C. trachomatis infection is the most common bacterial sexually transmitted infection, and infection in women can lead to pelvic inflammatory disease and infertility. No licensed vaccine exists to prevent C. trachomatis infection, and investigations of the natural immune response may inform the design of targeted vaccines for C. trachomatis. Our study fills a gap in knowledge regarding the epitope specificity of antibody responses that are elicited in response to C. trachomatis infection in women. We identified several new B cell epitopes for C. trachomatis antigens and confirmed B cell epitopes that have been identified by other methods. Our finding that women produce antibodies to the VD4-MOMP regardless of infection outcome provides insight into vaccine development, suggesting that vaccines targeting VD4-MOMP may need to elicit higher-titer antibody responses than natural infection imparts or that additional vaccine targets should be pursued in the future.
KEYWORDS: Chlamydia, VLP, affinity selection, antibody, bacteriophage, biopanning, epitope, vaccine, virus-like particle
ABSTRACT
Chlamydia trachomatis is an obligate intracellular bacterium. C. trachomatis infection is the most prevalent bacterial sexually transmitted infection and can lead to pelvic inflammatory disease and infertility in women. There is no licensed vaccine for C. trachomatis prevention, in part due to gaps in our knowledge of C. trachomatis-specific immune responses elicited during human infections. Previous investigations of the antibody response to C. trachomatis have identified immunodominant antigens and antibodies that can neutralize infection in cell culture. However, epitope-specific responses to C. trachomatis are not well characterized, and the impact of these antibodies on infection outcome is unknown. We recently developed a technology called deep sequence-coupled biopanning that uses bacteriophage virus-like particles to display peptides from antigens and affinity select against human serum IgG. Here, we used this technology to map C. trachomatis-specific antibodies in groups of women with defined outcomes following C. trachomatis infection: (i) C. trachomatis negative upon presentation for treatment (“spontaneous resolvers”), (ii) C. trachomatis negative at a 3-month follow-up visit after treatment (“nonreinfected”), and (iii) C. trachomatis positive at a 3-month follow-up after treatment (“reinfected”). This analysis yielded immunodominant epitopes that had been previously described but also identified new epitopes targeted by human antibody responses to C. trachomatis. We focused on human antibody responses to the C. trachomatis variable domain 4 serovar-conserved region of the major outer membrane protein (VD4-MOMP), a previously described immunodominant epitope. All three groups of women produced IgG to the VD4-MOMP, suggesting that detection of serum antibodies to VD4-MOMP in women with urogenital C. trachomatis infection is not associated with protection against reinfection.
IMPORTANCE C. trachomatis infection is the most common bacterial sexually transmitted infection, and infection in women can lead to pelvic inflammatory disease and infertility. No licensed vaccine exists to prevent C. trachomatis infection, and investigations of the natural immune response may inform the design of targeted vaccines for C. trachomatis. Our study fills a gap in knowledge regarding the epitope specificity of antibody responses that are elicited in response to C. trachomatis infection in women. We identified several new B cell epitopes for C. trachomatis antigens and confirmed B cell epitopes that have been identified by other methods. Our finding that women produce antibodies to the VD4-MOMP regardless of infection outcome provides insight into vaccine development, suggesting that vaccines targeting VD4-MOMP may need to elicit higher-titer antibody responses than natural infection imparts or that additional vaccine targets should be pursued in the future.
INTRODUCTION
In the United States, over 1.75 million Chlamydia trachomatis infections were reported in 2018 (1) with associated annual direct medical costs exceeding $500 million/year (2). The majority of C. trachomatis infections are in young women and are commonly asymptomatic, which may lead to a delay in infected women receiving curative antibiotic treatment. However, approximately 5 to 15% of C. trachomatis-infected women experience serious sequelae caused by ascension of C. trachomatis into the upper genital tract, including pelvic inflammatory disease (PID), ectopic pregnancy, and infertility (3). The World Health Organization (WHO) and the National Institute of Allergy and Infectious Diseases (NIAID) recently published recommendations for the development of a prophylactic vaccine against C. trachomatis (4, 5). These included a call for a better understanding of the specificity of the immune response to C. trachomatis during urogenital infection.
Animal models of C. trachomatis infection suggest that antibodies may play a role in protection from C. trachomatis reinfection, but are not required for C. trachomatis clearance. Animal models show that antibodies to particular antigens and epitopes may provide protection (6–15). Rabbit IgG specific for serovar L2 major outer membrane protein (MOMP) (6), and some MOMP monoclonal antibodies have in vitro neutralizing activity. In addition to neutralizing activity, in vitro and in vivo studies utilizing the mouse Chlamydia muridarum model show that polyclonal antibodies to C. muridarum elementary bodies (EBs) may provide protection by facilitating phagocytosis and killing of Chlamydia by gamma interferon (IFN-γ)-stimulated neutrophils (7). The specificity of these antibodies has not been determined. In addition, spontaneous resolution of C. trachomatis in ocular infections of nonhuman primates is associated with antibodies specific for C. trachomatis proteins PmpD, HSP60, CPAF, and Pgp3 (8). Together, these studies suggest that neutralization and opsonization leading to phagocytic killing are possible functions of C. trachomatis-specific antibodies, but the specificity of the antibodies is likely important.
In humans, the role of antibodies in protecting against urogenital C. trachomatis infection remains to be fully elucidated. To date, the only identified correlate of protection against C. trachomatis infection in humans is IFN-γ-producing CD4+ T cells, which have been shown to be associated with decreased frequency of C. trachomatis incidence and reinfection (9–11). Anti-C. trachomatis IgG and IgA are inversely correlated with cervical C. trachomatis burden, suggesting that antibodies may control C. trachomatis at the initial site of infection, but these antibodies are insufficient to prevent the ascension of C. trachomatis into the upper genital tract in women (12). In addition, women are commonly reinfected with the same C. trachomatis serovar, suggesting that serovar-specific anti-MOMP antibodies elicited during urogenital infection do not provide complete protective immunity (13). Finally, total serum C. trachomatis antibody levels are positively associated with the severity of tubal damage in infertile women (14). Specific antibody responses are associated with morbidity in women. Among women with chlamydia PID, those with IgG to C. trachomatis protein PmpA were less likely to become pregnant and less likely to report a live birth, while women with IgG to PmpI were more likely to have an upper genital tract infection (15). Tubal infertility is associated with IgG to C. trachomatis proteins HSP60, CT557, and CT443 (16). In addition, IgG3 to EBs is associated with lower rates of pregnancy and live births but higher rates of ectopic pregnancy (17). Whether these antibodies directly cause morbidity or are merely correlated with repeated infection is unclear. Together, these studies highlight the need for a more comprehensive assessment of antibody specificity during C. trachomatis infection.
