Abstract
Five sponge specimens belonging to the genera Spongilla and Ciocalypta were collected from Little Rann of Kutch (in Gujarat, India) and analysed for associated microbiomes. Critical analysis was done with respect to members of the phylum Planctomycetes, using two different strategies; 1. Culture-independent metagenomic approach and 2. culture-dependent anaerobic enrichment for anammox-planctomycetes. The 16S rRNA gene (V1-V3 region) amplicon metagenome analysis revealed significant divergence in bacterial diversity, including Planctomycetes among the sponges analysed. Community metagenomics revealed a total of 376 Operational Taxonomic Units (OTUs) belonging to 41 different phyla. OTUs belonging to Proteobacteria was the most abundant (38%) among the sponge analysed. The KEGG annotation predicted a total of 6909 KEGG orthologs (KOs); most of the KOs are associated with membrane transport, xenobiotic degradation, production of secondary metabolites, amino acid metabolism and carbohydrate metabolism. In the culture-dependent study, FISH analysis confirmed the association of anammox-planctomycetes with sponges. Partial 16S rRNA gene sequences of two planctomycetes (JC545, JC543) were cladding with those of uncultured Phycisphaerae class. The other three putative anammox bacteria (JC541, JC542, JC544) formed a different clade with “Candidatus Brocadia anammoxidans”. These three putative bacteria believably represent new species/genus related to “Candidatus Brocadia”.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02449-1) contains supplementary material, which is available to authorized users.
Keywords: Sponges, Culture independent, Metagenome, Culture dependent, Planctomycetes, Anammox bacteria
Introduction
An unprecedented amount of information through high-throughput sequencing methods revealed the structural and functional diversity of microbial communities, particularly those associated with sponges (Thomas et al. 2016). Sponges have highly diverse and specific symbiont communities. Due to the filter-feeding activities of sponges, they maintain a constant influx of seawater microorganisms. Approximately 40% of sponge volume comprises of sponge-associated microorganisms having densities over 109 CFU of sponge tissue, several orders higher than those typical for seawater (Abdelmohsen et al. 2014). Sponge microbiome consists of all the three domains of life which play an essential role in shaping sponge evolution (Hentschel et al. 2003, 2012; Imhoff and Stohr 2003; Taylor et al. 2007). Sponge symbionts are capable of producing secondary metabolites and provide a chemical defence to the sponge. On the other hand, sponge-associated microbiota for the degradation and metabolism of xenobiotics are still overlooked (Perez et al. 2002). Sponge symbionts probably contribute to the host’s nutrition by involving themselves in various metabolic processes such as photosynthesis, nitrification, nitrogen fixation, and sulfate reduction (Wilkinson 1979; Hoffmann et al. 2006, 2009; Bayer et al. 2008; Mohamed et al. 2010; Weisz et al. 2007). Culture-independent, as well as culture-dependent studies revealed that sponges have diverse bacterial populations. Phylogenetic studies clearly showed more than taxa belonging to thirty different bacterial phyla (Schleifer 2004; Webster et al. 2008; Sipkema et al. 2009), including the candidate phylum ‘Poribacteria’ (exclusively found within sponges; Fieseler et al. 2004; Lafi et al. 2009) are associated with sponges.
According to neoteric taxonomic refurbish, phylum Planctomycetes is divaricated into three main classes: Planctomycetia, Phycisphaerae, and Candidatus “Brocadiae.” Candidatus “Brocadiae” which still has a Candidatus status due to lack of axenic culture, consists of anammox bacteria (Dedysh et al. 2020; Wiegand et al. 2018, 2020). Anammox-planctomycetes are a unique group of anaerobic bacteria that play a pivotal role in the nitrogen cycle removing toxic ammonia (NH3) from the environment through a unique process in which ammonia is converted to dinitrogen gas using nitrite as the electron acceptor under anoxic conditions. Hydrazine is the common intermediate during the conversion of ammonia and nitrite to dinitrogen gas (van de Graaf et al. 1996, 1997; Strous et al. 2006; Kartal et al. 2013). The annamox-planctomycetes are also isolated from both marine as well as freshwater sponges. Association of anammox bacteria with sponges reveals the crucial role of Planctomycetes in the process of denitrification within the sponges (Mohamed et al. 2010).
During our survey on anammox-planctomycetes from India, we have collected five sponges from the Wild Ass Sanctuary in the Little Rann of Kutch, Gujarat, India. Little Rann of Kutch is a saline desert having an annual rainfall of less than 300 mm with temperatures ranging from 2 °C to 49.5 °C. The area present in this ecosystem is seasonally flooded, which makes it a wetland ecosystem. There is an occurrence of gradual variations in salinity due to the monsoon and also due to the mixing of ocean water and freshwater in the fringes of Little Rann of Kutch. This ecosystem harbours microbial diversity in a large extent (Patel et al. 2015). In the present study, sponges Spongilla sp. S9, Ciocalypta sp. S11 and Ciocalypta sp. S12 were investigated and analysed for the sponge-associated bacterial communities, focusing more on the diversity of Planctomycetes. Besides, functional composition of microbial community obtained from amplicon metagenome data was also predicted to recognise the ecological role of the associated bacterial communities and their biotechnological potential. Through culture-dependent study, we report the enrichment of five putative novel taxa of planctomycetes from sponges.
