Abstract
Amitriptyline is a tricyclic antidepressant commonly prescribed for major depressive disorders, as well as depressive symptoms associated with various neurological disorders. A possible correlation between the use of tricyclic antidepressants and the occurrence of Parkinson’s disease has been reported, but its underlying mechanism remains unknown. The accumulation of misfolded protein aggregates has been suggested to cause cellular toxicity and has been implicated in the common pathogenesis of neurodegenerative diseases. Here, we examined the effect of amitriptyline on protein clearance and its relevant mechanisms in neuronal cells. Amitriptyline exacerbated the accumulation of abnormal aggregates in both in vitro neuronal cells and in vivo mice brain by interfering with the (1) formation of aggresome-like aggregates and (2) autophagy-mediated clearance of aggregates. Amitriptyline upregulated LC3B-II, but LC3B-II levels did not increase further in the presence of NH4Cl, which suggests that amitriptyline inhibited autophagic flux rather than autophagy induction. Amitriptyline interfered with the fusion of autophagosome and lysosome through the activation of PI3K/Akt/mTOR pathway and Beclin 1 acetylation, and regulated lysosome positioning by increasing the interaction between proteins Arl8, SKIP, and kinesin. To the best of our knowledge, we are the first to demonstrate that amitriptyline interferes with autophagic flux by regulating the autophagosome maturation during autophagy in neuronal cells. The present study could provide neurobiological clue for the possible correlation between the amitriptyline use and the risk of developing neurodegenerative diseases.
Subject terms: Cellular neuroscience, Neurodegeneration
Introduction
Amitriptyline is a tricyclic antidepressant used to manage major depressive disorders, anxiety disorders, and other mental illnesses. Antidepressive activity of amitriptyline is associated with the increase in synaptic neurotransmission of serotonin and norepinephrine by blocking their neuronal reuptake from the synapse in the central nervous system1. Approximately, 40–80% of patients with neurodegenerative diseases suffer from depressive symptoms2–5. Amitriptyline could be used to treat depression and anxiety symptoms associated with neurodegenerative diseases including Parkinson’s disease (PD). Patients who received amitriptyline were reported to ameliorate PD-associated depressive symptoms, and delay the need for dopamine therapy as compared with patients who did not take the drug6,7. On the other hand, its chronic use has been demonstrated to be a risk factor for neurodegenerative diseases as evidenced by tricyclic antidepressants-induced increase in neuronal cell death in a dorsal root ganglion cell culture model8, PD-related neurotoxicity and movement disorders caused by its long-term use in an in vivo model9, and development of drug-induced PD by antidepressants10. Despite these evidences, there is critical knowledge gap in understanding the underlying mechanism by which amitriptyline causes neurotoxicity.
Protein quality control is crucial for maintaining cellular homeostasis and the functionality of eukaryotic cells. Reactive oxygen species are increased by environmental factors and intracellular metabolism, which cause the accumulation of abnormally high levels of damaged proteins in cells11,12. In addition, intracellular pathways responsible for protein homeostasis gradually decrease with aging13. Because neurons generally do not replicate, they are more vulnerable to the accumulation of toxic protein aggregates and damaged organelles than nonneuronal cells14,15. Therefore, the dysregulation of protein quality control is closely associated with an increased risk of developing neurodegenerative diseases16–18. The abnormal accumulation of misfolded proteins such as β-amyloid/tau, α-synuclein, and huntingtin is a characteristic pathology of Alzheimer’s disease, PD, and Huntington’s disease, respectively19–22.
There are two main intracellular protein clearance systems maintaining protein homeostasis: ubiquitin–proteasome system and autophagy. Autophagy is a degradation and recycling system targeting damaged or dysfunctional protein complexes and intracellular organelles by delivering them into lysosome, and is upregulated during stressful conditions including the accumulation of misfolded proteins23,24. In mammalian cells, there are three major types of autophagy: chaperone-mediated autophagy, microautophagy, and macroautophagy25. During chaperon-mediated autophagy, chaperones identify protein containing KFERQ-like sequence, associate them to the lysosomal membrane protein LAMP-2A, and translocate of the protein cargo into the lysosome. In microautophagy, the direct uptake of cargo protein is occurred through sequestration of the lysosomal membrane. Macroautophagy is the best studied, and has gained attention in the field of adult-onset neurodegeneration26. Macroautophagy, hereafter referred to as autophagy, begins with the formation of a double-membrane structure called phagophore from intracellular membrane components. During this process, microtubule-associated protein light chain 3 (LC3)-I is lipidated to LC3-II, then localized to the phagophore membrane. Autophagy adaptor p62/SQSTM1 (p62) recruits polyubiquitinated materials and binds to ubiquitinated cargoes as well as LC3, thereby targeting the autophagosome and facilitating clearance of ubiquitinated proteins. After the fusion of the autophagosome with lysosome, the substances are degraded by lysosomal enzymes27–29. The balance between autophagosome formation and lysosomal degradation is critical for autophagy-mediated protein clearance. Therefore, the dysregulation of autophagy induction and/or autophagic flux results in the abnormal accumulation of protein aggregates and cytotoxicity.
The purpose of this study is to evaluate whether amitriptyline causes abnormal accumulation of protein aggregates in neuronal cells and whether it dysregulates autophagy function. We also determined the specific step in the autophagy process regulated by amitriptyline, as well as its signaling mechanism. Because autophagy is primarily responsible in preventing abnormal protein accumulation in neuronal cells, this study demonstrating the association between the use of amitriptyline and the interference of autophagy-mediated protein clearance might provide new insight into the risk of neurodegenerative diseases.
Results
Amitriptyline increases protein aggregates in neuronal cells and mice brain
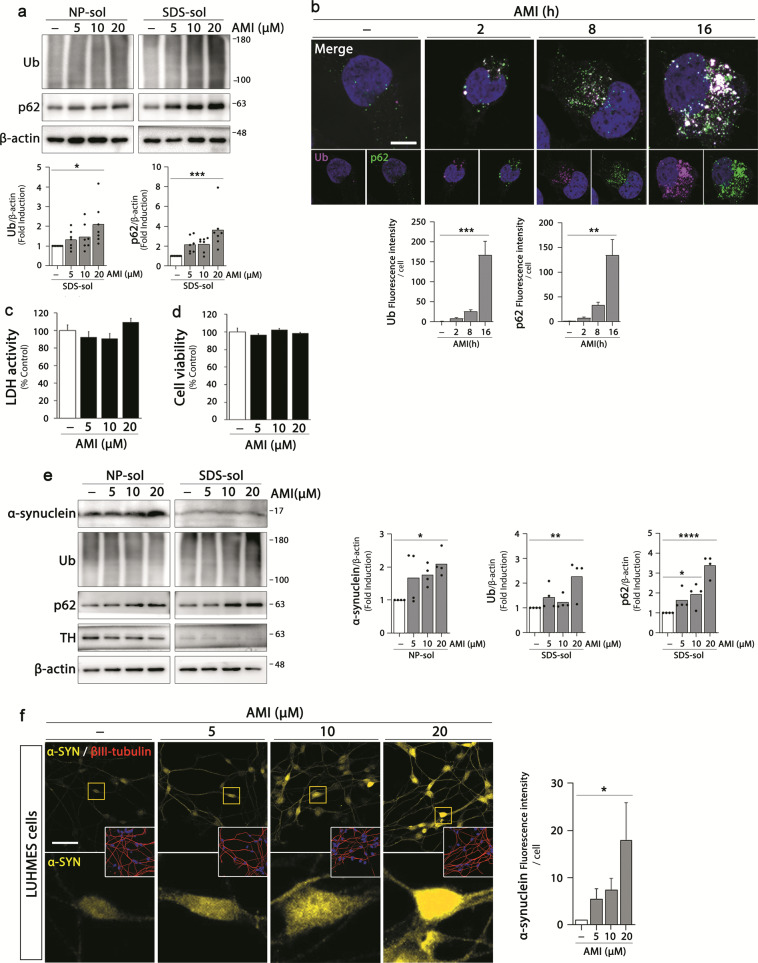
The effect of amitriptyline on the accumulation of protein aggregates in neuronal cells was examined by using all-trans retinoic acid-induced differentiated SH-SY5Y cells. We differentiated SH-SY5Y cells for 3 days and then treated with amitriptyline at a concentration range of 5–20 μM for additional 24 h, which concentrations were often used in in vitro studies on amitriptyline effects9,30,31. To evaluate the effect of amitriptyline on aggregates accumulation, immunoblot analyses with ubiquitin and p62 were performed on NP- and sodium dodecyl sulfate (SDS)-soluble cell fractions, respectively (Fig. 1a). The accumulation of both ubiquitin polymer (oligomer) and p62 tended to increase with increasing concentration of amitriptyline in SDS-soluble fractions of cells, and a significant increase was shown at 20 μM. We also performed immunostaining of amitriptyline-treated SH-SY5Y cells with ubiquitin and p62 antibodies and analyzed the intracellular levels and distribution pattern under confocal microscope. Ubiquitin and p62 immunoreactivities were significantly increased and largely colocalized with each other after 16 h (Fig. 1b). Amitriptyline-induced increase in ubiquitin and p62 was also detected at 8 h, but not significant. Moreover, there was less colocalization of ubiquitin and p62, which indicated that amitriptyline-induced protein aggregates were ubiquitinated and linked to p62 over time. Amitriptyline was not cytotoxic to differentiated SH-SY5Y cells in the given concentration range (Fig. 1c, d).
