Abstract
Environmental exposures have long been known to impact public health and safety. For example, exposures to airborne particulates, heavy metals in water, or certain industrial chemicals can contribute to aging and to risk of developing cancer and other diseases. Environmental factors can impact health in a variety of ways, but a key concern is DNA damage, which can lead to mutations that cause cancer. Cancer can take years to develop following chemical exposure; however, one way to predict carcinogenicity in a more practical time frame is by studying the chemical’s ability to induce DNA damage. The comet assay (or single-cell gel electrophoresis assay) has been used successfully for genotoxicity testing. The comet assay allows for the detection of DNA strand breaks via analysis of DNA migration during electrophoresis. Previously, the Engelward laboratory, in collaboration with the Bhatia laboratory, developed the CometChip for measurements of DNA damage and repair. The CometChip is a high- throughput comet assay that improves user reproducibility and significantly shortens total assay time. Here, we describe how the high-throughput CometChip platform can be used to measure DNA damage in established cell lines, animal models, and human samples. We also discuss technical challenges associated with these studies and provide recommendations on how to achieve optimal results for researchers interested in adopting this assay.
Keywords: CometChip, comet assay, DNA damage, DNA repair
INTRODUCTION
Our DNA is constantly exposed to exogenous and endogenous agents in our environment, resulting in structural changes such as base modifications or breaks to the DNA backbone (Figure 1).1–5 For example, reactive oxygen and nitrogen species (RONS) released during an inflammatory response can damage proteins and DNA.6,7 DNA damage can be toxic and mutagenic8 when the replication fork encounters unrepaired lesions. For instance, if an unrepaired 8-oxoguanine lesion (induced by ROS damage) is present during replication, the damaged guanine can now be paired effectively with an adenine, leading to G to T transversions.9,10 Over time, the accumulation of mutations drives the development of cancer.
Figure 1.
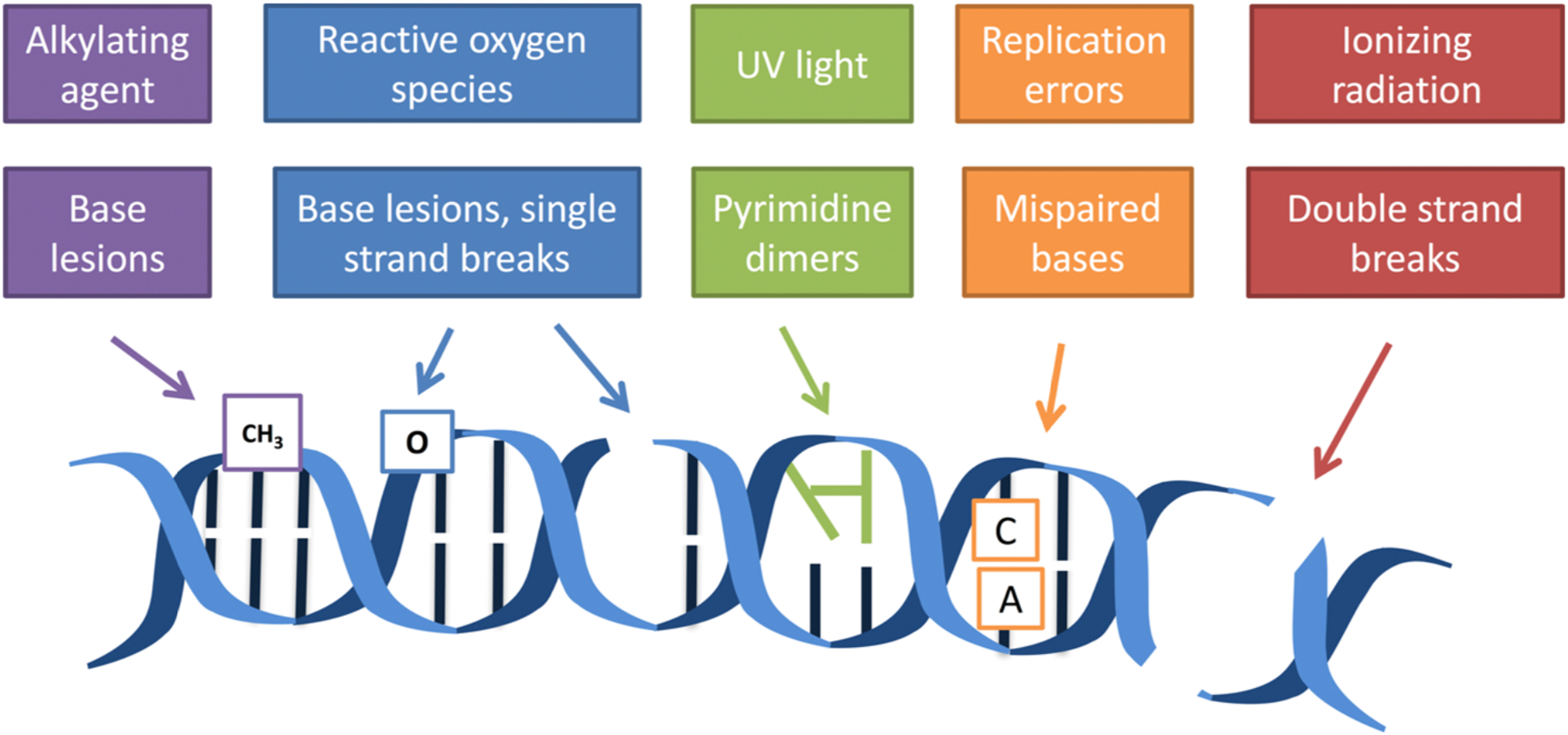
DNA is damaged by both external and internal agents. The lower boxes are examples of some of the classes of damage that can result from the exposures listed in the upper boxes.
Cancer can take years to develop after initial exposure to a carcinogen, making it important to utilize predictive assays that can rapidly assess carcinogenicity when early intervention is still possible. Since it is known that structural changes to DNA can lead to mutations and eventually cancer, studies quantifying DNA damage have been used for this purpose. Genotoxicity testing can play important roles in regulatory decisions relevant to public health and drug safety. Studying DNA damage and repair also has applications outside of disease prevention. For example, many chemotherapeutic agents rely on modulating DNA damage and repair pathways to kill tumor cells, so genotoxicity assays could lead to more effective pharmaceuticals and could also be valuable for personalized medicine (wherein drug selection and dose can be optimized according to the susceptibility of tumor cells to drug-induced genotoxicity).11–13
Understanding how DNA damage drives disease is an important step in improving public health, particularly from environmental exposures that may be chronic and that may impact many communities at once. External factors such as where people live, what they eat, and other lifestyle aspects can all contribute to the risk of developing disease. In addition, internal factors such as interindividual variations in DNA repair capacity play a role. Studying the mechanisms of how exposures impact DNA damage and disease, as well as learning about how different people in a population respond to genotoxic environmental chemicals, will help to promote public health while also providing avenues to protect those most at risk.
In this perspective, we describe how the CometChip (a high-throughput comet assay) has been used to perform studies of DNA damage and repair. We anticipate this tool will be valuable for researchers who routinely analyze large sample sizes (e.g., multichemical screens or interindividual variation studies). We begin by summarizing use of the established comet assay method and describing how a platform modification (the microwell array) simplifies sample handling while retaining the comet assay’s utility for studying various classes of DNA damage. We then discuss technical considerations for designing CometChip experiments, with a focus on the cell loading step that is unique to this platform, and we provide additional guidance for achieving consistent results. While many published studies that utilize the CometChip focus on analysis of cultured cell lines, we will also discuss how the CometChip enables high-throughput studies of in vivo samples collected from tissues and blood as well as challenges associated with performing these studies. Finally, we conclude by comparing this platform to other methods for DNA damage analysis.
THE COMET ASSAY
DNA damage measurements can be performed on mammalian cells using the comet assay.14–17 The comet assay is based on the principle that physical breaks to DNA result in loss of DNA supercoiling and fragmentation. When electrophoresed, these DNA loops and fragments migrate further through an agarose matrix compared to undamaged DNA. The degree to which the DNA migrates can be visualized by staining the DNA with a fluorescent dye and quantifying the amount of DNA in the comet tail (Figure 2). The comet assay allows for the detection of single-strand breaks, double-strand breaks, damage that is converted to strand breaks under assay conditions (all of which potentiate DNA migration), as well as interstrand cross-links, which can be detected by their ability to deter DNA migration.
Figure 2.
