Abstract
Recently, it was reported that tetra-peptides cyclized via lactam bond between the amino terminus and a glutamic residue in position 4 (termed here N-lock) can nucleate helix formation in longer peptides. We applied such strategy to derive N-locked covalent BH3 peptides that were designed to selectively target the anti-apoptotic protein Bfl-1. The resulting agents were soluble in aqueous buffer and displayed a remarkable (low nanomolar) affinity for Bfl-1 and cellular activity. The crystal structure of the complex between such N-locked covalent peptide and Bfl-1 provided insights on the geometry of the N-locking strategy and of the covalent bond between the agent and Bfl-1.
Keywords: N-locked peptide, stapled alpha helix, protein-protein interactions, PPIs, Bcl-2 family proteins, Bfl-1, covalent inhibitors
1. Introduction
Protein-protein interactions (PPIs) represent in principle a large class of viable therapeutic targets. Nonetheless, to date only one PPI drug is currently approved for clinical use is Venetoclax (ABT199) (Souers et al., 2013). Hence, recent years have witnessed increased efforts in devising novel strategies to design potent and selective PPI antagonists. These included the use of short linear peptides and their subsequent optimizations using structure-based approaches, and recent examples suggest that the derivation and optimization of short linear peptides into potent, cell permeable, pharmacologically active agents is possible (Baggio et al., 2018; Baggio, Udompholkul, Barile, & Pellecchia, 2017). Nonetheless, improving potency, selectivity and overall drug likeness (i.e. cell permeability) of these agents remain a major challenge. More recently, introduction of covalent warheads on peptide or peptide mimetics to target Cys (Baggio et al., 2018; Barile et al., 2017; de Araujo, Lim, Good, Skerlj, & Fairlie, 2017; Harvey et al., 2018; Huhn, Guerra, Harvey, Bird, & Walensky, 2016; Stebbins et al., 2013), and more recently also Lys and Tyr residues (Baggio et al., 2018), are emerging strategies to increase affinity and selectivity of these agents. A large number of therapeutically viable PPIs are mediated by an alpha helix (Bullock, Jochim, & Arora, 2011; Sawyer, Watkins, & Arora, 2017), suggesting that helical peptides could in principle also be used as starting point for the design of new therapeutic agents. However, isolated alpha helical peptides in solution tend to be disordered, with only a small population forming the bioactive alpha helical conformation. Long disordered peptides are susceptible to degradation and are not cell permeable, thus severely limiting their utility as pharmacological tools or therapeutics. During the past two decades, general strategies to stabilize alpha-helices have been reported over the years that are based on formation of cyclic peptides that include the synthesis of specific covalent bonds between side-chains of natural or un-natural amino acids spaced by 3, 4 or 7 residues (Bird, Crannell, & Walensky, 2011; Jayatunga, Thompson, & Hamilton, 2014; Lau, de Andrade, Wu, & Spring, 2015; Walensky & Bird, 2014), or by using hydrogen bond mimics (Cabezas & Satterthwait, 1999; Wang, Liao, & Arora, 2005). These strategies allowed the stabilization of short alpha helical peptides that in turn (and in principle) render them more cell permeable and resistant to proteases in plasma (Bird, Gavathiotis, LaBelle, Katz, & Walensky, 2014; Cohen et al., 2012; Henchey et al., 2010; LaBelle et al., 2012; Wang et al., 2005). Several studies have been proposed with the hydrocarbon stapled and lactam stabilized helices over the past decade (Bird et al., 2014; Cohen et al., 2012; Findeisen et al., 2017; Huhn et al., 2016; LaBelle et al., 2012; Walensky & Bird, 2014; H. Wu et al., 2018). These culminated in a clinical candidate from Aileron ALRN-6924, a p53 derived stapled alpha-helical peptide (https://www.cancer.gov/about-cancer/treatment/clinical-trials/intervention/mdm2mdmx-inhibitor-alrn-6924), suggesting that helix stabilized agents can produce potential therapeutic agents. Thus, additional and perhaps complementary helical stabilizing strategies are worthy of further investigations.
In folded proteins, an N-cap is defined as the first residue of the helix in which its NH group forms an hydrogen bond with the side chain of the residue in position i+4. Likewise, the side chain of the N-cap residue can form hydrogen bonding with the amide proton of residue in position 4. Such arrangement is also called a capping box (Fig 1A). Consequently, hydrogen bond acceptors are usually found at the N-cap and at the residue 4 of alpha-helices to stabilize/catalyze its formation. Hence, a simple strategy consisted of introducing a lactam bond in the peptide between the N-terminal backbone amine and the side chain carboxylate of a Glu (or a homoGlu) residue in position 4 of the helix (Fig 1B) (H. N. Hoang et al., 2011; Huy N. Hoang, Wu, Beyer, Hill, & Fairlie, 2017). However, based on NMR J-coupling measurements in solution, this arrangement has been shown to lead to an alpha-turn rather than a canonical alpha-helical turn (H. N. Hoang et al., 2011). It was also shown, however, that this approach, we termed here N-lock, propagates the alpha helical formation when inserted into longer peptides, especially if the Glu residue is replaced by a homoGlu (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). While these studies suggested that N-locked agents could result in stable alpha-helical binding agents, no binding studies on such constrained peptides have been reported thus far. Recently, we and others also proposed to introduce warheads into helical peptides to target Cys (Barile et al., 2017; Stebbins et al., 2013) or Lys residues (Baggio et al., 2018), resulting in very potent and selective agents. Hence, as an application, we derived N-locked covalent BH3 peptides that can target the anti-apoptotic protein Bfl-1. We have also obtained the crystal structure of the complex between such N-locked covalent peptide and Bfl-1, which provided insights on the geometry of the N-locking strategy and on the covalent bond between the agents. These studies further confirm previous finding on the use of this simple N-locking strategy as possibly a valid alternative or perhaps complementary method to hydrocarbon and/or lactam based alpha helix stapling, especially when combined with covalent targeting.
Fig 1. The capping box and the N-locking strategy.

A) The capping box: Hydrogen bonds in with capping box are highlighted in green, while other backbone to backbone hydrogen bonds are in yellow. B) Predicted geometry of a N-locked peptide, assuming an idealized alpha helical conformation, in which the side chain of the Glu in position 4 is cyclized via a lactam bind with the N-cap residue. An additional hydrogen bond could be formed between the lactam oxygen and the backbone of Glu 4 (green). C) Superposition of the 2D [1H, 1H] NOESY (black and cyan) and 2D [1H, 1H] TOCSY (red) spectra for the N-locked peptide [NH-ARAE]-CONH2 collected at 700 MHz 1H frequency, at 15 °C with a mixing time of 500 ms and 64 scans on a 500 μM sample in water. Selected resonance assignments and observed NOEs are labeled (sequential HN-HN NOEs are observed but not labeled). On the right, the idealized helical structure of the N-locked agent is reported, which is however not entirely compatible with the observed 3JαHN of ~ 10 Hz observed for residue in position 2 (see Fig 2).
2. Materials and Methods
2.1. Peptide synthesis
All reagents and anhydrous solvents were obtained from commercial sources, including Fmoc (fluorenylmethyloxycarbonyl) protected amino acids, allyl (allyl ester) protected Glu, and ivDde (1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3-methylbutyl) protected L-Dap (L-2,3-diaminopropionic acid), and resins for solid phase synthesis. Some of the reported agents were synthesized by Innopep, while others were synthesized in house by standard microwave-assisted Fmoc-peptide synthesis protocols on a rink amide resin using a Liberty Blue Peptide Synthesizer (CEM). The synthesis of N-locked peptides was accomplished using standard solid phase protocols with a rink-amide resin, using an allyl-protected glutamic acid in position 4. Selective allyl removal from the glutamic acid in position 4 was accomplished with a solution of chloroform (35 mL x g of resin), acetic acid (0.5 mL x g of resin), N-methylmorpholine (2 mL x g of resin), and 1 equivalent of Pd(PPh3)4, for 2 h. Subsequently, the resin was subjected to washing steps including three washes with DMF/DCM, two washes with 0.5% DIPEA (N,N-Diisopropylethylamine) in DMF, two additional washes with DMF, three additional washes with a solution 0.5% w/v of sodium dimethyl-dithio-carbamide in DMF, and a final wash with DMF/DCM. Fmoc removal was subsequently accomplished with 20% piperidine in DMF (2 times, each for 30 min), followed by washing with DMF/DCM. For the N-locking cyclization we used 1.1 equivalent of PyBOP (benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate), 2.2 equivalents of DIPEA in 1 mL DMF (overnight), and final wash with DMF/DCM. Incorporation of the L-Dap-2chloroacetamide was accomplished introducing an ivDde protected L-Dap at the N-terminus of the peptide. After cyclization, selective removal of ivDde protective group from L-Dap was accomplished with 4% NH2NH2 that allowed subsequent coupling with ClCH2COCl as we recently reported.(Barile et al., 2017; Stebbins et al., 2013) Peptides were purified via HPLC (>95%) and further characterized using HRMS and NMR spectroscopy.
