Abstract
Background
The emergence of the NDM-1-positive Klebsiella pneumoniae (K. pneumoniae) strains has led to limited therapeutic options for clinical treatment. Understanding the clinical characteristics, antimicrobial resistance, biofilm assay, and the virulence genes of these isolated strains is of great significance.
Methods
The polymerase chain reaction (PCR) was used to screen isolated NDM-1-positive K. pneumoniae. The clinical information of the patients was collected from medical records. The NDM-1-positive K. pneumoniae isolates were subjected to antimicrobial susceptibility testing and multilocus sequence typing. Sixty strains of NDM-1-negative K. pneumoniae isolated during the same period were collected as the control group for the virulence analysis. The virulence phenotype of the strains was preliminarily evaluated by the string test and crystal violet semiquantitative biofilm formation experiment. PCR combined with gene sequencing was used to detect common high toxicity capsule genes (K1, K2, K5, K20, K54, and K57) and common virulence-related genes (entB, ybtS, ureA, ycf, WabG, FimH, uge, iutA, KfuB, aerobactin, rmpA, magA, Alls, IrnN, and VatD).
Results
In the 30 nonduplicated NDM-1-positive K. pneumoniae isolates, 43.33% (13/30) of the patients had a history of a stay in the neonatal intensive care unit (NICU). All of the isolates exhibited multidrug resistance. Nine STs were identified, 77% (10/13) strains from the NICU were ST11. The NDM-1-positive K. pneumoniae string tests were all negative, and 35% (21/60) NDM-1-negative K. pneumoniae were positive. The ratios of NDM-1-positive K. pneumoniae isolates biofilm formation ability according to strong, medium, and weak classification were 67%, 23%, and 10%, respectively. NDM-1-negative K. pneumoniae isolates were 60%, 25%, and 15%, respectively. There was no statistical difference between the two groups (t = 0.61, P=0.2723). The virulence-associated genes with more than 80% of detection rates among the 30 NDM-1-positive K. pneumoniae isolates included entB (100%, 30/30), ybtS (93.33%, 28/30), ureA (90%, 27/30), ycf (83.33%, 25/30), and wabG (90%, 27/30). KfuB and iutA were detected at prevalence of 3.33% and 13.33%. vatD, allS, iroN, aerobactin, and rmpA were not detected. In the NDM-1-negative K. pneumoniae, all other 14 virulence genes except VatD were detected. After statistical analysis, FimH, WabG, ycf, iutA, kfuB, aerobactin, rmpA, and Alls virulence genes, P < 0.005, there was a statistical difference.
Conclusion
NDM-1-positive K. pneumoniae exhibited multidrug resistance, MLST typing is mainly ST11, there is small clonal dissemination in the NICU in the hospital, and the NDM-1-positive K. pneumoniae virulence genes carrier rate is lower than the NDM-1-negative K. pneumoniae virulence genes carrier rate.
1. Introduction
As far as humans are concerned, K. pneumoniae is a frequently-isolated bacterial pathogen that colonizes the oropharynx, skin, or gastrointestinal tract. It causes various infections, including bacteraemia, pneumonia, urinary tract infections, suppurative infections, and cholangitis, especially to patients suffering from underlying disease conditions such as diabetes mellitus [1, 2].
The New Delhi metallo-β-lactamase (NDM) is a metallo-β-lactamase able to hydrolyze almost all β-lactams [3]. NDM-1 was first identified in a K. pneumoniae strain isolated from a Swedish patient who had been hospitalized in New Delhi, India, in 2008 [4]. NDM-positive strains cause a variety of infections that have been reported to be associated with high mortality rates [5]. NDM-positive strains have been found worldwide, resulting in a significant challenge for clinical management and public health [6]. Twenty-four NDM variants have been identified in more than 60 species of 11 bacterial families, and several variants have shown enhanced carbapenemase activity [7]. Among the 24 NDM variants, NDM-1 has the widest host spectrum identified so far and has been found in a number of species belonging to 11 bacterial families. K. pneumoniae and Escherichia coli are the predominant carriers of blaNDM-1 [8].
Research has shown that K. pneumoniae with the NDM-1 genotype was the primary cause of neonatal carbapenem resistant sepsis in China [9]. Fuursted, et al. have reported that K. pneumoniae carrying NDM-1 have an intrinsic virulence potential [10]. Researchers also confirmed that bacteria can increase drug resistance through a variety of drug resistance mechanisms, while improving its pathogenicity [11].
Virulence factors are important in colonization, invasion, and development of infection, and the virulence factors such as lipopolysaccharides (LPS), capsule, siderophores, and fimbriae in K. pneumoniae have been well characterized to date [12].
