Abstract
Purpose
Cervical cancer (CC) is the fourth most common cancer with high death rate in females. The study aims to detect the mechanism of long non-coding RNA (LncRNA) PCAT1 on radiosensitivity of CC.
Methods
The expression of PCAT1, miR-128 and GOLM1 in CC tissues and cells was measured by qRT-PCR. Different doses of X-ray were used for radiation treatment of CC cells and 6 Gy was chosen to perform the following experiments. The proliferation, migration and invasion of CC cells were measured by MTT assay, wound healing assay and transwell assay, respectively. The target relationships among PCAT1, miR-128 and GOLM1 were predicted by StarBase and TargetScan and verified by luciferase reporter assay. The protein level of GOLM1 was determined by Western blot. The xenograft tumor model was constructed in nude mice to verify the effect of PCAT1 on radiosensitivity of CC in vivo.
Results
The PCAT1 expression was upregulated in CC tissues and cells. PCAT1 silencing enhances radiosensitivity of CC cells on proliferation, migration and invasion. MiR-128 was the target of PCAT1 and was negatively regulated by PCAT1. Upregulation of miR-128 enhances radiosensitivity of CC cells on proliferation, migration and invasion. GOLM1 was a target of miR-128 and was negatively regulated by miR-128. Upregulation of GOLM1 and downregulation of miR-128 both reversed the enhanced effect of PCAT1 knockdown on radiosensitivity of CC cells, which partly promoted the proliferation, migration and invasion of CC cells.
Conclusion
Silencing of PCAT1 enhanced radiosensitivity of CC via targeting miR-128/GOLM1, which provided a new idea for treating CC.
Keywords: cervical cancer, lncPCAT1, miR-128, GOLM1, radiosensitivity
Introduction
Cervical cancer (CC) is the most common gynecological malignancy caused by malignant proliferation of the uterine cervix cells.1 Currently, the therapeutic approaches for CC including surgery, cytotoxic chemotherapy and radiotherapy.2 However, the therapeutic effect of radiotherapy remains unsatisfactory due to resistance.3 Therefore, it is urgent to further explore the potential mechanisms of radioresistance and improve radiosensitivity in patients with CC.
Long non-coding RNAs (LncRNAs) are a class of non-protein-coding transcripts that longer than 200 nucleotides, exerting the pathological functions through their interactions with miRNAs, mRNAs and proteins.3 The abnormal expression of lncRNAs in tumors is closely related to tumorigenesis and progression.4,5 In addition, growing evidence has shown that lncRNA is relevant to the radiation resistance of cancer.6–8 LncRNA FAM201A mediates the radiosensitivity of esophageal squamous cell cancer by regulating ATM and mTOR expression via miR-101.4 LncRNA LINC00483 knockdown enhances the inhibiting effect of X-ray radiation on lung adenocarcinoma (LAD) cells.5 Overexpression of LncRNA HOTAIR leads to radioresistance of CC via promoting HIF-1α expression.6 It has been demonstrated that the expression of LncRNA PCAT1 is downregulated in CC.7 Knockdown of LncRNA PCAT1 in glioma stem cells promotes radiation sensitivity.8 However, the function of PCAT1 on radiosensitivity of CC needs to be further explored.
MicroRNAs (miRNAs) are a class of small non-coding RNAs with lengths of 20 to 22 nucleotides, which play important roles in the progression of cancers (including proliferation, migration and invasion) and the resistance to radiation therapy.9–11 MiR-144 promotes the radiosensitivity of LAD cells.5 MiRNA-200c enhances the radiosensitivity significantly in esophageal squamous cancer both in vitro and in vivo.11 Upregulation of miR-128 significantly increases radiosensitivity of GSC and glioblastoma lines.12 In addition, a recent study conducted by Zhang et al has demonstrated that miR-128 negatively regulated by PVT1 can attenuate the progression of CC.13 However, whether miR-128 is regulated by PCAT1 to enhance radiosensitivity of CC remains to be further explored.
Golgi membrane protein 1 (GOLM1), an identified Golgi-associated protein,14 is widely expressed in normal epithelial cells.15 Aruna has suggested that high expression of GOLM1 correlates with malignant clinicopathological characteristics of patients in non-small cell lung cancer.16 Chen et al have proved that miR-200a suppresses cell proliferation, invasion and migration via targeting GOLM1 in melanoma.17 A previous research has indicated that down-regulation of GOLM1 enhances the chemosensitivity of CC to methotrexate.18 However, the regulatory mechanism of PCAT1 involving GOLM1 in CC is still not entirely clear.
In this study, we measured the LncRNA PCAT1 expression in CC tissues and cells, and evaluated the effect of PCAT1 knockdown on radiosensitivity of CC cells. Besides, we analyzed the relationships among PCAT1, miR-128 and GOLM1. Our results may provide a basis for guiding radiation therapy of CC.
Materials and Methods
Patient Samples
Total 63 paired CC tissues and adjacent normal tissues were obtained from CC patients from March 2017 to February 2018. All patients had not received any treatment, including surgery, neoadjuvant chemotherapy, radiation therapy and other treatments before surgical resection. The clinicopathological features of all patients were collected including age, FIGO stage, lymph node metastasis and depth of cervical invasion. The study was permitted by the ethics committee of Yantaishan Hospital (approval ID: YSLZ2020026) and informed consents were obtained from all patients.
Cell Culture
The CC cell lines HeLa, SiHa, C33A, CaSki and human cervical epithelial cell line H8 were procured from American Type Culture Collection (ATCC; Manassas, VA, USA). HeLa, SiHa, C33A and CaSki cells were cultured in Eagle’s Minimum Essential Medium (EMEM, Sigma Aldrich, Shanghai, China) containing 10% fetal bovine serum (FBS, Sigma Aldrich, China). H8 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium containing 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. All the cells were cultured in an incubator with 5% CO2 at 37°C.
Quantitative Real-Time Reverse Transcriptase PCR (qRT-PCR)
Total RNA was isolated from cells or tissues by TRIzol Reagent (Thermo Fisher Scientific, Shanghai, China). The cDNA was synthesized with SuperScript First StrandcDNA System (Invitrogen, Carlsbad, CA, US). Then, the cDNA product was subjected to qPCR with SYBR Green kit under the following cycling conditions: 95°C for 10 min; 40 cycles of 95°C for 1 min, 63°C for 2 min, 72°C for 1 min; final 72°C for 10 min. The relative expression of PCAT1, miR-128 and GOLM1 was calculated by the 2−ΔΔCT method. GAPDH and U6 expression were used as the internal control. The sequences of primer are shown in Table 1.