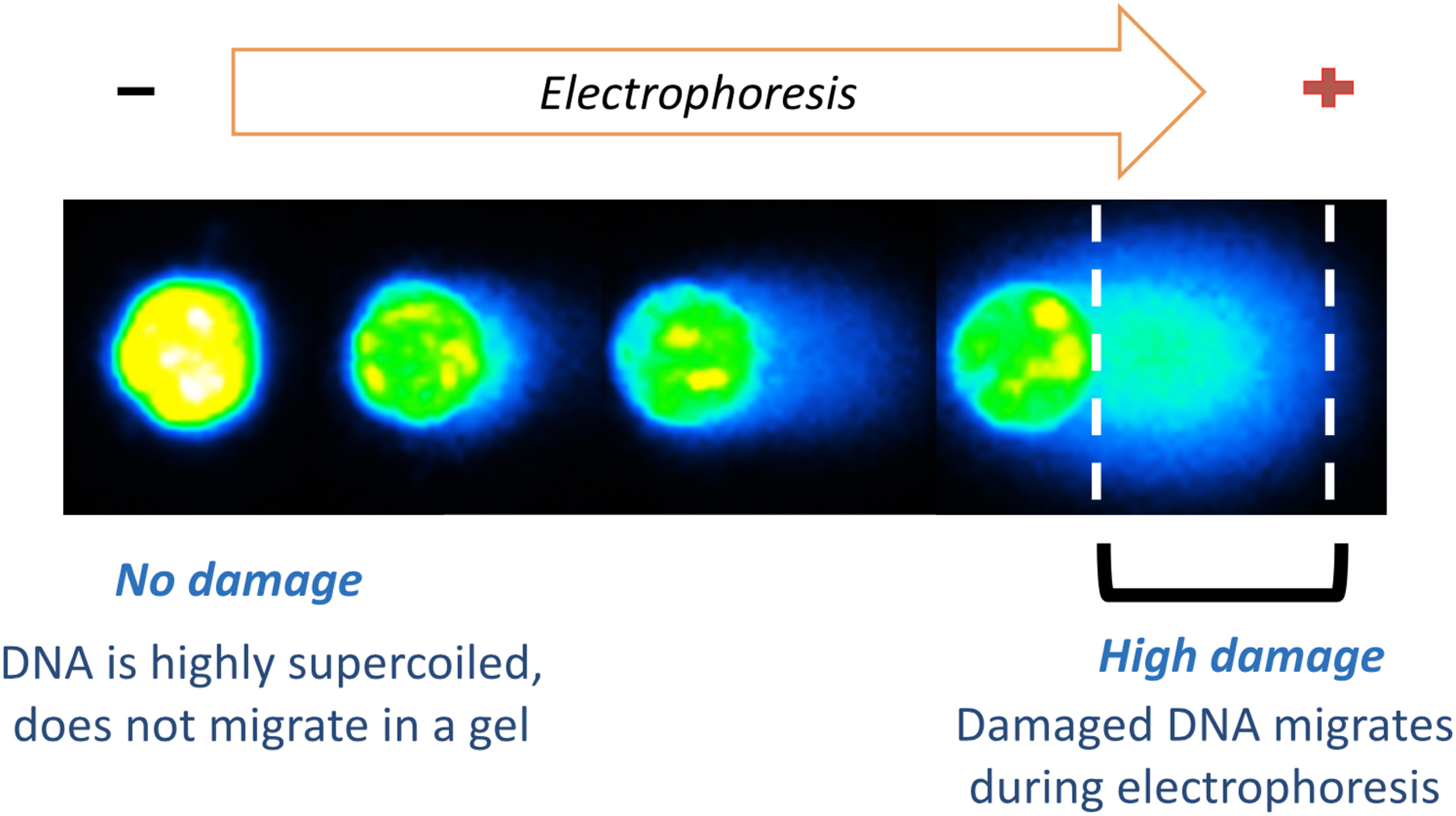
Undamaged DNA remains highly supercoiled and does not migrate significantly during electrophoresis (left). DNA damage leads to release of DNA supercoiling and fragmentation, enabling migration and the formation of the comet tail.
The traditional comet assay is performed using glass slides. Specifically, cells are suspended in agarose and allowed to adhere to the slide. The slide is then submerged into a high-salt, high-pH buffer (pH 10) containing detergent to lyse the cell membranes and denature proteins, leaving behind a nucleoid of DNA. Following cell lysis, the DNA is denatured in a higher pH buffer (pH >13) causing the DNA to unwind. DNA damage is revealed during electrophoresis when loops and fragments resulting from structural damage are pulled through the agarose. The DNA is stained with a fluorescent dye, and comets are scored to determine the extent of damage.
The comet assay has been used extensively over the years to evaluate the DNA damaging effects of chemicals (both in vitro and in vivo),18–22 to study DNA repair,23–25 and to evaluate environmental exposures in human samples and biomonitoring studies.26–29 However, use of this assay can be limited by throughput. First, the comet assay is traditionally performed on glass slides, with each experimental condition requiring its own slide. This makes handling large experiments and testing multiple conditions tedious and cumbersome. Second, when the cells are suspended in agarose, they are randomly embedded into the matrix. This can lead to overlapping comets, which complicates scoring. Finally, comets traditionally have been imaged one at a time, which is time-consuming. Together, these characteristics make the comet assay difficult to use in studies with large sample sizes, such as dose-response experiments in multichemical screens, or in studies where background noise may vary, for example, in samples collected from human subjects.
THE COMETCHIP
To minimize sample-to-sample variation and to reduce comet assay labor, the CometChip was developed by the Engelward laboratory and Bhatia laboratory at MIT.30–32 The CometChip is based on the same underlying principles as the traditional comet assay (Figure 3). The high-throughput capability is achieved by capturing cells within an array of agarose microwells. This eliminates overlapping comets, which enables more analyzable comets per mm2, and allows the assay to be performed in a 96-well format. In addition, cells are on the same plane, so dozens of comets can be analyzed from a single image. Combined with automated analysis, the CometChip assay is faster than the traditional comet assay by at least an order of magnitude.
Figure 3.
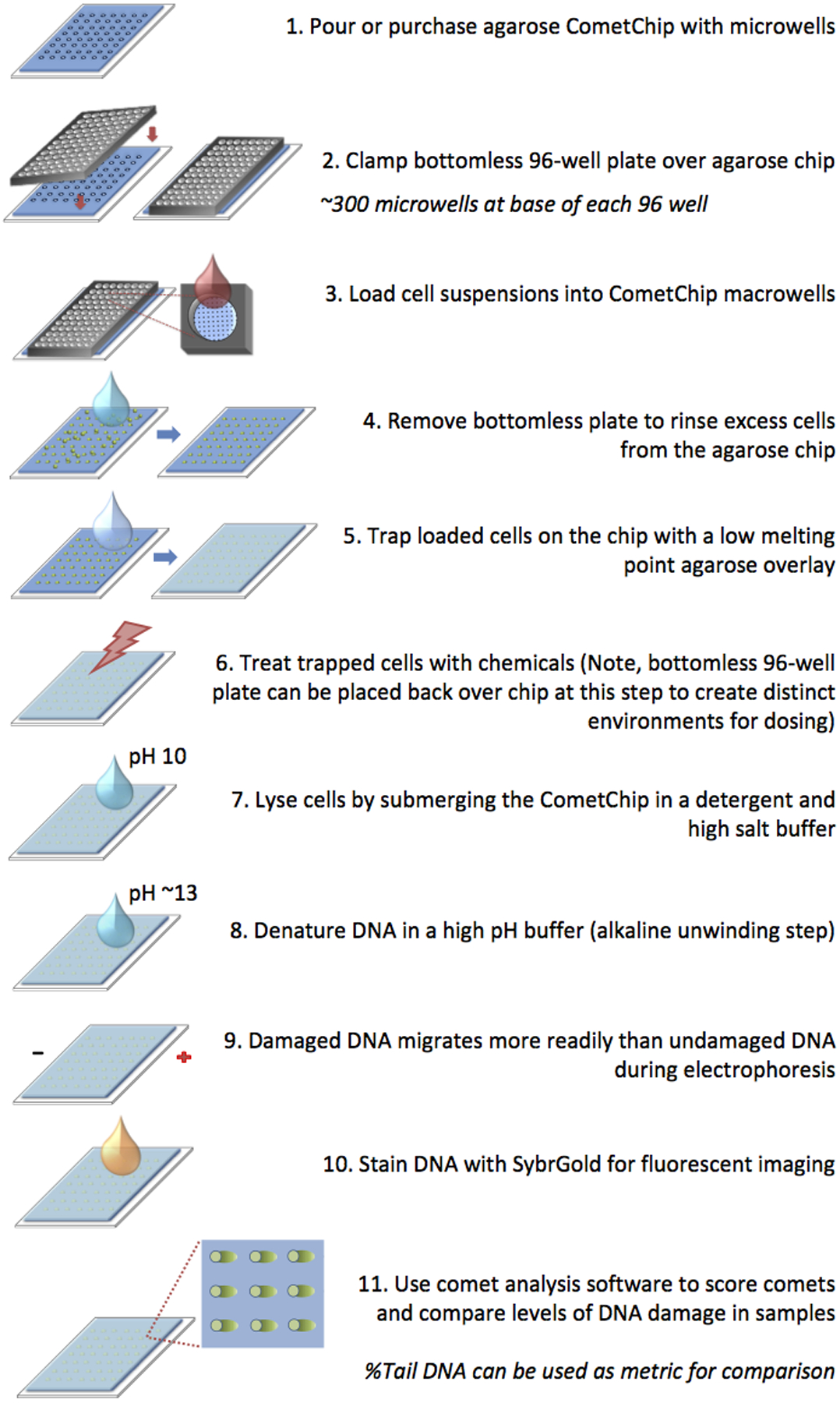
CometChip protocol involves loading cells into microwells, removing excess cells, and trapping cells with low melting point agarose. Shown here, cells are treated after being loaded into the CometChip. Cells can also be treated prior to loading into the CometChip (not shown).
Since the CometChip is a high-throughput version of the comet assay, it can be used to detect the same classes of DNA damage (Table 1), following exposure to a variety of DNA damaging agents (see below for a list of applications).
Table 1.