Materials and methods
Sample collection
During our survey for Planctomycetes from the wetland ecosystem, we collected five sponges from puddles at the Joghad Eco Park (part of Little Rann of Kutch). The GPS of the sampling site is N 22°43′26″, E 70°28′40″. The salinity, pH, and temperature of sampling sites were measured through conductivity meter, pH meter and thermometer, respectively. Sponges were identified to the genus through the world Porifera Database (https://www.marinespecies.org/porifera/). Three sponges belonged to the genus Spongilla (Spongilla sp. S8, Spongilla sp. S9, and Spongilla sp. S10), while two belonged to the genus Ciocalypta (Ciocalypta sp. S11, and Ciocalypta sp. S12). Spongilla spp. were collected from three puddles having at a depth of about 20 cm with continuous flow of surface water from different resources and the samples were collected at a distance of about 20 m between puddle. While Ciocalypta sp. S11 and Ciocalypta sp. S12 were collected from the salt marsh puddles on the road side at a distance of about 100 m between the puddles. Salinity, pH and temperature of the puddle water ranged between 1–5%, 7–9 and 26–30 °C, respectively. The collected sponges were processed separately for metagenome and cultured diversity analyses (Fig. 1). For metagenome-based diversity analysis, they were preserved on-site in formalin (4% formaldehyde in 1 × phosphate buffer saline). For the enrichment, they were directly inoculated on-site into the specific medium to avoid the loss of anammox bacterial diversity.
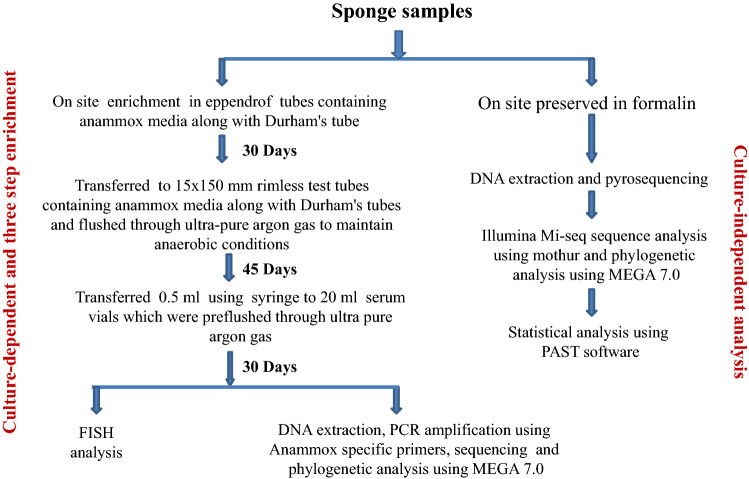
Fig. 1.
Methodology of culture-dependent and -independent studies applied to assess the bacterial diversity of sponge samples
Culture-independent study
Extraction of metagenomic DNA from sponge samples
Three hundred mg of sponge sample was macerated in liquid nitrogen with the help of a mortar and pestle. Metagenomic DNA was extracted from the macerated sponge sample using MO BIOs’ Power Soil DNA Isolation kit (QIAGEN make) and processed according to the kit’s protocols with a few modifications. Sponge tissue was lysed by vortexing for 40 min. The extracted DNA was stored at − 20 °C until further analysis. DNA samples were sent for Illumina Mi-Seq (2 × 300 bp) sequencing to Research and Testing Laboratory (RTL) LLC (Lubbock, TX, USA). The samples underwent 16S rRNA gene amplification for V1-V3 regions using universal primers 28F (5′ GAGTTTGATCNTGGCTCAG 3′) and 519R (5′ GTNTTACNGCGGCKGCTG 3′).
DNA processing and assessment of taxonomic diversity
The raw sequence reads obtained after Illumina paired-end sequencing was processed using mothur software (Schloss 2009). Two sets of data from each sample were joined to form contigs and further processed to reduce the sequencing and PCR errors. Sequences shorter than 350 nucleotides with homopolymers longer than eight nucleotides and all reads containing ambiguous base calls or incorrect primer sequences were removed (Kozich et al. 2013). The improved sequences were further processed to remove redundant sequences. Next, the processed sequences were aligned against the SILVA v132 database (Pruesse et al. 2007; https://www.mothur.org/wiki/Alignment_database). Putative chimeric sequences were detected and removed via Chimera Uchime, an integrated algorithm in mother (Edgar et al. 2011). All taxonomic classification was performed using the Bayesian classifier (Qiong et al. 2007), using a Greengenes dataset (gg_13_8_99; https://www.mothur.org/wiki/Greengenes-formatted_databases). Data have been normalized to obtain seven taxonomic levels for each sequence at 80% Naïve Bayesian bootstrap cut-off with 1000 iterations. Operational taxonomic units (OTUs; at 97–98% sequence similarity) were calculated using the mothur platform. The undesirable sequences belonging to mitochondria, chloroplast, Archaea, and eukaryota were removed. The constructed OTU table was corrected to comply with the current official nomenclature (List of Prokaryotic names with Standing in Nomenclature: LPSN-www.bacterio.net). Rarefaction analysis was carried out using a single rarefaction function of mothur, and alpha diversity was estimated by calculating the diversity indices like Inverse Simpson, Shannon diversity (H), and observed richness subsampling. Statistical analysis was carried out using PAST v3.18 (Hammer et al. 2001). Statistical tests like ANOVA and Tukey’s posthoc tests were used to detect remarkable differences in diversity indices between sponge samples. To further examine the community relationship among all three sponge samples, non-metric multidimensional scaling (NMDS) was performed using PAST software taking Bay-Curtis distance into consideration. The number of shared OTUs among all the sponge samples was calculated and represented using a Venn diagram (subsampled OTU table) by mothur.
