Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that causes life-threatening meningitis primarily in immunocompromised individuals. In order to survive and proliferate during infection, C. neoformans must adapt to a variety of stresses it encounters within the host. Patient outcome depends on the interaction between the pathogen and the host. Understanding the mechanisms that C. neoformans uses to facilitate adaptation to the host and promote pathogenesis is necessary to better predict disease severity and establish proper treatment. Several virulence phenotypes have been characterized in C. neoformans, but the field still lacks a complete understanding of how genotype and phenotype contribute to clinical outcome. Furthermore, while it is known that C. neoformans genotype impacts patient outcome, the mechanisms remain unknown. This lack of understanding may be due to the genetic heterogeneity of C. neoformans and the extensive phenotypic variation observed between and within isolates during infection. In this review, we summarize the current understanding of how the various genotypes and phenotypes observed in C. neoformans correlate with human disease progression in the context of patient outcome and recurrence. We also postulate the mechanisms underlying the genetic and phenotypic changes that occur in vivo to promote rapid adaptation in the host.
Keywords: Cryptococcus neoformans, genotype, microevolution, phenotype
Introduction
When it was first described in 1894, Cryptococcus neoformans was assumed to be a single species. More than a century of work, however, has revealed that what is now referred to as the Cryptococcus sp. complex has notable heterogeneity in pathogenesis, genetics, epidemiology, ecology, and biochemistry. Instead of the single species described in 1894, we now know that the Cryptococcus sp. complex contains at least seven distinct species or two major species complexes [1,2]. The Cryptococcus sp. are able to cause disease in both immunocompromised and immunocompetent individuals, although disease is much more commonly observed in the immunocompromised. Individuals at greatest risk of developing disease are those living with HIV/AIDS, patients taking corticosteroids, patients undergoing cancer chemotherapy, solid-organ transplant recipients, or individuals with other causes of immune deficiency [3]. Infection occurs when a person inhales a Cryptococcus spore into the lungs. If this initial infection is not eradicated by the immune response, or controlled in the lungs via granuloma formation, the infection can progress to result in fungal pneumonia. Systemic dissemination can result in skin lesions, but the most common disease presentation is dissemination to the brain and subsequent cryptococcal meningitis [4]. If left untreated, cryptococcal meningitis is fatal. Recent research has indicated the important role Cryptococcus sp. genetics and cell phenotype play during interactions with the host to promote disease. This review highlights the need to define the interplay between genetics and phenotype to understand and prevent C. neoformans disease.
Genetic variation affects patient disease outcomes
How has our understanding of Cryptococcus genetics evolved?
The first indication that Cryptococcus was not a single species was the discovery that there were strain-specific differences to reactive antibodies. Yeast cells were found to consistently fit within one of four distinct serotypes: A, B, C, or D, based on antigenic differences in capsular polysaccharides [5,6]. Later work revealed that A and D serotypes had the ability to mate as did B and C, suggesting that Cryptococcus contained two species [7–9]. This theory was supported by additional work that showed the A, D and B, C serotypes had distinct ecology, epidemiology, biochemistry, and genomic structure [10–17]. Serotypes A, D were renamed C. neoformans var. neoformans and serotypes B, C named C. neoformans var. gattii. The next shift in nomenclature came as a result of the molecular age. Due to variations in the URA5 gene that further differentiated serotypes A and D, serotype A was renamed C. neoformans var. grubii [18].
C. neoformans var. gattii was raised to the species level and eventually renamed C. gattii [10,19]. Additional molecular characterization of C. gattii populations revealed a substructure within the population and led to further subdivision into five species: C. tetragatii, C deuterogattii, C. decagattii, C. bacillisporus, and C. gattii [1]. This further subdivision of C. gattii has not been universally accepted by the research community with some researchers opting to use ‘C. gattii species complex’ to describe the complex population structure within C. gattii [2,20]. Concomitant with the renaming of the C. gattii species complex in 2015, C. neoformans var. grubii was renamed C. neoformans to reflect the fact that 95% of cryptococcal infections are caused by these strains, and C. neoformans var. neoformans was renamed C. deneoformans because these are the historical serotype D strains [1]. This name change was more universally accepted by the research community, and is used in this review, although ‘C. neoformans species complex’ is also commonly used (Figure 1A) [2].
