Abstract
Background:
The heart of the cell cycle regulatory machine is a group of enzymes named cyclin-dependent kinases (Cdks). The active form of these enzymes includes a kinase and its partner, a cyclin. The regulation of cyclin-Cdk complexes is provided by Cdk inhibitors (CKIs) such as Cip/Kip family comprising p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2. The hypermethylation and deacetylation of Cip/Kip gene family seem to be frequent in numerous cancers. It has been indicated that increased expression of DNMTs and HDACs contributes to cancer induction. Previously, we reported the effect of DNA demethylating agents and histone deacetylase inhibitors on histone deacetylase 1, DNA methyltransferase 1, and CIP/KIP family in colon cancer. The current study was designed to evaluate the effect of zebularine in comparison to and in combination with trichostatin A (TSA) on p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA methyltransferases (DNMT1, 3a and 3b) and histone deacetylases (HDAC1, 2, and 3) genes expression, cell growth inhibition and apoptosis induction in colon cancer LS 180 cell line.
Materials and Methods:
The colon cancer LS 180 cell line was cultured and treated with zebularine and TSA. To determine cell viability, apoptosis, and the relative expression level of the genes, MTT assay, cell apoptosis assay, and qRT-PCR were done respectively.
Results:
Both compounds significantly inhibited cell growth, and induced apoptosis. Furthermore, both compounds increased p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 significantly. Additionally, zebularine and TSA decreased DNMTs and HDACs gene expression respectively. Conclusion: The zebularine and TSA can reactivate the CIP/KIP family through inhibition of DNMTs and HDACs genes activity.
Key Words: Zebularine, trichostatin, TSGs, colon cancer
Introduction
The mammalian cell cycle refers to the determined intervals during which the cells prepare to duplicate their genome equally between two new daughter cells. This cycle is divided into four consecutive phases (G1, G2, S, and M phase) and comprises a series of regulated events controlling mitosis and proliferation to produce two daughter cells from a single cell, cell division is tightly controlled to ensure precise genome duplication (Matson et al., 2017). During the G1 phase, the cell accumulates necessary metabolites for DNA replication. Subsequently, the cell enters the S phase during which DNA is replicated. G2 phase is essential to ensure accurate DNA replication and finally, the cell is divided during M phase (Qie et al., 2016). The heart of the cell cycle regulatory machine is a group of enzymes named cyclin-dependent kinases (Cdks). The active form of these enzymes includes a kinase and its partner, a cyclin, together form a complex to derive cell cycle progression. The change in the kinase or cyclin components drives the cell from one phase to another phase of the cycle. A sequence of kinase subunits, Cdk4, Cdk6, Cdk2, and Cdk2, along with a sequence of cyclins, cyclin D, E, A, and B, are expressed during the cell progresses from G1 phase to mitosis (Park et al., 2008). Furthermore, the regulation of cyclin-Cdk complexes is provided by Cdk inhibitors (CKIs). The CKI gene families are divided into two groups based on structure and evolutionary origins consisting INK4 gene family which encodes p16INK4a, p15INK4b, p18INK4c, and p19INK4d and Cip/Kip family comprising p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 (Besson et al., 2008). The hypermethylation of the INK4 gene family (Wong et al., 2000; Chim et al., 2001) and Cip/Kip gene family (Yoshino et al., 2007; Zohny et al., 2017) seems to be frequent in numerous cancers. Another mechanism by which chromatin is inactivated is histone deacetylation reported in the hundreds of various cancers (Fang et al., 2002; Kikuchi et al., 2002). The Cip/Kip family could be inactivated by this pathway as reported in MCF-7 breast cancer (Varshochi et al., 2005), pancreatic cancer (Jiao et al., 2014), colon cancer (Chen et al., 2009), thyroid cancer (Weinlander et al., 2014), and gastric cancer (Sun et al., 2014). Primary regulators of INK4 and Cip/Kip family genes include DNA histone deacetylases (HDACs) and DNA methyltransferases (DNMTs). It has been indicated that increased expression of DNMTs and HDACs contribute to cancer induction through methylation- and deacetylation-mediated gene inactivation in various cancers (Patra et al., 2001). The over-expression of DNMTs (DNMT1, 3A, and 3B) has been shown in uterine cancer (Li et al., 2003), breast cancer (Girault et al., 2003), hepatocellular carcinoma (HCC) (Nagai et al., 2003), colorectal and stomach cancer (Kanai et al., 2001). Furthermore, high HDACs (HDACs 1, 2 and 3) expression levels are found in breast cancer (Müller et al., 2013), ovarian cancer (Khabele., 2014), bladder cancer (Poyet et al., 2014), and renal cancer (Fritzsche et al., 2008). DNA methyltransferase inhibitors (DNMTIs) can reactivate hypermethylated genes. It has been shown that DNMT-inhibiting cytosine nucleoside analogs, decitabine, azacitidine, and zebularine, have a significant effect on several cancers such as myeloid leukemia (AML) (Flotho et al., 2009), breast cancer (Chen et al., 2012), gastric cancer (Tan et al., 2013), colorectal cancer (His et al., 2005), endometrial cancer (Cui et al., 2010), lung cancer (Luszczek et al., 2010), colorectal cancer (Xiong et al., 2009), and prostate cancer (Walton et al., 2008). The HDACIs (carboxylic acids, hydroxamic acids, benzamides, and cyclic tetrapeptides) are other compounds that can restore silenced tumor suppressor genes (TSGs). They inhibit Class I (HDAC1, 2, 3, and 8) and class II HDACs (HDAC4, 5, 6, 7, 9, and 10) in various cancers including colon cancer, lung cancer, breast cancer, gastric cancer, and pancreatic cancer cells (Chueh et al., 2015). Previously, we reported the effect of DNA demethylating agent 5-aza-2′-deoxycytidine (decitabine, 5AZA-CdR) and histone deacetylase inhibitors valproic acid (VPA) and trichostatin A (TSA) on histone deacetylase 1, DNA methyltransferase 1, and Cip/Kip family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer SW480 cell line (Sanaei et al., 2018). The current study was designed to evaluate the effect of zebularine in comparison to and in combination with trichostatin A on p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA methyltransferases (DNMT1, 3a, and 3b) and histone deacetylases (HDAC1, 2, and 3) genes expression, cell growth inhibition and apoptosis induction in colon cancer LS 180 cell line.
