Abstract
Background
The development of molecularly tailored therapeutic agents such as the BCR/ABL-active tyrosine kinase inhibitors (TKi) resulted in an excellent treatment option for chronic myeloid leukemia (CML) patients. However, following TKi discontinuation, disease relapses in 40–60% of patients, an occurrence very likely due to the persistence of leukemic stem cells that are scarcely sensitive to TKi. Nevertheless, TKi are still the only current treatment option for CML patients.
Objective
The aim of this study was to compare the effects of TKi belonging to different generations, imatinib and ponatinib (first and third generation, respectively), on progenitor/stem cell expansion potential and markers.
Patients and Methods
We used stabilized CML cell lines (KCL22, K562 and LAMA-84 cells), taking advantage of the previous demonstration of ours that cell lines contain cell subsets endowed with progenitor/stem cell properties. Primary cells explanted from CML patients were also used. The effects of TKi on the expression of stem cell related genes were compared by quantitative PCR. Flow cytometry was performed to evaluate aldehyde-dehydrogenase (ALDH) activity and the expression of cluster of differentiation (CD) cell surface hematopoietic stem cell markers. Progenitor/stem cell potential was estimated by serial colony formation ability (CFA) assay.
Results
Ponatinib was more effective than imatinib for the reduction of cells with ALDH activity and progenitor/stem cell potential of CML patient-derived cells and cell lines. Furthermore, ponatinib was more effective than imatinib in reducing the percentage of CD26-expressing cells in primary CML cells, whereas imatinib and ponatinib showed similar efficacy on KCL22 cells. Both drugs strongly upregulated NANOG and SOX2 in CML cell lines, but in KCL22 cells this upregulation was significantly lower with ponatinib than with imatinib, an outcome compatible with a lower level of enrichment of the stem cell compartment upon ponatinib treatment.
Conclusion
Ponatinib seems to target CML progenitor/stem cells better than imatinib.
Electronic supplementary material
The online version of this article (10.1007/s11523-020-00741-x) contains supplementary material, which is available to authorized users.
Introduction
The pathogenesis of chronic myeloid leukemia (CML) is centered on the expression of the BCR/ABL oncoprotein, a constitutively active tyrosine kinase [1]. The clinical course of untreated CML typically includes an initial chronic phase (CP) lasting 3–5 years, an accelerated phase (AP) lasting 6–18 months, and a final, short, blast crisis (BC) with poor prognosis. The introduction of imatinib-mesylate, the prototypical tyrosine kinase inhibitor (TKi) active on BCR/ABL, signaled a new era in the treatment of CML, allowing up to 90% of CP-CML patients to survive after 20 years of treatment [2]. However, imatinib and subsequent second- (dasatinib, bosutinib, and nilotinib) and third- (ponatinib) generation TKi are not very effective in preventing the relapse of disease, as shown in particular by the outcome of TKi discontinuation protocols in CP patients. Several studies showed indeed that 40–60% of even well responding (sustained deep molecular remission) patients who have stopped therapy undergo relapse of disease (in 80% of cases, within the first 6 months) and require the restart of treatment, while others maintain treatment-free remission, in some cases despite the persistence of detectable molecular disease [3–7]. Based on available data, it is likely that relapse after TKi discontinuation is due to the persistence of leukemic stem cells (LSC), which apparently are relatively resistant to TKi [8–11]. However, while the identification of new treatments capable of targeting CML progenitor/stem cells seems necessary when aiming for eradication of disease [12, 13], TKi are still the only current treatment option for CML patients.
In CML, LSC are located within the CD34 +/CD38 − cell fraction, a phenotype which is, however, not exclusive to LSC of CML [14]. Therefore, different markers have been tested for being capable to discriminate LSC of CML from normal hematopoietic stem cells (HSC). Along this line, CD26 (dipeptidyl-peptidase IV) has been identified as a potential marker for the quantification and isolation of LSC in bone marrow (BM) samples of CML patients [15]. Indeed, while other antigens such as CD90 and IL-1RAP are co-expressed by LSC of CML and acute myeloid leukemia as well as by HSC, CD26 is consistently expressed in CP-CML patients, but it is not in HSC or stem cells of other myeloid neoplasms [15, 16]. Importantly, the concentration of CD26 + LSC correlates with resistance to TKi and identifies TKi-resistant sub-clones [17].
Stem cells from a variety of tissues exhibit high levels of aldehyde dehydrogenase (ALDH) activity, which is therefore considered a stem cell feature [18, 19]. HSC in particular are characterized by high ALDH activity [20–22], and cells with high ALDH activity and low side scattering are self-renewing and multipotent [21]. Based on all of the above, high ALDH activity has been increasingly used as a selectable marker for cell populations enriched in tumor-initiating stem-like cells in multiple types of cancers, including leukemias [23, 24].
Another set of potentially useful stem cell markers is represented by those defining the self-renewal and pluripotency of embryonic stem cells (ESC), including the transcription factors OCT4, SOX2, and NANOG [25–28]. The expression of OCT4 is indeed considered restricted to pluripotent stem cells, its expression being down-regulated in the course of differentiation, to reach undetectability in adult normal tissues [29]. SOX2, another transcriptional regulator of pluripotency and self-renewal in ESC [30, 31], has also been found expressed in a number of tumors [32–34]. Like OCT4 and SOX2, NANOG has been reported to be expressed in many tumors, and its knockdown to inhibit tumor development [35]. This set of markers also includes c-MYC, a proto-oncogene important for the regulation of the stem cell compartment [36, 37].
In this study, we compared the effects of TKi belonging to different generations, imatinib and ponatinib, on the expression of progenitor/stem cell markers and potential in both CML cell lines, taking advantage of our previous demonstration that leukemic cell lines contain cell subsets endowed with progenitor/stem cell properties [9, 12, 13, 38, 39], as well as patient-derived cells.
Methods
Cells and Culture Conditions
K562 [40] and KCL22 [41] (kindly supplied by Dr A.M. Vannucchi, Azienda Ospedaliero/Universitaria Careggi, Florence, Italy), LAMA-84 [42], and the imatinib-resistant LAMA-84R [43, 44] (kindly supplied by Dr C. Gambacorti-Passerini, University of Milano-Bicocca & San Gerardo Hospital, Monza, Italy) blast-crisis CML cells and Kasumi-1 [45] and NB4 [46] acute myeloid leukemia cells (kindly supplied by Dr C. Chomienne Université Denis Diderot, Paris, France) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 50 U/mL penicillin, and 50 mg/mL streptomycin (all from EuroClone, Paignton, UK), and incubated at 37 °C in a water-saturated atmosphere containing 95% air (21% O2) and 5% CO2. CML cell lines were routinely tested for mycoplasma contamination, BCR/ABL expression, and annually for their identity (Promega PowerPlex Fusion System kit; BMR Genomics, Padova, Italy). Cell cultures were renewed every 3 months by thawing a new vial of frozen cells.