Fig. 1. Effect of amitriptyline on protein accumulation in neuronal cells.
a SH-SY5Y cells were differentiated with 10 μM retinoic acid and treated with amitriptyline at concentrations of 5, 10, or 20 μM for 24 h. Ubiquitin and p62 protein levels in the NP- and SDS-soluble fractions were analyzed by western blotting. Protein levels in the SDS-fraction were quantified by densitometric analysis (n = 7). b SH-SY5Y cells were treated with amitriptyline at 20 μM for 2, 8, and 16 h and ubiquitin (pink) and p62 (green) levels were analyzed by immunofluorescence. DAPI staining (blue) represented the nuclei. Scale bar corresponds to 10 μm. Quantification of ubiquitin and p62 levels in b is shown in the below panel. (n = 15–20) c, d SH-SY5Y cells were treated amitriptyline at 5, 10, and 20 μM for 24 h, and the extent of cell death was assessed by LDH assay (c) and MTT assay (d) (mean ± S.E.M., n = 11–12). e LUHMES cells were differentiated as described in “Materials and methods” and treated with amitriptyline at concentrations of 5, 10, or 20 μM for 24 h. Alpha-synuclein, ubiquitin, p62, and tyrosine hydroxylase levels in the NP- and SDS-soluble fractions were analyzed by western blotting. Protein levels in the NP-soluble (α-synuclein) and SDS-soluble (ubiquitin and p62) fractions were quantified by densitometric analysis (n = 4). f LUHMES cells were treated with amitriptyline at 5, 10, or 20 μM for 24 h, and immunostained with α-synuclein (yellow) and βIII-tubulin (red) antibodies. DAPI staining (blue) represented the nuclei. Scale bar corresponds to 50 μm. Quantification of α-synuclein immunoreactivity in f was shown in the right panel. (n = 100–150) *P < 0.05, **P < 0.01, ***<0.001, ****P < 0.0001. AMI amitriptyline, NP-sol NP-soluble fraction, SDS-sol SDS-soluble fraction, Ub ubiquitin, α-SYN α-synuclein, TH tyrosine hydroxylase.
To verify the effect of amitriptyline on the protein accumulation in more pathophysiologically relevant in vitro model, we examined the amitriptyline effects in differentiated Lund human mesencephalic (LUHMES) cells. LUHMES cells are human embryonic neuronal precursor cells and differentiated into human dopaminergic neurons, which characterized by increased expression of dopaminergic neuronal markers32,33. Because LUHMES cells express α-synuclein and the accumulation of endogenous α-synuclein aggregate is associated with PD pathology33, we also evaluated whether accumulation of α-synuclein was induced by amitriptyline. LUHMES cells were differentiated for 5 days and treated with amitriptyline for additional 24 h. The expression of dopaminergic neuronal marker was confirmed by immunoblotting against tyrosine hydroxylase (Fig. 1e). Similar to SH-SY5Y cells, significant increase in ubiquitin and p62 proteins was detected in SDS-soluble fractions of amitriptyline (20 μM)-treated LUHMES cells (significant increase in p62 was shown even at 10 μM). Moreover, amitriptyline caused accumulation of α-synuclein in NP-soluble fractions. Confocal images also showed amitriptyline-induced accumulation of α-synuclein in LUHMES cells (Fig. 1f) and primary ventral mesenchymal (VM) dopaminergic neurons (Supplementary Fig. S1a). Taken together, these data indicate that amitriptyline induces accumulation of abnormal proteins in neuronal cells.
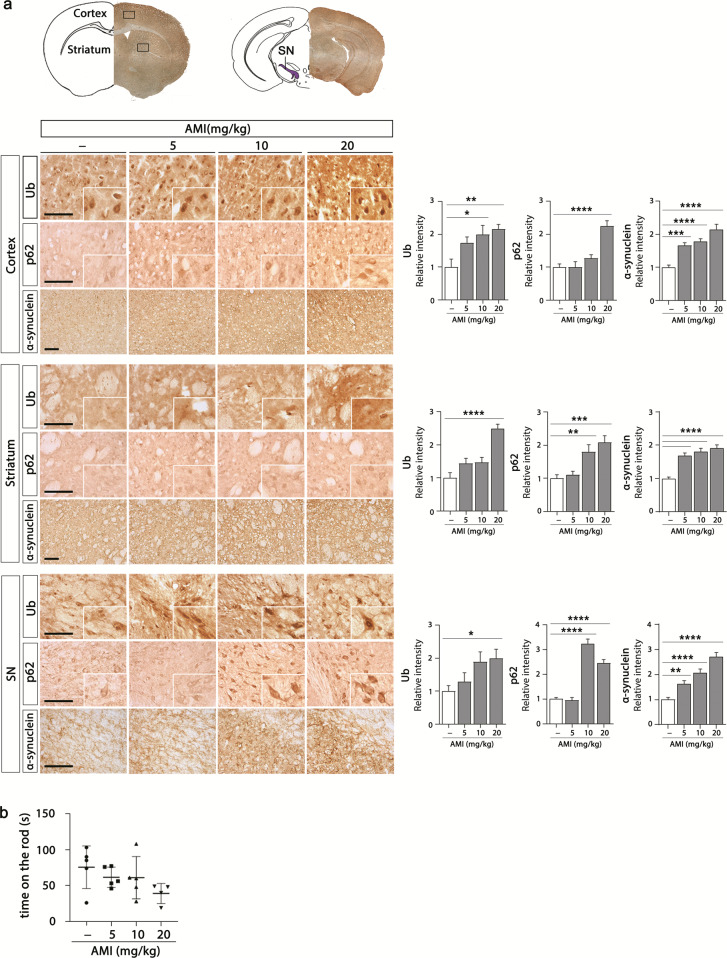
Next, we examined the effect of chronic exposure to amitriptyline on the accumulation of protein aggregates in the brain of mice. We injected amitriptyline (i.p. daily for 30 days; 5, 10, and 20 mg/kg) to male C57BL/6 mice, and evaluated ubiquitin, p62, and α-synuclein levels of the cortex, striatum, and substantia nigra (SN) by immunohistochemistry. As shown in Fig. 2a, the significantly increased accumulation of ubiquitin, p62, and α-synuclein was detected in amitriptyline-treated mice brain. Significantly increase in α-synuclein was shown by relatively low doses of amitriptyline (even at 5 mg/kg). To verify whether these conditions caused motor dysfunction, we examined motor coordination of mice by using rotarod test. The exposure of amitriptyline under this condition tended to decrease the motor function of the mice (Fig. 2b). These results clearly show that amitriptyline causes the accumulation of protein aggregates in in vivo models like as in vitro.
Fig. 2. Effect of amitriptyline on protein accumulation in mice brain.
a Mice were injected with amitriptyline (5, 10, and 20 mg/kg, i.p., n = 5) for 30 d, and immunohistochemistry against anti-ubiquitin, anti-p62, and anti-α-synuclein was performed in the cortex, striatum, and SN of mice brain. Immunoreactivities were quantified by densitometric analysis. Scale bar corresponds to 100 μm. b Motor dysfunction was assessed by the rotarod test. The latency to fall from a rotating rod was automatically recorded (n = 4–5). *P < 0.05, **P < 0.01, ***<0.001, ****P < 0.0001. AMI amitriptyline, SN subsantia nigra, Ub ubiquitin.
Amitriptyline aggravates a MG132-induced accumulation of aggregates
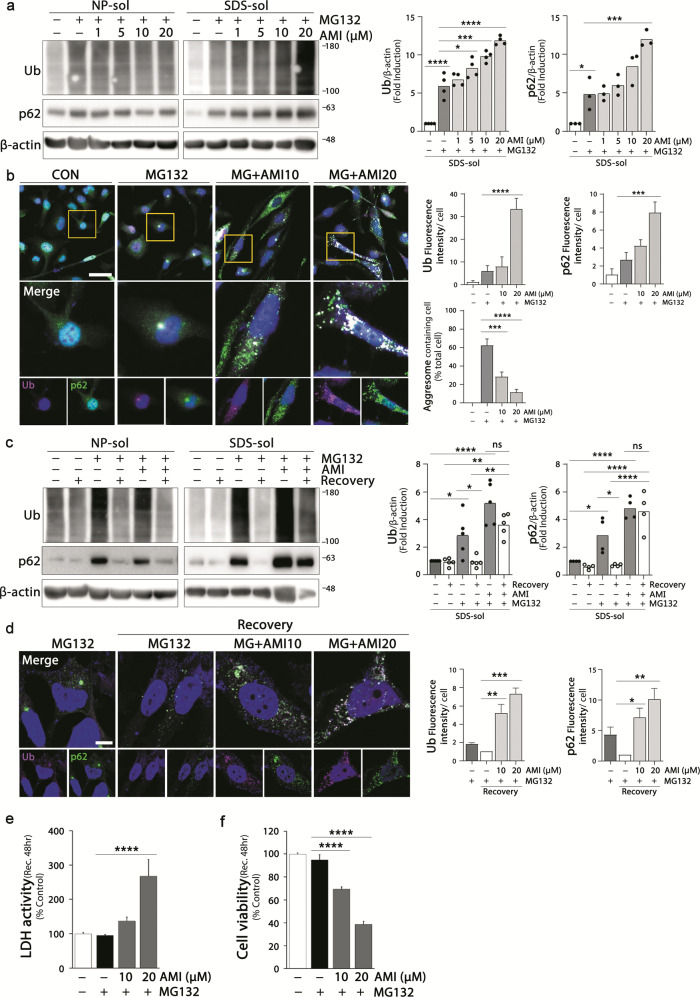
Next we would like to verify whether amitriptyline exacerbates the accumulation of already existing protein aggregates in neuronal cells. We used a previously established in vitro experimental model that represents the accumulation of aggresome-like aggregates and following autophagy-mediated clearance of aggregates34. Significant increases in protein aggregates were detected in differentiated SH-SY5Y cells by sequential treatment with proteasome inhibitor MG132 (treated with 0.1 μM MG132 for 16 h, incubated with fresh media for additional 8 h, and then treated with 1 μM MG132 for 16 h). MG132-induced increases in ubiquitin and p62 levels in SDS-soluble fractions were exacerbated by amitriptyline (Fig. 3a). Amitriptyline itself induced accumulation of protein aggregates in neuronal cells (Fig. 1a and Supplementary Fig. S2a), and significantly aggravated the accumulation in MG132-treated cells.
