Abstract
Stimulation of complex chaperone activity may be a viable means of therapy for neurodegenerative diseases. These chaperons execute reactivation of thermally and chemically aggregated protein substrates by cooperating with their partner co-chaperons. We optimized the expression and purification conditions of Tid1-L chaperone. Expression of Tid1-L in E. coli resulted in the formation of inclusion bodies which was further purified to soluble active form using 8 M urea and Ni-NTA column. Also, we investigated the events of the reactivation and disaggregation using aggregated G6PDH, luciferase and insulin as substrates. Incubation of aggregated/denatured enzymes with mortalin but not with Tid1 and/or Mge1 resulted in the initiation of the disaggregation reaction albeit very insignificantly. Under the same conditions coincubating the samples with chaperon and its assisted partners Tid1-L and nucleotide exchange factor Mge1 led to (40%) increase in enzyme activity of G6PDH. Similarly, luciferase activity was synergistically enhanced in the presence of mortlain/Tid1-L/Mge1 chaperones machinery. Chaperone-dependent disaggregation of thermally aggregated insulin showed that addition of Hsp70 and Hsp40 chaperones resulted in fast-track of renaissance reaction and inhibition of amyloid. The present study results conclude the quality of cell-control involves interaction of chaperon Hsp70 and its co-chaperones leading to complex formation with chemically/thermally aggregated substrate eventually causing its reactivation and disaggregation.
Keywords: Chaperones, Refolding, Amyloid, Mortalin, Insulin
1. Introduction
HSP70 (Heat Shock Proteins) are family of proteins which establish different cellular processes including protein folding, protein complex assembly and protein degradation (Hartl et al., 2011, Bukau and Horwich, 1998). The Human Hsp70 family comprises of eight members, six of whom are located in cytosol, one in the lumen of the ER and one in the mitochondrial matrix (mortalin, HspA9) (Bhattacharyya et al., 1995). Mortalin is related to a variety of diseases (Khan et al., 2017, Syken et al., 1999). Different studies showed the involvement of mortalin in numerous pathophysiological circumstances such as Alzheimer’s, Parkinson’s (Cook et al., 2016) and cancers (Na et al., 2016). Recently, mortalin autoantibody was documented a new serological marker for liver chirrosis (Lu et al., 2015). Numerous Hsp70 of co-chaperons are exists in mammalian mitochondria termed as Hsp40 (J-proteins), encourages the ATPase which appears in orthological proteins, GrpE and further controls the mortality rate through the factor of nucleotide amendment (DaSilva and Borges, 2011).
Tumorous imaginal Disk 1 (TID1) is a member of the molecular chaperone family DnaJ / Hsp40. Hsp40 family proteins share a DnaJ domain that functions to boost Hsp70′s ATPase activity through protein refolding and transportation. Mortalin is the only mitochondrial member of the Hsp70 family in mammalian cells, and was proposed to function as cochaperone with TID1 (Syken et al., 1999). There are two splice forms of TID1mRNA which encode long and short TID1 isoforms that function as DnaJ cochaperones in the cytosol (Schilling et al., 1998) and are processed into mature forms after import into the mitochondrial matrix. Several studies have shown that Tid1 acts as a regulator in intracellular signaling pathways associated with cell survival, cell proliferation and cellular response that is stress-induced. Tid1 also plays a role both in the apoptotic and antiapoptotic processes (Sarkar et al., 2001). Tid1 has also been suggested to control intracellular signaling for modulation of the Ras signaling pathway through its association with the RasGAP protein (Lo et al., 2004).