Table 1.
Primer Sequences Used in the Study
| Name of Primer | Sequences |
|---|---|
| LncRNA PCAT1 forward | 5ʹ-TTGCCTACTTGGGCAAATTC-3’ |
| LncRNA PCAT1 reverse | 5ʹ-TCCCCATTGTGTTCCAAGAT-3’ |
| miR-128 forward | 5ʹ-TTGGCCTTATTTGTGAGCTG-3’ |
| miR-128 reverse | 5ʹ-AAAAAGAAGCCAGGAAGCAG-3’ |
| GOLM1 forward | 5ʹ-CCGGAGCCTCGAAAAGAGATT-3’ |
| GOLM1 reverse | 5ʹ-ATGATCCGTGTCTGGAGGTC-3ʹ€€ |
| U6 forward | 5ʹ-CTCGCTTCGGCAGCACA-3’ |
| U6 reverse | 5ʹ-AACGCTTCACGAATTTGGT-3’ |
| GAPDH forward | 5ʹ-CTGGGCTACACTGAGCACC-3’ |
| GAPDH reverse | 5ʹ-AGTGGTCGTTGAGGGCAATG-3’ |
Cell Transfection and Radiation Treatment
ShRNAs targeting PCAT1 (sh-PCAT1-1, sh-PCAT1-2) and negative control (sh-NC), miR-128 inhibitor, miR-128 mimics and their negative controls (inhibitor NC and mimics NC), overexpression of GOLM1 (pcDNA-GOLM1) and negative control (pcDNA-NC) were all purchased from GenePharma (Shanghai, China). The transfection was conducted by Lipofectamine 3000 (Thermo Fisher Scientific, China) in light of the instructions. After transfection for 48 h, CC cells were irradiated with X-ray using an X-RAD 225Cx (Precision X-ray Inc, Blandford, Connecticut, US).
MTT Assay
CC cells were seeded into 96-well plates (0.2 × 104 cells/well). After 48h of culturing, 20 μL MTT (Sigma Aldrich, St. Louis, Missouri, US) was supplemented into each well, and cells were cultured for 2h. The absorbance values were detected by a microplate reader (Molecular Devices, Shanghai, China) at the wavelength of 490nm.
Wound Healing Assay
For migration analysis of CC cells, cells were seeded in 12-well plates and grown until the bottom of the plate was covered. Then, a scratch was made by a 200 μL pipette tip. To remove the detached cells, the cells were washed 3 times with PBS. After 24 h of culturing, photos were taken under a microscope (Olympus Corporation, Guangdong, China) and analyzed with Image J (NIH Image, Bethesda, MD, US).
Transwell Assay
For invasion analysis of CC cells, 1×105 cells were seeded into the matrigel-coated upper chamber. Then, 600 μL DMEM was added into the lower chamber. After 24h of culturing, the invasion cells were stained with 0.5% crystal violet. The invasion ability was evaluated by counting invasion cells under a microscope (Olympus Corporation, China, 40×10) at five randomly selected views.
Dual-Luciferase Reporter (DLR) Assay
Starbase software (http://starbase.sysu.edu.cn/) was utilized to predict the target relationship of PCAT1 and miR-128, whereas TargetScan software (http://www.targetscan.org) was used to predict the target relationship of miR-128 and GOLM1. Luciferase reporter assay was conducted to further verify the binding relationships between miR-128 and PCAT1/GOLM1. The fragments of PCAT1/GOLM1 containing the mutant (MUT) or wide type (WT) putative miR-128 binding site were inserted into the luciferase vector pGL2 (Promega Corporation, Madison, WI, US) to establish the recombinant reporter plasmids. CaSki and Hela cells (2 × 105 cells/well) were co-transfected with 40 nM of miR-128 mimics or mimics NC, and 20ng recombinant plasmids of either PCAT1/GOLM1 WT or PCAT1/GOLM1 MUT using Lipofectamine 3000 (Thermo Fisher Scientific, China). Following 48 h of incubation at 37°C, Renilla and firefly luciferase activities were analyzed using a dual-luciferase reporter gene assay system (Promega Corporation), according to the manufacturer’s protocol. The activity of firefly luciferase was normalized to the activity of Renilla luciferase.
Western Blot Assay
Cells were lysed with RIPA containing protease inhibitors to obtain the total protein (Thermo Fisher Scientific). The protein was separated by 10% SDS-PAGE and then transferred into PVDF membrane. The membrane was incubated with the primary antibody GOLM1 (1:1000, Sigma Aldrich, US) at the temperature of 4°C overnight. Then, membrane was incubated with secondary antibody (1:5000, Sigma Aldrich, US) under room temperature for 40 min. The bands were visualized with ECL detection reagents (Amersham Biosciences, Sweden).
Tumor Xenograft Experiments
Total 60 BALB/c nude mice (aged 4–5 weeks, weighing 22–24 g, GuangDong Experimental Animal Centre) were randomly assigned into 4 groups (10 mice/group): sh-NC, sh-PCAT1, sh-NC+6 Gy and sh-PCAT1 + 6 Gy group. The mice were anesthetized with 50 mg/kg pentobarbital sodium and subcutaneously injected with 100 μL CaSki cells that transfected with sh-NC or sh-PCAT1. Mice were irradiated with 6 Gy of X-ray per week and routinely checked. The tumor length (A) and width (B) were measured weekly, and the tumor volume was calculated using the following formula: V=1/2 AB2. Mice were anesthetized with 50 mg/kg pentobarbital sodium and sacrificed by cervical dislocation at 5 weeks after transplantation, and the tumors were collected and weighed. This study was approved by the Institutional Research Committee of Yantaishan Hospital (approval ID: YSLZ2020026). All animal experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical Analysis
Statistical analysis was performed with SPSS 23.0 (SPSS Inc., Chicago, IL, US). The two-tailed t test was used for comparison between two groups, and one-way ANOVA was used for comparison among multiple groups. Data were shown as means ± SD. Pearson’s correlation analysis was used to determine the correlations between the expression levels of miR-128 and PCAT1/GOLM1 in CC tissues. Statistical significance was considered when P < 0.05. All experiments were conducted in triplicate in at least three independent experiments.
