Abstract
Objective:
This study aims to explore the roles of long non-coding RNAs (lncRNAs) for predicting survival in hepatocellular carcinoma (HCC) patients with cirrhosis.
Methods:
A set of lncRNAs differentially expressed between HCC patients with or without cirrhosis was identified using expression profiles of The Cancer Genome Atlas database, and these lncRNAs were screened for their risk scoring system to predict recurrence-free survival (RFS) or overall survival (OS). Predictive ability of risk scoring systems was confirmed using uni- and multivariate Cox analyses while adjusting for clinical features. Predictive lncRNAs were analyzed by functional enrichment analysis.
Results:
Our screen identified 22 lncRNAs that were upregulated in the presence of cirrhosis and 59 that were downregulated. To predict OS of HCC patients with cirrhosis, a risk scoring system was developed with four lncRNAs (LINC02086, LINC00880, LINC01549 and AC136475.3); to predict RFS in these patients, the risk scoring system contained five lncRNAs (SH3RF3-AS1, AC104117.3, AC136475.3, LINC00239 and MRPL23-AS1). All risk scoring systems were associated with an area under the receiver operating characteristic curve > 0.7. Based on uni- and multivariate Cox analyses, the risk scoring system could serve as a significant independent predictor for OS in HCC patients with cirrhosis. Functional enrichment analysis suggested that the lncRNAs in the risk scoring systems are involved primarily in the pathway of Wnt signal and cytokine-cytokine receptor interaction.
Conclusion:
Risk scoring systems based on lncRNAs can effectively predict OS of HCC patients with cirrhosis. The system should be further developed and validated in larger, preferably multi-site patient populations.
Key Words: Hepatocellular carcinoma, cirrhosis, long non-coding RNA, survival
Introduction
Liver cancer remains the sixth most frequent cancer globally, with approximately 841,000 new diagnosed cases and 782,000 deaths annually. Hepatocellular carcinoma (HCC) is the most frequent common liver cancer, accounting for 75–85% of liver cancers (Bray et al., 2018; Forner et al., 2018; Kulik and El-Serag, 2019). Despite significant improvements in diagnosing and treating HCC, its heterogeneity means that it continues to be associated with relatively low rates of recurrence-free survival (RFS) and overall survival (OS) (Bruix et al., 2014; Fong and Tanabe, 2014). Currently no biomarkers have proven effective for predicting prognosis, reflecting the complicated nature of the disease. Further efforts are needed to identify biomarkers that can predict prognosis and guide treatment.
Long non-coding RNAs (lncRNAs), located in the cytoplasm and nucleus of eukaryotic cells, are non-coding RNAs longer than 200 nt (Ponting et al., 2009). These molecules play critical roles in the progression and occurrence of malignant cancers (Huarte, 2015; Schmitt and Chang, 2016; Huang et al., 2018; Wu et al., 2018a). For example, lncRNA-TUBB2A and KRTAP5-AS1 compete with endogenous RNAs to modulate the function of claudin-4, affecting prognosis of patients with gastric cancer (Song et al., 2017). The lncRNA lnc-EGFR stimulates T-regulatory cell differentiation, thereby promoting HCC immune evasion (Jiang et al., 2017), while lncRNA-6195 inhibits α-enolase activity and thus HCC progression (Yu et al., 2018).
An important risk factor for developing HCC is hepatic cirrhosis, which is characterized by the appearance of regenerative nodules surrounded by fibrous bands. Cirrhosis occurs secondary to chronic liver injury, and it can lead to portal hypertension or HCC(Schuppan and Afdhal, 2008). Since the clinical characteristics and prognosis of HCC patients can differ depending on whether cirrhosis is present (Grazi et al., 2003; Gassmann et al., 2010; Wang et al., 2013; Techathuvanan et al., 2015), we wanted to examine whether some lncRNAs are differentially expressed in the presence or absence of cirrhosis, and whether these might influence risk of recurrence or death. Using expression profile data in The Cancer Genome Atlas (TCGA) database, we developed a risk scoring system based on lncRNA levels and showed that it could predict survival of HCC patients with cirrhosis.
Materials and Methods
Datasets on HCC patients
Expression profiles of lncRNAs and mRNAs from HCC patients, together with the corresponding clinical information, were taken from TCGA (version 09-14-2017 for HCC) via the UCSC Xena server (https://xenabrowser.net/datapages/). Patient data were included in the present study if their HCC was confirmed by histology, complete lncRNA and mRNA expression profiles were available, cirrhosis status was known, and sufficient data were available to determine OS and RFS. After applying these criteria, 77 HCC patients with cirrhosis and 130 without cirrhosis were selected (Table 1). This study was prepared in accordance with TCGA guidelines (https://cancergenome.nih.gov/publications/publication-guidelines). No ethics approval was required for this study since the data came from TCGA.
Table 1.