Most of the chaperone systems are inefficient in successful dissolving of protein aggregates in vitro. Only one study states, for the DnaK system, that a molar excess of the DnaK system can effectively reactivate previously heat-aggregated RNA polymerase (Skowyra et al., 1990). In the presence of a large (100–1000-fold) molar excess of the DnaK or Hsp70 chaperone systems, negligible quantities of pre-aggregate enzymes such as luciferase are recovered (Schroder et al., 1993). In comparison, substoichiometric rates of the DnaK system are adequate to efficiently solubilize and reactivate a wide range of previously aggregated protein substrates in the presence of the E. coli chaperone ClpB (Goloubinoff et al., 1999). Synergistic behavior of chaperones in the refolding and disaggregation of denatured proteins for various proteins has been previously reported (Glover and Lindquist, 1998). In addition, nature appears to have strongly selected molecular chaperones to protect cells from the harmful effects of protein aggregates (Dobson, 2003). Genetic overexpression of Hsp70 in mouse models of type 1 spinocerebellar ataxia (Cummings et al., 2001) and Parkinson's disease has been shown to suppress amyloid-related phenotypes (Klucken et al., 2004). The formation of amyloid fibrils, induced by partial protein unfolding and enrichment in beta structure, has been studied earlier (Jahn and Radford, 2008). Several molecular chaperones, and even non-chaperone proteins have been implicated in the defenses against amyloid (Luo et al., 2014, Landreh et al., 2015).
Thus, the current paper was planned to optimize the condition of expression and purification of mitochondrial chaperone Tid1-L (HSP 40) and their synergistic role along with mortalin (HSP 70) for protein refolding and disaggregation studies. In addition, their functions in insulin aggregation and amyloid formation were also examined using diverse biophysical and imaging methods.
2. Materials and methods
2.1. Materials
Resin and superdex-200 column nickel-nitrilotriacetic acid (Ni-NTA) were purchased from the Qiagen and Gene script, respectively. All other analytical grade chemicals and were purchased from Sigma. Purchased from Promega, WI, USA, NdeI, Hind III enzymes (Promega, WI, USA), and T4 DNA ligase.
2.2. Methods
2.2.1. Purification of (HSP 70) human mortalin
Mortalin was expressed and purified with our previous method (Khan et al., 2017). Briefly, after successful expression conditions (18 °C, 0.5 mM IPTG for 16 h) in E. coli, bacterial lysate was loaded on Ni-NTA column and protein was eluted at 165 mM IPTG concentration. Further, minor contaminants were removed using Superdex- 200 gel filtration column. Purity of purified mortalin was checked on 4–15% gradient SDS-PAGE.
2.2.2. Expression of Tid1-L (HSP 40) in E. coli
The protein was changed after codon optimization to generate a His-tag at the N - terminal end and the recombinant plasmid pET-28a was E. coli BL21 (DE3)-pLysS. The transformed cells in the pET28a: Tidl-L and permitted to grow at 37 °C with broth media of lysogenic which involves 35 µg/mL of chloramphenicol and 50 µg/mL of kanamycin till 600 nm OD. Tidl-L expression was persuaded via 1 mM of IPTG and culture was permitted to ripe at 18 °C temp overnight. Further, cells were harvested for 1/2 h via centrifugation at 5 k-rpm and suspended pellet were gained by running on SDS-PAGE.
2.2.3. Purification of Tid1-L from inclusion bodies
The bacterial pellet obtained after centrifugation at 10, 000 × g for 10 min at 4 °C was used for the purification of tid1-L from inclusion bodies. The pellet was re-suspended in denaturing 15 ml Lysis Buffers (100 mM NaH2PO4, 10 mM Tris•Cl, 8 M urea, pH to 8.0, 1 mg/ml lysozyme, 0.1% NP-40 W/V and 1X PI cocktail). The cells were incubated at room temperature for 30 min followed by centrifugation at 10,000 × g for 30 min at 4 °C. The supernatant containing protein of interest was mixed with equilibrated Ni-NTA agarose resin (1 ml) and kept on shaker for 1 h. The resin was loaded onto a column to allow the unbound protein to pass through the column under the influence of gravitational force. Ni-NTA slurry with bound protein was washed with 10 column volumes of wash buffer (100 mM NaH2PO4, 10 mM Tris•Cl, 8 M urea, pH 6.3) and protein was eluted with 3 ml elution buffer (100 mM NaH2PO4, 10 mM Tris•Cl, 8 M urea, pH 5.9 & 4.5). The protein was analyzed by SDS-PAGE to check the purity of the protein. Further, after dialysis of tid1-L in a gradient concentration of urea (6, 3, 1 & 0 M), the protein tend to aggregate in 0 M urea and soluble fraction was reloaded on second Ni-NTA column and protein was eluted in pH gradient manner. The protein concentration was measured by Bradford method (Bradford, 1976).
