Abstract
The unprecedented global pandemic known as SARS-CoV-2 has exercised to its limits nearly all aspects of modern viral diagnostics. In doing so, it has illuminated both the advantages and limitations of current technologies. Tremendous effort has been put forth to expand our capacity to diagnose this deadly virus. In this work, we put forth key observations in the functionality of current methods for SARS-CoV-2 diagnostic testing. These methods include nucleic acid amplification–, CRISPR-, sequencing-, antigen-, and antibody-based detection methods. Additionally, we include analysis of equally critical aspects of COVID-19 diagnostics, including sample collection and preparation, testing models, and commercial response. We emphasize the integrated nature of assays, wherein issues in sample collection and preparation could impact the overall performance in a clinical setting.
Keywords: COVID-19, RT-PCR, CRISPR, Sequencing, Serological testing, SARS-CoV-2
Introduction
In December 2019, health officials in Wuhan, PRC, reported a disease outbreak involving a cluster of cases of “pneumonia of unknown cause.” Since then, the 2019 coronavirus disease (COVID-19 or SARS-CoV-2) outbreak has been characterized as a pandemic, spread to 188 countries worldwide, causing 9,30,000 fatalities and more than 29 million confirmed cases (as of September 15, 2020). The pandemic has proven to be a significant challenge to our ability to reduce the global spread of the SARS-CoV-2. Given the global scale of infections due to the novel virus and the lack of approved therapeutics and vaccines, the COVID-19 pandemic has drawn comparisons to the deadly 1918 Spanish Flu pandemic. A key difference is current advances in molecular diagnostic technology that has enabled us to rapidly characterize the novel virus, identify infectious (including asymptomatic) patients, and potentially isolate them to control the disease spread. However, initial delays in assay design and supply-chain bottlenecks prevented the deployment of accurate diagnostic tests at scale globally. This was found to be a critical gap in arresting the spread of this devastating disease worldwide. Nevertheless, recent developments have led to broader deployment of diagnostic tests, albeit ones that require at least a dedicated sample collection setup.
Nucleic acid (NA) amplification tests (e.g., polymerase chain reaction (PCR)) and immunoassays (e.g., enzyme-linked immunosorbent assay (ELISA)) are among the most utilized tools in today’s COVID-19 diagnostic testing landscape. This review paper examines current molecular diagnostic tools (Fig. 1), such as amplification-based (including CRISPR-Cas based), antibody and antigen tests, and sequencing, utilized for the detection of SARS-CoV-2. We discuss the capabilities offered by each of these molecular diagnostic tools and challenges encountered in utilizing them in the current pandemic, and provide an assessment of future directions in research that could help remedy some of the identified shortcomings. We also report how each of these methods plays a complementary role suited for different stages of the pandemic.
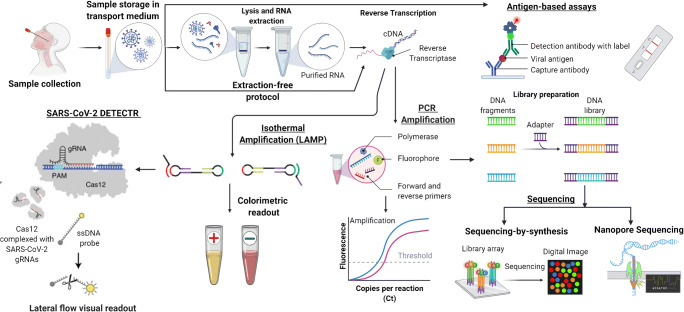
Fig. 1.
Overview of COVID-19 diagnostic workflow—samples are collected and stored in a transport medium, lysed, RNA extracted, reverse transcribed to complementary DNA (cDNA), and then amplified (via PCR or isothermal amplification). The amplified viral sequence is detected/quantified using fluorescent dyes or colorimetric readout. CRISPR-Cas-based detection (SARS-CoV-2 DETECTR) works by the activation of Cas12 due to the presence of a target RNA sequence. The activated Cas12 subsequently cleaves reporter labels generating a fluorescent signal. The sequencing workflow converts the cDNA into a form compatible with the sequencer (library preparation) and then determines the cDNA sequence via digital images (sequencing-by-synthesis) or using electrical signals (Nanopore sequencing). Antigen-based lateral flow assays detect the SARS-CoV-2 antigen using an immunoassay format. Viral antigen forms a sandwich bound by capture and detection antibodies. The presence of the labeled detection antibody indicates the presence of antigen in the sample (image created with BioRender.com)
Several review papers in the literature describe diagnostic approaches for COVID-19 detection. This paper underscores the importance of the integrated nature of these diagnostic tools, wherein improvements in sample collection and preparation are needed to complement advances towards sensitive and accurate diagnostic methods. This knowledge is relevant to the current scenario wherein differences in analytical sensitivity and clinical sensitivity have hampered the effectiveness of COVID-19 diagnostics. This situation reflects the challenges faced to develop and thoroughly assess the assays, reagents, and sample handling processes required for a reliable test. Also, this situation generates the need to mobilize a large-scale production pipeline to meet the enormous demand for assays for a new, largely unknown pathogen, SARS-CoV-2.
Furthermore, this paper discusses the challenges in ramping up testing. As an example, we discuss the problem of false-negatives that has impacted COVID-19 testing and how improvements in sample collection could help remedy the situation. We underscore the lack of research into ensuring repeatability of respiratory sample collection. We also discuss various clinical samples that are utilized for diagnostics and how viral loads (amount of viral proteins or viral nucleic acids in a sample) vary with disease onset in the sample types. We compare and contrast the utility of each sample type from the perspective of sensitivity to utilization in self-sampling formats.
In addition, we also discuss sample preparation aspects that are relevant to wider utilization and point-of-care (POC) deployment of COVID-19 diagnostic tests (PCR, isothermal amplification, and sequencing—including library preparation). Finally, we identify environmental sampling, surveillance opportunities, and advanced detection technologies being developed for commercial applications.
Diagnostic methods
Nucleic acid amplification–based methods
Nucleic acid (NA) amplification and its subsequent detection are the most widely used method for the diagnosis of viral agents. These methods have been extensively reported for the detection of past outbreaks caused by coronaviruses such as MERS and SARS-CoV [1]. In particular, reverse transcription polymerase chain reaction (RT-PCR) is the current standard for the detection of active SARS-CoV-2 infections [2]. RT-PCR broadly involves four steps—lysis of SARS-CoV-2 in the sample, purification of the viral RNA, reverse transcription to complementary DNA (cDNA), and amplification of specific regions of the cDNA, and finally, optical detection of the amplified cDNA. The development of RT-PCR assays for a novel virus such as SARS-CoV-2 involves sequencing the genome and design of primers and probes. A variety of RT-PCR assays utilizing different primer/probe sets have been developed. These assays utilize different primer/probe sets targeting different regions of the SARS-CoV-2 genome. The selection of the primer/probe set affects the sensitivity of the assay [3–5]. For instance, a study from the University of Washington reported E gene [6] and N2 gene primer/probe sets (US Centers for Disease Control and Prevention) to have better sensitivity for SARS-CoV-2 detection assay [7]. A recent report by Anantharajah et al. evaluated RT-PCR assays utilizing WHO-recommended primer/probe sets on clinical samples. They found substantial differences in the assay sensitivity depending on the choice of primer and probe [8]. The difference was more pronounced for samples with low viral load (median cycle threshold, Ct > 30). A recent study by Jung et al. compared the performance of seven primer-probe sets for the N gene and three primer-probe sets for the Orf1 gene for the detection of SARS-CoV-2 RNA. The study evaluated the specificity and sensitivity of amplification at three different reaction temperatures (55, 58, and 60 °C). The primer-probe sets from the Japan NIID (NIID_2019-nCOV_N) and China CDC (ORF1ab) were reported to exhibit the best performance without any non-specific amplification or cross-reactivity for other coronaviruses (hCoV-229E, hCoV-OC43, and MERS-CoV RNA) [9].
As an alternative to RT-PCR, other amplification-based formats such as digital PCR (dPCR) and isothermal amplification–based assays (RT-LAMP) are being utilized for COVID-19 diagnostics. Digital droplet PCR involves partitioning the sample into multiple droplets, with each droplet serving as a reactor for independent PCR. dPCR utilizes endpoint amplification for direct quantification of viral load, is less susceptible to amplification inhibitors in the sample matrix, and, unlike qPCR, does not require calibration curves. dPCR has been shown to have higher sensitivity and lower false-negative results when compared with RT-PCR [10, 11]. Thus, dPCR could be helpful in scenarios wherein low viral load or RNA degradation has resulted in false-negative results using RT-PCR [12]. In addition, dPCR assays could be used to identify insufficient samples by quantifying a reference gene from human RNA as a control [13].
Isothermal amplification–based methods such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are well suited for rapid POC detection. In contrast to RT-PCR, isothermal amplification is carried out at a constant temperature, does not require thermal cycling, and can be carried out using minimal instrumentation. In addition, due to the higher amplification efficiency, the test is rapid in comparison to RT-PCR. Abbott’s ID NOW POC platform for COVID-19 is based on isothermal amplification and has the shortest turnaround time among FDA-authorized diagnostic products [14].
In comparison to SARS-CoV, the increased transmissibility of SARS-CoV-2 has been attributed to asymptomatic carriers and presymptomatic patients [15]. The viral load of asymptomatic or presymptomatic cases has been found similar to that of symptomatic COVID-19 patients, suggesting comparable transmissibility [16]. There is increasing research suggesting the need for large-scale population-wide testing to identify and isolate asymptomatic and presymptomatic COVID-19 cases [17–20]. Centralized and high-throughput, automated RT-PCR assay platforms [21] are well suited to test a large number of samples in a cost-effective and timely manner. Recently, a shortage of commercial reagents for NA extraction and amplification has hindered efforts to scale up the number of COVID-19 tests. Alternative reagents, buffers, and protocols have been explored to remedy such shortages [22] and will need to be utilized to support mass testing strategies. These include open-source reagents and extraction-free RT-PCR protocols. It is important to validate such alternative reagents and protocols in clinical settings on automated assay platforms before use. For instance, due to the proprietary nature of reagents used in commercial RT-PCR platforms, it is critical to evaluate the effect of alternative reagents on the sensitivity and specificity of high-throughput, automated assays. The pooling of samples is an alternative method for scaling COVID-19 tests and could be useful for population-wide screening of COVID-19 cases. The strategy has been recently utilized to screen millions of people for active infections in Wuhan, China. These involve the pooling of multiple samples into a single RT-PCR. Various approaches (such as array testing [23]) have been proposed for the pooling of samples for COVID-19 testing [23–26]. Optimum pool size and approach depend on factors such as the prevalence of the COVID-19 in the population, and the sensitivity of the test utilized [23, 27]. dPCR, which has shown to have higher sensitivity in comparison to RT-PCR, has been proposed as an alternative for testing pooled samples [28].
Despite its wide use, amplification-based COVID-19 assays have faced numerous practical challenges [2]. These include vulnerabilities in the preanalytical and analytical aspects of the assay [29]. Analytical issues include those attributed to variation in viral load or timing of sample collection in relation to disease progression [29]. A study by Kucirka et al. notes that the probability of false-negatives using RT-PCR (from nasopharyngeal (NPS) or oropharyngeal (OPS) swabs) decreases from the day of infection (100%) to symptom onset (38%). The lowest false-negatives are observed at 3 days after symptom onset before increasing again [30].
Preanalytical issues, such as inadequate respiratory sample collection or improper sample handling, could result in false-negative results [29]. In comparison to the significant advances in analytical aspects such as automated operation and sensitivity of RT-PCR assays, respiratory sample collection using swabs is a manual process and prone to errors (see “SARS-CoV-2 clinical sample collection and preparation”). Issues in the collection of respiratory samples can manifest as an inadequate quantity of human respiratory epithelial cells in a sample. Such issues can be avoided by including a primer/probe set to detect the presence of the human RNase P gene in the collected samples. However, quantification of human RNase P in respiratory samples (rather than a qualitative presence) might be necessary to identify inadequate biological sampling [13]. Besides, the faulty design of the primer/probe set for the human RNase P gene in the RT-PCR assay has the potential to cause false-negative results [31, 32].
Proper interpretation of RT-PCR assays as a proxy for infectivity is another aspect that needs attention. Recently, there have been multiple reports of recurrent positive RT-PCR results in COVID-19 patients, even after the resolution of symptoms [33, 34]. A positive RT-PCR test does not necessarily indicate a patient is infectious. For instance, Wölfel and team were unable to isolate SARS-CoV-2 in culture from respiratory patient samples 8 days after symptom onset, despite positive RT-PCR results [34]. Amplification-based tests detect the viral RNA (including degraded viral RNA) and not necessarily the presence of an infectious virus. More work is necessary to understand the implications of positive RT-PCR results and the potential for disease transmission. This is important from the perspective of relying on RT-PCR tests as a criterion for determining return to work conditions for healthcare personnel and other essential workers [35]. Assays that utilize droplet dPCR to measure SARS-CoV-2 RNA in its intact form could be valuable to detect potentially infectious COVID-19 samples [36].
CRISPR-based diagnostics
RT-PCR is the most widely used method for the detection of viral pathogens, including the SARS-CoV-2 virus. However, given the current challenges with RT-PCR, alternative techniques involving CRISPR-Cas and isothermal amplification are being explored. For instance, there are severe limitations associated with the availability, costs, and the need for trained personnel to run RT-PCR tests. Diagnostic capabilities with lower cost, faster turnaround times, and portability are critical, given the global scope and magnitude of the pandemic.