The objectives of this study were to finely map the specificity of human antibodies elicited in response to urogenital C. trachomatis infection, empirically identify C. trachomatis B cell epitopes, and determine the association of antibodies with C. trachomatis infection outcomes (18, 19). We used a technology we developed called deep sequence-coupled biopanning (DSCB) to perform a global assessment of antibody responses to specific epitopes of 24 C. trachomatis antigens in sera from female patients with a history of C. trachomatis infection and different infection outcomes. We then performed a more in-depth assessment of the human antibody response to MOMP, due to its importance as a vaccine target (20).
RESULTS
Participant characteristics.
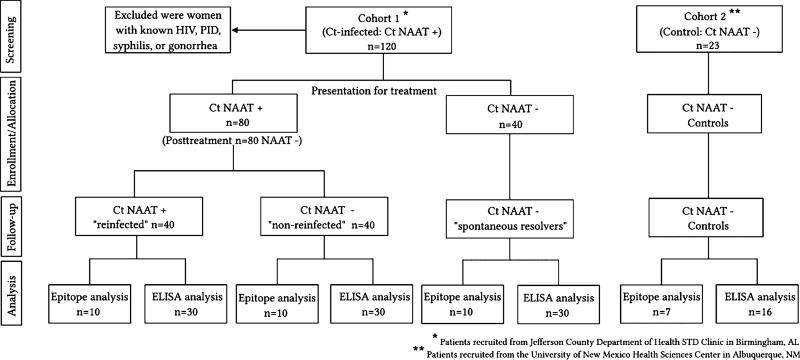
We evaluated sera from 120 women in cohort 1 who had one of three defined outcomes following a C. trachomatis infection: “spontaneous resolvers” (n = 40), “nonreinfected” (n = 40), or “reinfected” (n = 40). Of the 120 women evaluated in cohort 1, the median age at enrollment was 22 years (range, 16 to 41), and most were African-American race (99.2%) and non-Hispanic ethnicity (97.5%). Prior C. trachomatis infection (by self-report and/or review of medical records) was documented in 57.1%, and concomitant vaginal infections at enrollment were not uncommon: bacterial vaginosis in 25%, candidiasis in 10.8%, and trichomoniasis in 5.0%. For initial epitope mapping experiments, we utilized sera from 10 women from each outcome group of cohort 1 along with sera from 7 women in cohort 2. Sera from the remaining 30 cohort 1 participants from each outcome group and the remaining 16 cohort 2 participants were utilized to confirm findings by enzyme-linked immunosorbent assay (ELISA). A consort diagram graphically representing the assignment of patients to each group is shown in Fig. 1.
FIG 1.
Consort diagram. Labeling: Ct, C. trachomatis.
DSCB identifies MOMP as the most commonly selected antigen.
After constructing a bacteriophage MS2 virus-like particle (VLP) library displaying overlapping 6- to 10-amino-acid (aa) peptides from 24 C. trachomatis antigens, we used DSCB (see Fig. S1 in the supplemental material) to assess the specific epitopes recognized by antibodies in 10 patients from each outcome group in cohort 1 and 7 patients from cohort 2. Table 1 shows the 24 C. trachomatis antigens in decreasing order of total number of selections across all cohort 1 patients and also lists the most commonly selected epitope(s) from each individual antigen. MOMP was the most commonly selected antigen from our MS2-VLP C. trachomatis antigen fragment library (MS2-VLP-Ct-AFL). Two epitopes from MOMP were commonly selected by cohort 1 patients, not differing by the outcome group: CD4/VD3 (aa 233 to 250) and VD4 conserved region (aa 314 to 329). Indeed, 26/30 (86.7%) and 21/30 (70%) cohort 1 patients selected the CD4/VD3 region and VD4 conserved region, respectively (Table 1). Since the CD4/VD3 and VD4 regions were highly selected via DSCB compared to the other antigens, we subsequently focused on investigating these antibody responses.
TABLE 1.
Antigens included in the bacteriophage MS2 virus-like particle C. trachomatis antigen fragment library for DSCB and most commonly selected epitope(s) from DSCB
|
C. trachomatis
|
Epitope(s) most selected | Amino acid position(s) | No. of individual selections | No. of patients selecting | Infection outcomea |
|||
|---|---|---|---|---|---|---|---|---|
| Antigen | No. | SR | NR | R | ||||
| Major outer membrane protein (MOMP) | CT681 | EFTINKPKGYVGKEFPLD | 233–250 | 80 | 26/30 | 7 | 9 | 10 |
| FDTTTLNPTIAGAGDV | 314–329 | 21/30 | 6 | 10 | 5 | |||
| 60-kDa cysteine-rich OMP (OmcB) | CT443 | ATGPKQDSCFGRMY | 86–99 | 34 | 19/30 | 6 | 7 | 6 |
| Tarp | CT456 | NIYESIGGSRTSGPEN | 171–184, 219–234, 271–284b | 30 | 14/30 | 3 | 6 | 5 |
| Solute protein-binding family (metal ion transport protein) | CT067 | MNSCSSSRGNQPADESIY | 25–42 | 28 | 17/30 | 7 | 6 | 4 |
| Putative outer membrane protein C (PmpC) | CT414 | LEGSQGDTADTGT | 647–659 | 24 | 11/30 | 5 | 5 | 1 |
| Hypothetical protein | CT147 | WHNQYQM | 1423–1429 | 17 | 15/30 | 7 | 2 | 6 |
| CT875 | CT875 | DPLGRRTPNYQSKNP | 229–243 | 17 | 11/30 | 3 | 3 | 5 |
| Putative outer membrane protein D (PmpD) | CT812 | ASEDGDLSPE | 764–773 | 15 | 10/30 | 4 | 4 | 2 |
| Hypothetical protein pCHL1p5 (plasmid) (Pgp3) | pCT03 | RTSITNTGLT | 208–217 | 14 | 9/30 | 5 | 2 | 2 |
| Inclusion membrane protein A | CT119 | ESSDLCSQIR | 247–256 | 13 | 10/30 | 4 | 2 | 4 |
| Hypothetical protein | CT143 | QAYNCATHRNG | 123–133 | 13 | 9/30 | 3 | 4 | 2 |
| L31 ribosomal protein | CT022 | CGSTYQTDKT | 25–34 | 12 | 11/30 | 5 | 3 | 3 |
| CHLPN 76-kDa homolog | CT622 | LKQEHTGLTD | 283–292 | 12 | 9/30 | 5 | 2 | 2 |
| Hypothetical protein | CT695 | RTTSSSGVSED | 57–67 | 11 | 9/30 | 3 | 3 | 3 |
| Aspartyl tRNA synthetase | CT542 | NQALDHLRRL | 403–412 | 11 | 9/30 | 4 | 3 | 2 |
| Hypothetical protein | CT795 | LIYPPV | 105–110 | 9 | 9/30 | 3 | 1 | 5 |
| CQQEAEEDC | 27–35 | 7/30 | 5 | 1 | 1 | |||
| Arginine binding protein (ArtJ) | CT381 | TGCLKEGGDS | 22–31 | 9 | 9/30 | 4 | 3 | 2 |
| HSP60 | CT110 | EDEQIGARIV | 436–445 | 9 | 7/30 | 4 | 1 | 2 |
| Hypothetical protein | CT101 | SMLKQHELDA | 65–74 | 8 | 8/30 | 2 | 4 | 2 |
| Thio-specific antioxidant (TSA) peroxidase | CT603 | VINDLPLGRSI | 138–149 | 8 | 7/30 | 2 | 2 | 3 |
| Recombination protein | CT240 | ISQTSPCNF | 66–74 | 8 | 7/30 | 5 | 2 | 0 |
| SPITGKHLS | 114–122 | 7/30 | 2 | 2 | 3 | |||
| DO serine protease | CT823 | LNQVLKNSKGENV | 466–477 | 8 | 6/30 | 3 | 1 | 2 |
| ATP-dependent zinc metalloprotease | CT841 | LSKEVRNAIT | 118–127 | 7 | 7/30 | 3 | 4 | 0 |
| PGRFDRRV | 584–591 | 7/30 | 3 | 3 | 1 | |||
| FKBP-type peptidyl-prolyl cis-trans isomerase | CT541 | ALTDNQKLS | 40–48 | 5 | 5/30 | 2 | 1 | 2 |
| IKGMQSEI | 73–80 | 5/30 | 2 | 1 | 2 | |||
| GMQGMKEGE | 193–201 | 3/30 | 2 | 0 | 1 | |||
SR, spontaneous resolver; NR, nonreinfected; R, reinfected.