Clinicopathological Characteristics of 207 HCC Patients with or without Cirrhosis
| Clinicopathological characteristics | Patients(n=207) |
||
|---|---|---|---|
| n | % | ||
| Age | ≤60 | 96 | 46.4 |
| >60 | 111 | 53.6 | |
| BMI | <25 | 94 | 45.4 |
| ≥25 | 103 | 49.8 | |
| Not reported | 10 | 4.8 | |
| Race | Non-Asian | 121 | 58.5 |
| Asian | 80 | 38.6 | |
| Not reported | 6 | 2.9 | |
| AFP | ≤20ng/mL | 109 | 52.7 |
| >20ng/ml | 76 | 36.7 | |
| Not reported | 22 | 10.6 | |
| Gender | Female | 66 | 31.9 |
| Male | 141 | 68.1 | |
| Hepatitis B or C | NO | 94 | 45.4 |
| YES | 104 | 50.3 | |
| Not reported | 9 | 4.3 | |
| Alcohol consumption | NO | 148 | 71.5 |
| YES | 50 | 24.2 | |
| Not reported | 9 | 4.3 | |
| Cirrhosis status | NO | 130 | 62.8 |
| YES | 77 | 37.2 | |
| Histologic grade | G1-2 | 133 | 64.2 |
| G3-4 | 72 | 34.8 | |
| Not reported | 2 | 1.0 | |
| New tumor event | NO | 97 | 46.9 |
| YES | 101 | 48.8 | |
| Not reported | 9 | 4.3 | |
| Pathologic stage* | Stage I+II | 155 | 74.9 |
| Stage III+IV | 41 | 19.8 | |
| Not reported | 11 | 5.3 | |
| Cancer status | Tumor free | 116 | 56 |
| With tumor | 84 | 40.6 | |
| Not reported | 7 | 3.4 | |
| Family cancer history | NO | 111 | 53.6 |
| YES | 68 | 32.9 | |
| Not reported | 28 | 13.5 | |
| Residual tumor | R0 | 192 | 92.7 |
| non-R0 | 13 | 6.3 | |
| Not reported | 2 | 1.0 | |
| Vascular invasion | Negative | 138 | 66.7 |
| Positive | 60 | 29 | |
| Not reported | 9 | 4.3 | |
BMI, Body mass Index; AFP, Alpha fetoprotein; *TNM staging systems.
Identification of lncRNAs differentially expressed between HCC patients with or without cirrhosis
After removal of lncRNAs showing zero expression in more than 50% of patients, the “edgeR” package in R (Robinson et al., 2010) was used to identify lncRNAs differentially expressed between patients with or without cirrhosis, defined as those associated with |log2fold change (log2FC)| > 1 and false discovery rate (FDR) < 0.05. Volcano and cluster heat maps were created by “gplots” and “heatmap” packages in R.
LncRNA–based risk scoring systems
Firstly, the normalized expression values of multiple samples of the same patient were averaged. Then, univariate Cox analysis was performed to screen differentially expressed lncRNAs for a significant relationship with OS or RFS. Those lncRNAs associated with p < 0.05 were included in subsequent multivariate Cox regression, and the best model was selected using the method of backward stepwise. A risk scoring system was defined using a linear combination of the lncRNA expression levels, each multiplied by a regression coefficient β:
Risk score = (β1 * lncRNA1 level) + (β2 * lncRNA2 level) + (β3 * lncRNA3 level) + (β4 * …level) + ...
Using this formula, a risk score was calculated for each patient, and the ability of this score to predict survival was evaluated by time-dependent receiver operating characteristic (ROC) curves in three years. Patients were divided into groups at high or low risk based on the median risk score, and shown on a non-cluster heat map. Kaplan–Meier survival curves were compared between patients at high or low risk. All these analyses were performed using R/Bioconductor (version 3.4.4).
Validation of the prognostic significance of risk scoring systems
Uni- or multivariate analyses were used to verify associations between risk score and survival. If these analyses did not return any significant results, stratified analyses were conducted to identify factors that might be affecting the results, based on the chi-squared test. All these analyses were carried out using SPSS 16.0 (IBM, Chicago, IL, USA), and the threshold for significance was a two-sided p < 0.05.
Co-expression and functional analyses of lncRNA-related mRNAs
Based on data from the 207 HCC patients, mRNAs whose expression co-varied with that of lncRNAs in the risk scoring system were identified based on a two-sided |Pearson correlation coefficient | > 0.40 and a z-test p < 0.01(Fan and Liu, 2016). These mRNAs were analyzed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment using the “clusterProfiler” package in R (Yu et al., 2012). Differences in enrichment were considered significant when associated with p < 0.05.
Results
LncRNAs differentially expressed in the presence or absence of cirrhosis in HCC patients
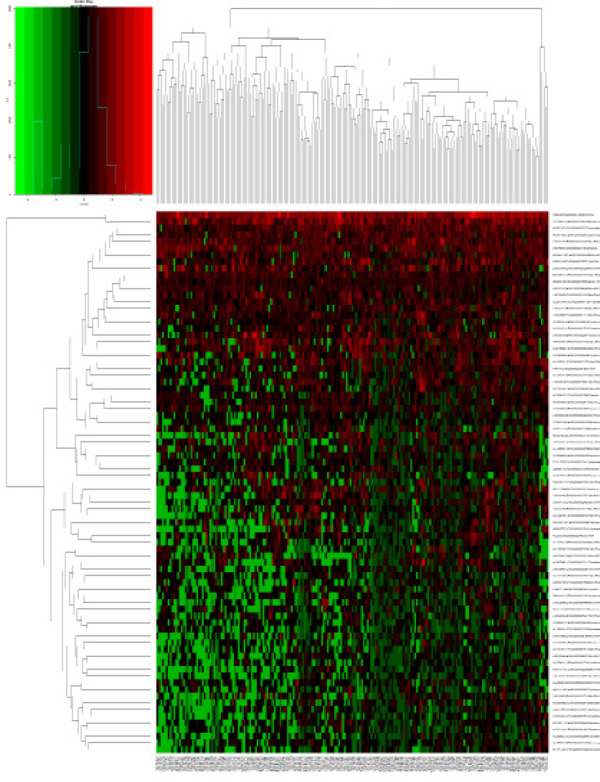
Our analysis included the lncRNA expression profiles in 77 HCC patients with cirrhosis and 130 HCC without cirrhosis obtained from the TCGA database. A total of 81 differentially expressed lncRNAs were identified, 22 (27.16%) of which were up-regulated in cirrhosis and 59 (72.84%) down-regulated. Table 2 shows the first 20 up- and down-regulated lncRNAs, together with the corresponding values for log2FC, p, and FDR. Figure 1 shows all differentially expressed lncRNAs plotted according to -log10 (FDR) and log2FC, while Figure 2 shows a heatmap indicating the relative specificity of differentially expressed lncRNAs.
Table 2.