Patient-derived CML cells (see Table 1 for patients’ characteristics) were collected following informed consent and under approval of the Ethical Committee of AOUC (authorization no. 520/10, October 18, 2010, renewed with no. 2015/0032965, November 4, 2015). Mononuclear cells (MCs) from BM of CML patients were isolated by Ficoll-Hypaque gradient (Cedarlane Laboratories, ON, Canada). CML BMMCs were cultured in IMDM supplemented with 20% FBS, 2 mM glutamine, 50 U/mL penicillin, 50 mg/mL streptomycin (all from EuroClone, Paignton, UK), and 50 ng/mL Flt-3 ligand (cat. no. 300-19), 20 ng/mL TPo (cat. no. 300-18), 50 ng/mL stem cell factor (SCF) (cat. no. 300-07) and 10 ng/mL interleukin-3 (IL-3) (cat. no. 200-03) (all from PeproTech, Rocky Hill, NJ, USA).
Table 1.
Patients’ characteristics
| Patient ID | Age at diagnosis | CML phase | Sokal risk | BCR/ABL transcript | ELN criteria response |
|---|---|---|---|---|---|
| 23 | 42 | ECP | Intermediate | b3a2 (p210) e1a2 (p190) | Optimal response to nilotinib |
| 33 | 59 | ECP | Intermediate | b3a2 (p210) | Optimal response to nilotinib |
| 46 | 75 | ECP | Low | b3a2 (p210) | Optimal response to imatinib |
| 47 | 65 | ECP | High | b3a2 (p210) | Optimal response to dasatinib |
CML chronic myeloid leukemia, ECP early chronic phase, ELN European Leukemia Net (2013)
Experiments were performed with cells harvested from maintenance cultures by sub-culturing viable cells (5 × 105/mL) in fresh medium for 24 h before seeding for the experiments (3 × 105/mL). Cells were counted in a hemocytometer and cell viability was assessed by trypan blue exclusion test.
IC50 Determination in Cytotoxicity Experiments
Exponentially growing KCL22, LAMA-84, and K562 cells from routine cultures were seeded in fresh medium at 3 × 105 cells/mL. Cells were treated with dimethyl-sulfoxide (DMSO; vehicle) or increasing doses of TKi at the beginning of incubation (treatment at time 0). Half maximal inhibitory concentration (IC50) values were determined by counting viable cells following trypan blue staining after 72 h of treatment and processing data with GraphPad Prism software (La Jolla, CA, USA).
Drugs
Imatinib (cat. no. 202180, Santa Cruz, Biotechnology, Dallas, TX, USA) [47], dasatinib (cat. no. HY-10181) [48], nilotinib (cat. no. HY-10159) [49], ponatinib (cat. no. HY-12047) [50], bosutinib (cat. no. HY-10158) [51], and XMD8-92 (cat. no. HY-14443) [52, 53], from MedChemExpress (LLC, Princeton, NJ, USA), were all dissolved in DMSO.
Immunophenotype Determination
To analyze cell surface markers, 105 cells were stained with anti-human-CD26-PE (#12-0269), -CD90(Thy-1)-PE-Cyanine7 (#25-0909) from Affymetrix eBioscience (San Diego, CA, USA) [13], or with anti-human CD38-APC (#21270386), -CD34-FITC (#21270343) from ImmunoTools, JPT Peptide Technologies GmbH (Berlin, Germany) for 30 min at 4 °C in the dark. Expression of cell surface markers was measured with a FACSCanto flow-cytometer (Becton–Dickinson, Franklin Lakes, NJ, USA). Background signal was established in the same populations by staining with matched isotype control.
ALDH Activity Measurement
ALDH enzymatic activity was quantified by flow cytometry using the Aldefluor kit (#01700, Stem Cell Technologies, Vancouver, BC, Canada) [54] according to the manufacturer’s instructions. Non-viable cells were excluded from analysis following staining with 7-actino-aminomycin-D (7-AAD; BD Biosciences, San Jose, CA, USA). Diethylaminobenzaldehyde (DEAB), a specific inhibitor of ALDH, was used to determine background fluorescence.
RNA Extraction and Quantitative PCR
Total RNA was extracted using Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. Residual DNA was removed by Deoxyribonuclease I (Roche Diagnostics, Lewes, UK). The quality of extracted RNA was evaluated in a 2% agarose gel. RNA 2 µg were retro-transcribed into cDNA using the ImProm-IITM Reverse Transcription System kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. Quantitative polymerase chain reaction (q-PCR) was performed as described previously [55] and carried out using Green GoTaq q-PCR Master Mix (Promega). Primers (Qiagen, Hilden, Germany) for c-MYC, NANOG, SOX2, OCT4, KLF4, β-actin, and GAPDH are described in Supplementary Table S1 (see electronic supplementary material [ESM]).
Colony Formation Ability (CFA) Assay
K562 (300 cells/dish) and KCL22 (750 cells/dish) cells as well as CML BMMCs (10,000 cells/dish) were suspended in IMDM containing 2% FBS, 100 U/mL penicillin/streptomycin. Cells were plated in methylcellulose-containing medium (StemCell Technologies #04230 for CML cell lines, #04435 for CML BMMCs) in 35-mm dishes and treated with drugs from time 0 to the end of primary culture (day 7). Cells recovered from day-7 cultures were washed to remove drugs and replated weekly (passages II–III) in methylcellulose-containing medium. Colonies were scored on day 7 after each passage.
Statistical Analysis
Data represent means ± standard error of the mean (SEM) or means ± standard deviation (SD) of values obtained from at least three independent experiments. The exact number of experiments performed and used for statistical analysis is indicated in each figure legend. Variance between groups that were statistically compared was similar. p Values were calculated using Student’s t test (two groups) or one-way analysis of variance (more than two groups; multiple comparison using Bonferroni’s correction). A two-tailed value of p < 0.05 was considered statistically significant.
Results
Effects of Imatinib and Ponatinib on Stem Cell Markers in CML Cells
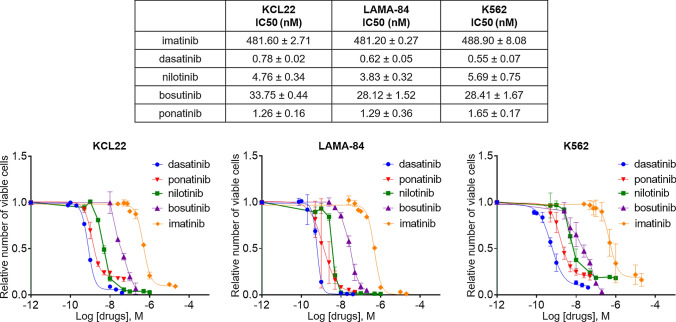
We determined first the IC50 values relative to the effects of several TKi on viable cell number in cultures of three CML cells lines (Fig. 1). Imatinib showed the highest IC50 value, and the IC50 of ponatinib and dasatinib were lower than those of nilotinib and bosutinib. Thus, the data show a good progression of efficacy going from first to second to third TKi generation. The IC50 values were similar in all cell lines, suggesting that the use of a panel of CML cell lines to assess the overall effects of TKi is adequate. Based on these results and on the fact that ponatinib is a third-generation TKi, we chose to compare the effects of imatinib with those of ponatinib on a number of progenitor/stem cell features.