Multiple amino acid positions are indicated for a single peptide sequence because this is a repeated sequence in the Tarp antigen.
Schematic of DSCB technology workflow. Download FIG S1, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CD4/VD3 and VD4 region of MOMP are commonly selected epitopes regardless of outcome group.
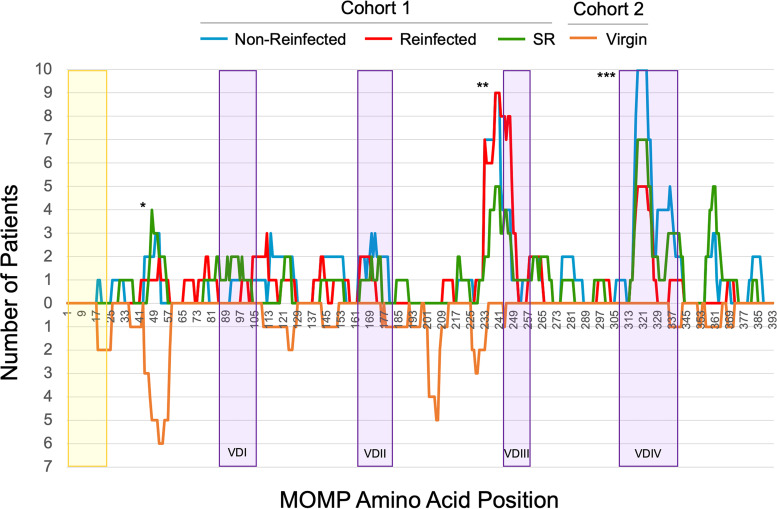
In order to investigate MOMP-specific epitopes in the context of specific outcome groups, we determined the number of patients in each outcome group that selected epitopes of MOMP (Fig. 2). CD4/VD3 (aa 225 to 253, labeled “**”) and VD4 (aa 313 to 328, labeled “***”), including a serovar conserved region (aa 319 to 325, LNPTIAG), were commonly selected by cohort 1 patients, regardless of outcome. There were 27 of 30 cohort 1 patients (10 nonreinfected, 7 spontaneous resolvers, and 10 reinfected patients) whose serum antibodies identified peptides within the CD4/VD3 region, and 23 of 30 cohort 1 patients (7 nonreinfected, 10 spontaneous resolvers, and 6 reinfected patients) whose serum antibodies identified peptides within the VD4 conserved epitope. Cohort 2 patient sera (n = 7) showed no substantial increased selection of the CD4/VD3 or VD4 regions observed for these patients. Similar graphs were generated for the other 23 antigens (see Fig. S2 to S10 in the supplemental material).
FIG 2.
DSCB results for MOMP from sera of female patients with variable clinical outcomes. The numbers of patients selecting peptides at each amino acid of the MOMP at ≥10-fold above the starting library, with experimental patient groups pointing in the positive y axis and virgin controls pointing in the negative y axis, are shown. The yellow shaded region represents the signal sequence of MOMP. The purple shaded regions represent the four variable domains of MOMP (VDI, VDII, VDIII, and VDIV). Labeling: *, a region with nonspecific antibody binding; **, a commonly selected region (CD4/VD3); ***, another commonly selected region, the VD4 conserved region.
DSCB results for the OmcB (CT443) from sera of female patients with variable infection outcomes. Download FIG S2, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MOMP epitopes are represented by overlapping peptide sequences.
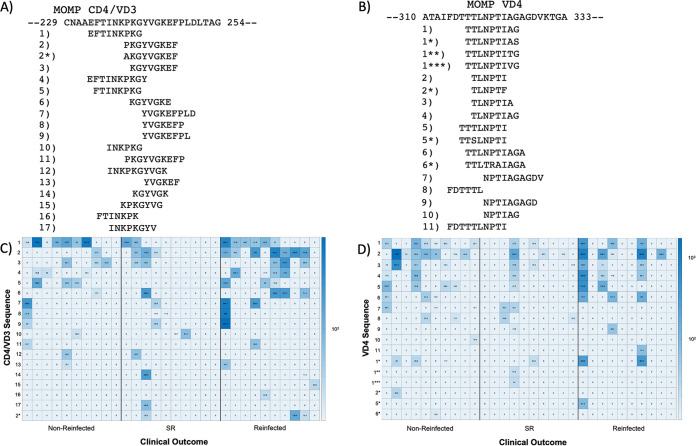
The MS2-VLP-Ct-AFL was constructed using multiple overlapping 6 to 10 amino acid peptide sequences, allowing for fine-mapping of antibody specificity. Notably, peptide sequences from the CD4/VD3 and VD4 conserved region of MOMP were identified by sera from a majority of cohort 1 patients (26 of 30 patients and 21 of 30 patients, respectively; Table 1). Figure 3 shows the peptides identified for both CD4/VD3 and VD4. The CD4/VD3 peak was composed of 17 different total peptides with two distinct core epitopes (FTINKPK and KGYVGKEF) that overlapped to comprise the CD4/VD3 region (Fig. 3A). The top ranked peptides for the FTINKPK and KGYVGKEF core epitopes, regardless of the fold change increase over the starting library, were numbers 1 and 2, respectively (Fig. 3C). The VD4 peak consisted of 11 different peptides encompassing a single core epitope (NPTI) and flanking sequence selections (Fig. 3B). For the VD4 region, three individual peptide sequences (1, 2, and 3) made up over half of the total peptides identified by the patient serum antibodies (Fig. 3D). Number 1, along with unexpected amino acid substitutions, is the highest ranked peptide selection. However, number 2 comprises the three highest fold changes across all selections (Fig. 3D) and is the top ranked peptide according to fold change, with 12 of 21 patient serum antibodies that recognize VD4 MOMP identifying this particular peptide as their highest fold change selection.