Fig. 3. Effect of amitriptyline on MG132-induced accumulation of aggregates.
a, b Ubiquitin and p62 levels in differentiated SH-SY5Y cells treated with amitriptyline and MG132 were analyzed by western blotting. SH-SY5Y cells were treated with MG132 in a two-step process (pro-incubated with a low dose of 0.1 μM for 16 h and changed to fresh normal media; after 8 h, incubated with a high dose of 1 μM for additional 16 h). Amitriptyline was applied 1 h prior to MG132 high-dose treatment. Quantification of ubiquitin and p62 in the SDS-fractions in a is shown in the right panel (n = 3–4). The expression and localization of ubiquitin (magenta) and p62 (green) in SH-SY5Y cells treated with MG132 and amitriptyline were analyzed by immunofluorescence (b). Scale bar corresponds to 50 μm. Quantification of ubiquitin and p62 immunofluorescence intensity in b is shown in the right panel. Percentage of aggresome (spots larger than 2 μm in diameter) in total cell is shown in the right panel (n = 60–100) c SH-SY5Y cells were treated with 1 μM MG132 in a two-step process and 20 μM amitriptyline as described in (a) and incubated with fresh media for an additional 24 h after MG132 treatment. Amitriptyline groups were treated amitriptyline in fresh media. Ubiquitin and p62 protein levels were analyzed by western blot. Protein levels in the SDS-fraction were quantified by densitometric analysis (n = 4–5). d The ubiquitin and p62 aggregates in MG132 and amitriptyline co-treated cells after the recovery period was evaluated by immunofluorescence. Scale bar corresponds to 10 μm. (n = 25–30) Quantification of ubiquitin and p62 immunofluorescence intensity in (d) is shown in the right panel. e, f SH-SY5Y cells were pretreated with amitriptyline for 1 h and treated MG132 in a two-step process, followed by a recovery period for 48 h. Cell death was assessed by LDH assay (e) and MTT assay (f) (mean ± S.E.M.; n = 7–8). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. AMI amitriptyline, CON control, NP-sol NP-soluble fraction, SDS-sol SDS-soluble fraction, Ub ubiquitin.
Immunofluorescence images also showed that amitriptyline increased the MG132-induced accumulation of ubiquitin and p62 and their colocalization with increasing concentration, which was significant at 20 μM (Fig. 3b). In addition, amitriptyline significantly attenuated aggresome formation in MG132-treated cells. Clear and large immunospots (spots larger than 2 μm in diameter) of ubiquitin and p62, indicative of the formation of aggresome-like aggregates34, were detected in the cytosol of MG132-treated cells. However, in cells co-treated with amitriptyline and MG132, smaller and more scattered aggregates were distributed in the cytosol, instead of clear perinuclear aggresomes.
Because aggresome formation is necessary for the autophagy-mediated protein clearance, these findings suggest the possibility that amitriptyline interferes with autophagy-mediated protein clearance. To verify this, we examined the effect of amitriptyline on the clearance of MG132-induced protein aggregate in SH-SY5Y cells. After sequential treatment with the proteasome inhibitor MG132 to induce autophagy, the differentiated SH-SY5Y cells were subjected to recovery through fresh media for 24 h, and the protein aggregates levels were evaluated. During the recovery period, amitriptyline was added at the same concentration without MG132. As shown in Fig. 3c, d, almost all the ubiquitin and p62 proteins were removed in MG132-treated control cells after the recovery period, but not in cells treated with amitriptyline and MG132. Immunofluorescence showed that MG132-induced aggresome-like aggregates disappeared after the recovery period. In cells co-treated with amitriptyline and MG132, ubiquitin and p62 in the form of scattered spots throughout the cells were still detected after the recovery period. These results indicate that amitriptyline significantly interrupted the protein clearance system in MG132-treated cells even at 10 μM, and consequently, ubiquitinated proteins bound with p62 were not removed, but accumulated in the cytoplasm.
To evaluate whether disrupted protein clearance makes cells more vulnerable to MG132 toxicity, we examined the effect of amitriptyline on the viability of MG132-treated SH-SY5Y cells after a recovery period of 48 h (Fig. 3e, f). The viability of MG132-treated cells slightly decreased compared with that of control cells, whereas that of cells treated with 20 μM of amitriptyline and MG132 decreased to 38.55% compared with that of control cells (38.55 ± 2.80%, P < 0.0001 vs. untreated control cells; Fig. 3f). These results suggest that amitriptyline aggravates the accumulation of abnormal protein aggregates in the SH-SY5Y cells by disturbing the protein clearance system.
Amitriptyline interferes with autophagy turnover
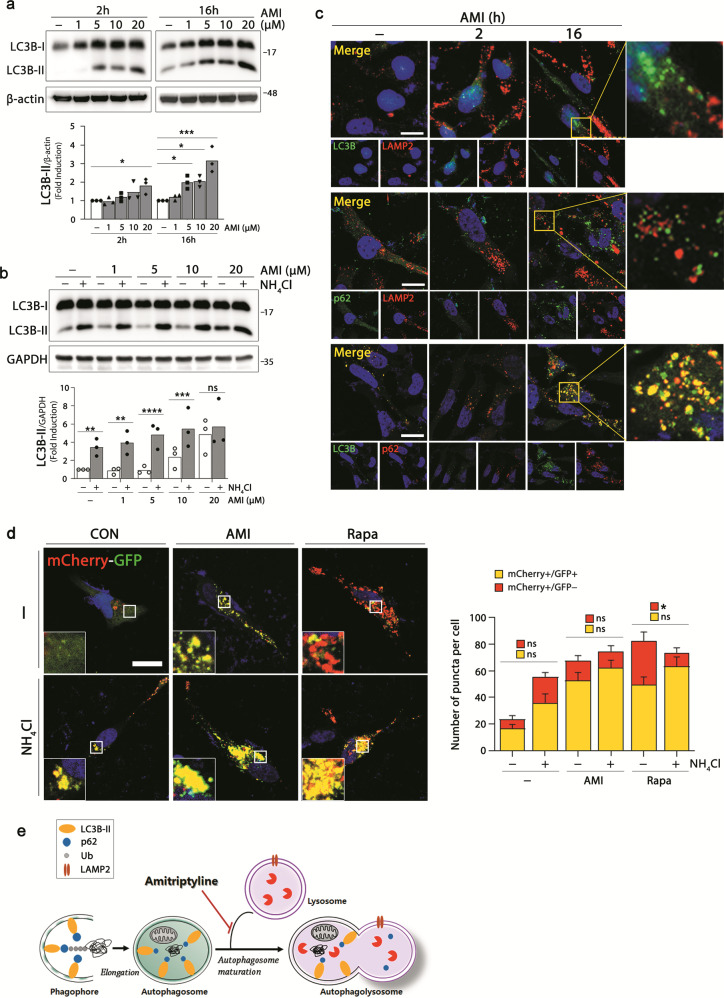
We next asked which stage of the autophagy process was affected by amitriptyline and therefore relevant to abnormal protein clearance in amitriptyline-treated cells. To identify the step in the autophagy process affected by amitriptyline, we first examined the effect of amitriptyline on the LC3-II shift, a marker for autophagosomal formation (Fig. 4a). The increase in LC3B-II was detected with increasing concentration of amitriptyline as early as 2 h after treatment. A significant increase in LC3B-II was induced at 20 μM after 2 h of amitriptyline treatment, but more significant increases were detected even at low concentrations (5 and 10 μM) after 16 h. Because these increases can also be caused by the blockade of the autophagy process, we evaluated autophagic flux by measuring LC3B-II turnover. We compared the LC3B-II levels in amitriptyline-treated SH-SY5Y cells in the presence or absence of NH4Cl. NH4Cl inhibits the fusion of autophagosome and lysosome, as well as the degradation of a group of autophagosomal cargoes, by preventing the acidification in the lysosome35. Therefore, it is used to monitor autophagic flux. As shown in Fig. 4b, the presence of NH4Cl significantly increased LC3B-II levels, without significant differences between control and amitriptyline-treated cells. Higher concentration of amitriptyline (20 μM) itself caused LC3B-II accumulation, and there were no significant differences in the LC3B-II levels of cells treated with 20 μM of amitriptyline in terms of the absence and the presence of NH4Cl. These results suggest that the increase in LC3B-II by amitriptyline could be due to the blockade of autophagic flux.