Materials and Methods
Materials
The human colon cancer LS 180 cell line was provided from the National Cell Bank of Iran-Pasteur Institute and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with fetal bovine serum 10% and antibiotics in a humidified atmosphere of 5% CO2 in air at 37oC. Zebularine, TSA, and 3 (4,5 dimethyl 2 thiazolyl) 2, 5 diphenyl 2H tetrazolium bromide (MTT) were obtained from Sigma–Aldrich (Sigma–Aldrich, Louis, MI, USA) and the Annexin V and also propidium iodide (PI) apoptosis kit from Life Technologies. Total RNA extraction kit (TRIZOL reagent) and Real-time polymerase chain reaction (PCR) kits (qPCR MasterMix Plus for SYBR Green I dNTP) were purchased from Applied Biosystems Inc. (Foster, CA, USA) and Dimethyl sulfoxide (DMSO) from Merck Co. (Darmstadt, Germany).
Cell culture and cell viability
To determine cell viability, colon cancer LS 180 cell line was cultured in DMEM supplemented with 10% FBS and antibiotics and incubated overnight and then seeded into 96-well plates (4 × 105 cells per well). Next day, the culture medium was replaced by experimental medium containing various concentration of zebularine (0, 5, 10, 25, 50 and 100 μM) and TSA (0, 1, 2.5, 5, 10, and 25 μM) for 24 and 48 h, except control groups which treated with DMSO at a concentration of 0.05 %. Subsequently, the colon cancer LS 180 cell viability was assessed by MTT assay according to Standard protocols. First, the MTT solution was added for 4 h at 37oC and then the MTT solution was changed with DMSO. To dissolve all of the crystals, the solution was shaken for 10 min. Finally, the optical density was detected by a microplate reader at a wavelength of 570 nM. Each experiment was repeated three times (triplicates).
Cell apoptosis assay
For apoptotic cell investigation, the colon cancer LS 180 cell line were cultured at a density of 4 × 105 cells/well and treated with zebularine (50 μM) and TSA (2.5 μM), individually and combined, for 24 and 48 h. After treatment, the cells were harvested by trypsinization, washed twice in PBS and resuspended in Binding buffer (1x). Next, annexin V-FITC and PI were added to obtain the apoptotic cells. After incubation at room temperature, 15 min in the dark, the apoptotic cells were counted by FACScan flow cytometry (Becton Dickinson, Heidelberg, Germany).
Real-time Quantitative Reverse Transcription Polymerase
Chain Reaction (qRT-PCR)
The qRT-PCR was done to investigate the relative expression level of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA methyltransferases (DNMT1, 3a, and 3b) and histone deacetylases (HDAC1, 2, and 3) genes. The colon cancer LS 180 cell line was treated with zebularine (50 μM) and TSA (2.5 μM), individually and combined, for 24 and 48 h, based on IC5o values, and total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer protocol and treated by RNasefree DNase (Qiagen). Other processes were done as we did previously (Sanaei et al., 2018). The primer sequences of the genes are indicated in Table 1. GAPDH was used as an endogenous control. Data were analyzed using the comparative Ct (ΔΔct) method.
Table 1.
The Primer Sequences of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA Methyltransferases (DNMT1, 3a, and 3b) and Histone Deacetylases (HDAC1, 2, and 3) Genes
| Primer name | Primer sequences (5' to 3') | References |
|---|---|---|
| DNMT1 Forward | GAG GAA GCT GCT AAG GAC TAG TTC | Sanaei et al., 2018 |
| DNMT1 Reverse | ACT CCA CAA TTT GAT CAC TAA ATC | Sanaei et al., 2018 |
| DNMT3a Forward | GGA GGC TGA GAA GAA AGC CAA GGT | Sanaei et al., 2018 |
| DNMT3a Reverse | TTT GCC GTC TCC GAA CCA CAT GAC | Sanaei et al., 2018 |
| DNMT3b Forward | TAC ACA GAC GTG TCC AAC ATG GGC | Sanaei et al., 2018 |
| DNMT3b Reverse | GGA TGC CTT CAG GAA TCA CAC CTC | Sanaei et al., 2018 |
| P21 Forward | AGG CGC CAT GTC AGA ACC GGC TGG | Sanaei et al., 2019 |
| P21 Reverse | GGA AGG TAG AGC TTG GGC AGG C | Sanaei et al., 2019 |
| P 27 Forward | ATG TCA AAC GTG CGA GTG TCT AAC | Sanaei et al., 2019 |
| P 27 Reverse | TTA CGT TTG ACG TCT TCT GAG GCC A | Sanaei et al., 2019 |
| P 57 Forward | GCGGCGATCAAGAAGCTGTC | Sanaei et al., 2019 |
| P 57 Reverse | CCGGTTGCTGCTACATGAAC | Sanaei et al., 2019 |
| GAPDH Forward | TCCCATCACCATCTTCCA | Sanaei et al., 2019 |
| GAPDH Reverse | CATCACGCCACAGTTTCC | Sanaei et al., 2019 |
| HDAC1 Forward | AACCTGCCTATGCTGATGCT | Jin et al., 2008 |
| HDAC1 Reverse | CAGGCAATTCGTTTGTCAGA | Jin et al., 2008 |
| HDAC2 Forward | GGGAATACTTTCCTGGCACA | Jin et al., 2008 |
| HDAC2 Reverse | ACGGATTGTGTAGCCACCTC | Jin et al., 2008 |
| HDAC3 Forward | TGGCTTCTGCTATGTCAACG | Jin et al., 2008 |
| HDAC3 Reverse | GCACGTGGGTTGGTAGAAGT | Jin et al., 2008 |
Results
Result of cell viability by the MTT assay
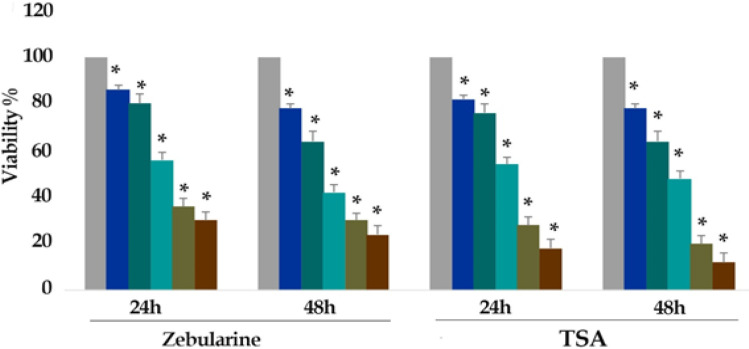
The viability of the LS 180 cell line was assessed by MTT assay. As mentioned above, the cells were treated with zebularine (0, 5, 10, 25, 50 and 100 μM) and TSA (0, 1, 2.5, 5, 10, and 25 μM) for 24 and 48 h and the cell viability was determined based on the activity of cellular enzymes to reduce the tetrazolium salt MTT resulting in a dark-blue formazan. To determine the number of viable cells, the product was dissolved in DMSO. The result of the assay indicated that both compounds significant cell growth inhibition with all used concentrations at different periods as shown in Figure 1 (P< 0.001).