Variations of the alkaline CometChip
| Type of Damage | Comet Assay Modifications | Description |
|---|---|---|
| Single-strand breaks | No modifications are necessary to detect strand breaks (and damage converted to strand breaks) under standard assay conditions. | Cells can be lysed immediately to assess levels of spontaneous damage or exposed to a DNA damaging agent and assessed following exposure. |
| Base Lesions | Enzyme incubations are performed after cell lysis and prior to electrophoresis. DNA nicks are created at sites of base lesions . | Base damage caused by ROS or UV exposure may not be readily detected using the comet assay. However, the presence of a damaged base can be revealed using enzymes that cleave the DNA at sites of damage (e.g., using DNA repair enzymes).17 |
| Bulky Adducts | Co-exposing cells to HU/AraC traps NER repair intermediates,33,34 creating a persis- tent strand break that can be detected using the comet assay.35 | DNA adducts, bases to which chemical moieties have been added, can be formed directly (e.g., cisplatin),36 or indirectly following metabolic activity (e.g., benzo[a]pyrene).37 Due to the short half-life of single-strand breaks during nucleotide excision repair (NER), the standard comet assay is not very sensitive to the presence of bulky lesions. Assay sensitivity can be improved by trapping NER intermediates with hydroxyurea (HU) and 1-β-D-arabinofuranosyl cytosine (AraC). |
| Cross-links | Prior to electrophoresis, exposing the DNA to irradiation38 (or another DNA damaging agent)39,40 creates additional breaks in the DNA. Migration of this damage is inhibited by the presence of cross-links. | Cross-links, such as those caused by chemotherapeutics (e.g., Mytomicin C)2,5 deter DNA migration, instead of enabling DNA migration. To reveal whether a compound induces cross-links, cells are exposed to agents that induce single-strand breaks, and the extent to which migration is deterred reveals the presence of interstrand cross-links.41,42 |
Examples of CometChip applications:
Genotoxic effects of RONS in mouse embryonic fibroblasts (MEFs) exposed to S-Nitrosoglutathione (GSNO), 43 AS52 cells treated with alkylanilines, 44 and TK6 cells exposed to H2O2 45
Alkylation damage in Chinese hamster ovary (CHO) cells 48 and in HepaRG and HepG2 cells 49
DNA damage associated with asthma in BEAS-2B and A549 cells50,51
Damage induced by influenza in infected cells 52
Oxidative DNA damage induced by live cell imaging 53
Cross-links induced by an intestinal microbe (colibactin) 38
Damage in blood samples collected from turtles 54
Strand breaks in MEFs resulting from co-exposure to Bisphenol A and camptothecin 55
Response of ovarian cancer cells to a poly (ADP-ribose) polymerase (PARP) inhibitor 56
COMETCHIP TERMINOLOGY
In Vitro vs In Vivo Comet.
In this perspective, we refer to experiments performed using cultured cell lines as in vitro CometChip experiments. In this context, we will focus on experiments where cellular responses to DNA damage are assessed in live cells (rather than assessment of cell extracts, which is more akin to a biochemical assay, as opposed to a cell- based assay). The term in vivo CometChip will be used to describe experiments where the tissues or liquid biopsies are collected from animals or humans, and the comet assay is performed on the cells isolated from these samples.
Alkaline vs Neutral Comet Assay.
There are two main versions of the comet assay: neutral and alkaline.57 The neutral comet assay is more specific to double-strand breaks, while the alkaline assay has been used to detect single-strand breaks, abasic sites, and alkali-sensitive sites. In addition, there is a modified alkaline comet assay wherein enzymes are used to reveal the presence of specific classes of DNA damage. For the remaining discussion, we will focus on CometChip applications using the more commonly used alkaline comet assay (although, we have shown it is also possible to study DSBs under neutral conditions using the CometChip).32
%Tail DNA.
Various metrics have been used to analyze DNA damage levels using the comet assay (e.g., %Tail DNA, tail length, and olive tail moment).58 For the alkaline CometChip assay, we generally use %Tail DNA as a metric for quantifying damage. %Tail DNA is defined during image analysis as the intensity of the comet tail divided by the total cell fluorescence. This analysis method is self-calibrating, making this metric effective even when there are several cells per comet, as described below.
Detection of Base Lesions vs Downstream Intermediates.
While changes to base structure are not detectable using the comet assay (as they are not a break in the phosphodiester backbone), enzymatic processes that convert base lesions into downstream DNA repair intermediates lead to the formation of detectable damage (see Table 1 for additional details on enzymatic CometChip experiments). Specifically, for base excision repair (BER), after removal of the damaged bases, an abasic site and, subsequently, a single-strand break is formed. These are detectable lesions. Thus, oxidative base damage can be studied using the comet assay, since damaged bases are processed by the BER pathway (Figure 4).
Figure 4.
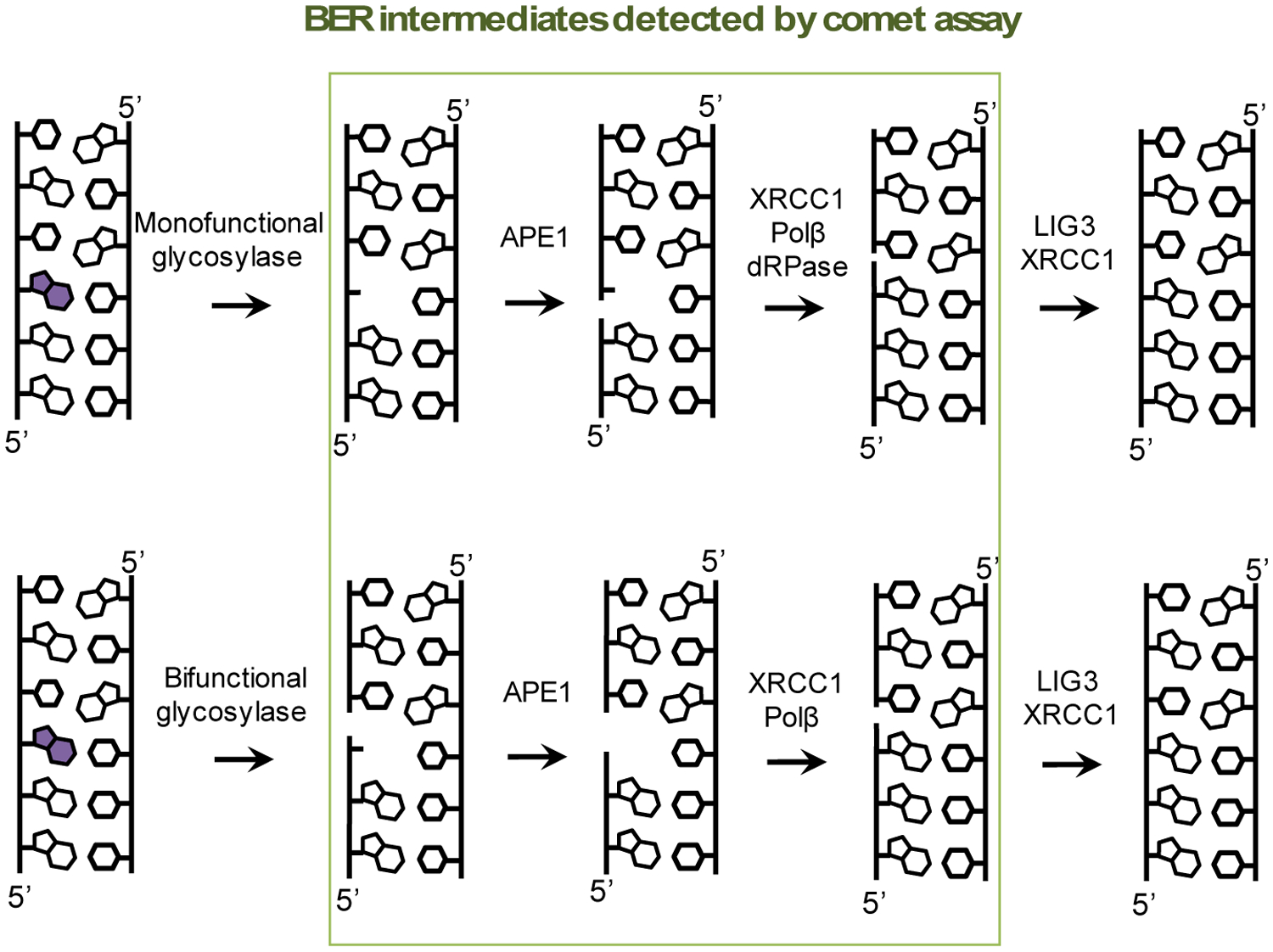
Base excision repair (BER) intermediates can be detected using the alkaline comet assay. During BER, DNA repair enzymes create abasic sites and single-strand breaks (boxed in green) that can be measured. Illustrated here is a simplified version of BER and is one of several pathways.
Microwells vs Macrowells.
A CometChip macrowell is defined as the area of an individual well in a standard 96-well plate. Macrowells are created when a bottomless 96-well plate is clamped over the agarose chip. Each macrowell creates an individual environment, allowing the CometChip to be used in a 96-well format during experiments. The base of each macrowell contains ~300 microwells. Most experiments are performed using microwells that are ~30–50 μm in diameter. Note that during loading, multiple cells may settle into each individual microwell (Figure 5). Previous studies show that multicell comets are readily analyzable using in-house or commercial software, and they yield comparable results compared to single-cell comets.30
Figure 5.