Fig. 4. Inhibition of autophagy turnover by amitriptyline.
a Differentiated SH-SY5Y cells were treated with amitriptyline (1, 5, 10, and 20 μM) for 2 and 16 h, and analyzed LC3B-I and LC3B-II levels by western blotting. LC3B-II levels were quantified by densitometric analysis (n = 3). b Cells were pretreated with 5 mM NH4Cl for 1 h, followed by amitriptyline (1, 5, 10, and 20 μM) for 24 h, and LC3B-I and II levels were analyzed by western blotting. Quantification of LC3B-II in (b) is shown in the below panel (n = 3). c Cells were treated with 20 μM amitriptyline for 2 or 16 h, and performed immunofluorescence against LC3B (green) and LAMP2 (red) in the first line, p62 (green) and LAMP2 (red) in the second line, and LC3B (green) and p62 (red) in the third line. Scale bar corresponds to 20 μm. d Cells were transfected with mCherry-GFP-LC3 plasmid and differentiated for 3 days. Cells were pretreated with 5 mM NH4Cl for 1 h, and treated with amitriptyline (20 μM) or rapamycin (2.5 μM) for additional 16 h. Scale bar corresponds to 50 μm. Quantification of mRFP+/GFP+ and mRFP+/GFP− puncta per cell is shown in right panel (n = 8–12). e Schematic diagram predicted effect of amitriptyline on autophagolysosome formation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. AMI amitriptyline. AMI amitriptyline, CON control, Rapa rapamycin, Ub ubiquitin.
From these results, we examined whether amitriptyline interfered with the fusion of autophagosome and lysosome to form autophagolysosome. Cells were treated with amitriptyline, and the distribution patterns of autophagosome marker LC3B, autophagosomal substrate marker p62, and lysosome marker LAMP2 were evaluated by immunofluorescence analysis (Fig. 4c). Both LC3B and LAMP2 levels were increased by amitriptyline in a time-dependent manner, but most were not colocalized. Similar result was detected in immunofluorescence image against p62 and LAMP2. But large portion of LC3B and p62 immunoreactivities were colocalized in amitriptyline-treated cells. To confirm the effect of amitriptyline on the autophagolysosome formation, we conducted additional experiment using a tandem tagged mCherry-GFP-LC3B plasmid. mCherry-GFP-LC3B is widely used to monitor autophagic flux; mCherry and green fluorescent protein (GFP) retain each fluorescence in autophagosome, but GFP fluorescence is quenched in the low pH of autophagolysosome36. SH-SY5Y cells transfected with mCherry-GFP-LC3B plasmids were treated with amitriptyline in the presence or absence with NH4Cl. As shown in Fig. 4d, both mCherry and GFP showed up in amitriptyline-treated cells, and NH4Cl had no significant effect on the number of autophagosome (yellow puncta) among the total puncta in amitriptyline-treated cells, which indicates defective formation of autophagolysosome by amitriptyline. On the other hand, rapamycin, a positive regulator of the autophagy induction, increased the quenching of GFP (suggestive of the maturation of autophagosome), which was significantly interrupted by NH4Cl (Fig. 4d). These results demonstrate that amitriptyline does not inhibit autophagosome formation, but inhibits autophagic flux by interfering with the fusion of autophagosome and lysosome (Fig. 4e).
Amitriptyline regulates lysosome positioning
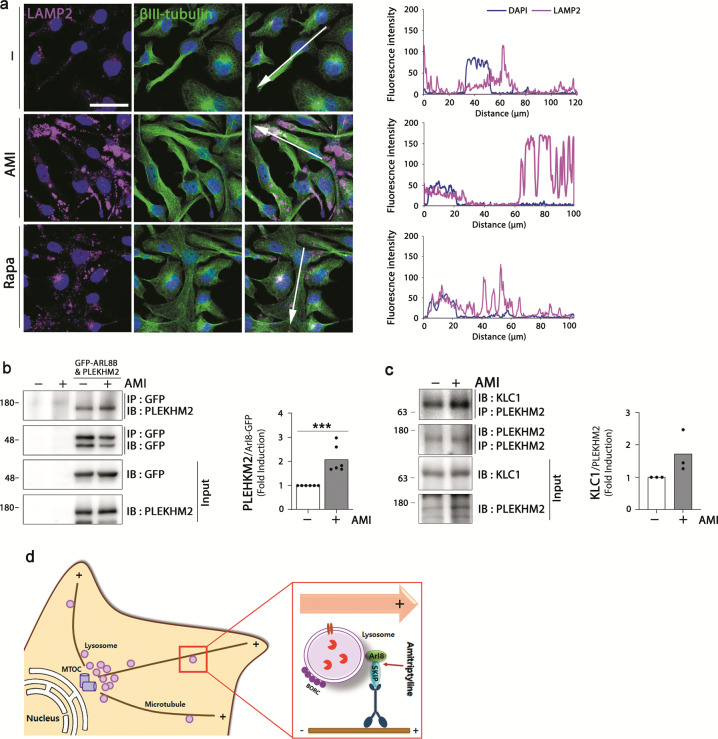
Lysosomes are highly dynamic organelles and their spatiotemporal characteristics could affect autophagolysosome formation and cargo degradation37. It is of interesting that immunofluorescence images of amitriptyline-treated SH-SY5Y cells showed that LAMP2 was mostly located at the distal end of cells compared with that of the control (Fig. 4c). To verify this, we compared the cytosolic localization of LAMP2 between cells treated with amitriptyline and positive autophagy regulator rapamycin (Fig. 5a). Large portion of LAMP2 immunoreactivity was detected in the peripheral part of amitriptyline-treated cells, but most of LAMP2 was detected in the perinuclear location of rapamycin-treated cells. Because SKIP (SifA and kinesin-interacting protein, also called PLEKHM2) and small GTPase Arl8 (Arf-like G protein) interact with kinesin and are relevant to the anterograde movement of the lysosomes38, we then determined whether the increased binding of Arl8, kinesin-1, and SKIP was involved in the peripheral distribution of lysosomes in amitriptyline-treated cells. We overexpressed HEK-293T cells with GFP-Arl8 and SKIP and performed immunoprecipitation for Arl8 and SKIP in cell lysates treated with 20 μM of amitriptyline. As shown in Fig. 5b, the interaction between SKIP and Arl8 was significantly increased by amitriptyline. Interaction between kinesin LC-1 (KLC1) and SKIP was also showed tendency to increase after treatment with amitriptyline (Fig. 5c). These results suggest that amitriptyline induces the translocation of lysosome to the plus-end direction of the cells by increasing the interactions between Arl8, kinesin, and SKIP (Fig. 5d).
Fig. 5. Effect of amitriptyline on lysosome distribution.
a Differentiated SH-SY5Y cells were treated with amitriptyline (20 μM) or rapamycin (2.5 μM) for 24 h and analyzed LAMP2 and βIII-tubulin localization by immunofluorescence. Scale bar corresponds to 50 μm. Quantification of LAMP2 and DAPI intensity along the arrow line in a is shown in the right panel. b HEK-293T cells were transiently transfected with Arl8-GFP and PLEKHM2 and treated with 20 μM amitriptyline for 24 h. The binding of Arl8 and SKIP/PLEKHM2 in cells was analyzed by immunoprecipitation with anti-GFP and immunoblotting with anti-PLEKHM2. Quantification of PLEKHM2 per Arl8-GFP is shown in the right panel (n = 6). c HEK-293T cells were treated with amitriptyline for 24 h and the binding of KLC1 and PLEKHM2 in cells was analyzed by immunoprecipitation. Quantification of KLC1 per PLEKHM2 is shown in the right panel (n = 3). d Schematic diagram showing the effect of amitriptyline on lysosome positioning. ***P < 0.001. AMI amitriptyline, IP immunoprecipitation, IB Immunoblotting, Rapa rapamycin.
Amitriptyline activates PI3K/Akt/mTOR pathway
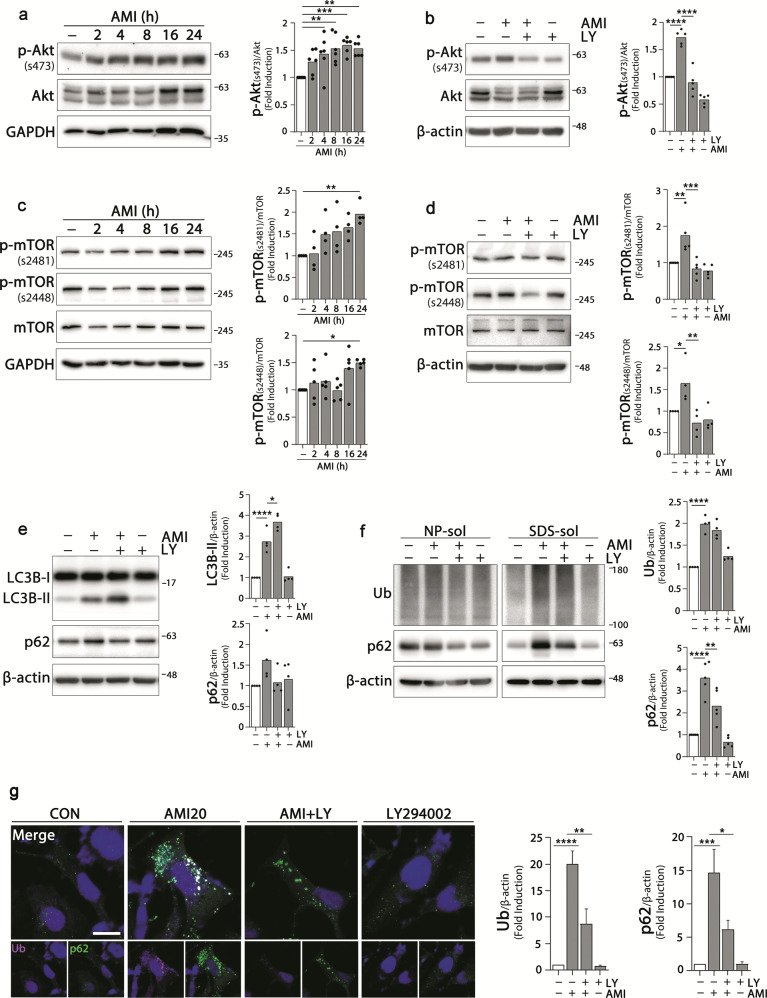
PI3K/Akt/mTOR signaling pathway is a well-known upstream signaling pathway of autophagy39. Activated Akt phosphorylates and activates mTOR, which leads to the inhibition of autophagy through the phosphorylation of multiple autophagy-related proteins and blocks the elimination of aggregates. In this study, as shown in Fig. 6a–d, the levels of phospho-Akt(s473) were increased in cells treated with amitriptyline, which resulted in the increase of downstream signaling molecules such as p-mTOR(s2481) and p-mTOR(s2448). Pretreatment with PI3K inhibitor LY294002 blocked the activation of Akt-mTOR signals induced by amitriptyline. Activated mTOR phosphorylates the UNC-51 like kinase (ULK) at serine 758 in human cell and consequently inhibits autophagy40. Amitriptyline increases p-ULK(s758), which was inhibited by LY294002 treatment (Supplementary Fig. 3a).