2.2. Circular dichroism and NMR spectroscopy.
Circular dichroism (CD) measurement were conducted with a JASCO spectrometer with aqueous solutions containing 250 μM of each peptide and quartz cell with 1 mm path and at room temperature. The parameters used were: scanning speed of 100 nm/min, response of 2 s, bandwidth 1 nm, 4 accumulations. The spectra were recorded in a continuous mode from 190 to 260 nm. NMR spectra were acquired on a 700 MHz Bruker Avance spectrometer equipped with a TCI cryoprobe. All NMR data were processed and analyzed using TOPSPIN2.1 (Bruker Biospin, Billerica, MA, USA). 1D 1H NMR spectra were collected at 500 μM peptides in water containing 10% D2O + 1% d6-DMSO, at T = 15 °C. Resonance assignment were obtained by a combination of 2D [1H, 1H] TOCSY and 2D [1H, 1H] NOESY experiments collected at various mixing times, with peptides at 500 μM in the same aqueous conditions at T = 15° C.
2.3. DELFIA (Dissociation Enhanced Lanthanide Fluorescence Immuno-Assay)
The assay was performed as we described recently (Barile et al., 2017). Briefly, in one hundred microliters of a 600 ng/ml solution of biotin-BID (Biotin- aminohexanoic acid -EDIIRNIARHLAQVGDSMDR-NH2) were added to each well of 96-well streptavidin-coated plates (Perkin-Elmer). After 2 hours incubation and three washing steps, 11 μl of a solution of 6His-hBcl-xL (8.5 nM final concentration in the assay well) or 6His-hBfl-1-ΔTm (15 nM final concentration in the assay well) and a serial dilution of the test peptide(s) were added to the assay plate (in triplicates). Subsequently, 89 μl of 1.56 nM solution of Eu-N1-labeled anti-6xHis Antibody (Perkin-Elmer) was added to capture the His-tagged protein (2 h followed by one washing step). Finally, 200 μl of the enhancement solution (Perkin-Elmer) was added and fluorescence measured (excitation wavelength: 340 nm; emission wavelength: 615 nm). Each well also contained a final DMSO concentration equal to 1%. Fluorescence readings were normalized to control wells (receiving only DMSO to a final concentration of 1%) and reported as percent inhibition (IC50 values were calculated by Prism5, GraphPad). The reported standard errors were calculated from replicates. Specifically, two independent measurements, each in duplicates, were conducted, except for 138C5 for which only one experiment was measured (in duplicate).
2.4. Protein expression, X-ray crystallography, and molecular modeling
6His-hBfl-1-ΔTm and 6His-hBcl-xL-ΔTm were expressed and purified as we described recently (Barile et al., 2017) and used directly for the DELFIA assays, while untagged hBfl-1-ΔTm was used for crystallization studies. To obtain the co-crystals of 138C7 and hBfl-1-ΔTm, the peptide was first dissolved in DMSO to 50 mM final concentration. Subsequently, 1.2 μl of the 50 mM 138C7 peptide solution was added to 100 μl of 9.9 mg/ml hBfl-1-ΔTm to make a 1:1 complex of protein and peptide. The crystal was grown at 20 °C using a MRC 3-well plate in a 100 + 100 nl drop with reservoir as follows: 0.1 M sodium acetate pH 4.6, 16 % (w/v) PEG smear high. A few days after crystallization setup, rod-shaped crystals grew with dimensions up to approximately 0.03 × 0.03 × 0.2 μm. 1.5 μl cryoprotectant containing: 18 % w/v PEG smear high, 0.1 M sodium acetate pH 4.6, 25 % v/v glycerol, 2 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) and 150 mM sodium chloride was added to the drop, and the crystal was cryo-cooled in liquid N2. Data were collected at 100K at station ID29 of the ESRF, Grenoble, France (λ = 0.9753 Å) equipped with a Pilatus 6M detector. Data were processed using XDS (Kabsch, 2010), Aimless (Evans & Murshudov, 2013), and Autoproc (Vonrhein et al., 2011) programs. The structure was determined by Phaser (McCoy et al., 2007) using the PDB entry 2VM6 as the molecular replacement mode. Two disulfide-linked Bfl-1 molecules (chain A and chain B) are present in the asymmetric unit. Initial refinement to 3.0 Å revealed clear presence of peptide bound covalently to Cys55 in both molecules. However, the density was not of high enough quality to unambiguously trace the peptide. Therefore, to improve the electron density maps, the data was reprocessed using Autoproc (Vonrhein et al., 2011) and the weak data was removed by application of an anisotropic resolution cutoff using STARANISO (Cambridge, United Kingdom, Global Phasing Ltd.). In summary, weak reflections were removed in certain directions and the data cut-off was made using the strength of average reflections in an area, rather than a strict spherical resolution cut-off. This means that the final refinement was made with data between 30 and 2.57 Å, but the data were cut irregularly between 2.57, 3.33 and 3.33 Å in different directions. The structure was refined in Refmac5 (Murshudov et al., 2011) and Buster (version 2.11.7 Cambridge, United Kingdom: Global Phasing Ltd.). Throughout refinement non-crystallographic symmetry (NCS) restraints were applied between the two copies of the molecule in the asymmetric unit, to make convergence of the refinement easier, and final refinement was performed in Refmac5. Model building was completed in Coot, (Emsley, Lohkamp, Scott, & Cowtan, 2010) and the bound 138C7 peptides were built with modified amino acids in position 1, 3 and 4, with restraints for these amino acids and linkers were created using Jligand (Lebedev et al., 2012). Molecular modeling figures were prepared using Chimera (UCSF).
2.5. Cell apoptosis assay
SKMEL28 is a melanoma cell line with high Bfl-1 expression (Placzek et al., 2010). SKMEL28 cells were grown in EMEM media (cell line and media were purchased from ATCC) supplemented with 10% fetal bovine serum and 1% Pen/Strep (supplements were purchased from GIBCO, ThermoFisher). At the day of the experiment, cells were trypsinized and plated in 96-well plate at cell density 2,500 cells per well. Cells were incubated o/n to attached, and the following day media was aspirated and replaced with serum-free media containing 1% Pen/Strep and 1x IncuCyte® Cytotox Green Reagent (Sartorious) for counting dead cells. Compounds were tested at the indicated concentrations in triplicates, and the plate was imaged using IncuCyte S3 (Sartorious). IncuCyte software 2019A was used to measure Total Green Object Integrated Intensity. GraphPad Prism version 7 was used to plot and measure significance between groups.