Figure 1.
In vitro Effects of Zebularine (0, 5, 10, 25, 50 and 100 μM) and TSA (0, 1, 2.5, 5, 10, and 25 μM) on LS 180 Cells Viability Determined by MTT Assay at 24 and 48 h. As shown in figure 1, from right to the left, the first column of each group belongs to the control group. Values are means of three experiments in triplicate. Asterisks (*) demonstrate significant differences between treated and untreated control groups
The IC50 values were determined with approximately 50 and 2.5 μM for zebularine and TSA respectively.
Result of cell apoptosis assay
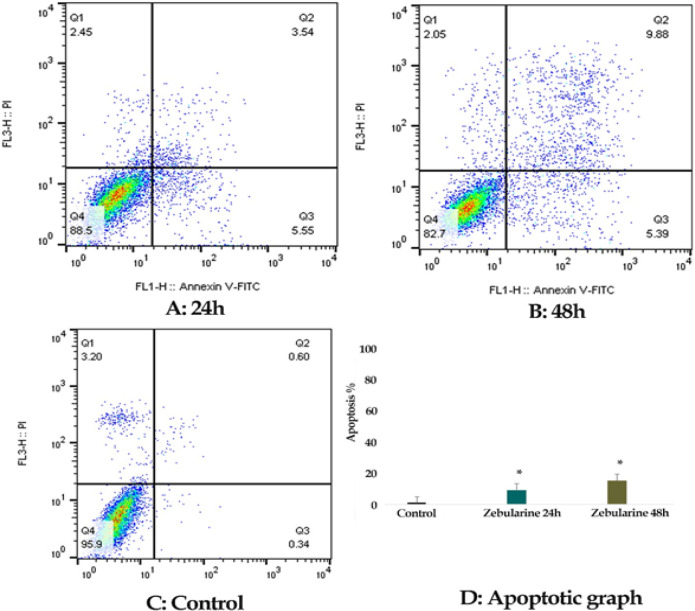
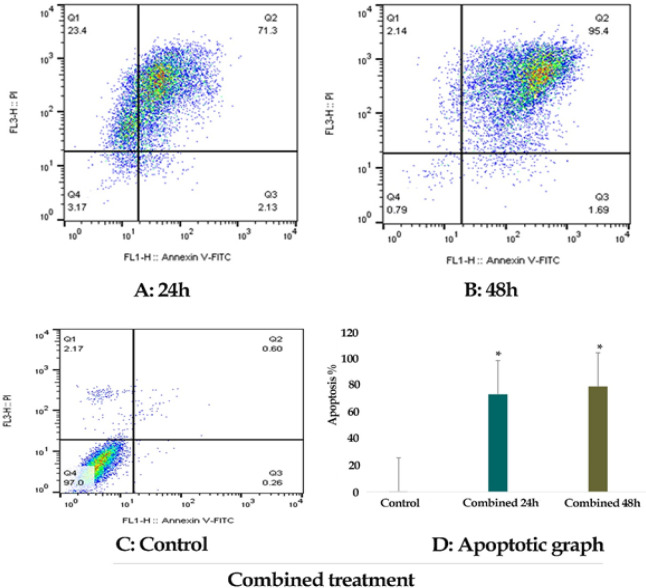
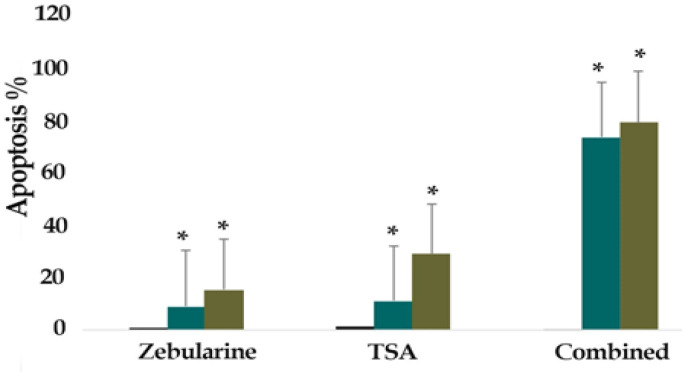
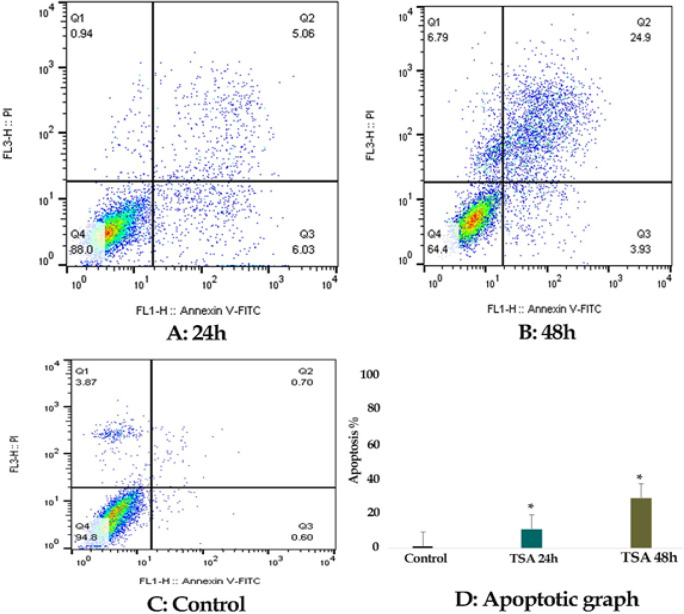
The percentage of LS 180 apoptotic cells was evaluated by staining with annexin V-FITC AND PI. The cells were treated with zebularine (50 μM) and TSA (2.5 μM) individually and combined for 24 and 48 h and then stained using annexin-V-(FITC) and PI. The result indicated that both compounds induced significant apoptosis as alone and combined, Figures 2-4. Further, TSA had a more significant effect in comparison to zebularine. Maximal apoptotic effect was observed with combined treatment after 48 h and minimal apoptosis was seen with zebularine at 24 h, Figure 5. The percentage of apoptosis has been shown in Table 2.