FIG 3.
CD4/VD3 region and VD4 region of MOMP are represented by overlapping peptides. The CtsvD sequences for a portion of the CD4/VD3 region (A) and the VD4 region (B) are listed at the top with patient sequences selections below with a corresponding letter, in decreasing order of selection number. Peptides with unexpected amino acid substitutions were also selected, as noted by the asterisks listed. Heat maps show selections made by individual patients in each group, with higher fold changes represented by a darker color for CD4/VD3 (C) and VD4 (D).
Antibody response toward the VD4 conserved region of MOMP does not differ among patient outcome groups.
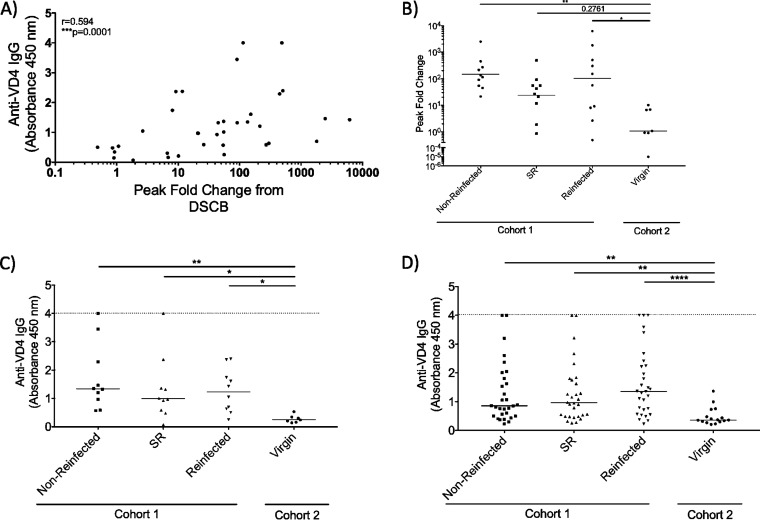
Since the VD4 conserved region of MOMP has been pursued as a vaccine target (20), we were interested in investigating the antibody response to this specific region in our study population. Using ELISA against a VD4 synthetic peptide corresponding to aa 313 to 325, we found that the DSCB-identified peak fold changes correlated with peptide-specific ELISA optical density at 450 nm (OD450) values against this same peptide (Fig. 4A). We next compared the cohort 1 patient groups with a favorable outcome (spontaneous resolver and nonreinfected) versus the group who were reinfected with C. trachomatis by analyzing peak fold change and anti-VD4 IgG separately. There was no difference in the geometric mean peak fold change between the cohort 1 outcome groups, although nonreinfected and reinfected patients had peak fold changes above that of cohort 2 patients (Fig. 4B). In addition, we found that all cohort 1 patient groups had anti-VD4 IgG levels above that of cohort 2 but did not differ among cohort 1 patient groups (Fig. 4C). Finally, we independently verified this finding utilizing sera from each of the 30 remaining cohort 1 outcome groups not previously used in DSCB, assessing these sera for anti-VD4 IgG by ELISA. Again, we found that all cohort 1 outcome groups had anti-VD4 IgG levels that were higher than cohort 2 patients but were not statistically different from one another (Fig. 4D).
FIG 4.
Patients with different clinical outcomes have comparable antibody responses toward the VD4 region. (A) Correlation of peak fold changes against the VD4 conserved region and ELISA readings from virgin patients and all clinical outcome groups. (B) The peak fold change for each patient was determined within the VD4 region and geometric mean between patient groups were compared. (C) OD450 determined by bait-peptide ELISA to measure patient antibody responses against the VD4 conserved region (FDTTTLNPTIAGAGDVK). Patient sera utilized from DSCB experiments, with duplicates performed and values averaged after subtraction of background. (D) OD450 determined via bait-peptide ELISA to measure additional patient antibody responses against the VD4 conserved region. A nonparametric Kruskal-Wallis multiple-comparison test was used for ELISA and peak fold change analysis. A nonparametric Spearman test was used for correlation analysis. ****, P < 0.0001; ***, P < 0.0001; **, P < 0.01; *, P < 0.05.
IgG1 and IgG3 subclasses produced in response to the VD4 conserved region of MOMP are not different among patient groups, regardless of outcome group.
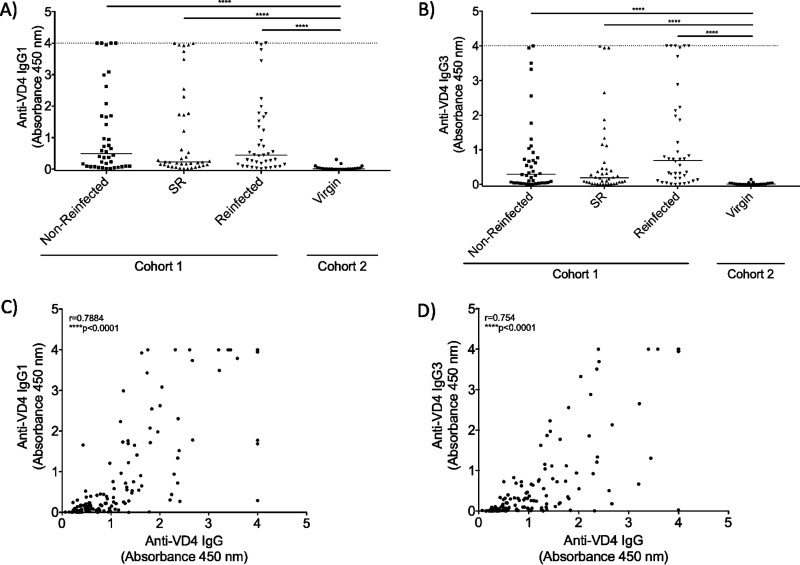
Patients with active C. trachomatis infection are seropositive for IgG1 and IgG3 in an EB ELISA (21). Since EB ELISA can detect antibodies to all EB-expressed antigens and epitopes, we wanted to know whether IgG1 and IgG3 antibodies are both produced to the VD4 MOMP. To test this, we performed ELISAs against the same synthetic VD4 MOMP peptide mentioned above (aa 313 to 325) and used secondary antibodies specific for either IgG1 or IgG3. We found that all cohort 1 outcome groups had a magnitude of OD readings higher than that of cohort 2 patients (supporting the specificity of the assay), but were not significantly different from one another (Fig. 5A and B). The magnitude of the OD readings for total IgG correlated with both IgG1 and IgG3 independently (Fig. 5C and D), suggesting that no patient group preferentially produced IgG1 or IgG3 toward the VD4 conserved region.