Differentially Expressed lncRNAs in HCC Ppatients between with Cirrhosis and without Cirrhosis
| Top 20 up-regulated lncRNAs |
Top 20 down-regulated lncRNAs |
||||||
|---|---|---|---|---|---|---|---|
| logFC | P-Value | FDR | logFC | P-Value | FDR | ||
| MEG3 | 1.911879 | 2.84E-09 | 2.02E-06 | AP000439.3 | -5.46056 | 7.87E-14 | 2.24E-10 |
| LINC01549 | 1.197689 | 0.000727 | 0.037371 | AC139749.1 | -3.78362 | 4.00E-12 | 5.70E-09 |
| C5orf66 | 1.025117 | 1.57E-05 | 0.002357 | AC002398.2 | -3.23053 | 4.09E-11 | 4.67E-08 |
| AC091182.1 | 1.341751 | 6.41E-09 | 4.06E-06 | AC023906.5 | -2.39771 | 6.05E-11 | 5.75E-08 |
| MEG9 | 2.83443 | 4.49E-14 | 2.24E-10 | LINC01474 | -2.24764 | 2.67E-10 | 2.18E-07 |
| BX649601.1 | 1.124886 | 1.16E-07 | 5.09E-05 | AC139769.2 | -3.10429 | 2.62E-08 | 1.49E-05 |
| LINC01214 | 1.051004 | 2.18E-06 | 0.000485 | AC104031.1 | -1.95623 | 5.05E-08 | 2.62E-05 |
| HOXA11-AS | 1.473062 | 3.43E-05 | 0.004256 | AC069431.1 | -1.89819 | 2.27E-07 | 8.40E-05 |
| AC098869.2 | 2.185602 | 2.85E-12 | 5.43E-09 | TH2LCRR | -1.58174 | 2.36E-07 | 8.40E-05 |
| LINC01133 | 1.321311 | 7.40E-05 | 0.007962 | AL359853.1 | -2.33083 | 3.69E-07 | 0.000117 |
| LINC01882 | 1.620611 | 0.00011 | 0.010802 | Z93930.3 | -1.21647 | 5.25E-07 | 0.000158 |
| AL117190.1 | 2.177359 | 7.04E-08 | 3.35E-05 | SH3RF3-AS1 | -1.45216 | 6.98E-07 | 0.000199 |
| LINC02086 | 1.774533 | 9.62E-06 | 0.001567 | AC148476.1 | -2.04117 | 9.03E-07 | 0.000245 |
| Z99289.1 | 1.010418 | 0.000601 | 0.032977 | AC007099.1 | -2.74984 | 1.22E-06 | 0.000304 |
| HOXA-AS3 | 1.220775 | 0.000419 | 0.028584 | AP006285.1 | -1.77935 | 1.23E-06 | 0.000304 |
| TRBV11-2 | 1.045662 | 0.001038 | 0.047752 | LINC02334 | -2.29279 | 2.05E-06 | 0.000485 |
| LINC00487 | 1.137215 | 0.000192 | 0.016844 | MIR9-3HG | -1.90608 | 2.24E-06 | 0.000485 |
| AC122108.2 | 1.167812 | 7.02E-05 | 0.007735 | LINC01970 | -1.30815 | 2.29E-06 | 0.000485 |
| AC073115.1 | 1.884735 | 1.37E-07 | 5.57E-05 | AL445493.2 | -1.97111 | 2.40E-06 | 0.000489 |
| AC104051.2 | 1.141559 | 0.000478 | 0.029645 | ADIRF-AS1 | -1.12681 | 2.62E-06 | 0.000516 |
logFC, log2fold change; FDR, false discovery rate
Figure 1.
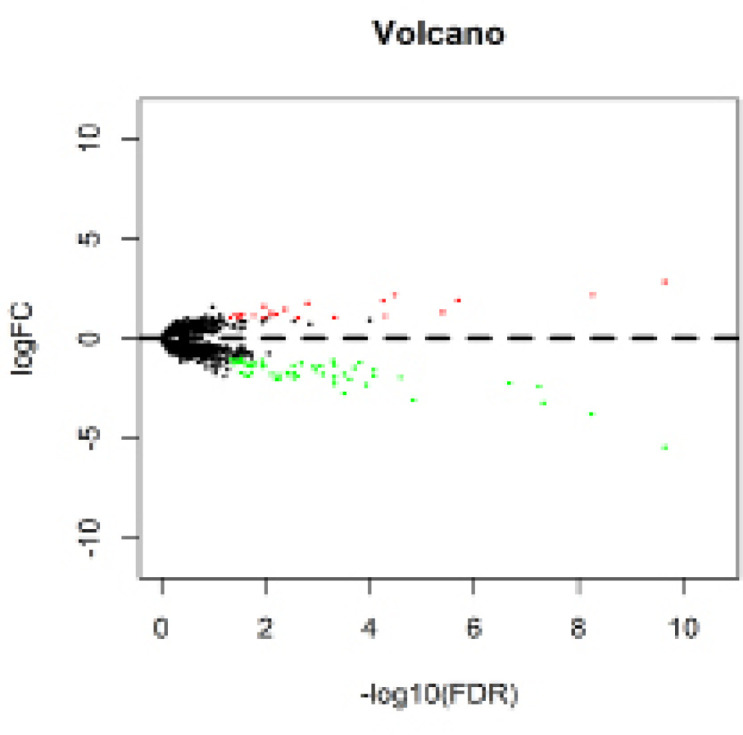
Volcano Map of the Differentially Expressed lncRNAs in HCC Patients between with Cirrhosis or without Cirrhosis. Red spots represent up-regulated genes, and green spots represent down-regulated genes
Figure 2.