Figure 2.
The Apoptotic Effect of Zebularine (50 μM) on LS 180 Cell versus Control Groups at Different Periods (24 and 48h). The cells were treated with this agent for 24 and 48h and then the apoptotic effect was evaluated by flow cytometric analysis. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean
Figure 4.
The Apoptotic Effect of Zebularine (50 μM) in Combination with TSA (2.5 μM) for 24 and 48 h on LS 180 Cell versus Control Groups at Different Periods (24 and 48h). The cells were treated with this agent for 24 and 48h and then the apoptotic effect was evaluated by flow cytometric analysis. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean
Figure 5.
The Comparative Apoptotic Effects of Zebularine (50 μM) in Comparison to and in Combination with TSA (2.5 μM) on LS 180 cells. Asterisks (*) indicate significant differences between the treated and untreated control groups. As demonstrated above, TSA had a more significant apoptotic effect on LCL-PI 11 cells in comparison to zebularine. The combined treatment had a maximum effect on apoptosis
Table 2.
Percentage of Apoptosis in the Groups Treated with Zebularine and TSA, as alone and Combined, at Different Periods
| Drug | Dose/ μM | Duration/ h | Apoptosis % | P-value |
|---|---|---|---|---|
| Zebularine | 25 | 24 | 9.09 | P < 0.001 |
| 25 | 48 | 15.27 | P < 0.001 | |
| TSA | 5 | 24 | 11.09 | P < 0.001 |
| 5 | 48 | 28.83 | P < 0.001 | |
| Zebularine/TSA | 25/5 | 24 | 73.43 | P < 0.001 |
| 25/5 | 48 | 79.09 | P < 0.001 |
Result of determination of genes expression
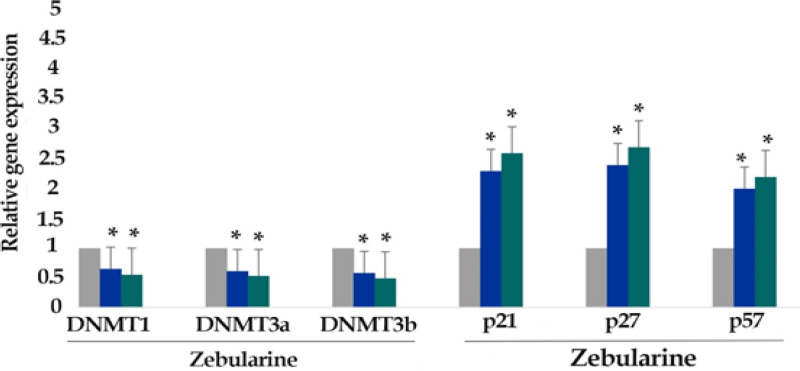
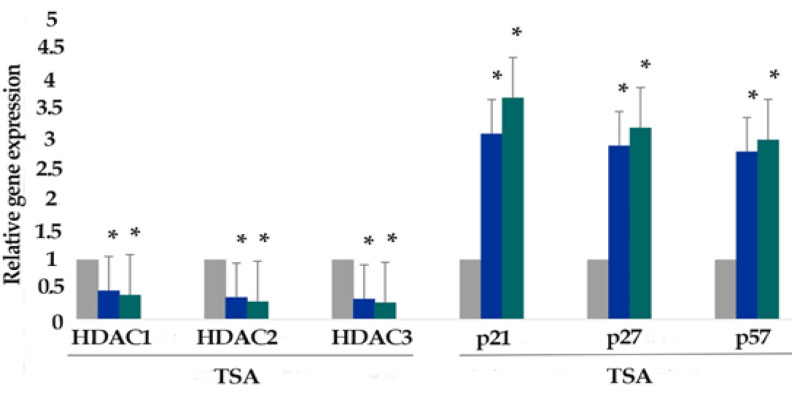
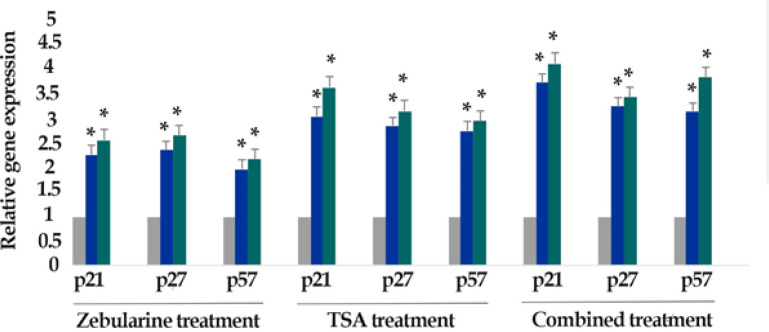
The effect of zebularine (50 μM) and TSA (2.5 μM), alone and combined, on the relative expression level of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA methyltransferases (DNMT1, 3a, and 3b) and histone deacetylases (HDAC1, 2, and 3) genes was determined by quantitative real-time RT-PCR analysis. The finding indicated that zebularine decreased DNA methyltransferases (DNMT1, 3a and 3b), decreased DNA methyltransferases (DNMT1, 3a and 3b) and histone deacetylases (HDAC1, 2, and 3), and both compounds increased p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 genes expression significantly in LS 180 cell line after 24 and 48 h, Figure 6 and Figure 7. Additionally, TSA had a more significant effect on the up-regulation of p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 in comparison to zebularine. Further, the maximum expression of p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 genes was observed with combined treatment as demonstrated in Figure 8. The relative expression level of the genes has been indicated in Table 3 and 4.