Fig. 6. Activation of PI3K/Akt/mTOR pathway by amitriptyline.
a, c Differentiated SH-SY5Y cells were treated with 20 μM amitriptyline for 2, 4, 8, 16, and 24 h, and the expression levels of p-Akt(S473), Akt, p-mTOR(S2481), p-mTOR(S2448), and mTOR were analyzed by western blotting. Quantification of each protein in (a, c) is shown in the right panel (n = 4–6). b, d Cell were pretreated with 10 μM PI3K inhibitor LY294002 for 1 h and treated with 20 μM amitriptyline for additional 24 h, and analyzed the protein levels as in (a, c) by western blotting. Quantification of each protein in b, d is shown in the right panel (n = 5). e LC3B and p62 levels in SH-SY5Y cells treated with amitriptyline in the presence or absence with LY294002 as b, d were analyzed by western blotting, and the quantification of protein is shown in the right panel (n = 4). f SH-SY5Y cells were treated LY294002 and amitriptyline, and the ubiquitin and p62 protein levels in NP- and SDS-soluble fractions were analyzed by western blotting. Protein levels in the SDS-fraction were quantified by densitometric analysis (n = 4–5). g Cells were treated with LY294002 and amitriptyline, and analyzed ubiquitin and p62 levels and localization by immunofluorescence. Scale bar corresponds to 20 μm. Quantification of ubiquitin and p62 immunoreactivities in (g) is shown in the right panel (5 independent sample, n = 15–25 cells) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. AMI amitriptyline, CON control, LY LY294002, NP-sol NP-soluble fraction, SDS-sol SDS-soluble fraction, Ub ubiquitin.
To verify the role of the activation of PI3K/Akt/mTOR in the blockade of autophagic flux and in the accumulation of protein aggregates in amitriptyline-treated SH-SY5Y cells, we treated cells with LY294002 and amitriptyline, and measured the levels of LC3B-II, p62, and ubiquitin. As shown in Fig. 6e, f, greater increases in LC3B-II levels and the attenuated accumulation of ubiquitin and p62 were detected in LY294002 co-treated cells compared with amitriptyline-treated cells. Immunofluorescence image also showed that amitriptyline-induced increase in ubiquitin and p62 immunoreactivities was significantly attenuated by LY294002 (Fig. 6g). These results suggest that the activation of the PI3K/Akt/mTOR pathway is responsible for the impaired autophagic flux observed in amitriptyline-treated cells.
Amitriptyline induces Beclin 1 acetylation and interaction to Rubicon
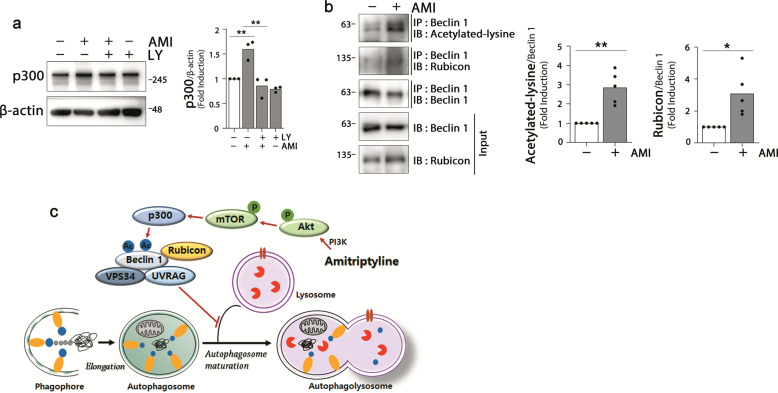
It has been suggested that Akt plays a crucial role in p300 regulation; Akt-induced phosphorylation of p300 is critical for the transactivation of p30041, and Akt/protein kinase B(PKB) is a positive regulator of the p300 expression42. To verify the involvement of Akt-linked p300 regulation in the amitriptyline-induced autophagy dysfunction, we would like to determine whether amitriptyline upregulates or activates p300, and Akt is responsible for the regulation. p300 was upregulated in amitriptyline-treated cells but not in cells co-treated with LY294002, indicating that amitriptyline activates Akt and subsequently, positively regulates p300 (Fig. 7a).
Fig. 7. Effect of amitriptyline on Beclin 1 acetylation and Rubicon binding.
a Differentiated SH-SY5Y cells were pretreated with 10 μM LY294002 for 1 h, followed by 20 μM amitriptyline for 24 h. p300 protein levels were analyzed by western blotting, and quantified by densitometric analysis (n = 3). b Cells were treated with 20 μM amitriptyline for 24 h, and analyzed protein interaction by immunoprecipitation with anti-Beclin 1 and immunoblotting with anti-Beclin 1, anti-acetylated-lysine, and anti-Rubicon, and quantified by densitometric analysis (n = 5). c Schematic diagram representing amitriptyline-mediated inhibition of autophagosome maturation and related signaling pathway. *P < 0.05, **P < 0.01. AMI amitriptyline, LY LY294002, IP immunoprecipitation, IB immunoblotting.
Recent studies have shown that Beclin 1 can be acetylated by p300 acetyltransferase, and acetylated Beclin 1 promotes the recruitment of Rubicon and inhibits autophagosome maturation and endocytic trafficking43,44. To verify this, we examined the effect of amitriptyline on the acetylation of Beclin 1. The amount of the acetylated-lysine protein binding to Beclin 1 was measured by immunoprecipitation of SH-SY5Y cell lysates treated with amitriptyline for 24 h. As shown in Fig. 7b, the acetylated Beclin 1 level was significantly increased in amitriptyline-treated cells. Simultaneously, a significant increase in Rubicon binding with Beclin 1 was also detected. These results suggest that amitriptyline induces Beclin 1 acetylation and Rubicon binding, and interferes with the fusion of autophagosome and lysosome (Fig. 7c).
Discussion
In the present study, we showed that amitriptyline increases the accumulation of abnormal protein aggregates in neuronal cells and in the brain. Amitriptyline blocks autophagic flux by regulating the intracellular distribution of lysosomes, thereby resulting in the inhibition of autophagosome maturation.
Autophagy is a catabolic pathway that degrades unfolded proteins and damaged organelles. Throughout our lives, our bodies are exposed to a variety of stress factors both externally and internally, which result in protein modifications and accumulation of misfolded proteins. As the protein clearance system that controls protein quality becomes dysfunctional with aging, more and more protein aggregates are accumulated45,46. Interestingly, recent studies have demonstrated that basal autophagy is implicated in the regulation of neurogenesis in mature brain47, which might play a potential role in neurodegenerative pathologies. Therefore, autophagy dysfunction is closely related to the pathogenesis of neurodegenerative diseases48–51, and drugs targeting autophagy induction have a potential to prevent or ameliorate neurodegenerative diseases52,53. Conversely, it is also speculated that drugs or environments that inhibit autophagy might be risk factors for the development of neurodegenerative diseases54.
Amitriptyline is a TCA drug prescribed for regulating depressive symptoms. Conflicting results have been reported regarding its effects on autophagy function and on the development of neurodegenerative diseases. Several studies have shown that amitriptyline increases LC3B-II and Beclin 1 in neuronal cells55, augments the degradation of long-lived proteins through class-III PI3 kinase-dependent pathway56, and prevents neuronal degeneration in rotenone-induced PD mice model57. On the other hand, some studies have demonstrated that amitriptyline induces dopaminergic cell damage and causes PD-associated neurotoxicity9, and blocks the proliferation and angiogenesis in vascular endothelial cells by inhibiting autophagic flux58.
In this study, we verified the effect of amitriptyline on autophagy-mediated protein clearance and its relevant signaling pathways in neuronal cells. Clinically, amitriptyline is usually prescribed 75–150 mg/day, which corresponds to 0.18–0.5 µM levels in plasma after 6 weeks of treatment59. In animal study using mice, most pharmacological dose of amitriptyline to relief depressive symptoms is ranged 10–30 mg/kg (i.p.), and intraperitoneal LD50 (mouse) is 65 mg/kg (SAFETY DATA SHEET, Cayman). In this study, we injected amitriptyline (i.p. daily for 30 days; 5, 10, and 20 mg/kg) to male C57BL/6 mice, and showed tendency to increase in protein accumulation in the brain as low as 10 mg/kg, which was more significant in mice treated with 20 mg/kg. In vitro neuronal cell culture system, amitriptyline does not cause cytotoxicity up to 20 μM (Fig. 1) and IC50 for cell proliferation is reported to be 35.59 ± 3.48 μM60. To evaluate whether amitriptyline accumulation through long-term and/or high-dose use could be associated with the increased risk of neurodegenerative disease, we used relatively high concentrations in this study.