2.2.4. Refolding of aggregated G6PDH and luciferase
Based on Iosefson (2012) studies, the current study technique was implemented. Briefly, accumulation of G6PDH (0.85 µM) was carried out in 100 mM tris (pH 7.4) buffer containing 100 mM KCl, 10 mM MgCl2, and 10 mM DTT for 10 min at 52 °C. Outstanding activity afterwards the step of denaturation was < 3%. Further, to measure refolding activity, aggregates were incubated in ATP regenerating solution (0.2 M glycine betaine monohydrate, 10 µg/ml pyruvate kinase, 5 mM phosphoenolpyruvate, and 4 mM ATP). Thereafter, mortalin (8 µM) along with (0.01–1 μM) of the tid1-L, and 1 μM of the Mge1 were added to the G6PDH aggregates. This technique is performed with the previous study (Hansen et al., 2002).
Firefly luciferase was aggregated under thermal condition. Briefly, 200 mM luciferase stock was diluted 300-fold with PBS containing BSA (1 mg/ml) and then incubated at 45 °C for 15 min. Aggregated luciferase (1 mM) was diluted 20-fold with refolding buffer containing (25 mM Hepes, pH 7.6, 150 mM KCl, 10 mM MgCl2, 4 mM ATP, 10 mM DTT, 1 mM EDTA and 0.1 mg/ml BSA) along with chaperone (mortalin, 1 mM) and its co-chaperones (1 mM of each Tid1-L and Mge1). At RT, mixture was gestated and aliquoted with luminescence assay system.
2.2.5. Preparation of amyloid (Insulin)
Stock solution of insulin (Sigma) was prepared in glycine-HCl buffer (50 mM, pH-2.0). The concentration of insulin was determined using an extinction coefficient of 1.0 for 1 mg/mL at 276 nm (Khan et al., 2015). Stock solution of chaperones (mortalin/tid1-L) were prepared in sodium phosphate buffer pH 7.4 and various concentration of these chaperones were added to the fibrillated insulin. Listed conc is the mortalin conc, and the tid1-L was held at 100-fold lesser value. For example, treatment with 200 nM mortalin/tid1-L involved final concentrations of 200 nM mortalin and 2 nM Tid1-L.
2.2.6. Measurements of ThT fluorescence
ThT Fluorescence assay was measured on Jasco FP-750 spectrofluorometer (Japan). Stock of ThT dye was prepared in milli Q water and their concentration was estimated using its coefficient of molar extinction at ε412 nm = 36,000 M−1 cm−1 (Khan et al., 2018). 100 μl samples were aliquoted from the thermal fibrillated insulin in the absence and presence of mortalin/tid1-L and combined with ThT to achieve a 1:1 protein to dye ratio. The spectra were recorded at 450–650 nm at 440 nm, when excited. All measurements were in triplicate.
2.2.7. Electron microscopy
At the end of the spectroscopic measurements, electron microscopic analyses were performed. For this, 20 µl aliquots were removed from each sample and placed on 200-mesh copper grids covered with carbon stabilized Formvar film for 1 min, washed twice with distilled water, and treated with 3% uranyl acetate for 1 min. Images were captured on transmission electron microscope (TEM) JEOL operating at an accelerating voltage of 200 Kv with magnifications x10000.
2.3. Statistical analysis
All data were presented as mean ± standard deviation from 3 independent determinations. The statistical analysis was made by performing one-way ANOVA for 3 independent determinations. Significance of results was determined as p ≤ 0.05.
3. Results
3.1. Purification of mortalin (HSP 70) chaperone
Cloned mortalin was expressed in E. coli, containing name and further filtered Ni-NTA and gel filtration column in a 2-step chromatography process. SDS-PAGE profiling demonstrates homogeneity and mortalin purity (Fig. 1). Their expression method and purification has been published earlier in our lab (Khan et al., 2017).
Fig. 1.

SDS-PAGE (4–15% gradient) profile of purified mortalin. Lane 1- contain protein standard marker, lane 2- purified fraction of mortalin after superdex-200 chromatography.