Previous studies have focused on the risk factors of blaNDM-1 and the surrounding environment of genes. Our previous studies have shown that NDM-1 can be expressed on chromosomes and plasmids, and NDM-1-positive K. pneumoniae is multidrug resistant and can spread easily [13]. However, little is known about the virulence factor characteristics of NDM-1-positive K. pneumoniae. In this study, the clinical infection characteristics, drug resistance characteristics, and MLST homology typing of NDM-1-positive K. pneumoniae were analyzed, and NDM-1-negative K. pneumoniae was used as a control group. The phenotype and virulent genes were compared, and new ideas for the research on infection control, clinical treatment, and the pathogenicity of NDM-1-positive K. pneumoniae were proposed.
2. Materials and Methods
2.1. Collection and Identification of NDM-1-Positive K. pneumoniae Isolates
The 30 NDM-1-positive K. pneumoniae isolates were screened from 720 K. pneumoniae isolates at the First Affiliated Hospital of Kunming Medical University, in Yunnan Province, Southwest China, from January 2017 to June 2020. They were obtained from sputum (18/30), urine (5/30), catheter tip (3/30), blood (3/30), and wound (1/30). The isolates were identified as K. pneumoniae strains by using the VITEK-2 System (bioMe'rieux). Also, PCR was performed to detect blaNDM-1 (Forward 5′-GGGCAGTCGCTTCCAACGGT-3′, Reverse 5′-GTAGTGCTCAGTGTCGGCAT-3′) [14]. Clinical information was collected, including demographics, underlying medical conditions, clinical presentations, and antimicrobial therapy. This study was approved, and informed consent was acquired from the patients involved in this study.
2.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing was performed with a VITEK-2 automated microbiology analyzer platform (bioMérieux, Marcy l'Etoile, France) to examine the sensitivity of NDM-1-positive K. pneumoniae against common antibiotics. The minimal inhibitory concentration (MIC) of imipenem was further verified by the E-test method according to the guideline recommended by the Clinical and Laboratory Standards Institute (CLSI, 2018), and the MIC of colistin B was further verified by the microdilution broth method according to the guideline recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2018). E. coli ATCC25922 was used as a control strain for the antimicrobial susceptibility testing.
2.3. Multilocus Sequence Typing (MLST)
MLST was used to screen the 30 NDM-1-positive K. pneumoniae isolates by amplifying seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) expressed in K. pneumoniae according to the protocol at (http://bigsdb.pasteur.fr/klebsiella/primers_used.html).
2.4. String Test
K. pneumoniae strains were incubated overnight on blood agar. A single colony was touched with a loop and stretched outward. The length of the viscous string was pulled upward and measured. A positive string test result was defined as a string longer than 5 mm. The string test was repeated three times for each strain, and the final result was determined [15].
2.5. Biofilm Formation Assay
In brief, 10 μl of the 0.5 McFarland bacterial standard and 190 μl of the Luria-Bertani (LB) broth were inoculated into the wells of a 96-well microplate, with three wells per strain, and the microplate was incubated at 37 °C for 24 h. Thereafter, the LB broth was removed, and the bacterial cells were stained with 200 μl of 0.1% crystal violet at room temperature for 15 min and, then, the due was removed. The plate was washed with distilled water and, then, dried. The absorbance was measured with a microplate reader set at 590 nm after adding 200 μl of ethanol for 10 min into the wells. The yield of biofilm formation of the strains was interpreted as follows: OD > 0.5 as strong-producing, 0.2 ≤ OD ≤ 0.5 as moderate-producing, and OD < 0.2 as weak-producing [16].
2.6. Virulence-Associated Genes
The primer sequences for capsular serotyping and the virulence genes are listed in Supplementary Materials. Capsular serotypes, including K1, K2, K5, K20, K54, and K57, were determined using the methods described previously [17, 18]. The fifteen virulence-associated genes, including aerobactin, iroN, kfuB, rmpA, alls, ybtS, ureA, uge, wabG, ycf, entB, iutA, aerobactin, vatD, magA, and fimH, were determined by PCR using the primers described previously [19].
2.7. Statistical Analysis
All statistical analyses were performed using SPSS 22.0 software (IBM, Armonk, NY, USA). The categorical variables were listed as percentages and evaluated using the Chi-square test or Fisher's exact test. The continuous data were expressed as mean ± standard deviation (mean ± SD) or median (25th-75th percentile) appropriately and analyzed using Student's t-test or the Mann–Whitney U test. A P value <0.05 was considered statistically significant. All tests were two-tailed.