Heatmap Based on the Differentially Expressed lncRNAs in HCC Patients between with Cirrhosis or without Cirrhosis
Risk scoring system based on lncRNA expression and OS prediction
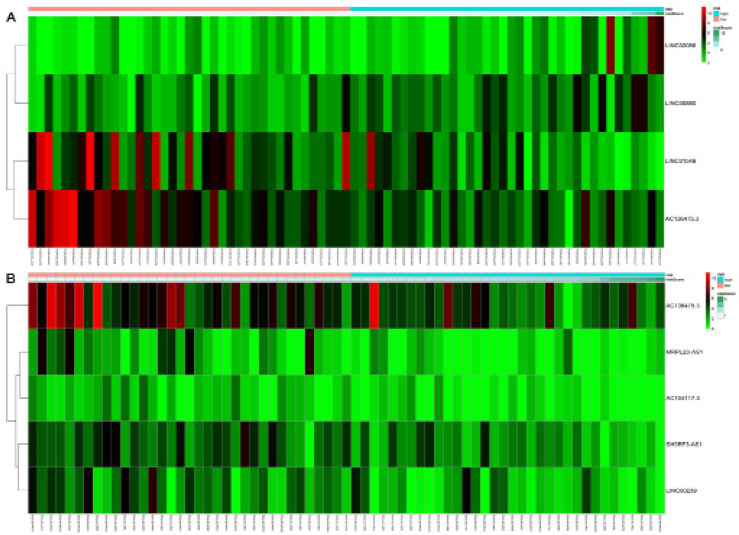
Univariate Cox analysis of patients with cirrhosis identified five lncRNAs (LINC02086, LINC00880, LINC01549, AC136475.3 and HOXA-AS3) significantly associated with OS (p < 0.05). Of these, multivariate regression analyses identified the first four as independent prognostic indicators of OS (Table 3). The resulting risk scoring system was
Table 3.
Four lncRNAs Correlated with OS of HCC Patients with Cirrhosis in the Best Statistical Model
| lncRNA | β | HR | z | P-value |
|---|---|---|---|---|
| LINC02086 | 0.335 | 1.398 | 2.51 | 0.012 |
| LINC00880 | 0.372 | 1.451 | 2.42 | 0.015 |
| LINC01549 | -0.161 | 0.851 | -1.39 | 0.165 |
| AC136475.3 | -0.309 | 0.734 | -1.82 | 0.068 |
HR, Hazard ratio
Risk score = (0.335 * LINC02086) + (0.372 * LINC00880) + (-0.161 * LINC01549) + (-0.309 * AC136475.3).
In this scoring system, higher expression of LINC02086 and LINC00880 predicted worse OS (β > 0), while higher expression of AC136475.3 and LINC01549 predicted better OS (β < 0).
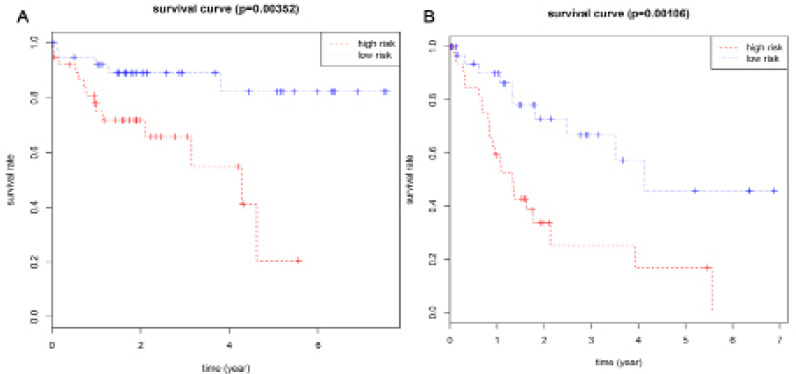
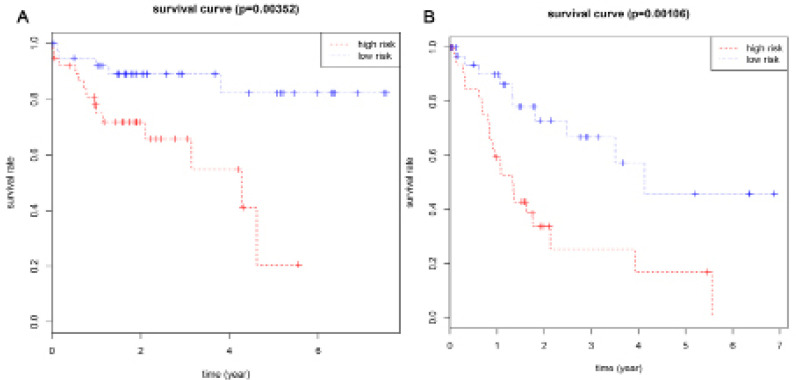
Based on their risk scores, patients with cirrhosis were classified as being at low or high risk of poor OS using the median risk score as a cut-off (Figure 3A). Kaplan–Meier curves showed that high-risk patients showed significantly lower OS rates at 3 years (65.8% vs 89.1%) and 5 years (20.6% vs 82.2%) (Figure 4A). The area under the ROC curve (AUC) for the risk scoring system was 0.818 (Figure 5A).
Figure 3.
The Non-Cluster Risk Heat Map of Risk Score for OS(A) and RFS (B) in HCC Patients with Cirrhosis. The risk score rises gradually from left to right
Figure 4.
Kaplan-Meier Survival Curves for OS (A) and RFS (B) in HCC Patients with Cirrhosis According to the Risk Cutoff Point
Figure 5.
ROC Curves Analysis of the Risk Score for OS(A) and RFS (B) in HCC Patients with Cirrhosis
After obtaining these promising results, we wished to check whether risk scoring based on lncRNAs could predict OS independently of clinicodemographic characteristics of the patients. This analysis is important given the heterogeneity of clinical presentation of HCC, and the range of factors that can influence prognosis. First we conducted univariate Cox regression to identify which clinical features were significantly associated with OS, and then we subjected this subset of factors to multivariate analysis.
Univariate analysis of patients with cirrhosis identified risk score and age as significantly associated with OS, but not body mass index (BMI), ethnicity, alpha fetoprotein (AFP), gender, hepatitis, alcohol consumption, cancer status, histology grade, new tumor event, pathology stage, family cancer history, residual tumor, or vascular invasion. Multivariate Cox regression confirmed that age was an independent predictor of poor OS [hazard ratio (HR) 2.86, 95%CI 1.09-7.56], as was risk score (HR 4.08, 95%CI 1.43-11.68) (Table 4).