3.2. Expression and purification of HSP 40 (tid1-L)
SDS-PAGE tid1-L expression analysis showed more protein in the pellet than supernatant (Fig. 2a), indicating inclusion bodies of tid1-L types under experimental conditions. It may be because of the inability of the prokaryotic machinery in E. coli to fold tid1-L, a eukaryotic protein. His tag containing tid1-L was purified using 8 M urea from inclusion bodies followed by purification with Ni-NTA. A pH gradient of 5.9 & 4.5 was used to eluted protein. Therefore, eluted protein was dialyzed to remove urea and to get tid1-L refolded. Nevertheless, as shown in the SDS-PAGE profile (Fig. 2b, Lane-4), the protein appears to aggregate in 0 M urea dialyzed sample and showed accumulation (higher aggregate); Supernatant was further reloaded onto Ni-NTA affinity resin to resolve protein contamination and aggregation. After eluting at pH 4.5, on SDS - PAGE (Fig. 2b, Lane-9), tid1-L showed single band, suggesting tid1-L purity.
Fig. 2a.
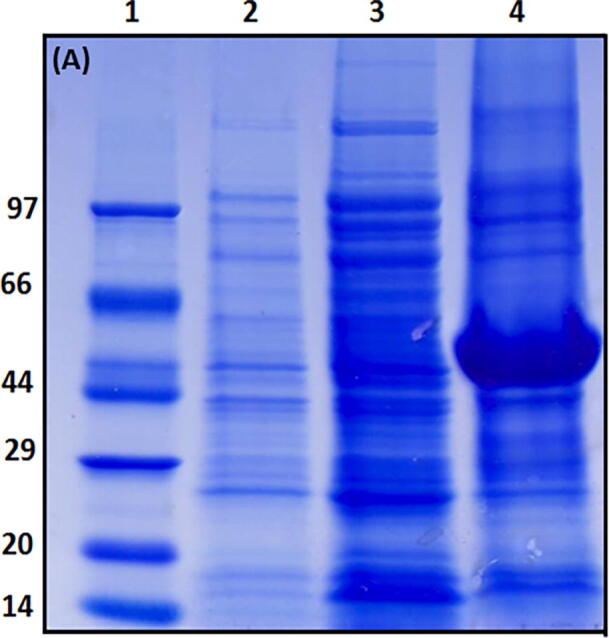
SDS-PAGE of expression of Tid1-L (HSP 40) in E. coli: Lane 1- Protein standard marker, Lane 2- Uninduced tid1-L, Lane 3- total lysate (supernatant), Lane 4- pellet.
Fig. 2b.

SDS-PAGE showing purification steps of Tid1-L: Lane 1 & 2- Elution of Tid1-L at pH 5.9 and 4.5 respectively. Lane 3 & 4- contain dialysed sample in 1 M and 0 M urea. Lane 6- Total lysate. Lane 7-Flow through. Lane 8 & 9- Second Ni-NTA eluted fractions at pH 5.9 & 4.5 respectively. Lane 10- Protein standard marker.
Analysis of expression of tid1-L showed formation of inclusion bodies. Earlier literature also identified tid1-L bodies for inclusion (Iosefson et al., 2012). The formation of inclusion bodies may be due to the fact that the folding of tid1-L, a eukaryotic protein, in the E. coli folding system was not efficiently folded by the prokaryotic machinery (Goder et al., 2004, Opekarova and Tanner, 2003).
3.3. Aggregate-reactivation activity of mortalin and Tid1-L
In the present study we tested glucose-6-phosphate dehydrogenase (G6PDH) and firefly luciferase reactivation using the mortalin and Tid1-L chaperone method. Large aggregates were formed from chemically denatured G6PDH using 10 mM urea and 40 mM DTT (Barnett et al., 2005), and firefly luciferase with thermal aggregation (Barnett et al., 2000). As illustrated in Fig. 3a, Fig. 3b, mortalin (HSP70) does not reactivate the aggregated G6PDH by itself 3a and 3b. However, in the presence of its partner co-chaperone Tid1-L (HSP 40), the percentage of G6PDH 's remaining activity increases as Tid1-L concentrations grow. In addition, the reactivation activity of the preformed G6PDH aggregate increased more significantly (2-fold) with the addition of another mitochondrial co-chaperone Mge1 (Fig. 3b). Our finding showed clearly the disaggregation of misfolded proteins (G6PDH) by mitochondrial mortalin chaperones in combination with its partner tid1-L and Mge1. The role of mortalin reactivation and other ClpB chaperones for similar substrate has been reported earlier (Barnett et al., 2005, Iosefson et al., 2012, Nagy et al., 2010).