3. Results
3.1. Design, synthesis, and characterization of constrained (N-locked) alpha helical peptides
Water soluble alpha helical peptides can be designed by properly placing side-chain stabilizing interactions including hydrophobic and salt bridges on the helix surface. Synthetic approaches however are more effective and these can include hydrogen bond surrogates, (Bullock et al., 2011; Cabezas & Satterthwait, 1999; Henchey et al., 2010; Patgiri, Jochim, & Arora, 2008; Sawyer et al., 2017; Wang et al., 2005) or tethering approaches that include hydrocarbon stapling, side chain lactams, and other approaches (Bird et al., 2011; Bird et al., 2014; Cohen et al., 2012; Jayatunga et al., 2014; Lau et al., 2015; Nguyen, Luong, & Kim, 2016; Walensky & Bird, 2014; Walensky et al., 2004; Wu, Hoang, Liu, & Fairlie, 2018). In natural alpha helices, the backbone is further stabilized by intramolecular hydrogen bonding between the carboxyl oxygen of a given residue i and the amide hydrogen of the residue located in the sequence at position i+4 (Fig 1A). This arrangement however leaves the amide hydrogen of the first 4 amino acids non-hydrogen bonded via the backbone (Fig 1A). Rather, the helix can be stabilized by hydrogen bonding between such amides and the side chain hydrogen bond acceptor atoms of capping residues (Asn, Ser, Thr, Asp, or Glu) at positions 1 and 4 (Fig 1A), and such arrangement, termed capping box is often found in native protein structures (Harper & Rose, 1993; Jimenez, Munoz, Rico, & Serrano, 1994; Presta & Rose, 1988). Based on this simple observation, it was reported that a straightforward way to stabilize a linear peptide into a helix could be via a direct lactam bond between the N-terminus amine and the side chain of a glutamic acid placed in position 4 (Fig 1B) (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). While simple molecular mechanics would predict that such arrangement is compatible to an ideal alpha helical peptide geometry (Fig 1B), more detailed studies using molecular dynamics and experimental CD amd NMR data in solution suggested that the peptide is slightly distorted and assumes an alpha-turn conformation (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). Most notably, this is supported by a 3JαHN coupling constant that, in particular, in position 2 assumes values close to 10 Hz, hence incompatible with an idealized alpha-helical geometry. Synthetically, the cyclization of model peptides was accomplished using orthogonal deprotection of allyl-protected Glu on the side chain in position 4. Because the allyl protecting group is stable in 20% piperidine but is cleaved by Pd(PPh3)4 in chloroform, it allows the carboxylate of the allyl-protected Glu in position 4 to be selectively unmasked in the solid phase without affecting the side-chain protecting groups of other residues, facilitating subsequent site-specific modifications on the resin. Hence, after selective deprotection of the glutamic acid in position 4 and of the N-terminal amine, subsequent reaction of the free amine provided the desired N-locked peptides. As proof of concept, we prepared a model N-locked peptide of sequence [NH-ARAE]-CONH2, where the underlined residues indicated lactam bond between the N-terminus and the side chain of the glutamic acid in position 4. Remarkably, the 2D [1H, 1H] NOESY spectrum of such peptide in water displayed a pattern of sequential HN-HN and Hα-HN NOEs typical of a helical conformation (Fig 1C). While it is difficult to unambiguously discriminate between an alpha helix, an alpha-turn, or a 3–10 helix, based solely on these NOEs, additional NOEs for example between the −CONH1 proton and the β-hydrogens of Glu 4 would favor of the alpha conformation as the predicted geometry (Fig 1C). Noteworthy is also the observation of strong NOEs between the amide proton of Ala 1 and the Hγ1,2 of Glu 4, as expected from the intended cyclization (Fig 1C). However, as mentioned above, 3JαHN coupling measurements are more in agreement with an alpha-turn as previously reported (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017) (Fig 1D), with a characteristic large value (~ 10 Hz) for the residue in position 2 of the N-lock. Not surprisingly, no NOEs were observed with the corresponding linear tetra-peptide (not shown). However, it was recently reported that the cyclic tetrapeptide [NH-ARAhE]-CONH2, while also assuming an alpha-turn in solution, it can nucleate the formation of a more idealized alpha-helical conformation when placed in longer peptides (Huy N. Hoang et al., 2017). Hence, two additional longer N-locked peptides ([NH-AIAE]ALR-CONH2 and [NH-AIAE]ALRRIGDAF-CONH2) were synthesized and characterized using biophysical methods, side by side with their corresponding linear counterparts. Remarkably, and in agreement with the 2D NOESY experiments (Fig 1C), we found that the CD spectrum of the N-locked peptide [NH-ARAE]-CONH2 in water was typical of a helical structure, while, and again not surprisingly, the CD of the linear corresponding agents suggested a random coil conformation (Fig 2A). In agreement, amide proton chemical shifts in the 1D 1H NMR spectrum of the cyclic peptide were more spread, perhaps due to the helical conformation and hydrogen bonding, compared to the linear peptides (Fig 2A–C). However, J-coupling measurements still supported the formation of an alpha turn with a 3JαHN ~ 10 Hz for the residue in position 2 for all agents (Fig. 2) which make it difficult to ascertain the exact geometry of the agents in solution. While a pattern of strong sequential HN-HN can be clearly observed in the 2D [1H,1H] NOESY spectrum of the N-locked agent (Fig 2C), typical of a stable helical conformation (Fig 2C), the J coupling values in position 2 would suggest that at the least for first turn of the helix is not adopting the canonical alpha helix geometry. Because it is difficult to evaluate whether such stabilization (and concomitant helical distortion) can result useful in binding peptides, we decided to carry out more detailed binding and structural studies as reported below.
Fig 2. N-locked model peptides.

A) Comparison between the CD and NMR spectra of the linear Ac-ARAE-CONH2 (blue) with the corresponding N-locked peptide [NH-ARAE]-CONH2 (red); B) Comparison between the CD and NMR spectra of the linear Ac-AIAEALR-CONH2 (blue) with the corresponding N-locked peptide [NH-AIAE]ALR-CONH2 (red); C) Comparison between the CD and NMR spectra of the linear Ac-AIEALRRIGDAF-CONH2 (blue) with the corresponding N-locked peptide [NH-AIAE]ALRRIGDAF-CONH2 (red). Panel C) also displays the amide region of a 2D [1H, 1H] NOESY spectrum collected at 700 MHz 1H frequency, at 15 °C with a mixing time of 200 ms and 80 scans on a 500 μM sample of the N-locked peptide [NH-AIAE]ALRRIGDAF-CONH2. Backbone resonance assignments are indicated. Sequential HN-HN NOEs are clearly observed. In the 1D 1H NMR spectra for each agent the 3JαHN coupling constant for residue in position 2 is also reported.
3.2. Design, synthesis and structural characterization of N-locked covalent BIM derived Bfl-1 targeting peptides.
To explore the potential of the N-locking strategy in the design of PPIs that are mediated by an alpha helix, we further prepared a number of longer peptides derived from the pro-apoptotic protein BIM and evaluated their binding properties to anti-apoptotic Bcl-2 family proteins, using a biochemical displacement assay based on the DELFIA (Dissociation Enhanced Lanthanide Fluorescence Immuno-Assay) platform (Rega, Reed, & Pellecchia, 2007). Briefly, a biotinylated Bid BH3 peptide was adsorbed on streptavidin coated plates while histidine tagged recombinant proteins (anti-apoptotic Bcl-2 family proteins hBfl-1 and hBcl-xL were used in this study) were subsequently conjugated with a Eu-tagged anti-His antibody (Perkin Elmer, see Methods). Exposing the complex to test peptides resulted in the displacement of the Eu-tagged-anti-His antibody/Bcl-2-protein complex from the BH3-peptide, and after washing steps the displacement was detected by a decrease in fluorescence signal (Rega et al., 2007).
We and others have recently reported on BIM derived alpha helical peptides that target covalently Cys55 in Bfl-1 (Barile et al., 2017),(Huhn et al., 2016), a member of the Bcl-2 family protein for which there are currently no viable pharmacological inhibitors. Targeting Cys55 can also be used to engineer selectivity on the resulting agents, in the attempt of obtaining peptides that do not target Bcl-xL. Hence, to further investigate the suitability of the N-locking strategy to deliver potent, Bfl-1 selective, covalent binding peptides, we designed a series of N-locked BIM covalent peptides that are predicted to target Cys55 hBfl-1 (Table 1).
Table 1.