The microbial composition of sponge samples was characterized and the results are displayed as bacterial phyla abundance. The bacterial families and genera with abundances greater than 0.5% in at least one of the samples were also identified. Remarkable differences between sponge samples were assessed using the Kruskal–Wallis test and Dunn’s posthoc test (non-normal data). The heatmap showing relative abundance were constructed using Morpheus web-based tool (https://software.broadinstitute.org/morpheus). KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation of the OTU table obtained from mothur was performed using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) to predict the functional composition of microbial community obtained from amplicon metagenome data (Langille et al. 2013).
Culture-dependent study
For a culture-dependent study, we used three-step enrichment strategy (Fig. 1). The enrichments were followed by fluorescence in situ hybridization (FISH) to screen for the anammox-planctomycetes and finally the DNA was extracted, PCR amplified and 16S rRNA gene was sequenced to identify the anammox-planctomycetes (Fig. 1).
Enrichment strategy
All the sponge samples were inoculated independently, on-site in 2 ml microcentrifuge (Eppendorf) tubes along with a Durham tube containing oligotrophic anammox bacterial mineral salts medium (Egli et al. 2001) containing [mM] NaNO2 (6), KHCO3 (25), (NH4)2SO4 (3), K2HPO4 (1), CaCl2.5H20 (0.5), MgCl2.7H20 (0.5), 2 ml of trace element solution-1 and 1 ml trace element solution-2 per liter. (Trace element solution 1 contained (g.l−1) Na2EDTA·2H2O (10) and FeSO4.7H2O (5). Trace element solution 2 contained (g.l−1) Na2EDTA·2H2O (15), ZnSO4·7H2O (0.43), CoCl2·6H2O (0.24), MnCl2·4H2O (1), CuSO4·5H2O (0.25), NaMoO4·2H2O (0.22), NiCl2·6H2O (0.2), Na2SeO3 (0.08) and H3BO4 (0.014). Durham tube was introduced to detect nitrogen gas production due to the activity of anammox bacteria. The inoculated microcentrifuge tubes were sealed with para-film. After 30 days of anaerobic/anoxic incubation, enrichment cultures were transferred into pre-flushed (with ultra-pure argon gas to maintain anaerobic conditions) rimless test tubes (15 × 150 mm; Borosil make) containing 5 ml of anammox medium and Durham tubes (Borosil make) and tightly closed by butyl rubber stoppers and incubated at room temperature for 45 days. One ml of culture from these test tubes were drawn and inoculated into serum vials (20 ml) containing 15 ml of anammox medium pre-flushed with ultra-pure argon gas (Fig. 1) to maintain anaerobic condition.
Fluorescence in situ hybridization (FISH)
To corroborate the presence of Planctomycetes in the anaerobically grown enrichment cultures, FISH was performed according to Parsley et al. (2010) with slight modifications. Samples were treated with Triton × 100 (0.04%) for 30 min at 37 °C. FISH was performed using CY3 labelled Planctomycetes-specific probes, Pla46 (5′-GACTTGCATGCCTAATCC-3′) (Parsley et al. 2010) supplied by BIOMERS, Germany. A probe concentration of 6 ng.µl−1 was used for overnight hybridization at 58 °C (Schmid et al. 2005). Samples were washed with SSC buffer (0.2 ×) and observed under a confocal microscope (Carl Zeiss LSM880).
DNA extraction, PCR amplification and identification of bacteria
Genomic DNA was extracted from 20 ml of enrichment cultures using Nucleo-pore gDNA Fungal/ Bacterial Mini Kit (GENETIX make) and processed according to the manufacturer’s protocols. To identify anammox bacteria from enrichment cultures, the Planctomycetes16S rRNA gene amplification was performed using group-specific oligonucleotide primers Pla352f (5′-GGCTGCAGTCGAGRATCT-3′) and Pla920r (5′-TGTGTGAGCCCCCGTCAA-3′). The PCR mixture (50 µl) contained 14 µl of Milli-Q, 25 µl of master-mix (Takara), 2.5 µl of each primer (100 ng.μl−1) and 6 µl of genomic DNA (30 ng.µl−1). PCR conditions were standardized through gradient PCR (50 °C to 60 °C). Maximum amplification was obtained at 54 °C. The reaction cycle parameters included an initial denaturation step consisting of 10 min at 95 °C, followed by 54 s of annealing at 54 °C, and 1:40 s of elongation at 72 °C for 35 cycles and a final extension of 15 min at 72 °C. Further sequencing of the amplified 16S rRNA gene (568 base pair) was carried out by M/S. 1st Base, Malaysia. The 16S rRNA gene sequences were aligned and phylogenetic analyses were done using Molecular Evolutionary Genetic Analysis-7 (MEGA-7) (https://www.megasoftware.net/) (Kumar et al. 2016). The evolutionary distance was calculated using the Tamura 3-parameter model (Tamura 1992) in pairwise deletion procedure. Phylogenetic trees were constructed in MEGA-7 software using the maximum-likelihood (ML) method (Felsenstein 1985). Further analysis of the phylogenetic tree was carried out on available online software, Interactive Tree of Life (Ciccarelli et al. 2006) (https://itol.embl.de/itol.cgi/).