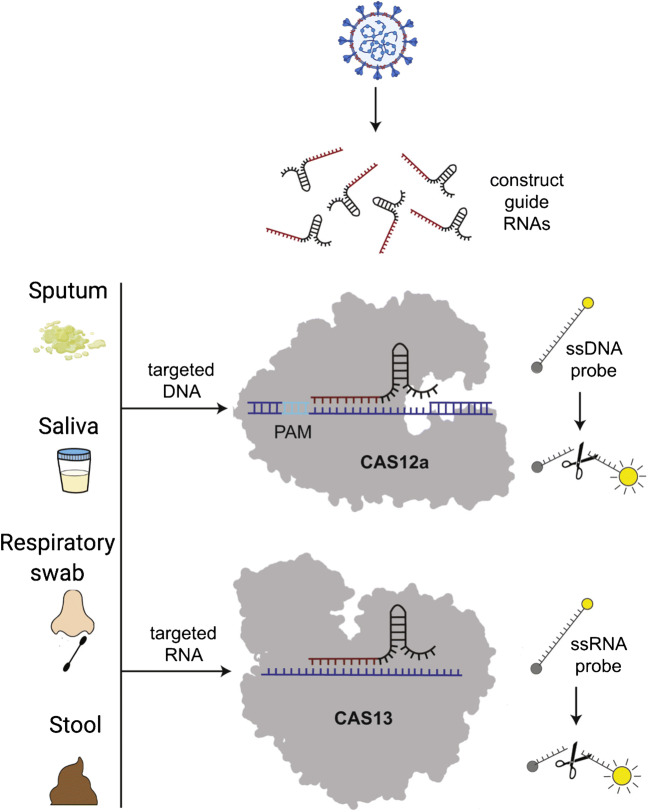
CRISPR, well known as a gene-editing tool, has been leveraged for rapid, POC detection of viral pathogens [38, 39]. Figure 2 describes the schematic for a molecular diagnostics assay utilizing the CRISPR/Cas system. The CRISPR-Cas adaptive immune system consists of a guide RNA (gRNA) and a CRISPR-associated nuclease (Cas). The gRNA consists of a nucleotide sequence, called CRISPR RNA (crRNA), which is complementary to the target sequence. The Cas13 (a type VI CRISPR-Cas) nuclease is activated by the presence of an RNA sequence (target RNA) complementary to the crRNA. The Cas13 then performs a targeted cleavage and also triggers a non-specific RNase activity, leading to degradation of nearby RNA (regardless of the presence of a complementary sequence). This “collateral cleavage” activity of Cas13 is utilized to cut RNA reporter labels, which then release a fluorescent signal. Other CRISPR-Cas systems such as Cas9 or Cas12, which target DNA, have also been utilized for the detection of viral targets [40, 41].
Fig. 2.
Schematic of a CRISPR/Cas-based molecular diagnostic test. Adapted from [37] with permission
CRISPR-based diagnostics (CRISPR-Dx) have been utilized for the detection of viral targets, including dengue virus, Zika virus, and lentiviruses [39]. These assays enable multiplexing and increased sensitivity in comparison to stand-alone isothermal amplification methods. CRISP-Dx combines conventional NA amplification with the CRISPR-Cas13 system to achieve better sensitivity and specificity. The specificity is achieved through the activation of Cas13 via the binding of crRNA to target RNA. In addition, sensitivity is achieved by signal amplification due to the “collateral cleavage” of reporter labels [42]. Chiu and team reported a rapid lateral flow–based assay (SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR)) for detection of SARS-CoV-2 [43]. The assay can detect the virus from respiratory swab samples with sensitivity comparable to that of the US Centers for Disease Control and Prevention (CDC) SARS-CoV-2 real-time RT-PCR assay in 30–40 min. The assay combines reverse transcription loop-mediated isothermal application (RT-LAMP), CRISPR-Cas12-based detection, and a readout on a lateral flow strip. The current protocol can be incorporated into a POC instrument, which would involve microfluidic automation to achieve accurate metering and mixing of reagents along with an integrated isothermal heater [44]. However, the RNA extraction from swab samples is performed using a CDC-recommended protocol that involves the use of a conventional spin column (Qiagen DSP Viral RNA Mini Kit) and a MagNA Pure 24 instrument. These steps involve bulky equipment and are not suitable for POC use. Other recent reports have demonstrated the utility of CRISPR-Cas for COVID-19 diagnostics in clinical samples [45–47]. However, RNA extraction is performed manually, proving to a critical limitation to its POC use. The automation of this key RNA extraction step using POC sample preparation methods and integration with coupled isothermal RT-LAMP+CRISPR–based lateral assay readouts will enable a fast, field-deployable platform that can be used in current and potential future pandemics. Another advantage of CRISPR-based assays is the use of lyophilized reagents and the capability to run assays directly on raw samples. Gootenberg et al. developed a multiplexed CRISPR-Cas13 assay (SHERLOCKv2—specific high-sensitivity enzymatic reporter unlocking-version 2) capable of detecting four targets in a single reaction [41]. This multiplexing is achieved by using distinct cleavage preferences of different Cas13 enzymes on homopolymer reporters. In addition, they utilized lyophilized Cas13 reagents on a lateral flow strip format. The cost of a single paper–based SHERLOCK test was estimated to be as low as USD 0.61 [48]. The multiplexing capability reported in this work is relevant to the current pandemic due to reports of co-infection among SARS-CoV-2-infected patients [49, 50]. Multiplexed assays could be useful to identify potential false-negative assay results. For instance, the multiplexed assay could be used to detect infection from alternative respiratory pathogens with clinical symptoms similar to COVID-19 [51, 52].
Building on the capabilities of SHERLOCKv2, Myhrvold et al. developed a protocol (HUDSON) that can detect multiple viral targets directly from clinical samples, including saliva with minimal sample processing, which is relevant to the current pandemic since saliva is being explored as an alternative sample for COVID-19 testing [42]. Despite a lower observed SARS-CoV-2 viral load in comparison with NPS and OPS, saliva samples have several advantages for POC screening and self-administered tests (see “COVID-19 testing models”) [53, 54]. HUDSON uses heat to lyse viral particles and chemical reduction to inactivate RNases subsequently. This approach obviates the need for centrifuges used in column-based extraction and can be used in POC platforms [55]. A protocol utilizing CRISPR-Cas13 SHERLOCK assay has been reported for the detection of SARS-CoV-2 [56]. Curti et al. reported a CRISPR-Cas12-based method to detect synthetic SARS-CoV-2 RNA fragments from spiked saliva [57]. However, the limit of detection was much lower (105 copies/μL) in comparison to SARS-CoV-2 RNA–spiked buffer samples (10 copies/μL). Recently, SHINE (SHERLOCK and HUDSON Integration to Navigate Epidemics), a diagnostic assay combining SHERLOCK and HUDSON, was utilized to detect SARS-CoV-2 from unextracted clinical specimens (NP swabs and saliva) [58].
CRISPR-Dx for COVID-19 testing has shown to have comparable accuracy to CDC-recommended quantitative RT-PCR tests. In addition, these tests are simpler, are less expensive, and have a faster turnaround time. The integration of these assays with sample preparation on an automated POC format utilizing lyophilized reagents is needed, which will greatly enhance the clinical utility of CRISP-Dx for the COVID-19 pandemic as an alternative to RT-PCR. Given their lower cost, faster turnaround time, and higher sensitivity, CRISPR-Dx could also be used in population-wide diagnostic screening or POC settings.
Sequencing for COVID-19 pandemic management
Next-generation sequencing (NGS) plays a critical role in identifying and monitoring a viral outbreak. Unbiased NGS is the key first step in identifying a novel viral strain [59]. The sequencing data has also enabled the determination of possible origins [60] and transmission patterns of viral pathogens, such as SARS-CoV-2 [61]. Additionally, the identification of the sequence is the first step towards the design of primers and probes for NA-based RT-PCR tests. Sequencing will continue to give us important information about how the virus is evolving, guiding the development of potential vaccines and therapies [62]. Recently, sequencing has been approved as a clinical diagnostic test (under FDA Emergency Use Authorization guidelines) for COVID-19 [63].
Sequencing approaches can be broadly divided into short-read and long-read technologies [64]. Short-read technology generates sequence data from NA fragments shorter than 1000 base pairs. Illumina sequencing, a short-read technology, is based on a sequencing-by-synthesis approach. The workflow involves three steps: library preparation (fragmentation of the DNA to be sequenced and ligation of adapters), the amplification of DNA fragments, and finally, sequencing of the amplified DNA strands based on the sequencing-by-synthesis approach. The collective fluorescent signal resulting from synthesizing a large number of amplified identical DNA strands allows the inference of nucleotide identity. Long-read technologies include single-molecule real-time sequencing (PacBio) and Nanopore sequencing (Oxford Nanopore), which are capable of generating sequencing data from high molecular weight NA (> 1000 base pairs). Nanopore sequencing works by measuring the electric current across an engineered protein nanopore, as a single strand of NA translocates through it.
NGS is an unbiased approach to the identification of novel infectious agents since it does not require a priori knowledge of the pathogen. NGS was used to identify the novel pathogen (2019-nCoV, now known as SARS-CoV-2) in bronchoalveolar lavage fluid (BLF) from three patients in Wuhan, China, exhibiting symptoms of severe pneumonia [59]. Sequencing was utilized after conventional PCR using a multiplexed RT-PCR panel failed to identify the presence of known pathogens (including human coronaviruses). NGS was able to identify the genome sequence of the causative pathogen in BLF from nine patients in Wuhan, exhibiting similar clinical symptoms [65]. The sequence was shown to be 99.98% identical to each other, which confirmed the presence of a novel, single virus. When similar symptoms were detected in patients in Australia [66], India [67], and Cambodia [68], sequencing was used to confirm that the symptoms were indeed caused by the same pathogen.
Timely availability of viral genome sequences is a key step in the design of RT-PCR-based assays for novel viruses such as SARS-CoV-2 [6]. Early on, SARS-related sequencing information allowed for the release of primers and probes for COVID-19 [69]. Since then, improved COVID-19 primers have been proposed [70], and a bait capture hybridization probe sequence has been released [71]. NGS is also an important tool that can provide insights into the temporal or geographic traits of pathogens. For instance, sequencing was an important tool in understanding the nature of early undetected community transmission of COVID-19 in the USA [72, 73]. In the past, researchers have tracked the evolution and spread of the Zika virus in the Americas [74, 75], and of the Ebola virus in West Africa [76]. In the case of COVID-19, sharing of this genomic data has already been very helpful [77], with existing consortiums employed. Early reports showed 99.9% sequence homology [78], but, more recently, higher sequence diversity has been reported [79, 80]. Recent research has shown that mutations in the SARS-CoV-2 spike protein led to the emergence of a more transmissible form of the virus [81, 82]. Understanding of SARS-CoV-2 mutations is also important to understand the effectiveness of future vaccines and therapeutics [83]. Recently, the CDC launched a nationwide genomics consortium to utilize real-time sequencing data to support such efforts [84].
Metagenomic NGS has been proposed as a diagnostic tool for detecting a broad spectrum of pathogens [75], including SARS-CoV-2 in clinical samples [85]. These include direct RNA sequencing of SARS-CoV-2, avoiding inherent amplification bias associated with RT-PCR [86, 87]. However, clinical samples with low viral titers will require targeted sequencing approaches [75], such as amplicon-based [88], hybridization-based capture [71, 89], and CRISPR-Cas-based enrichment [90]. In addition to its ability to detect co-infections, NGS can offer better sensitivity compared with RT-PCR [85]. The ability of NGS to detect small variations in the viral genome that could guide clinical decisions is also notable [85].
In comparison to RT-PCR, NGS has not been used extensively as a diagnostic tool in the current pandemic. This is possibly due to complex sample and library preparation protocols, expensive platforms requiring expertise to operate and analyze results, and relatively long turnaround time. The greatest difficulty is overcoming other signals in the sample: typically, < 1% reads are non-human, and NGS is particularly prone to contamination [91]. NGS has also traditionally been slower (a standard Illumina instrument takes > 18 h [92]) and more expensive than other methods.
In large-scale testing, NGS is not currently competitive with PCR-based methods; however, there is ample evidence to imply that it could become a viable testing method in the future. NGS diagnostic tests can be scaled using approaches utilizing reverse transcription (RT) primers to barcode up to 19,200 patient samples in a single sequencing run [93]. Viral genomes have been recovered directly from clinical samples [94], including testing of bacterial, fungal, viral, and parasitic RNA from multiple body fluids and tissues for multiplexed pathogen detection [91, 95]. Additionally, chemiluminescence enzyme immunoassay (CLIA)–certified laboratories are increasingly utilizing NGS for diagnosis of encephalitis, meningitis [96, 97], sepsis, and pneumonia [92]. In one test specific to COVID-19, nanopore sequencing was used to identify SARS-CoV-2 from NPS within 8 h [88]. Nanopore sequencing has a lower raw-read accuracy in comparison to sequencing-by-synthesis approaches. However, Nanopore sequencers such as the MinION are inexpensive, have faster turnaround time, and are portable. Further advances in automation and integration of sample and library preparation (see “Sample preparation”) will enable the widespread adoption of its capabilities. Table 1, summarizes the various approaches we have described in this section on the utilization of sequencing for COVID-19 pandemic management.
Table 1.