Bfl-1 targeting agents and biochemical assays data.
| Compd ID | Sequences | DELFIA IC50 (nM)* | |
|---|---|---|---|
| hBfl-1 | hBcl-xL | ||
| hBIM-BH3 | Ac-IWIAQELRRIGDEFNAYYARR-CONH2 | 18 ± 2 | 5.0 ± 1 |
| 135P1 | Ac-λIAAALRRIGDAFNAAY-CONH2 | 3.0 ± 0.1 | 280 ± 180 |
| 138C5 | [NH-λIAE]ALRRIGDAFNAAY-CONH2 | 2.9 ± 0.1 | 1750 ± 500 |
| 138C7 | [NH-λI-Aib-E]ALRRIGDAFNAAY-CONH2 | 1.1 ± 0.1 | 1500 ± 200 |
| 138C8 | [NH-λI-Aib-E]-Aib-LRRIGD-Aib-FNA-Aib-Y-CONH2 | 3.1 ± 0.1 | >10 μM |
λ = L-Dap-2-chloroacetamide, Aib = 2-aminobutyric acid,
2 h incubation, see methods.
To introduce a proper electrophile, the N-terminal residue was replaced with an L-Dap-(2-chloroacetamide) as we recently reported (Barile et al., 2017). As expected, the linear BIM BH3 peptide was very potent for both hBfl-1 and hBcl-xL, with IC50 values of 18 nM and 5 nM, respectively (Table 1). Likewise, for hBfl-1, the shorter covalent peptides resulted even more potent than the BIM BH3 peptide, while the same agents resulted dramatically less active for Bcl-xL (Table 1). Interestingly, comparison of the displacement ability of the linear covalent agent 135P1 with its N-locked corresponding agent 138C5 for both proteins, suggested that not only the N-locked sequence preserved the affinity of the linear agent for hBfl-1, but resulted more selective (Fig 3A). Introduction of further aminobutyric acid residues in lieu of Ala residues in the N-locked peptide again preserved the affinity for Bfl-1, while progressively rendered the resulting agents less active for hBcl-xL (Table 1). These observations are in agreement with the anticipated covalent bond formation between the peptides and hBfl-1, but not with hBcl-xl. To verify that the remarkable affinity of the compounds was due to a specific covalent interaction with hBfl-1, we used SDS gel electrophoresis. Given the relative large molecular weight of the helical peptides, a stable covalent adduct with hBfl-1 would manifest itself in the SDS gel electrophoresis in a significant band shift. In agreement, significant band shifts can be observed with hBfl-1, when exposed to each of the 3 agents (Fig 3B). On the contrary, no shift was observed when hBcl-xL was exposed to the same agents (Fig 3B). Also, similar to the previously mentioned shorter peptide, the increased amide proton chemical shift dispersion (Fig 3C) in the N-locked peptide compared to the linear one, confirmed the ability of the N-lock strategy to propagate the helix throughout the length of the peptide. Likewise, a pattern of sequential HN-HN NOEs can be clearly observed in the 2D [1H,1H] NOESY spectrum of the N-locked agent (Fig 3D), typical of a stable helical conformation. Strong NOEs between the amide proton of residue 1 (Dap) and the Hγ1,2 of Glu 4 are also observed that further confirm the intended cyclization of the peptide (Fig 3D).
Fig 3. Comparative characterizations of N-locked covalent peptides targeting Bfl-1.

A) Chemical structure of 138C5 (Table 1). Biochemical dose response curves representing the ability of each tested agent to displace the binding of a reference BH3 peptide from Bfl-1 or Bcl-xL (2 h incubation, see methods) are also reported. The curves represent the dose response curves obtained with the BIM peptide (black), with a linear covalent Bfl-1 antagonist 135P1 (blue), and with its N-locked corresponding agent, 138C5 (red) (Table 1 and structure shown above). Data were collected in duplicates and experiments were repeated twice, all other experimental details for the DELFIA assay are reported in the methods section. B) SDS gel electrophoresis were performed incubating either hBfl-1-ΔTM or hBcl-xL-ΔTM (10 μM) with the indicated covalent agents (each at 100 μM concentration). Gel shifts can be clearly appreciated when the Bfl-1 is exposed to the covalent agents, while no shifts are observed when the compounds are exposed to Bcl-xL. No DTT was present in the protein buffer. C) Superposition of the 1D 1H NMR spectra of the linear covalent agent 135P1 (blue) and its corresponding N-locking equivalent, 138C5 (red) (Table 1). The spectra were recorded at 700 MHz 1H frequency with equimolar samples of each peptide (500 μM) at 15 °C in water. The 3JαHN coupling constant for residue Ile 2 of the N-locked agent is also reported. D) 2D [1H,1H] NOESY and CD spectra of the 138C5 recorded with a 500 μM peptide sample in water at 15 °C. A number sequential NOEs are clearly observed. NOEs between the amide proton of residue 1 (Dap) and the Hγ1,2 of Glu 4 are highlighted (confirming the cyclic structure) as well as the NOEs between the amide proton of Ile2 and Hβ1,2 of Dap1.
However, neither NMR studies nor CD (Fig 3D) can easily discriminate between the alpha or the 3–10 helical or the presence of the N-terminal alpha-turn. Indeed, the 3JαHN coupling constant for residue Ile 2 still supports the formation of an alpha turn (3JαHN ~ 10 Hz) for the agent (Fig. 3C) which makes it difficult to ascertain the exact geometry of the peptide in solution. Hence, to further elucidate the binding conformation of the N-locked covalent agent to Bfl-1, we also determined the crystal structure of 138C7 in complex with hBfl-1-ΔTm. A summary of structural parameters is reported in Table 2. The general structure of the complex is similar to previously determined Bfl-1 in complex with BH3 peptides (Fig 4A), with a monomer of hBfl-1-ΔTm superimposing with of that from PDB code 2VM6 at 0.8 Å. The geometry of the N-locked and the covalent bond between the 138C7 peptide and Cys55 of hBfl-1-ΔTm can be delineated by a contiguous electron density (Fig 4B), in agreement with all other data that strongly supported covalent adduct formation between these two molecules. The general peptide-hBfl-1 interactions are well maintained in the crystal structure. Here, the hBfl-1-ΔTm peptide‐binding pocket is lined with hydrophobic regions that are well conserved in Bcl‐2 proteins, allowing for interactions with the conserved residues on the amphipathic helix of BH3 proteins, such as BIM (Table 1). The 138C7 N-locked covalent peptide maintained these hydrophobic interactions, which includes Ile2, Leu6, Ile9, Phe13, and Tyr17 of 138C7, in addition to a charge interaction between 138C7 Asp11 and Arg88 of hBfl-1. Moreover, the geometry of the N-locked moiety closely resembles that of the ideal helical peptide (Fig 1C), with the lactam carbonyl oxygen forming a hydrogen bond with the backbone amide of residue Glu in position 4 (Fig 4C). Also, a potential hydrogen bond can be observed between the carbonyl oxygen on the side chain of the Dap residue 1 and the backbone amide proton of residue 2. Hence, in this arrangement, three out of the first four amide protons of the helix are involved in intramolecular hydrogen bonding (Fig 4C).
Table 2. Crystallization summary table.
Figures in parentheses are for the highest resolution shell. Other relevant quality indicators can be easily extracted from the PDB file header.
| Refinement of data done in two steps | Data processed in XDS, scaled in Aimless and used for initial refinement | Data included in the final refinement after anisotropic removal of weak data (STARANISO) |
|---|---|---|
| Resolution (Å) | 49.3 – 3.00 (3.18 –3.00) | 61.7 – 2.57 (2.94 – 2.57) |
| Wavelength (Å) | 0.97625 | 0.97625 |
| Space group | P31 2 1 | P31 2 1 |
| Unit cell (Å) | a=b=113.8,c=79.1 | a=b=113.75, c=79.2 |
| Completeness (%) | 99.6 (99.8) | 55.5 (8.4) |
| Redundancy | 3.6 (3.8) | 3.6 (3.2) |
| No. of observations / unique reflections | 43 475 / 12 088 | 38 750 / 10 669 |
| <I/σ(I)> | 11.6 (1.0) | 13.3 (1.9) |
| CC(1/2) (%) | 99.8 (68.8) | 99.9 (69.2) |
| Rmerge (I) (%) | 4.6 (132.0) | 4.3 (68.5) |
| Rcryst (F) (%) | 19.9 (27.4) | |
| Rfree (F) (%) | 26.6 (53.2) | |
| No. of non-hydrogen atoms | 2704 | |
| No. of water molecules | 10 | |
| r.m.s. deviations from ideal geometry: Bond lengths (Å) | 0.015 | |
| Bond angles (deg) | 1.9 | |
| Mean B-factor protein chain, A, B (Å2) | 74.0, 110.9 | |
| Mean B-factor peptide C, D (Å2) | 44.0, 78.1 | |
| Mean B-factor (water, Å2) | 71.7 | |
| Ramachandran plot quality# | ||
| Favored regions (%) | 95.7 | |
| Allowed regions (%) | 4.3 | |
| Outliers (%) | 0 | |
Calculated using a local Molprobity server (Chen et al., 2010).