Table 4.
Univariate and Multivariate Cox Regression Analysis for OS in HCC Patients with Cirrhosis
| Univariate Cox regression |
Multivariate Cox regression |
|||||||
|---|---|---|---|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI | ||
| Risk score (high/low) | 0.02 | 7.11 | 1.42 | 35.54 | 0.01 | 4.08 | 1.43 | 11.68 |
| Age (>60/≤60) | 0.02 | 7.34 | 1.3 | 41.57 | 0.03 | 2.86 | 1.09 | 7.56 |
| BMI | 0.21 | |||||||
| <25 | Reference | |||||||
| ≥25 | 0.48 | 0.07 | 3.13 | |||||
| Not reported | 25.56 | 0.37 | 1750.1 | |||||
| Race | 0.26 | |||||||
| Non-Asian | Reference | |||||||
| Asian | 0.25 | 0.05 | 1.31 | |||||
| AFP | 0.34 | |||||||
| ≤20ng/mL | Reference | |||||||
| >20ng/ml | 2.8 | 0.66 | 11.81 | |||||
| Not reported | 0.44 | 0.01 | 15.3 | |||||
| Gender(Male/Female) | 0.44 | 2.15 | 0.3 | 15.11 | ||||
| Hepatitis B or C | 0.1 | |||||||
| No | Reference | |||||||
| Yes | 0.17 | 0.03 | 0.87 | |||||
| Not reported | 0.28 | 0.01 | 10.28 | |||||
| Alcohol consumption | ||||||||
| No | 0.12 | Reference | ||||||
| Yes | 0.12 | 0.01 | 1.74 | |||||
| Histologic grade | 0.06 | |||||||
| G1-2 | Reference | |||||||
| G3-4 | 9.6 | 1.49 | 61.93 | |||||
| New tumor event | 0.79 | 0.69 | 0.05 | 10.32 | ||||
| Pathologic stage* | 0.7 | |||||||
| Stage I+II | Reference | |||||||
| Stage III+IV | 2.3 | 0.33 | 16.02 | |||||
| Cancer status | 0.37 | |||||||
| Tumor free | Reference | |||||||
| With tumor | 2.11 | 0.12 | 35.83 | |||||
| Not reported | 0.11 | 0 | 8.67 | |||||
| Family cancer history | 0.25 | |||||||
| No | Reference | |||||||
| Yes | 3.11 | 0.46 | 20.93 | |||||
| Not reported | 8.42 | 0.48 | 147.11 | |||||
| Residual tumor | 0.7 | |||||||
| R0 | 0.66 | 0.08 | 5.71 | |||||
| Vascular invasion | 0.21 | |||||||
| Negative | Reference | |||||||
| Positive | 3.61 | 0.87 | 14.91 | |||||
BMI, Body mass index; AFP, Alpha fetoprotein; HR, Hazard ratio; CI, Confidence interval; *TNM staging systems.
Risk scoring system based on lncRNA expression and RFS prediction
Univariate analysis identified the following 11 lncRNAs as significantly correlated with RFS of patients with cirrhosis: SH3RF3-AS1, AC104117.3, AC136475.3, LINC00239, MRPL23-AS1, LINC00494, LINC01970, MEG3, Z93930.3, MIR9-3HG and TRBV11-2. Multivariate analysis showed the first five to be independent prognostic indicators of RFS (Table 5). The resulting risk scoring system was
Table 5.
Five lncRNAs Correlated with RFS of HCC Patients with Cirrhosis in the Best Statistical Model
| lncRNA | β | HR | z | P-value |
|---|---|---|---|---|
| SH3RF3-AS1 | -0.273 | 0.7611 | -2.12 | 0.034 |
| MRPL23-AS1 | -0.2463 | 0.7817 | -1.76 | 0.078 |
| LINC00239 | -0.2425 | 0.7847 | -1.99 | 0.047 |
| AC136475.3 | -0.1312 | 0.877 | -1.46 | 0.143 |
| AC104117.3 | -0.3609 | 0.6971 | -1.73 | 0.083 |
HR, Hazard ratio
Risk score = (-0.2730 * SH3RF3-AS1) + (-0.2463 * MRPL23-AS1) + (-0.2425 * LINC00239) + (-0.1312 * AC136475.3) + (-0.3609 * AC104117.3), in which higher expression of all five lncRNAs was associated with better RFS (β < 0).
Based on their risk scores, patients with cirrhosis were classified as being at low or high risk of poor RFS using the median risk score as cut-off (Figure 3B). Kaplan–Meier curves showed that high-risk patients showed significantly lower RFS rates at 3 years (25.3% vs 66.8%) and 5 years (16.9% vs 57.3%) (Figure 4B). The AUC of the risk scoring system was 0.819 (Figure 5B).
As we did above for OS, we wished to check whether risk scoring based on lncRNAs could predict RFS independently of clinicodemographic characteristics of the patients. For patients with cirrhosis, our analysis showed that AFP and vascular invasion, but not risk score, correlated significantly with RFS in univariate analysis (Table 6). Frequencies of patients with certain clinicodemographic characteristics were stratified into groups at low or high risk of RFS; this analysis identified significant associations of the risk score with AFP, new tumor event and cancer status (Table 7).
Table 6.