Figure 6.
The Relative Expression Level of DNA Methyltransferases (DNMT1, 3a, and 3b), p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 in the LS 180 cell line treated with zebularine (50 μM) versus untreated control groups at different periods (24 and 48h). The first column of each group belongs to the untreated control group and the others belong to the treated cells with zebularine. Asterisks (*) indicate significant differences between the treated and untreated groups
Figure 7.
The Relative Expression Level of Histone Deacetylases (HDAC1, 2, and 3), p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 in the LS 180 cell line treated with TSA (2.5 μM) versus untreated control groups at different periods (24 and 48h). The first column of each group belongs to the untreated control group and the others belong to the treated cells with zebularine. Asterisks (*) indicate significant differences between the treated and untreated groups
Figure 8.
The Relative Expression Level of p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 in the LS 180 cell line treated with combined compounds versus untreated control groups at different periods (24 and 48h). The first column of each group belongs to the untreated control group and the others belong to the treated cells with zebularine in combination with TSA. Asterisks (*) indicate significant differences between the treated and untreated groups
Table 3.
The Relative Expression Level of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNMTs, HDAC1, HDAC2, and HDAC3 Genes
| Cell line | Gene | Drug | Dose (μM) | Duration (h) | Expression | P-value |
|---|---|---|---|---|---|---|
| LS 180 | DNMT1 | Zebularine | 2.5 μM | 24 | 0.65 | 0.001 |
| LS 180 | DNMT1 | Zebularine | 25 μM | 48 | 0.55 | 0.001 |
| LS 180 | DNMT3a | Zebularine | 25 μM | 24 | 0.61 | 0.001 |
| LS 180 | DNMT3a | Zebularine | 25 μM | 48 | 0.53 | 0.001 |
| LS 180 | DNMT3b | Zebularine | 25 μM | 24 | 0.58 | 0.001 |
| LS 180 | DNMT3b | Zebularine | 25 μM | 48 | 0.49 | 0.01 |
| LS 180 | P21 | Zebularine | 25 μM | 24 | 2.3 | 0.004 |
| LS 180 | P21 | Zebularine | 25 μM | 48 | 2.6 | 0.001 |
| LS 180 | P27 | Zebularine | 25 μM | 24 | 2.4 | 0.001 |
| LS 180 | P27 | Zebularine | 25 μM | 48 | 2.7 | 0.001 |
| LS 180 | P57 | Zebularine | 25 μM | 24 | 2 | 0.001 |
| LS 180 | P57 | Zebularine | 25 μM | 48 | 2.2 | 0.001 |
| LS 180 | HDAC1 | Trichostatin A | 5 μM | 24 | 0.48 | 0.001 |
| LS 180 | HDAC1 | Trichostatin A | 5 μM | 48 | 0.41 | 0.004 |
| LS 180 | HDAC2 | Trichostatin A | 5 μM | 24 | 0.37 | 0.002 |
| LS 180 | HDAC2 | Trichostatin A | 5 μM | 48 | 0.3 | 0.01 |
| LS 180 | HDAC3 | Trichostatin A | 5 μM | 24 | 0.34 | 0.007 |
| LS 180 | HDAC3 | Trichostatin A | 5 μM | 48 | 0.28 | 0.001 |
| LS 180 | P21 | Trichostatin A | 5 μM | 24 | 3.1 | 0.001 |
| LS 180 | P21 | Trichostatin A | 5 μM | 48 | 3.7 | 0.001 |
| LS 180 | P27 | Trichostatin A | 5 μM | 24 | 2.9 | 0.001 |
| LS 180 | P27 | Trichostatin A | 5 μM | 48 | 3.2 | 0.001 |
| LS 180 | P57 | Trichostatin A | 5 μM | 24 | 2.8 | 0.001 |
| LS 180 | P57 | Trichostatin A | 5 μM | 48 | 3 | 0.001 |
Table 4.
The Relative Expression Level of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 Genes with Combined Treatment
| Cell line | Gene | Drug | Dose (μM) | Duration (h) | Expression | P-value |
|---|---|---|---|---|---|---|
| LS 180 | P21 | Zebularine/TSA | 25/5 μM | 24 | 3.8 | 0.001 |
| LS 180 | P21 | Zebularine/TSA | 25/5 μM | 48 | 4.2 | 0.001 |
| LS 180 | P27 | Zebularine/TSA | 25/5 μM | 24 | 3.3 | 0.001 |
| LS 180 | P27 | Zebularine/TSA | 25/5 μM | 48 | 3.5 | 0.001 |
| LS 180 | P57 | Zebularine/TSA | 25/5 μM | 24 | 3.2 | 0.001 |
| LS 180 | P57 | Zebularine/TSA | 25/5 μM | 48 | 3.9 | 0.001 |
Figure 3.