Results and discussion
Metagenomic DNA extraction, sequencing and quality control
DNA was extracted successfully from all the five formalin-preserved sponges for further study. However, metagenomic DNA extracted from Spongilla sp. S8 and Spongilla sp. S10 failed when subjected to amplicon sequencing on the Illumina Mi-Seq platform, though the genomic DNA quality was good. The metagenomic sequences of the sponge samples, Spongilla sp. S9, Ciocalypta sp. S11 and Ciocalypta sp. S12 were submitted to the NCBI-SRA (Sequence Read Archive) under accession numbers SRR6347343, SRR6347342, and SRR6347344 respectively.
Bacterial abundance and diversity analysis
A total of 10161operational taxonomic units (OTUs) belonging to 41 different phyla (376 different species) were obtained from the three sponge samples, Spongilla sp.S9 (growing at pH 8 and salinity 1%), Ciocalypta sp. S11 (pH 8; salinity 5%) and Ciocalypta sp. S12 (pH 9; salinity 1.5%) with 98% cut-off and 9148 OTUs with 97% cut-off. There were no significant differences in the abundance of bacterial diversity obtained using both the cut-off’s (Table S1); hence 98% cut-off was used for further analysis. The abundance here refers to the percentage of sequence reads in the final OTU table. The unclassified OTUs observed were 5.8% in 97% cut-off and 5.9% in 98% cut-off. In this analysis, OTU represents a cluster of a particular group of sequences. Thus, some sequences representing OTUs constructed at 97% were not classifiable by the RDP classifier and were labelled as unclassified. At 98% cut-off, these longer unclassifiable sequences were placed in separate OTUs with fewer sequences and shorter abundant classifiable sequences placed in other OTUs, consequently increasing identifiable sequences. Rarefaction curves were generated (Fig. S1) for each sample at a sequence similarity cut-off of 98%, which indicated that the majority of taxonomic diversity was covered in all the three samples. The curves showed saturation indicating proper sampling to continue with further analysis.
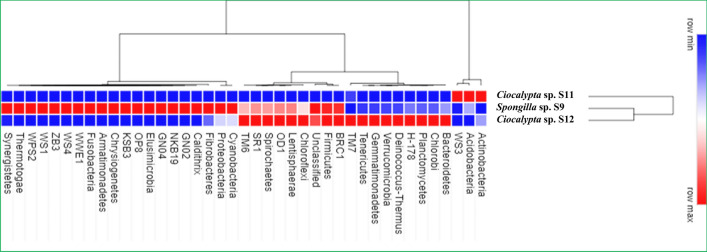
Bacterial abundance or distributions at the level of phylum (Fig. 2) and classes (Fig. S2) are shown in the form of heat map. Overall, 8792 OTUs (at 98% sequence similarity cut-off) belonging to 41 different bacterial phyla (23 known bacterial phyla, 17 "Candidatus" phyla, and one unclassified phylum) were obtained from the three sponge samples. Detected OTUs belong to the phyla Actinobacteria, Firmicutes, Bacteroidetes, Chloroflexi, Planctomycetes, Acidobacteria, and Gemmatimonodetes accounting for 40% of the observed OTUs. Seventeen OTUs belong to the Candidatus phyla which include BRC1, GN02, GN04, H-178, KSB3, NKB19, OD1, OP8, SR1, TM6, TM7, WPS2, WS1, WS3, WS4, WWE1 and ZB31. Members of the phylum “Candidatus Poribacteria” which are widely associated with many marine sponges (Schmitt et al. 2012) were not detected in our study. Majority of the OTUs belong to unclassified taxa and taxa belonging to the phylum Aquificae, Caldiserica, Dictyoglomi, Nitrospira, and Thermodesulfobacteria were absent in all the sponges studied (Fig. 2). The most abundant OTUs belong to the phylum Proteobacteria with 11810 sequences distributed across all the five classes (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria). OTUs belonging to the class Alphaproteobacteria are the most dominant in all the three sponge (Fig. S2). Among the phylum Planctomycetes, OTUs belonging to the Class Phycisphaerae and Planctomycetia are dominant in Ciocalypta sp. S12 (Fig. S2). At the family level, OTUs belonging to Rhodobacteraceae (27–48%) are dominant in all the sponge samples. The dominant families were similar in Spongilla sp. S9 and Ciocalypta sp. S12. However, Spongilla sp. S9 showed a higher ratio of unique families compared to Ciocalypta sp. S12 and Ciocalypta sp. S11 (Data not shown).
Fig. 2.