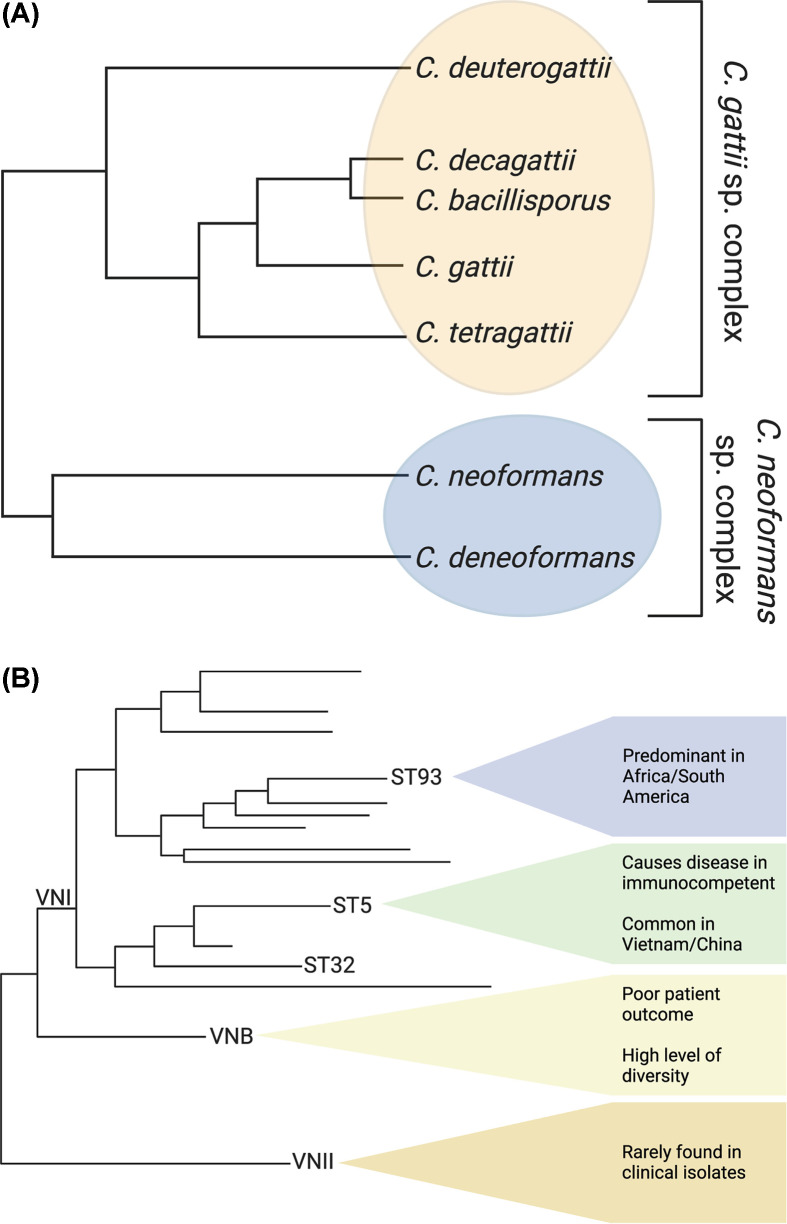
Figure 1. Pathogenic Cryptococcus species.
Relatedness of the (A) Cryptococcus species complex and (B) subpopulations within C. neoformans. (A) There are five proposed species in the C. gattii species complex (yellow), and two species in the C. neoformans species complex (blue) [1,2,20]. (B) There are three evolutionarily distinct subpopulations within C. neoformans, as defined by molecular type: VNI, VNII, and VNB. VNII is very rarely found in clinical isolates. VNB is more diverse, was initially discovered in Botswana, and patients infected with these strains tend to have poor clinical outcomes. VNI causes the most cases of cryptococcosis and has three evolutionarily distinct subpopulations; ST93 and ST5 are in separate subpopulations [21,54,55].