The utility of sequencing for various COVID-19 pandemic management requirements via pathogen discovery, clinical diagnosis, and outbreak surveillance
| Phases | Methodology | Ref. |
|---|---|---|
| Pathogen discovery | Identified a novel pathogen from patient samples in Wuhan (named 2019-nCoV, now known as SARS-CoV-2). Reports utilize targeted enrichment and sequencing using combinations of Sanger, BGI, Nanopore, & Illumina platforms | [59, 65, 98, 99] |
| Based on SARS-CoV-2 genomic data, insight into mutation in the spike protein (receptor-binding domain) involved in attachment to host cells (to human receptor ACE2) that helped explain the ability of the virus to infect human cells | [100] | |
| Comparative analysis of sequence data to deduce possible origins of SARS-CoV-2 | [60] | |
| Sanger, Illumina, & Nanopore sequencing on clinical samples from first patient/s diagnosed with COVID-19 in the USA, Australia, and India respectively | [66, 67, 101] | |
| Sequence data to design primers and probes for COVID-19 | [69] | |
| Clinical diagnosis | Amplicon-based target sequencing using Nanopore sequencer to detect SARS-CoV-2 and other respiratory organisms in clinical samples | [85, 88] |
| Target (amplicon-based and hybridization-based capture) sequencing using BGI sequencer to detect SARS-CoV-2 and other respiratory organisms in clinical samples | [89] | |
| Direct RNA sequencing of SARS-CoV-2 (from clinical specimens grown in cell culture) using Nanopore sequencer | [86, 87] | |
| Proposed scaled testing protocol using RT primers to barcode up to 19,200 patient samples in a single sequencing run | [93] | |
| Outbreak surveillance | Analysis of sequencing data from clinical samples (using Illumina sequencer) unveiled undetected community transmission of COVID-19 in the state of Washington | [72] |
| Analysis of sequencing data from clinical samples (using Illumina sequencer) unveiled multiple routes of introduction of COVID-19 into the state of California | [73] | |
| Amplicon-based targeted sequencing from clinical samples (using Nanopore sequencer) unveiled multiple routes of introduction of COVID-19 into the Netherlands | [102] | |
| Platforms and global consortiums utilizing genomic data to track real-time spread and evolution of pathogens including COVID-19 | [84, 103, 104] |
Antigen detection
Given the increasing global need for COVID-19 tests, rapid and inexpensive assays are required to supplement current NA amplification–based assays. These include antigen-based tests and often in the form of an immunoassay. In SARS-CoV-2 antigen detection, targets could be the nucleocapsid, spike, envelope, or membrane proteins of the virus [105–107]. Viral antigens bind specifically to a corresponding antibody, and the event can be detected using optical, magnetic, electrochemical, and surface plasmon resonance–based techniques, among other methods (see “Nanomaterials for POC COVID-19 diagnostics”) [108]. One of the challenges in developing effective antigen tests is the lack of antibodies for specific proteins of the SARS-CoV-2 virus. As an alternative to antibodies, SELEX (systematic evolution of ligands by exponential enrichment)-based strategies are useful in identifying affinity ligands such as aptamers specific to SARS-CoV-2. Recently, aptamers target the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein [109]. Aptamers are significantly easier and cheaper to produce in comparison to antibodies. Such efforts will enable the development of reliable and more accessible antigen-based assays.
The viral load is found to be variable in COVID-19 patients [110, 111], depending on factors such as time between disease onset and sample collection, type and quality of the sample, disease severity, and patient age [112]. Zou et al. reported Ct values in the range of 19–40 in upper respiratory specimens of infected patients [110]. Antigen tests are less sensitive than RT-PCR and could be less reliable in the clinical diagnosis of COVID-19 patients with low viral load. Such challenges have affected the clinical sensitivity of antigen tests for influenza and other respiratory viruses [112]. Recent studies on four different commercial antigen tests demonstrated a wide range of sensitivities from 16.7 to 85% (with 100% specificity) in COVID-19 clinical samples [113]. Other studies have shown sensitivity as high as 93.9% (CI 95%: 86.5–97.4). However, the reported sensitivities were lower and more variable (72.2%, CI 95%: 49.1–87.5) in samples with low viral load (Ct value > 25.1) [114]. Table 2 lists recent work utilizing the detection of SARS-CoV-2 antigens along with the sensitivity/limit of detection (LOD). Despite these limitations, rapid antigen tests can be used as a simple and inexpensive screening tool for active COVID-19 infections. For instance, rapid antigen tests have been proposed as a screening tool for COVID-19 at airports and border checkpoints. Sample collection remains a significant challenge in antigen testing. More ideal POC sample types, such as saliva, are less invasive, and their adoption is expected to accelerate the use of antigen tests as a much-needed screening tool. A recent evaluation reported a low sensitivity of 11.7% for self-collected COVID-19-positive saliva samples using antigen tests [115]. The wide variation in the sensitivities of rapid antigen tests needs to be evaluated and understood [120]. Based on the current understanding of the sensitivity challenges associated with antigen tests for influenza, we believe advances in respiratory sample collection, antigen preconcentration, and detection modalities will drive the adoption of antigen-based screening [122].
Table 2.
Antigen-based detection methods utilized for COVID-19 diagnostics
| Ref. | Detection method | Antigen | Sample type | Num. of samples | Days since symptom onset (days) | Sensitivity (%)/LOD | Specificity (%) |
|---|---|---|---|---|---|---|---|
| [106] | Fluorescence immunochromatographic assay | Nucleocapsid protein | NPS and urine | 239 | 3 | 68 | 100 |
| [115] | Fluorescence immunochromatographic assay | Nucleocapsid protein | Saliva | 103 | 9 | 11.7 | N/A |
| [116] | Chemiluminescence enzyme immunoassay | Nucleocapsid protein | NPS | 313 | N/A | 55.2 | 99.6 |
| [117] | Fluorescence immunochromatographic assay | Nucleocapsid protein (Genscript Cat #Z03488 & Genemedi GMP-V-2019nCoV-N002) | Non-clinical samples (in PBS buffer) | N/A | N/A | Genemedi − 0.65 ng/mL, Genscript − 3.03 ng/mL | N/A |
| [118] | Fluorescence immunochromatographic assay | SARS-CoV-2 antigen | NPS | 19 | N/A | N/A (low) | N/A |
| [114] | Fluorescence immunochromatographic assay | SARS-CoV-2 antigen | NPS and OPS | 127 | < 7 for 93.7% of samples | 93.9 | 100 |
| [119] | GICA | Nucleoprotein | NPS | 138 | N/A | 50 | 100 |
| [120] | GICA | Nucleoprotein | NPS | 148 | Median: 4, mean: 6.6, range: 0–34 | 30.2 | 100 |
| [121] | GICA | Nucleoprotein | NPS | 328 | N/A | 57.6 | 99.5 |
Antibody-based tests
Antibody-based tests detect antibodies generated as a result of a SARS-CoV-2 infection. Antibodies are proteins generated by the immune system in response to an infection. The level of antibody response can vary with age, gender, and presence of comorbidities [123, 124]. Immunoglobulin G, M, and A (IgG, IgM, and IgA) are being used as potential markers for COVID-19. IgM is shown to be an indicator of early-stage infection, while higher IgG levels are observed during late stages or post-recovery [124–128]. COVID-19 antibody can be detected based on existing methods (Table 3) such as colloidal gold-based immunochromatographic assay (GICA) [129–132], magnetic chemiluminescence enzyme immunoassay (CLIA) [123–125], and enzyme-linked immunosorbent assay (ELISA) [126, 133–136]. Pan et al. [137] and Li et al. [130] reported the detection of SARS-CoV-2 antibodies from whole blood. The sensitivity and specificity were found to be consistent with tests performed on serum samples [130].
Table 3.
Antibody-based tests utilized for COVID-19 diagnostics
| Ref. | Detection method | Antibody | Sample type | Num. of samples | Seroconversion (days) | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| [129] | GICA | IgG and IgM | Serum/whole blood | 134 | 7 | 96.81 | N/A |
| [130] | GICA | IgG and IgM | Serum/whole blood | 525 | N/A | 88.662 | 90.632 |
| [131] | GICA | IgG and IgM | Serum | 814 | 5 | 86.892 | 99.392 |
| [132] | GICA | IgG and IgM | Serum | 179 | 8 | 95.102 | 912 |
| [125] | CLIA | IgG and IgM | Serum | 285 | 13 | 1001 | N/A |
| [123] | CLIA | IgG and IgM | Serum | 159 | 14 | 91.142 | 802 |
| [124] | CLIA | IgG and IgM for nucleocapsid protein | Serum | 222 | 4 | 81.52 | 96.62 |
| [133] | ELISA | IgG and IgM | Serum | 238 | 11 | 81.32 | N/A |
| [134] | ELISA | IgG and IgM for nucleocapsid and spike protein | Serum | 214 | 10 | 82.2 | N/A |
| [135] | ELISA | IgG and IgM | Serum | 15 | 5 | N/A | N/A |
| [136] | ELISA | IgA, IgM, and IgG | Serum | 208 | 5 | 85.4 | N/A |
| [126] | ELISA | IgG, IgA for spike protein | Serum | 61 | N/A | N/A | N/A |
1Highest sensitivity among samples tested
2Sensitivity reported as a mean of all samples tested
N/A, data not reported or not relevant in the context of the referenced publication
Antibody tests are useful in epidemiological studies to assess the number of asymptomatic cases in the population [138, 139]. However, cross-reactivity of antibody is observed between human coronaviruses, such as SARS-CoV-2, SARS-CoV, and MERS-CoV. Antibody-based rapid diagnostic tests are performed on serum, plasma, or whole blood (fingerstick) POC settings. However, false-positives could lead to an inflated estimate of COVID-19 prevalence [139]. In addition, the temporal antibody response could vary depending on the individuals tested. Also, by design, antibody-based rapid diagnostic tests do not detect the presence of SARS-CoV-2 neutralizing antibodies. Tests that detect antibodies that have a high affinity for SARS-CoV-2 virus are more likely to indicate the presence of neutralizing antibodies. More research is needed to understand if a positive antibody test is indicative of immunity against future SARS-CoV-2 infections.
Despite these limitations, given the low cost and ease of use, with improvements in clinical sensitivity and specificity, antibody tests could be used for mass-screening of COVID-19 prevalence. It could also play a role in the evaluation of vaccines in the future [140].
COVID-19 testing models
Monitoring the prominence of infection throughout a pandemic is of critical importance in all stages of a pandemic. Each stage of a pandemic, as defined by the World Health Organization, has unique testing needs [141]. In early and peak phases, rapid turnaround times and scale-up are of great importance. In the post-peak phases of a pandemic, easily accessible, low-complexity, inexpensive, and high-throughput monitoring is needed. Current virus testing technologies and methods have proven effective in various capacities. However, each method has unique strengths and limitations, which must be considered and uniquely paired with the pandemic phase, population, available resources, and virus characteristics if an optimal response is to be achieved. While testing continues inside hospitals and clinics, other unique methods are proving to be highly effective testing models for the SARS-CoV-2 pandemic. Several unique testing models have been incorporated in the current pandemic response and will undoubtedly play a role in future pandemic response planning. Here, we describe these methods and their relation to diagnostic testing, and provide insight into their application.
The testing of infectious viruses presents several unique challenges. Traditionally, a patient visits a hospital for evaluation and diagnostic testing. The limitations of this method include increased risk of exposure due to travel and interaction with healthcare workers and social concentration of infected and non-infected persons. Other concerns include overloading healthcare workers and facilities, as well as the financial cost of in-person testing. Several unique testing models are being utilized to reduce exposure and resource consumption. Testing can be categorized by the location of the sample collection and analysis as well as if a healthcare worker aided in this process. The combination of these possibilities results in several unique testing models.
Drive-thru models have distinct advantages over traditional testing and have proven effective in rapid community testing [142]. While this model is limited in the number of communities worldwide that could utilize it, many urban and rural areas of the world could incorporate such a method. This method minimizes interaction while enabling more oversight on proper sample collection. The need for reporting of self-illness has been well argued [143, 144]. Self-testing models involve the collection and possibly the analysis of a sample at home by the patient. Recent work by Tu et al. has shown the clinical utility of patient self-collected swabs from the tongue, nasal, and mid-turbinate for COVID-19 diagnostic testing [145, 146]. The FDA has recently authorized at-home self-collection of saliva and nasal swabs for COVID-19 diagnostics [147]. The advantages of this method include expanded testing and reduced risk of nosocomial transmission in overcrowded healthcare settings. The primary limitation is the adaption of established testing methods to a home collection model. Additionally, accuracy in sample collection and processing in self-testing could be of concern. To address this concern, a home testing method where a healthcare worker visits a patient’s home to collect samples has found success in treating patients [148]. Healthcare workers can aid in sample collection and analysis to reduce the risk for error at the expense of increased risk of exposure. Several companies, including LabCorp, LetsGetChecked, and Nurx, have presented home-based POC collection devices for testing. It is encouraging to note that recent studies have reported equivalent sensitivity in self-collected samples [142, 149].
Point-of-care diagnostic methods with an emphasis on portability and speed are seeing a large increase in development; these tests are predominantly immunoassay based [107]. Aside from the assay itself, sample collection methods are also seeing progress. Spectrum Solutions LLC, a developer of biosample collection devices, developed the SDNA-1000 for collecting saliva biosamples, which is currently being used in SARS-CoV-2 testing and analysis. Such technologies enable self-testing models to become highly feasible. Yang et al. proposed a home-based testing model wherein the patient collects and processes their sample and then images the result from a paper-based assay using a cell phone for subsequent cloud upload and evaluation [150]. The integration of POC diagnostic assay with self-collection of samples will reduce the risk associated with the shipping of infectious samples and avoid potential issues due to sampling degradation during transport.