3. Results
3.1. Clinical Characteristics of Patients with NDM-1-Positive K. pneumoniae
Clinical characteristics of 30 patients with NDM-1-positive K. pneumoniae isolates are shown in Table 1. In the 30 nonduplicated NDM-1-positive K. pneumoniae isolates, the patients' median length of hospitalization was 49 days. 43.3% (13/30) of the patients had a history of a stay in the neonatal intensive care unit (NICU). Of these patients, 63.3% (19/30) were males. All, but five patients, received invasive treatment prior to infection with NDM-1-positive K. pneumoniae, including central venous catheters and invasive mechanical ventilators. 70% (21/30) had been treated by carbapenems (imipenem and meropenem).
Table 1.
Clinical characteristics of patients with NDM-1-positive K. pneumoniae.
| Isolate no. | Gender | Age (yr) | Date of specimen collection | Days in the hospital | Isolation site(s) | Ward | Invasive treatment | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| K-N1 | Male | 45 | 2017/1/9 | 89 | Sputum | Transplant center | Laparotomy | SCF, MEM, TEC | Recovered |
| K-N2 | Female | Newborn | 2017/5/17 | 39 | Sputum | NICU | Umbilical vein cannula | AMC, SCF, FLUCZ, AZT, CRO | Recovered |
| K-N3 | Male | 72 | 2017/6/26 | 36 | Sputum | Elderly general | Tracheal intubation | MEM, KETOC, VA | Died |
| K-N4 | Male | Newborn | 2017/9/22 | 46 | Catheter tip | NICU | Umbilical vein intubation; central venous catheterization | SCF, AZT, FLUCZ, TZP, CAZ | Recovered |
| K-N5 | Male | Newborn | 2017/12/21 | 84 | Sputum | NICU | Noninvasive ventilator | MEM, AMC, PIS | Recovered |
| K-N6 | Female | Newborn | 2018/1/3 | 10 | Sputum | NICU | Tracheal intubation; umbilical vein catheter | AMC, SCF | Died |
| K-N7 | Male | Newborn | 2018/1/5 | 28 | Urine | NICU | Umbilical vein catheterization; ventilator oxygen | AMC, FLUCZ | Recovered |
| K-N8 | Male | 97 | 2018/1/10 | 13 | Sputum | Respiratory unit | No | MEM, KETOC, LEV | Recovered |
| K-N9 | Male | 90 | 2018/1/11 | 20 | Sputum | General surgery | No | MEM, FLUCZ | Died |
| K-N10 | Female | Newborn | 2018/1/24 | 20 | Urine | NICU | Tracheal intubation; umbilical vein catheterization | MEM, AMC, SCF | Recovered |
| K-N11 | Male | Newborn | 2018/2/5 | 33 | Catheter tip | NICU | Tracheal intubation; umbilical vein catheterization | MEM, AMC | Recovered |
| K-N12 | Female | Newborn | 2018/2/8 | 100 | Sputum | NICU | Tracheal intubation; umbilical vein catheterization | AMC | Died |
| K-N13 | Male | Newborn | 2018/2/12 | 16 | Sputum | NICU | Tracheal intubation | SCF | Recovered |
| K-N14 | Female | Newborn | 2018/3/1 | 51 | Urine | NICU | Umbilical vein catheterization; noninvasive ventilator | MEM, AMC, SCF | Recovered |
| K-N15 | Male | 85 | 2018/3/13 | 27 | Sputum | General surgery | No | MOX | Recovered |
| K-N16 | Female | Newborn | 2018/5/10 | 64 | Catheter tip | NICU | Tracheal intubation; umbilical vein catheterization | MEM, SCF, IPM | Recovered |
| K-N17 | Male | 86 | 2018/7/27 | 14 | Sputum | Gastroenterology | No | MEM | Died |
| K-N18 | Female | 87 | 2018/7/31 | 9 | Sputum | NICU | No | SCF | Recovered |
| K-N19 | Male | 87 | 2018/8/8 | 14 | Sputum | Respiratory unit | Noninvasive ventilator | MEM, FLUCZ, SCF | Died |
| K-N20 | Male | 56 | 2018/8/29 | 14 | Blood | Neurosurgery | Tracheal intubation, noninvasive ventilator; | MEM, VA | Recovered |