Fig. 1.
TKi IC50 determination in CML cell lines. KCL22, LAMA-84, and K562 cells (3 × 105/mL) were cultured for 72 h in RPMI medium supplemented with 10% FBS in the presence of the indicated drugs or their solvent DMSO. Viable cells were counted by trypan-blue exclusion and IC50 were calculated by the GraphPad software. IC50 values are indicated in the Table. Graphs show TKi dose/response curves after 72 h of treatment or vehicle. Values are mean ± SD of data from three independent experiments normalized to matched vehicle-treated sample. CML chronic myeloid leukemia, DMSO dimethyl-sulfoxide, FBS fetal bovine serum, IC50 half maximal inhibitory concentration, TKi tyrosine kinase inhibitors
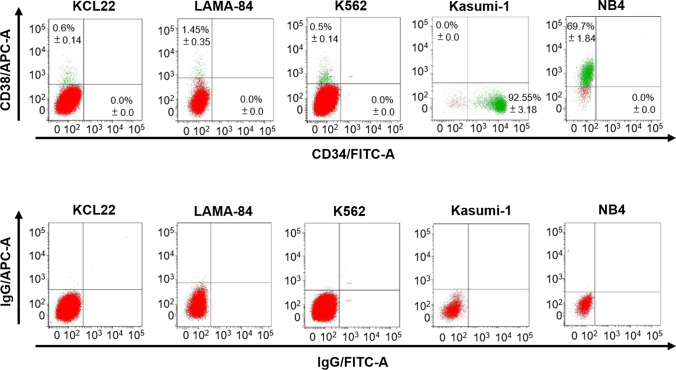
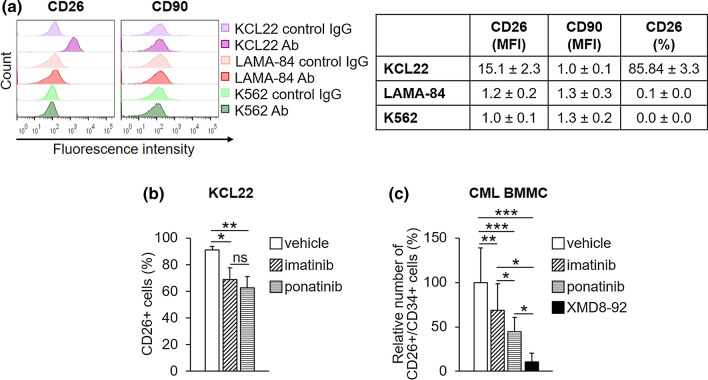
CML stem cells, besides being CD34 +/CD38 −/CD90 + , express CD26, a CML-specific stem cell marker [15, 56–58]. With the aim of comparing the effects of ponatinib with those of imatinib on CML progenitor/stem cells, we performed a characterization of the above cell surface molecules in a number of CML cell lines (Figs. 2, 3a; Table 2). All three CML cell lines tested were CD34-negative, a feature previously reported for K562 cells [40, 59], and shown to include a small subpopulation of CD38 + cells (Fig. 2), at variance with previous data showing that K562 cells do not express CD38 [59]. The acute myeloid leukemia cell lines Kasumi-1 and NB4 were used as positive controls for the expression of CD34 or CD38, respectively [59–61]. All three CML cell lines were found to be negative for CD90 (Fig. 3a). CD26 was expressed at high levels in KCL22, but not in K562 and LAMA-84 cells, as previously reported by us and others [13, 62] (Fig. 3a). The pattern of CD expression in LAMA-84R (imatinib-resistant) cells was identical to that of LAMA-84 cells (not shown). Based on all of the above, CD26 emerged as the only cell surface marker suitable to monitor progenitor/stem cells for subsequent experiments. Using imatinib and ponatinib at around two-fold the IC50 concentrations, we found that the two drugs are similarly effective in reducing the percentage of CD26-expressing cells (Fig. 3b). The same results were obtained when analyzing mean fluorescence intensity (MFI) values (Supplementary Fig. S1a, see ESM). Importantly, when the effects of the two drugs were tested on primary cells explanted from CML patients, we found that ponatinib was significantly more effective than imatinib in reducing the percentage of CD26 +/CD34 + cells (Fig. 3c, Supplementary Fig. S1b [see ESM]). XMD8-92, a small-molecule inhibitor of ERK5 that we previously showed to suppress progenitor/stem cell potential of primary CML cells [13], was used as a positive control.
Fig. 2.
Evaluation of the expression of the CD34 and CD38 cell surface markers in CML cell lines. The expression of CD34 and CD38 in routinely cultured cells was assessed by flow cytometry. Dot plots from one representative experiment are shown. The background signal was measured by cell staining with matched-isotype IgG controls. Values represent mean ± SD of data from three independent experiments. Kasumi-1 and NB4 cells were used as positive controls for CD34 and CD38, respectively
Fig. 3.
Effects of TKi on CD26-expressing CML cells. a The expression of CD90 and CD26 cell surface antigens in routinely cultured cells was assessed by flow cytometry using specific antibodies (Ab). Cell mean fluorescence intensity (MFI), calculated using matched-isotype IgG as negative control (control IgG), and the percentages of CD26-expressing cells obtained from the same experiments are reported in the table. Values represent mean ± SD of data obtained from three independent experiments. b KCL22 cells (3 × 105/mL) were treated with DMSO (vehicle), 1 μM imatinib or 4 nM ponatinib at time 0 and incubated for 72 h. Histograms relative to the average percentages of KCL22 cells expressing CD26 from three independent experiments (mean ± SD) are shown. *p < 0.05, **p < 0.01; ns not significant. c Patient-derived CML cells (3 × 105/mL) were treated with DMSO (vehicle), 1 μM imatinib, 4 nM ponatinib or 10 μM XMD8-92 at time 0 and incubated for 72 h. Histograms relative to the average percentages of CD26/CD34-expressing cells from three patients (mean ± SD) are shown. See Supplementary Fig. S1b for single patient data in the electronic supplementary material. CML chronic myeloid leukemia, DMSO dimethyl-sulfoxide, TKi tyrosine kinase inhibitors; *p < 0.05, **p < 0.01, ***p < 0.001
Table 2.