SARS-CoV-2 clinical sample collection and preparation
Though all the test methods are fundamentally different, the quality of the sample is crucial for successful detection. This is contingent on the detectable presence of the biomolecules (viral RNA, antigens, antibody, etc.) in the sample. Thus, the type of sample (e.g., blood, serum, sputum, nasal swabs, fecal matter, urine), collection methods, and sample pre-processing all play a vital role in detection regardless of the technique. For instance, the sensitivity of RT-PCR tests is dependent on the type of swab used [151]. Increasingly, inaccuracies in RT-PCR tests have been attributed to sample collection [13, 152] and handling. Upper respiratory samples are the primary specimen used for COVID-19 diagnostic testing [153]. In comparison to advances in amplification-based tests, methods for the collection of respiratory samples need improvement. Given the scale of testing required, improper sampling by untrained personnel has contributed to false-negative results [152]. There have been efforts to develop a robot for respiratory swab sampling [21]. However, the device was not tested in vivo or clinical settings. Alternative samples like saliva or nasal swabs are simpler, less invasive, and could potentially be used to avoid improper sampling associated with respiratory swab samples.
Sample preparation
Sample preparation (or preprocessing) refers to the sequence of steps required to convert a clinical sample into a form compatible with downstream analysis and detection such as amplification or sequencing. Sample preparation has been a major bottleneck in widespread testing during the COVID-19 pandemic. The availability of RNA extraction kits and lysis reagents and low-throughput RNA extraction protocols have resulted in increased turnaround times and delays. A rapid surge in demand for tests is threatening to overwhelm the diagnostic capabilities. A review by Esbin et al. describes alternative COVID-19 diagnostic protocols involving lysis reagents and direct addition of samples (extraction-free protocols) into the amplification reaction mix [22]. There have been recent advances in extraction-free protocols for COVID-19 amplification [154–159] and sequencing-based tests [160]. However, such protocols involve trade-offs between lower sensitivity and simplicity [22, 161, 162]. Automated extraction platforms utilizing liquid handling robots are useful for high-throughput, centralized COVID-19 diagnostic testing. In addition, decentralized point-of-care (POC) diagnostic tests also have a role to fulfill. Integrated POC platforms with “Sample-in Answer-out” capability will enable COVID-19 diagnostic capability in doctor’s office and clinics. Such platforms will ensure reduced turnaround time, repeatability, and potentially lower costs by reducing reagent consumption. These will involve automation of sample preparation utilizing microfluidics and their integration with the detection assay. Such methods have been extensively reviewed [44, 163, 164] and will need to be validated with COVID-19 clinical samples.
Similar approaches are required to push increased utilization of sequencing to support the COVID-19 pandemic. For instance, the complexity and labor-intensive nature of sample and library preparation workflow could limit the widespread adoption of sequencing [165–167]. Efforts towards the automation and integration of sample and library preparation steps will enable increased adoption of sequencing platforms for COVID-19 pandemic management. Expanding sequencing capabilities will enable us to track mutations of the SARS-CoV-2 virus in real-time with potential implications to the efficacy of future COVID-19 vaccines and therapeutics.
It is important to treat a diagnostic assay as an integrated unit, with each step of the workflow from sample collection and preprocessing to the final detection and analysis, impacting the overall sensitivity.
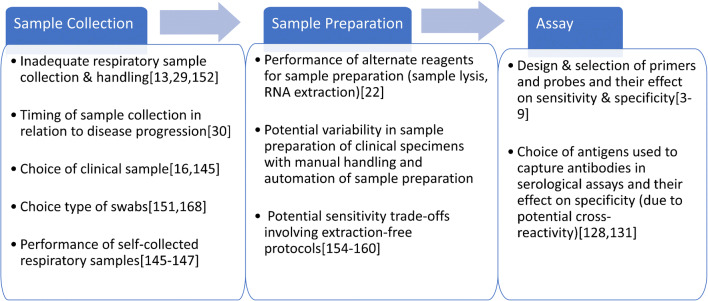
We have summarized some of the key factors impacting each step in the diagnostic workflow (from sample collection to detection) with reference to relevant literature (Fig. 3).
Fig. 3.
Key factors that could potentially impact each step of the workflow for COVID-19 diagnostic assays
SARS-CoV-2 clinical sample types
Nasopharyngeal/oropharyngeal sample
Nasopharyngeal/oropharyngeal swabs are generally used as the collection mode for upper respiratory samples in early detection of SARS-CoV-2 by PCR. In particular, flocked swabs are especially useful when the total volume of the sample matrix is low. These special types of nylon swabs help in collecting and retaining more analyte than traditional spun-fiber swabs made from cotton, polyester, or rayon [168]. Swabs are generally taken from the nasopharynx or oropharynx [169], with a nasopharyngeal sample being more sensitive than oropharyngeal [16, 170, 171]. Usually, a healthcare worker collects these samples, but it has been shown that the collection can be done at home by the patient [172, 173]. Self-sampling reduces the risks to healthcare workers (see “COVID-19 testing models”). Following swab collection, swabs are placed into a viral RNA extraction medium and subsequently amplified (e.g., by RT-PCR) [169].
Depending on the disease progression and the number of days passed since the onset of symptoms, the viral load varies considerably [169]. In such cases, swabs from different sources can be combined to increase the probability of detection.
Sputum
Sputum is mucus produced by the act of coughing up and spitting out the material produced in the respiratory tract (the trachea and bronchi) [174]. It has been shown that, for COVID-19, sputum contains more viral RNA than NPS, resulting in a higher chance of detection [170, 175, 176]. Sputum collection is a much simpler process than swab sampling and can easily be done by the patient. This reduces the chances of healthcare providers getting infected while collecting samples. This process involves rinsing the mouth with water, followed by expectorating of deep cough sputum directly into a sterile, leak-proof, collection cup. However, not every patient produces sputum. Alternatively, sputum can be clinically induced [177]. Han et al. showed that SARS-CoV-2 could be detected from such samples [178].
Saliva
Saliva is an alternative sample that can be easily be collected by the patient at home. Zheng et al. showed that saliva had a higher detection rate (88.09%) for SARS-CoV-2 in comparison to throat swabs (45.24%) and NPS (76.19%) [171]. While most studies have found saliva to be as sensitive as NPS for COVID-19 diagnostic tests [54, 179, 180], few have reported slightly lower sensitivity [181].
Bronchoalveolar lavage
BLF is an excellent sample for COVID-19 diagnostic with a high detection rate (93%) compared to sputum (73%) and NPS (63%) [170]. It is generally collected from patients with severe illness or undergoing mechanical ventilation. Collecting BLF involves the instillation of sterile normal saline into a subsegment of the lung, followed by suction and collection [182]. Due to the complexity involved and the aerosol-generating nature of the procedure, BLF is more useful in determining the recovery of admitted patients.
Stool
Stool samples can be collected and tested concurrently with other samples to both monitor disease progression and limit the instance of false-positives. Like many other coronaviruses, large viral loads of SARS-CoV-2 can be found in stool [170, 183, 184]. However, it should be noted that stool samples tend to be among the most difficult samples to process for viral detection. Stool samples are difficult to process due to the presence of many PCR inhibitors, such as bile, polysaccharides, hemoglobin, and bilirubin [185]. Processing of stool samples often requires highly trained technicians, and results are highly susceptible to user error. Therefore, the viral detection rate could be variable [183, 186–188]. Viral loads can be found in stool samples at early onset through the convalescent stage of illness. Viral loads have been found in stool samples from as early as the onset of symptoms to as late as 33 days after onset of symptoms [175, 186].
Stool samples may provide a non-invasive alternative to NPS and OPS. The collection of stool samples is simple and can often be performed at home. Sample collection involves the capture of a stool sample in a clean, dry container and transfer of the sample into a sterile specimen cup.
Serum and whole blood
Serum and whole blood are primarily used in antibody tests for tracking disease progression, epidemiological studies, and patient immunity. These have been discussed in the section “Antibody-based tests.”
Table 4 lists the various clinical samples used in COVID-19 diagnostics and important factors such as peak viral load, time to peak viral load, and average detection rate. These factors should enable guidance on their utility for different testing models such as at-home or POC testing.
Table 4.
Types of SARS-CoV-2 clinical samples and the associated time to peak viral/antigen load
| Sample type | Self-sampling (yes/no) | Viral load (high/medium/low) | Approx. time to peak viral/antigen load | Average detection rate | Ref. |
|---|---|---|---|---|---|
| NPS | No, medical facility preferred | High (Ct: 16.9–38.4, average: 24.3) | 0–7 days after symptom onset | 63–65% | [16, 170–173] |
| Sputum | Yes, can be done with minimal instruction | Medium–high (Ct: 18.4–38.8, average: 31.3) | 3–7 days after symptom onset | 70–95% | [170, 171, 175, 189] |
| Saliva | Yes, can be done with minimal instruction | High (Ct: 24.31–30.31, average: 27.41) | 3–7 days after symptom onset | 88–90% | [110, 171, 179, 190, 191] |
| BLF | No, medical facility required | Medium (Ct: 26.4–36.2, average: 31.1) | N/A | 92% | [170] |
| Stool | Yes, can be done with minimal instruction | Medium (Ct: 22.3–38.4, average: 31.4) | 6–14 days after symptom onset | 50–89% | [170, 183, 186–188, 192] |
| Serum | Yes, at-home sampling kits available | Low (Ct: 34.1–35.4, average: 34.6) | Viral presence rare. 3–14 days for antigen detection | 1% viral RNA detection | [170, 193] |
Environmental sampling and surveillance
COVID-19 is primarily spread via human-to-human transmission, though environmental transmission has also been reported [194]. Unlike other enveloped viruses, SARS-CoV-2 are less susceptible to environmental stresses [194]. Depending on the environment, temperature, and type of surfaces, it can survive up to 14 days [195]. Hence, environmental surveillance for SARS-CoV-2 is useful in COVID-19 containment and preventing transmission.
Air sampling
Air sampling can be done using an air sampling pump [196] (e.g., SKC BioLite+, Zefon Bio-Pump® Plus). These pumps can sample dust/particulates, vapors/gases, bioaerosols, or environmental air samples. They use cassettes and filters to collect samples for further analysis. Air samplers are efficient, adaptable, and easy to use. But for preserving the viability of samples, quick and subsequent handling of sampling is needed. Air samplers are used for confined spaces such as hospital rooms and apartments.
Sewage water sampling
Researchers are now focusing on the sampling of sewage water for COVID-19 surveillance [197–199]. This can be used to detect the presence of SARS-CoV-2 in certain populations and also to quantify the scale of infection [200].
Sampling from surfaces
The persistence of SARS-CoV-2 on surfaces is important to understand the risk of infection from fomites. For instance, sampling from high-touch surfaces will help uncover modes of transmission and also the effectiveness of disinfection protocols [201]. WHO has provided guidelines on potential sampling sites for such studies at COVID-19 healthcare facilities [202]. Premoistened swabs are used to collect samples from such surfaces with RT-PCR used to detect the presence of SARS-CoV-2 [203–205]. Viral cultures could demonstrate the viability of SARS-CoV-2 on such surfaces. Surface sampling using swabs is labor-intensive, and a large number of samples are required in surveillance studies.
Nanomaterials for POC COVID-19 diagnostics
In April this year, the National Institutes of Health (NIH) launched the Rapid Acceleration of Diagnostics-Radical (RADx-rad) initiative to accelerate the development of novel approaches to COVID-19 diagnostics [206]. This initiative is also aimed at the repurposing of current diagnostic techniques towards COVID-19 and future pandemics. The initiative plans to enable rapid COVID-19 detection platforms for home-based and POC use. In this section, we review some of the recent work on advanced techniques utilizing nanostructured materials, developed for the detection of active COVID-19 infections at POC. These include platforms utilizing field-effect transistors (FET), plasmonics, and breath-based detection of SARS-CoV-2 virus, associated antigens, or RNA.