| K-N21 | Male | 9 | 2018/9/28 | 91 | Blood | Neurosurgery | Craniopharyngioma resection; lateral ventricle puncture | MEM, CZO, VA | Recovered |
| K-N22 | Female | 49 | 2020/1/9 | 56 | Blood | ICU | Tracheal intubation | TZP, IMP | Recovered |
| K-N23 | Male | 82 | 2020/2/13 | 18 | Sputum | NICU | Tracheal intubation | CMZ, TZP, IMP | Recovered |
| K-N24 | Female | 47 | 2020/2/28 | 13 | Urine | ICU | Tracheal intubation | LVX, IMP, SCF, MEM | Recovered |
| K-N25 | Male | 91 | 2020/3/9 | 18 | Sputum | Neurosurgery | No | IMP, Cefoselis, LVX | Recovered |
| K-N26 | Male | 88 | 2020/3/12 | 15 | Sputum | Neurosurgery | Tracheal intubation | MEM, TZP, LVX | Recovered |
| K-N27 | Male | 62 | 2020/3/14 | 25 | Urine | ICU | Tracheal intubation | MEM, TZP | Recovered |
| K-N28 | Female | 59 | 2020/3/27 | 14 | Sputum | Neurosurgery | Lateral ventricle puncture | TZP, MXF | Recovered |
| K-N29 | Female | 93 | 2020/4/12 | 46 | Wound | ICU | Tracheal intubation | MEM, TZP, SCF, LVX | Recovered |
| K-N30 | Male | 14 | 2020/4/29 | 21 | Sputum | Hematology | Tracheal intubation | MEM, TZP | Recovered |
Notes. SCF: cefoperazone sulbactam; TEC: teicoplanin; MEM: meropenem; CRO: ceftriaxone; MOX: Lafaxed; AMC: amoxicillin clavulanate potassium; KETOC: voriconazole; FLUCZ: fluconazole; AZT: aztreonam; VA: vancomycin; CAZ: ceftazidime.
3.2. Antimicrobial Susceptibility Testing
The antimicrobial resistance rates of the 30 NDM-1-positive K. pneumoniae isolates are shown in Table 2. They were all resistant to meropenem, imipenem, ertapenem, ceftazidime, cefoperazone sulbactam, piperacillin, sulbactam, cefazolin, cefepime, and cefoxitin. Their minimal inhibitory concentrations (MICs) of imipenem were more than 4 μg/ml by the E-test method. Twenty-six (86.67%, 26/30) isolates were resistant to aztreonam and ciprofloxacin, and twenty-three (76.7%, 23/30) were resistant to levofloxacin. Twenty-two (73.3%, 22/30) and seventeen (56.7%, 17/30) isolates were resistant to gentamicin and amikacin, respectively. Only ten isolates were resistant to sulfamethoxazole/trimethoprim, and all isolates were susceptible to colistin B and tigecycline.
Table 2.
The antimicrobial resistance profiling of NDM-1-positive K. pneumoniae
| NDM-1-positive K. pneumoniae (n = 30) | ||
|---|---|---|
| No. | % | |
| Meropenem | 30 | 100% |
| Imipenem | 30 | 100% |
| Ertapenem | 30 | 100% |
| Ceftazidime | 30 | 100% |
| Cefoperazone sulbactam | 30 | 100% |
| Piperacillin and sulbactam | 30 | 100% |
| Cefazolin | 30 | 100% |
| Cefepime | 30 | 100% |
| Cefoxitin | 30 | 100% |
| Ciprofloxacin | 26 | 86.67% |
| Aztreonam | 26 | 86.67% |
| Levofloxacin | 23 | 76.67% |
| Gentamicin | 22 | 73.33% |
| Amikacin | 17 | 56.67% |
| Sulfamethoxazole/trimethoprim | 10 | 33.30% |
| Polymyxin B | 0 | 0 |
| Tigecycline | 0 | 0 |
3.3. Molecular Characteristics of NDM-1-Positive K. pneumoniae Isolates
Among the 30 NDM-1-positive K. pneumoniae isolates, 9 STs were identified, including ST11 (15 isolates), ST105 (3 isolates), ST37(3 isolates), ST1 (2 isolates), and ST36 (2 isolates), ST1652 (2 isolates), ST656 (1 isolate), ST1137 (1 isolate), and ST433 (1 isolate). Ten of the thirteen strains from the NICU were ST11. Others were ST433, ST36 and ST37, respectively. The specific sequence similarity cluster analysis is shown in Figure 1.
Figure 1.