Univariate Cox Regression Analysis for RFS in HCC Patients with Cirrhosis
| Variables | P-value | HR | 95% CI | |
|---|---|---|---|---|
| Risk score (high/low) |
0.92 | 0.93 | 0.23 | 3.79 |
| Age(>60/≤60) | 0.11 | 0.33 | 0.08 | 1.3 |
| BMI | 0.94 | |||
| <25 | Reference | |||
| ≥25 | 0.72 | 0.1 | 4.95 | |
| Not reported | 390.28 | 0 | 2.21E+173 | |
| Race | 0.35 | |||
| Non-Asian | Reference | |||
| Asian | 0.22 | 0.03 | 1.68 | |
| Not reported | 23.5 | 0 | 3.52E+226 | |
| AFP | 0.03 | |||
| ≤20ng/mL | Reference | |||
| >20ng/ml | 4.48 | 0.45 | 44.66 | |
| Not reported | 45.46 | 2.18 | 947.68 | |
| Gender (Male/Female) | 0.52 | 2.6 | 0.14 | 47.19 |
| Hepatitis B or C | 0.3 | |||
| No | Reference | |||
| Yes | 0.35 | 0.07 | 1.61 | |
| Not reported | 0.13 | 0.01 | 2.63 | |
| Alcohol consumption (Yes/No) | 0.41 | 0.42 | 0.05 | 3.27 |
| Histologic grade | 0.32 | |||
| G1-2 | Reference | |||
| G3-4 | 5.29 | 0.61 | 45.68 | |
| Not reported | 67.46 | 0 | 1.27E+173 | |
| New tumor event | 0.66 | |||
| No | 152118.71 | 0 | 3.36E+28 | |
| Yes | Reference | |||
| Pathologic stage* | 0.1 | |||
| Stage I+II | Reference | |||
| Stage III+IV | 5.34 | 1.16 | 24.58 | |
| Not reported | 0.27 | 0 | 4.97E+170 | |
| Cancer status | 0.1 | |||
| Tumor free | Reference | |||
| With tumor | 6.89 | 1.17 | 40.49 | |
| Not reported | 1.46 | 0 | . | |
| Family cancer history | 0.23 | |||
| No | Reference | |||
| Yes | 1.21 | 0.09 | 15.49 | |
| Not reported | 0.11 | 0.01 | 1.38 | |
| Residual tumor | 0.71 | 0.58 | 0.04 | 9.56 |
| Vascular invasion | <0.05 | |||
| Negative | Reference | |||
| Positive | 3.4 | 0.59 | 19.61 | |
| Not reported | 0.07 | 0 | 6.845 | |
Note: BMI, Body mass index; AFP, Alpha fetoprotein; HR, Hazard ratio; CI, Confidence interval; *TNM staging systems.
Table 7.
Stratified Analyses for the Risk Score of RFS in HCC Patients with Cirrhosis Using Chi-Square Test
| Risk score |
P-value | ||
|---|---|---|---|
| Variables | low-risk (n) | high-risk (n) | |
| Age | 0.19 | ||
| ≤60 | 23 | 17 | |
| >60 | 12 | 17 | |
| BMI | 0.9 | ||
| <25 | 17 | 17 | |
| ≥25 | 16 | 17 | |
| Race | 0.81 | ||
| Non-Asian | 15 | 16 | |
| Asian | 19 | 18 | |
| AFP | 0 | ||
| ≤20ng/mL | 16 | 26 | |
| >20ng/ml | 18 | 5 | |
| Gender | 0.28 | ||
| Female | 10 | 6 | |
| Male | 25 | 28 | |
| Hepatitis B or C | 0.82 | ||
| No | 9 | 9 | |
| Yes | 26 | 23 | |
| Alcohol consumption | 0.85 | ||
| No | 28 | 25 | |
| Yes | 7 | 7 | |
| Histologic grade | 0.39 | ||
| G1-2 | 22 | 24 | |
| G3-4 | 13 | 9 | |
| New tumor event | 0 | ||
| No | 25 | 11 | |
| Yes | 10 | 23 | |
| Pathologic stage* | 1 | ||
| Stage I+II | 29 | 29 | |
| Stage III+IV | 4 | 4 | |
| Cancer status | 0 | ||
| Tumor free | 27 | 13 | |
| With tumor | 7 | 21 | |
| Family cancer history | 0.49 | ||
| No | 20 | 21 | |
| Yes | 10 | 7 | |
| Residual tumor | 0.64 | ||
| R0 | 33 | 30 | |
| non-R0 | 2 | 4 | |
| Vascular invasion | 0.08 | ||
| Negative | 27 | 19 | |
| Positive | 7 | 13 | |
BMI, Body mass index; AFP, Alpha fetoprotein; *TNM staging systems
Functional analysis of mRNAs related to differentially expressed lncRNAs
Correlation in expression levels between the lncRNAs in our risk scoring systems and mRNAs from RNA-seq data was analyzed using Pearson correlation, and several related mRNAs were identified (Supplementary Tables 1-2). These related mRNAs were analyzed using the KEGG signal pathway databases (Figure 6). Functional enrichment analysis showed that mRNAs strongly related to the lncRNAs in our risk scoring systems were involved mainly in the pathway of Wnt signal (Supplementary Figure 1) and cytokine-cytokine receptor interaction (Supplementary Figure 2).
Figure 6.