Results
PCAT1 is Highly Expressed in CC Tissues and Cells
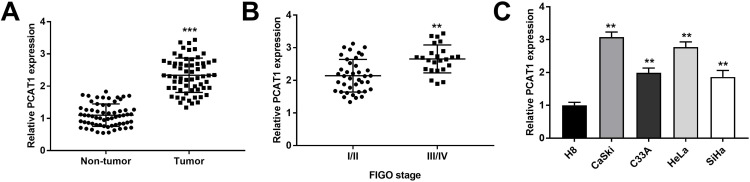
We measured the PCAT1 expression in CC and non-tumor tissues from 63 patients. The results found that PCAT1 expression was significantly higher in tumor tissues than that in non-tumor tissues (P < 0.001, Figure 1A). In line with this result, PCAT1 was highly expressed in stage III/IV of CC in contrast to that in stage I/II (P < 0.01, Figure 1B). Besides, the PCAT1 expression was proved to be positively correlated with FIGO stage, lymph node metastasis and depth of cervical invasion (Table 2). In addition, the expression of PACT1 in the CC cell lines was detected. The results showed that PCAT1 expression in CC cell lines (CaSki, C33A, SiHa and HeLa) was up-regulated compared with normal cell line (P < 0.01, Figure 1C).
Figure 1.
PCAT1 is highly expressed in CC tissues and cells. (A) The PCAT1 expression in CC tissues and non-tumor tissues (n =63) were measured by qRT-PCR. ***P < 0.001 vs the non-tumor group. (B) The PCAT1 expression in CC tissues at different FIGO stages was measured by qRT-PCR. **P < 0.01 vs the stage I/II group. (C) The PCAT1 expression in CC cell lines (CaSki, C33A, SiHa and HeLa) and normal cell line (H8) was measured by qRT-PCR. **P < 0.01 vs the H8 group.
Table 2.
Correlation Between lncRNA PCAT1 Expression and Clinicopathological Parameters of Cervical Cancer Patients
| Clinical Parameters | Cases (n = 63) | PCAT1 Expression Level | P | |
|---|---|---|---|---|
| Low (n = 34) | High (n = 29) | |||
| Age (years) | ||||
| ≤40 | 25 | 16 | 9 | 0.195 |
| >40 | 38 | 18 | 20 | |
| FIGO stage | ||||
| I/II | 39 | 27 | 12 | 0.0019** |
| III–IV | 24 | 7 | 17 | |
| Histology | 0.923 | |||
| Squamous | 33 | 18 | 15 | |
| Adenocarcinoma | 30 | 16 | 14 | |
| Lymph node metastasis | ||||
| No | 36 | 24 | 12 | 0.0195* |
| Yes | 27 | 10 | 17 | |
| Depth of cervical invasion | ||||
| <2/3 | 34 | 23 | 11 | 0.0183* |
| ≥2/3 | 29 | 11 | 18 | |
Note: *P, **P < 0.05.
PCAT1 Knockdown Enhances Radiosensitivity of CC Cells on Proliferation, Migration and Invasion in vitro
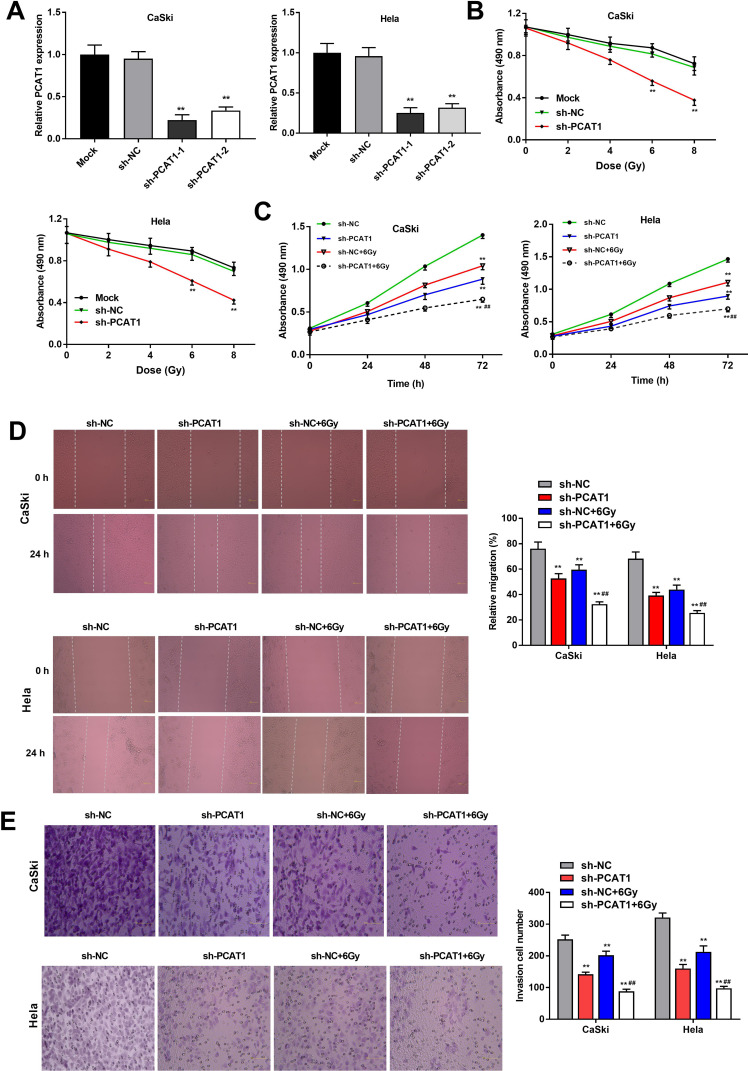
qRT-PCR was performed to detect the transfection efficiency of sh-PCAT1-1/-2/NC. The results demonstrated that PCAT1 expression was significantly downregulated after transfection of sh-PCAT1-1/-2 in CC cells (P < 0.01, Figure 2A). MTT assay showed that in a time-dependent manner, the viability of CC cells was decreased in the sh-PCAT1 group compared to the sh-NC group (P < 0.01, Figure 2B). Silencing of PCAT1 and radiation treatment (6 Gy) could, respectively, inhibit the viability of CC cells and the viability of CC cells could be further suppressed in the sh-PCAT1 + 6 Gy group (P < 0.01, Figure 2C). Similarly, wound healing assay and transwell assay indicated that the migration ability and invasion ability of CC cells in sh-PCAT1 group and sh-NC + 6 Gy group were significantly repressed compared with sh-NC group and further decreased in the sh-PCAT1 + 6 Gy group (P < 0.01, Figure 2D and E).