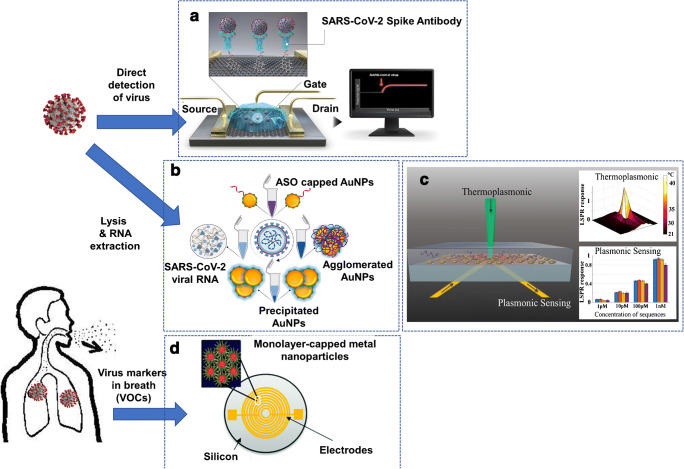
FET biosensors consist of an electrode (gate) functionalized with receptors or probe molecules to specifically bind with the analyte of interest. The binding of the target analyte causes a change in electrostatic surface potential (electrostatic gating effect), resulting in a change in the current flowing between the source and drain [211]. Seo et al. developed a graphene-based FET sensor for the detection of SARS-CoV-2 in clinical samples (Fig. 4a) [207]. The sensor consists of graphene sheets conjugated with the SARS-CoV-2 spike antibody (receptor) that specifically binds with the SARS-CoV-2 spike protein (analyte). The binding causes a real-time change in the current response, indicating the presence of SARS-CoV-2 spike protein or SARS-CoV-2 virus in the sample. The platform was shown to be highly specific (exhibiting no response to MERS-CoV spike proteins) and was able to detect clinical samples with SARS-CoV-2 viral load as low as 242 copies/mL. Researchers from the University of Maryland, Baltimore, developed a colorimetric assay utilizing thiol-modified antisense oligonucleotides (ASO) capped on the surface of gold nanoparticles (AuNPs) for the detection of isolated SARS-CoV-2 RNA (Fig. 4b) [208]. The ASO-capped AuNPs agglomerate in the presence of SARS-CoV-2 viral RNA, causing a change in surface plasmon resonance (SPR) of the agglomerates in solution. Further addition of RNase H causes the ASO-capped AuNPs to precipitate, resulting in further amplification of the SPR signal and enabling a visual detection of SARS-CoV-2 RNA in 10 min. Qiu and co-workers developed a dual-functional plasmonic biosensor incorporating localized surface plasmon resonance (LSPR) and plasmonic photothermal (PPT) effect for the detection of SARS-CoV-2 (Fig. 4c) [209]. The biosensor consists of two-dimensional gold nanoislands (AuNIs) functionalized with complementary DNA (cDNA) receptors that hybridize with the target SARS-CoV-2 RNA sequence. The hybridization event leads to an LSPR phase change (response) that can be detected. In addition, when the AuNIs are illuminated at the plasmonic resonance frequency, a localized thermoplasmonic heat is generated, promoting the selective hybridization of the target SARS-CoV-2 RNA sequence to cDNA receptors. Biosensors based on electrochemical sensing [212, 213], quartz crystal microbalance [214], and magnetoresistance [215] have been utilized in the past for viral detection [108]. These techniques can, in principle, be applied to COVID-19 detection at POC. Haick et al. developed an interesting alternative to COVID-19 diagnostics involving the detection of volatile organic compounds (VOCs) from the exhaled breath of patients (Fig. 4d) [210]. The chemi-resistive sensor array consists of AuNPs linked to organic ligands that swell or shrink upon exposure to various VOCs, causing a change in the electric resistance. Although the technique will need to be validated in a clinical study with a larger sample size, it could prove very useful as a rapid screening tool for COVID-19.
Fig. 4.
Schematic of novel COVID-19 diagnostics platforms utilizing nanostructured materials for potential POC use. a A graphene-based FET sensor for SARS-CoV-2 detection. The device consists of the SARS-CoV-2 spike antibody conjugated to graphene sheets. The specific binding of spike antibody to SARS-CoV-2 spike protein causes a change in current (response) between source and drain. (modified with permission from [207]. Copyright © American Chemical Society) b A colorimetric assay based on SPR utilizing thiol-modified antisense oligonucleotides capped on the surface of gold nanoparticles for the detection of isolated SARS-CoV-2 RNA. (modified with permission from [208]. Copyright © American Chemical Society) c A dual-functional LSPR biosensor utilizing gold nanoislands for sensitive detection of target SARS-CoV-2 RNA sequence from a multigene mixture. (modified with permission from [209]. Copyright © American Chemical Society) d A chemi-resistive sensor utilizing array consisting of AuNPs for detection of VOCs from the exhaled breath of COVID-19 patients (reproduced from [210] with permission from the Royal Society of Chemistry)
Commercial tests for COVID-19 diagnostics
Rapid and reliable commercial screening of SARS-CoV-2 is critical to limit the 2019 COVID-19 pandemic. Research labs and medical device companies across the world made major shifts in manufacturing desperately needed test kits at a rate that matches the rapidly increasing demand during the 2019–2020 pandemic. To produce effective COVID-19 tests, companies must overcome the challenges associated with scalability, accuracy, cost, testing supply-demand, and sample collection. The USA alone has performed over 5 million COVID-19 tests in the first 3 months of testing [216]. Reducing storage, transportation, and testing time are of utmost importance. COVID-19 tests generally fall into two categories, virus RNA testing (using mostly RT-PCR) to diagnose current infections or serological tests for anti-SARS-CoV-2 antibodies to determine if the patient was exposed.
China first released the COVID-19 genome on Jan 11, 2020, and the Malaysian Institute for Medical Research successfully produced “primers and probes” for SARS-CoV-2 the same day [217]. Because of the early and rapid deployment of tests, South Korea stands out for its response to the COVID-19 pandemic. South Korean company, Seegene, was one of the first commercial companies to provide large-scale testing with the development of the Allplex 2019-nCoV Assay, which was deployed in an impressive 3 weeks within the release of the COVID-19 genome. Allplex™ 2019-nCoV Assay is a multiplex real-time RT-PCR assay that targets three specific genes for SARS-CoV-2, which include RdRP and N genes and E gene for all Sarbecovirus, including SARS-CoV-2. In the USA, the CDC was the first to produce a one-step process that tests samples on a 96-well plate and completes a full test in ~ 45 min using rRT-PCR [218].
A major issue that hindered PCR testing is the supply chain for reagents and swabs. Samples for COVID-19 PCR are collected from the nasal, nasopharyngeal, or oropharyngeal areas using specialized synthetic fiber swabs with plastic shafts. Guidelines set by the US Centers for Disease Control and Prevention (CDC) state that the sample be suspended in viral transport media and stored between 2 and 8 °C for up to 72 h and after 72 h must be stored at − 70 °C, which poses challenges for widespread sample collection [219]. Collected samples are combined with primers and polymerase and then passed through a thermocycler for amplification and optical detection.
Considering the scale of required tests and the highly infectious nature of COVID-19, sample processing is well suited for automated devices such as Roche’s (Basel, Switzerland) rapid test polymerase chain reaction detection devices. In an 8-h period, these devices can reportedly process 384 and 1058 patient samples for the Cobas 6800 and 8800, respectively [220]. Point-of-care devices remove the need for storage and transportation, saving substantial testing time. Bosch (Waiblingen, Germany) has developed a portable point-of-care RT-PCR device called Vivalytic, which is fully automatic and can complete a full in situ test in 2.5 h.
Serological testing [221] provides surveillance of SARS-CoV-2 antibodies identifying who has been in contact with the virus and information on immunity. The development of antibodies typically takes 2 weeks after infection, and for this reason, serological testing is not the preferred method for diagnosing infection. Common commercialized serological tests include rapid diagnostic tests (RDTs) and enzyme-linked immunosorbent assay (ELISA). RDTs are typically point-of-care qualitative lateral flow assays where samples are collected from a finger prick, saliva sample, or nasal swab [52]. ELISA tests whole blood, plasma, or serum and is typically performed in a laboratory. Patient samples are incubated with an immobilized pathogen in which antibodies in the sample can form bonds. A secondary immunolabel with a fluorescent signature is used to quantifiably and qualitatively identify the pathogen [222, 223].
Conclusion and future direction
The 2019 SARS-CoV-2 pandemic brought to light the weaknesses of many health organizations worldwide in meeting the challenges of a global pandemic. The global financial and health costs associated with this pandemic are sure to total trillions of dollars. Global pandemics will likely continue to be a significant threat in the modern world, and the pandemic response strategy is worthy of a thorough evaluation. Viral diagnostic testing has been observed to be one of the areas in the SARS-CoV-2 pandemic response in need of assessment and improvement. Assessment of the global response with a focus on the diagnostic methods and instruments utilized will provide recognition on what should be done to better prepare for future pandemics. We reviewed aspects of viral diagnostic tools relevant in the SARS-CoV-2 pandemic and provided insight into their current state, including advantages, limitations, and scalability, and then provided direction on future work that would enable diagnostics to be better prepared for a future pandemic response.
RT-PCR remains the gold standard in SARS-CoV-2 infection testing. RT-PCR has encountered limitations in sensitivity. Improved primer and probe design, along with better control over preanalytical variables such as sample collection and handling, is expected to improve performance in clinical settings. Other NA amplification techniques, including dPCR and isothermal amplification, are being used to a lesser degree and have distinct advantages in sensitivity and turnaround time, respectively. CRISPR-Dx shows promise in achieving higher sensitivity and specificity over NA amplification alone. Additionally, CRISPR-based technologies appear to be more conducive to multi-target reactions, raw body sample compatibility, and POC applications.
In addition to being the first step in the identification of a novel virus strain, sequencing can provide insight into the evolution, transmission, and unique characteristics of a virus. While not commonly used as a diagnostic method due to higher turnaround time and costs over traditional methods, the FDA has recently granted EUA to a high-throughput SARS-CoV-2 clinical testing (Illumina COVIDSeq) [63]. The advent of nanopore sequencers combined with advances in integrated sample and library preparation will enable the widespread adoption of its sequencing for COVID-19 pandemic management.
Although antibody and antigen detection methods were initially limited by low sensitivity and specificity as well as a longer development cycle in comparison to RT-PCR, they promise a cost-effective approach to population-wide mass-screening and are amenable to POC use.
Commercial testing has relied on RT-PCR and serological testing, aiming to diagnose a current infection and previous exposure, respectively. The commercial response was limited by supply chain issues rather than by technology constraints. RT-PCR-based assays were available within a few weeks of genome discovery, while serological testing methods became available within a few months.
Beyond the viral diagnostic technologies themselves, the methodologies involving the collection and analysis of samples have played and will continue a critical role in effectively diagnosing viral infections during a pandemic. Methods, including self- and drive-thru testing, are expected to limit transmission while reducing overall per-patient testing costs. As diagnostic testing technologies develop, it will be important to incorporate the unique user needs and requirements associated with pandemic response methodologies.
Finally, biological sample collection and preparation remain essential regardless of the sample detection method. The type of sample, viral load, collection method, illness phase, and preprocessing are all factors that impact the sensitivity of the diagnostic method and must be appropriately matched for ideal results. Improvements to preanalytical vulnerabilities (such as ensuring repeatable sample collection and processing) should move in tandem with advances in the analytical performance of an assay to ensure the optimum clinical performance of a COVID-19 assay.
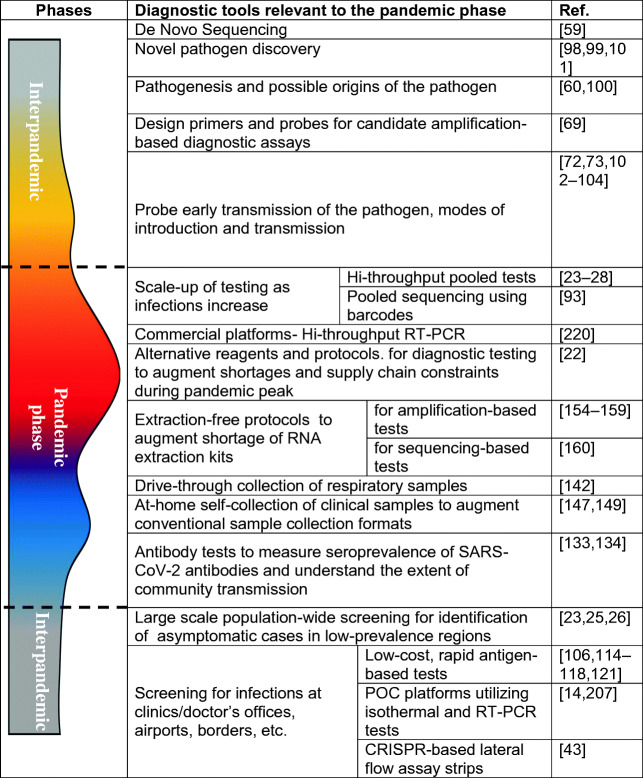
Table 5 provides a summary of various techniques reviewed in this work and their potential role in disease control and pandemic management.
Table 5.
The complementary role played by various COVID-19 diagnostic tools in different phases of the pandemic
In conclusion, given the global spread and scale of COVID-19 infections, the diagnostic ecosystem has encountered various bottlenecks. In spite of these challenges, various diagnostic tools will continue to play a critical and complementary role in the management of various stages of the COVID-19 pandemic.
Abbreviations
- ASO
Thiol-modified antisense oligonucleotides
- AuNIs
Two-dimensional gold nanoislands
- AuNPs
Gold nanoparticles
- BLF
Bronchoalveolar lavage fluid
- Cas
CRISPR-associated nuclease
- CDC
US Centers for Disease Control and Prevention
- cDNA
Complementary DNA
- CLIA
Chemiluminescence enzyme immunoassay
- COVID-19
Coronavirus disease 2019
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CRISPR-Dx
CRISPR-based diagnostics
- crRNA
CRISPR RNA
- Ct
Cycle threshold
- DETECTR
SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter
- dPCR
Digital PCR
- ELISA
Enzyme-linked immunosorbent assay
- FET
Field-effect transistor
- GICA
Colloidal gold-based immunochromatographic assay
- gRNA
Guide RNA
- hCoV-229E
Human coronavirus 229E
- hCoV-OC43
Human coronavirus OC43
- HUDSON
Heating unextracted diagnostic samples to obliterate nucleases
- LAMP
Loop-mediated isothermal amplification
- LOD
Limit of detection
- LSPR
Localized surface plasmon resonance
- MERS-CoV
Middle East respiratory syndrome coronavirus
- NA
Nucleic acid
- NGS
Next-generation sequencing
- NIH
National Institutes of Health
- NPS
Nasopharyngeal swabs
- OPS
Oropharyngeal swabs
- PCR
Polymerase chain reaction
- POC
Point of care
- PPT
Plasmonic photothermal
- RADx-rad
Rapid Acceleration of Diagnostics-Radical
- RBD
Receptor-binding domain
- RT-LAMP
Reverse transcription loop-mediated isothermal amplification
- RT-PCR
Reverse transcription polymerase chain reaction
- SARS-CoV
Severe acute respiratory syndrome coronavirus (virus identified in 2003)
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SELEX
Systematic evolution of ligands by exponential enrichment
- SHERLOCK
Specific high-sensitivity enzymatic reporter unlocking
- SHINE
SHERLOCK and HUDSON Integration to Navigate Epidemics
- SPR
Surface plasmon resonance
- VOCs
Volatile organic compounds
Compliance with ethical standards
Conflict of interest
Harikrishnan Jayamohan is a former employee of Roche Sequencing Solutions Inc. and owns shares in Roche Holding AG. All other authors have no conflicts to declare.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Harikrishnan Jayamohan and Christopher J. Lambert contributed equally to this work.