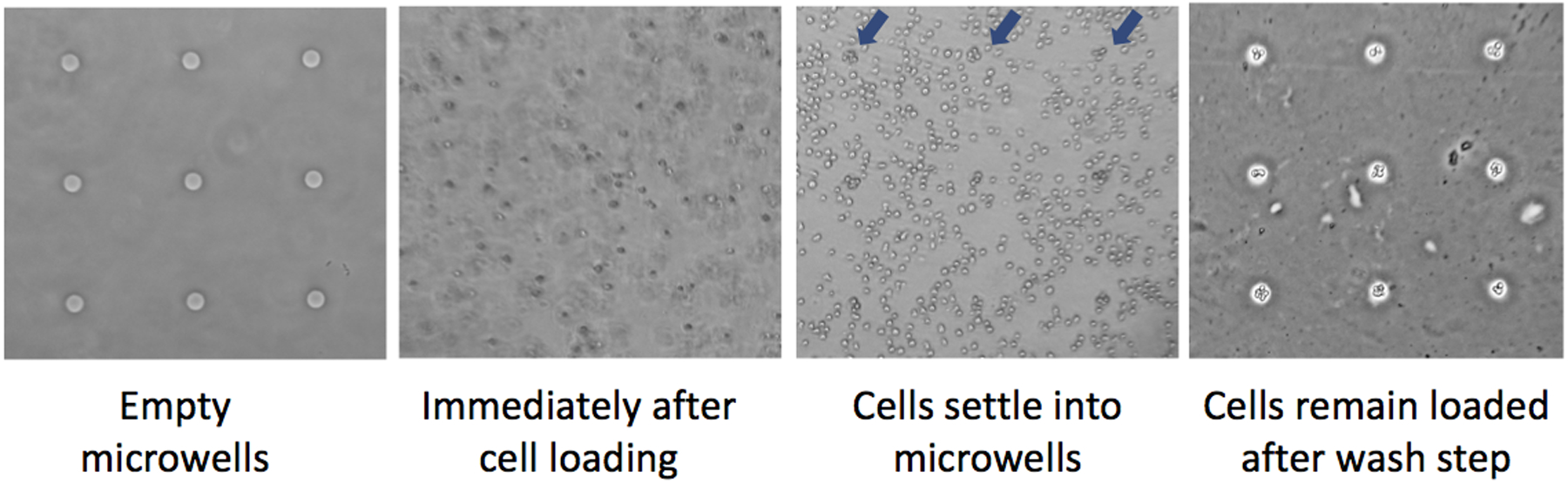
At the base of each macrowell on a 96-well plate are ~300 microwells in a grid. Pictured here are microwells that are ~40 μm in diameter and spaced 240 μm apart. Cells are loaded into each macrowell, and they settle into the microwells over time. Loading efficiency can be checked using a microscope by visualizing a grid of loaded microwells among the off-grid/excess cells (examples of loaded wells are marked by the blue arrows). Following the wash step, excess cells are removed, leaving behind an array of patterned cells. For these images, 40,000 TK6 cells were added to each macrowell in a volume of 80 μL. Cells remain trapped in wells following 20 min of loading time. Note that loading time can be optimized for different cell types, but can be as low as 5 min.
COMETCHIP REPRODUCIBILITY AND VARIATION
It is known that many factors contribute to comet assay variability (e.g., cell line, electrophoresis conditions, and comet scoring).35,59–61 Different approaches have been used to address these factors. For example, to minimize electrophoresis variation, Cassano et al.59 developed a tank to provide a greater level of control over electrophoresis conditions. The CometChip also decreases the variation introduced during electrophoresis by condensing what originally required multiple slides run across many electrophoresis tanks into a single chip where all 96 wells are run simultaneously. Previous studies of the CometChip30,45,32 have demonstrated that condensing samples within a single chip does in fact reduce the amount of slide-to-slide variation reported with the traditional comet assay. For example, when Weingeist et al.32 compared slide-to-slide variability in traditional comet analysis vs macrowell-to- macrowell variability in neutral CometChip analysis, a coefficient of variation (CV) of ~20% was observed using the traditional comet assay, while a CV of ~5% was observed using the CometChip. In addition, comet-to-comet variability decreased, with fewer comets required for analysis.
The comet scoring process may introduce operator bias if the user manually deletes comets or adjusts head/tail cutoffs following image analysis. CometChip decreases user bias by removing operator intervention from the comet scoring process. An in-house MATLAB CometChip analysis program, described in the Supporting Information of Wood et al., is available upon request.30 We have also observed that while absolute levels of %Tail DNA varied when CometChip results were analyzed using two separate programs (Trevigen’s comet analysis software vs in-house Matlab analysis), the trends in DNA damage remained similar.
IN VITRO COMETCHIP EXPERIMENTS: TECHNICAL DETAILS
Creating Agarose Microwell Arrays.
CometChips with 30 μm-diameter microwells are commercially available from Trevigen (catalog number: 4260-096-01). Reusable polydimethylsiloxane (PDMS) stamps can also be created in-house, and this fabrication process is described by Weingeist et al.32 The stamp contains a grid of microposts that are set into molten agarose, creating an array of microwells for cell loading. Note that the PDMS stamps can be secured to a rigid surface (e.g., a glass plate). This allows for a fixed distance to be maintained between the microposts and the Gelbond film, while the agarose is cooling, creating chips with uniform thickness (~0.3 mm).
To pour a CometChip, a Gelbond film (trimmed to the size of a standard 96-well plate) is placed hydrophilic side up into a rectangular tray (e.g., the lids of Nunc Rectangular Dishes [Thermo Scientific, 267060]). (Note that if using these lids for CometChip fabrication, the feet at the bottom corners of each lid should be cut away with a razor to create a flat surface). Prior to placing the Gelbond film into the tray, it is recommended that ~3 mL of molten agarose be added to the center of the tray followed by pressing the hydrophobic side of the Gelbond into the agarose. This helps secure the Gelbond film to the tray for the remaining steps.
Once the film is secure, molten 1% normal melting point agarose is poured over the Gelbond. When using the Nunc lids, ~12 mL of agarose is sufficient to cover the entire area. After molten agarose is added, the PDMS stamp is immediately set into the agarose over the Gelbond. To avoid bubbles forming when the stamp is placed into the agarose, we suggest setting the mold down at an angle (similar to how coverslips are placed at an angle). This can be achieved by setting down the long edge of the stamp first and slowly lowering the stamp as if it is on a hinge. We also recommend that when placing the stamp into the agarose, a small gap should be left at the edge of the tray, keeping it accessible to future PBS additions (which will aid in the mold removal step). The stamp should be centered so the edges of the PDMS are not directly touching the sides of the tray. Leaving this space will also aid in the mold removal step.
After the agarose has cooled, ~5 mL of PBS is added to the sides of the tray, and the stamp is lifted at an angle in one continuous motion. Before continuing to the cell loading step, we recommend viewing the CometChip under a microscope to ensure that microwells have not been torn during the stamp removal step (as torn wells may decrease cell loading efficiency and contribute to abnormally shaped comets). We have observed that torn wells may result from leaving insufficient space for PBS addition prior to the stamp removal step. Failing to lift the stamp in a single fluid motion from the agarose may also damage microwells. The CometChip can be removed from the tray (taking care to handle the chip using the Gelbond film so that the agarose layer is not disturbed). There may be agarose on the bottom of the chip from the agarose used to secure the Gelbond to the tray, and this can be removed gently with a scalpel or Kimwipe. When storing CometChips for short-term use, chips can be covered in PBS and placed in the fridge.
Sample Loading Considerations.
To perform the CometChip assay, suspended cells are added above the agarose microwell array, and cells enter the microwells by gravity (Figure 5). While straightforward, several factors affect the efficiency of cells entering the wells as well as the likelihood that cells are dislodged during the CometChip rinse step.
Cell Type.
Both adherent and suspension cells can be analyzed using the CometChip. Compared to suspension cells, adherent cells tend to stick together and can aggregate during the loading process. Cell aggregation can prevent cells from entering the microwells. In addition, it is possible that long incubation during cell loading can enable formation of aggregates that are then readily washed away during the rinse step. Nevertheless, adherent cells have been successfully studied by optimizing cell loading time (see Loading Time section). In addition, cells vary in size, which further influences loading efficiency (see section below about microwell size).
Loading Time.
We have observed that most cell types are sufficiently loaded into microwells within 5–20 min at 37 °C. The minimum loading time needed should be determined for each cell type at the start of the experiment by visually checking microwell loading under a microscope every 5 min. It is important to visually confirm cells are properly loaded before moving on to the next step of the protocol.
Microwell Size.
Prior to beginning an experiment, it may be beneficial to optimize cell loading by testing different-sized microwells. In general, we have observed that smaller cells are trapped more efficiently by smaller diameter microwells, and the minimum microwell size possible should be selected for a cell type. (Note that CometChips sold by Trevigen have a microwell diameter of ~30 μm. The Engelward laboratory has also had success using chips made in-house with microwell diameters of 40 or 50 μm.)
Cell Loading Density.
For most CometChip experiments, efficient cell loading (where the majority of the microwells result in analyzable comets) can be achieved by loading at least 10,000 cells per microwell. This cell number can be delivered to the macrowell in volumes ranging from 30 to 200 μL. Volume may further impact loading time as additional cell media in the macrowell results in a higher distance for the cells to drop before they settle into the microwells. Though, we expect that 10–20 min loading times will be sufficient for any volume in the 30–200 μL range.
Rinsing off-Grid Cells.