Figure 2.
PCAT1 silencing enhances radiosensitivity of CC cells. (A) The expression of PCAT1 in CaSki and Hela cells was measured by qRT-PCR. (B) The viability of CC cells at different doses of X-ray radiation was analyzed by MTT assay. (C) The viability of CC cells transfected with sh-PCAT1/NC under X-ray (6 Gy) was measured by MTT assay. (D) The migration ability of CC cells was measured by wound healing assay. (E) The invasion number of CC cells was measured by transwell assay. **P < 0.01 vs sh-NC group. ##P < 0.01 vs sh-NC + 6 Gy group.
PCAT1 Targets miR-128
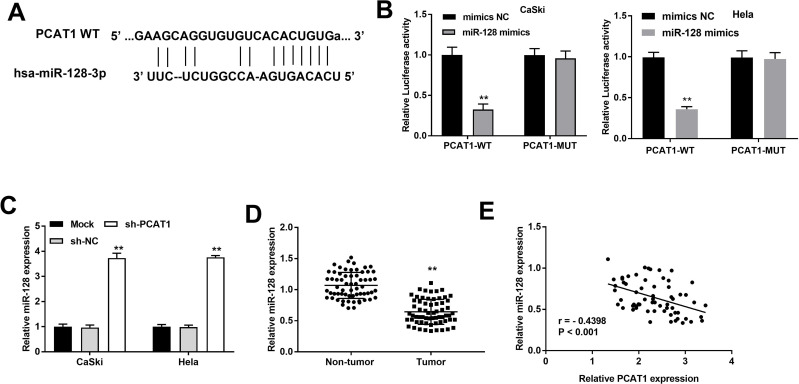
To further explore the mechanism of PCAT1, StarBase software was used to predict the putative target of PCAT1. We found that miR-128 is one of the potential targets of PCAT1 (Figure 3A). To further determine the relationship between PCAT1 and miR-128, DLR assay was performed both in CaSki and Hela cells. We discovered the luciferase activity in the PCAT1-WT/miR-128 mimics group was obviously decreased in contrast to that in PCAT1-WT/mimics NC group (P < 0.01, Figure 3B). The results of qRT-PCR demonstrated that the expression of miR-128 was increased markedly in CaSki and Hela cells after transfection of sh-PCAT1 (P < 0.01, Figure 3C), whereas was decreased in tumor tissues compared to that in non-tumor tissues (P < 0.01, Figure 3D). Additionally, a negative correlation between PCAT1 and miR-128 was exhibited in CC tissues (P < 0.001, r = −0.4398; Figure 3E).
Figure 3.
The expression of PCAT1 is negatively correlated with miR-128. (A) The binding site between PCAT1 and miR-128 was predicted by StarBase. (B) The luciferase activity of CC cells co-transfected with miR-128 mimic or mimics NC and luciferase reporters containing PCAT1 WT or PCAT1 MUT was determined by luciferase reporter gene assay. **P < 0.01 vs mimics NC group. (C) The expression of miR-128 in CC cells was detected by qRT-PCR assay. **P < 0.01 vs sh-NC group. (D) The expression of miR-128 in CC and non-tumor tissues was measured by qRT-PCR. **P < 0.01 vs non-tumor group. (E) The correlation between PCAT1 and miR-128. P < 0.001.
Upregulation of miR-128 Enhances Radiosensitivity of CC Cells on Proliferation, Migration and Invasion in vitro
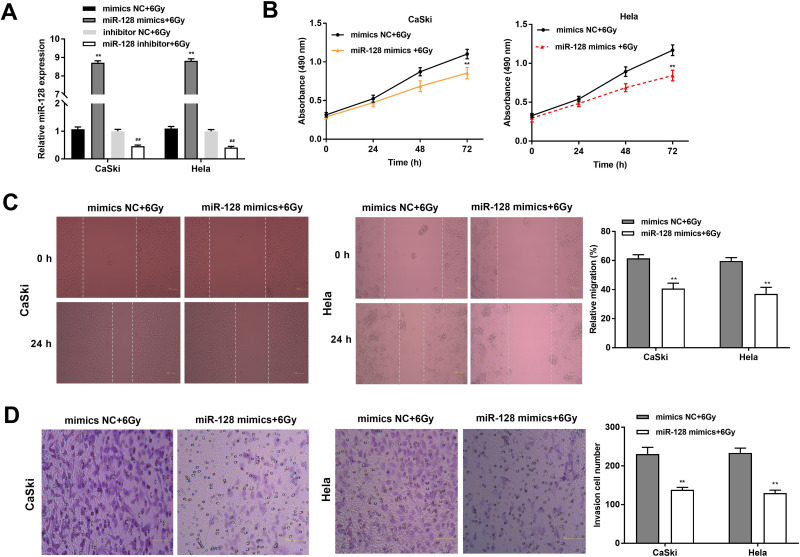
The expression of miR-128 was detected under radiation treatment (6 Gy) after transfection of miR-128 mimics, miR-128 inhibitor and miR-NC. The results disclosed that miR-128 expression was increased in the miR-128 mimics + 6 Gy group compared with the mimics NC + 6 Gy group and decreased in the miR-128 inhibitor + 6 Gy group compared to that in the inhibitor NC + 6 Gy group (P < 0.01, Figure 4A). MTT assay revealed that the viability of CC cells in the miR-128 mimics + 6 Gy group was decreased dramatically in contrast to the mimics NC + 6 Gy group (P < 0.01, Figure 4B). In line with this result, wound healing assay and transwell assay uncovered that the migration ability and invasion ability of CC cells in the miR-128 mimics + 6 Gy group was obviously decreased in comparison to the mimics NC + 6 Gy group (P < 0.01, Figure 4C and D).
Figure 4.