Characteristics of CML cell lines used in the text
| Cell line | Cell type | BCR-ABL junction type | Derivation | ALDH activity | CD34 | CD38 | CD26 | CD90 |
|---|---|---|---|---|---|---|---|---|
| K562 | BC CML | e14-a2 (b3-a2) | Pleural effusion of a 53-year-old woman with CML in terminal BC [40] | ++ | − | +/− | − | − |
| LAMA-84 | BC CML | e14-a2 (b3-a2) | Peripheral blood of a 29-year-old woman with CML after onset of myeloid-megakaryocytic BC [42] | ++ | − | +/− | − | − |
| KCL22 | BC CML | e13-a2 (b2-a2) | Pleural effusion of a 32-year-old woman with CML [41] | ± | − | +/− | ++ | − |
CML chronic myeloid leukemia, BC blast crisis; ++ high expression, ± low expression, − no expression
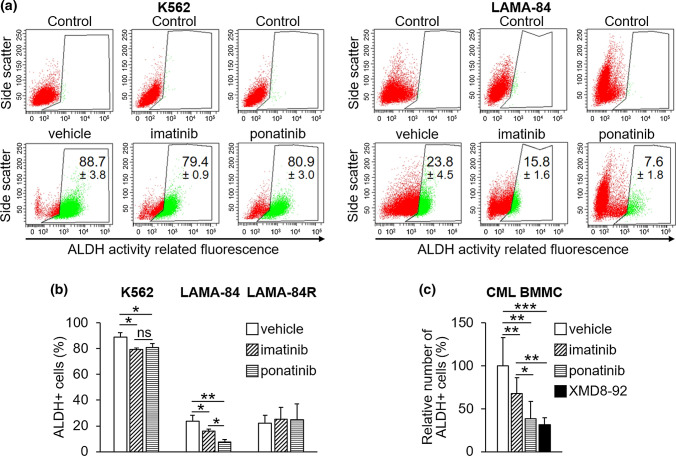
We then tested the effects of imatinib and ponatinib on cells endowed with ALDH activity using K562, LAMA-84, and LAMA-84R cells that, differently from KCL22 cells, show high levels of ALDH activity (Supplementary Fig S2a [see ESM]; Table 2), in agreement with data reported previously for K562 cells [63, 64]. Both drugs similarly reduced the percentages (Fig. 4a, b) and MFI values (Supplementary Fig. S2b [see ESM]) of ALDH-expressing cells in K562 cells, while ponatinib was significantly more effective than imatinib in LAMA-84 cells. Neither imatinib nor ponatinib was capable of reducing the ALDH activity of LAMA-84R cells (Fig. 4b, Supplementary Fig. S2c). The enhanced efficacy of ponatinib with respect to imatinib was confirmed on primary CML cells (Fig. 4c, Supplementary Fig. S2d [see ESM]).
Fig. 4.
Effects of TKi on CML cells endowed with ALDH activity. a K562 and LAMA-84 cells (3 × 105/mL) were treated with DMSO (vehicle), 1 μM imatinib or 4 nM ponatinib at time 0 and incubated for 72 h. The enzymatic activity of ALDH in viable cells was assessed by flow cytometry. The gating for ALDH activity-negative cells (control) was performed in the presence of the ALDH inhibitor DEAB (upper plots). Percentages of ALDH activity-positive viable cells averages (± SD) from three independent experiments are show in representative plots. b Data from a are plotted in the graphs together with statistical analysis; *p < 0.05, **p < 0.01; ns not significant. Data obtained from similar experiments carried out with LAMA-84R are also shown. See Supplementary Fig. S2c for representative dot plots from one out of three experiments performed with LAMA-84R (electronic supplementary material). c Patient-derived CML cells (3 × 105/mL) were treated with DMSO (vehicle), 1 μM imatinib, 4 nM ponatinib, or 10 μM XMD8-92 at time 0 and incubated for 72 h. The percentages of ALDH activity-positive viable cells from three patients (mean ± SD) were determined by flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001. Data obtained for individual patients are shown in Supplementary Fig. S2d (see electronic supplementary material). ALDH aldehyde-dehydrogenase, CML chronic myeloid leukemia, DMSO dimethyl-sulfoxide, TKi tyrosine kinase inhibitors
Effects of Imatinib and Ponatinib on the Expression of Stem Cell–Related Genes in CML Cells
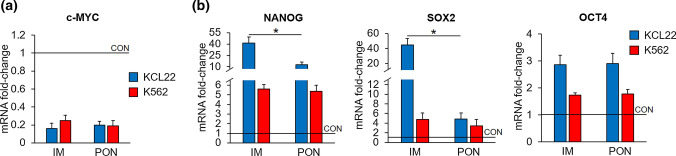
Both imatinib and ponatinib markedly reduced the expression of c-MYC, a proto-oncogene important for the regulation of stem cell compartments [36, 37], in either KCL22 or K562 cells (Fig. 5a). Moreover, both drugs upregulated the expression of the NANOG, SOX2, and OCT4 stem cell–related genes [65–67] with respect to control cells (Supplementary Fig. S3a [see ESM]), an effect likely due to the scarce sensitivity of LSC to TKi. Interestingly, in KCL22 cells, the upregulation of NANOG and SOX2 expression under ponatinib treatment was significantly lower than that induced by imatinib (Fig. 5b). Furthermore, no statistically significant difference as for KLF4 expression was found between imatinib- and ponatinib-treated cells (Supplementary Fig. S3a, b [see ESM]). In these experiments, cells were exposed to higher concentrations of both drugs (around 4-fold IC50) with respect to the experiments of Figs. 3 and 4, due to the fact that cells were incubated for shorter times.
Fig. 5.