Heat map showing Phylum level comparison of bacterial community structure obtained from amplicon metagenome of three sponge samples, Spongilla sp. S9, Ciocalypta sp. S11---- and Ciocalypta sp. S12. The relative abundance is represented as the percentage of each phylum to the total effective bacterial sequences of each sample. The relative abundance is represented as the percentage of each phylum to the total effective bacterial sequences of each sample. The heat map was hierarchically clustered in two dimensions. The horizontal axis refers to relative abundance of each phylum and vertical axis represents the sponge sample
Among the 4828 OTUs used for constructing Venn diagram by mothur, no OTU represented core microbiome of the three sponge samples analysed (Fig. S3). Spongilla sp. S9 and Ciocalypta sp. S12 shared about 80 OTUs, mostly belonging to Proteobacteria. Spongilla sp. S9 contains more unique OTUs (3072), accounting for 70% of total OTUs, followed by Ciocalypta sp. S12 (700 OTUs, 15.6%) and finally, Ciocalypta sp. S11 contained 583 unique OTUs (11.8%). Ciocalypta sp. S12 and Ciocalypta sp. S11 shared only 2 OTUs (Fig. S3). This finding suggests that environmental parameters like salinity, temperature, and pH affect the microbiomes along with host specificity. Ciocalypta sp. S11, isolated from a pond with relatively high salinity (5%) and temperature (30 °C) and had a pH of 8 shared no OTUs with Spongilla sp. S9 and shared just two OTUs with Ciocalypta sp. S12, which were both isolated from puddles with low salinity (1–1.5%, respectively), low temperature (26 °C and 28 °C) and different pH (7–9, respectively). In contrast, Spongilla sp. S9 and Ciocalypta sp. S12, which were isolated from puddles with similar salinity and temperature shared 80 OTUs.
To further examine the community relationship among all three sponge samples, non-metric multi-dimensional scaling (nMDS) was performed using PAST software. The Bay-Curtis distance was taken into consideration. The nMDS analysis positioned them at altogether different places (Fig. S4). This indicated that the three sponge samples have distinct bacterial diversity. It is mostly affected by environmental parameters (especially salinity) and host specificity as well. Various alpha diversity indices (Mean and Standard deviation) like Shannon (H), Inverse Simpson, and Observed Richness were calculated using mothur by rarefying the OTU table to 4828 OTUs. Alpha diversity indices were similar between the cut-off percentages (97–98% sequence similarity) for the three samples analysed. Shannon diversity index varied between the sponge samples, with the highest diversity found in Spongilla sp. S9 (H = 8.2 ± 0.0) and the lowest in Spongilla sp. S11 (4.8 ± 0.0). Observed Richness and Inverse Simpson indices were also higher for Spongilla sp. S9, followed by Ciiocalypta sp. S12 and lowest for Ciiocalypta S11 (Table S1). The Spongilla sp. S9 showed the most diverse bacterial community (Shannon's diversity, which considers how evenly the OTUs are distributed), followed by Ciocalypta sp. S12 and Ciocalypta sp. S11. Although the sponges analysed in the present study were collected from not very far-off puddles, they showed significant variation in bacterial composition, which may be attributed to the variation of environmental parameters (pH and salinity and temperature) of the niches from where these were collected.
Planctomycetes diversity among sponges
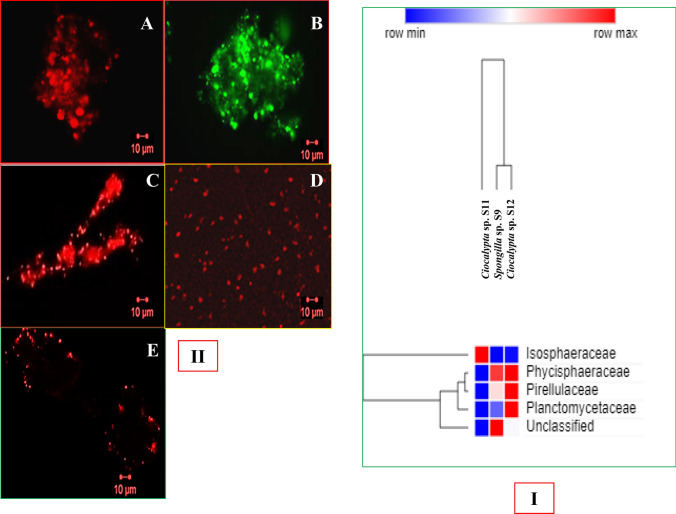
Planctomycetes in all three sponge samples were analysed using the abundance values obtained by amplicon sequencing. Planctomycetes, although not a dominant phylum, was observed in all the three samples ranging from 1.0 to 4.3% (Fig. 2). The sponge sample Ciocalypta sp. S12 contains more OYUs (4.3%) belonging to Planctomycetes, followed by Ciocalypta sp. S11 (1.1%) and Spongilla sp. S9 (1.2%). Family level distribution showed a different diversity among the three sponges. Ciocalypta sp. S11 contained OTUs belonging to Isosphaeraceae (unclassified at genus level), whereas Ciocalypta sp. S12 and Spongilla sp. S9 showed similar distribution with different dominant families (Fig. 3I). Spongilla sp. S9 is dominated by OTUs belonging to the family Phycisphaeraceae (unclassified) followed by OTUs of the family Pirellulaceae (unclassified), Planctomycetaceae (Planctomyces sp.), and Isosphaeraceae, whereas in Ciocalypta sp. S12, Pirellulaceae family is dominant, followed by Planctomycetaceae, Phycisphaeraceae and Isosphaeraceae. Overall, most of the Planctomycetes were unclassified at the family level, indicating the need to explore further diversity (Fig. 3I). Thus, culture-independent study, intimate and impel us to cultivate unclassified planctomycetes along with anammox bacteria.