Fig. 3a.

Describes the time dependent G6PDH activity in the presence of 8 μM mortalin assisted by different concentration of Tid1-L and 1 μM Mge1 co-chaperone.
Fig. 3b.
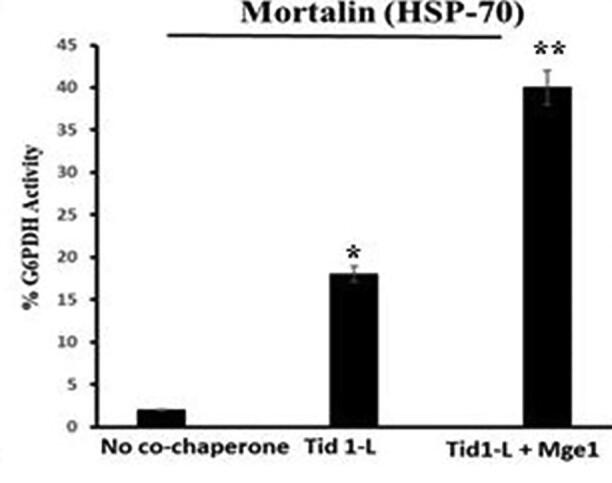
Yield of G6PDH refolding (after 6 h) obtained by mortalin and their co-chaperones. A stable G6PDH aggregates were reactivated at 30 °C in the presence of mortalin (8 μM), either alone or assisted by the indicated co-chaperone Tid1-L (1 μM) and Mge1 (1 μM). *Statistically different from the control group, p ≤ 0.05 and **statistically different from the Tid1-L, p ≤ 0.05.
Reactivation of another firefly luciferase substratum has also been determined in the absence and presence of the co-chaperone mortalin system (Fig. 4). The results showed negligible luciferase activity that mortalin and tid1-L display only in the absence of assisted partner Mge1. The growth in luciferase activity with mortalin was confirmed through co-chaperons (i.e., Tidl-L and Mge1). The luciferase disaggregation was earlier studies in yeast, bacteria, chaperons and co-chaperons (Zhang et al., 2012, Zolkiewski, 1999, Motohashi et al., 1999).
Fig. 4.

Reactivation of Luciferase activity. Aggregated luciferase (1 mM) was diluted 20-fold with refolding buffer along with chaperone (mortalin, 1 mM) and its co-chaperones (1 mM of each Tid1-L and Mge1). Remaining activity of luciferase was measured as described in methodology. Experimental data represent the average ± s.d.
3.4. ThT fluorescence measurements
ThT fluorescence assays were performed to determine the capacity of chaperone co-chaperone system for aggregation inhibition. 100 μl samples were aliquoted from the fibrillated insulin samples (65 °C) in the absence and presence of mortalin / tid1-L and combined with ThT to achieve a 1:1 protein-to-dye ratio. Insulin fibrillated at 65 °C displayed an enhanced ThT signal indicating the presence of aggregated structures, and similar results were observed when Tid1-L alone (Fig. 5) or mortalin alone (data not shown) was treated with insulin. The combination of chaperone and co-chaperone, however, significantly reduced the ThT fluorescence signal whose magnitude depended on the mortalin and Tid1 concentrations. As illustrated in Fig. 5A combined amyloid inhibition of 400 nM mortalin and corresponding concentration of its 100-fold lower concentration co-chaperone was achieved. Mortalin concentrations of 100 nM and 200 nM have also been significantly effective in keeping insulin fibrillation triggered by heat at bay. Fig. 5b, illustrated ThT fluorescence intensity of thermal aggregated insulin after 72 h in the absence and presence of mortalin and tid1 chaperones, decrease in ThT fluorescence intensity indicating inhibition of insulin amyloid by these chaperones. Inhibition of Aβ amyloid by peptide and chaperones has been reported earlier (Luo et al., 2014, Landreh et al., 2015).
