Dear Editor,
We propose a nontoxic, sustainable alternative to conventional surface disinfection, possibly useful in fighting the present COVID‐19 pandemics. The global spread of COVID‐19 has increased awareness of how the SARS‐CoV‐2 virus is transmitted on surfaces. 1 Person to person contagion can occur through contact with contaminated surfaces. To limit this contagion pathway, regular surface disinfection is recommended. Research indicates that this virus can remain viable for 4 to 72 hours on plastic, copper, and steel, and up to 7 days on surgical mask material, 2 creating increased transmission risk in social and medical environments. Presently, the application of ethanol in combination with sodium hypochlorite or hydrogen peroxide or the use of ultraviolet surface irradiation effectively inactivates the virus. However, the practical application of these methods, as well as other antiviral protocols, is hindered by their toxic impact on human health. 3 It is vital to develop surfaces, fabrics, and other materials that could inherently inhibit viral spread while concurrently being safe for humans.
One such material is silicon nitride (Si3N4), an FDA‐cleared bioceramic, which may be used in the human body. It has superior antibacterial behavior and has been proven safe for long‐term use in humans. It possesses a unique surface biochemistry that inhibits bacterial infections by long‐term elution of nitrogen (promptly converted into ammonia) in minute concentrations that, unlike bacteria and viruses, mammalian cells can easily metabolize. 4 Within 1 minute, influenza A and enterovirus were completely inactivated by Si3N4 bioceramic particles suspended in water. 5
In this study, we exposed SARS‐CoV‐2 virions to the above bioceramic as well as to aluminum nitride (AlN) micrometric powders suspended in water. The nitrogen‐based ceramic, AlN, undergoes surface hydrolysis analogous to that of Si3N4 when in such a solution. We used two controls, namely, a copper (Cu) particle suspension (a positive control, known to strongly inactivate pathogens and viruses 6 ) and a negative control expected to have no effect, H2O. The supernatant virions were then inoculated into VeroE6/TMPRSS2 cells. We expected comparable antiviral behavior for Si3N4 and AlN, as these nitride compounds share the chemical similarity of N atoms with strong electronegativity.
Figure 1A shows results for TCID50 assay in case of virions exposed to Si3N4, AlN, and Cu powders in 15 wt.% at 1‐minute inactivation time. Compared with the water‐exposed negative control (sham sample), these three powders produced equally effective inactivation of SARS‐CoV‐2 virions (>99%). We then examined fragmentation of viral RNA upon 1‐minute contact with the powders by means of RT‐PCR experiments on the virions N‐gene sequence (Figure 1B). Unlike the case of powder‐unexposed control supernatant (sham sample), the viral RNA underwent nearly complete fragmentation when exposed to Cu, and was significantly damaged after both AlN and Si3N4 contact. Viral RNA on pelleted powders, after 1‐minute exposure, was not detectable for any of the three powders (Figure 1B). Experiments repeated at 10‐minute exposure revealed substantial RNA cleavage for all powders tested (see Supporting Information).
FIGURE 1.

(A) TCID50/50 μL and % reduction plots by TCID50 assay (based on the Reed‐Muench method). (B) RT‐PCR tests to evaluate viral RNA using two sets of N gene primers; a comparison is given using evaluations of supernatants and powders with viral RNA from virions simply suspended in water. (C) Fluorescence micrographs inoculated VeroE6/TMPRSS2 cells after staining: red, green, and blue stains visualize viral protein, F‐actin, and cell nuclei, respectively. (D) Quantification of fluorescence microscopy data given as % infected cells on total cells, namely, the percent fraction of red‐stained cells with respect to the total number of blue‐stained nuclei, and the percent fraction of viable cells on total cells, namely, the percent fraction of green‐stained cells with respect to the total number of blue‐stained nuclei. Labels in inset specify statistics (unpaired two‐tailed Student's test with n = 3)
Figure 1C shows immunofluorescence imaging results on inoculated cells. The envelope antibody of the anti‐SARS coronavirus stained red; viable cell F‐actin (phalloidin‐stained) green; and cell nuclei (DAPI‐stained) blue. Micrographs showing fluorescence in Figure 1C compare the sham (negative) control VeroE6/TMPRSS2 cell population with populations inoculated with supernatant virions exposed to Si3N4, AlN, and Cu (see labels). The synthesis of viral protein, visualized by red‐fluorescent signals, imaged the sham sample cells extensively infected by the virus. As expected, as‐cultured VeroE6/TMPRSS2 cells unexposed to virions (mock sample) showed no red staining. A striking result was that cells inoculated with supernatant treated with Si3N4 and, to a lesser extent, with AlN, were viable and showed a low fraction of infected cells. On the other hand, cells infected with Cu‐treated viral supernatant were essentially dead (see complete lack of F‐actin), clearly indicating that it was free copper ions in the cells having toxic effects, and not viral infection, that caused cell death. 7 We confirmed this using in situ Raman spectroscopy (see Supporting Information). In a quantitative plot of fluorescence microscopy results (Figure 1D), ∼35% fraction of cells in the sham sample (negative control) were infected. Comparatively, cells inoculated with Si3N4 supernatants showed only 2% infection and with AlN supernatants showed 8% infection (see Supporting Information for experimental procedures).