Fig 4. Structural analysis of the N-locked covalent agents 138C7 in complex with hBfl1-ΔTM.

A) Ribbon representation of the crystal structure of hBfl-1-ΔTM (light blue ribbons) in complex with the N-locked covalent agent 138C7 (Table 1; red ribbon, and stick side chains). The side chain of Bfl-1 residue Cys 55 is also displayed. The N-lock and the covalent bind between the N-cap residue and Cys are highlighted in the zoomed insert. B) Blue electron density (2m|Fo|-D|Fc|) map contoured at 1σ level for 138C7 peptide chain C and Cys55 in hBfl-1. C) Close-up view of the geometry of the N-locking moiety, highlighting the observed intramolecular hydrogen bonding.
To preliminarily ascertain that the agents may induce apoptosis in cancer cells, we selected the melanoma SKMEL28 cell line that presents high Bfl-1 expression (Placzek et al., 2010). Hence, SKMEL28 cells (in 10% fetal bovine serum) were incubated with test agents or the potent Bcl-2 antagonist ABT199 (that is inactive against Bfl-1), and apoptosis was monitored using the IncuCyte® Cytotox Green Reagent (Sartorious) (Fig 5). Unlike ABT199, both N-locked agents 138C5 and 138C8 induced significant apoptosis in the low micromolar range (Fig 5).
Fig 5. N-locked covalent Bfl-1 compounds induce apoptosis in SKMEL28 melanoma cell line.
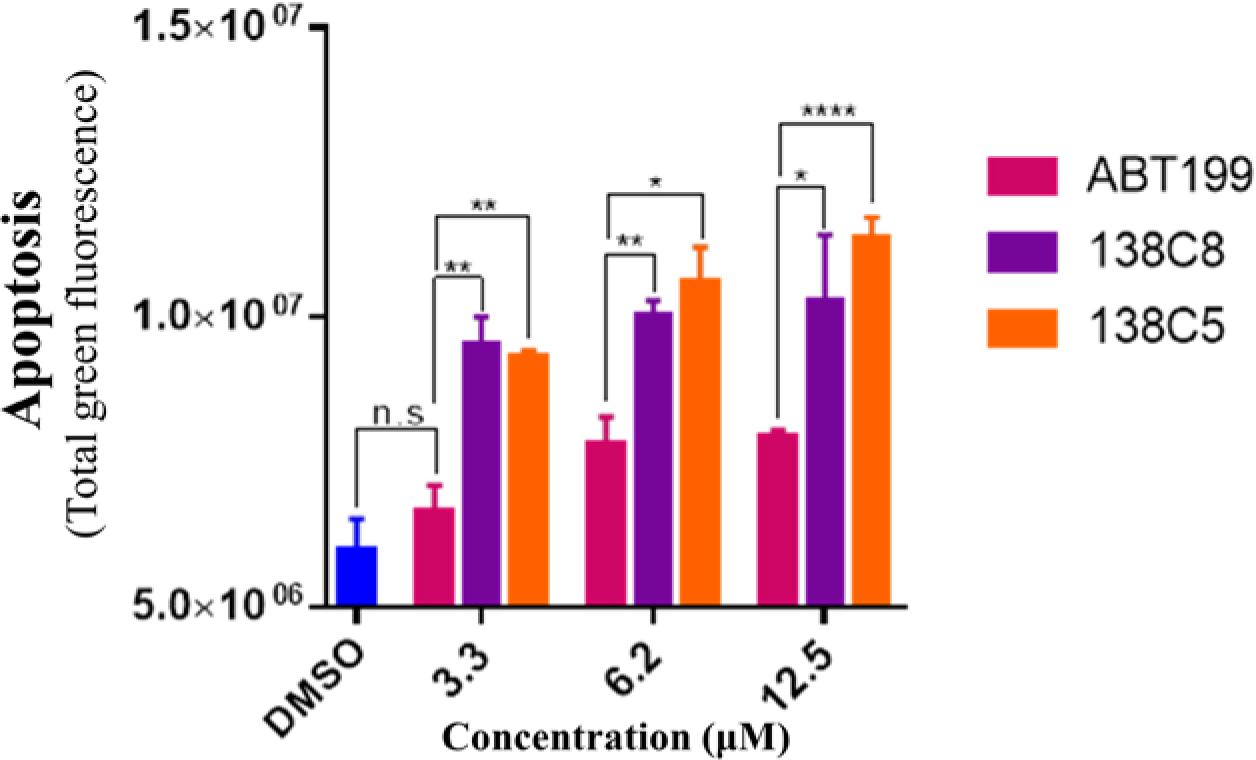
SKMEL28 cells were treated with different compounds at various concentrations. Late apoptotic marker, IncuCyte® Cytotox Green was imaged and measured using IncuCyte S3 live-cell analysis system. The histogram represents data collected at 30 h post-treatment. Experiments were performed in triplicates and error bars indicate SD. ****, P < 0.0001; ***, P = 0.0002; **, P < 0.002; *, P = 0.02; n.s., not significant.
4. Discussion
During the past two decades, applications of rational biophysical approaches combined with fragment- and/or structure-based design strategies have resulted in the design of a variety of novel PPIs antagonists, culminating in 2016 in the approval of Venetoclax (ABT199) (Souers et al., 2013), a Bcl-2 antagonist derived using a combination of NMR-based fragment screening (Bruncko et al., 2007; Liu et al., 2000; Oltersdorf et al., 2005; Oost et al., 2004; Tse et al., 2008) and iterative structure-based optimizations. These studies were aimed at obtaining a potent (single digit nanomolar) Bcl-2 antagonist that was also voided of Bcl-xL activity, deemed responsible for the observed toxicity of earlier, less selective, agents (Souers et al., 2013). Alternative strategies to Bcl-2 family proteins inhibitor design have been proposed, including the use of constrained BIM derived stapled peptides. Several recent studies revealed that both side chain lactam and hydrocarbon stapling can produce stable, cell penetrating peptides, with high binding affinity for the intended anti-apoptotic Bcl-2 targets (Bird et al., 2011; Bird et al., 2014; Cohen et al., 2012; LaBelle et al., 2012). While ultimately successful, each approach has also shown significant limitations, and obtaining effective, soluble, stapled alpha helices often required a systematic scanning of the positioning and the composition of the side-chain staples (Cohen et al., 2012; Okamoto et al., 2013). Hydrocarbon stapling can also result in peptides with limited aqueous solubility (Barile et al., 2017), while others found that introduction of lactam or hydrocarbon stapling severely affected the affinity of the peptides for the intended Bcl-2 targets (Okamoto et al., 2013). Given the potential of alpha helices as pharmacological tools or even therapeutics (Bullock et al., 2011), we sought here to test a simpler strategy to induce alpha helix formation in short peptides. The N-locking strategy used here is based on simple theoretical geometrical considerations and experimental studies that were recently reported (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). In alpha helices in proteins, an N-cap is defined as the first residue of the helix. Due to the geometry of the alpha helix, the amide hydrogen of the N-cap and those of the next 3 residues are not hydrogen bonded to backbone carbonyl oxygen atoms in alpha helices (Fig 1A). Rather, hydrogen bond acceptors are usually found in the side chain of amino acids in positions 1 (at the N-cap) and 4 of alpha-helices that often are found to stabilize the helix via a so-called capping box (Fig 1A). It is believed that capping boxes, perhaps similar to what was observed with hydrogen bond surrogate stabilized helices (Cabezas & Satterthwait, 1999; Wang et al., 2005), can nucleate the folding of alpha helices (Fig 1A). Based on previous observations with N-locked model peptides, our strategy consisted of introducing a simple N-locking cyclisation involving a lactam bond between the N-terminal backbone amine and the side chain carboxylate of a Glu residue placed in position 4 of the helix (Fig 1B). Based on simple molecular mechanics modeling, such arrangement could also bring the carbonyl oxygen of the lactam closer to the amide proton of residue in position 4, hence presumably resembling the stabilization provided by a capping box (Fig 1). However, and as reported recently (Huy N. Hoang et al., 2017), such an arranged introduces a distortion in the helical turn that results in the formation of an alpha-turn rather than a typical alpha helical turn. This manifests experimentally primarily in a large 3JαHN coupling (~ 10 Hz) in the residue in position 2, while values smaller than 5 Hz would be expected in an idealized alpha helical geometry (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). However, a recent work suggested that even if the N-lock in solution can adopt an alpha turn, it can nucleate alpha helical peptides when incorporated into longer sequences (H. N. Hoang et al., 2011; Huy N. Hoang et al., 2017). However, experimental studies on the binding ability of such agents to their targets have not been reported, which is the focus of our work.