Fig. 5.
ThT fluorescence of aggregated insulin in the absence and presence of chaperones (a) Kinetics of aggregation of insulin (b) bar graph representing ThT fluorescence intensity of aggregated insulin at 65 °C after 72 h alone and in the presence of mortlain/tid1 chaperones. Indicated concentration represent mortalin while tid1-L was taken 100 fold less.
3.5. Electron microscopy
The results of spectroscopic measurements have been confirmed and authenticated using the transmission electron microscopy (TEM) Fig. 6 by physical evaluation of insulin samples. Panel-1 matches native insulin which shows no aggregation. Insulin incubated at 65 °C for 72 h shows abundant aggregation in the form of fibrils, protofibrils, amorphous aggregates and mature amyloids which indicate the favorable fibrillation condition of the test protein under study (panel 2). There was a significant reduction in the formation and existence between aggregated protein when test protein was co-incubated under thermal stress with varying doses of moratlin and tid1 combinations (as defined in the Methods section). Panel 3–4 corresponds to concentrations of 300 nm and 400 nm of mortalin and its co-chaperon concentration of tid1-L. However, neither chaperone alone nor co-chaperone alone could have a significant impact on the prevention of insulin aggregation –although some positive impact could be seen (panel 5), indicating that the chaperone system 's effectiveness in preventing unwanted misfolding and protein aggregation in the physiological system also requires a co-chaperone.
Fig. 6.
Electron micrographs of control and aggregated insulin samples under various concentrations of chaperones and co-chaperones. Tid1-L was added 100-fold lower concentration of mortalin.
4. Discussion
Cellular thermotolerance is a crucial factor and is carried out by proteins belonging to the family of chaperones. Cellular chaperones such as mortalin (Hsp70) collaborate with their respective co-chaperons such as tid1 (Hsp40) chaperones and allow the thermally aggregated protein substrates such as insulin to reactivate. Reactivation studies of thermally aggregated proteins suggested that chaperones from the Hsp100 family alone do not achieve sufficient disaggregation but need co-chaperone proteins in this step as well. For instance, In E. coli Hsp70 (DnaK) and its co-chaperons comply with ClpB when several aggregated proteins are disaggregated (Mogk, et al., 1999). Similarly, after thermal stress, in addition to Hsp78, mitochondrial Hsp70 and its co-chaperons are necessary for cellular quality control and organelle reactivation as described in yeast studies (Schmitt et al., 1996, Germaniuk et al., 2002).
Chaperone Hsp70 and its co-chaperon Hsp40 were successfully expressed and purified to homogeneity in the present communication, as described in detail in the results section before being subjected to thermally aggregated protein reactivation studies. The chaperone structures were also used to demonstrate their rescue ability against chemically denatured glucose-6-phosphate dehydrogenase (G6PDH) and firefly luciferase heat denatures. After the introduction of chaperon and its co-chaperone, significant recovery of enzyme activity indicated the potential for reactivation of the chaperon system and the cellular quality control performed by these molecular chaperones. An important observation was found when adding another co-chaperon mge1 which led to overwhelming activity recovery of the test enzyme under chemical stress indication that there is a complex structure and an intricate mechanism for reactivating the stressed biomolecules involving not just one or two chaperones but an array of them. Complex chaperone-cochaperone systems thus perform quality control of the cells. Interestingly, firefly luciferase showed similar behaviour when undergoing thermal stress and then incubated with chaperones. Overall the results suggest cellular biomolecules synergistic and cooperative mechanism to alleviate any unfavourable stress.