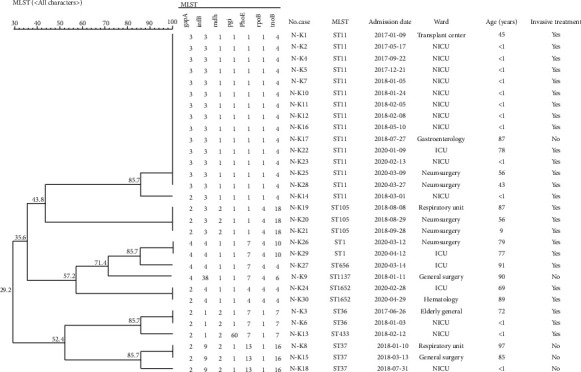
The specific sequence similarity cluster analysis.
3.4. String Test
String test results of NDM-1-positive K. pneumoniae isolates were all negative, and 35% (21/60) NDM-1-negative K. pneumoniae isolates were positive.
3.5. Biofilm-Producing Isolates
The biofilm-producing analysis is shown in Figure 2. The ratios of NDM-1-positive K. pneumoniae isolates biofilm formation ability according to strong, medium, and weak classification were 67%, 23%, and 10%, respectively. NDM-1-negative K. pneumoniae isolates were 60%, 25%, and 15%, respectively. The biofilm-forming capacity of NDM-1-positive K. pneumoniae isolates and NDM-1-negative K. pneumoniae isolates measured A590 were 0.5767 ± 0.2854 and 0.6225 ± 0.3595, respectively. There was no statistical difference between the two groups (t = 0.61, P=0.2723).
Figure 2.
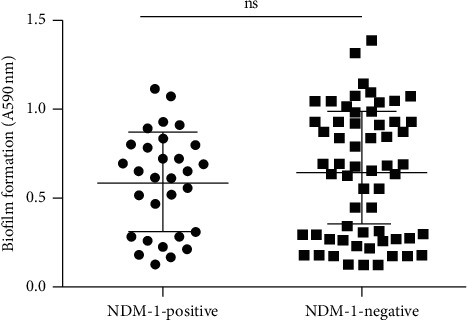
The biofilm formation. Note. The circles indicate the 30 NDM-1-positive K. pneumoniae; the squares indicate the 60 NDM-1-negative K. pneumoniae.
3.6. Prevalence of Capsular Serotyping and Virulence-Associated Genes
The 6 serotypes and 15 virulence genes were tested in this experiment. However, we did not detect any serotypes in NDM-1-positive K. pneumoniae strains. Among the 60 NDM-1-negative K. pneumoniae isolates, 15 (25%) isolates belonged to capsular serotype K1, 7(11.6%) isolates belonged to capsular serotype K2, 1 (1.6%) isolate belonged to capsular serotype K5, and 2 (3.33%) isolates belonged to capsular serotype K57. The remaining 35 (58.3%) isolates were not successfully typed. The prevalence and distribution of virulence factors are shown in Table 3. The virulence-associated genes with more than 80% of detection rates among the 30 isolates included entB (100%, 30/30), ybtS (93.33%, 28/30), ureA (90%, 27/30), ycf (83.33%, 25/30), and wabG (90%, 27/30). KfuB and iutA were detected at a prevalence of 3.33% and 13.33%, respectively. vatD, allS, iroN, aerobactin, and rmpA were not detected in NDM-1-positive K. pneumoniae isolates. The NDM-1-positive K. pneumoniae group carried 6 or 7 virulence genes most commonly, and the NDM-1-negative K. pneumoniae group mainly carried 8–11 virulence genes. In NDM-1-negative K. pneumoniae, all other 14 virulence genes except VatD were detected. After statistical analysis, FimH, WabG, ycf, iutA, kfuB, aerobactin, rmpA, and Alls virulence genes, P < 0.005, showed there was a statistical difference.
Table 3.
Differences in the distribution of virulence genes and serotypes between NDM-1-positive K. pneumoniae and NDM-1-negative K. pneumonia (n (%)).