We and others recently reported on covalent alpha helical BH3 peptides targeting a unique Cys that is present only on the surface of hBfl-1 and not on other anti-apoptotic Bcl-2 proteins such as hBcl-xL (Barile et al., 2017; D. de Araujo et al., 2018; de Araujo et al., 2017; Huhn et al., 2016; Jenson, Ryan, Grant, Letai, & Keating, 2017). In these previous studies, various hBfl-1 targeting covalent peptides were rendered more stable by a variety of approaches including the introduction of non-natural amino acids (Barile et al., 2017), introduction of hydrocarbon stapling (de Araujo et al., 2017; Huhn et al., 2016; Jenson et al., 2017) or simultaneous introduction of one hydrocarbon staple and one side chain to side chain lactam bond (D. de Araujo et al., 2018). Hence, we applied the N-locking strategy and introduced a Dap-2Cl-acetoamide at the N-terminal residue to verify whether such agents could covalently, and selectively, target hBfl-1 Cys55. Using a biochemical displacement assay we confirmed that these agents were particularly potent (single digit nanomolar) against hBfl-1 compared to hBcl-xL (Table 1), owing to their covalent interaction with Bfl-1 Cys55, as confirmed by SDS gel shift assays (Fig 3A, B). Interestingly, comparing the linear agent 135P1 with its corresponding N-locked equivalent, 138C5, we noticed that while the affinity for hBfl-1 was similar between the two agents, the N-locked peptide was significantly less active against hBcl-xL. This was a pleasant surprise, given that deriving hBfl-1 antagonists that are void of hBcl-xL inhibitory properties is deemed desirable to avoid toxicity in vivo. Further introduction of helix inducing Aib residues (Toniolo et al., 1993) in lieu of Ala residues in 138C5 led to agents 138C7 and 138C8 that were increasingly less potent for hBcl-xl, but retained their ability to effectively displace a reference Bid peptide from hBfl-1 in the low nanomolar range (Table 1). It is possible that, the N-lock introduced conformational distortions in the side chains of residues 1 to 4 of the helix, as suggested by the large 3JαHN coupling constant values of the residues in position 2, which negatively impacted the affinity for hBcl-xL, but not hBfl-1. In agreement with this hypothesis, others have also observed that subtle changes in the peptide sequence and/or the introduction of a lactam staple in BH3 peptides targeting Bcl-xL can result in large losses in binding affinity (Okamoto et al., 2013). This appears to be a rather common feature of constrained helices, in that these agents more often than not tend to lose affinity for the intended target compared to the linear corresponding peptides (Barile et al., 2017; Baxter et al., 2017; Okamoto et al., 2013). While the N-locking strategy seems to suffer from this same nuisance with respect to hBcl-xL, most likely due to the alpha turn at the N-terminal sequence, the data with hBfl-1 suggest the contrary: the N-locked covalent agent was equipotent as its linear equivalent, and even more potent than the linear native and much longer BIM peptide (Fig 3A). Hence, it is most likely that the covalent bond compensates for any losses of binding due to the distortions of the helix, hence making the covalent agent even more selective for the intended target.
To ascertain that the agents are active in cell, a preliminary cell based apoptosis assay against the Bfl-1 expressing melanoma cell line SKMEL28 was conducted. Here, we could observe that ABT199 (potent Bcl-2 antagonist but inactive against Bfl-1 (Barile et al., 2017),) was inactive at the concentrations tested (Fig 5) in agreement with this cell line being dependent on Bfl-1 and not on Bcl-2 for survival. On the contrary, both N-locked agents 138C5 and 138C8 induced significant cell death in the low micromolar range in the same preliminary assay (Fig 5).
Detailed additional cell mechanistic studies, including studies on the passive diffusion of helical constrained peptides have been reported and involve a number of systematic side chain optimizations, including modifying the nature and positioning of the staple(s), the number and nature of charged amino acids, and solubility of the agents (Bird et al., 2011; Cohen et al., 2012; Walensky & Bird, 2014; Walensky et al., 2004). Hence further systematic refinements of the side chains of the N-locked agents in Table 1 will be likely needed and subsequently systematically characterize the cell efficacy of the resulting agents, which fall outside the scope of this current structural/biochemical study.
As it is difficult to accurately discriminate between the alpha helical, the 3–10 helical confirmations and/or distortions introduced in the terminal aminoacids (alpha-turn) only using NMR and CD in solution, we obtained the crystal structure of Bfl-1 in complex with the N-locked covalent peptide 138C7 (Fig 4). Contiguous electron density could be observed between the peptide and the protein, in particular between the Dap residue of 138C7 and the side chain of hBfl-1 Cys55, further corroborating the anticipated covalent intermolecular interaction between these residues (Fig 4B). In addition, however, the structure also largely confirmed an idealized alpha helical geometry also for the N-locked moiety (Fig 4C). For example, the peptide geometry in the crystal structure seems further stabilized by a hydrogen bond between the oxygen on side chain of the residue Dap1 and the amide hydrogen of Ile 2 (Fig 4C). This geometry brings the amide proton of Ile 2 close to the Hβ1,2 of Dap1, and, in agreement, strong NOEs were observed in solution in the 2D [1H, 1H] NOESY spectrum of 138C5 between these protons (Fig 3D). These data suggest that while the 3JαHN coupling of the N-locked covalent peptide (10 Hz) suggests an alpha turn in solution, the interconversion of the alpha turn into the alpha helical conformation is not disallowed. Such needed conformational change would likely cause losses in binding compared to a linear peptide, but such losses seem to be overturned by introducing a covalent moiety. A favorable consequence of these observations is that the N-locked covalent agents will result more selective for their intended target (Table 1).
Recently, various efforts have been reported in deriving covalent helix stabilized hBfl-1 targeting BH3 peptides, including i, i+4 (Huhn et al., 2016) and i, i+7 hydrocarbon stapled covalent peptides (Harvey et al., 2018). However, and as in most cases with stapling strategies, direct comparisons between their linear counterparts was not provided, which makes it difficult to evaluate the advantages of the proposed stapling strategies. Nonetheless, hydrocarbon stapling produced relatively large solvent exposed hydrophobic surfaces that could interfere with some of the native intermolecular interactions, and that could lead to peptide aggregation, limited solubility, and non-specific interactions. On the contrary, compared to these peptides, the N-locked positioned all the side chains approximately in the same orientation as those of the linear peptide, a feature that could result particularly advantageous compared to these other stapling strategies. In addition, and also based on our own previous experience with a stapled Bfl-1 covalent BIM peptide (Barile et al., 2017), solubility can be a major drawback with hydrocarbon stapling. The N-locking strategy, on the contrary, is expected not to significantly diminish the solubility of the originating linear peptide, as it can be observed comparing the 1D 1H NMR spectra of both the shorter N-locked peptides (Fig 2), and those of 138C5 and its corresponding linear 135P1 (Fig 3C). We believe this is a critically important advantage of the N-locking approach compared to hydrocarbon-stapled strategies.