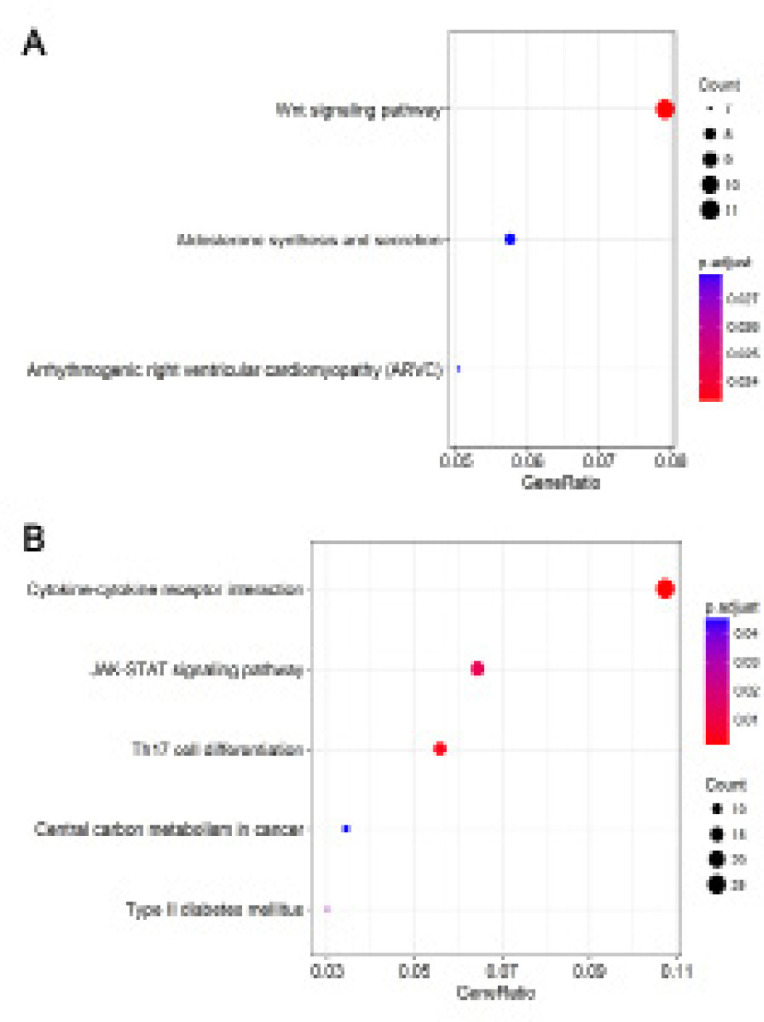
The Top 10 Significantly Enriched Enriched KEGG Pathways of the Highly Related Genes of the lncRNAs of the Risk Scoring Systems for OS(A) and RFS (B) in HCC Patients with Cirrhosis
Discussion
HCC is associated with high morbidity and poor prognosis, and the factors that contribute to poor outcomes are likely to vary substantially in the presence or absence of cirrhosis (Grazi et al., 2003; Gassmann et al., 2010; Wang et al., 2013; Techathuvanan et al., 2015). Therefore we developed an lncRNA-based scoring system to assess a patient’s risk of poor OS or RFS in the presence of cirrhosis. Our results establish the potential of four or five lncRNAs to serve as prognostic biomarkers with respective AUCs of 0.818 and 0.819, suggesting reasonable predictive power.
After adjusting for other clinical variables, uni- and/or multivariate Cox analyses showed the lncRNA-based scoring system to be significantly associated with OS of HCC patients with cirrhosis, suggesting the lncRNA-based scoring system can serve as a significant independent predictor. However, the system for predicting RFS was not significantly associated with RFS in univariate analysis. Therefore, we stratified patients by low or high risk score and found the following factors to influence risk: AFP, new tumor event and cancer status. Our results suggest that these factors should be taken into account when predicting RFS in HCC patients with cirrhosis. Furthermore, other clinical factors also influenced the survival of HCC patients in our sample. Our study showed that age was an independent factor for the prediction of OS in HCC patients with cirrhosis, which was similar with previous studies (Wang et al., 2013; Techathuvanan et al., 2015; Liu et al., 2018). To gain insight into the prognostic significance of our risk scoring systems, we analyzed the molecular functions of genes highly related to these lncRNAs. We found that the lncRNAs associated with prognosis in HCC are involved mainly in Wnt signaling and cytokine-cytokine receptor interactions.
Several previous studies constructed risk scoring systems to predict the prognosis of HCC patients (Gu et al., 2018; Liao et al., 2018; Ma et al., 2018; Shi et al., 2018; Sui et al., 2018; Wu et al., 2018b; Zhao et al., 2018; Bai et al., 2019; Yan et al., 2019; Ye et al., 2019; Zhang et al., 2019). However, all these risk scoring systems are based on lncRNAs differentially expressed between HCC and normal samples, while our systems are based on lncRNAs differentially expressed between HCC patients with or without cirrhosis. Therefore, our systems may be more specific for HCC involving cirrhosis. One previous study identified a four-lncRNA signature as a prognostic indicator in cirrhotic HCC (Ma and Deng, 2019), with no overlap with our four or five-lncRNA signatures. Those authors extracted their data from a different expression profile dataset, and they used only stratified analyses based on clinical characteristics to validate the prognostic significance of their risk scoring system. In contrast, we applied multivariate Cox regression, which may be more rigorous. This may help explain why none of the lncRNAs in our risk scoring systems occurs in the previously published risk scoring systems for HCC prognosis. The importance of developing prognostic tools for HCC with cirrhosis is underscored by the observation of several microRNAs differentially expressed between patients with or without cirrhosis, particularly mir-149 (Mei et al., 2018), miR-24 and miR-27a (Salvi et al., 2013). These differences may help explain the different prognosis in the two types of HCC. Together, these studies suggest that non-coding RNAs may hold the key to unlocking new strategies in the diagnosis, treatment and management of HCC in the presence of cirrhosis.
Our results should be interpreted with caution in light of several limitations. First, we could not include type of HCC treatment in our multivariate Cox regression for lack of data, and this variable may affect OS and RFS. Second, some Cox analyses may have been biased because certain clinical data were unavailable for some patients. Third, although the AUC was 0.819 for the system to score risk of poor RFS in HCC patients with cirrhosis, univariate Cox analysis showed that the risk score was not a significant independent predictive factor of HCC prognosis. This model, in particular, should be re-examined carefully with a larger dataset. Last but not least, we could not divide the samples into training and test dataset, which may result in some bias. Indeed, future work should validate all our findings in larger, preferably multi-site patient populations.
In conclusion, we have defined lncRNA-based risk scoring systems that can effectively predict OS in HCC patients with cirrhosis. Our work highlights the potential usefulness of lncRNAs as prognostic biomarkers, and it provides several leads for the development of novel therapies.