FIG 5.
IgG subclasses produced against the VD4 region are the same among patients with different clinical outcomes. The OD450 was measured via bait-peptide ELISA to determine anti-VD4 IgG1 (A) and IgG3 (B) antibody responses utilizing patient sera in duplicate. Correlation of IgG and IgG1 (C) and IgG and IgG3 (D) for each patient. A nonparametric Kruskal-Wallis multiple-comparison test was used for ELISA and peak fold change analysis. A nonparametric Spearman test was used for correlation analysis. ****, P < 0.0001.
DISCUSSION
The WHO and NIAID have highlighted the need to elucidate the role of antibodies during urogenital C. trachomatis infection and to more comprehensively screen for potential targets in C. trachomatis vaccine development (4, 5). In the present study, we utilized DSCB to investigate the antibody response elicited against C. trachomatis in women whose C. trachomatis infection had different outcomes. The primary human antibody response we observed was toward MOMP, with both the CD4/VD3 region and the VD4 region being highly selected (Table 1). However, we found no difference in peak fold change (Fig. 3A), the magnitude of the antibody response (Fig. 3B and D), or IgG1 versus IgG3 subclass production (Fig. 4A and B) toward the VD4 conserved region between C. trachomatis-infected women based on infection outcome.
A variety of approaches have been used to investigate immunodominant epitopes of C. trachomatis antigens. DSCB is unique in that it utilizes a high-throughput, relatively unbiased method to identify epitope-specific antibody responses from human serum samples (18, 19). This allows users to screen a large number of antigens from various patient groups, without relying on B cell epitope prediction algorithms, blind peptide ELISAs, recombinant peptides, or the use of monoclonal antibodies. Other recent studies have carried out investigations to identify immunodominant antigens (22) and immunodominant epitopes of C. trachomatis antigens, with the primary goal being the development of C. trachomatis species-specific diagnostics (23, 24). One approach used a B cell epitope prediction algorithm coupled with confirmation by ELISA to identify the immunodominant sequences of a number of C. trachomatis antigens (23, 24). Among the sequences identified in that study were MOMP, Tarp, PmpD, IncA, and CT875. Notably, the same immunodominant epitopes were also identified in our studies. However, our approach additionally identified immunodominant epitopes of CT143, HSP60, and PmpC that differed from those previously found (23, 24). This highlights one advantage of using DSCB, which does not rely on B cell epitope prediction algorithms and provides an unbiased screening approach. However, DSCB has limitations as well: we only included 24 C. trachomatis antigens in our library, and our overlapping peptides were 6 to 10 aa in length, making it unlikely that we captured discontinuous or complex conformational epitopes. Despite the short length of the peptides displayed by our library, the DSCB technique consistently selected regions containing overlapping peptide epitopes, indicating that it can efficiently identify linear epitopes on the surface of C. trachomatis antigens, similar to what we previously showed in a study in which we mapped the epitopes targeted by human antibody responses to dengue virus (25). The MS2 bacteriophage VLP display platform displays short peptides in a constrained β-hairpin loop on the surface of the VLPs, likely providing the scaffolding necessary for antibodies to bind these short peptides and allowing us to identify epitope-specific antibody responses. Our VLP platform does not provide information on the appropriateness of peptides as potential T cell epitopes in a vaccine formulation. We also note that with short peptides there are likely nonspecific interactions and cross-reactive antibodies that can be identified. This was controlled for with our virgin serum samples.
MOMP is of primary interest in vaccine development, with the first MOMP-targeted vaccine candidates emerging in 1992 (20, 25). The VD4 conserved region of MOMP is known to be an immunodominant epitope and monoclonal antibodies toward MOMP and/or this region have neutralizing activity (24, 26, 27). In addition, the vaccine candidate CTH522 targeting the VD4 region of MOMP induces neutralizing antibodies in mice and humans (28). CTH522 is a recombinant fusion protein of the VD4 region of MOMP and flanking sequences from C. trachomatis serovars D, E, F, and G with the addition of Cationic Adjuvant Formulation no. 1 (CAF01) (28). CTH522-elicited antibodies have functionality in in vitro neutralization assays and in vivo challenge with C. trachomatis intravaginally at 6 weeks and 1 year postvaccination and also through passive transfer of antibodies to Rag1 knockout mice, which prevented the establishment of an infection in approximately half of mice (28). Initial studies of the immune response of the 15 women receiving CTH522:CAF01 in a phase I clinical trial showed promising immunogenicity, with induction of neutralizing antibody titers after three intramuscular injections, mucosal specific IgG and IgA, and induction of cell-mediated immunity as measured by CTH522 stimulated IFN-γ release (29). Whether neutralizing antibodies are sufficient to provide protection in urogenital C. trachomatis infection in women remains to be seen.
In contrast to the protective value of VD4-directed antibodies seen with CTH522 in animal models, our findings suggest that, in women, antibody responses generated during urogenital C. trachomatis infection toward the VD4 conserved region are not associated with C. trachomatis infection outcomes. Our study, along with the observation that repeat infection with the same serovar of C. trachomatis is common in females (13), support the hypothesis that the detection of MOMP-specific antibodies and the magnitude of the antibody response are not associated with protection against C. trachomatis outcomes. MOMP VD4 was the most commonly identified peptide among our patients and was the highest ranked peptide by fold change, supporting it as the immunodominant B cell epitope during a urogenital infection in women. However, there is evidence from other pathogens, such as HIV, that targeting immunodominant regions of antigens may not provide protection (30, 31). Instead, pathogens may “hide” regions, known as cryptic epitopes, which are normally not very immunogenic but may be protective (30). Targeting cryptic epitopes is of interest for vaccine development against pathogens, such as group A streptococcus, and could be a promising new direction in C. trachomatis vaccine development (32). Together, our present study is of particular interest in the context of vaccine development. Although further functional responses of specific antibodies, including those against the MOMP VD4 region, are necessary to fully understand the role of specific antibodies in C. trachomatis infection and protection, our data suggest that naturally occurring antibodies to the VD4 conserved region of MOMP are not associated with C. trachomatis infection outcomes.
MATERIALS AND METHODS
Ethics statement.
