD1 (Figure 6C–M). These results indicated that miR-1283 could inhibit glioma progression by decreasing CHD1 expression.
Down-Regulation of circ-TTBK2 Decreases CHD1 Expression by Regulating miR-1283
To explore whether circTTBK2 could regulate CHD1 expression, T98G and U251 cells were transfected with si-NC, si-circ-TTBK2#3, si-circ-TTBK2#3 + anti-NC or anti-miR-1283. As shown in Figure 7A and B, the mRNA and protein levels of CHD1 were markedly decreased via knockdown of circ-TTBK2 in the two cell lines, which were restored via down-regulation of miR-1283. Moreover, there was a positive correlation between the levels of circ-TTBK2 and CHD1 in glioma tissues (Figure 7C). These data suggested that circ-TTBK2 could positively regulate CHD1 expression by miR-1283.
Knockdown of circ-TTBK2 Decreases Tumor Growth
To explore the role of circ-TTBK2 in vivo, U251 cells stably transfected with sh-circ-TTBK2 or sh-NC were used to establish the xenograft models, named as sh-circ-TTBK2 or sh-NC group, respectively. As shown in Figure 8A and B, tumor volume and weight were obviously decreased in sh-circ-TTBK2 group in comparison to the sh-NC group. Furthermore, the abundances of circ-TTBK2, miR-1283 and CHD1 were detected in the tumor tissues. The results showed that the abundances of circ-TTBK2 and CHD1 were remarkably down-regulated and miR-1283 expression was markedly up-regulated in the sh-circ-TTBK2 group (Figure 8C–F). These findings indicated that inhibition of circ-TTBK2 obviously reduced xenograft tumor growth.
Discussion
Glioma is a common brain malignancy, and large challenges remain on the treatment of glioma.23 circRNAs are widely expressed and play important roles in the development of tumors, including glioma.24 A previous study reported that circ-TTBK2 played an oncogenic role in glioma by promoting cell proliferation, migration and invasion via functioning as a ceRNA for miR-217.10 In this paper, we also assessed the effect of circ-TTBK2 on glioma cell proliferation, migration and invasion. Moreover, we were the first to assess the function of circ-TTBK2 on glycolysis and find a new ceRNA network of circ-TTBK2/miR-1283/CHD1 in glioma.
Here we examined the expression of circ-TTBK2 in glioma and performed the experiments using Actinomycin D and RNase R, which showed that circ-TTBK2 was highly and stably expressed in glioma. Moreover, we found the anti-glioma role of circ-TTBK2 silence, revealed via suppression of cell proliferation, migration and invasion. This is also in agreement with the previous study.10 Furthermore, glycolysis is also an important feature in glioma development.5 By detecting glucose consumption, lactate production and HK2 protein expression, this study indicated that silence of circ-TTBK2 inhibited glycolysis of glioma. These results implied that circ-TTBK2 could play as a promising target for the development and treatment of glioma.
The ceRNA network is responsible for elucidating the mechanism of circRNAs in glioma progression.25 A former report has confirmed that circ-TTBK2 could act as a ceRNA for miR-217 to regulate HNF1β.10 To find a novel ceRNA network mediated via circ-TTBK2, we validated that circ-TTBK2 could function as a sponge for miR-1283, which plays an inhibitive role in glioma via suppressing cell proliferation and invasion.15 Similarly, here we also found that miR-1283 overexpression suppressed glioma progression via decreasing cell proliferation, migration, invasion and glycolysis. Moreover, knockdown of miR-1283 could attenuate the role of silence of circ-TTBK2 in glioma progression, indicating that circ-TTBK2 regulated glioma progression via sponging miR-1283. In addition, the former work suggested that ATF4 was required for miR-1283 to participate in glioma development.15 To explore the new mechanism, here we were the first to validate CHD1 as a target of miR-1283. Previous studies indicated that CHD1 could exhibit an oncogenic role in breast cancer and glioma.17,26 Similarly, this research validated that CHD1 abolished the suppressive effect of miR-1283 by a carcinogenic role in glioma. Additionally, by detecting the function of circ-TTBK2 on CHD1 expression, we found that circ-TTBK2 knockdown reduced CHD1 level by mediating miR-1283, implying the ceRNA network of circ-TTBK2/miR-1283/CHD1 in glioma cells. Besides, the subcutaneous xenograft model using U251 cells is helpful for understanding the potential mechanism of glioma in vivo.27–29 Here we using the xenograft model further confirmed the anti-tumor role of circ-TTBK2 inhibition in glioma via regulating the miR-1283/CHD1 axis in vivo.
In conclusion, inhibition of circ-TTBK2 could repress glioma progression in vitro and in vivo, possibly via acting as a ceRNA for miR-1283 to target CHD1. This study provides a new insight into the pathogenesis of glioma, and indicates that circ-TTBK2 can be used as a target for the treatment of glioma.
Funding Statement
There is no funding to report.