The Apoptotic Effect of TSA (2.5 μM) on LS 180 Cell versus Control Groups at Different Periods (24 and 48h). The cells were treated with this agent for 24 and 48h and then the apoptotic effect was evaluated by flow cytometric analysis. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean
Discussion
Epigenetic regulation such as DNA methylation and histone modification is the mechanism by which gene is activated or inactivated in the mammalian cells. This mechanism is more specified genetic information and involved in gene repression. Recent studies have identified a variety of regulatory proteins comprising histone-modifying enzymes, DNA methyltransferases, chromatin remodeling factors, and methyl-CpG binding proteins. Abnormalities and changes in the epigenetic states such as DNA hypermethylation and histone deacetylation represent several diseases, especially tumorigenesis. However, promoter hypermethylation and histone deacetylation play a significant role in cancer through transcriptional silencing of TSGs. Meanwhile, the DNA demethylating agents and histone deacetylase inhibitors can induce re-expression of silenced TSGs causing growth arrest and apoptosis (Baylin., 2007; Kelly et al., 2003). In this study, we observe that zebularine and TSA can up-regulate p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 and down-regulate DNA methyltransferases (DNMT1, 3a, and 3b) and histone deacetylases (HDAC1, 2, and 3) gene expression resulting in cell growth inhibition and apoptosis induction. Further, the effect of TSA was stronger than that of zebularine. Maximal expression of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 and also cell apoptosis was seen with combined treatment. Inconsistent with our result, it has been reported that zebularine can increase the level of p21Cip1/Waf1/Sdi1 in SW48 cells (Flis et al., 2014). The other members of DNMTIs act by a similar pathway. In vitro studies have shown that 5-aza increases p21WAF1 in colon cancer Colo-320 and SW1116 (Fang et al., 2004; Chen et al., 2004). The re-activation of Cip/Kip family by DNMTIs has been shown in several cancers such as myeloma cell line WL2 (Chim et al., 2005), human pancreatic cancer (Wang et al., 2013), and gastric cancer (Pellegrini et al., 2010). In addition to Cip/Kip, zebularine can up-regulate ink4 p15INK4b and p16INK4a in colon cancer Caco-2 cell (Berner et al., 2010), and also p53 in colon cancer (Yang et al., 2013) and other cancer such as HCC (Nakamura et al., 2013). In contrast, some studies have shown that zebularine is not effective on the up-regulation of p21 and p27 in colon cancer HCT15, SW48, and HT-29 colon cancer (Cheng et al., 2004).
In the current study, we indicated that TSA can up-regulate p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 genes significantly. As we reported, other studies have been demonstrated that TSA up-regulates p21Cip1/Waf1/Sdi1 in colon cancer SW480 and HT-29 cell lines (Spurling et al., 2008), p27Kip1 in colon cancer HT-29 M6 (Mayo et al., 2007). The same pathway, Cip/Kip up-regulation, has been demonstrated in various cancers including, lung cancer (Platta et al., 2007), HCC Hep3B cells (Svechnikova et al., 2007), and human gastric cancer cell lines, OCUM-8 and MKN-74 (Zhang et al., 2006). We observed that cell apoptosis was increased after combined treatment. Other researchers have demonstrated that apoptosis induction is greatly enhanced in the presence of combined treatment, HDACIs in combination with DNMTIs, in human lung, thoracic, breast, leukemia and colon cancer cell lines (Zhu et al., 2003; Belinsky et al., 2003), and breast cancer (Primeau et al., 2003). After the evaluation of gene expression, we performed a further investigation and found that zebularine and TSA induce this effect through inhibition of DNMTs and HDACs respectively. Such inhibitory effect on DNMTs has been shown in T24 bladder cancer, HCT15, SW48, and HT-29 colon cancer, CFPAC-1 pancreatic cancer, PC3 prostate cancer, CALU-1 lung cancer cells [48], cholangiocarcinoma (CCA) TFK-1 and HuCCT1 cell lines (Nakamura et al., 2015). Additionally, the similar inhibitory effect of TSA on HDAC Class I has been demonstrated in other cancers comprising gastric MKN-7, MKN-28 and Ho-1-N-1, and oral Ho-1-N-1 cell lines (Suzuki et al., 2000). It should be noted that the inhibition of HDAC Class I is not the only mechanism of TSA. It can inhibit other HDACs including class I, II, and IV (Rikiishi., 2011). Summery, we indicated that zebularine and TSA down-regulate DNA methyltransferases (DNMT1, 3a, and 3b) and histone deacetylases (HDAC1, 2, and 3) by which up-regulate p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 gene expression resulting in LS 180 cell growth inhibition and apoptosis induction.
In conclusion, our findings demonstrated that zebularine and TSA can epigenetically down-regulate DNMTs and HDACs gene expression by which re-activate the Cip/Kip family gene in colon cancer LS 180 cell line cells resulting in cell growth inhibition, and apoptosis induction. Thus, this result suggests a dependence of the p21Cip1/Waf1/Sdi1, and p27Kip1 gene silencing through histone deacetylation and DNA hypermethylation by a mechanism that involves the up-regulation of histone deacetylases and DNA methyltransferases.
Acknowledgments
This article was supported by the adjutancy of research of Jahrom University of Medical Sciences, Iran.
Conflict of interest
The authors report no conflict of interest.