Cohort 1 samples were originally collected under approvals from the University of Alabama at Birmingham Institutional Review Board (IRB) and also the Jefferson County Department of Health (JCDH) in Birmingham, AL, and the University of New Mexico Health Sciences Center received deidentified samples under exempt status approved by their Institutional Review Board. Cohort 2 samples were collected, analyzed, and deidentified under approval from the University of New Mexico Health Sciences Center Human Research Review Committee.
Study population.
Patient serum samples were obtained from two study populations. One population (cohort 1) consisted of mostly African-American female patients 16 years of age or older presenting to the JCDH STD Clinic in Birmingham, AL, for treatment of a positive routine C. trachomatis nucleic acid amplification test (NAAT) who participated in a study on immune responses to C. trachomatis infection. Interested female patients provided written consent and were enrolled, at which time a cervical swab was collected for C. trachomatis NAAT (Hologic Aptima Combo 2 [AC2]; Hologic, Inc., Marlborough, MA). Blood was also obtained for isolation of serum. Women with known gonorrhea, syphilis, or HIV were excluded, as were women with PID. Participants returned for a 3-month follow-up visit for repeat cervical C. trachomatis NAAT to evaluate for reinfection (9, 33). Women were grouped according to defined outcomes following initial C. trachomatis infection: (i) women who originally were C. trachomatis positive but were C. trachomatis negative upon presentation for treatment (“spontaneous resolvers”), (ii) women with a lack of C. trachomatis reinfection at a 3-month follow-up visit following initial treatment (“nonreinfected”), and (iii) women with C. trachomatis reinfection at a 3-month follow-up following initial treatment (“reinfected”) (Table 2). Cohort 2 was derived from a second study population comprised of Hispanic and non-Hispanic white women aged 18 to 40 years presenting for routine gynecologic examinations in Albuquerque, NM. A subset of these women who self-reported no history of sexual activity (i.e., virginal women) was designated for our study as cohort 2 and were used as C. trachomatis-negative controls. Study participants enrolled in cohort 2 signed informed consent, and cervical swab and blood samples were obtained.
TABLE 2.
C. trachomatis nucleic acid amplification test status of cohort 1 outcome group patients
| Patient group (n) |
C. trachomatis NAAT status |
||
|---|---|---|---|
| Screening | Enrollment | 3-mo follow-up | |
| Nonreinfected (10) | + | + | – |
| Reinfected (10) | + | + | + |
| Spontaneous resolvers (10) | + | – | – |
Generation of a bacteriophage MS2 VLP C. trachomatis serovar D antigen fragment library.
A C. trachomatis antigen fragment library was generated as previously described (18). The library (MS2-VLP-Ct-AFL) contains overlapping peptides of 6 to 10 amino acids corresponding to 24 C. trachomatis antigens displayed on bacteriophage MS2 VLPs. These peptides are displayed as fusions with the MS2 coat protein and are displayed in a surface-exposed, β-hairpin loop which provides for display in a constrained manner. C. trachomatis antigens (Table 1) were chosen based on a literature review showing that they either: (i) are predicted or confirmed to be expressed on the surface of EBs and/or (ii) have been shown to elicit antibody responses in humans, nonhuman primates, or mouse models of C. trachomatis infection. The amino acid sequence of each antigen was based on the sequence of C. trachomatis serovar D. The library contained 26,657 unique peptides representing 13,335 amino acids from the C. trachomatis antigens of interest.
Deep sequence-coupled biopanning.
Deep sequence-coupled biopanning (DSCB) was performed with patient serum as previously described (18). Isolated patient serum IgG (500 ng) was mixed with MS2-VLP-Ct-AFL (40 μg) in a 100-μl total volume in 1× phosphate-buffered saline (PBS) and incubated at 4°C overnight with rotation. VLP/antibody complexes were then isolated using Dynabeads (Invitrogen), and complexes were extensively washed and then eluted with acidic glycine. Encapsidated MS2 RNA was isolated by RNA extraction kit and cDNA generated by RT-PCR and previously described primers (18). PCR products were prepared for Ion Torrent deep sequencing, and FASTQ files were processed and analyzed as previously described (19). Data were normalized to account for the relative abundance of each peptide in the starting VLP library.
Synthetic peptide ELISA.
ELISAs were carried out using Immulon 2 HB 96-well flat-bottom microtiter plates (Thermo Scientific). All incubations were done at 25°C with rocking unless otherwise noted; washes were in 1× PBS or 1× PBS plus 0.05% Tween 20 (PBS-T), and 100-μl volumes were used. Wells were coated with carrier protein (1 μg of streptavidin [Invitrogen]) in PBS at 4°C overnight. Plates were washed three times with PBS and succinimidyl 6-(β-maleimidopropionamido)-hexanoate (SMPH; 2 μg/100 μl) was added. Plates were incubated for 1 h and washed three times with PBS, and FDTTTLNPTIAGAGDVKGGGC peptide (GenScript Biotech Corporation) was added at 2 μg/well. Plates were incubated for 2 h and washed three times with 1× PBS. Blocking was carried out overnight at 4°C with 300 μl of 0.5% nonfat dry milk in PBS-T (blocking reagent). Patient sera (diluted 1:700 for total IgG or 1:100 for IgG subclasses in blocking reagent) were added after three washes with PBS-T. Plates were incubated for 2 h and washed five times with PBS-T, and secondary antibody was added. Total IgG was assessed with peroxidase-conjugated AffiniPure goat anti-human IgG H+L (Jackson ImmunoResearch) at 1:5,000 in blocking reagent, IgG1 with mouse anti-human IgG1 Hinge-HRP (Southern Biotech), and IgG3 with mouse anti-human IgG3 Hinge-HRP (Southern Biotech). Plates were incubated for 45 min, washed five times with PBS-T, and five times with PBS. Then, 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added, and plates were incubated for 8 min. Then, 100 μl of 1% HCl was added, and the OD450 was determined using a microplate reader. Background OD450 values of empty wells were averaged and subtracted. The reported OD450 values are the means of two technical replicates.
Statistical analysis.
Statistical analysis of data was done using Prism 7 for Macintosh. For ELISA data and peak fold change analysis utilizing patient serum samples, a nonparametric multiple comparisons Kruskal-Wallis test was performed, followed by a Dunn’s multiple-comparison test. For correlation data, analysis was performed by using nonparametric Spearman correlation tests.
Data availability.
The data and code used in the manuscript are available upon request by contacting the corresponding author.