Effects of TKi on the expression of stem cell–related genes in CML cell lines. Cells (3 × 105/mL) were treated with DMSO (CON), 2 μM imatinib (IM) or 8 nM ponatinib (PON) at time 0 and incubated for 24 h and c-MYC (a), NANOG, SOX2, and OCT4 (b) mRNA were measured by Q-PCR. Data were normalized with respect to the mean of GAPDH and β-actin mRNA levels and expressed as fold-change with respect to the values obtained for CON samples. Values represent mean ± SEM of three independent experiments, each carried out in triplicate. *p < 0.05; CML chronic myeloid leukemia, DMSO dimethyl-sulfoxide, GAPDH glyceraldehyde 3-phosphate dehydrogenase, Q-PCR quantitative polymerase chain reaction, SEM standard error of the mean, TKi tyrosine kinase inhibitors
Effects of Imatinib and Ponatinib on Progenitor/Stem Cell Potential of CML Cells
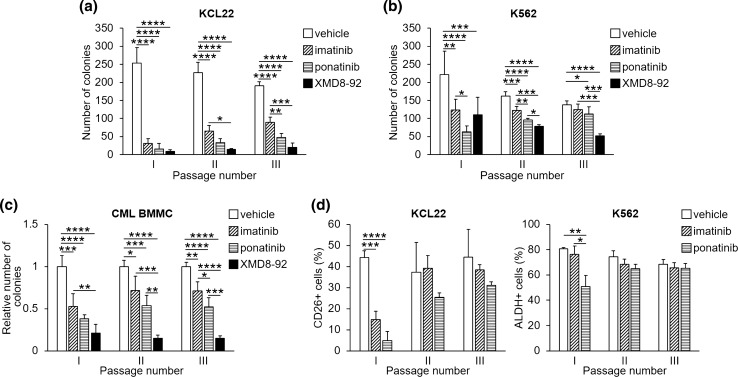
In order to evaluate functionally, rather than phenotypically, the effects of imatinib and ponatinib on CML progenitor/stem cell potential, cells were subjected to serial CFA assay (Fig. 6) [12, 13]. Both drugs strikingly (KCL22, Fig. 6a) or markedly (K562, Fig. 6b) reduced CFA in primary semisolid cultures (passage I), an effect which was progressively quenched as cells were replated to secondary and tertiary cultures (passages II–III). XMD8-92, used as a positive control for its suppressive effect on CML progenitor/stem cell potential (see above and [13]), maintained or even enhanced the inhibitory effect on CFA throughout the subsequent replatings. Of note, ponatinib did progressively better than imatinib along the subsequent passages, an outcome compatible with an enhanced effect of ponatinib on progenitors of relatively higher hierarchical level. Differences between imatinib and ponatinib were significant for KCL22 cells from passage III and for K562 cells from passage I. Ponatinib, differently from imatinib, maintained an inhibitory effect on CFA until passage III for K562 cells. Serial CFA assays performed with primary CML cells yielded results similar to those obtained for CML cell lines (Fig. 6c, Supplementary Fig. S4 [see ESM]). Indeed, both drugs markedly reduced CFA from passage I, the effect being progressively quenched along subsequent passages. However, at passage III, ponatinib was more effective than imatinib (Fig. 6c). Again, XMD8-92 was used as a positive control [13].
Fig. 6.
Effects of TKi on serial CFA of CML cells. Cells were incubated in semisolid medium and treated with DMSO (vehicle), 1 μM imatinib, 4 nM ponatinib or 10 μM XMD8-92 from time 0 to the end of primary culture (day 7; passage I). Colony cells were rescued and washed to remove drugs and replated (passage II); colony cells from drug-free, day-7 passage II cultures were then replated and incubated for 7 further days (passage III). Colonies were scored on day 7 after each passage. a, b Values are mean ± SD of data from three independent experiments each performed in duplicate. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. c Values are mean ± SD of data, normalized for vehicle-treated control, from three patients, each CFA assay being performed in duplicate. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data obtained for individual patients are shown in Supplementary Fig. S4 (see electronic supplementary material). d Expression of CD26 or ALDH activity, respectively, in KCL22 (left) or K562 (right) viable cells rescued from each passage of serial CFA assays shown in a and b. The percentages of cells expressing CD26 or ALDH activity as determined by flow cytometry from three independent experiments (mean ± SD) are shown. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ALDH aldehyde-dehydrogenase, CFA colony formation ability, CML chronic myeloid leukemia, DMSO dimethyl-sulfoxide, TKi tyrosine kinase inhibitors
In order to evaluate the correspondence between functional and phenotypical features of CML stem/progenitor cells in relation to their sensitivity to treatment, the effects of TKi on CD26 expression or ALDH activity were evaluated at each passage of serial CFA assays (Fig. 6d). Both drugs markedly reduced the percentage of CD26-expressing KCL22 cells at passage I, but this effect was no longer detected from passage II onward (Fig. 6d, left). It is useful to point out that control vehicle-treated KCL22 cultures contained fewer CD26 + cells at the end of passage I (around 45%) than at time 0 (Fig. 3a, table), a fact possibly due to the suppression in the course of culture of a sizeable subset of CD26 + cells of relatively low hierarchical level. Likewise, a significant reduction of the percentage of K562 cells expressing ALDH activity under ponatinib treatment was observed at passage I only, while imatinib did not show any effect at any passage (Fig. 6d, right). Taken together, these results seem to indicate that an LSC-like phenotype was selected progressively in culture across subsequent passages, paralleled by a loss of sensitivity to TKi.
Discussion
The development of molecularly tailored therapeutic agents such as BCR/ABL-active TKi led to the availability of an excellent treatment for CML. However, although capable of efficiently inducing remission of disease, neither imatinib nor later-generation TKi are very effective in preventing relapse upon treatment discontinuation. This is due to a number of reasons, including a relatively low efficacy of TKi on the LSC of CML [11, 68]. Despite the intensive research to develop non-TKi-based strategies capable of targeting TKi-insensitive LSC, TKi are still the only current treatment option for CML patients. It is to be noted, however, that a sizeable percentage of patients do not undergo relapse following TKi treatment discontinuation, indicating that in some cases TKi are actually effective on the CML stem/progenitor cell compartment(s). This prompted us to evaluate the effects of imatinib and ponatinib on a number of stem/progenitor cell features. In this study, ponatinib was more effective than imatinib in reducing cells with ALDH activity as well as the progenitor/stem cell potential of CML cell lines and patient-derived cells. In the latter, ponatinib did better that imatinib in reducing the number of CD26 + cells. Furthermore, ponatinib was less effective than imatinib in increasing the expression of NANOG and SOX2 stem cell–related genes.
When the effects of TKi on cells endowed with ALDH activity were tested, ponatinib was more effective than imatinib in reducing the percentage of ALDH activity-positive cells in primary CML cells and LAMA-84 cells. Imatinib and ponatinib exhibited comparable efficacy in K562 cells. The effect of imatinib on the K562 cell fraction expressing ALDH had been previously reported [69], while that of ponatinib on K562 cells and of imatinib or ponatinib on LAMA-84 cells have, to our knowledge, never been described before. It is worth pointing out that LAMA-84R cells, selected for resistance to imatinib, were equally insensitive to ponatinib as for the maintenance of ALDH-positive cells in culture (Fig. 4b). This effect was expected, as the mechanism of induction of LAMA-84R resistance to imatinib relies on BCR/ABL amplification [43]. The ALDH experiments seem to indicate, on one hand, that imatinib is actually capable of hitting, at least partially, the more immature fraction of CML cell population, and on the other hand, that in this respect the later-generation ponatinib is more effective than imatinib.
CD26 is nowadays considered the only CML-specific marker of LSC, and may be exploited as a prognostic marker of relapse [15]. In our study, ponatinib was more effective than imatinib in reducing the percentages of CD34 +/CD26 + primary CML cells, whereas imatinib and ponatinib showed similar efficacy in cultures of KCL22 cells. Other interesting observations came from the CFA assay performed with KCL22, where the effects of TKi progressively decreased, indicating that the CD26-positive population is heterogeneous and composed of TKi-sensitive and TKi-insensitive cell subsets. The existence of this duality seems to mirror the clinical observation that the percentage of CD26 + CML cells is reduced to low or undetectable levels in CML patients who respond to imatinib therapy and increases in peripheral blood and BM in the course of relapse of disease [17]. In this respect, when compared with CD26-positive cells, the ALDH-positive cells were overall more TKi resistant, indicating that the latter subpopulation includes relatively more immature progenitor/stem cells.