Interestingly, the dominant clades VNI, VNII, and VNB observed in C. neoformans have not changed significantly as the field transitioned from the early microsatellite genotyping technologies to modern sequence-based technologies such as multilocus sequence typing (MLST) and whole-genome sequencing (WGS). The vast majority of clinical isolates are in the VNI clade (Figure 1B). The VNI and VNII clades are both globally distributed and tend to have predominantly clonal population structures [21–23]. In contrast, the VNB clade is primarily found in sub-Saharan Africa and South America and is highly diverse, suggesting ongoing or recent recombination [21,24]. To further classify major sequence types (STs) within the C. neoformans clades, a standardized, globally accepted MLST scheme for C. neoformans that uses seven unlinked genetic loci (CAP59, GPD1, LAC1, PLB1, SOD1, URA5, IGS1) sequenced individually or derived from WGS was developed [25]. Four of these genes are believed to be under neutral selective pressure, while three (CAP59, LAC1, and PLB1) code for Cryptococcus virulence factors. This globally accepted method to identify the ST cluster of strains has begun to reveal the vast genetic diversity within C. neoformans and has also led to fine-scale studies associating C. neoformans genetics with geographic location and human disease.
Are there differences in Cryptococcus sp. disease presentation?
The Cryptococcus sp. have dramatic differences in their ecology and disease epidemiology. C. neoformans and C. deneoformans predominantly cause disease in immunocompromised individuals across the globe, and thus are thought to be ubiquitous in the environment. C. neoformans causes 95% of all cryptococcal meningitis globally, and 99% of cryptococcosis cases in HIV/AIDS patients (Figure 2A) [4]. C. neoformans is known to be a disease of immunocompromised patients, but approximately 25% of patients with C. neoformans have no immunocompromising conditions (Figure 2B) [26–32]. In contrast, infections caused by strains in the C. gattii species complex are more commonly seen in immunocompetent individuals (∼64%) (Figure 2B) [30,31,33–36]. C. gattii infections had previously only been observed in tropical and subtropical regions of the world, leading to the assumption that C. gattii was ecologically restricted to these regions. However, an outbreak on Vancouver Island in Canada in 1999, that spread across North America, showed this assumption was incorrect [37]. Genetic analysis of the C. gattii outbreak strains revealed introgression of a ‘virulent’ South American genotype into previously uncharacterized ‘native’ North American genotypes, suggesting C. gattii was present previously in North America but was not associated with disease [38].
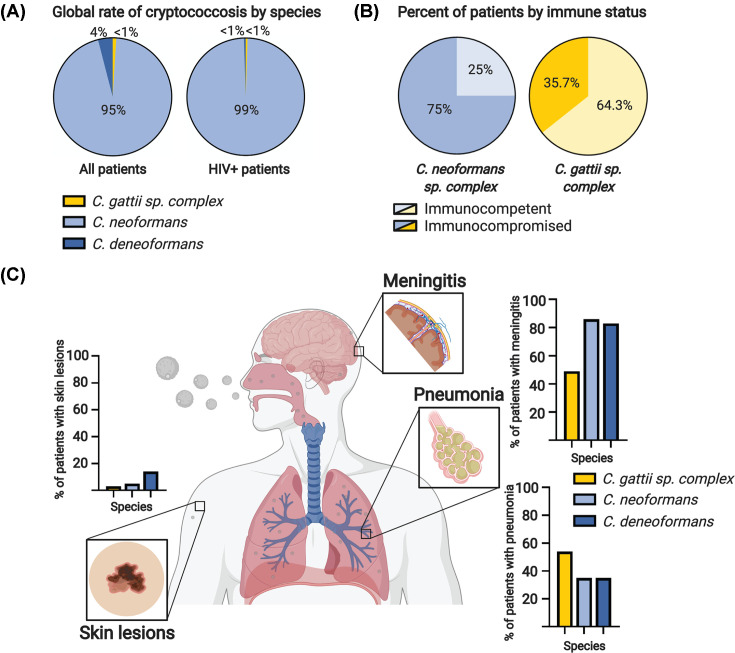
Figure 2. Epidemiology and pathobiology of the Cryptococcus species complex.