DSCB results for the Tarp (CT456) from sera of female patients with variable clinical outcomes. Amino acid positions from ca. 200 to 300 are a highly repetitive region in which the selected peptides cannot be resolved by our analysis pipeline, resulting in no apparent selection in this region. Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the solute protein-binding family (CT067) from sera of female patients with variable clinical outcomes. Download FIG S4, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpC (CT414) from sera of female patients with variable clinical outcomes. Download FIG S5, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT147 from sera of female patients with variable clinical outcomes. Download FIG S6, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT875 from sera of female patients with variable clinical outcomes. Download FIG S7, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpD (CT812) from sera of female patients with variable clinical outcomes. Download FIG S8, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the Pgp3 (pCT03) from sera of female patients with variable clinical outcomes. Download FIG S9, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the inclusion membrane protein A (CT119) from sera of female patients with variable clinical outcomes. Download FIG S10, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This study has been supported by the National Institutes of Health, National Center for Advancing Translational Sciences (grants KL2TR001448 and UL1TR001449 [supporting K.M.F.]) and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grants U19 AI113187 [principal investigator, C.M.W.] and R01AI093692 [principal investigator, W.M.G.]). This research was partially supported by UNM Comprehensive Cancer Center Support Grant NCI P30CA118100 and the Analytical and Translational Genomics shared resource. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. A.L.C. was supported by a Stephanie Ruby Predoctoral Travel Fellowship from the Molecular Genetics and Microbiology Department at UNM HSC.
The authors had the following specific contributor roles as defined by CASRI: conceptualization (K.M.F. and B.C.), data curation (A.L.C. and K.M.F.), formal analysis (A.L.C.), funding acquisition (B.C., K.M.F., and C.M.W.), investigation (A.L.C., A.C.L., and S.B.C.), methodology (A.L.C., A.C.L., S.B.C., B.C., D.S.P., K.M.F., WMG, and C.M.W.), project administration (K.M.F.), resources (C.M.W., W.M.G., and D.S.P.), supervision (B.C. and K.M.F.), visualization (A.L.C.), writing—original draft (A.L.C.), and writing—review and editing (A.L.C., A.C.L., S.B.C., C.M.W., W.M.G., D.S.P., B.C., and K.M.F.).
REFERENCES
- 1.CDC. 2018. Sexually transmitted disease surveillance, 2018. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Owusu-Edusei K, Chesson HW, Gift TL, Tao G, Mahajan R, Ocfemia MC, Kent CK. 2013. The estimated direct medical cost of selected sexually transmitted infections in the United States, 2008. Sex Transm Dis 40:197–201. doi: 10.1097/OLQ.0b013e318285c6d2. [DOI] [PubMed] [Google Scholar]
- 3.Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. 2010. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis 201(Suppl 2):S134–S155. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- 4.Zhong G, Brunham RC, de la Maza LM, Darville T, Deal C. 2019. National Institute of Allergy and Infectious Diseases workshop report: “Chlamydia vaccines: the way forward.” Vaccine 37:7346–7354. doi: 10.1016/j.vaccine.2017.10.075. [DOI] [PubMed] [Google Scholar]
- 5.Broutet N, Fruth U, Deal C, Gottlieb SL, Rees H, participants of the 2013 STI Vaccine Technical Consultation. 2014. Vaccines against sexually transmitted infections: the way forward. Vaccine 32:1630–1637. doi: 10.1016/j.vaccine.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell HD, Perry LJ. 1982. Neutralization of Chlamydia trachomatis infectivity with antibodies to the major outer membrane protein. Infect Immun 38:745–754. doi: 10.1128/IAI.38.2.745-754.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naglak EK, Morrison SG, Morrison RP. 2017. Neutrophils are central to antibody-mediated protection against genital Chlamydia. Infect Immun 85:e00409-17. doi: 10.1128/IAI.00409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kari L, Bakios LE, Goheen MM, Bess LN, Watkins HS, Southern TR, Song L, Whitmire WM, Olivares-Zavaleta N, Caldwell HD. 2013. Antibody signature of spontaneous clearance of Chlamydia trachomatis ocular infection and partial resistance against re-challenge in a nonhuman primate trachoma model. PLoS Negl Trop Dis 7:e2248. doi: 10.1371/journal.pntd.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakshi RK, Gupta K, Jordan SJ, Chi X, Lensing SY, Press CG, Geisler WM. 2018. An adaptive Chlamydia trachomatis-specific IFN-γ-producing CD4+ T cell response is associated with protection against Chlamydia reinfection in women. Front Immunol 9:1981. doi: 10.3389/fimmu.2018.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell AN, Zheng X, O’Connell CM, Wiesenfeld HC, Hillier SL, Taylor BD, Picard MD, Flechtner JB, Zhong W, Frazer LC, Darville T. 2016. Identification of Chlamydia trachomatis antigens recognized by T cells from highly exposed women who limit or resist genital tract infection. J Infect Dis 214:1884–1892. doi: 10.1093/infdis/jiw485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barral R, Desai R, Zheng X, Frazer LC, Sucato GS, Haggerty CL, O’Connell CM, Zurenski MA, Darville T. 2014. Frequency of Chlamydia trachomatis-specific T cell interferon-gamma and interleukin-17 responses in CD4-enriched peripheral blood mononuclear cells of sexually active adolescent females. J Reprod Immunol 103:29–37. doi: 10.1016/j.jri.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darville T, Albritton HL, Zhong W, Dong L, O’Connell CM, Poston TB, Quayle AJ, Goonetilleke N, Wiesenfeld HC, Hillier SL, Zheng X. 2019. Anti-chlamydia IgG and IgA are insufficient to prevent endometrial chlamydia infection in women, and increased anti-chlamydia IgG is associated with enhanced risk for incident infection. Am J Reprod Immunol 81:e13103. doi: 10.1111/aji.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batteiger BE, Tu W, Ofner S, Van Der Pol B, Stothard DR, Orr DP, Katz BP, Fortenberry JD. 2010. Repeated Chlamydia trachomatis genital infections in adolescent women. J Infect Dis 201:42–51. doi: 10.1086/648734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Hakim EA, Gordon UD, Akande VA. 2010. The relationship between serum Chlamydia antibody levels and severity of disease in infertile women with tubal damage. Arch Gynecol Obstet 281:727–733. doi: 10.1007/s00404-009-1225-7. [DOI] [PubMed] [Google Scholar]