Upregulation of miR-128 enhances radiosensitivity of CC cells on proliferation, migration and invasion. (A) The expression of miR-128 after transfection of miR-128 mimics/NC or miR-128 inhibitor/NC was measured by qRT-PCR. **P < 0.01 vs mimics NC group, ##P < 0.01 vs inhibitor NC group. (B) The viability of CC cells transfected with miR-128 mimics/NC under X-ray (6 Gy) was measured by MTT assay. (C) The migration ability of CC cells was measured by wound healing assay. (D) The invasion number of CC cells was measured by transwell assay. **P < 0.01 vs mimics NC group.
MiR-128 Targets GOLM1
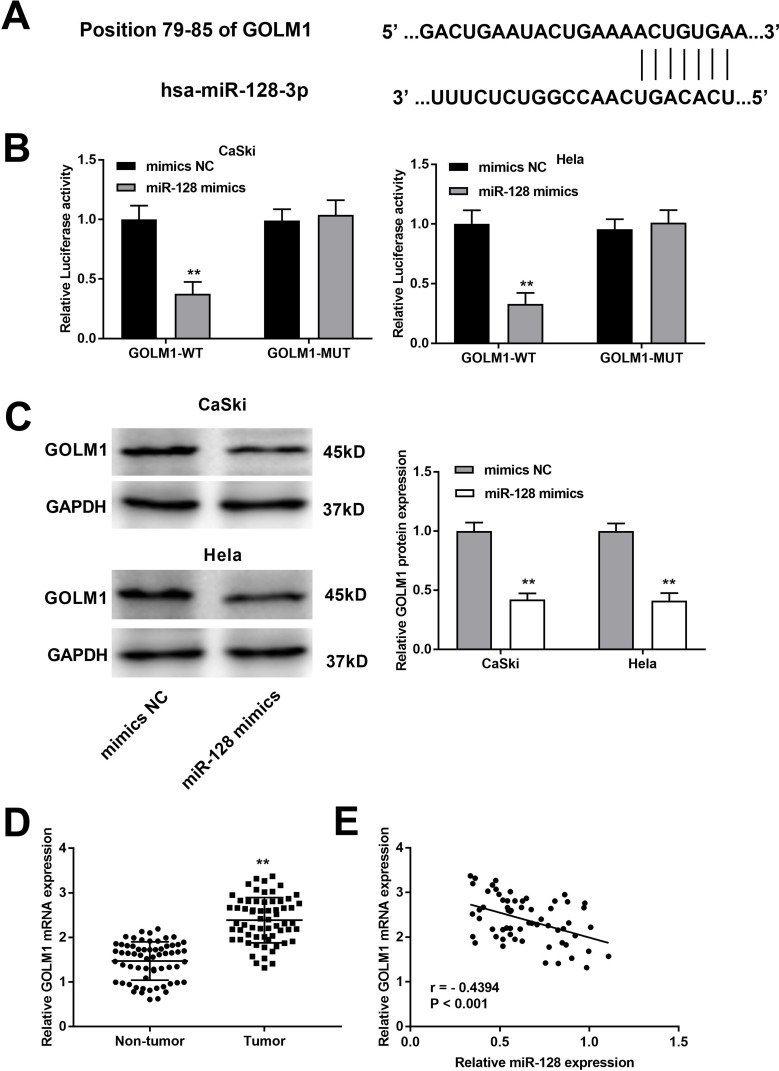
The complementary binding site of miR-128 and GOLM1 was predicted by TargeScan (Figure 5A). Through dual-luciferase reporter assay both in CaSki and Hela cells, decreased luciferase activities were exhibited in the GOLM1-WT/miR-128 mimics groups in contrast to the GOLM1-WT/mimics NC groups, which proved GOLM1 was a target gene of miR-128 (P < 0.01, Figure 5B). Western blot assay indicated that upregulation of miR-128 reduced the protein level of GOLM1 in CaSki cells and Hela cells (P < 0.01, Figure 5C). Furthermore, we found that GOLM1 expression was upregulated in tumor tissues in contrast to that in non-tumor tissues (P < 0.01, Figure 5D). The results of Pearson’s correlation analysis showed that miR-128 expression was negatively correlated with GOLM1 in CC tissues (P < 0.001, r = −0.4394; Figure 5E).
Figure 5.
MiR-128 targets GOLM1. (A) The binding site between miR-28 and GOLM1 was predicted by TargetScan. (B) The luciferase activity of CC cells co-transfected with miR-128 mimic or NC mimic and luciferase reporters containing GOLM1 WT or GOLM1 MUT was determined by luciferase reporter gene assay. **P < 0.01 vs mimics NC group. (C) The protein level of GOLM1 was measured by Western blot assay. **P < 0.01 vs mimics NC group. (D) The expression of GOLM1 in CC and non-tumor tissues was measured by qRT-PCR. **P < 0.01 vs the non-tumor group. (E) The correlation between miR-128 and GOLM1. P < 0.001.