Acknowledgments
This work was supported by The Young and Middle-aged Teacher Basic Ability Enhancement Project of Guangxi University (2018KY0111), Innovation Project of Guangxi Graduate Education (YCBZ2019036)- Guangxi First-class Discipline Project for Basic medicine Sciences (GXFCDP-BMS-2018) and the National Natural Science Foundation of China (81860504). Guangxi Natural Science Foundation (2016GXNSFCB380003 and 2015GXNSFCA139014).
Appendix
Supplementary Table 1. LncRNA-mRNA co-expression analysis for OS in HCC with cirrhosis.
Supplementary Table 2. LncRNA-mRNA co-expression analysis for RFS in HCC with cirrhosis.
Supplementary Figure 1. Wnt signaling pathway
Supplementary Figure 2. The pathway of cytokine-cytokine receptor interaction
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234:18837–48. doi: 10.1002/jcp.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2018;0:1–31. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–55. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Liu B. Identification of a RNA-Seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163–72. doi: 10.12659/MSM.902615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- Gassmann P, Spieker T, Haier J, et al. Prognostic impact of underlying liver fibrosis and cirrhosis after curative resection of hepatocellular carcinoma. World J Surg. 2010;34:2442–51. doi: 10.1007/s00268-010-0655-5. [DOI] [PubMed] [Google Scholar]
- Grazi GL, Cescon M, Ravaioli M, et al. Liver resection for hepatocellular carcinoma in cirrhotics and noncirrhotics Evaluation of clinicopathologic features and comparison of risk factors for long-term survival and tumour recurrence in a single centre. Aliment Pharmacol Ther. 2003;17:119–29. doi: 10.1046/j.1365-2036.17.s2.9.x. [DOI] [PubMed] [Google Scholar]
- Gu J, Zhang X, Miao R, et al. A three-long non-coding RNA-expression-based risk score system can better predict both overall and recurrence-free survival in patients with small hepatocellular carcinoma. Aging (Albany NY) 2018;10:1627–39. doi: 10.18632/aging.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Xiang B, Liu Y, et al. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi: 10.1016/j.canlet.2018.08.024. [DOI] [PubMed] [Google Scholar]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–91. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Yang C, Huang R, et al. Identification of potential prognostic long non-coding RNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cell Physiol Biochem. 2018;48:1854–69. doi: 10.1159/000492507. [DOI] [PubMed] [Google Scholar]
- Liu H, Cen D, Yu Y, et al. Does fibrosis have an impact on survival of patients with hepatocellular carcinoma: evidence from the SEER database? BMC Cancer. 2018;18:1125. doi: 10.1186/s12885-018-4996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Deng C. Identification of a novel four-lncRNA signature as a prognostic indicator in cirrhotic hepatocellular carcinoma. Peer J. 2019;7:e7413. doi: 10.7717/peerj.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Luo T, Dong D, et al. Characterization of long non-coding RNAs to reveal potential prognostic biomarkers in hepatocellular carcinoma. Gene. 2018;663:148–56. doi: 10.1016/j.gene.2018.04.053. [DOI] [PubMed] [Google Scholar]
- Mei Y, You Y, Xia J, et al. Identifying differentially expressed MicroRNAs between cirrhotic and non-cirrhotic hepatocellular carcinoma and exploring their functions using bioinformatic analysis. Cell Physiol Biochem. 2018;48:1443–56. doi: 10.1159/000492254. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi A, Abeni E, Portolani N, et al. Human hepatocellular carcinoma cell-specific miRNAs reveal the differential expression of miR-24 and miR-27a in cirrhotic/non-cirrhotic HCC. Int J Oncol. 2013;42:391–402. doi: 10.3892/ijo.2012.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–51. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YM, Li YY, Lin JY, et al. The discovery of a novel eight-mRNA-lncRNA signature predicting survival of hepatocellular carcinoma patients. J Cell Biochem. 2018;0:1–10. doi: 10.1002/jcb.28028. [DOI] [PubMed] [Google Scholar]
- Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Miao Y, Han J, et al. Systematic analyses of a novel lncRNA-associated signature as the prognostic biomarker for Hepatocellular Carcinoma. Cancer Med. 2018;0:1–12. doi: 10.1002/cam4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techathuvanan K, Srisajjakul S, Pongpaibul A, et al. Comparison between disease free survival of hepatocellular carcinoma after hepatic resection in chronic hepatitis B patients with or without cirrhosis. J Med Assoc Thai. 2015;98:334–42. [PubMed] [Google Scholar]
- Wang Q, Fiel MI, Blank S, et al. Impact of liver fibrosis on prognosis following liver resection for hepatitis B-associated hepatocellular carcinoma. Br J Cancer. 2013;109:573–81. doi: 10.1038/bjc.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Pan C, Wei X, et al. lncRNA KRAL reverses 5-fluorouracil resistance in hepatocellular carcinoma cells by acting as a ceRNA against miR-141. Cell Commun Signal. 2018a;16 doi: 10.1186/s12964-018-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang PS, Wang BG, et al. Genomewide identification of a novel six-LncRNA signature to improve prognosis prediction in resectable hepatocellular carcinoma. Cancer Med. 2018b;7:6219–33. doi: 10.1002/cam4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhou C, Guo K, et al. A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma. J Cell Biochem. 2019;120:213–23. doi: 10.1002/jcb.27321. [DOI] [PubMed] [Google Scholar]
- Ye J, Zhang J, Lv Y, et al. Integrated analysis of a competing endogenous RNA network reveals key long noncoding RNAs as potential prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:13810–25. doi: 10.1002/jcb.28655. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Li N, Huang Z, et al. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. 2018;9:1184. doi: 10.1038/s41419-018-1231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ouyang Y, Huang Y, et al. Comprehensive bioinformatics analysis reveals potential lncRNA biomarkers for overall survival in patients with hepatocellular carcinoma: an on-line individual risk calculator based on TCGA cohort. Cancer Cell Int. 2019;19:174. doi: 10.1186/s12935-019-0890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QJ, Zhang J, Xu L, et al. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J Gastroenterol. 2018;24:3426–39. doi: 10.3748/wjg.v24.i30.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]