The effects of TKi on other phenotypic features were also addressed by evaluating the effects of imatinib and ponatinib on the expression of stem cell–related genes, including NANOG, SOX2, c-MYC, and KLF4. Either drug strongly upregulated NANOG and SOX2 in KCL22 and K562 cells, but in KCL22 cells this upregulation was significantly lower with ponatinib than with imatinib. The fact that the increase of NANOG and SOX2 expression was less pronounced with ponatinib than with imatinib cannot be attributed, in our opinion, to a reduced effect of ponatinib on stem cell pool, rather to the possibility that ponatinib, while determining, like imatinib, an enrichment of less immature cells, is in addition better suited than imatinib to suppress the NANOG +/SOX2 + cell subset. While the increased expression of NANOG and SOX2 by imatinib has been previously reported [70, 71], the effects of ponatinib shown here are novel. Both drugs, on the contrary, markedly reduced the expression of c-MYC. This is in agreement with the overall antiproliferative effect of TKi and with the fact that c-MYC plays multiple roles, including the control of stem cell clonal expansion, while NANOG and SOX2 are upregulated in association with the maintenance of cancer stem cell pool [26, 72]. The enhanced NANOG and SOX2 expression is also in keeping with the preferential effects of TKi on relatively less immature CML cell subsets, resulting in an apparent enrichment of stem cell phenotype [11]. Finally, in a previous study of ours carried out with non-CML cells [70], NANOG was markedly down-regulated and c-MYC markedly upregulated upon G1-S transition. Our conclusion was that NANOG preserves slowly or non-cycling stem cells, while c-MYC parallels stem cell clonal expansion. In the same study, as far as KLF4 is concerned, changes of NANOG and KLF4 expression appeared inversely proportional, their sum being constant across the different experimental variants, suggesting that c-MYC expression (and thereby G1-S transition) be regulated by the NANOG/KLF4 ratio. Along this line, in the present study, no statistically significant difference in expression levels of KLF4 was found between imatinib- and ponatinib-treated cells.
Additional interesting information came from CFA assays, which showed that, in both CML cell lines and patient-derived cells, ponatinib was, overall, more effective than imatinib in reducing progenitor/stem cell potential. This effect was progressively reduced across subsequent replatings, in keeping with the notion that TKi get progressively less effective as the hierarchical level of progenitor increases. This conclusion is further supported by the finding that the progressive fading off of TKi effect was paralleled by the consistent effectiveness of XMD8-92, which we previously showed to exhibit an enhanced inhibitory action on the relatively more immature CML cells and on the maintenance of the CML stem cell compartment. Besides providing interesting pieces of evidence with respect to the effect of imatinib versus ponatinib, our experiments indicated that serial CFA assays are adequate to test the effects of drugs on leukemia progenitor/stem cell subsets [8, 9, 12, 13, 38, 73].
Finally, the fact that the results obtained with primary cells were very similar to those obtained with stabilized CML cell lines suggests that the structure of the CML stem/progenitor cell compartment within stabilized cell lines derived from patients in blast crisis is rather similar to that of primary cells explanted from patients in the chronic phase.
Conclusion
This study shows that ponatinib exhibited similar or superior effects to those of imatinib when tested on a panel of CML progenitor/stem cell features. These in vitro results are well in keeping with the lower risk of relapse upon TKi discontinuation in patients treated with ponatinib [74]. However, in clinical practice, the superior effects of ponatinib over imatinib need to be balanced with the enhanced risk of adverse cardiovascular events consequent to ponatinib treatment [75–77]. We addressed in detail elsewhere the question of the phenotypical heterogeneity of the CML stem cell compartment in terms of the sensitivity or resistance to TKi [78–80]. Nevertheless, this work provides a sizeable contribution to the discussion of whether or not TKi are capable of targeting the CML stem cell compartment. Finally, the experimental approach exploited here may be useful in the future for rapid and cost-effective preclinical screening of the capacity of treatments, including those with drugs that do not target BCR/ABL, to reduce or suppress cell subsets endowed with progenitor/stem cell properties.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open access funding provided by Universitá degli Studi di Firenze within the CRUI-CARE Agreement.
Declarations
Funding
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (Grants IG5220, IG13466, and IG23607 to Persio Dello Sbarba; grants IG15282 and IG21349 to Elisabetta Rovida) and Istituto Toscano Tumori (Persio Dello Sbarba).
Conflicts of Interest
Antonella Gozzini and Barbara Scappini received honoraria from Novartis and Incyte for lectures at conferences on issues outside those of this work. Overall, the authors have no conflict of interest to declare.
Ethics approval
Patient-derived CML cells were collected following informed consent and under approval of the Ethical Committee of AOUC (authorization no. 520/10, October 18, 2010, renewed with no. 2015/0032965, November 4, 2015).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Authors’ contributions
Not applicable.
Footnotes
Ignazia Tusa and Giulia Cheloni contributed equally to the work.
Persio Dello Sbarba and Elisabetta Rovida are co-last authors.
Contributor Information
Persio Dello Sbarba, Email: persio@unifi.it.
Elisabetta Rovida, Email: erovida@unifi.it.