Following cell loading, it is necessary to wash away off-grid cells prior to the low melting point overlay, as they can interfere with comet scoring processes (step 4 in Figure 3). Note that overwashing the chip can result in loss of cells within the microwells, and we recommend visually checking the agarose chip under a microscope to confirm loading efficiency following the wash step. This is perhaps the most technically challenging step in the protocol, because shear force is difficult to control and depends on the bore size of the pipet, the force of liquid ejection applied to the chip, and the rate of movement of the pipet across the chip. Analysis of rinsing in Engelward laboratory has shown that for a full 96-well-plate- sized CometChip, a single wash step ejecting 5 mL of PBS across the top of the chip from a 5 mL serological pipet in the course of 1–2 s is sufficient for many cell types. The CometChip should be held at a 45° angle during the rinse step so that excess cells and PBS runoff the bottom of the chip (Video S1). Following the rinse step, PBS can be gently aspirated off the chip from between the macrowells (Video S2).
Chemical Exposures: off-Chip vs on-Chip.
DNA repair can happen very rapidly. For example, we have observed during CometChip analysis that repair of oxidative damage is complete within approximately 30 min after exposure.31 As a result, if cells are treated in a tissue culture dish and subsequently trypsinized and loaded into the CometChip (defined here as an off-chip exposure experiment), then repair will have mostly come to completion prior to lysis. In other words, DNA repair that occurs during cell handling can make it difficult to detect DNA damage. One way to overcome this limitation is to load cells into the agarose microwells before exposing them to DNA damaging agents (defined here as an on-chip exposure experiment). On-chip experiments decrease assay handling time between chemical exposure and cell lysis, allowing for rapidly repaired damage to be captured. This can be advantageous when the half time of repair is about the same as cell trypsinization and loading step times. Indeed, we have observed that rapid repair leads to drastically different dose-response results for off-chip vs on-chip conditions in the case of H2O2 and SIN-1.62
It is noteworthy that on-chip exposures may not be ideal for all chemical agents. A potentially confounding factor is that some chemicals are difficult to rinse from the agarose chip. Although the pore size for the agarose is relatively large compared to the size of chemical DNA damaging agents, for particulates, it remains a possibility that agarose will interfere with the ability of the particles to reach the cells. In those situations, off-chip exposure experiments may be preferred. In addition, for some cell types, extended culture on an agarose chip may be toxic, as many adherent cell types require attachment for normal behavior. In these cases, off-chip experiments may be necessary to retain cell viability over the course of long-term chemical exposures. Note that the Engelward laboratory has explored incorporating collagen into the agarose for culture of some adherent cell types.63
IN VIVO COMETCHIP EXPERIMENTS
Rather than study cell lines, tissue and liquid biopsy samples can also be collected from animals and people. This analysis technique, the in vivo comet assay, is gaining popularity due to great interest in learning about the existing levels of DNA damage in animals and in people. For example, peripheral blood mononuclear cells (PBMCs) can be isolated from human blood samples and either directly analyzed for DNA damage levels or cultured for further experiments, and many researchers have used the traditional comet assay to reveal the impacts of smoking, diet, and other variables on steady-state levels of DNA damage in human lymphocytes (e.g., see refs 27, 64, and 65). In addition, tissues can be minced and filtered to collect cells for DNA damage analysis.
Analysis of Spontaneous/Steady-State DNA Damage.
While in vivo techniques are not yet well characterized using the CometChip, they involve methods (e.g., cell loading, washing, and encapsulation on the agarose chip) similar to in vitro experiments, and so this avenue is promising. The Engelward laboratory has observed that PBMCs (suspension cells) load efficiently into the CometChip microwell array, and DNA damage can be readily studied in primary cells from people. We have also observed that minced tissue collected from mice can be successfully loaded onto the CometChip for analysis (discussed below). Studies are ongoing to further explore the efficacy of the in vivo CometChip for analysis of steady-state DNA damage for various cell types. Note that background DNA damage levels may vary depending on cell type, and we have observed a range of baseline damage levels across various tissues. It is also important to consider that the process of collecting tissues and isolating cells takes time and that rapidly repaired damage may be difficult to capture if tissue processing time exceeds repair time.
Loading Samples Isolated from Tissues.
Preliminary studies in the Engelward laboratory show that cells derived from minced tissues (fresh or flash frozen) can be studied using CometChip. One reason for efficacy could be that while individual cells can load into the microwells, larger fragments of tissue will be excluded and removed during the rinse step, such that the CometChip acts as a sieve. It may also be the case that using a cell strainer to first remove larger clusters may help to improve microwell loading. In addition, efficient loading may be achieved using larger microwells to capture cell clusters (CometChip is effective for analyzing both single cells and clusters of several cells, as described above).30 For in vivo studies, keeping minced tissue and filtered cells on ice helps to keep background levels low.
SPHEROID COMETCHIP EXPERIMENTS
There is growing interest in using three-dimensional cell culture models to perform in vitro experiments as a way to better mimic conditions found in tissues in vivo. For example, spheroids can be cultured, treated with chemicals of interest, and then disaggregated into single cell suspensions for comet analysis.66,67 It is also possible to create hepatocyte spheroids directly within the CometChip microwells and perform comet analysis on intact samples.62 In this section, we describe challenges of analyzing spheroids using the CometChip.
Loading Disaggregated Spheroids.
We previously discussed that 10,000 cells per macrowell is the approximate minimum number of cells necessary to achieve efficient microwell loading on the CometChip. Since individual spheroids harbor a relatively low number of cells, when disaggregating spheroids, one needs to consider whether there is a sufficient cell number in the resulting cell suspension for efficient microwell loading. For example, when culturing spheroids using commercially available ultralow attachment plates (96- or 384-well format), it may be necessary to pool multiple spheroids to achieve this minimum loading requirement.
Analysis of Rapidly Repaired Damage.
When a spheroid is disaggregated for CometChip analysis, it is trypsinized and resuspended in cell media. This can take 5–10 min (depending on cell type and spheroid size). Next, the single cell suspension is loaded onto the CometChip, which can take another 10–20 min. Combined with other CometChip steps (Figure 3), it can take 30–40 min before cells can be lysed. During this time, DNA damage (such as oxidative damage caused by RONS) can be fully repaired by cells (see discussion of on-chip vs off-chip CometChip experiments above).
Recently, we have developed a method for intact spheroid comet analysis that eliminates the disaggregation step.62 Specifically, intact spheroids that have been cultured in the agarose microwells can be electrophoresed to reveal DNA damage levels. It is noteworthy that when analyzing intact spheroids (~100 μm in diameter), we observed a loss in assay sensitivity, which we suspect is due to incomplete DNA migration. This effect was also reported by Wood et al.30 when larger diameter CometChip microwells were tested. However, despite a decrease in assay sensitivity, it was still possible to use intact spheroid-comet analysis to detect dose responses following chemical treatment with H2O2 and SIN-1. These dose responses were not detected using off-chip CometChip treatment methods.
USE OF COMETCHIP FOR WHOLE BLOOD ANALYSIS
The ability to analyze baseline levels of DNA damage in patient-derived samples quickly and accurately could be a useful tool and provide new insights into individual responses from environmental exposures. In epidemiology, whole blood samples are a simple and minimally invasive option and have been used to study DNA damage along with the traditional comet assay.68–70 Compared to in vitro and in vivo applications, whole blood analysis on human samples has not been studied as extensively on the CometChip platform. While limited data are currently available on using CometChip for baseline DNA damage analysis of whole blood, we anticipate that these samples will load into the microwell array using similar parameters as those used for suspension cell lines. However, since whole blood contains red blood cells and these cells lack DNA, we also anticipate it will be advantageous and essential to lyse the red blood cells prior to loading to ensure they do not settle into the microwells and prevent other DNA-containing cell types from loading.
POTENTIAL TECHNICAL CHALLENGES AND ALTERNATIVE METHODS FOR DNA DAMAGE ANALYSIS
While there are advantages to using CometChip for DNA damage studies, certain challenges may also arise. For example, during CometChip loading, multiple cells are trapped per microwell, resulting in an average %Tail DNA value. This is a disadvantage in experiments that focus on single cell behavior. Nevertheless, with adjustments to microwell size, one can achieve single cell loading.30 Additionally, we have observed that not all cell types load easily into the circular microwells due to cell morphology (e.g., long, cylindrical cardiomyocytes). In these scenarios, testing various microwell sizes may help solve the issue.
In certain cases, when cell loading remains a limiting factor, it may also be desirable to use a different platform for DNA damage analysis. For example, other methods have been developed to improve comet assay throughput and reliability. A recent review summarizes these technologies,71 which utilize similar concepts of condensing multiple samples onto surfaces such as glass slides72 or a GelBond film.73 For these high-throughput comet assay systems, droplets of agarose containing cell suspensions are applied to the supporting material (e.g., in a 96-well format), and comets in each droplet are analyzed as a separate gel. Note that these arrays are different than the arrayed CometChip microwells described above, as the cells in the agarose droplets are dispersed randomly, rather than in a regularly spaced grid. An advantage of the microwell CometChip arrays is that they are compatible with automated and unbiased scoring of up to ~300 non-overlapping and in-focus comets per single well of a 96-well plate. In addition, as discussed above, a bottomless 96-well plate can be clamped directly over the CometChip array, allowing the CometChip to be used like a standard 96-well plate when loading multiple sample types, performing chemical exposures, or performing downstream enzyme digestion experiments. This may be desirable for experimental and handling purposes.
Another challenge of CometChip analysis is that when treating cells on-chip, there is a possibility that chemicals become trapped in the agarose chip. It is also possible that rinsing the chip with a high volume of buffer to dilute the chemical following exposure may assist in clearing the test compound. Another option is to perform off-chip chemical exposures prior to cell loading. Note, however, that repair may be missed when off-chip experiments are performed (discussed above).