- 15.Taylor BD, Darville T, Tan C, Bavoil PM, Ness RB, Haggerty CL. 2011. The role of Chlamydia trachomatis polymorphic membrane proteins in inflammation and sequelae among women with pelvic inflammatory disease. Infect Dis Obstet Gynecol 2011:989762. doi: 10.1155/2011/989762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budrys NM, Gong S, Rodgers AK, Wang J, Louden C, Shain R, Schenken RS, Zhong G. 2012. Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet Gynecol 119:1009–1016. doi: 10.1097/AOG.0b013e3182519326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner AZ, Diamond MP, Legro RS, Schlaff WD, Barnhart KT, Casson PR, Christman GM, Alvero R, Hansen KR, Geisler WM, Thomas T, Santoro N, Zhang H, Eisenberg E, Reproductive Medicine Network. 2015. Chlamydia trachomatis immunoglobulin G3 seropositivity is a predictor of reproductive outcomes in infertile women with patent fallopian tubes. Fertil Steril 104:1522–1526. doi: 10.1016/j.fertnstert.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frietze KM, Pascale JM, Moreno B, Chackerian B, Peabody DS. 2017. Pathogen-specific deep sequence-coupled biopanning: a method for surveying human antibody responses. PLoS One 12:e0171511. doi: 10.1371/journal.pone.0171511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frietze KM, Roden RB, Lee JH, Shi Y, Peabody DS, Chackerian B. 2016. Identification of anti-CA125 antibody responses in ovarian cancer patients by a novel deep sequence-coupled biopanning platform. Cancer Immunol Res 4:157–164. doi: 10.1158/2326-6066.CIR-15-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips S, Quigley BL, Timms P. 2019. Seventy years of Chlamydia vaccine research: limitations of the past and directions for the future. Front Microbiol 10:70. doi: 10.3389/fmicb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler WM, Morrison SG, Doemland ML, Iqbal SM, Su J, Mancevski A, Hook EW, III, Morrison RP. 2012. Immunoglobulin-specific responses to Chlamydia elementary bodies in individuals with and at risk for genital chlamydial infection. J Infect Dis 206:1836–1843. doi: 10.1093/infdis/jis621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong G. 2010. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol 185:1670–1680. doi: 10.4049/jimmunol.1001240. [DOI] [PubMed] [Google Scholar]
- 23.Rahman KS, Darville T, Russell AN, O’Connell CM, Wiesenfeld HC, Hillier SL, Lee DE, Kaltenboeck B. 2018. Comprehensive molecular serology of human Chlamydia trachomatis infections by enzyme-linked immunosorbent assays. mSphere 3:e00253-18. doi: 10.1128/mSphere.00253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman KS, Darville T, Russell AN, O’Connell CM, Wiesenfeld HC, Hillier SL, Chowdhury EU, Juan Y-C, Kaltenboeck B. 2018. Discovery of human-specific immunodominant Chlamydia trachomatis B cell epitopes. mSphere 3:e00246-18. doi: 10.1128/mSphere.00246-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuffrey M, Alexander F, Conlan W, Woods C, Ward M. 1992. Heterotypic protection of mice against chlamydial salpingitis and colonization of the lower genital tract with a human serovar F isolate of Chlamydia trachomatis by prior immunization with recombinant serovar L1 major outer-membrane protein. J Gen Microbiol 138(Pt 8):1707–1715. doi: 10.1099/00221287-138-8-1707. [DOI] [PubMed] [Google Scholar]
- 26.Peterson EM, Cheng X, Markoff BA, Fielder TJ, de la Maza LM. 1991. Functional and structural mapping of Chlamydia trachomatis species-specific major outer membrane protein epitopes by use of neutralizing monoclonal antibodies. Infect Immun 59:4147–4153. doi: 10.1128/IAI.59.11.4147-4153.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YX, Stewart S, Joseph T, Taylor HR, Caldwell HD. 1987. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J Immunol 138:575–581. [PubMed] [Google Scholar]
- 28.Olsen AW, Lorenzen EK, Rosenkrands I, Follmann F, Andersen P. 2017. Protective effect of vaccine promoted neutralizing antibodies against the intracellular pathogen Chlamydia trachomatis. Front Immunol 8:1652. doi: 10.3389/fimmu.2017.01652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham S, Juel HB, Bang P, Cheeseman HM, Dohn RB, Cole T, Kristiansen MP, Korsholm KS, Lewis D, Olsen AW, McFarlane LR, Day S, Knudsen S, Moen K, Ruhwald M, Kromann I, Andersen P, Shattock RJ, Follmann F. 2019. Safety and immunogenicity of the chlamydia vaccine candidate CTH522 adjuvanted with CAF01 liposomes or aluminium hydroxide: a first-in-human, randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect Dis 19:1091–1100. doi: 10.1016/S1473-3099(19)30279-8. [DOI] [PubMed] [Google Scholar]
- 30.Good MF, Yanow SK. 2016. Cryptic epitope for antibodies should not be forgotten in vaccine design. Expert Rev Vaccines 15:675–676. doi: 10.1586/14760584.2016.1154791. [DOI] [PubMed] [Google Scholar]
- 31.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 32.Ozberk V, Pandey M, Good MF. 2018. Contribution of cryptic epitopes in designing a group A streptococcal vaccine. Hum Vaccin Immunother 14:2034–2052. doi: 10.1080/21645515.2018.1462427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta K, Bakshi RK, Van Der Pol B, Daniel G, Brown L, Press CG, Gorwitz R, Papp J, Lee JY, Geisler WM. 2018. Repeated Chlamydia trachomatis infections are associated with lower bacterial loads. Epidemiol Infect doi: 10.1017/S0950268818002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of DSCB technology workflow. Download FIG S1, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the OmcB (CT443) from sera of female patients with variable infection outcomes. Download FIG S2, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the Tarp (CT456) from sera of female patients with variable clinical outcomes. Amino acid positions from ca. 200 to 300 are a highly repetitive region in which the selected peptides cannot be resolved by our analysis pipeline, resulting in no apparent selection in this region. Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the solute protein-binding family (CT067) from sera of female patients with variable clinical outcomes. Download FIG S4, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpC (CT414) from sera of female patients with variable clinical outcomes. Download FIG S5, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT147 from sera of female patients with variable clinical outcomes. Download FIG S6, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT875 from sera of female patients with variable clinical outcomes. Download FIG S7, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpD (CT812) from sera of female patients with variable clinical outcomes. Download FIG S8, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the Pgp3 (pCT03) from sera of female patients with variable clinical outcomes. Download FIG S9, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the inclusion membrane protein A (CT119) from sera of female patients with variable clinical outcomes. Download FIG S10, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data and code used in the manuscript are available upon request by contacting the corresponding author.
DSCB results for the Tarp (CT456) from sera of female patients with variable clinical outcomes. Amino acid positions from ca. 200 to 300 are a highly repetitive region in which the selected peptides cannot be resolved by our analysis pipeline, resulting in no apparent selection in this region. Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the solute protein-binding family (CT067) from sera of female patients with variable clinical outcomes. Download FIG S4, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpC (CT414) from sera of female patients with variable clinical outcomes. Download FIG S5, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT147 from sera of female patients with variable clinical outcomes. Download FIG S6, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the CT875 from sera of female patients with variable clinical outcomes. Download FIG S7, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the PmpD (CT812) from sera of female patients with variable clinical outcomes. Download FIG S8, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the Pgp3 (pCT03) from sera of female patients with variable clinical outcomes. Download FIG S9, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DSCB results for the inclusion membrane protein A (CT119) from sera of female patients with variable clinical outcomes. Download FIG S10, TIF file, 2.8 MB (2.9MB, tif) .
Copyright © 2020 Collar et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.