Autophagy is a complex and dynamic process; therefore, it is often difficult to distinguish between induction and inhibition. Autophagy begins with the nucleation of an isolated membrane that elongates to envelop target proteins into a double-membrane vacuole autophagosome and matures by the formation of autolysosomes61. The increased LC3-II level is most commonly used as an autophagy induction marker, but it also suggests impaired autophagic flux62. LC3-II levels are increased by amitriptyline, which may be due to the inhibition of autophagy turnover in SH-SY5Y cells. These are evidenced by our significant observations: (1) there was no significant difference in LC3B-II levels in amitriptyline-treated cells in terms of the presence or absence of NH4Cl (Fig. 4b); (2) autophagosome marker LC3B and p62 were colocalized, but not LC3B/or p62 and lysosome marker LAMP2, suggestive of disrupted autophagolysosome formation (Fig. 4c); and (3) no quenching of GFP was observed in cells transfected with mCherry-GFP-LC3B plasmid, suggesting defective formation of autophagolysosome (Fig. 4d). All these findings suggest that amitriptyline inhibits autophagy turnover rather than autophagy initiation.
The immunoreactivity of LAMP2 was increased by amitriptyline but was not colocalized with LC3B (Fig. 4). Interestingly, almost all LAMP2 were detected at the distal end of the cells (Fig. 5). The location of lysosome could be affected by environmental circumstances, such as pH and nutrition, and kinesin or dynein direct the lysosome through the microtubule to the plus or minus direction63. The Arf-like Arl8 is a small G protein that is a major regulator of components related to endocytic and secretory pathways64. Arl8 recruits kinesin-1 to lysosomes by binding to SKIP and migrates lysosomes to the cell periphery38,65,66, which suggests that Arl8 and SKIP constitute a core link between lysosomal membranes and kinesin-1. In this study, we demonstrated for the first time that the increased interaction between Arl8, SKIP, and kinesin could be responsible for the transport of the lysosomes at the distal end of amitriptyline-treated cells.
The PI3K/Akt/mTOR is one of the typical pathways modulating autophagy function. Akt is phosphorylated by PI3K, followed by the phosphorylation of mTOR, thus resulting in autophagy inhibition39. Several studies have shown that amitriptyline increases the phosphorylation of Akt67,68. Consistent with this, we found that amitriptyline activated Akt and mTOR, and the protein accumulation due to amitriptyline was attenuated by pre-treatment with PI3K inhibitor LY294002. mTOR activation plays a crucial role in the regulation of autophagy induction69,70, but evidences for the involvement of mTOR in the regulation of autophagic flux are conflicting71,72. Studies have shown that mTOR directly phosphorylates acetyltransferase p300 to regulate autophagy73. The fusion of autophagosome and lysosome to form autophagolysosome is critical for autophagy-mediated protein degradation. Beclin 1, a mammalian ortholog of yeast Atg6, interacts with several cofactors (e.g., Atg14L, UVRAG, Bif-1, Rubicon, Ambra1, HMGB1, nPIST, VMP1, SLAM, IP3R, PINK, and survivin) and regulates autophagy74. When Beclin 1 forms the Beclin 1–VPS34–UVRAG complex, autophagy is activated by autophagosome maturation. On the other hand, Beclin 1 can be acetylated by p300 acetyltransferase, and the Beclin 1–VSP34–UVRAG complex then binds to Rubicon, eventually interfering with the fusion of autophagosome and lysosome44,75,76. In this study, acetylated Beclin 1 was increased in amitriptyline-treated cells, and its interaction with Rubicon was also increased (Fig. 7).
In summary, our study shows that amitriptyline causes an accumulation of protein aggregates in neuronal cells through autophagy dysregulation. Amitriptyline does not block the initiation of autophagy, but blocks the autophagy turnover. Amitriptyline inhibits autophagic flux through (1) the activation of PI3K/Akt/mTOR pathway, (2) the activation of p300 and Beclin-1 acetylation, and (3) the increased interaction of Arl8b, SKIP, and kinesin, which moves the lysosome to the periphery of the cells and thus interferes with autophagosome fusion. The accelerated accumulation of abnormal aggregates could directly increase the risk of neuronal cell death in both healthy persons and those with already abnormal accumulation of protein aggregates. Therefore, our findings suggest that long-term and/or high-dose administration of amitriptyline might be associated with the increased risk of neurodegenerative disease.
Materials and methods
Antibodies and reagents
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, and trypsin/EDTA were purchased from Corning (Corning, NY, USA). Alexa Fluor®-conjugated secondary antibody, 4'6-Diamidino-2-Phenylindole, Dilactate (DAPI), and ProLong™ Gold antifade reagent were purchased from Invitrogen (Carlsbad, CA, USA). MG132 was from Enzo Life Sciences (Farmingdale, NY, USA). The protease and phosphatase inhibitor cocktails were obtained from Roche (Mannheim, Germany). The following antibodies were used: anti-LAMP-2 (H4B4, sc-18822), anti-p62 (D-3, sc-28359), anti-ubiquitin (P4D1, sc-8017), anti-ARL8A/B (H-8, sc-398635), anti-BECN1 (E-8, sc-48341), anti-phospho-p70 S6 kinase α (A-6, sc-8416), anti-p300 (F-4, sc-48343), anti-GFP (B-2, sc-9996), anti-TH (F-11, sc-25269), all obtained from Santa Cruz Biotechnology, Inc. (Santa cruz, CA, USA); anti-mTOR (2972S), anti-phospho-mTOR (Ser2448, 2971S), anti-phospho-mTOR (Ser2481, 2974S), anti-Akt1 (C73H10, 2938S), phospho-Akt (Ser473, 9271L), anti-beclin-1 (3738), anti-acetylated-lysine (9441S) from Cell Signaling Technology (Danvers, MA, USA); and anti-LC3 were purchased from MBL International (PM046, Woburn, MA, USA) and Novus Biologicals (NB600–1384, Littleton, CO, USA). Anti-GAPDH (G9545) and anti-β-actin (A1978) were from Sigma-Aldrich (St. Louis, MO, USA) and anti-α synuclein was from GeneTex (Irvine, CA, USA). Anti-phospho-ULK1(Ser758, PA5-78251), goat anti-Rabbit IgG (H + L) secondary antibody HRP (31460), and goat anti-Mouse IgG (H + L) secondary antibody HRP (31430) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All other chemicals including amitriptyline were of reagent grade and were purchased from Sigma-Aldrich.
Cell culture
Human neuroblastoma SH-SY5Y cells and HEK-293T cells were grown in DMEM containing 10% FBS, 100 IU/L penicillin, and 10 μg/mL streptomycin at 37 °C in a humidified atmosphere with 5% CO2 and 95% air. The cells were plated on polystyrene culture dishes at a density of0.055 × 105 cells/well in a 48-well culture plate, 0.19 × 105 cells/well in a 6-well culture plate, or 0.1 × 106 cells in a 60-mm dish. SH-SY5Y cells were differentiated by incubation for 3 days with 10 μM all-trans retinoic acid and 1% FBS in DMEM. LUHMES cells were cultured in precoated dishes with 50 μg/mL poly-l-ornithine hydrobromide and 1 μg/mL fibronectin (Sigma-Aldrich). Cells were maintained in DMEM/F12 containing 1× N2 supplement and 40 ng/mL recombinant basic fibroblast growth factor (bFGF) (Thermo Fisher). For differentiation, cells were changed to the differentiation medium consisting with DMEM/F12, 1× N2 supplement, 1 mM dibutyryl cAMP, 1 μg/mL tetracyclin (Sigma-Aldrich), and 2 ng/mL recombinant human GDNF (R&D systems, Minneapolis, MN, USA). After 5 days of differentiation, cells were treated with amitriptyline in fresh proliferation medium. For primary cultures of cortical neurons, E18 C57BL/6 mice were dissected, as described previously77. Cortex tissues were incubated in StemPro® Accutase® Cell Dissociation Reagent (Gibco, Grand Island, NY, USA) for 10 min at 37 °C and washed twice with Hank’s Balanced Salt Solution. Dissociated tissues were triturated and resuspended in neurobasal medium (Gibco) with 1% of l-glutamine (100×; Gibco), 2% of B27 supplement (50×; Gibco), and 1% of penicillin–streptomycin solution (100×; Corning, NY, USA). Cells were plated at an appropriate density on 60 mm dish precoated with poly-d-lysine (Sigma-Aldrich). After incubation for 4 h, cells were changed with fresh media and maintained in a humidified CO2 incubator. At day in vitro 11, amitriptyline were treated for 24 h.
Transient transfection
SH-SY5Y cells were transiently transfected with pDEST-CMV mcherry-GFP-LC3B (Addgene, Watertown, MA, USA) using polyethylenimine (PEI) reagents (Polyscience, Warrington, PA, USA) according to the manufacturer’s instructions. Transient transfection of HEK-293T cells with pDEST47-ARL8B-GFP (Addgene) and pCMV3-PLEKHM2 plasmids (Sino biological, Beijing, China) was performed using PEI reagents. pDEST47-ARL8B-GFP was deposited by the Richard Kahn Lab (Emory University Department of Biochemistry, Atlanta).