While these comet-based methods have both been shown to be effective for analysis of DNA damage and repair, other technologies with their own advantages and disadvantages have also been developed and reviewed extensively in the literature.13,74–77 In the remainder of this section, we will briefly highlight a few popular methods discussed in these reviews:
The micronucleus assay quantifies chromosomal damage in cells and is a standard test in the regulatory approval process.78,79 This method can be adapted for high- throughput experiments through the use of flow cytometry.80,81 However, compared to the comet assay, the micronucleus assay does not provide information about the types of DNA damage present (e.g., oxidative damage, bulky lesions) or information about DNA repair.
An alternative approach for DNA damage analysis is using immunofluorescence methods to measure the extent to which chromatin is modified near a DNA DSB (γ-H2AX staining).82–84 While the comet assay measures a physical break, this method detects a signaling event (the phosphorylation of the H2AX histone), which may be influenced by additional factors.
Analytical methods have also been developed that utilize mass spectrometry to accurately measure DNA adducts following exposures, and these studies can provide valuable insight into a chemical’s mechanism of action.85–87 Compared to comet analysis, mass spectrometry is often more expensive and technically challenging. In addition, comet data are relatively straightforward to interpret for new users and provide a simple way to study repair kinetics.
Similar to the alkaline comet assay, the Fluorimetric Detection of Alkaline DNA Unwinding (FADU) quantifies DNA strand breaks following DNA unwinding in an alkaline buffer. Following unwinding, the extent of DNA damage is determined by measuring how much double-stranded DNA remains in the sample (e.g., the smaller the fluorescence signal, the more DNA strand breaks were present).88 This method can also be automated. Similar to the CometChip, this method can analyze multiple cells per condition, providing an average amount of damage for cells in a well. DNA repair studies are also possible using this method, and the platform may provide an alternative way to capture rapidly repaired intermediates.
CONCLUSIONS
DNA damage levels are relevant in a variety of contexts. For example, DNA damage analysis can be used as a downstream predictor of carcinogenicity in safety assessments or drug screens. In addition, DNA damaging agents are often used to treat cancer, and knowing about DNA repair in a person’s tumor could help guide treatment. Here, we have summarized use of the CometChip platform for genotoxicity studies. We have discussed how this high-throughput tool can be used to study environmental exposures relevant to human health in different sample types, and we have provided technical considerations for adopting the assay. CometChip has been successfully used for many studies of cultured cells and is currently being applied to animal models and human population studies in vivo. Given its throughput and ease of use, it is anticipated that the CometChip will become a broadly applicable platform for researchers interested in DNA damage and repair.
Supplementary Material
Video S1 describes the wash step of the CometChip assay
Video S2 describes the aspiration step of the CometChip assay
ACKNOWLEDGMENT
We thank funding by the Training Grant in Environmental Toxicology (T32-ES007020), the MIT Nitric Oxide Program Project Grant (P01-CA026731), the National Institute of Environmental Health Sciences Superfund Basic Research Program, National Institute of Health (P42 ES027707), and the National Institute of Environmental Health Sciences Core Center Grant, National Institute of Health (P30-ES002109).
Funding Sources
Training Grant in Environmental Toxicology (T32-ES007020). MIT Nitric Oxide Program Project Grant (P01-CA026731). National Institute of Environmental Health Sciences Superfund Basic Research Program, National Institute of Health (P42 ES027707). National Institute of Environmental Health Sciences Core Center Grant, National Institutes of Health (P30-ES002109)
ABBREVIATIONS
- PDMS
polydimethylsiloxane
- LMP
low melting point
- ROS
reactive oxygen species
- RONS
reactive oxygen and nitrogen species
- PBMCs
peripheral blood mononuclear cells
- BER
base excision repair
- MEFs
mouse embryonic fibroblasts
- GSNO
S- nitrosoglutathione
- CHO
Chinese hamster ovary
- PARP
poly(ADP-ribose) polymerase
- NER
Nucleotide excision repair
- HU
hydroxyurea
- AraC
1-B-D-arabinofuranosyl cytosine
Footnotes
The authors declare the following competing financial interest(s): Bevin P. Engelward is a co-inventor for the CometChip, which has been patented and licensed to Trevigen.
REFERENCES
- (1).Fu D, Calvo JA, and Samson LD (2012) Balancing Repair and Tolerance of DNA Damage Caused by Alkylating Agents. Nat. Rev. Cancer 12 (2), 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Deans AJ, and West SC (2011) DNA Interstrand Crosslink Repair and Cancer. Nat. Rev. Cancer 11 (7), 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chatterjee N, and Walker GC (2017) Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen 58 (5), 235– 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Duclos S, Doublié S, and Wallace SS (2012) Consequences and Repair of Oxidative DNA Damage. Issues in Toxicology: The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens. No. 13, 115–159. [Google Scholar]
- (5).Schar̈ OD (2005) DNA Interstrand Crosslinks: Natural and Drug-Induced DNA Adducts That Induce Unique Cellular Responses. ChemBioChem 6 (1), 27–32. [DOI] [PubMed] [Google Scholar]
- (6).Dedon PC, and Tannenbaum SR (2004) Reactive Nitrogen Species in the Chemical Biology of Inflammation. Arch. Biochem. Biophys 423 (1), 12–22. [DOI] [PubMed] [Google Scholar]
- (7).Kay J, Thadhani E, Samson L, and Engelward B (2019) Inflammation-Induced DNA Damage, Mutations and Cancer. DNA Repair 83, 102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fedeles BI, and Essigmann JM (2018) Impact of DNA Lesion Repair, Replication and Formation on the Mutational Spectra of Environmental Carcinogens: Aflatoxin B 1 as a Case Study. DNA Repair 71, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Grollman AP, and Moriya M (1993) Mutagenesis by 8- Oxoguanine: An Enemy Within. Trends Genet. 9 (7), 246–249. [DOI] [PubMed] [Google Scholar]
- (10).Shibutani S, Takeshita M, and Grollman AP (1991) Insertion of Specific Bases during DNA Synthesis Past the Oxidation- Damaged Base 8-OxodG. Nature 349, 431–434. [DOI] [PubMed] [Google Scholar]
- (11).McKenna DJ, McKeown SR, and McKelvey-Martin VJ (2008) Potential Use of the Comet Assay in the Clinical Management of Cancer. Mutagenesis 23 (3), 183–190. [DOI] [PubMed] [Google Scholar]
- (12).Gavande NS, Vandervere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, and Turchi JJ (2016) DNA Repair Targeted Therapy: The Past or Future of Cancer Treatment? Pharmacol. Ther 160, 65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Nagel ZD, Engelward BP, Brenner DJ, Begley TJ, Sobol RW, Bielas JH, Stambrook PJ, Wei Q, Hu JJ, Terry MB, et al. (2017) Towards Precision Prevention: Technologies for Identifying Healthy Individuals with High Risk of Disease. Mutat. Res., Fundam. Mol. Mech. Mutagen 800–802, 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Olive PL, and Banat́h JP (2006) The Comet Assay: A Method to Measure DNA Damage in Individual Cells. Nat. Protoc 1 (1), 23–29. [DOI] [PubMed] [Google Scholar]
- (15).Ostling O, and Johanson KJ (1984) Microelectrophoretic Study of Radiation-Induced DNA Damages in Individual Mammalian Cells. Biochem. Biophys. Res. Commun 123 (1), 291–298. [DOI] [PubMed] [Google Scholar]
- (16).Singh NP, McCoy MT, Tice RR, and Schneider EL (1988) A Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp. Cell Res 175 (1), 184–191. [DOI] [PubMed] [Google Scholar]
- (17).Collins AR (2004) The Comet Assay for DNA Damage and Repair: Principles, Applications, and Limitations. Mol. Biotechnol 26 (3), 249–261. [DOI] [PubMed] [Google Scholar]
- (18).Araldi RP, de Melo TC, Mendes TB, de Sa Junior PL, Nozima BHN, Ito ET, de Carvalho RF, de Souza EB, and de Cassia Stocco R (2015) Using the Comet and Micronucleus Assays for Genotoxicity Studies: A Review. Biomed. Pharmacother 72, 74–82. [DOI] [PubMed] [Google Scholar]