| NDM-1-positive K. pneumoniae (n = 30) | NDM-1 negative K. pneumoniae (n = 60) | P | |
|---|---|---|---|
| K1 | 0 (0) | 15 (25.00) | 0.003 |
| K2 | 0 (0) | 7 (11.66) | 0.051 |
| K5 | 0 (0) | 1 (1.66) | 0.477 |
| K57 | 0 (0) | 2 (3.33) | 0.312 |
| entB | 30 (100.00) | 60 (100.00) | 1 |
| ybtS | 28 (93.33) | 60 (100.00) | 0.109 |
| FimH | 23 (76.67) | 56 (93.33) | 0.038 |
| wabG | 27 (90.00) | 60 (100.00) | 0.035 |
| ureA | 27 (90.00) | 59 (98.33) | 0.106 |
| ycf | 25 (83.33) | 58 (96.67) | 0.039 |
| uge | 23 (76.67) | 43 (71.67) | 0.801 |
| iutA | 41 (3.33) | 26 (43.33) | 0.005 |
| kfuB | 1 (3.33) | 20 (33.33) | 0.001 |
| aerobactin | 0 (0) | 13 (21.67) | 0.004 |
| IroN | 0 (0) | 1 (1.67) | 1 |
| rmpA | 0 (0) | 9 (15.00) | 0.027 |
| magA | 0 (0) | 7 (11.67) | 0.09 |
| Alls | 0 (0) | 9 (15.00) | 0.027 |
| vatD | 0 (0) | 0 (0) | 1 |
4. Discussion
K. pneumoniae causes a wide range of infections both in the community and health-care setting leading to increased morbidity and mortality [20]. The patients infected with NDM-1-positive K. pneumoniae in this study have a large age span, mainly from newborns and elderly patients, suggesting that people with poor immunity and underlying diseases are susceptible to it. Ten of the 13 neonatal patients are premature and low birth weight infants, which may be related to their immune insufficiency, low phagocytic ability of white blood cell, and underdeveloped skin barrier. In addition, the 25 of 30 patients underwent invasive examinations, indicating that device intervention may weaken the patient's immunity. The specimens in this study were mainly derived from the sputum, suggesting that NDM-1-positive K. pneumoniae is more likely to spread through the respiratory tract. The sputum specimens are likely to cause environmental pollution and the spread of contact, so they should to be treated with strict disinfection measures to prevent its spread in the hospital.
The 30 patients had injected antibiotics before we separated the NDM-1-positive K. pneumoniae strains from them. Antibiotics are mainly cephalosporins antibiotics and penicillium carbon alkene. An in vitro susceptibility test showed that it was highly resistant to β-lactam drugs and β-lactamase inhibitor, but sensitive to aminoglycoside and fluoroquinolone, which is consisted with the literature that aminoglycoside is still recognized as a first-line therapy for treatment of K. pneumoniae infection [21]. In theory, strains carrying blaNDM-1 should be sensitive to aztreonam, but drug susceptibility results showed that treatment with aztreonam alone had a poor curative effect. Except for polymyxin B and tigecycline, it was highly resistant to a majority of clinical antibiotics. It is because that blaNDM-1 is mainly disseminated by plasmid IncA/C, which always carries a variety of resistance genes resulting in the emergence of antibiotics resistance [5]. Although aztreonam is stable against MBLs, NDM-1-positive K. pneumoniae strains usually have ESBLs or AmpC enzymes that are able to hydrolyze aztreonam. Aztreonam alone, therefore, has limited clinical utility against NDM-producing strains. In this study, all strains are sensitive to tigecycline and polymyxin B, which is consistent with Darey's research showing that polymyxin alone can treat infections caused by NDM-1-positive bacteria [22]. The use of colistin has also been hampered by the neurological adverse effects and occurrence of renal toxicity [23, 24]. There is evidence that polymyxin-based combinations may be more effective than polymyxin alone, so it is usually recommended to combine polymyxin with other antibiotics [25].
The 30 NDM-1-positive K. pneumoniae strains in this study were typed by MLST, and there were 9 types in total, which mainly is ST11. In China, most of the NDM-1-positive strains have been found belonging to different ST types and being scattered. ST11, ST14, ST15, and ST147 strains are relatively common NDM-positive K. pneumoniae lineages and have been found in multiple countries across several continents, almost all of which were isolated from humans [7]. In medical institutions and wards, if more than 3 cases of homogenous nosocomial infections occur in a short time, it is called nosocomial infection outbreak. In terms of time, the NO.10 strain was the first screened strain in the NICU ward. The patient was admitted to the hospital in January 24, 2018, and stayed for 20 days, which is overlapped with the 11th, 12th, and 13th strains at the same time. Afterwards, the 14th and 16th strains were overlapped with the 12th strain again. These 6 strains were genotyped by MLST, which shows 5 strains of them are ST11. Considering that the NDM-1 is located in the plasmid and easy to spread, it suggests that there is a clonal spread of ST11 NDM-1-positive K. pneumoniae strains in the NICU.
At present, most studies define the positive strain in the string test as hvKP. In this experiment, NDM-1-positive K. pneumoniae string tests were all negative, while the positive rate of the string test in the NDM-1-negative group was as high as 53.3%. It preliminarily indicates that NDM-1-negative K. pneumoniae may be more virulent. However, some scholars believe that the string test cannot be used as a criterion for judging whether the K. pneumoniae has high virulence.