Assessment of cell viability: lactate dehydrogenase assay
The extent of cell death was measured from the amount of lactate dehydrogenase released into the culture medium. Fifty microlitre of cell culture medium were incubated at room temperature (RT) with 100 mM potassium phosphate buffered saline (PBS) containing 23 mM pyruvate (pH 7.4) and 0.26 mM NADH with a total volume of 200 μL. The rate of NAD + formation was measured at 340 nm for 5 min at 25-s intervals using a microplate spectrophotometer (VersaMax; Molecular Devices, San Jose, CA, USA).
Assessment of cell viability: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells were plated in a 48-well tissue culture plate and incubated with yellow MTT solution at a final concentration 0.5 mg/mL for 4 h. Then, purple formazan crystals were dissolved in DMSO. The absorbance of the solubilized formazan product was measured at 570 nm using a microplate spectrophotometer (VersaMax; Molecular Devices).
Cell fractionations and western blot analysis
Cells were lysed in NP buffer (150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, 25 mM Tris–Cl; pH 7.5) with protease and phosphatase inhibitor cocktails (Roche, Basel, Switzerland) for 60 min at −80 °C. Cell lysates were centrifuged at 15,000 rpm for 60 min at 4 °C, and the supernatant proteins in the NP-soluble fraction were collected. The pellet was further lysed in SDS buffer (2% SDS, 50 mM Tris–Cl; pH 7.5) with protease and phosphatase inhibitor cocktails (Roche, Basel, Switzerland), sonicated for homogenization, and boiled at 100 °C for 5 min. The SDS-soluble fraction was centrifugated at 15,000 rpm for 60 min at 4 °C, and the supernatant were collected. Protein samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted as previously described34.
Western blot analysis
Cells were washed with ice-cold PBS, harvested, and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris–Cl; pH 7.5) with complete protease and phosphatase inhibitor cocktails for 30 min at –20 °C. Cell lysates were centrifuged at 15,000 rpm for 30 min at 4 °C. Equal amounts of protein were separated by 8–12% SDS polyacrylamide gels and transferred onto polyvinylidene difluoride-nitrocellulose membranes (Millipore). The membranes were blocked with 5% skim milk in TBST and were incubated at 4 °C overnight with specific primary antibodies and then with HRP-conjugated horseradish peroxide-conjugated secondary antibody. Specific bands were detected by enhanced chemiluminescence (ECL; Millipore, Billerica, MA, USA) and analyzed by a luminescent image analyzer LAS-4000 (Fujifilm, Tokyo, Japan). The relative intensities of each band were measured using Image J software.
Immunoprecipitation
Cell lysates were incubated with A/G agarose beads (Santa Cruz, sc-2003) for 1 h to eliminate non-specific binding proteins. After the preclearing step, the lysates were centrifugated at 2500 rpm for 5 min, and the supernatant were incubated with the indicated antibody for 1 h and with A/G agarose beads overnight. The beads were washed with IP buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol; pH 7.4) 5 times and boiled with 2× loading dye for 10 min. The sample is loaded on SDS-PAGE for further analysis.
Immunofluorescence confocal microscopy
SH-SY5Y cells were plated on poly-d-lysine-coated cover slips in 12-well culture plates. After the individual experiments, the cells were washed twice with PBS and fixed for 15 min in cold 4% paraformaldehyde and 4% sucrose in PBS (pH 7.4). After fixation, the cells were permeabilized with 0.25% triton-X-100 in PBS for 10 min and then blocked with PBS containing 1% bovine serum albumin (BSA), 1% normal donkey serum, and 3% FBS for 30 min at room RT to reduce non-specific binding. They were then incubated overnight with primary antibodies in PBS containing 1% BSA at 4 °C and with Alexa Fluor®-conjugated secondary antibodies for 50 min at RT, and DAPI was added for 10 min.
Animal treatment
All experiments involving animals were approved by the Institutional Animal Care and Use Committee at CHA University (IACUC 170116). Male C57BL/6 mice, 6 weeks of age, were housed under a 12:12 light–dark lighting schedule with free access to food and water. The RT was 22 °C. The mice were injected intraperitoneally with sterile PBS (100 μL each, n = 5) or amitriptyline (5, 10, or 20 mg/kg in a volume of 100 μL PBS each, n = 5) daily for 30 days. After the last injection, the mice were euthanized with Zoletil 50 (Virbac) and transcardially perfused with PBS, heparin, and 4% PFA in PBS (pH 7.4).
Rotarod test
Motor function of the mice was assessed using a rotarod system (Rota Rod-R V2.0, B.S. Technolab). Before the experiment, mice were trained on the rod (diameter, 3.5 cm) with an increasing speed 4 rpm for 3 min. After the training, the speed of rod increased from 4 to 40 rpm and retention time of each mouse on the rod was recorded, ≤300 s. Each animal was tested twice times and the final time records were used for statistical analysis.
Immunohistochemistry
After perfusion, fixation was performed in 4% paraformaldehyde for more than 24 h and cryo-protected in 30% sucrose/0.1 M PBS solution for another 24 h followed by the freezing of tissues in optimal cutting temperature compound. Afterwards, 20 μm PFA-fixed cryo-sections were serially cut in the coronal plane for unbiased stereology, and brain sections were incubated for 30 min in 1% BSA, 0.2% Triton X, 0.3% H2O2, and 2% normal goat serum (NGS) in 0.05 M PBS. The sections were then incubated with mouse anti-p62/SQSTM1 or mouse anti-ubiquitin, diluted in 2% NGS in 0.05 M PBS overnight at 4 °C. After two rinses in 0.05 M PBS, the sections were incubated with secondary biotinylated goat anti-mouse (Vector Laboratories, Burlingame, CA) for 1 h and with ABC solution (Vector Laboratories) in 0.05 M PBS for 1 h and developed with 0.025% 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO).
Statistical analysis
Data were analyzed and expressed as mean ± standard error of the mean. Statistical significance was determined by one-way ANOVA, followed by Bonferroni post hoc tests using Prism software (GraphPad Software; La Jolla, CA, USA). P < 0.05 was considered significant for all statistical analysis.
Supplementary information
Acknowledgements
This work was supported by the Ministry of Science, ICT & Future Planning (NRF-2018R1A2B6004008, NRF-2016R1C1B1015991, and 2019M3A9A8067070) through the National Research Foundation of Korea.
Author contributions
Y.K., Y.B., A.K., and H.J.C. conceived and designed the study. Y.K. and Y.B. performed the in vitro experiments and Y.K. and S.H.M. performed the vivo experiments. Y.K., Y.B., S.H.M., and H.J.C. performed the data analysis/interpretation. Y.K., Y.B., S.H.M., A.K., and H.J.C. wrote the paper draft. Y.B. and H.J.C. obtained the research funding. All authors have read and approved the final paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by B. Joseph
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03085-6).