PCAT1 Knockdown Enhances Radiosensitivity of CC Cells on Proliferation, Migration and Invasion by Regulating miR-128/GOLM1 in vitro
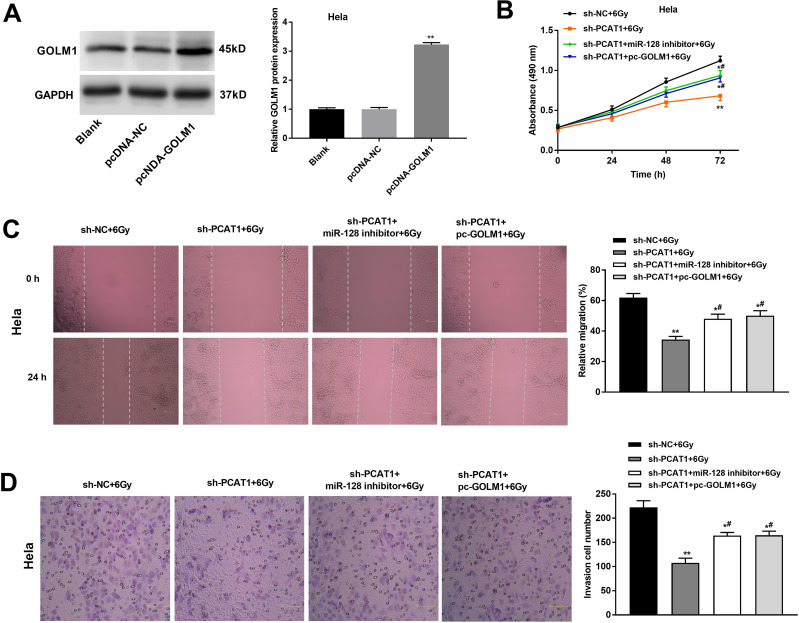
After transfection of pcDNA-GOLM1/NC, we investigated the protein level of GOLM1 in CC cells by Western blot assay. The results showed that the protein level of GOLM1 was upregulated in the pcDNA-GOLM1 group in comparison to the pcDNA-NC group (P < 0.01, Figure 6A). The results of MTT assay showed that the viability of CC cells in the sh-PCAT1 + 6 Gy group was significantly lower than that in the sh-NC + 6 Gy group, while this situation was partly reversed in the sh-PCAT1 + miR-128 inhibitor + 6 Gy group and sh-PCAT1 + pcDNA-GOLM1 + 6 Gy group (P < 0.05, Figure 6B). Wound healing assay and transwell assay showed the similar results: the migration ability and invasion ability of CC cells in the sh-PCAT1 + 6 Gy group was dramatically decreased compared with the sh-NC + 6 Gy group. Downregulation of miR-128 and upregulation of GOLM1 both reversed the enhanced effect of PCAT1 knockdown on radiosensitivity of CC cells, which partly promoted the migration ability and invasion ability of CC cells (P < 0.05, Figure 6C and D).
Figure 6.
PCAT1 knockdown enhances radiosensitivity of CC cells on proliferation, migration and invasion by regulating miR-128/GOLM1 in vitro. (A) The protein level of GOLM1 was measured by Western blot assay. **P < 0.01 vs the pcDNA-NC group. (B) The viability of Hela cells was determined by MTT assay. (C) The migration ability of CC cells was measured by wound healing assay. (D) The invasion number of CC cells was measured by transwell assay. *P < 0.05; **P < 0.01 vs sh-NC + 6 Gy group. #P < 0.05 vs sh-PCAT1 + 6 Gy group.
PCAT1 Knockdown Enhances Radiosensitivity of CC by Regulating miR-128/GOLM1 in vivo
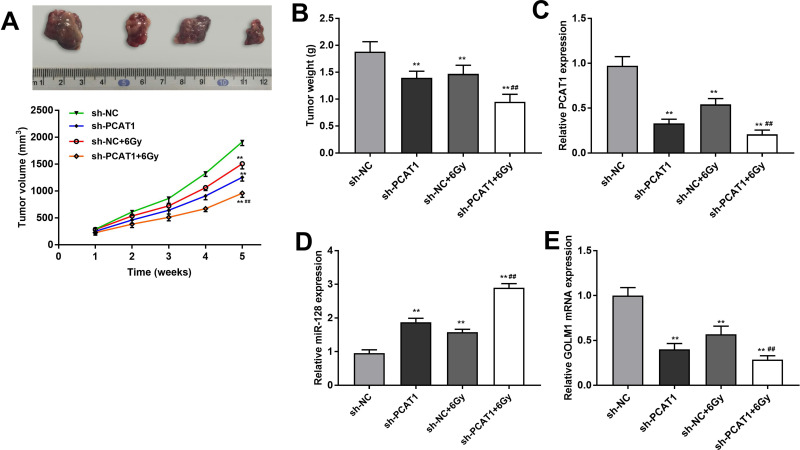
In order to evaluate the inhibitory effect of PCAT1 on the tumor growth of CC in vivo, CaSki cells transfected with sh-PCAT1/NC were inoculated into mice subcutaneously. The tumor volume and weight were obviously smaller or lighter in sh-PCAT1 group and sh-NC + 6 Gy group than that in the sh-NC group, while this situation was further declined in the sh-PCAT1 + 6 Gy group (P < 0.01, Figure 7A and B). The results implied that the antitumor effect of radiation was significantly strengthened by PCAT1 knockdown. In addition, the expression of PCAT1, miR-128 and GOLM1 was detected in tumor tissues under radiation treatment (6 Gy) after injection of sh-PCAT1/NC into mice. The results showed that both silencing of PCAT1 and treatment with radiation could downregulate the expression of PCAT1 and GOLM1 and upregulate the expression of miR-128. The expression of PCAT1 and GOLM1 was further reduced in the sh-PCAT1 + 6 Gy group compared with the sh-NC + 6 Gy group. On the contrary, the expression of miR-128 was dramatically increased in the sh-PCAT1 + 6 Gy group in contrast to that in the sh-NC + 6 Gy group (P < 0.01, Figure 7C–E).
Figure 7.
PCAT1 knockdown enhances radiosensitivity of CC by regulating miR-128/GOLM1 in vivo. (A) The volume of tumor tissues after injection of sh-PCAT1/NC under X-ray (6 Gy). (B) The weight of tumor tissues after injection of sh-PCAT1/NC under X-ray (6 Gy). (C) The expression of PCAT1 in CC tissues was detected by qRT-PCR. (D) The expression of miR-128 in CC tissues was detected by qRT-PCR. (E) The expression of GOLM1 in CC tissues was detected by qRT-PCR. **P < 0.01 vs sh-NC group. ##P < 0.01 vs sh-NC + 6 Gy group.
Discussion
Among females, CC is the fourth most common cancer and also the fourth leading cause of death.19 LncRNAs are participated in the progression of several cancers, including CC.7 Song et al have displayed that the expression of OIP5-AS1 is dramatically upregulated in CC cells, which is significantly related to lymph node metastasis and tumor stage.20 Hu et al have uncovered that CAR10 expression in CC tissues and cells is markedly upregulated and upregulation of CAR10 is closely related to lymph node metastasis.21 In line with these results, in our study, we demonstrated that PCAT1 expression was significantly higher in tumor tissues and PCAT1 upregulation was positively correlated with tumor stage, lymph node metastasis and depth of cervical invasion. Hence, PCAT1 might act as a pathogenic factor in CC.