References
- 1.Shen M, Zhou Y, Ye J, Abdullah Al-maskri AA, Kang Y, Zeng S, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020. 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed]
- 2.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020. 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed]
- 3.Vogels CBFF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol. 2020) 2020.03.30.20048108. 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed]
- 4.John Hopkins Center for Health Security, Comparison of national RT-PCR primers, probes, and protocols for SARS-CoV-2 diagnostics, Centerforhealthsecurity.Org. 2020;5. https://www.centerforhealthsecurity.org/resources/COVID-19/COVID-19-fact-sheets/200410-RT-PCR.pdf.
- 5.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020:0–28. 10.1021/acsnano.0c02624. [DOI] [PubMed]
- 6.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed]
- 7.Nalla AK, Casto AM, Casto AM, Huang MLW, Perchetti GA, Sampoleo R, et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol. 2020. 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed]
- 8.Anantharajah A, Helaers R, Olive N, Defour J-P, Olive N, Kabera F, et al. How to choose the right real-time RT-PCR primer sets for the SARS-CoV-2 genome detection? Res Sq. 2020. 10.21203/rs.3.rs-36512/v1. [DOI] [PMC free article] [PubMed]
- 9.Jung YJ, Park G-S, Moon JH, Ku KB, Beak S-H, Kim SISS-JJ, et al. Comparative analysis of primer-probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2). ACS Infect Dis. 2020. 10.1021/acsinfecdis.0c00464. [DOI] [PubMed]
- 10.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020:1–30. 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed]
- 11.Dong L, Zhou J, Niu C, Wang Q, Pan Y, Wang X, et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. MedRxiv. 2020. 10.1101/2020.03.14.20036129. [DOI] [PMC free article] [PubMed]
- 12.Lu R, Wang J, Li M, Wang Y, Dong J, Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. MedRxiv. 2020) 2020.03.24.20042689. 10.1101/2020.03.24.20042689.
- 13.Kinloch NN, Ritchie G, Brumme CJ, Dong W, Dong W, Lawson T, et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. MedRxiv. 2020) 2020.05.05.20091728. 10.1101/2020.05.05.20091728.
- 14.Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol. 2020. 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed]
- 15.Gandhi M, Yokoe DS, Havlir DV. Asymptomatic transmission, the achilles’ heel of current strategies to control Covid-19. N Engl J Med. 2020. 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed]
- 16.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen H-L, Peiris M, Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano G, Blanchini F, Bruno R, Colaneri P, Di Filippo A, Di Matteo A, et al. Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy. Nat Med. 2020. 10.1038/s41591-020-0883-7. [DOI] [PMC free article] [PubMed]
- 18.Peto J. Covid-19 mass testing facilities could end the epidemic rapidly. BMJ. 2020. 10.1136/bmj.m1163. [DOI] [PubMed]
- 19.Balilla J. Assessment of COVID-19 mass testing: the case of South Korea. SSRN Electron J. 2020. 10.2139/ssrn.3556346.
- 20.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection. Ann Intern Med. 2020. 10.7326/m20-3012. [DOI] [PubMed]
- 21.Craney AR, Velu P, Satlin MJ, Fauntleroy KA, Callan K, Robertson A, et al. Comparison of two high-throughput reverse transcription-polymerase chain reaction systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol. 2020. 10.1128/JCM.00890-20. [DOI] [PMC free article] [PubMed]
- 22.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdun CM, Fuchs T, Harar P, Elbrächter D, Fischer DS, Berner J, et al. Group testing for SARS-CoV-2 allows for up to 10-fold efficiency increase across realistic scenarios and testing strategies. MedRxiv. 2020. 10.1101/2020.04.30.20085290. [DOI] [PMC free article] [PubMed]
- 24.Shani-Narkiss H, Gilday OD, Yayon N, Landau ID. Efficient and practical sample pooling high-throughput PCR diagnosis of COVID-19. MedRxiv. 2020. 10.1101/2020.04.06.20052159.
- 25.Aragón-Caqueo D, Fernández-Salinas J, Laroze D. Optimization of group size in pool testing strategy for SARS-CoV-2: a simple mathematical model. J Med Virol. 2020. 10.1002/jmv.25929. [DOI] [PMC free article] [PubMed]
- 26.Narayanan K, Frost I, Heidarzadeh A, Tseng KK, Banerjee S, John J, et al. Pooling RT-PCR or NGS samples has the potential to cost-effectively generate estimates of COVID-19 prevalence in resource limited environments. MedRxiv. 2020. 10.1101/2020.04.03.20051995.
- 27.Keeler E, Goldman D, Trish E. Getting Americans back to work (and school) with pooled testing – USC Schaeffer; n.d..
- 28.Martin A, Storto A, Andre B, Mallory A, Dangla R, Visseaux B, Gossner O. High-sensitivity COVID-19 group testing by digital PCR, ArXiv Prepr. ArXiv2006.02908; 2020. [DOI] [PMC free article] [PubMed]
- 29.Lippi G, Simundic A-M, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020. 10.1515/cclm-2020-0285. [DOI] [PubMed]
- 30.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020. 10.7326/m20-1495. [DOI] [PMC free article] [PubMed]
- 31.Dekker RJ, Ensink WA, van Leeuwen S, Rauwerda H, Breit TM. Overhauling a faulty control in the CDC-recommended SARS-CoV-2 RT-PCR test. BioRxiv. 2020. 10.1101/2020.06.12.147819.
- 32.Rosebrock AP. Patient DNA cross-reactivity of the CDC SARS-CoV-2 extraction control leads to an inherent potential for false negative results. BioRxiv. 2020. 10.1101/2020.05.13.094839.
- 33.Widders A, Broom A, Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health. 2020. 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed]
- 34.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020:1–10. 10.1038/s41586-020-2196-x. [DOI] [PubMed]
- 35.Centers for Disease Control and Prevention, Criteria for return to work for healthcare personnel with suspected or confirmed COVID-19 (Interim Guidance); 2020.
- 36.Mello CJ, Kamitaki N, de Rivera H, McCarroll SA. Absolute quantification and degradation evaluation of SARS-CoV-2 RNA by droplet digital PCR. MedRxiv. 2020. 10.1101/2020.06.24.20139584.
- 37.Chiu C. Cutting-edge infectious disease diagnostics with CRISPR. Cell Host Microbe. 2018;23:702–704. doi: 10.1016/j.chom.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L, Collins JJ. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 41.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed]
- 44.Jayamohan H, Romanov V, Li H, Son J, Samuel R, Nelson J, Gale BK. Advances in microfluidics and lab-on-a-chip technologies, 2016. 10.1016/B978-0-12-802971-8.00011-0.
- 45.Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q, et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull. 2020. 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed]
- 46.Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W, et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens Bioelectron. 2020. 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed]
- 47.Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, Pain A, Alofi FS, Alsomali A, Hashem AM, Khogeer A, Almontashiri NAM, Abedalthagafi M, Hassan N, Mahfouz MM. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah N. Higher co-infection rates in COVID19, Medium.Com. 2020.
- 50.Zhu X, Ge Y, Wu T, Zhao K, Chen Y, Wu B, et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020. 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed]
- 51.Bordi L, Nicastri E, Scorzolini L, Di Caro A, Capobianchi MR, Castilletti C, Lalle E. On behalf of Inmi Covid-Study Group, Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Euro Surveill. 2020;25:2–5. doi: 10.2807/1560-7917.ES.2020.25.8.2000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roche Molecular Systems, Fact sheet for healthcare providers-cobas® SARS-CoV-2; 2020.
- 53.To KKW, Yip CCY, Lai CYW, Wong CKH, Ho DTY, Pang PKP, Ng ACK, Leung KH, Poon RWS, Chan KH, Cheng VCC, Hung IFN, Yuen KY. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect. 2019;25:372–378. doi: 10.1016/j.cmi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 54.To KK-W, Tsang OT-Y, Yip CC-Y, Chan K-H, Wu T-C, Chan JM-C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020:4–6. 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed]
- 55.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F, Abudayyeh OO, Gootenberg JS, Sciences C, Mathers L. A protocol for detection of COVID-19 using CRISPR diagnostics. Cambridge: Broad Inst. MIT Harvard; 2020. pp. 1–8. [Google Scholar]
- 57.Lucia C, Federico P-B, Alejandra GC, Curti L, Pereyra-Bonnet F, Gimenez C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. BioRxiv. 2020. 10.1101/2020.02.29.971127.
- 58.Arizti-Sanz J, Freije CA, Stanton AC, Boehm CK, Petros BA, Siddiqui S, et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. BioRxiv Prepr Serv Biol. 2020. 10.1101/2020.05.28.119131. [DOI] [PMC free article] [PubMed]
- 59.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Sen Tan K, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao H, Lu X, Chen Q, Xu K, Chen Y, Cheng L, et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. SSRN Electron J. 2020. 10.2139/ssrn.3578153.
- 63.U.S. Food and Drug Administration, Coronavirus (COVID-19) update: FDA authorizes first next generation sequence test for diagnosing COVID-19; 2020.
- 64.Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet. 2016. 10.1146/annurev-genom-083115-022413. [DOI] [PubMed]
- 65.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caly L, Druce J, Roberts J, Bond K, Tran T, Kostecki R, et al. Isolation and rapid sharing of the 2019 novel coronavirus (SAR-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust. 2020:2–5. 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed]
- 67.Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, Majumdar TD, Shete-Aich A, Basu A, Abraham P. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151(2 & 3):200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manning JE, Bohl JA, Lay S, Chea S, Sovann L, Sengdoeurn Y, et al. Rapid metagenomic characterization of a case of imported COVID-19 in Cambodia. BioRxiv. 2020. 10.1101/2020.03.02.968818.
- 69.World Health Organization, Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR; 2020. https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf.
- 70.Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by the ARTIC Network’s multiplex PCR. BioRxiv. 2020. 10.1101/2020.03.10.985150. [DOI] [PMC free article] [PubMed]
- 71.Nasir JA, Speicher DJ, Kozak RA, Poinar HN, Miller MS, Mcarthur AG. Rapid design of a bait capture platform for culture-and amplification-free next-generation sequencing of SARS-CoV-2. Biorxiv Org. 2020. 10.20944/preprints202002.0385.v1.
- 72.Bedford T, Greninger AL, Roychoudhury P. Cryptic transmission of SARS-CoV-2 in Washington State Trevor. MedRxiv. 2020. 10.1101/2020.04.02.20051417. [DOI] [PMC free article] [PubMed]
- 73.Deng X, Gu W, Federman S, du Plessis L, Pybus OG, Faria N, Wang C, Yu G, Bushnell B, Pan C-Y, Guevara H, Sotomayor-Gonzalez A, Zorn K, Gopez A, Servellita V, Hsu E, Miller S, Bedford T, Greninger AL, Roychoudhury P, Starita LM, Famulare M, Chu HY, Shendure J, Jerome KR, Anderson C, Gangavarapu K, Zeller M, Spencer E, Andersen KG, MacCannell D, Paden CR, Li Y, Zhang J, Tong S, Armstrong G, Morrow S, Willis M, Matyas BT, Mase S, Kasirye O, Park M, Masinde G, Chan C, Yu AT, Chai SJ, Villarino E, Bonin B, Wadford DA, Chiu CY. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grubaugh ND, Ladner JT, Lemey P, Pybus OG, Rambaut A, Holmes EC, Andersen KG. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng X, Achari A, Federman S, Yu G, Somasekar S, Bártolo I, Yagi S, Mbala-Kingebeni P, Kapetshi J, Ahuka-Mundeke S, Muyembe-Tamfum JJ, Ahmed AA, Ganesh V, Tamhankar M, Patterson JL, Ndembi N, Mbanya D, Kaptue L, McArthur C, Muñoz-Medina JE, Gonzalez-Bonilla CR, López S, Arias CF, Arevalo S, Miller S, Stone M, Busch M, Hsieh K, Messenger S, Wadford DA, Rodgers M, Cloherty G, Faria NR, Thézé J, Pybus OG, Neto Z, Morais J, Taveira N, Hackett JR, Chiu CY. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat Microbiol. 2020;5:443–454. doi: 10.1038/s41564-019-0637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, Ouédraogo N, Afrough B, Bah A, Baum JHJ, Becker-Ziaja B, Boettcher JP, Cabeza-Cabrerizo M, Camino-Sánchez Á, Carter LL, Doerrbecker J, Enkirch T, Garciá-Dorival I, Hetzelt N, Hinzmann J, Holm T, Kafetzopoulou LE, Koropogui M, Kosgey A, Kuisma E, Logue CH, Mazzarelli A, Meisel S, Mertens M, Michel J, Ngabo D, Nitzsche K, Pallasch E, Patrono LV, Portmann J, Repits JG, Rickett NY, Sachse A, Singethan K, Vitoriano I, Yemanaberhan RL, Zekeng EG, Racine T, Bello A, Sall AA, Faye O, Faye O, Magassouba N, Williams CV, Amburgey V, Winona L, Davis E, Gerlach J, Washington F, Monteil V, Jourdain M, Bererd M, Camara A, Somlare H, Camara A, Gerard M, Bado G, Baillet B, Delaune D, Nebie KY, Diarra A, Savane Y, Pallawo RB, Gutierrez GJ, Milhano N, Roger I, Williams CJ, Yattara F, Lewandowski K, Taylor J, Rachwal P, Turner DJ, Pollakis G, Hiscox JA, Matthews DA, O’Shea MK, Johnston AMD, Wilson D, Hutley E, Smit E, Di Caro A, Wolfel R, Stoecker K, Fleischmann E, Gabriel M, Weller SA, Koivogui L, Diallo B, Keita S, Rambaut A, Formenty P, Gunther S, Carroll MW. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020:1–5. 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed]
- 78.World Health Organization (WHO), Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19), WHO-China Jt. Mission Coronavirus Dis. 2019. 1. 2020;40.