Disclosure of interest
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. doi: 10.1016/S0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- 2.Lenting K, Verhaak R, Ter Laan M, Wesseling P, Leenders W. Glioma: experimental models and reality. Acta Neuropathol. 2017;133(2):263–282. doi: 10.1007/s00401-017-1671-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16(12):1575–1584. doi: 10.1093/neuonc/nou147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CA, Chang CY, Hsueh KW, et al. Migration/invasion of malignant gliomas and implications for therapeutic treatment. Int J Mol Sci. 2018;19(4):1115. doi: 10.3390/ijms19041115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbin Z, Spielman D, Recht L. A metabolic therapy for malignant glioma requires a clinical measure. Curr Oncol Rep. 2017;19(12):84. doi: 10.1007/s11912-017-0637-y [DOI] [PubMed] [Google Scholar]
- 6.Nie JH, Li TX, Zhang XQ, Liu J. Roles of non-coding RNAs in normal human brain development, brain tumor, and neuropsychiatric disorders. Noncoding RNA. 2019;5(2):36. doi: 10.3390/ncrna5020036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Zhao K, Huang N, Zhang N. Circular RNAs and human glioma. Cancer Biol Med. 2019;16(1):11–23. doi: 10.20892/j.issn.2095-3941.2018.0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor LM, McMillan PJ, Liachko NF, et al. Pathological phosphorylation of tau and TDP-43 by TTBK1 and TTBK2 drives neurodegeneration. Mol Neurodegener. 2018;13(1):7. doi: 10.1186/s13024-018-0237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor LM, McMillan PJ, Kraemer BC, Liachko NF. Tau tubulin kinases in proteinopathy. FEBS J. 2019;286(13):2434–2446. doi: 10.1111/febs.14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyer S, Fleming J, Meng W, Singh R, Haque SJ, Chakravarti A. The role of miRNAs in angiogenesis, invasion and metabolism and their therapeutic implications in gliomas. Cancers. 2017;9(7):85. doi: 10.3390/cancers9070085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Yuan J, Xu Q, Chen R, Chen L, Fang M. miRNA-1283 regulates the PERK/ATF4 pathway in vascular injury by targeting ATF4. PLoS One. 2016;11(8):e0159171. doi: 10.1371/journal.pone.0159171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latchana N, Abrams ZB, Howard JH, et al. Plasma microRNA levels following resection of metastatic melanoma. Bioinform Biol Insights. 2017;11:1177932217694837. doi: 10.1177/1177932217694837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S, Lim W, Bazer FW, Whang KY, Song G. Quercetin inhibits proliferation of endometriosis regulating cyclin D1 and its target microRNAs in vitro and in vivo. J Nutr Biochem. 2019;63:87–100. doi: 10.1016/j.jnutbio.2018.09.024 [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Zhang Y, Su H, Shi H, Xiong Q, Su Z. Overexpression of miR-1283 inhibits cell proliferation and invasion of glioma cells by targeting ATF4. Oncol Res. 2019;27(3):325–334. doi: 10.3727/096504018X15251282086836 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Zhou J, Li J, Serafim RB, et al. Human CHD1 is required for early DNA-damage signaling and is uniquely regulated by its N terminus. Nucleic Acids Res. 2018;46(8):3891–3905. doi: 10.1093/nar/gky128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Gu W, Yu C. MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif. 2019;e12700. doi:doi: 10.1111/cpr.12700 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 19.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–572. doi: 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- 20.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–430. doi: 10.1038/nrclinonc.2016.26 [DOI] [PubMed] [Google Scholar]
- 21.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lis P, Dylag M, Niedzwiecka K, et al. The HK2 dependent “Warburg Effect” and mitochondrial oxidative phosphorylation in cancer: targets for effective therapy with 3-bromopyruvate. Molecules. 2016;21(12):1730. doi: 10.3390/molecules21121730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins S, Sontheimer H. Unique biology of gliomas: challenges and opportunities. Trends Neurosci. 2012;35(9):546–556. doi: 10.1016/j.tins.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Yang S, Wang H, et al. The progress of circular RNAs in various tumors. Am J Transl Res. 2018;10(6):1571–1582. [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Y, Jiaoming L, Xiang W, et al. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol. 2018;137(3):493–502. doi: 10.1007/s11060-018-2757-0 [DOI] [PubMed] [Google Scholar]
- 26.Tan S, Ding K, Li R, et al. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res. 2014;16(2):R40. doi: 10.1186/bcr3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang M, Qiao L, Li B, et al. Suppression of SIRT6 by miR-33a facilitates tumor growth of glioma through apoptosis and oxidative stress resistance. Oncol Rep. 2017;38(2):1251–1258. doi: 10.3892/or.2017.5780 [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018. doi: 10.3727/096504018X15178768577951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Yang Y, Wu M, et al. β-asarone induces apoptosis and cell cycle arrest of human glioma U251 cells via suppression of HnRNP A2/B1-mediated pathway in vitro and in vivo. Molecules. 2018;23(5):1072. doi:doi: 10.3390/molecules23051072 [DOI] [PMC free article] [PubMed] [Google Scholar]