Prostate cancer-associated transcript 1 (PCAT1) is first described as a prostate-specific regulator in 2011 and contributes to cancer progression by promoting cell proliferation.22 Huang et al have demonstrated that overexpression of PCAT1 markedly increases proliferation of ESCC cells, thus promoting tumor growth of ESCC.23 Prensner et al have disclosed that lncRNA PCAT1 expression is increased in prostate cancer and promotes invasion and metastasis of prostate cancer cells.22 Sur et al have indicated that knockdown of PCAT1 in head and neck cancer (HNSCC) cells inhibits tumor growth and induces cell apoptosis.24 The results in our study concur with previous researches. In the current study, the data showed that silencing of PCAT1 inhibited proliferation, migration and invasion of CC cells. The results implied that PCAT1 knockdown attenuated the development of CC in vitro.
Guidelines from the NCCN (version 1.2020) recommend radiation therapy forms the basis of treatment of patients with CC.25 Radioresistance is one of the most important reasons for the failure or ineffectiveness of CC radiation therapy.26,27 Previous studies have found that the abnormal expression of LncRNAs has an important effect on the radioresistance of cancer.6–8 The radiosensitivity of LAD is enhanced by knockdown of lncRNA LINC00483.5 LncRNA RI silencing increases radiosensitivity of colorectal cancer (CRC).28 Zhang et al have demonstrated that PCAT1 knockdown strengthens the promoting effect of radiation on apoptosis of glioma stem cells.8 Similarly, the current study showed that silencing of PCAT1 markedly enhanced the inhibiting effect of radiation therapy on CC cells. Besides, PCAT1 silencing further reduced the volume and weight of CC tissues in radiation-treated mice. The results implied that PCAT1 knockdown enhanced radiosensitivity of CC in vivo and vitro.
MiRNAs are involved in pathological processes of many cancers including breast cancer, colorectal cancer, lung cancer, CC and others.29–32 As a member of miRNAs family, miR-128 plays a vital role in cancers. MiR-128 is downregulated in CC cells and miR-128 upregulation suppresses proliferation, invasion and metastasis in CC cells.33 Overexpression of miR-128 enhanced the chemosensitivity of oxaliplatin in colorectal cancer.34 MiR-128 upregulation significantly increases radiosensitivity of GSC and glioblastoma lines.12 In line with the above results, in our study, we found that the expression of miR-128 was downregulated in CC tissues and cells. Overexpression of miR-128 significantly increased the inhibitory effect of radiation therapy on CC cells. Besides, miR-128 was proved to be the target of PCAT1 and negatively regulated by PCAT1. Downregulation of miR-128 reversed the enhanced effect of PCAT1 knockdown on radiosensitivity of CC cells. The results suggested PCAT1 knockdown enhanced radiosensitivity of CC cells by regulating miR-128.
Researches have revealed that GOLM1 plays a vital role in many cancers, including hepatocellular carcinoma, prostate cancer and CC.35–37 GOLM1 silencing inhibits the migration and invasion of U251 cells and U87 cells in glioma.38 GOLM1 siRNA reverses the promoting effect of miR-143 inhibitor on cell migration of CC.37 Besides, downregulation of GOLM1 has been reported to enhance the methotrexate chemosensitivity of CC.18 In the present study, GOLM1 was highly expressed in CC tissues. GOLM1 was identified to be the direct target gene of miR-128 and was negatively modulated by miR-128. In addition, upregulation of GOLM1 reversed the enhanced effect of PCAT1 knockdown on radiosensitivity of CC cells. In a word, the results implied that PCAT1 knockdown enhanced radiosensitivity of CC cells by regulating miR-128/GOLM1.
In conclusion, the present study displayed that PCAT1 knockdown could enhance radiosensitivity of CC, thus inhibiting the growth of CC both in vivo and in vitro. However, a limitation may exist in this study. We did not deeply investigate the detailed regulatory mechanism between PCAT1 and miR-128/GOLM1. Further studies to elucidate this issue will be performed in the future. Nevertheless, we also hope these findings will provide a new strategy for treating CC.
Funding Statement
There is no funding to report.
Data Sharing Statement
All data are included in this article.
Ethics Approval and Informed Consent
The protocols of this study were reviewed and approved by ethical committee in Yantaishan Hospital (approval ID: YSLZ2020026). Each patient in this study obtained the written informed consent. And animal experiment obtained the approval of ethical committee in our hospital (approval ID: YSLZ2020026) and performed in the Animal Experimental Center of our hospital.
Consent for Publication
All participants agreed to publish the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors disclosed that they have no competing interests.
References
- 1.Daniel A, Ganesan S, Joseph L. Biochemical assessment of human uterine cervix by micro-Raman mapping. Photodiagnosis Photodyn Ther. 2017;17:65–74. doi: 10.1016/j.pdpdt.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 2.Lin C-L, Lee C-H, Chen C-M, et al. Protodioscin induces apoptosis through ROS-mediated endoplasmic reticulum stress via the JNK/p38 activation pathways in human cervical cancer cells. Cell Physiol Biochem. 2018;46(1):322–334. doi: 10.1159/000488433 [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Shen L, Chen X, et al. High expression of PKM2 as a poor prognosis indicator is associated with radiation resistance in cervical cancer. Histol Histopathol. 2015;30(11):1313–1320. [DOI] [PubMed] [Google Scholar]
- 4.Chen M, Liu P, Chen Y, et al. Long noncoding RNA FAM201A mediates the radiosensitivity of esophageal squamous cell cancer by regulating ATM and mTOR expression via miR-101. Front Genet. 2018;9:611. doi: 10.3389/fgene.2018.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang QS, Li B, Xu G, et al. Long noncoding RNA LINC00483/microRNA‐144 regulates radiosensitivity and epithelial–mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol. 2019;234(7):11805–11821. doi: 10.1002/jcp.27886 [DOI] [PubMed] [Google Scholar]