- 79.Chatterjee S. An overview of mutations occurring within the coronavirus-2 genome: mutations data reporting on SARS-CoV-2. SSRN Electron J. 2020. 10.2139/ssrn.3632241.
- 80.van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020. 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed]
- 81.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed]
- 82.Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, Tenoever BR, et al. The spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. BioRxiv. 2020. 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed]
- 83.Robson B. COVID-19 coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput Biol Med. 2020. 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed]
- 84.Centers for Disease Control and Prevention, SARS-CoV-2 sequencing for public health emergency response, epidemiology, and surveillance, 2020.
- 85.Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z, Gu J, Xiang B, Liu J, Jiang W, Shen G, Zhao W, Men D, Deng Z, Yu L, Wei W, Li Y, Liu T, Deng Z, Li Y, Liu T. Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small. 2020;16:2002169. doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim D, Lee J, Yang J, Kim JW, Kim VN, Chang H, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020:1–29. 10.1017/CBO9781107415324.004. [DOI] [PMC free article] [PubMed]
- 87.Taiaroa G, Rawlinson D, Featherstone L, Pitt M, Caly L, Druce J, et al. Direct RNA sequencing and early evolution of SARS-CoV-2. BioRxiv. 2020. 10.1101/2020.03.05.976167.
- 88.Moore SC, Penrice-Randal R, Alruwaili M, Dong X, Pullan ST, Carter D, et al. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. MedRxiv. 2020. 10.1101/2020.03.05.20032011.
- 89.Xiao M, Liu X, Ji J, Li M, Li J, Yang L, Sun W, Ren P, Yang G, Zhao J, Liang T, Ren H, Chen T, Zhong H, Song W, Wang Y, Deng Z, Zhao Y, Ou Z, Wang D, Cai J, Cheng X, Feng T, Wu H, Gong Y, Yang H, Wang J, Xu X, Zhu S, Chen F, Zhang Y, Chen W, Li Y, Li J. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med. 2020;12:57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsai Y-C, Greenberg D, Powell J, Hoijer I, Ameur A, Strahl M, et al. Amplification-free, CRISPR-Cas9 targeted enrichment and SMRT sequencing of repeat-expansion disease causative genomic regions. BioRxiv. 2017;203919. 10.1101/203919.
- 91.Thézé J, Li T, du Plessis L, Bouquet J, Kraemer MUG, Somasekar S, Yu G, de Cesare M, Balmaseda A, Kuan G, Harris E, Wu CH, Ansari MA, Bowden R, Faria NR, Yagi S, Messenger S, Brooks T, Stone M, Bloch EM, Busch M, Muñoz-Medina JE, González-Bonilla CR, Wolinsky S, López S, Arias CF, Bonsall D, Chiu CY, Pybus OG. Genomic epidemiology reconstructs the introduction and spread of Zika virus in Central America and Mexico. Cell Host Microbe. 2018;23:855–864.e7. doi: 10.1016/j.chom.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hossain A, Reis AC, Rahman S, Salis HM. A massively parallel COVID-19 diagnostic assay for simultaneous testing of 19200 patient samples. Google Docs. 2020.
- 94.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, De Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–1266. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One. 2013;8. 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed]
- 96.Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, Stryke D, Pham E, Fung B, Bolosky WJ, Ingebrigtsen D, Lorizio W, Paff SM, Leake JA, Pesano R, DeBiasi R, Dominguez S, Chiu CY. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 98.Zhou P, Lou Yang X, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Di Jiang R, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, Sun Z, Liu F, Wu K, Zhong B, Mei Y, Zhang W, Chen Y, Li Y, Shi M, Lan K, Liu Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:1–9. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox LA, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Munnink BBO, Nieuwenhuijse DF, Stein M, O’Toole Á, Haverkate M, Mollers M, et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med. 2020. 10.1038/s41591-020-0997-y. [DOI] [PubMed]
- 103.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nextstrain-real-time tracking of pathogen evolution; (n.d.). [DOI] [PMC free article] [PubMed]
- 105.Li M, Jin R, Peng Y, Wang C, Ren W, Lv F, et al. Generation of antibodies against COVID-19 virus for development of diagnostic tools. MedRxiv. 2020. 10.1101/2020.02.20.20025999.
- 106.Diao B, Wen K, Chen JJJ, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. MedRxiv. 2020. 10.1101/2020.03.07.20032524.
- 107.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;2. 10.1038/d41587-020-00010-2. [DOI] [PubMed]
- 108.Poschenrieder A, Thaler M, Junker R, Luppa PB. Recent advances in immunodiagnostics based on biosensor technologies—from central laboratory to the point of care. Anal Bioanal Chem. 2019;411:7607–7621. doi: 10.1007/s00216-019-01915-x. [DOI] [PubMed] [Google Scholar]
- 109.Song Y, Song J, Wei X, Huang M, Sun M, Zhu L, Lin B, Shen H, Zhu Z, Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 110.To KKW, Tsang OTY, Leung WS, Tam AR, Wu TC, Lung DC, Yip CCY, Cai JP, Chan JMC, Chik TSH, Lau DPL, Choi CYC, Chen LL, Chan WM, Chan KH, Ip JD, Ng ACK, Poon RWS, Luo CT, Cheng VCC, Chan JFW, Hung IFN, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, Peiris M, Poon LLM, Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bruning AHL, Leeflang MMG, Vos JMBW, Spijker R, De Jong MD, Wolthers KC, Pajkrt D. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin Infect Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, et al. Head-to-head comparison of four antigen-based rapid detection tests for the diagnosis of SARS-CoV-2 in respiratory samples. Biorxiv. 2020. 10.1101/2020.05.27.119255. [DOI] [PMC free article] [PubMed]
- 114.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020. 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed]
- 115.Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, Horiuchi M, Kato K, Imoto Y, Iwata M, Mimura S, Ito T, Tamura K, Kato Y, Ikeda M, Imai K, Tabata S, Miyoshi K, Mizuno T, Murahara N, Horiuchi M, Kato K, Imoto Y, Iwata M, Mimura S, Ito T, Tamura K, Kato Y. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription–loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58:e01438–e01420. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int J Infect Dis. 2020. 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed]
- 117.Grant BD, Anderson CE, Williford JR, Alonzo LF, Glukhova VA, Boyle DS, Weigl BH, Nichols KP. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 118.Lin Q, Wen D, Wu J, Liu L, Wu W, Fang X, Kong J. Microfluidic immunoassays for sensitive and simultaneous detection of IgG/IgM/antigen of SARS-CoV-2 within 15 min. Anal Chem. 2020;92:9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- 119.Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso A-M, Le Goff J, Delaugerre C. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58:e00977–e00920. doi: 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, Van den Wijngaert S, Monteil V, Melin P, Stoffels K, Yin N, Mileto D, Delaunoy S, Magein H, Lagrou K, Bouzet J, Serrano G, Wautier M, Leclipteux T, Van Ranst M, Vandenberg O, Gulbis B, Brancart F, Bry F, Cantinieaux B, Corazza F, Cotton F, Dresselhuis M, Mahadeb B, Roels O, Vanderlinden J. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front Med. 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Green DA, George KS. Rapid antigen tests for influenza: rationale and significance of the FDA reclassification. J Clin Microbiol. 2018;56:1–10. doi: 10.1128/JCM.00711-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur J Clin Microbiol Infect Dis. 2020) 2020.03.27.20045153. 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed]
- 124.Zhang B, Zhou X, Zhu C, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B, Wang J. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Long Q, Deng H, Chen J, Hu J, Liu B, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. MedRxiv. 2020. 10.1101/2020.03.18.20038018.
- 126.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020) 2020.03.18.20038059. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed]
- 127.Zhang J, Zhang X, Liu J, Ban Y, Li N, Wu Y, Liu Y, Ye R, Liu J, Li X, Li L, Qin X, Zheng R. Serological detection of 2019-nCoV respond to the epidemic: a useful complement to nucleic acid testing. Int Immunopharmacol. 2020;88:1–13. doi: 10.1016/j.intimp.2020.106861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lv H, Wu NC, Tsang OT-Y, Yuan M, Perera RAPM, Leung WS, So RTY, Chan JMC, Yip GK, Chik TSH, Wang Y, Choi CYC, Lin Y, Ng WW, Zhao J, Poon LLM, Peiris JSM, Wilson IA, Mok CKP. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31:107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yunbao Pan YL, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Inf Secur. 2020) 2020.03.13.20035428. 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed]
- 130.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020:0–1. 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed]
- 131.Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19). MedRxiv. 2020. 10.1101/2020.03.17.20036954.
- 132.L. Ying, L. Yue-ping, D. Bo, R. Feifei, W. Yue, D. Jinya, H. Qianchuan, G. Hospital, C.T. Command, G. Hospital, C. Theater, Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2; 2020.
- 133.Liu L, Liu W, Zheng Y, Jiang X, Kou G, Ding J, Wang Q, Huang Q, Ding Y, Ni W, Wu W, Tang S, Tan L, Hu Z, Xu W, Zhang Y, Zhang B, Tang Z, Zhang X, Li H, Rao Z, Jiang H, Ren X, Wang S, Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020. 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed]
- 135.Zhang W, Du R-HH, Li B, Zheng X-SS, Lou Yang X-L, Hu B, Wang Y-YY, Xiao G-FF, Yan B, Shi Z-LL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020. 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed]
- 137.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Inf Secur. 2020. 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed]
- 138.Bendavid E, Mulaney B, Sood N, Shah S, Ling E, Bromley-Dulfano R, et al. COVID-19 antibody seroprevalence in Santa Clara County, California. MedRxiv. 2020. 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed]
- 139.Mallapaty S. Antibody tests suggest that coronavirus infections vastly exceed official counts. Nature. 2020. 10.1038/d41586-020-01095-0. [DOI] [PubMed]
- 140.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395:1101–1102. doi: 10.1016/s0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karcher F, McClean J. Pandemic influenza preparedness and response planning. Eur J Pub Health. 2010;20:610–611. doi: 10.1093/eurpub/ckq123. [DOI] [Google Scholar]
- 142.Kwon KT, Ko JH, Shin H, Sung M, Kim JY. Drive-through screening center for COVID-19: a safe and efficient screening system against massive community outbreak. J Korean Med Sci. 2020. 10.3346/jkms.2020.35.e123. [DOI] [PMC free article] [PubMed]
- 143.McCullough PA, Eidt J, Rangaswami J, Lerma E, Tumlin J, Wheelan K, Katz N, Lepor NE, Vijay K, Soman S, Singh B, McCullough SP, McCullough HB, Palazzuoli A, Ruocco GM, Ronco C. Urgent need for individual mobile phone and institutional reporting of at home, hospitalized, and intensive care unit cases of SARS-CoV-2 (COVID-19) infection. Rev Cardiovasc Med. 2020;21:1–7. doi: 10.31083/j.rcm.2020.01.42. [DOI] [PubMed] [Google Scholar]
- 144.Mahase E. Coronavirus: home testing pilot launched in London to cut hospital visits and ambulance use. BMJ. 2020;368:m621. doi: 10.1136/bmj.m621. [DOI] [PubMed] [Google Scholar]
- 145.Tu Y-P, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, et al. Patient-collected tongue, nasal, and mid-turbinate swabs for SARS-CoV-2 yield equivalent sensitivity to health care worker collected nasopharyngeal swabs. MedRxiv. 2020. 10.1101/2020.04.01.20050005.
- 146.Tu Y-P, Jennings R, Hart B, Cangelosi GA, Wood RC, Wehber K, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020. 10.1056/nejmc2016321. [DOI] [PMC free article] [PubMed]
- 147.U.S. Food and Drug Administration, Coronavirus (COVID-19) update: FDA authorizes first standalone at-home sample collection kit that can be used with certain authorized tests; 2020.
- 148.Bryson-Cahn C, Duchin J, Makarewicz VA, Kay M, Rietberg K, Napolitano N, et al. A novel approach for a novel pathogen: using a home assessment team to evaluate patients for 2019 novel coronavirus (SARS-CoV-2). Clin Infect Dis. 2020. 10.1093/cid/ciaa256. [DOI] [PMC free article] [PubMed]
- 149.Wehrhahn MC, Robson J, Brown S, Bursle E, Byrne S, New D, et al. Self-collection: an appropriate alternative during the SARS-CoV-2 pandemic. J Clin Virol. 2020. 10.1016/j.jcv.2020.104417. [DOI] [PMC free article] [PubMed]
- 150.Yang T, Wang Y-CC, Shen C-FF, Cheng C-MM. Point-of-care RNA-based diagnostic device for Covid-19. Diagnostics. 2020;10:9–11. doi: 10.3390/diagnostics10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.M. Nairz, R. Bellmann-Weiler, M. Ladstätter, F. Schüllner, M. Zimmermann, A.-M. Koller, S. Blunder, H. Naschberger, W. Klotz, M. Herold, Overcoming limitations in the availability of swabs systems used for SARS-CoV-2 laboratory diagnostics; 2020. [DOI] [PMC free article] [PubMed]
- 152.Piras A, Rizzo D, Uzzau S, De Riu G, Rubino S, Bussu F. Inappropriate nasopharyngeal sampling for SARS-CoV-2 detection is a relevant cause of false-negative reports. Otolaryngol Head Neck Surg. 2020. 10.1177/0194599820931793. [DOI] [PubMed]
- 153.Centers for Disease Control and Prevention, Interim guidelines for collecting, handling, and testing clinical specimens from persons under investigation (PUIs) for coronavirus disease 2019 COVID-19; 2020.