Fig. 3.
I Heat map depicting the abundance and distribution of Planctomycetes at family level from amplicon sequence data of three sponge samples. The heat map was hierarchically clustered in two dimensions. The horizontal axis refers to relative abundance and vertical axis represents the sponge sample. II Fluorescence micrograph of Planctomycetes bacteria from five sponges enriched samples using CY3 labeled Pla46 probe. a JC541, b JC542, c JC543, d JC544, e JC544. Scale is of 10 µm
Functional profile prediction
The metabolic capacity of the microbiomes inferred from amplicon genomic libraries will provide a better understanding of the role of the microbiome in host metabolism and disease (Steinert et al. 2019). Functional metabolic profiling by using PICRUSt, a predictive approach, provide insights into potential functional aspects of sponge-associated bacterial communities. The KEGG annotation predicted a total of 6909 KEGG Orthologs (KOs; i.e., sets of homologous sequences) mapping to 328 different KEGG pathways (Fig. S5). Out of the total predicted KOs, 4448 were shared in microbiomes of all the three sponges. Most of the KOs are associated with membrane transport, followed by amino acid metabolism, carbohydrate metabolism and energy metabolism. Besides, the occurrence of different classes of energy metabolism (e.g., carbon or nitrogen metabolism) or symbiosis factors (e.g., eukaryotic-like proteins, ankyrin repeats, or tetratricopeptide repeats), KOs related to synthesis of secondary metabolites and bioactive natural products were predicted. Sponges are acknowledged widely often to synthesize secondary metabolites and bioactive natural products and the same may be contributed by the associated microbiome. All the three sponge microbiomes showed functional genes related to xenobiotic degradation (1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT), aminobenzoate, atrazine, benzoate, caprolactam, Polycyclic aromatic hydrocarbons). The functionally related genes for biosynthesis of secondary metabolites like carotenoids, neomycin, streptomycin, vancomycin, penicillin and cephalosporin were predicted in the samples. Such a vast secondary metabolite repertoire may play pivotal roles in microbiome community assembly, host-symbiont signalling and host defence. They also showed genes related to beta-Lactam resistance. They also showed KOs mapped to terpenoid backbone biosynthesis; terpenoids represent the most extensive natural products group and are a highly diverse class of organic chemicals. Terpene synthases have also been shown to be widely distributed in bacteria and represent a potentially important source for discovering novel natural products (Fig. S5).
Phylogenetic analysis of anammox-planctomycetes from the culture-dependent study
Five putative new species in the enrichment cultures, represented by bacterial isolates, JC541, JC542, JC543, JC544 and JC545, were identified from Spongilla sp. S8, Spongilla sp. S9, Spongilla sp. S10, Ciocalypta sp. S11 and Ciocalypta sp. S12, respectively, through 16S rRNA gene sequence analysis. The 16S rRNA gene sequences of JC541, JC542, JC543, JC544 and JC545, were deposited with European Molecular Biology Laboratory (EMBL) accession numbers LT907934, LT907935, LT907936, LT907938 and LT907937, respectively.
On-site inoculation of the sample in anammox-planctomycetes medium (in microcentrifuge tube having Durham's tube inside) ensured positive enrichment of anammox-planctomycetes (in co-cultured communities) and thus enhanced chances of recovering them in culture and minimizing the probability of losing. A three-step enrichment strategy (Fig. 1) proved quite useful in cultivating different Planctomycetes bacteria. The relative abundance of planctomycetes bacteria in samples was 1–4.3% (from metagenome data), which could be enhanced to 60–70% (deduced from microscopic observation during FISH) in the enrichment cultures. Positive fluorescence (Fig. 3II) observed with all the five co-cultured communities using CY3 labeled Pla46 probes for hybridization confirms the presence of anammox-planctomycetes cells (which are growing either as clusters or in isolation) in all the enrichments. Further, sequential sub-culturing in the test tubes and serum vials in anaerobic conditions helped increase the number of Planctomycetes and reduce relative abundance of other bacteria.