References
- 1.Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Saußele S, Richter J, Hochhaus A, Mahon FX. The concept of treatment-free remission in chronic myeloid leukemia. Leukemia. 2016;30:1638–1647. doi: 10.1038/leu.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hehlmann R, Müller MC, Lauseker M, Hanfstein B, Fabarius A, Schreiber A, et al. Deep molecular response is reached by the majority of patients treated with imatinib, predicts survival, and is achieved more quickly by optimized high-dose imatinib: results from the randomized CML-study IV. J Clin Oncol. 2014;32:415–423. doi: 10.1200/JCO.2013.49.9020. [DOI] [PubMed] [Google Scholar]
- 5.Rea D, Nicolini FE, Tulliez M, Guilhot F, Guilhot J, Guerci-Bresler A, et al. Discontinuation of dasatinib or nilotinib in chronic myeloid leukemia: interim analysis of the STOP 2G-TKI study. Blood. 2017;129:846–854. doi: 10.1182/blood-2016-09-742205. [DOI] [PubMed] [Google Scholar]
- 6.Hughes TP, Ross DM. Moving treatment-free remission into mainstream clinical practice in CML. Blood. 2016;128:17–23. doi: 10.1182/blood-2016-01-694265. [DOI] [PubMed] [Google Scholar]
- 7.Francois-Xavier M. Treatment-free remission in CML: who, how, and why? Hematol Am Soc Hematol Educ Program. 2017;2017:102–109. doi: 10.1182/asheducation-2017.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham SM, Jørgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 9.Giuntoli S, Rovida E, Barbetti V, Cipolleschi MG, Olivotto M, Dello Sbarba P. Hypoxia suppresses BCR/Abl and selects Imatinib-insensitive progenitors within clonal CML population. Leukemia. 2006;20:1291–1293. doi: 10.1038/sj.leu.2404224. [DOI] [PubMed] [Google Scholar]
- 10.Ghiaur G, Gerber J, Jones RJ. Concise review: cancer stem cells and minimal residual disease. Stem Cells. 2012;30:89–93. doi: 10.1002/stem.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129:1595–1606. doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 12.Cheloni G, Tanturli M, Tusa I, Ho DeSouza N, Shan Y, Gozzini A, et al. Targeting chronic myeloid leukemia stem cells with the hypoxia-inducible factor inhibitor acriflavine. Blood. 2017;130:655–665. doi: 10.1182/blood-2016-10-745588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusa I, Cheloni G, Poteti M, Gozzini A, DeSouza NH, Shan Y, et al. Targeting the extracellular signal-regulated kinase 5 pathway to suppress human chronic myeloid leukemia stem cells. Stem Cell Reports. 2018;11:929–943. doi: 10.1016/j.stemcr.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisterer W, Jiang X, Christ O, Glimm H, Lee KH, Pang E, et al. Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia. 2005;19:435–441. doi: 10.1038/sj.leu.2403649. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann H, Sadovnik I, Cerny-Reiterer S, Rulicke T, Stefanzl G, Willmann M, et al. Dipeptidylpeptidase IV CD26 defines leukemic stem cells LSC in chronic myeloid leukemia. Blood. 2014;123:3951–3962. doi: 10.1182/blood-2013-10-536078. [DOI] [PubMed] [Google Scholar]
- 16.Jiang X, Distinguishing CML. LSCs from HSCs using CD26. Blood. 2014;123:3851–3852. doi: 10.1182/blood-2014-05-574293. [DOI] [PubMed] [Google Scholar]
- 17.Warfvinge R, Geironson L, Sommarin MNE, Lang S, Karlsson C, Roschupkina T, et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 2017;129:2384–2394. doi: 10.1182/blood-2016-07-728873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, Weiss ML, Rao MS. In search of “stemness”. Exp Hematol. 2004;32:585–598. doi: 10.1016/j.exphem.2004.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douville J, Beaulieu R, Balicki D. ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev. 2008;23:23. doi: 10.1089/scd.2008.0055. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 21.Fallon P, Gentry T, Balber AE, Boulware D, Janssen WE, Smilee R, et al. Mobilized peripheral blood SSCloALDHbr cells have the phenotypic and functional properties of primitive haematopoietic cells and their number correlates with engraftment following autologous transplantation. Br J Haematol. 2003;122:99–108. doi: 10.1046/j.1365-2141.2003.04357.x. [DOI] [PubMed] [Google Scholar]
- 22.Hess DA, Meyerrose TE, Wirthlin L, Craft TP, Herrbrich PE, Creer MH, et al. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood. 2004;104:1648–1655. doi: 10.1182/blood-2004-02-0448. [DOI] [PubMed] [Google Scholar]
- 23.Fleischman AG. ALDH marks leukemia stem cell. Blood. 2012;119:3376–3377. doi: 10.1182/blood-2012-02-406751. [DOI] [PubMed] [Google Scholar]
- 24.Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B, et al. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195:52–60. doi: 10.1016/j.cbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 28.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 29.Nichols J, Pesce M, Scholer HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 30.Chen YC, Hsu HS, Chen YW, Tsai YH, How CK, Wang CY, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE. 2008;3:e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;280:1931–1938. doi: 10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- 32.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3 ⁄ 4 and Sox2 regulate Oct-3 ⁄ 4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 33.Phi JH, Park SH, Kim SK, Paek SH, Kim JH, Lee YJ, et al. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol. 2008;32:103–112. doi: 10.1097/PAS.0b013e31812f6ba6. [DOI] [PubMed] [Google Scholar]
- 34.Sholl LM, Long KB, Hornick JL. Sox2 Expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 2010;18:55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- 35.Ezeh UI, Turek PJ, Reijo RA, Amander TC. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 36.Sharma N, Magistroni V, Piazza R, Citterio S, Mezzatesta C, Khandelwal P, et al. BCR/ABL1 and BCR are under the transcriptional control of the MYC oncogene. Mol Cancer. 2015;14:132. doi: 10.1186/s12943-015-0407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham SA, Hopcroft LEM, Carrick E, Drotar ME, Dunn K, Williamson AJ, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giuntoli S, Tanturli M, Di Gesualdo F, Barbetti V, Rovida E, Dello Sbarba P. Glucose availability in hypoxia regulates the selection of chronic myeloid leukemia progenitor subsets with different resistance to imatinib-mesylate. Haematologica. 2011;96:204–212. doi: 10.3324/haematol.2010.029082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giuntoli S, Rovida E, Gozzini A, Barbetti V, Cipolleschi MG, Olivotto M, et al. Severe hypoxia defines heterogeneity and selects highly immature progenitors within clonal erythroleukemia cells. Stem Cells. 2007;25:1119–1125. doi: 10.1634/stemcells.2006-0637. [DOI] [PubMed] [Google Scholar]
- 40.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 41.Kubonishi I, Miyoshi I. Establishment of a Ph1 chromosome-positive cell line from chronic myelogenous leukemia in blast crisis. Int J Cell Cloning. 1983;1:105–117. doi: 10.1002/stem.5530010205. [DOI] [PubMed] [Google Scholar]
- 42.Seigneurin D, Champelovier P, Mouchiroud G, Berthier R, Leroux D, Prenant M, et al. Human chronic myeloid leukemic cell line with positive Philadelphia chromosome exhibits megakaryocytic and erythroid characteristics. Exp Hematol. 1987;15:822–1987. [PubMed] [Google Scholar]
- 43.le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, et al. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–1766. [PubMed] [Google Scholar]
- 44.Redaelli S, Boschelli F, Perini P, Pirola A, Viltadi M, Gambacorti-Passerini C. Synergistic activity of the Src/Abl inhibitor bosutinib in combination with imatinib. Leukemia. 2010;24:1223–1227. doi: 10.1038/leu.2010.79. [DOI] [PubMed] [Google Scholar]