Our work revealed two pivotal aspects of Si3N4 surface chemistry that likely play fundamental roles in inactivating SARS‐CoV‐2: (a) protonation of the amino groups creates Si3N4 surface sites Si–NH3 + that resemble N‐terminals of lysine, C–NH3 +, the cell side viral receptor; and, (b) hydrolytically eluted ammonia from the Si3N4 surface as a strong virucidal compound. Figure 2 (center) draws the interaction between virus and bioceramic surface in aqueous environment. At pH 7.4, positively charged viral envelope/membrane proteins are strongly attracted to the Si3N4 surface (see Supporting Information). The left panel depicts similarity between protonated amine and the lysine N‐terminal. As is the case with hepatitis B and influenza A, 5 , 8 an extremely effective “competitive binding” effect on SARS‐CoV‐2 occurs. Once in contact with the virus, eluted ammonia gas penetrates the virions and cuts through the RNA backbone 9 (see Figure 2, right panel). The combination of RT‐PCR results and fluorescence microscopy suggest that SARS‐CoV‐2 inactivation takes place through a sequence of events: virions are first electrically trapped, locked by “competitive binding,” and then killed by “ammonia poisoning.” Such a scenario could be referred to as “catch and kill.”
FIGURE 2.
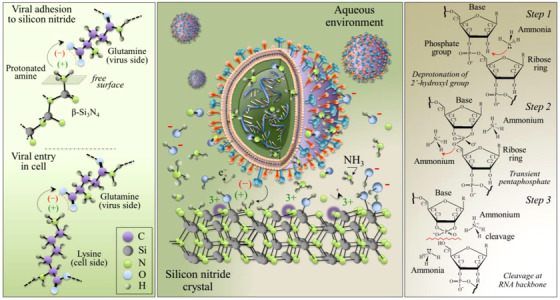
The “catch and kill” mechanism. Central panel: Draft of the electrochemical interaction between Si3N4 surface and SARS‐CoV‐2 virions (envelope and membrane proteins are electrostatically attracted at the negatively charged Si3N4 surface while protonated amines, which resemble cell lysine N‐terminal receptors, link with the spike protein and lock the virions; once the virion is “caught” and locked on the Si3N4 surface, eluted NH3 gas freely penetrates envelope proteins and “kills” it). Left panel: Draft of electrochemical “binding competitive” interactions between protonated amine groups on the surface of Si3N4 and lysine N‐terminals in cells. Right panel: RNA cleavage by ammonia species occurs in three successive steps including the deprotonation of backbone 2′‐hydroxyls, the formation of a transient pentaphosphate group, and the final RNA cleavage by alkaline transesterification
Results confirm SARS‐CoV‐2 inactivation was almost instantaneous upon contact with Cu, AlN, and Si3N4, but only the latter compound proved completely safe to host cells. The bioceramic, Si3N4, is thus a primary candidate to replace toxic and allergenic compounds in long‐term environmental sanitation. 10 The use of micron‐sized Si3N4 particles in disinfectant sprays or their direct embedment in personal protective equipment fabrics (facemasks, surgical drapes, and other garments) in hospitals could limit viral transmission for both health workers and patients. As neither anion‐ nor cation‐side surface chemistry of Si3N4 will affect human health, even in the long term, this bioceramic has potential as an invaluable tool in fighting the SARS‐CoV‐2 pandemic.
Supporting information
Supporting information
Contributor Information
Giuseppe Pezzotti, Email: pezzotti@kit.ac.jp.
Osam Mazda, Email: mazda@koto.kpu-m.ac.jp.
REFERENCES
- 1. Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Warnes SL, Little ZR, Keevil CW. Human coronavirus 229E remains infectious on common touch surface materials. MBio. 2015;6:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chin A, Chu J, Perera M, et al. Stability of SARS‐CoV‐2 in different environmental conditions. medRxivorg. 2020;5247 10.1101/2020.03.15.20036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pezzotti G, Nitride S. A bioceramic with a gift. ACS Appl Mater Interfaces. 2019;11:26619‐26636. [DOI] [PubMed] [Google Scholar]
- 5. Pezzotti G, et al. Sci Rep. 2020. under review. [Google Scholar]
- 6. Grass G, Rensing C, Solioz M. Metallic copper as an antimicrobial surface. Appl Environ Microbiol. 2011;77:1541‐1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balamurugan K, Schaffner W. Copper homeostasis in eukaryotes: teetering on a tightrope. Biochim Biophys Acta – Mol Cell Res. 2006;1763:737‐746. [DOI] [PubMed] [Google Scholar]
- 8. Ye X, et al. Sci Rep. 2016;6:1‐11.28442746 [Google Scholar]
- 9. Decrey L, Kazama S, Udert KM, Kohn T. Ammonia as an in situ sanitizer: inactivation kinetics and mechanisms of the ssRNA virus MS2 by NH3 . Environ Sci Technol. 2015;49:1060‐1067. [DOI] [PubMed] [Google Scholar]
- 10. Scholar PG. Eur J Pharm Med Res. 2018;5:232‐237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