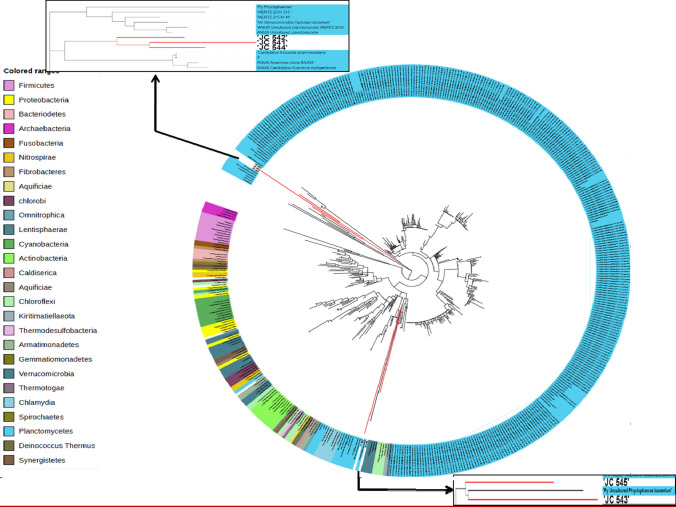
The16S rRNA gene sequences having a sequence length greater than 500 bp representing different orders of Planctomycetes were obtained from NCBI. The five 16S rRNA gene sequences from the enrichment cultures of the present study were used for phylogenetic analysis using the Maximum Likelihood (ML) method of MEGA-7. 16S rRNA gene sequences obtained from the isolates JC541, JC542, and JC544 cladded with CTUs of "Candidatus Brocadia anammoxidans”, but distinctly out-grouped (Fig. 4), suggesting that they represent new taxa. In contrast, the sequences of isolates, JC543, and JC545 have formed a separate clade with the uncultured Phycisphaerae class. Since the enrichment cultivation conditions mostly favour the growth of anammox bacteria, we suspect the two isolates (JC543 and JC545) representing putative novel taxa of Phycisphaerae might also have anammox property, which is under investigation since, so far, no members of Phycisphaerae are reported as anammox bacteria.
Fig. 4.
Phylogenetic tree constructed using the partial sequences of 16S r RNA gene of isolates JC 541, JC542, JC543, JC544 and JC545 along with the sequences belonging to phylum Planctomycetes obtained from amplicon metagenome of three sponge samples. Type strains of different members belonging to different phyla were also taken for phylogenetic analysis. Tree was constructed by the neighbour-joining method using MEGA-7 software. The constructed tree was visualized and coloured using online available software Interactive Tree Of Life (iTOL)
To the best of our knowledge, this is the first report of bacterial diversity of the two mini-sponge genera, Ciocalypta and Spongilla (except Spongillala custris; Gernert et al. 2005), and also the sponges from the Little Rann of Kutch. The diversity of Planctomycetes determined through metagenome analysis also differed in the three sponges, with members of the families Planctomycetaceae and Phycisphaeraceae dominating. The success of getting five putative novel taxa of anammox bacteria into cultivation in enrichment cultures in this study might be due to the three-step enrichment strategy used, coupled to inoculation into the enrichment medium immediately after sample collection. There is a need to understand their role in the nitrogen cycling and detoxification of ammonia in sponges, which may also have practical implications for aquaculture ponds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Chintalapati Venkata Ramana thanks to the Department of Biotechnology and CSIR Government of India for the award of Tata Innovative Fellowship and financial assistance, respectively. Vaddavalli Radha thanks to University Grants Commission for the award of Women-Post Doctoral Fellowship. Gaurav Kumar thanks to Department of Biotechnology (DBT) for providing financial assistance in the form of fellowship, and Uppadda Jagadeeshwari thanks Technical Education Quality Improvement Program (TEQIP) of World Bank, given to IST, JNTU Hyderabad. Ministry of Earth Sciences (MoES) and TEQIP are acknowledged for providing financial assistance to JNT University Hyderabad. DST and UGC are acknowledged for providing infrastructural facilities under FIST and SAP-DRS programs to Department of Plant Sciences, University of Hyderabad.
Compliance with ethical standards
Conflict of interest
Authors express no conflict of interest.
Footnotes
Gaurav Kumar, Vaddavalli Radha and Uppadda Jagadeeshwari contributed equally and considered as first authors.
Contributor Information
Chintalapati Sasikala, Email: sasi449@yahoo.ie.
Chintalapati Venkata Ramana, Email: cvr449@gmail.com.