- 45.Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- 46.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 47.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 48.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 49.Weisberg E, Manley PW, Breitenstein W, Brüggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 50.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;6:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boschelli DH, Wang YD, Ye F, Wu B, Zhang N, Dutia M, et al. Synthesis and Src kinase inhibitory activity of a series of 4-phenylamino-3-quinolinecarbonitriles. J Med Chem. 2001;44:822–833. doi: 10.1021/jm000420z. [DOI] [PubMed] [Google Scholar]
- 52.Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, et al. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18:258–267. doi: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin EC, Amantea CM, Nomanbhoy TK, Weissig H, Ishiyama J, Hu Y, et al. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc Natl Acad Sci USA. 2016;113:11865–11870. doi: 10.1073/pnas.1609019113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;567:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbetti V, Tusa I, Cipolleschi MG, Rovida E, Dello Sbarba P. AML1/ETO sensitizes via TRAIL acute myeloid leukemia cells to the pro-apoptotic effects of hypoxia. Cell Death Dis. 2013;4:e536. doi: 10.1038/cddis.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Culen M, Borsky M, Nemethova V, Razga F, Smejkal J, Jurcek T, et al. Quantitative assessment of the CD26 + leukemic stem cell compartment in chronic myeloid leukemia: patient-subgroups, prognostic impact, and technical aspects. Oncotarget. 2016;7:33016–33024. doi: 10.18632/oncotarget.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerber JM, Gucwa JL, Esopi D, Gurel M, Haffner MC, Vala M, et al. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget. 2013;4:715–728. doi: 10.18632/oncotarget.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bocchia M, Sicuranza A, Abruzzese E, Iurlo A, Sirianni S, Gozzini A, et al. Residual Peripheral Blood CD26(+) leukemic stem cells in chronic myeloid leukemia patients during TKI therapy and during treatment-free remission. Front Oncol. 2018;30:8–194. doi: 10.3389/fonc.2018.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue T, Swain A, Nakanishi Y, Sugiyama D. Multicolor analysis of cell surface marker of human leukemia cell lines using flow cytometry. Anticancer Res. 2014;34:4539–4550. [PubMed] [Google Scholar]
- 60.Jensen HA, Yourish HB, Bunaciu RP, Varner JD, Yen A. Induced myelomonocytic differentiation in leukemia cells is accompanied by noncanonical transcription factor expression. FEBS Open Bio. 2015;5:789–800. doi: 10.1016/j.fob.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darwish NH, Sudha T, Godugu K, Elbaz O, Abdelghaffar HA, Hassan EE, et al. Acute myeloid leukemia stem cell markers in prognosis and targeted therapy: potential impact of BMI-1, TIM-3 and CLL-1. Oncotarget. 2016;7:57811–57820. doi: 10.18632/oncotarget.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landberg N, von Palffy S, Askmyr M, Lilljebjörn H, Sandén C, Rissler M, et al. CD36 defines primitive chronic myeloid leukemia cells less responsive to imatinib but vulnerable to antibody-based therapeutic targeting. Haematologica. 2018;103:447–455. doi: 10.3324/haematol.2017.169946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anorma C, Hedhli J, Bearrood TE, Pino NW, Gardner SH, Inabaet H, et al. Surveillance of cancer stem cell plasticity using an isoform-selective fluorescent probe for aldehyde dehydrogenase 1A1. ACS Cent Sci. 2018;4:1045–1055. doi: 10.1021/acscentsci.8b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bliss SA, Paul S, Pobiarzyn PW, Ayer S, Sinha G, Pant S, et al. Evaluation of a developmental hierarchy for breast cancer cells to assess risk-based patient selection for targeted treatment. Sci Rep. 2018;8:367. doi: 10.1038/s41598-017-18834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stivarou T, Cipolleschi MG, Damico M, Mannini A, Mini E, Rovida E, et al. The complex metabolic network gearing the G1/S transition in leukemic stem cells: hints to a rational use of antineoplastic agents. Oncotarget. 2015;6:31985–31996. doi: 10.18632/oncotarget.5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yaghmaie M, Yeung CC. Molecular mechanisms of resistance to tyrosine kinase inhibitors. Curr Hematol Malig Rep. 2019;14:395–404. doi: 10.1007/s11899-019-00543-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhang M, Luo Z, Liu H, Croce CM, Jr, Burke TR, Bottaro DP. Synergistic anti-leukemic activity of imatinib in combination with a small molecule Grb2 SH2 domain binding antagonist. Leukemia. 2014;28:948–951. doi: 10.1038/leu.2013.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bono S, Dello Sbarba P, Lulli M. Imatinib-mesylate enhances the maintenance of chronic myeloid leukemia stem cell potential in the absence of glucose. Stem Cell Res. 2018;28:33–38. doi: 10.1016/j.scr.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 71.Chorzalska A, Kim JF, Roder K, Tepper A, Ahsan N, Rao RSP, et al. Long-term exposure to imatinib mesylate downregulates hippo pathway and activates YAP in a model of chronic myelogenous leukemia. Stem Cells Dev. 2017;26:656–677. doi: 10.1089/scd.2016.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marzi I, Cipolleschi MG, D’Amico M, Stivarou T, Rovida E, Vinci MC, et al. The involvement of a Nanog, Klf4 and c-Myc transcriptional circuitry in the intertwining between neoplastic progression and reprogramming. Cell Cycle. 2013;12:353–364. doi: 10.4161/cc.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puissant A, Dufies M, Fenouille N, Ben Sahra I, Jacquel A, Robert G, et al. Imatinib triggers mesenchymal-like conversion of CML cells associated with increased aggressiveness. J Mol Cell Biol. 2012;4:207–220. doi: 10.1093/jmcb/mjs010. [DOI] [PubMed] [Google Scholar]
- 74.Cortes JE, Kim DW, Pinilla-Ibarz J, le Coutre PD, Paquette R, Chuah C, et al. Ponatinib efficacy and safety in Philadelphia chromosome-positive leukemia: final 5-year results of the phase 2 PACE trial. Blood. 2018;132:393–404. doi: 10.1182/blood-2016-09-739086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caocci G, Mulas O, Abruzzese E, Luciano L, Iurlo A, Attolico I, et al. Arterial occlusive events in chronic myeloid leukemia patients treated with ponatinib in the real-life practice are predicted by the Systematic Coronary Risk Evaluation (SCORE) chart. Hematol Oncol. 2019;37:296–302. doi: 10.1002/hon.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Latifi Y, Moccetti F, Wu M, Xie A, Packwood W, Qi Y, et al. Thrombotic microangiopathy as a cause of cardiovascular toxicity from the BCR-ABL1 tyrosine kinase inhibitor ponatinib. Blood. 2019;133:1597–1606. doi: 10.1182/blood-2018-10-881557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dorer DJ, Knickerbocker RK, Baccarani M, Cortes JE, Hochhaus A, Talpaz M, et al. Impact of dose intensity of ponatinib on selected adverse events: multivariate analyses from a pooled population of clinical trial patients. Leuk Res. 2016;48:84–91. doi: 10.1016/j.leukres.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 78.Rovida E, Peppicelli S, Bono S, Bianchini F, Tusa I, Cheloni G, et al. The metabolically-modulated stem cell niche: a dynamic scenario regulating cancer cell phenotype and resistance to therapy. Cell Cycle. 2014;13:3169–3175. doi: 10.4161/15384101.2014.964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cipolleschi MG, Marzi I, Rovida E, Dello Sbarba P. Chronic myeloid leukemia and hepatoblastoma: two cancer models to link metabolism to stem cells. Front Oncol. 2016;6:95. doi: 10.3389/fonc.2016.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, Li S. Leukemia Stem Cells in Hematologic Malignancies. 1st ed. Springer Nature Singapore Pte Ltd; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.