References
- 1.Richelson E. Pharmacology of antidepressants–characteristics of the ideal drug. Mayo Clin. Proc. 1994;69:1069–1081. doi: 10.1016/S0025-6196(12)61375-5. [DOI] [PubMed] [Google Scholar]
- 2.Beghi E, et al. The epidemiology of ALS and the role of population-based registries. Biochim. Biophys. Acta. 2006;1762:1150–1157. doi: 10.1016/j.bbadis.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Grimm A, Eckert A. Brain aging and neurodegeneration: from a mitochondrial point of view. J. Neurochem. 2017;143:418–431. doi: 10.1111/jnc.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castilla-Puentes RC, Habeych ME. Subtypes of depression among patients with Alzheimer’s disease and other dementias. Alzheimers Dement. 2010;6:63–69. doi: 10.1016/j.jalz.2009.04.1232. [DOI] [PubMed] [Google Scholar]
- 5.Ferentinos P, et al. Prevalence of major depression in ALS: comparison of a semi-structured interview and four self-report measures. Amyotroph. Lateral Scler. 2011;12:297–302. doi: 10.3109/17482968.2011.556744. [DOI] [PubMed] [Google Scholar]
- 6.Baquero M, Martin N. Depressive symptoms in neurodegenerative diseases. World J. Clin. Cases. 2015;3:682–693. doi: 10.12998/wjcc.v3.i8.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reijnders JS, et al. A systematic review of prevalence studies of depression in Parkinson’s disease. Mov. Disord. 2008;23:183–189. doi: 10.1002/mds.21803. [DOI] [PubMed] [Google Scholar]
- 8.Haller I, et al. Differential neurotoxicity of tricyclic antidepressants and novel derivatives in vitro in a dorsal root ganglion cell culture model. Eur. J. Anaesthesiol. 2007;24:702–708. doi: 10.1017/S0265021507000154. [DOI] [PubMed] [Google Scholar]
- 9.Lee MY, et al. Tricyclic antidepressants amitriptyline and desipramine induced neurotoxicity associated with Parkinson’s disease. Mol. Cells. 2015;38:734–740. doi: 10.14348/molcells.2015.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Sendon J, Mena MA, de Yebenes JG. Drug-induced Parkinsonism. Expert Opin. Drug Saf. 2013;12:487–496. doi: 10.1517/14740338.2013.787065. [DOI] [PubMed] [Google Scholar]
- 11.Hipkiss AR. Accumulation of altered proteins and ageing: causes and effects. Exp. Gerontol. 2006;41:464–473. doi: 10.1016/j.exger.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Press M, et al. Protein aggregates and proteostasis in aging: amylin and beta-cell function. Mech. Ageing Dev. 2019;177:46–54. doi: 10.1016/j.mad.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik S, Cuervo AM. Proteostasis and aging. Nat. Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 14.Remondelli P, Renna M. The endoplasmic reticulum unfolded protein response in neurodegenerative disorders and its potential therapeutic significance. Front. Mol. Neurosci. 2017;10:187. doi: 10.3389/fnmol.2017.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iram A, Naeem A. Protein folding, misfolding, aggregation and their implications in human diseases: discovering therapeutic ways to amyloid-associated diseases. Cell Biochem. Biophys. 2014;70:51–61. doi: 10.1007/s12013-014-9904-9. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Polo RA, et al. Is the modulation of autophagy the future in the treatment of neurodegenerative diseases? Curr. Top. Med. Chem. 2015;15:2152–2174. doi: 10.2174/1568026615666150610130645. [DOI] [PubMed] [Google Scholar]
- 17.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein DC, Bento CF, Deretic V. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J. Exp. Med. 2015;212:979–990. doi: 10.1084/jem.20150956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abramov, A. Y. et al. Interaction of misfolded proteins and mitochondria in neurodegenerative disorders. Biochem. Soc. Trans.10.1042/BST20170024 (2017). [DOI] [PubMed]
- 20.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 21.Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 22.Rakhit R, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 23.Meijer AJ. Autophagy research: lessons from metabolism. Autophagy. 2009;5:3–5. doi: 10.4161/auto.5.1.7207. [DOI] [PubMed] [Google Scholar]
- 24.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menzies FM, Moreau K, Rubinsztein DC. Protein misfolding disorders and macroautophagy. Curr. Opin. Cell Biol. 2011;23:190–197. doi: 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budini M, Buratti E, Morselli E, Criollo A. Autophagy and its impact on neurodegenerative diseases: new roles for TDP-43 and C9orf72. Front. Mol. Neurosci. 2017;10:170. doi: 10.3389/fnmol.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, et al. Impaired autophagy promotes bile acid-induced hepatic injury and accumulation of ubiquitinated proteins. Biochem. Biophys. Res. Commun. 2018;495:1541–1547. doi: 10.1016/j.bbrc.2017.11.202. [DOI] [PubMed] [Google Scholar]
- 29.Moloudizargari M, et al. Autophagy, its mechanisms and regulation: Implications in neurodegenerative diseases. Ageing Res. Rev. 2017;40:64–74. doi: 10.1016/j.arr.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Hisaoka-Nakashima K, et al. Amitriptyline induces brain-derived neurotrophic factor (BDNF) mRNA expression through ERK-dependent modulation of multiple BDNF mRNA variants in primary cultured rat cortical astrocytes and microglia. Brain Res. 2016;1634:57–67. doi: 10.1016/j.brainres.2015.12.057. [DOI] [PubMed] [Google Scholar]
- 31.Farrelly LA, et al. Therapeutic concentrations of valproate but not amitriptyline increase neuropeptide Y (NPY) expression in the human SH-SY5Y neuroblastoma cell line. Regul. Pept. 2013;186:123–130. doi: 10.1016/j.regpep.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Lotharius J, et al. Effect of mutant alpha-synuclein on dopamine homeostasis in a new human mesencephalic cell line. J. Biol. Chem. 2002;277:38884–38894. doi: 10.1074/jbc.M205518200. [DOI] [PubMed] [Google Scholar]
- 33.Marreiros R, et al. Disruption of cellular proteostasis by H1N1 influenza A virus causes alpha-synuclein aggregation. Proc. Natl Acad. Sci. USA. 2020;117:6741–6751. doi: 10.1073/pnas.1906466117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bang Y, Kang BY, Choi HJ. Preconditioning stimulus of proteasome inhibitor enhances aggresome formation and autophagy in differentiated SH-SY5Y cells. Neurosci. Lett. 2014;566:263–268. doi: 10.1016/j.neulet.2014.02.056. [DOI] [PubMed] [Google Scholar]
- 35.Orhon, I. & Reggiori, F. Assays to monitor autophagy progression in cell cultures. Cells10.3390/cells6030020 (2017). [DOI] [PMC free article] [PubMed]
- 36.Yoshii, S. R. & Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci.10.3390/ijms18091865 (2017). [DOI] [PMC free article] [PubMed]
- 37.Cabukusta B, Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic. 2018;19:761–769. doi: 10.1111/tra.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 2011;21:1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, et al. Molecular basis for the regulation of transcriptional coactivator p300 in myogenic differentiation. Sci. Rep. 2015;5:13727. doi: 10.1038/srep13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esteves AR, et al. The role of Beclin-1 acetylation on autophagic flux in Alzheimer’s disease. Mol. Neurobiol. 2019;56:5654–5670. doi: 10.1007/s12035-019-1483-8. [DOI] [PubMed] [Google Scholar]
- 44.Sun T, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat. Commun. 2015;6:7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowotny K, Jung T, Grune T, Hohn A. Accumulation of modified proteins and aggregate formation in aging. Exp. Gerontol. 2014;57:122–131. doi: 10.1016/j.exger.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Lim J, Bang Y, Choi HJ. Abnormal hippocampal neurogenesis in Parkinson’s disease: relevance to a new therapeutic target for depression with Parkinson’s disease. Arch. Pharm. Res. 2018;41:943–954. doi: 10.1007/s12272-018-1063-x. [DOI] [PubMed] [Google Scholar]
- 48.Loos B, Klionsky DJ, Wong E. Augmenting brain metabolism to increase macro- and chaperone-mediated autophagy for decreasing neuronal proteotoxicity and aging. Prog. Neurobiol. 2017;156:90–106. doi: 10.1016/j.pneurobio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Tammineni P, Cai Q. Defective retrograde transport impairs autophagic clearance in Alzheimer disease neurons. Autophagy. 2017;13:982–984. doi: 10.1080/15548627.2017.1291114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kegel KB, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anglade P, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 52.Levine B, Packer M, Codogno P. Development of autophagy inducers in clinical medicine. J. Clin. Investig. 2015;125:14–24. doi: 10.1172/JCI73938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kroemer G. Autophagy: a druggable process that is deregulated in aging and human disease. J. Clin. Investig. 2015;125:1–4. doi: 10.1172/JCI78652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin HW, Chung SJ. Drug-induced Parkinsonism. J. Clin. Neurol. 2012;8:15–21. doi: 10.3988/jcn.2012.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcocer-Gomez E, et al. Antidepressants induce autophagy dependent-NLRP3-inflammasome inhibition in major depressive disorder. Pharmacol. Res. 2017;121:114–121. doi: 10.1016/j.phrs.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 56.Zschocke J, et al. Antidepressant drugs diversely affect autophagy pathways in astrocytes and neurons–dissociation from cholesterol homeostasis. Neuropsychopharmacology. 2011;36:1754–1768. doi: 10.1038/npp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kandil EA, Abdelkader NF, El-Sayeh BM, Saleh S. Imipramine and amitriptyline ameliorate the rotenone model of Parkinson’s disease in rats. Neuroscience. 2016;332:26–37. doi: 10.1016/j.neuroscience.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Guan Y, et al. Tricyclic antidepressant amitriptyline inhibits autophagic flux and prevents tube formation in vascular endothelial cells. Basic Clin. Pharm. Toxicol. 2019;124:370–384. doi: 10.1111/bcpt.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziegler VE, Clayton PJ, Biggs JT. A comparison study of amitriptyline and nortriptyline with plasma levels. Arch. Gen. Psychiatry. 1977;34:607–612. doi: 10.1001/archpsyc.1977.01770170117012. [DOI] [PubMed] [Google Scholar]
- 60.Solek P, et al. Neuronal life or death linked to depression treatment: the interplay between drugs and their stress-related outcomes relate to single or combined drug therapies. Apoptosis. 2019;24:773–784. doi: 10.1007/s10495-019-01557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016;129:4329–4339. doi: 10.1242/jcs.196287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 65.Khatter D, Sindhwani A, Sharma M. Arf-like GTPase Arl8: moving from the periphery to the center of lysosomal biology. Cell Logist. 2015;5:e1086501. doi: 10.1080/21592799.2015.1086501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 2006;344:1186–1191. doi: 10.1016/j.bbrc.2006.03.221. [DOI] [PubMed] [Google Scholar]
- 67.Olianas MC, Dedoni S, Onali P. LPA1 mediates antidepressant-induced ERK1/2 signaling and protection from oxidative stress in glial cells. J. Pharm. Exp. Ther. 2016;359:340–353. doi: 10.1124/jpet.116.236455. [DOI] [PubMed] [Google Scholar]
- 68.Chadwick W, et al. Amitriptyline-mediated cognitive enhancement in aged 3xTg Alzheimer’s disease mice is associated with neurogenesis and neurotrophic activity. PLoS ONE. 2011;6:e21660. doi: 10.1371/journal.pone.0021660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, Z. et al. Balancing mTOR signaling and autophagy in the treatment of Parkinson’s disease. Int. J. Mol. Sci. 10.3390/ijms20030728 (2019). [DOI] [PMC free article] [PubMed]
- 70.Jung CH, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cina DP, et al. Inhibition of MTOR disrupts autophagic flux in podocytes. J. Am. Soc. Nephrol. 2012;23:412–420. doi: 10.1681/ASN.2011070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Senturk M, et al. Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat. Cell Biol. 2019;21:384–396. doi: 10.1038/s41556-019-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wan W, et al. mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol. Cell. 2017;68:323–335 e326. doi: 10.1016/j.molcel.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 74.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 77.Kaech S, Banker G. Culturing hippocampal neurons. Nat. Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.