References
- Abdelmohsen UR, Bayer K, Hentschel U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep. 2014;31:381–399. doi: 10.1039/c3np70111e. [DOI] [PubMed] [Google Scholar]
- Bayer K, Schmitt S, Hentschel U. Physiology, phylogeny and in situ evidence for bacterial and archaealnitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol. 2008;10:2942–2955. doi: 10.1111/j.1462-2920.2008.01582.x. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;312:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damste JSS, Ravin NV. Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov.and proposal of the orders Pirellulales ord. nov., Gemmatalesordnov.andIsosphaerales ord. nov. SystApplMicrobiol. 2020;43:126050. doi: 10.1016/j.syapm.2019.126050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli K, Fanger U, Alvarez PJJ, Siegrist H, van der Meer JR, Zehnder AJB. Enrichment and characterization of ananammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch Microbiol. 2001;175:198–207. doi: 10.1007/s002030100255. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fieseler L, Horn M, Wagner M, Hentschel U. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl Environ Microbiol. 2004;70:3724–3732. doi: 10.1128/AEM.70.6.3724-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernert C, Glockner FO, Krohne G, Hentschel U. Microbial diversity of the freshwater sponge Spongillalacustris. MicrobEcol. 2005;50:206–212. doi: 10.1007/s00248-004-0172-x. [DOI] [PubMed] [Google Scholar]
- Hammer O, Harper D, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 1:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm
- Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, et al. Microbial diversity of marine sponges. ProgMolSubcellBiol. 2003;37:59–88. doi: 10.1007/978-3-642-55519-0_3. [DOI] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10:641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Rapp HT, Reitner J. Monitoring microbial community composition by fluorescence in situ hybridization during cultivation of the marine cold-water sponge Geodia barretti. Mar Biotechnol. 2006;8:373–379. doi: 10.1007/s10126-006-5152-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, et al. Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol. 2009;11:2228–2243. doi: 10.1111/j.1462-2920.2009.01944.x. [DOI] [PubMed] [Google Scholar]
- Imhoff JF, Stohr R. Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea. ProgMolSubcellBiol. 2003;37:35–57. doi: 10.1007/978-3-642-55519-0_2. [DOI] [PubMed] [Google Scholar]
- Kartal B, de Almeida NM, Maalcke WJ, et al. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–461. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeqIllumina sequencing platform. Appl Environ Microbiol. 2013;17:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. Molecular evolutionary genetics analysis version 7.0 for bigger datasets. MolBiolEvol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Fuerst JA, Fieseler L, Engels C, Goh WWL, Hentschel U. Widespread distribution of Poribacteria in Demospongiae. Appl Environ Microbiol. 2009;75:5695–5699. doi: 10.1128/AEM.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes J, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NM, Saito K, Tal Y, Hill RT. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 2010;4:38–48. doi: 10.1038/ismej.2009.84. [DOI] [PubMed] [Google Scholar]
- Parsley LC, Newman MM, Liles MR. Fluorescence in situ hybridization of bacterial cell suspensions. Cold Spring HarbProtoc. 2010;9:pdb.prot5493. doi: 10.1101/pdb.prot5493. [DOI] [PubMed] [Google Scholar]
- Patel R, Mevada V, Prajapati D, Dudhagara P, Koringa P, Joshi CG. Metagenomic sequence of saline desert microbiota from wild ass sanctuary, Little Rann of Kutch, Gujarat, India. Genom Data. 2015;15:137–139. doi: 10.1016/j.gdata.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez T, Sarrazin L, Rebouillon P, Vacelet J. First evidences of surfactant biodegradation by marine sponges (Porifera): an experimental study with a linear alkylbenzenesulfonate. Hydrobiologia. 2002;489:225–233. doi: 10.1023/A:1023217218585. [DOI] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiong W, George MG, James MT, James RC. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. Microbial diversity: facts, problems and prospects. SystApplMicrobiol. 2004;27:3–9. doi: 10.1078/0723-2020-00245. [DOI] [PubMed] [Google Scholar]
- Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, Maas B, Dapena A, Van de Pas-Schoonen K, Van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, et al. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (Anammox) bacteria. Appl Environ Microbiol. 2005;4:1677–1684. doi: 10.1128/AEM.71.4.1677-1684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, Perez T, Rodrigo A, Schupp PJ, et al. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012;3:564–576. doi: 10.1038/ismej.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipkema D, Holmes B, Nichols SA, Blanch HW. Biological characterisation of Haliclona (gellius) sp.: sponge and associated microorganisms. MicrobEcol. 2009;58:903–920. doi: 10.1007/s00248-009-9534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert G, Wemheuer B, Janussen D, Erpenbeck D, Daniel R, Simon M, Brinkhoff T, Schupp PJ. Prokaryotic diversity and community patterns in Antarctic continental shelf sponges. Front Mar Sci. 2019;6:297. doi: 10.3389/fmars.2019.00297. [DOI] [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, BartolMavel D, Wincker P. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C content. MolBiolEvol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. MicrobiolMolBiol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, Olson JB, Erwin PM, López-Legentil S, Luter H, Chaves-Fonnegra A. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:1–2. doi: 10.1038/ncomms11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Graaf AA, de Bruijn P, Robertson LA, Jetten MSM, Kuenen JG. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiol. 1996;142:2187–2196. doi: 10.1099/13500872-142-8-2187. [DOI] [Google Scholar]
- Van de Graaf AA, de Bruijn P, Robertson LA, Jetten MSM, Kuenen JG. Metabolic pathway of anaerobic ammonium oxidation on the basis of 15N studies in a fluidized bed reactor. Microbiol. 1997;143:2415–2421. doi: 10.1099/00221287-143-7-2415. [DOI] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Negri AP. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2008;2:830–842. doi: 10.1038/ismej.2008.42. [DOI] [PubMed] [Google Scholar]
- Weisz JB, Hentschel U, Lindquist N, Martens CS. Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol. 2007;152:475–483. doi: 10.1007/s00227-007-0708-y. [DOI] [Google Scholar]
- Wiegand S, Jogler M, Jogler C. On the maverick Planctomycetes. FEMS Microbiol Rev. 2018;42:739–760. doi: 10.1093/femsre/fuy029. [DOI] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, et al. Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat Microbiol. 2020;5:126–140. doi: 10.1038/s41564-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR. Nutrient translocation from symbiotic cyanobacteria to coral reef sponges. In: Levi C, Boury-Esnault N, editors. Biologie des Spongiaires. Paris: C.N.R.S p; 1979. pp. 373–380. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.