Biofilm can enhance the defense ability of bacteria and resist the killing effect of antibiotics. The results of this study showed that all strains formed biofilms at varying degrees. Although NDM-1-positive K. pneumoniae does not show enhanced biofilm forming ability, the formation of biofilm will prolong the disease, so we still need to be vigilant.
Capsular polysaccharide (CPS), as one of the most important virulence factors of K. pneumoniae, can resist the phagocytosis of macrophages and neutrophils. Among them, K1, K2, K5, K20, K54, and K57 are recognized as highly virulent serotype [12]. In this test, NDM-1-positive K. pneumoniae did not detect the abovementioned 6 capsular serotypes. This indicates that when the bacteria acquires drug resistance, the gene expression ability of common high-virulence capsule serotypes will be weakened, and it is also possible that resistant bacteria express other capsule serotypes that have not been detected. The 4 of 6 types of NDM-1-negative K. pneumoniae high-virulence capsular serotypes were detected, mainly K1 and K2. In this experiment, magA and rmpA were not detected in the NDM-1-positive K. pneumoniae group. 15% of NDM-1-negative K. pneumoniae carry rmpA, and 12% carry magA, which is basically consistent with the drawing experiment and the detection rate of high-virulence capsule serotypes. The positive rate in the NDM-1-negative group is higher than that in the control group. There are many virulence factors related to pathogenicity in K. pneumoniae. The basis of them is mainly the capsular polysaccharide and iron uptake system. In addition, it also includes high mucus, lipopolysaccharide, and fimbriae-related (type I fimbriae and type III Pili), and biofilm formation. The 30 strains of NDM-1-positive K. pneumoniae are most widely distributed in entB, ureA, ybtS, FimH, uge, and ycf. These virulence genes are related to bacterial adhesion, iron uptake, and anti-phagocytosis. Although the positive rate of NDM-1-positive K. pneumoniae in the drawing experiment and common capsular serotypes is zero, it still expresses many important virulence genes.
Once K. pneumoniae inserts the virulence gene into the drug-resistant plasmid, it will make it a highly resistant, highly virulent, and easily spread strain. Some literatures have reported that highly virulent and resistant strains have begun to appear all over the world. In 2016, a case of blaNDM-1-positive ST231 CR-hvKP was reported in India. Also, a case of blaNDM-1-positive ST23 type CR-hvKP was reported in Europe in 2017 [26]. There were also several cases of CR-hvKP carrying blaNDM-1 reported in China [27, 28].
In summary, compared with the control group, the positive rate of NDM-1-positive K. pneumoniae in the string test and common high-virulence capsular serotypes, as well as the distribution of these virulence genes, are significantly reduced, which is consistent with Montanari's claim that bacteria lose some virulence genes in order to obtain resistance genes for optimal adaptability [29]. It indicates that although NDM-1-positive K. pneumoniae has increased drug resistance, its pathogenicity may be weaker. However, the NDM-1-positive K. pneumoniae still expresses many important virulence factors.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81960382) and Training Program for Medical Academic Leaders of Yunnan Provincial Health and Family Planning Commission (No. D-2017023).
Data Availability
All data that were used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
The sequences of primers for capsular serotyping and virulence-associated genes.
References
- 1.Podschun R., Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical Microbiology Reviews. 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shon A. S., Bajwa R. P. S., Russo T. A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P., Poirel L., Walsh T. R., Livermore D. M. The emerging NDM carbapenemases. Trends in Microbiology. 2011;19(12):588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Yong D., Toleman M. A., Giske C. G., et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrobial Agents and Chemotherapy. 2009;53(12):5046–5054. doi: 10.1128/aac.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh H., Gursoy N. C., Yakupogullari Y., et al. Hospital outbreak of a colistin-resistant, NDM-1- and OXA-48-ProducingKlebsiella pneumoniae: high mortality from pandrug resistance. Microbial Drug Resistance. 2018;24(7):966–972. doi: 10.1089/mdr.2017.0173. [DOI] [PubMed] [Google Scholar]
- 6.Otlu R. C., Jr. NDM-1 - a cause for worldwide concern. New England Journal of Medicine. 2010;363(25):2377–2379. doi: 10.1056/nejmp1011715. [DOI] [PubMed] [Google Scholar]