5. Conclusions
In conclusion, our preliminary studies suggested that the N-locking may be a viable strategy in designing inhibitors of PPIs that are mediated by alpha helices, yet their applications are likely more effective when a covalent agent is obtained, to compensate losses in binding due to helical distortions (the alpha turn) introduced at the N-terminus. For longer peptides, it may be worth exploring whether it may be beneficial to simultaneously combine the N-locking with other stapling strategies. Along the same lines, we are also attempting similar strategies to “lock” the C-terminus of helical peptides to obtain C-locked, and/or N-locked and C-locked peptides. We are currently exploring these possibilities.
Perhaps most importantly, our studies resulted in a novel series of N-locked covalent hBfl-1 antagonists with remarkable potency and selectivity against an undesirable counter target, hBcl-xL. Recent clinical studies with CLL (chronic lymphocytic leukemia) patients identified hBfl-1 as the strongest discriminating gene, with higher expression in chemotherapy resistant CLL patients, clearly suggesting that Bfl-1 represents a highly significant therapeutic target (Morales et al., 2005; Olsson et al., 2007). Similarly, it was found that Bfl-1 conferred resistance to CLL cells treated with ABT737 (the earlier Bcl-2/Bcl-xL antagonist, recently replaced by the FDA approved Bcl-2 selective Venetoclax/ABT199) (Al-Harbi et al., 2011; Yecies, Carlson, Deng, & Letai, 2010). More recent studies identified Bfl-1 to be overexpressed in lymphoma, both in cell lines and in primary cultures, after prolonged exposure to Venetoclax, confirming the role of Bfl-1 in acquired resistance to Bcl-2 antagonists (Esteve-Arenys & Roue, 2018). Likewise, we and others found that Bfl-1 is highly expressed in melanomas (Placzek et al., 2010), where its over-expression could be associated with resistance to chemotherapy and to the BRAF inhibitor Vemurafenib (Hind et al., 2015). Nonetheless, no small molecule pharmacological tools that target hBfl-1 potently and selectively have been reported to date. In this regard, we are confident that our agents, as also supported by our preliminary apoptosis assay data, could represent a significant step forward the identification of such agents.
Acknowledgements
Financial support was obtained in part by the NIH, with grants CA168517, CA242620 and NS107479 (to MP) and UC CRCC CRN-18-524906 and UCOP LFR-17-476732 grants to JJPP. MP holds the Daniel Hays Chair in Cancer Research at the School of Medicine at UCR. PU is a recipient of the 2017-2018 Pease Cancer Fellowship through the Division of Biomedical Sciences, School of Medicine at UCR. The funders provided support in the form of salaries for authors employed at UCR and materials and supplies used at UCR to carry out these studies, and also supported the expenses related to the studies conducted at SARomics Biostructures AB, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
Abbreviations:
- DELFIA
Dissociation-Enhanced Lanthanide Fluorescent Immunoassay
- DMF
Dimethylformamide
- DCM
Dichloromethane
- ivDde
1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3-methylbutyl
- DIPEA
N,N-diisopropylethylamine
- TCEP
(Tris(2-carboxyethyl)phosphine hydrochloride)
- CLL
chronic lymphocytic leukemia
Footnotes
Conflicts of interest
The authors employed at UCR declare no conflict of interests. MH is an employee of SARomics Biostructures AB and worked for hire by UCR to determine the structure reported in Fig 4. This does not alter our adherence to the journal policies on sharing data and materials.
Data Availability Statement
Our agents can be distributed in small amounts (1–5 mg) for research purposes upon request and signing of a standard material transfer agreement. The crystallographic coordinates of hBFl-1 in complex with 138C7 have been deposited in the Protein Data Bank under the PDB ID 6RJP.
References
- Al-Harbi S, Hill BT, Mazumder S, Singh K, Devecchio J, Choudhary G,…Almasan A (2011). An antiapoptotic BCL-2 family expression index predicts the response of chronic lymphocytic leukemia to ABT-737. Blood, 118(13), 3579–3590. doi: 10.1182/blood-2011-03-340364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio C, Gambini L, Udompholkul P, Salem AF, Aronson A, Dona A,…Pellecchia M (2018). Design of Potent pan-IAP and Lys-Covalent XIAP Selective Inhibitors Using a Thermodynamics Driven Approach. J Med Chem. doi: 10.1021/acs.jmedchem.8b00810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio C, Udompholkul P, Barile E, & Pellecchia M (2017). Enthalpy-Based Screening of Focused Combinatorial Libraries for the Identification of Potent and Selective Ligands. ACS Chem Biol, 12(12), 2981–2989. doi: 10.1021/acschembio.7b00717 [DOI] [PubMed] [Google Scholar]
- Barile E, Marconi GD, De SK, Baggio C, Gambini L, Salem AF,…Pellecchia M (2017). hBfl-1/hNOXA Interaction Studies Provide New Insights on the Role of Bfl-1 in Cancer Cell Resistance and for the Design of Novel Anticancer Agents. ACS Chem Biol, 12(2), 444–455. doi: 10.1021/acschembio.6b00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter D, Perry SR, Hill TA, Kok WM, Zaccai NR, Brady RL,…Mason JM (2017). Downsizing Proto-oncogene cFos to Short Helix-Constrained Peptides That Bind Jun. ACS Chem Biol, 12(8), 2051–2061. doi: 10.1021/acschembio.7b00303 [DOI] [PubMed] [Google Scholar]
- Bird GH, Crannell WC, & Walensky LD (2011). Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr Protoc Chem Biol, 3(3), 99–117. doi: 10.1002/9780470559277.ch110042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Gavathiotis E, LaBelle JL, Katz SG, & Walensky LD (2014). Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol, 9(3), 831–837. doi: 10.1021/cb4003305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A,…Elmore SW (2007). Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem, 50(4), 641–662. doi: 10.1021/jm061152t [DOI] [PubMed] [Google Scholar]
- Bullock BN, Jochim AL, & Arora PS (2011). Assessing helical protein interfaces for inhibitor design. J Am Chem Soc, 133(36), 14220–14223. doi: 10.1021/ja206074j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas E, & Satterthwait AC (1999). The Hydrogen Bond Mimic Approach: Solid-Phase Synthesis of a Peptide Stabilized as an α-Helix with a Hydrazone Link. Journal of the American Chemical Society, 121(16), 3862–3875. doi: 10.1021/ja983212t [DOI] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ,…Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr, 66(Pt 1), 12–21. doi: 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NA, Stewart ML, Gavathiotis E, Tepper JL, Bruekner SR, Koss B,…Walensky LD. (2012). A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol, 19(9), 1175–1186. doi: 10.1016/j.chembiol.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D. de Araujo A, Lim J, Wu K-C, Xiang Y, Good AC, Skerlj R, & Fairlie DP (2018). Bicyclic Helical Peptides as Dual Inhibitors Selective for Bcl2A1 and Mcl-1 Proteins. Journal of Medicinal Chemistry, 61(7), 2962–2972. doi: 10.1021/acs.jmedchem.8b00010 [DOI] [PubMed] [Google Scholar]
- de Araujo AD, Lim J, Good AC, Skerlj RT, & Fairlie DP (2017). Electrophilic Helical Peptides That Bond Covalently, Irreversibly, and Selectively in a Protein–Protein Interaction Site. ACS Medicinal Chemistry Letters, 8(1), 22–26. doi: 10.1021/acsmedchemlett.6b00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, & Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr, 66(Pt 4), 486–501. doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Arenys A, & Roue G (2018). BFL-1 expression determines the efficacy of venetoclax in MYC+/BCL2+ double hit lymphoma. Oncoscience, 5(3–4), 59–61. doi: 10.18632/oncoscience.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, & Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr, 69(Pt 7), 1204–1214. doi: 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen F, Campiglio M, Jo H, Abderemane-Ali F, Rumpf CH, Pope L,…Minor DL Jr. (2017). Stapled Voltage-Gated Calcium Channel (CaV) alpha-Interaction Domain (AID) Peptides Act As Selective Protein-Protein Interaction Inhibitors of CaV Function. ACS Chem Neurosci, 8(6), 1313–1326. doi: 10.1021/acschemneuro.6b00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ET, & Rose GD (1993). Helix stop signals in proteins and peptides: the capping box. Biochemistry, 32(30), 7605–7609. [DOI] [PubMed] [Google Scholar]