Furthermore, distilled chaperone, mortalin (Hsp70) and its partner Tidl1 (Hsp40) have been tested for their functions in the defence of thermally aggregated insulin and subsequent reactivation using multimethodological approach. Any protein's misfolding and aggregation precedes unfolding and anything that prevents a protein's unfolding will largely protect it against misfolding and aggregation. In the present study, insulin unfolding was effectively resisted by molecular chaperone mortalin helped by its co-chaperone when thermal insult was made to the test protein. Such results are also consistent with the above spectroscopic assessments of the preventive and protective effects of molecular chaperon Hsp70 and its co-chaperon Hsp40 against insulin amyloidogenesis induced by heat.
Furthermore, the results indicate that the molecular chaperon system acts rather early on sensing the physical or chemical stress to cellular molecules thus preventing any unfavorable transfer of its biomolecules, particularly proteins. This is important because a protein can be reactivated to its normal form during the early stages of protein aggregation, but late stages of aggregation may allow cells to kill and destabilize them which cost cells energetically. Finally, electron microscopy was employed after spectroscopic examination to observe the cooperative effects of the chaperone system physically. Once again, the results are very convincing and clearly show the cellular chaperones quality control mechanism.
In summary, our results demonstrate the importance of the cellular quality control achieved through cooperation between chaperones of the families Hsp70 (mortalin) and Hsp40 (Tidl) in the reactivation of various protein aggregates and it is important to establish the complex reality structure of different chaperones and supported partners.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology (KACST), Kingdom of Saudi Arabia (Award Number 12-MED2928-02).
Footnotes
Peer review under responsibility of King Saud University.
References
- Barnett M.E., Nagy M., Kedzierska S., Zolkiewski M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005;280:34940–34945. doi: 10.1074/jbc.M505653200. [DOI] [PubMed] [Google Scholar]
- Barnett M.E., Zolkiewska A., Zolkiewski M. Structure and activity of ClpB from Escherichia coli. Role of the amino-and -carboxyl-terminal domains. J. Biol. Chem. 2000;275:37565–37571. doi: 10.1074/jbc.M005211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T., Karnezis A.N., Murphy S.P., Hoang T., Freeman B.C., Phillip B. Cloning and subcellular localization of human mitochondrial hsp70. J. Biol. Chem. 1995;270:1705–1710. doi: 10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Cook T.J., Hoekstra J.G., Eaton D.L., Zhang J. Mortalin is expressed by astrocytes and decreased in the midbrain of Parkinson's disease patients. Brain Pathol. 2016;26(1):75–81. doi: 10.1111/bpa.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings C.J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H.T., Dillmann W.H., Zoghbi H.Y. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- da Silva K.P., Borges J.C. The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism. Protein Pept. Lett. 2011;18:132–142. doi: 10.2174/092986611794475057. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Germaniuk A., Liberek K., Marszalek J. A bichaperone (Hsp70-Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J. Biol. Chem. 2002;277:27801–27808. doi: 10.1074/jbc.M201756200. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lindquist Susan. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goder V., Junne T., Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P., Mogk A., Peres Ben-Zvi A., Tomoyasu T., Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U. S. A. 1999;23:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T., Schlichting B., Schonheit P. Glucose-6-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima: expression of the g6pd gene and characterization of an extremely thermophilic enzyme. FEMS Microbiol. Lett. 2002;216:249–253. doi: 10.1111/j.1574-6968.2002.tb11443.x. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Iosefson O., Sharon S., Goloubinoff Pierre, Azem A. Reactivation of protein aggregates by mortalin and Tid1—the human mitochondrial Hsp70 chaperone system. Cell Stress and Chaperones. 2012;17:57–66. doi: 10.1007/s12192-011-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.S., Ahmed A., Tabrez S. Optimization of expression and purification of human mortalin (Hsp70): Folding/unfolding analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017;187:98–103. doi: 10.1016/j.saa.2017.06.015. [DOI] [PubMed] [Google Scholar]