- 154.Smyrlaki I, Ekman M, Vondracek M, Papanicoloau N, Lentini A, Aarum J, et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCR. MedRxiv. 2020. 10.1101/2020.04.17.20067348. [DOI] [PMC free article] [PubMed]
- 155.Bruce E, Huang M-L, Perchetti G, Tighe S, Laaguiby P, Hoffman J, et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. BioRxiv. 2020. 10.1101/2020.03.20.001008. [DOI] [PMC free article] [PubMed]
- 156.Srivatsan S, Han PD, van Raay K, Wolf CR, McCulloch DJ, Kim AE, et al. Preliminary support for a “dry swab, extraction free” protocol for SARS-CoV-2 testing via RT-qPCR. BioRxiv. 2020. 10.1101/2020.04.22.056283.
- 157.Lalli MA, Chen X, Langmade SJ, Fronick CC, Sawyer CS, Burcea LC, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. MedRxiv. 2020. 10.1101/2020.05.07.20093542. [DOI] [PMC free article] [PubMed]
- 158.Byrnes SA, Gallagher R, Steadman A, Bennett C, Rivera R, Ortega C, et al. Multiplexed and extraction-free amplification for simplified SARS-CoV-2 RT-PCR tests. MedRxiv. 2020. 10.1101/2020.05.21.20106195. [DOI] [PubMed]
- 159.Ranoa DRE, Holland RL, Alnaji FG, Green KJ, Wang L, Brooke CB, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. BioRxiv. 2020. 10.1101/2020.06.18.159434.
- 160.Credle JJ, Robinson ML, Gunn J, Monaco D, Sie B, Tchir A, et al. Highly multiplexed oligonucleotide probe-ligation testing enables efficient extraction-free SARS-CoV-2 detection and viral genotyping. BioRxiv. 2020. 10.1101/2020.06.03.130591. [DOI] [PMC free article] [PubMed]
- 161.Bremner R, Pearson JD, Trcka D, Hyduk SJ, Aynaud M-M, Hernandez JJ, et al. Comparison of SARS-CoV-2 indirect and direct detection methods. BioRxiv. 2020. 10.1101/2020.05.12.092387. [DOI] [PMC free article] [PubMed]
- 162.Israeli O, Beth-Din A, Paran N, Stein D, Lazar S, Weiss S, et al. Evaluating the efficacy of RT-qPCR SARS-CoV-2 direct approaches in comparison to RNA extraction. Int J Infect Dis. 2020. 10.1016/j.ijid.2020.08.015. [DOI] [PMC free article] [PubMed]
- 163.Sonker M, Sahore V, Woolley AT. Recent advances in microfluidic sample preparation and separation techniques for molecular biomarker analysis: a critical review. Anal Chim Acta. 2017. 10.1016/j.aca.2017.07.043. [DOI] [PMC free article] [PubMed]
- 164.Zhu H, Fohlerová Z, Pekárek J, Basova E, Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens Bioelectron. 2020;153. 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed]
- 165.Jayamohan H, Sinha A, Krishnakumar R, Edwards HS, Younis TA, Trevithick JD, Patel KD, Bartsch MS. Library preparation for the Oxford Minion sequencer with “ASPIRE”: automated sample prep by indexed rotary exchange, in: 21st Int. Conf Miniaturized Syst Chem Life Sci MicroTAS 2017; 2020.
- 166.Snider A, Nilsson M, Dupal M, Toloue M, Tripathi A. A microfluidics workflow for sample preparation for next-generation DNA sequencing. SLAS Technol. 2019. 10.1177/2472630318796133. [DOI] [PubMed]
- 167.Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, et al. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv. 2020. 10.1016/j.biotechadv.2020.107537. [DOI] [PubMed]
- 168.Daley P, Castriciano S, Chernesky M, Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J Clin Microbiol. 2006. 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed]
- 169.Patel R, Babady E, Theel E, Storch G, Pinsky B, George K, et al. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of diagnostic testing for SARS–CoV-2/COVID-19. MBio. 11(2020):1–5. 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed]
- 170.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA - J Am Med Assoc. 2020:11–2. 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed]
- 171.Zheng S, Yu F, Fan J, Zou Q, Xie G, Yang X, et al. Saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. SSRN Electron J. 2020. 10.2139/ssrn.3543605. [DOI] [PMC free article] [PubMed]
- 172.Murray MA, Schulz LA, Furst JW, Homme JH, Jenkins SM, Uhl JR, Patel R, Cockerill FC, Myers JF, Pritt BS. Equal performance of self-collected and health care worker-collected pharyngeal swabs for group a streptococcus testing by PCR. J Clin Microbiol. 2015;53:573–578. doi: 10.1128/JCM.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dhiman N, Miller RM, Finley JL, Sztajnkrycer MD, Nestler DM, Boggust AJ, Jenkins SM, Smith TF, Wilson JW, Cockerill FR, Pritt BS. Effectiveness of patient-collected swabs for influenza testing. Mayo Clin Proc. 2012;87:548–554. doi: 10.1016/j.mayocp.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Walker HK, Hall WD, Hurst JW. Clinical methods: the history, physical, and laboratory examinations. 3. Boston: Butterworths; 1990. [PubMed] [Google Scholar]
- 175.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zheng S, Yu F, Fan J, Zou Q, Xie G, Yang X, et al. Saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. SSRN Electron J. 2020. 10.2139/ssrn.3543605. [DOI] [PMC free article] [PubMed]
- 177.Weiszhar Z, Horvath I. Induced sputum analysis: step by step. Breathe. 2013;9:301–306. doi: 10.1183/20734735.042912. [DOI] [Google Scholar]
- 178.Han H, Luo Q, Mo F, Long L, Zheng W. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect Dis. 2020;3099:30174. doi: 10.1016/s1473-3099(20)30174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, et al. University of Insubria COVID-19 Task Force, A. Baj, Saliva is a reliable tool to detect SARS-CoV-2. J Inf Secur. 2020:1–6. 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed]
- 180.Alizargar J, Etemadi Sh M, Aghamohammadi M, Hatefi S. Saliva samples as an alternative for novel coronavirus (COVID-19) diagnosis. J Formos Med Assoc. 2020. 10.1016/j.jfma.2020.04.030. [DOI] [PMC free article] [PubMed]
- 181.Griesemer SB, Van Slyke G, Ehrbar D, Strle K, Yildirim T, Centurioni DA, et al. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. MedRxiv. 2020. 10.1101/2020.06.16.20133041. [DOI] [PMC free article] [PubMed]
- 182.Patel PH, Antoine M, Ullah S. Bronchoalveolar lavage, in: StatPearls [internet] Treasure Island: StatPearls Publishing; 2019. [Google Scholar]
- 183.Xie C, Jiang L, Huang G, Pu H, Gong B, Lin H, Ma S, Chen X, Long B, Si G, Yu H, Jiang L, Yang X, Shi Y, Yang Z. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peiris JSM, Chu CM, Cheng VCC, Chan KS, Hung IFN, Poon LLM, Law KI, Tang BSF, Hon TYW, Chan CS, Chan KH, Ng JSC, Zheng BJ, Ng WL, Lai RWM, Guan Y, Yuen KY. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Feghoul L, Salmona M, Cherot J, Fahd M, Dalle JH, Vachon C, Perrod A, Bourgeois P, Scieux C, Baruchel A, Simon F, LeGoff J. Evaluation of a new device for simplifying and standardizing stool sample preparation for viral molecular testing with limited hands-on time. J Clin Microbiol. 2016;54:928–933. doi: 10.1128/JCM.02816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;1253:20–21. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA - J Am Med Assoc. 2020:1–7. 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed]
- 188.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020:1–3. 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed]
- 189.Peter JG, Theron G, Pooran A, Thomas J, Pascoe M, Dheda K. Comparison of two methods for acquisition of sputum samples for diagnosis of suspected tuberculosis in smear-negative or sputum-scarce people: a randomised controlled trial. Lancet Respir Med. 2013;1:471–478. doi: 10.1016/S2213-2600(13)70120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. MedRxiv. 2020. 10.1101/2020.04.16.20067835.
- 191.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, et al. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. MedRxiv. 2020. 10.1101/2020.04.11.20062372. [DOI] [PMC free article] [PubMed]
- 192.Cheung KS, Hung IF, Chan PP, Lung KC, Tso E, Liu R, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020. 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed]
- 193.National Institutes of Health, NIH begins study to quantify undetected cases of coronavirus infection; 2020.
- 194.Warnes SL, Little ZR, Keevil CW. Human coronavirus 229E remains infectious on common touch surface materials. MBio. 2015;6:1–10. doi: 10.1128/mBio.01697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen H-L, Chan MCW, Peiris M, Poon LLM. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA - J Am Med Assoc. 2020. 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed]
- 197.Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- 198.Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. MedRxiv. 2020. 10.1101/2020.05.19.20105999.
- 199.Daughton CG. Wastewater surveillance for population-wide Covid-19: the present and future. Sci Total Environ. 2020;139631. [DOI] [PMC free article] [PubMed]
- 200.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 201.Gutierrez-arroyo A, De Velasco-sada PG, Ruiz-bastián M, Alguacil-guillén M, González-donapetry P, Guedez-lópez GV, et al. Detection of SARS-CoV-2 on high-touch surfaces in a clinical microbiology laboratory. J Hosp Infect. 2020. 10.1016/j.jhin.2020.05.017. [DOI] [PMC free article] [PubMed]
- 202.WHO Global. Surface sampling of coronavirus disease (COVID-19): a practical “how to” protocol for health care and public health professionals, COVID-19 WHO surveilllance. Case Investig Epidemiol Protoc. 2020:1–26 https://apps.who.int/iris/handle/10665/331058.
- 203.Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA - J Am Med Assoc. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lee S, Lee D, Lee W, Kang B, Suk Y, Ryu B, et al. Detection of novel coronavirus on the surface of environmental materials contaminated by COVID-19 patients in the Republic of Korea. Osong Public Health Res Perspect. 2020:1–5. 10.24171/j.phrp.2020.11.3.03. [DOI] [PMC free article] [PubMed]
- 205.Ye G, Lin H, Chen L, Wang S, Zeng Z, Wang W, et al. Environmental contamination of SARS-CoV-2 in healthcare premises. J Inf Secur. 2020:2–6. 10.1016/j.jinf.2020.04.034. [DOI] [PMC free article] [PubMed]
- 206.Tromberg BJ, Schwetz TA, Pérez-Stable EJ, Hodes RJ, Woychik RP, Bright RA, et al. Rapid scaling up of Covid-19 diagnostic testing in the United States — the NIH RADx initiative. N Engl J Med. 2020. 10.1056/nejmsr2022263. [DOI] [PMC free article] [PubMed]
- 207.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 208.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020. 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed]
- 209.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 210.Shan B, Broza YY, Li W, Wang Y, Wu S, Liu Z, et al. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020. 10.1021/acsnano.0c05657. [DOI] [PubMed]
- 211.Lee JO, So HM, Jeon EK, Chang H, Won K, Kim YH. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal Bioanal Chem. 2008. 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed]
- 212.Layqah LA, Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tripathy S, Singh SG. Label-free electrochemical detection of DNA hybridization: a method for COVID-19 diagnosis. Trans Indian Natl Acad Eng. 2020;5:205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Klangprapan S, Choke-arpornchai B, Lieberzeit PA, Choowongkomon K. Sensing the classical swine fever virus with molecularly imprinted polymer on quartz crystal microbalance. Heliyon. 2020;6. 10.1016/j.heliyon.2020.e04137. [DOI] [PMC free article] [PubMed]
- 215.Su D, Wu K, Krishna VD, Klein T, Liu J, Feng Y, Perez AM, Cheeran MC-J, Wang J-P. Detection of influenza A virus in swine nasal swab samples with a wash-free magnetic bioassay and a handheld giant magnetoresistance sensing system. Front Microbiol. 2019;10:1077. doi: 10.3389/fmicb.2019.01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Centers for Disease Control and Prevention, Testing data in the U.S.; 2020.
- 217.Researchers develop first diagnostic test for novel coronavirus in China | German Center for Infection Research; n.d.
- 218.Centers for Disease Control and Prevention, CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel- for emergency use only; 2020.
- 219.Centers for Disease Control and Prevention, Centers for Disease Control and Prevention preparation of viral transport medium#: DSR-052-02, (2020). https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf. Accessed 29 Aug 2020.
- 220.Roche Molecular Solutions, Roche receives FDA emergency use authorization for cobas SARS-CoV-2 test to detect novel coronavirus; 2020.
- 221.Voller A, Bidwell DE, Bartlett A. Enzyme immunoassays in diagnostic medicine. Theory and practice. Bull World Health Organ. 1976;53(1):55–65. [PMC free article] [PubMed] [Google Scholar]
- 222.Cellex cleared to market antibody test for COVID-19 | North Carolina Biotechnology Center; n.d.
- 223.Hanna MA. FDA emergency use authorization letter; 2020.