- Harvey EP, Seo HS, Guerra RM, Bird GH, Dhe-Paganon S, & Walensky LD (2018). Crystal Structures of Anti-apoptotic BFL-1 and Its Complex with a Covalent Stapled Peptide Inhibitor. Structure, 26(1), 153–160 e154. doi: 10.1016/j.str.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchey LK, Kushal S, Dubey R, Chapman RN, Olenyuk BZ, & Arora PS (2010). Inhibition of hypoxia inducible factor 1-transcription coactivator interaction by a hydrogen bond surrogate alpha-helix. J Am Chem Soc, 132(3), 941–943. doi: 10.1021/ja9082864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind CK, Carter MJ, Harris CL, Chan HT, James S, & Cragg MS (2015). Role of the pro-survival molecule Bfl-1 in melanoma. Int J Biochem Cell Biol, 59, 94–102. doi: 10.1016/j.biocel.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Hoang HN, Driver RW, Beyer RL, Malde AK, Le GT, Abbenante G,…Fairlie DP (2011). Protein alpha-turns recreated in structurally stable small molecules. Angew Chem Int Ed Engl, 50(47), 11107–11111. doi: 10.1002/anie.201105119 [DOI] [PubMed] [Google Scholar]
- Hoang HN, Wu C, Beyer RL, Hill TA, & Fairlie DP (2017). Alpha Helix Nucleation by a Simple Cyclic Tetrapeptide. Australian Journal of Chemistry, 70(2), 213–219. doi: 10.1071/CH16591 [DOI] [Google Scholar]
- Huhn AJ, Guerra RM, Harvey EP, Bird GH, & Walensky LD (2016). Selective Covalent Targeting of Anti-Apoptotic BFL-1 by Cysteine-Reactive Stapled Peptide Inhibitors. Cell Chem Biol. doi: 10.1016/j.chembiol.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayatunga MK, Thompson S, & Hamilton AD (2014). alpha-Helix mimetics: outwards and upwards. Bioorg Med Chem Lett, 24(3), 717–724. doi: 10.1016/j.bmcl.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Jenson JM, Ryan JA, Grant RA, Letai A, & Keating AE (2017). Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1. Elife, 6. doi: 10.7554/eLife.25541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez MA, Munoz V, Rico M, & Serrano L (1994). Helix stop and start signals in peptides and proteins. The capping box does not necessarily prevent helix elongation. J Mol Biol, 242(4), 487–496. doi: 10.1006/jmbi.1994.1596 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr, 66(Pt 2), 125–132. doi: 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, …Walensky LD (2012). A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest, 122(6), 2018–2031. doi: 10.1172/JCI46231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YH, de Andrade P, Wu Y, & Spring DR (2015). Peptide stapling techniques based on different macrocyclisation chemistries. Chem Soc Rev, 44(1), 91–102. doi: 10.1039/c4cs00246f [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, & Murshudov GN (2012). JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr, 68(Pt 4), 431–440. doi: 10.1107/S090744491200251X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, …Fesik SW (2000). Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature, 408(6815), 1004–1008. doi: 10.1038/35050006 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, & Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr, 40(Pt 4), 658–674. doi: 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales AA, Olsson A, Celsing F, Osterborg A, Jondal M, & Osorio LM (2005). High expression of bfl-1 contributes to the apoptosis resistant phenotype in B-cell chronic lymphocytic leukemia. Int J Cancer, 113(5), 730–737. doi: 10.1002/ijc.20614 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, …Vagin AA (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr, 67(Pt 4), 355–367. doi: 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Luong HX, & Kim Y-W (2016). Helix Nucleation via Hydrocarbon Cross-link Mimicking N-capping Box. Bulletin of the Korean Chemical Society, 37(4), 566–570. doi: 10.1002/bkcs.10727 [DOI] [Google Scholar]
- Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, …Czabotar PE (2013). Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol, 8(2), 297–302. doi: 10.1021/cb3005403 [DOI] [PubMed] [Google Scholar]
- Olsson A, Norberg M, Okvist A, Derkow K, Choudhury A, Tobin G, …Osorio LM (2007). Upregulation of bfl-1 is a potential mechanism of chemoresistance in B-cell chronic lymphocytic leukaemia. Br J Cancer, 97(6), 769–777. doi: 10.1038/sj.bjc.6603951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA,…Rosenberg SH (2005). An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature, 435(7042), 677–681. doi: 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- Oost TK, Sun C, Armstrong RC, Al-Assaad AS, Betz SF, Deckwerth TL,…Fesik SW (2004). Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J Med Chem, 47(18), 4417–4426. doi: 10.1021/jm040037k [DOI] [PubMed] [Google Scholar]
- Patgiri A, Jochim AL, & Arora PS (2008). A hydrogen bond surrogate approach for stabilization of short peptide sequences in alpha-helical conformation. Acc Chem Res, 41(10), 1289–1300. doi: 10.1021/ar700264k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, & Pellecchia M (2010). A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis, 1, e40. doi: 10.1038/cddis.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta LG, & Rose GD (1988). Helix signals in proteins. Science, 240(4859), 1632–1641. [DOI] [PubMed] [Google Scholar]
- Rega MF, Reed JC, & Pellecchia M (2007). Robust lanthanide-based assays for the detection of anti-apoptotic Bcl-2-family protein antagonists. Bioorg Chem, 35(2), 113–120. doi: 10.1016/j.bioorg.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Sawyer N, Watkins AM, & Arora PS (2017). Protein Domain Mimics as Modulators of Protein-Protein Interactions. Acc Chem Res, 50(6), 1313–1322. doi: 10.1021/acs.accounts.7b00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J,…Elmore SW (2013). ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med, 19(2), 202–208. doi: 10.1038/nm.3048 [DOI] [PubMed] [Google Scholar]
- Stebbins JL, Santelli E, Feng Y, De SK, Purves A, Motamedchaboki K,…Pellecchia M (2013). Structure-based design of covalent Siah inhibitors. Chem Biol, 20(8), 973–982. doi: 10.1016/j.chembiol.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniolo C, Crisma M, Formaggio F, Valle G, Cavicchioni G, Precigoux G,…Kamphuis J (1993). Structures of peptides from alpha-amino acids methylated at the alpha-carbon. Biopolymers, 33(7), 1061–1072. doi: 10.1002/bip.360330708 [DOI] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S,…Elmore SW (2008). ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res, 68(9), 3421–3428. doi: 10.1158/0008-5472.CAN-07-5836 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W,…Bricogne G (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr, 67(Pt 4), 293–302. doi: 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, & Bird GH (2014). Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem, 57(15), 6275–6288. doi: 10.1021/jm4011675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD,…Korsmeyer SJ (2004). Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science, 305(5689), 1466–1470. doi: 10.1126/science.1099191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liao W, & Arora PS (2005). Enhanced metabolic stability and protein-binding properties of artificial alpha helices derived from a hydrogen-bond surrogate: application to Bcl-xL. Angew Chem Int Ed Engl, 44(40), 6525–6529. doi: 10.1002/anie.200501603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hoang HN, Liu L, & Fairlie DP (2018). Glucuronic acid as a helix-inducing linker in short peptides. Chem Commun (Camb), 54(17), 2162–2165. doi: 10.1039/c7cc09785a [DOI] [PubMed] [Google Scholar]
- Wu H, Acharyya A, Wu Y, Liu L, Jo H, Gai F, & DeGrado WF (2018). Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle. Chembiochem, 19(9), 902–906. doi: 10.1002/cbic.201800026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies D, Carlson NE, Deng J, & Letai A (2010). Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood, 115(16), 3304–3313. doi: 10.1182/blood-2009-07-233304 [DOI] [PMC free article] [PubMed] [Google Scholar]