References
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Rev Clin Oncol. 2005;2:1–4. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–93. [PubMed] [Google Scholar]
- Berner C, Aumüller E, Gnauck A, et al. Epigenetic control of estrogen receptor expression and tumor suppressor genes is modulated by bioactive food compounds. Ann Nutr Metab. 2010;57:183–9. doi: 10.1159/000321514. [DOI] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–69. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Chen M, Shabashvili D, Nawab A, et al. DNA methyltransferase inhibitor, zebularine, delays tumor growth and induces apoptosis in a genetically engineered mouse model of breast cancer. Mol Cancer Ther. 2012;11:370–82. doi: 10.1158/1535-7163.MCT-11-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fang J, Lu J, Qiu D. Regulation of histone acetylation on the expression of cell cycle-associated genes in human colon cancer cell lines. Zhonghua Yi Xue Za Zhi. 2004;84:312–7. [PubMed] [Google Scholar]
- Chen Y-X, Fang J-Y, Lu R, et al. Expression of p21WAF1 is related to acetylation of histone H3 in total chromatin in human colorectal cancer. World J Gastroenterol. 2007;13:2209. doi: 10.3748/wjg.v13.i15.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–8. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Chim C, Liang R, Fung T, et al. Infrequent epigenetic dysregulation of CIP/KIP family of cyclin-dependent kinase inhibitors in multiple myeloma. Leukemia. 2005;19:2352. doi: 10.1038/sj.leu.2403904. [DOI] [PubMed] [Google Scholar]
- Chim C, Liang R, Tam C, et al. Methylation of p15 and p16 genes in acute promyelocytic leukemia: potential diagnostic and prognostic significance. J Clin Oncol. 2001;19:2033–40. doi: 10.1200/JCO.2001.19.7.2033. [DOI] [PubMed] [Google Scholar]
- Chueh AC, Tse JW, Tögel L, et al. Mechanisms of histone deacetylase inhibitor-regulated gene expression in cancer cells. Antioxidant Redox Signal. 2015;23:66–84. doi: 10.1089/ars.2014.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Wen Z, Chen J, et al. 5AZA-CdR is a potent inhibitor of DNA methyltransferase 3B and induces apoptosis in human endometrial cancer cell lines with the up-regulation of hMLH1. Med Oncol. 2010;27:278–85. doi: 10.1007/s12032-009-9204-1. [DOI] [PubMed] [Google Scholar]
- Fang JY, Chen YX, Juan L, et al. Epigenetic modification regulates both expression of tumor-associated genes and cell cycle progressing in human colon cancer cell lines: Colo-320 and SW1116. Cell Res. 2004;14:217. doi: 10.1038/sj.cr.7290222. [DOI] [PubMed] [Google Scholar]
- Fang J-Y, Lu Y-Y. Effects of histone acetylation and DNA methylation on p21WAF1 regulation. World J Gastroenterol. 2002;8:400. doi: 10.3748/wjg.v8.i3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flis S, Gnyszka A, Flis K. DNA methyltransferase inhibitors improve the effect of chemotherapeutic agents in SW48 and HT-29 colorectal cancer cells. PLoS One. 2014;9:92305. doi: 10.1371/journal.pone.0092305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho C, Claus R, Batz C, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- Fritzsche FR, Weichert W, Röske A, et al. Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer. BMC Cancer. 2008;8:381. doi: 10.1186/1471-2407-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault I, Tozlu S, Lidereau R, et al. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–22. [PubMed] [Google Scholar]
- Hsi LC, Xi X, Wu Y, et al. The methyltransferase inhibitor 5-aza-2-deoxycytidine induces apoptosis via induction of 15-lipoxygenase-1 in colorectal cancer cells. Mol Cancer Ther. 2005;4:1740–6. doi: 10.1158/1535-7163.MCT-05-0218. [DOI] [PubMed] [Google Scholar]
- Jiao F, Hu H, Yuan C, et al. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int J Oncol. 2014;45:1523–30. doi: 10.3892/ijo.2014.2568. [DOI] [PubMed] [Google Scholar]
- Jin KL, Pak JH, Park J-Y, et al. Expression profile of histone deacetylases 1, 2 and 3 in ovarian cancer tissues. J Gynecol Oncol. 2008;19:185–90. doi: 10.3802/jgo.2008.19.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Kondo Y, et al. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer. 2001;91:205–12. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kelly WK, Richon VM, O’Connor O, et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin Cancer Res. 2003;9:3578–88. [PubMed] [Google Scholar]
- Khabele D. The therapeutic potential of class I selective histone deacetylase inhibitors in ovarian cancer. Front Oncol. 2014;4:111. doi: 10.3389/fonc.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Toyota M, Itoh F, et al. Inactivation of p57KIP2 by regional promoter hypermethylation and histone deacetylation in human tumors. Oncogene. 2002;21:2741. doi: 10.1038/sj.onc.1205376. [DOI] [PubMed] [Google Scholar]
- Li S, Chiang T-c, Richard-Davis G, et al. DNA hypomethylation and imbalanced expression of DNA methyltransferases (DNMT1, 3A, and 3B) in human uterine leiomyoma. Gynecol Oncol. 2003;90:123–30. doi: 10.1016/s0090-8258(03)00194-x. [DOI] [PubMed] [Google Scholar]
- Luszczek W, Cheriyath V, Mekhail TM, et al. Combinations of DNA methyltransferase and histone deacetylase inhibitors induce DNA damage in small cell lung cancer cells: correlation of resistance with IFN-stimulated gene expression. Mol Cancer Ther. 2010;9:2309–21. doi: 10.1158/1535-7163.MCT-10-0309. [DOI] [PubMed] [Google Scholar]
- Matson JP, Cook JG. Cell cycle proliferation decisions: the impact of single cell analyses. FEBS J. 2017;284:362–75. doi: 10.1111/febs.13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo C, Lloreta J, Real FX, et al. In vitro differentiation of HT-29 M6 mucus-secreting colon cancer cells involves a trychostatin A and p27KIP1-inducible transcriptional program of gene expression. J Cell Physiol. 2007;212:42–50. doi: 10.1002/jcp.20999. [DOI] [PubMed] [Google Scholar]