(A) C. neoformans is the most common species of Cryptococcus to cause infections globally, accounting for 95% of infections, compared with C. deneoformans (4%) and the C. gattii species complex (<1%) (left). In HIV+ patients, C. neoformans causes more than 99% of cases globally (right) [4]. (B) Infections by the C. gattii species complex most commonly occur in immunocompetent patients (64.3%), while infections by C. neoformans species complex primarily occur in patients with an immunocompromising condition (75%) [26–36]. (C) The most common clinical manifestations of cryptococcosis are skin lesions, pneumonia, and meningitis. Skin lesions, while rare, occur most often in patients infected with C. deneoformans, in approximately 14% of infections [41]. Patients with C. neoformans infections have skin lesions in 5% of cryptococcal meningitis cases [44]. Only 3% of C. gattii species complex cases have cutaneous cryptococcosis as a symptom [3]. The second most common manifestation of cryptococcosis is pneumonia. Pneumonia occurs most frequently in infections caused by C. gattii species complex infections, with 54% of C. gattii patients in the United States displaying lung involvement [36,45]. Species in the C. neoformans species complex manifest pulmonary cryptococcosis in 35% of clinical infections [41]. Cryptococcal meningitis occurs most frequently in the C. neoformans species complex, with over 80% of patients displaying meningitis symptoms [42]. Cryptococcal meningitis is less common in the C. gattii species complex with only 49% of patients in the United States developing meningitis [36].
Differences in disease progression between the Cryptococcus sp. are also observed in both patient populations and animal models. As mentioned, C. neoformans is known to be associated with cryptococcal meningitis in patients and readily disseminates to the brain after inhalation in mouse models of disease [39]. While not universal, C. deneoformans is often less virulent in animal models and less commonly observed as meningitis in clinical isolates worldwide (Figure 2C) [40–42]. C. deneoformans is more frequently found to cause disease in Europe, although many of these isolates have been shown to be hybrids with C. neoformans [43]. C. deneoformans also causes skin lesions more frequently; 14% of C. deneoformans patients manifest lesions, while skin lesions appear in 5% of C. neoformans patients and 3% of C. gattii patients (Figure 2C) [3,41,44]. The C. gattii species complex tends to cause a much more pronounced pneumonia in humans, and some studies have recapitulated this phenomenon in the animal model [45]. C. gattii manifests as pneumonia in 54% of patients in the United States (a percentage lower than patients in British Columbia), while the C. neoformans sp. complex causes pneumonia in 35% of cases (Figure 2C) [36,41,45]. Due to the known differences in Cryptococcus sp. disease potential and presentation and the fact that C. neoformans causes 95% of all cryptococcal meningitis worldwide, the following sections of this review will focus on C. neoformans.
How does C. neoformans genotype impact patient outcome?
Cryptococcal meningitis mortality rates differ across the globe [46]. Historically, these differences have been attributed to treatment regimens, type of care, level of patient immune function, and patient genetics [47]. It was not until 2012, when Wiesner et al. published the first study linking the genetics of C. neoformans with virulence in humans, that the importance of C. neoformans genetics in virulence began to surface [48]. Wiesner et al. performed MLST sequencing on clinical isolates of C. neoformans from Ugandan HIV/AIDS patients and showed the isolates grouped into three distinct, phylogenetically linked clonal clusters within the VNI clade. In one cluster centered on ST5 and ST63 only 38% of the patients died, compared with a 79% mortality rate in the other two clusters, centered on ST93 and ST77, respectively (Figure 1B) [48]. Surprisingly, the differences in patient mortality were not due to numbers of yeast cells cultured from patient samples but rather alterations in the host immune response to the different genotypes that were associated with capsule and melanin production [48]. These data indicated for the first time that the pathophysiology of cryptococcal meningitis was associated with pathogen genotype within C. neoformans.