- (19).Kang SH, Kwon JY, Lee JK, and Seo YR (2013) Recent Advances in In Vivo Genotoxicity Testing: Prediction of Carcinogenic Potential Using Comet and Micronucleus Assay in Animal Models. J. Cancer Prev 18 (4), 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Glei M, Schneider T, and rmann W (2016) Comet Assay: An Essential Tool in Toxicological Research. Arch. Toxicol 90 (10), 2315–2336. [DOI] [PubMed] [Google Scholar]
- (21).Brendler-Schwaab S, Hartmann A, Pfuhler S, and Speit G (2005) The in Vivo Comet Assay: Use and Status in Genotoxicity Testing. Mutagenesis 20 (4), 245–254. [DOI] [PubMed] [Google Scholar]
- (22).Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu J-C, and Sasaki YF (2000) Single Cell Gel/Comet Assay: Guidelines for in Vitro and in Vivo Genetic Toxicology Testing. Environ. Mol. Mutagen 35 (3), 206– 221. [DOI] [PubMed] [Google Scholar]
- (23).Gunter S, and Rothfuss A (2012) The Comet Assay: A Sensitive Genotoxicity Test for the Detection of DNA Damage and Repair DNA Repair Protocols, pp 79–90, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- (24).Collins AR (2014) Measuring Oxidative Damage to DNA and Its Repair with the Comet Assay. Biochim. Biophys. Acta, Gen. Subj 1840 (2), 794–800. [DOI] [PubMed] [Google Scholar]
- (25).Azqueta A, Slyskova J, Langie S.aS., O’Neill Gaivaõ I, and Collins A (2014) Comet Assay to Measure DNA Repair: Approach and Applications. Front Genet. 5,288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kassie F, Parzefall W, and Knasmuller S (2000) Single Cell Gel Electrophoresis Assay: A New Technique for Human Biomonitoring Studies. Mutat. Res., Rev. Mutat. Res 463 (1), 13–31. [DOI] [PubMed] [Google Scholar]
- (27).Dusinska M, and Collins AR (2008) The Comet Assay in Human Biomonitoring: Gene-Environment Interactions. Mutagenesis 23 (3), 191–205. [DOI] [PubMed] [Google Scholar]
- (28).Møller P, Hemmingsen JG, Jensen DM, Danielsen PH, Karottki DG, Jantzen K, Roursgaard M, Cao Y, Kermanizadeh A, Klingberg H, et al. (2015) Applications of the Comet Assay in Particle Toxicology: Air Pollution and Engineered Nanomaterials Exposure. Mutagenesis 30 (1), 67–83. [DOI] [PubMed] [Google Scholar]
- (29).Wasson GR, McKelvey-Martin VJ, and Downes CS (2008) The Use of the Comet Assay in the Study of Human Nutrition and Cancer. Mutagenesis 23 (3), 153–162. [DOI] [PubMed] [Google Scholar]
- (30).Wood DK, Weingeist DM, Bhatia SN, and Engelward BP (2010) Single Cell Trapping and DNA Damage Analysis Using Microwell Arrays. Proc. Natl. Acad. Sci. U. S. A 107 (22), 10008–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ge J, Prasongtanakij S, Wood DK, Weingeist DM, Fessler J, Navasummrit P, Ruchirawat M, and Engelward BP (2014) CometChip: A High-Throughput 96-Well Platform for Measuring DNA Damage in Microarrayed Human Cells. J. Visualized Exp 18 (92), e50607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Weingeist DM, Ge J, Wood DK, Mutamba JT, Huang Q, Rowland EA, Yaffe MB, Floyd S, and Engelward BP (2013) Single-Cell Microarray Enables High-Throughput Evaluation of DNA Double-Strand Breaks and DNA Repair Inhibitors. Cell Cycle 12 (6), 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Smerdon MJ (1986) Completion of Excision Repair in Human Cells: Relationship between Ligation and Nucleosome Formation. J. Biol. Chem 261 (1), 244–252. [PubMed] [Google Scholar]
- (34).Fram RJ, and Kufe DW (1985) Inhibition of DNA Excision Repair and the Repair of X-Ray-Induced DNA Damage by Cytosine Arabinoside and Hydroxyurea. Pharmacol. Ther 31, 165–176. [DOI] [PubMed] [Google Scholar]
- (35).Ngo LP, Owiti NA, Swartz C, Winters J, Su Y, Ge J, Xiong A, Han J, Recio L, Samson LD, and Engelward BP (2020) Sensitive CometChip Assay for Screening Potentially Carcinogenic DNA Adducts by Trapping DNA Repair Intermediates. Nucleic Acids Res. 48, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Rocha CRR, Silva MM, Quinet A, Cabral-Neto JB, and Menck CFM (2018) DNA Repair Pathways and Cisplatin Resistance: An Intimate Relationship. Clinics 73, e478s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Xue W, and Warshawsky D (2005) Metabolic Activation of Polycyclic and Heterocyclic Aromatic Hydrocarbons and DNA Damage: A Review. Toxicol. Appl. Pharmacol 206 (1), 73–93. [DOI] [PubMed] [Google Scholar]
- (38).Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, Brennan CA, Chun E, Ngo L, Samson LD, et al. (2019) The Human Gut Bacterial Genotoxin Colibactin Alkylates DNA. Science (Washington, DC, U. S.) 363 (6428), No. eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Li L, Wang W, Ding M, Luo G, and Liang Q (2016) Single- Cell-Arrayed Agarose Chip for in Situ Analysis of Cytotoxicity and Genotoxicity of DNA Cross-Linking Agents. Anal. Chem 88 (13), 6734–6742. [DOI] [PubMed] [Google Scholar]
- (40).Pfuhler S, and Wolf HU (1996) Detection of DNA- Crosslinking Agents With the Alkaline Comet Assay. Environ. Mol. Mutagen 27, 196–201. [DOI] [PubMed] [Google Scholar]
- (41).Olive PL, Wlodek D, Durand RE, and Banat́h JP (1992) Factors Influencing DNA Migration from Individual Cells Subjected to Gel Electrophoresis. Exp. Cell Res 198 (2), 259–267. [DOI] [PubMed] [Google Scholar]
- (42).Hartley JM, Spanswick VJ, Gander M, Giacomini G, Whelan J, Souhami RL, and Hartley J. a. Measurement of DNA Cross-Linking in Patients on Ifosfamide Therapy Using the Single Cell Gel Electrophoresis (Comet) Assay Measurement of DNA Cross- Linking in Patients on Ifosfamide Therapy Using the Single Cell Gel Electrophoresis. Clin. Cancer Res 1999, 5 (3), 507–512. [PubMed] [Google Scholar]
- (43).Parrish MC, Chaim IA, Nagel ZD, Tannenbaum SR, Samson LD, and Engelward BP (2018) Nitric Oxide Induced S- Nitrosation Causes Base Excision Repair Imbalance. DNA Repair 68, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chao M, Kim MY, Ye W, Ge J, Trudel LJ, Belanger CL, Skipper PL, Engelward BP, Tannenbaum SR, and Wogan GN (2012) Genotoxicity of 2,6- and 3,5-Dimethylaniline in Cultured Mammalian Cells: The Role of Reactive Oxygen Species. Toxicol. Sci 130 (1), 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ge J, Chow DN, Fessler JL, Weingeist DM, Wood DK, and Engelward BP (2015) Micropatterned Comet Assay Enables High Throughput and Sensitive DNA Damage Quantification. Mutagenesis 30 (1), 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Watson C, Ge J, Cohen J, Pyrgiotakis G, Engelward BP, and Demokritou P (2014) High-Throughput Screening Platform for Engineered Nanoparticle-Mediated Genotoxicity Using CometChip Technology. ACS Nano 8 (3), 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Sotiriou GA, Watson C, Murdaugh KM, Darrah TH, Pyrgiotakis G, Elder A, Brain JD, and Demokritou P (2014) Engineering Safer-by-Design, Transparent, Silica-Coated ZnO Nano- rods with Reduced DNA Damage Potentials. Environ. Sci.: Nano 1 (2), 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mutamba JT, Svilar D, Prasongtanakij S, Wang X-H, Lin Y-C, Dedon PC, Sobol RW, and Engelward BP (2011) XRCC1 and Base Excision Repair Balance in Response to Nitric Oxide. DNA Repair 10 (12), 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Seo JE, Tryndyak V, Wu Q, Dreval K, Pogribny I, Bryant M, Zhou T, Robison TW, Mei N, and Guo X (2019) Quantitative Comparison of in Vitro Genotoxicity between Metabolically Competent HepaRG Cells and HepG2 Cells Using the High-Throughput High-Content CometChip Assay. Arch. Toxicol 93, 1433–1448. [DOI] [PubMed] [Google Scholar]
- (50).Chan TK, Loh XY, Peh HY, Tan WNF, Tan WSD, Li N, Tay IJJ, Wong WSF, and Engelward BP (2016) House Dust Mite-Induced Asthma Causes Oxidative Damage and DNA Double-Strand Breaks in the Lungs. J. Allergy Clin. Immunol 138 (1), 84–96. [DOI] [PubMed] [Google Scholar]
- (51).Chan TK, Tan WSD, Peh HY, and Wong WSF (2017) Aeroallergens Induce Reactive Oxygen Species Production and DNA Damage and Dampen Antioxidant Responses in Bronchial Epithelial Cells. J. Immunol 199 (1), 39–47. [DOI] [PubMed] [Google Scholar]