- Müller BM, Jana L, Kasajima A, et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer-overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Nakamura A, Makino R, et al. Expression of DNA (5-cytosin)-methyltransferases (DNMTs) in hepatocellular carcinomas. Hepatol Res. 2003;26:186–91. doi: 10.1016/s1386-6346(03)00091-3. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Aizawa K, Nakabayashi K, et al. DNA methyltransferase inhibitor zebularine inhibits human hepatic carcinoma cells proliferation and induces apoptosis. PLoS One. 2013;8:e54036. doi: 10.1371/journal.pone.0054036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Nakabayashi K, Aung KH, et al. DNA methyltransferase inhibitor zebularine induces human cholangiocarcinoma cell death through alteration of DNA methylation status. PLoS One. 2015;10:e0120545. doi: 10.1371/journal.pone.0120545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med. 2016;94:1313–26. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M-T, Lee S-J. Cell cycle and cancer. J Biochem Mol Biol. 2003;36:60–5. doi: 10.5483/bmbrep.2003.36.1.060. [DOI] [PubMed] [Google Scholar]
- Patra SK, Patra A, Dahiya R. Histone deacetylase and DNA methyltransferase in human prostate cancer. BBRC. 2001;287:705–13. doi: 10.1006/bbrc.2001.5639. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Argibay P, Gomez D. Dietary factors, genetic and epigenetic influences in colorectal cancer. Exp Ther Med. 2010;1:241–50. doi: 10.3892/etm_00000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta CS, Greenblatt DY, Kunnimalaiyaan M, et al. The HDAC inhibitor trichostatin A inhibits growth of small cell lung cancer cells. J Surg Res. 2007;142:219–26. doi: 10.1016/j.jss.2006.12.555. [DOI] [PubMed] [Google Scholar]
- Poyet C, Jentsch B, Hermanns T, et al. Expression of histone deacetylases 1, 2 and 3 in urothelial bladder cancer. BMC Clin Pathol. 2014;14:10. doi: 10.1186/1472-6890-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primeau M, Gagnon J, Momparler RL. Synergistic antineoplastic action of DNA methylation inhibitor 5-AZA-2′-deoxycytidine and histone deacetylase inhibitor depsipeptide on human breast carcinoma cells. Int J Cancer. 2003;103:177–84. doi: 10.1002/ijc.10789. [DOI] [PubMed] [Google Scholar]
- Rikiishi H. Autophagic and apoptotic effects of HDAC inhibitors on cancer cells. Bio Med Res Int. 2011;5:1–9. doi: 10.1155/2011/830260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Effect of 5-aza-2’-deoxycytidine in comparison to valproic acid and trichostatin A on histone deacetylase 1, DNA methyltransferase 1, and CIP/KIP family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer SW480 cell line. Adv Biomed Res. 2019;8:52. doi: 10.4103/abr.abr_91_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Roustazadeh A, et al. Effect of genistein in comparison with trichostatin a on reactivation of DNMTs genes in hepatocellular carcinoma. J Clin Translat Hepatol. 2018;6:141. doi: 10.14218/JCTH.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Df, Zhang Yj, Tian Xq, et al. Inhibition of m TOR signalling potentiates the effects of trichostatin A in human gastric cancer cell lines by promoting histone acetylation. Cell Biol Int. 2014;38:50–63. doi: 10.1002/cbin.10179. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Yokozaki H, Kuniyasu H, et al. Effect of trichostatin A on cell growth and expression of cell cycle-and apoptosis-related molecules in human gastric and oral carcinoma cell lines. Int J Cancer. 2000;88:992–7. doi: 10.1002/1097-0215(20001215)88:6<992::aid-ijc24>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Svechnikova I, Ammerpohl O, Ekström TJ. p21waf1/Cip1 partially mediates apoptosis in hepatocellular carcinoma cells. BBRC. 2007;354:466–71. doi: 10.1016/j.bbrc.2006.12.222. [DOI] [PubMed] [Google Scholar]
- Tan W, Zhou W, Yu H-g, et al. The DNA methyltransferase inhibitor zebularine induces mitochondria-mediated apoptosis in gastric cancer cells in vitro and in vivo. BBRC. 2013;430:250–5. doi: 10.1016/j.bbrc.2012.10.143. [DOI] [PubMed] [Google Scholar]
- Varshochi R, Halim F, Sunters A, et al. ICI182, 780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor α from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J Biol Chem. 2005;280:3185–96. doi: 10.1074/jbc.M408063200. [DOI] [PubMed] [Google Scholar]
- Walton T, Li G, Seth R, et al. DNA demethylation and histone deacetylation inhibition co-operate to re-express estrogen receptor beta and induce apoptosis in prostate cancer cell-lines. Prostate. 2008;68:210–22. doi: 10.1002/pros.20673. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Jiang N, et al. Effect of inhibition of MEK pathway on 5-aza-deoxycytidine-suppressed pancreatic cancer cell proliferation. Genet Mol Res. 2013;12:5560–73. doi: 10.4238/2013.November.18.6. [DOI] [PubMed] [Google Scholar]
- Weinlander E, Somnay Y, Harrison AD, et al. The novel histone deacetylase inhibitor thailandepsin A inhibits anaplastic thyroid cancer growth. J Surg Res. 2014;190:191–7. doi: 10.1016/j.jss.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong IH, Ng MH, Huang DP, et al. Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood. 2000;95:1942–9. [PubMed] [Google Scholar]
- Xiong H, Chen ZF, Liang QC, et al. Inhibition of DNA methyltransferase induces G2 cell cycle arrest and apoptosis in human colorectal cancer cells via inhibition of JAK2/STAT3/STAT5 signalling. J Cell Mol Med. 2009;13:3668–79. doi: 10.1111/j.1582-4934.2009.00661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P-M, Lin Y-T, Shun C-T, et al. Zebularine inhibits tumorigenesis and stemness of colorectal cancer via p53-dependent endoplasmic reticulum stress. Sci Rep. 2013;3:3219. doi: 10.1038/srep03219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino A, Katayama Y, Ogino A, et al. Promoter hypermethylation profile of cell cycle regulator genes in pituitary adenomas. JNO. 2007;83:153–62. doi: 10.1007/s11060-006-9316-9. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yashiro M, Ren J, et al. Histone deacetylase inhibitor, trichostatin A, increases the chemosensitivity of anticancer drugs in gastric cancer cell lines. Oncol Rep. 2006;16:563–8. [PubMed] [Google Scholar]
- Zhu W-G, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anti-Cancer Agents. 2003;3:187–99. doi: 10.2174/1568011033482440. [DOI] [PubMed] [Google Scholar]
- Zohny SF, Baothman OA, El-Shinawi M, et al. The KIP/CIP family members p21^{Waf1/Cip1} and p57^{Kip2} as diagnostic markers for breast cancer. Cancer Biomark. 2017;18:413–23. doi: 10.3233/CBM-160308. [DOI] [PubMed] [Google Scholar]