- Khan M.S., Bhat S.A., Rehman M.T. Rutin attenuates negatively charged surfactant (SDS)-induced lysozyme aggregation/amyloid formation and its cytotoxicity. Int. J. Biol. Macromol. 2018;120:45–58. doi: 10.1016/j.ijbiomac.2018.07.112. [DOI] [PubMed] [Google Scholar]
- Klucken J., Shin Y., Masliah E., Hyman B., McLean P.J. Hsp70 reduces alpha-synuclein aggregation and toxicity. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- Lo J.F., Hayashi M., Woo-Kim S., Tian B., Huang J.F., Fearns C., Takayama S., Zapata J.M., Yang Y., Lee J.D. Tid1, a cochaperone of the heat shock 70 protein and the mammalian counterpart of the Drosophila tumor suppressor l (2) tid, is critical for early embryonic development and cell survival. Mol. Cell Biol. 2004;24(6):2226–2236. doi: 10.1128/MCB.24.6.2226-2236.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.J., Saxena N., Luk J.M., Kaul S.C., Wadhwa R. Circulating mortalin autoantibody—a new serological marker of liver cirrhosis. Cell Stress Chaperones. 2015;20(4):715–719. doi: 10.1007/s12192-015-0592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Tomoyasu T., Goloubinoff P., Rüdiger S., Röder D., Langen H., Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18(24):6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K., Watanabe Y., Yohda M., Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Y., Kaul S.C., Ryu J., Lee J.S., Ahn H.M., Kaul Z., Kalra R.S., Li L., Widodo N., Yun C.O., Wadhwa R. Stress chaperone mortalin contributes to epithelial-mesenchymal transition and cancer metastasis. Cancer Res. 2016;76(9):2754–2765. doi: 10.1158/0008-5472.CAN-15-2704. [DOI] [PubMed] [Google Scholar]
- Nagy M., Guenther I., Akoyev V., Barnett M.E., Zavodszky M.I. Synergistic cooperation between two ClpB isoforms in aggregate reactivation. J. Mol. Biol. 2010;396:697–707. doi: 10.1016/j.jmb.2009.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opekarova M., Tanner W. Specific lipid requirements of membrane proteins—a putative bottleneck in heterologous expression. BBA. 2003;1610:11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Pollack B.P., Lin K.T., Kotenko S.V., Cook J.R., Lewis A., Pestka S. hTid-1, a human DnaJ protein, modulates the interferon signaling pathway. J. Biol. Chem. 2001;276:49034–49042. doi: 10.1074/jbc.M103683200. [DOI] [PubMed] [Google Scholar]
- Schilling B., De-Medina T., Syken J., Vidal M., Munger K. A novel human DnaJ protein, hTid-1, a homolog of the Drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology. 1998;247:74–85. doi: 10.1006/viro.1998.9220. [DOI] [PubMed] [Google Scholar]
- Schmitt M., Neupert W., Langer T. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell. Biol. 1996;134(6):1375–1386. doi: 10.1083/jcb.134.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder H., Langer T., Hartl F.U., Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Georgopoulos C., Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62(5):939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- Syken J., De-Medina T., Münger K. TID1, a human homolog of the Drosophila tumor suppressor l (2) tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Ploetz E.A., Nagy M., Doyle S.M., Wickner S. Flexible connection of the N-terminal domain in ClpB modulates substrate binding and the aggregate reactivation efficiency. Proteins. 2012;80:2758–2768. doi: 10.1002/prot.24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- Jahn T.R., Radford S.E. Folding versus aggregation: polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008;469(1):100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Wärmländer S.K., Gräslund A., Abrahams J.P. Non-chaperone proteins can inhibit aggregation and cytotoxicity of Alzheimer amyloid β peptide. J. Biol. Chem. 2014;289:27766–27775. doi: 10.1074/jbc.M114.574947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreh M., Rising A., Presto J., Jörnvall H., Johansson J. Specific chaperones and regulatory domains in control of amyloid formation. J. Biol. Chem. 2015;290(44):26430–26436. doi: 10.1074/jbc.R115.653097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.S., Tabrez S., Rabbani N., Oves M., Shah A., Alsenaidy M.A., Al-Senaidy A.M. Physico-chemical stress induced amyloid formation in insulin: Amyloid characterization, cytotoxicity analysis against human neuroblastoma cell lines and its prevention using black seeds (Nigella sativa) Chin. J. Integr. Med. 2015 doi: 10.1007/s11655-015-2153-y. [DOI] [PubMed] [Google Scholar]