- (52).Li N, Parrish M, Chan TK, Yin L, Rai P, Yoshiyuki Y, Abolhassani N, Tan KB, Kiraly O, Chow VTK, et al. (2015) Influenza Infection Induces Host DNA Damage and Dynamic DNA Damage Responses during Tissue Regeneration. Cell. Mol. Life Sci 72 (15), 2973–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ge J, Wood DK, Weingeist DM, Prasongtanakij S, Navasumrit P, Ruchirawat M, and Engelward BP (2013) Standard Fluorescent Imaging of Live Cells Is Highly Genotoxic. Cytometry, Part A 83 (6), 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sykora P, Chiari Y, Heaton A, Moreno N, Glaberman S, and Sobol RW (2018) Application of the CometChip Platform to Assess DNA Damage in Field-Collected Blood Samples from Turtles. Environ. Mol. Mutagen 59 (4), 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sonavane M, Sykora P, Andrews JF, Sobol RW, and Gassman NR (2018) Camptothecin Efficacy to Poison Top1 Is Altered by Bisphenol A in Mouse Embryonic Fibroblasts. Chem. Res. Toxicol 31 (6), 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Fang P, Madden JA, Neums L, Moulder RK, Forrest ML, and Chien J (2018) Olaparib-Induced Adaptive Response Is Disrupted by FOXM1 Targeting That Enhances Sensitivity to PARP Inhibition. Mol. CancerRes 16(6),961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Fairbairn DW, Olive PL, and O’Neill KL (1995) The Comet Assay: A Comprehensive Review. Mutat. Res., Rev. Genet. Toxicol 339, 37–59. [DOI] [PubMed] [Google Scholar]
- (58).Kumaravel TS, Vilhar B, Faux SP, and Jha AN (2009) Comet Assay Measurements: A Perspective. Cell Biol. Toxicol 25 (1), 53–64. [DOI] [PubMed] [Google Scholar]
- (59).Cassano J, Roesslein M, Kaufmann R, Luethi T, Schicht O, Wick P, and Hirsch C (2019) A Novel Approach to Increase Robustness, Precision and High-Throughput Capacity of Single Cell Gel Electrophoresis. ALTEX 37, 1–15. [DOI] [PubMed] [Google Scholar]
- (60).Forchhammer L, Johansson C, Loft S, Möller L, Godschalk RWL, Langie SAS, Jones GDD, Kwok RWL, Collins AR, Azqueta A, et al. (2010) Variation in the Measurement of DNA Damage by Comet Assay Measured by the ECVAG Inter-Laboratory Validation Trial. Mutagenesis 25 (2), 113–123. [DOI] [PubMed] [Google Scholar]
- (61).Azqueta A, Meier S, Priestley C, Gutzkow KB, Brunborg G, Sallette J, Soussaline F, and Collins A (2011) The Influence of Scoring Method on Variability in Results Obtained with the Comet Assay. Mutagenesis 26 (3), 393–399. [DOI] [PubMed] [Google Scholar]
- (62).Chao C, Ngo LP, and Engelward BP (2020) SpheroidChip: Patterned Agarose Microwell Compartments Harboring HepG2 Spheroids Are Compatible with Genotoxicity Testing. ACS Biomater. Sci. Eng 6, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Ngo LP, Chan TK, Ge J, Samson LD, and Engelward BP (2019) Microcolony Size Distribution Assay Enables High- Throughput Cell Survival Quantitation. Cell Rep. 26 (6), 1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Azqueta A, Langie SAS, Boutet-Robinet E, Duthie S, Ladeira C, Møller P, Collins AR, and Godschalk RWL (2019) DNA Repair as a Human Biomonitoring Tool: Comet Assay Approaches. Mutat. Res., Rev. Mutat. Res 781, 71–87. [DOI] [PubMed] [Google Scholar]
- (65).Brevik A, Gaivao I, Medin T, Jørgenesen A, Piasek A, Elilasson J, Karlsen A, Blomhoff R, Veggan T, Duttaroy AK, and Collins AR (2011) Supplementation of a Western Diet with Golden Kiwifruits (Actinidia Chinensis Var.′Hort 16A′:) Effects on Biomarkers of Oxidation Damage and Antioxidant Protection. Nutr. J 10, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Olive PL, and Banath JP (1997) Multicell Spheroid Response to Drugs Predicted with the Comet Assay. Cancer Res. 57 (24), 5528–5533. [PubMed] [Google Scholar]
- (67).Elje E, Hesler M, Rundeń-Pran E, Mann P, Mariussen E, Wagner S, Dusinska M, and Kohl Y (2019) The Comet Assay Applied to HepG2 Liver Spheroids. Mutat. Res., Genet. Toxicol. Environ. Mutagen 845, 403033. [DOI] [PubMed] [Google Scholar]
- (68).Koppen G, De Prins S, Jacobs A, Nelen V, Schoeters G, and Langie SAS (2018) The Comet Assay in Human Biomonitoring: Cryopreservation of Whole Blood and Comparison with Isolated Mononuclear Cells. Mutagenesis 33 (1), 41–47. [DOI] [PubMed] [Google Scholar]
- (69).Chuang CH, and Hu ML (2004) Use of Whole Blood Directly for Single-Cell Gel Electrophoresis (Comet) Assay in Vivo and White Blood Cells for in Vitro Assay. Mutat. Res., Genet. Toxicol. Environ. Mutagen 564 (1), 75–82. [DOI] [PubMed] [Google Scholar]
- (70).Hininger I, Chollat-Namy A, Sauvaigo S, Osman M, Faure H, Cadet J, Favier A, and Roussel AM (2004) Assessment of DNA Damage by Comet Assay on Frozen Total Blood: Method and Evaluation in Smokers and Non-Smokers. Mutat. Res., Genet. Toxicol. Environ. Mutagen 558 (1–2), 75–80. [DOI] [PubMed] [Google Scholar]
- (71).Brunborg G, Jackson P, Shaposhnikov S, Dahl H, Azqueta A, Collins AR, and Gutzkow KB (2014) High Throughput Sample Processing and Automated Scoring. Front. Genet 5, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Yamani N. El, Collins AR, Rundeń-Pran E, Fjellsbø LM, Shaposhnikov S, Zienolddiny S, and Dusinska M (2017) In Vitro Genotoxicity Testing of Four Reference Metal Nanomaterials, Titanium Dioxide, Zinc Oxide, Cerium Oxide and Silver: Towards Reliable Hazard Assessment. Mutagenesis 32 (1), 117–126. [DOI] [PubMed] [Google Scholar]
- (73).Gutzkow KB, Langleite TM, Meier S, Graupner A, Collins AR, and Brunborg G (2013) High-Throughput Comet Assay Using 96 minigels. Mutagenesis 28 (3), 333–340. [DOI] [PubMed] [Google Scholar]
- (74).Nelson BC, Wright CW, Ibuki Y, Moreno-Villanueva M, Karlsson HL, Hendriks G, Sims CM, Singh N, and Doak SH (2017) Emerging Metrology for High-Throughput Nanomaterial Genotoxicology. Mutagenesis 32 (1), 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Figueroa-Gonzaĺez G, and Peŕez-Plasencia C (2017) Strategies for the Evaluation of DNA Damage and Repair Mechanisms in Cancer. Oncol. Lett 13 (6), 3982–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Collins AR, Annangi B, Rubio L, Marcos R, Dorn M, Merker C, Estrela-Lopis I, Cimpan MR, Ibrahim M, and Cimpan E (2017) High Throughput Toxicity Screening and Intracellular Detection of Nanomaterials. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology 9 (1), e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Gassman NR, and Holton NW (2019) Targets for Repair: Detecting and Quantifying DNA Damage with Fluorescence-Based Methodologies. Curr. Opin. Biotechnol 55, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Hayashi M (2016) The Micronucleus Test-Most Widely Used in Vivo Genotoxicity Test. Genes Environ. 38 (1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Kirsch-Volders M, Decordier I, Elhajouji A, Plas G, Aardema MJ, and Fenech M (2011) In Vitro Genotoxicity Testing Using the Micronucleus Assay in Cell Lines, Human Lymphocytes and 3D Human Skin Models. Mutagenesis 26 (1), 177–184. [DOI] [PubMed] [Google Scholar]
- (80).Bryce SM, Bemis JC, Avlasevich SL, and Dertinger SD (2007) In Vitro Micronucleus Assay Scored by Flow Cytometry Provides a Comprehensive Evaluation of Cytogenetic Damage and Cytotoxicity. Mutat. Res., Genet. Toxicol. Environ. Mutagen 630, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Nüsse M, and Kramer J (1984) Flow Cytometric Analysis of Micronuclei Found in Cells after Irradiation. Cytometry 5 (1), 20–25. [DOI] [PubMed] [Google Scholar]
- (82).Rogakou EP, Pilch DR, Orr AH, Ivanova VS, and Bonner WM (1998) DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem 273 (10), 5858– 5868. [DOI] [PubMed] [Google Scholar]
- (83).Kuo LJ, and Yang L-X (2008) Gamma-H2AX - a Novel Biomarker for DNA Double-Strand Breaks. In Vivo 22 (3), 305–309. [PubMed] [Google Scholar]
- (84).Rothkamm K, Barnard S, Moquet J, Ellender M, Rana Z, and Burdak-Rothkamm S (2015) DNA Damage Foci: Meaning and Significance. Environ. Mol. Mutagen 56 (6), 491–504. [DOI] [PubMed] [Google Scholar]
- (85).Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, and Dedon PC (2008) Quantification of DNA Damage Products Resulting from Deamination, Oxidation and Reaction with Products of Lipid Peroxidation by Liquid Chromatography Isotope Dilution Tandem Mass Spectrometry. Nat. Protoc 3 (8), 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Stornetta A, Villalta PW, Hecht SS, Sturla SJ, and Balbo S (2015) Screening for DNA Alkylation Mono and Cross-Linked Adducts with a Comprehensive LC-MS3 Adductomic Approach. Anal. Chem 87 (23), 11706–11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Cadet J, Douki T, Frelon S, Sauvaigo S, Pouget JP, and Ravanat JL (2002) Assessment of Oxidative Base Damage to Isolated and Cellular DNA by HPLC-MS/MS Measurement. Free Radical Biol. Med 33 (4), 441–449. [DOI] [PubMed] [Google Scholar]
- (88).Moreno-Villanueva M, Pfeiffer R, Sindlinger T, Leake A, Müller M, Kirkwood TBL, and rkle A (2009) A Modified and Automated Version of the “Fluorimetric Detection of Alkaline DNA Unwinding” Method to Quantify Formation and Repair of DNA Strand Breaks. BMC Biotechnol. 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 describes the wash step of the CometChip assay
Video S2 describes the aspiration step of the CometChip assay