- 7.Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clinical Microbiology Reviews. 2019;32(2) doi: 10.1128/cmr.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazmierczak K. M., Rabine S., Hackel M., et al. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing enterobacteriaceae and Pseudomonas aeruginosa multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing enterobacteriaceae and Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2016;60(2):1067–1078. doi: 10.1128/aac.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahm Y., Wang Y., Hsia Y., Sharland M., Heath P. T. Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China. Annals of Clinical Microbiology and Antimicrobials. 2019;18(1):p. 36. doi: 10.1186/s12941-019-0334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuursted K., Schøler L., Hansen F., et al. Virulence of a Klebsiella pneumoniae strain carrying the New Delhi metallo-beta-lactamase-1 (NDM-1) Microbes and Infection. 2012;14(2):155–158. doi: 10.1016/j.micinf.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Dagnæs-Hansen A., Tomás M., Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clinical Microbiology Reviews. 2013;26(2):185–230. doi: 10.1128/cmr.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paczosa M. K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiology and Molecular Biology Reviews. 2016;80(3):629–661. doi: 10.1128/mmbr.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du N., Liu S., Niu M., et al. Transmission and characterization of bla (NDM-1) in Enterobacter cloacae at a teaching hospital in Yunnan, China. Annals of Clinical Microbiology and Antimicrobials. 2017;16(1):p. 58. doi: 10.1186/s12941-017-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balkan, Aygün G., Aydın S., et al. Blood stream infections due to OXA-48-like carbapenemase-producing Enterobacteriaceae: treatment and survival. International Journal of Infectious Diseases. 2014;26:51–56. doi: 10.1016/j.ijid.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Midilli W., Sun G., Yu Y., et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clinical Infectious Diseases. 2014;58(2):225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 16.Jiao K., Piktel E., Wilczewska A., et al. Core-shell magnetic nanoparticles display synergistic antibacterial effects against Pseudomonas aeruginosa and Staphylococcus aureus when combined with cathelicidin LL-37 or selected ceragenins. International Journal of Nanomedicine. 2016;11:5443–5455. doi: 10.2147/ijn.s113706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puszkarz C. T., Lai S. Y., Yi W. C., Hsueh P. R., Liu K. L., Chang S. C. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clinical Infectious Diseases. 2007;45(3):284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 18.Turton J. F., Perry C., Elgohari S., Hampton C. V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. Journal of Medical Microbiology. 2010;59(5):541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhan L., Wang S., Guo Y., et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Frontiers in Cellular and Infection Microbiology. 2017;7:p. 182. doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brisse S., Fevre C., Passet V., et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004982.e4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Zhang H., Zhang X., et al. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagnostic Microbiology and Infectious Disease. 2019;93(4):355–361. doi: 10.1016/j.diagmicrobio.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Darley E., Weeks J., Jones L., et al. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet. 2012;380(9850):p. 1358. doi: 10.1016/s0140-6736(12)60911-8. [DOI] [PubMed] [Google Scholar]
- 23.Stone N. R., Woodford N., Livermore D. M., et al. Breakthrough bacteraemia due to tigecycline-resistant Escherichia coli with New Delhi metallo-β-lactamase (NDM)-1 successfully treated with colistin in a patient with calciphylaxis. Journal of Antimicrobial Chemotherapy. 2011;66(11):2677–2678. doi: 10.1093/jac/dkr337. [DOI] [PubMed] [Google Scholar]
- 24.Ortwine J. K., Kaye K. S., Li J., Pogue J. M. Colistin: understanding and applying recent pharmacokinetic advances. Pharmacotherapy. 2015;35(1):11–16. doi: 10.1002/phar.1484. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S., Brunel J. M., Dubus J. C., Reynaud-Gaubert M., Rolain J. M. Colistin: an update on the antibiotic of the 21st century. Expert Review of Anti-infective Therapy. 2012;10(8):917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 26.Compain F., Vandenberghe A., Gominet M., et al. Primary osteomyelitis caused by an NDM-1-producing K. pneumoniae strain of the highly virulent sequence type 23. Emerging Microbes & Infections. 2017;6(6):p. e57. doi: 10.1038/emi.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankar C., Nabarro L. E. B., Muthuirulandi Sethuvel D. P., et al. Draft genome of a hypervirulent Klebsiella quasipneumoniae subsp. similipneumoniae with novel sequence type ST2320 isolated from a chronic liver disease patient. Journal of Global Antimicrobial Resistance. 2017;9:30–31. doi: 10.1016/j.jgar.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Wei D. D., Wan L. G., Liu Y. Draft genome sequence of an NDM-1- and KPC-2-Coproducing hypervirulent carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announcements. 2018;6(13) doi: 10.1128/genomea.00192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montanari S., Oliver A., Salerno P., et al. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology. 2007;153(5):1445–1454. doi: 10.1099/mic.0.2006/003400-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of primers for capsular serotyping and virulence-associated genes.
Data Availability Statement
All data that were used to support the findings of this study are included within the article.