- 6.Li N, Meng D-D, Gao L, et al. Overexpression of HOTAIR leads to radioresistance of human cervical cancer via promoting HIF-1α expression. Radiat Oncol. 2018;13(1):210. doi: 10.1186/s13014-018-1153-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma TT, Zhou LQ, Xia JH, Shen Y, Yan Y, Zhu RH. LncRNA PCAT-1 regulates the proliferation, metastasis and invasion of cervical cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(7):1907–1913. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P, Liu Y, Fu C, et al. Knockdown of long non-coding RNA PCAT1 in glioma stem cells promotes radiation sensitivity. Med Mol Morphol. 2019;52(2):114–122. doi: 10.1007/s00795-018-0209-8 [DOI] [PubMed] [Google Scholar]
- 9.Pizzini S, Bisognin A, Mandruzzato S, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589. doi: 10.1186/1471-2164-14-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Zhao H, Xin Y, Fan L. MicroRNA-198 inhibits proliferation and induces apoptosis of lung cancer cells via targeting FGFR1. J Cell Biochem. 2014;115(5):987–995. doi: 10.1002/jcb.24742 [DOI] [PubMed] [Google Scholar]
- 11.Zheng R, Liu Y, Zhang X, Zhao P, Deng Q. miRNA-200c enhances radiosensitivity of esophageal cancer by cell cycle arrest and targeting P21. Biomed Pharmacother. 2017;90:517–523. [DOI] [PubMed] [Google Scholar]
- 12.Peruzzi P, Bronisz A, Nowicki MO, et al. MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol. 2013;15(9):1212–1224. doi: 10.1093/neuonc/not055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang D, Sun G, Zhang H, Wang X, Li Y. Long non-coding RNA PVT1 facilitates cervical cancer progression through negative modulation of miR-128-3p. Int J Clin Exp Pathol. 2017;10(4):4522–4529. [Google Scholar]
- 14.Kim H-J, Lv D, Zhang Y, Peng T, Ma X. Golgi phosphoprotein 2 in physiology and in diseases. Cell Biosci. 2012;2(1):31. doi: 10.1186/2045-3701-2-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Li L, Hu L, Peng T. Golgi phosphoprotein 2 (GOLPH2/GP73/GOLM1) interacts with secretory clusterin. Mol Biol Rep. 2011;38(3):1457–1462. doi: 10.1007/s11033-010-0251-7 [DOI] [PubMed] [Google Scholar]
- 16.Aruna L. Overexpression of golgi membrane protein 1 promotes non-small-cell carcinoma aggressiveness by regulating the matrix metallopeptidase 13. Am J Cancer Res. 2018;8(3):551–565. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Xu Y, Zhang X. Targeting GOLM1 by microRNA-200a in melanoma suppresses cell proliferation, invasion and migration via regulating PI3K/Akt signaling pathway and epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2019;23(16):6997–7007. [DOI] [PubMed] [Google Scholar]
- 18.Li RM, Man MN, Duan SJ, Li SX, Song WY. Down-expression of GOLM1 enhances the chemo-sensitivity of cervical cancer to methotrexate through modulation of the MMP13/EMT axis. Am J Cancer Res. 2018;8(6):964–980. [PMC free article] [PubMed] [Google Scholar]
- 19.Bray F, Jacques J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. [DOI] [PubMed] [Google Scholar]
- 20.Song L, Wang L, Pan X, Yang C. lncRNA OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit cell apoptosis through a mechanism involving miR-143-3p in cervical cancer. Braz J Med Biol Res. 2020;53(1):e8883. doi: 10.1590/1414-431x20198883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu T, Zhang Q, Gao L. LncRNA CAR10 upregulates PDPK1 to promote cervical cancer development by sponging miR-125b-5p. Biomed Res Int. 2020;2020:4351671. doi: 10.1155/2020/4351671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Wang Y, Chen J, Wang Y, Liu X. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10(7). doi: 10.1038/s41419-019-1745-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sur S, Nakanishi H, Steele R, Ray RB. Depletion of PCAT-1 in head and neck cancer cells inhibits tumor growth and induces apoptosis by modulating c-Myc-AKT1-p38 MAPK signalling pathways. BMC Cancer. 2019;19(1):019–5562. doi: 10.1186/s12885-019-5562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Rustum NR, Yashar CM, Bean S, et al. NCCN guidelines insights: cervical cancer, version 1.2020. J Natl Compr Canc Netw. 2020;18(6):660–666. doi: 10.6004/jnccn.2020.0027 [DOI] [PubMed] [Google Scholar]
- 26.Benedet J, Odicino F, Maisonneuve P, et al. Carcinoma of the cervix uteri. J Epidemiol Biostat. 2001. [PubMed] [Google Scholar]
- 27.Su WH, Chuang PC, Huang EY, Yang KD. Radiation-Induced Increase in cell migration and metastatic potential of cervical cancer cells operates via the K-ras pathway. Am J Pathol. 2012;180(2):0–871. doi: 10.1016/j.ajpath.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Zhang Q, Shen L, Chen S, Wang Z. Long noncoding RNA lnc-RI regulates DNA damage repair and radiation sensitivity of CRC cells through NHEJ pathway. Cell Biol Toxicol. 2020;3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang S, Yin J, Xu R. MiR-566 mediates cell migration and invasion in colon cancer cells by direct targeting of PSKH1. Cancer Cell Int. 2019;19(1). doi: 10.1186/s12935-019-1053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Wang Z, Wang Q, Cui Y, Luo S. Clinical significance of the expression of miRNA-21, miRNA-31 and miRNA-let7 in patients with lung cancer. Saudi J Biol Sci. 2019;26(4):777–781. doi: 10.1016/j.sjbs.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Li D, Fang L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947. doi: 10.1016/j.biopha.2019.108947 [DOI] [PubMed] [Google Scholar]
- 32.Park S, Kim J, Eom K, et al. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. Bmc Cancer. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Guo L, Liang Z, Liu Y, Yao Z. Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/beta-catenin pathway in cervical cancer cells. J Cancer. 2020;11(8):2201–2212. doi: 10.7150/jca.40319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi: 10.1186/s12943-019-0981-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Ai N, Liu W, Li ZG, Ji H, Li B, Yang G. High expression of GP73 in primary hepatocellular carcinoma and its function in the assessment of transcatheter arterial chemoembolization. Oncol Lett. 2017;14(4):3953–3958. doi: 10.3892/ol.2017.6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varambally S, Laxman B, Mehra R, et al. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia. 2008;10(11):1285–1294. doi: 10.1593/neo.08922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou M, Chen X, Wu J, et al. MicroRNA-143 regulates cell migration and invasion by targeting GOLM1 in cervical cancer. Oncol Lett. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding X, Deng G, Liu J, et al. GOLM1 silencing inhibits the proliferation and motility of human glioblastoma cells via the Wnt/β-catenin signaling pathway. Brain Res. 2019. [DOI] [PubMed] [Google Scholar]