There have been several additional studies linking human disease with sequencing type. Beale et al. genotyped clinical isolates from South African HIV/AIDS patients using MLST [49]. This population exhibited much higher diversity than the Ugandan population, yet the authors still found that VNB and ST32 isolates both had worse patient outcome that could not be explained by in vitro phenotypic differences [49]. A study in Brazil that combined analysis of environmental and clinical isolates, and HIV/AIDS and transplant patients, showed that STs other than ST93 had lower patient survival rates, but ST93 was also over-represented in the environmental isolates (Figure 1B) [23,24]. In a study of renal transplant recipients in Brazil, patients infected with VNII were more likely to survive (Figure 1B) [50]. A particularly interesting link between patient disease and genotype was found in Asia, where C. neoformans infections are observed in immunocompetent individuals [51]. In Vietnam, analysis of isolates from immunocompetent patients showed that almost all are in a closely related VNI cluster centered on ST5 (Figure 1B) [52–54]. ST5 caused 82% of infections in HIV uninfected patients, compared with only 35% in HIV infected patients [26]. Combined, these studies analyzing clinical isolates from across the globe indicate that C. neoformans genotype is responsible for variations in patient clinical outcome but also highlight the global genetic diversity present in C. neoformans. In addition, the genetic changes in C. neoformans are occurring on too fine a level for the underlying mechanism to be defined by MLST or current in vitro assays. This showcases the need for WGS studies and better understanding of the host–pathogen interaction to truly link C. neoformans genetics with patient clinical outcome.
The first WGS studies in C. neoformans linked to clinical outcomes have provided unexpected results. Day et al. performed WGS on eight of the ST5 isolates mentioned previously [26]. The isolates were compared with the C. neoformans H99 reference genome and SNPs and INDELs either specific to the Vietnamese ST5 or non-ST5 isolates that caused translation truncation (and therefore, likely impacted protein production) were identified. The variants specific to ST5 were associated with 19 genes. Only three coded for previously identified virulence proteins. The non-ST5 variants truncated the translation of 25 genes, of which only 4 were previously known virulence determinants [26]. The next WGS study that investigated the link between C. neoformans genotype and patient phenotype was published in 2019 and specifically explored differences between ST93 isolates causing changes to patient outcome. Gerstein et al. performed WGS on clinical isolates from patients in Uganda, where ST93 is the most frequently identified sequence type [55]. They identified non-synonymous INDELs and SNPs that were specific to ST93 and not located in the extreme telomeric or centromeric regions. They then performed a genome-wide association study (GWAS) to explore association of the variants with patient clinical data that included parameters such as clinical outcome, patient immune response, and isolate in vitro phenotype. The GWAS identified 145 variants associated with 40 genes, with only two previously known to be virulence determinants. Gerstein et al. identified that four of these genes were novel virulence factors by testing deletion strains in a mouse inhalation model [55]. Perhaps most intriguing, considering the evolutionary distance between ST5 and ST93 (Figure 1B), is that four genes (CNAG_ 05185, CNAG_05987, CNAG_02475, and CNAG_05185) were identical between the two studies and an additional three were only a single gene off (CNAG_07704, CNAG_04921, CNAG_01240 versus CNAG_07703, CNAG_04922, CNAG_01241 for Day et al. and Gerstein et al., respectively). While genomic studies looking at the specific genetic polymorphisms that underlie the clinical difference between C. neoformans STs are still in their infancy, a number of striking observations can be made. First, while the exact polymorphisms within distinct ST lineages may differ, the observation that the same genes or regions of the genome are under selective pressure suggests these genes are universally important for C. neoformans virulence in humans. Second, only a small proportion of the virulence determinants identified in the GWASs had been previously identified as C. neoformans virulence factors. These data suggest that previous in vitro and in vivo studies aimed at defining the minimum requirements for C. neoformans to be a pathogen, which are shared by all the Cryptococcus sp., may have missed the nuances of the host–pathogen interaction that impact clinical outcome in patients with cryptococcal meningitis. Thus, renewed effort needs to be placed on understanding the complexities of the C. neoformans–host interaction and how genetic polymorphisms impact these interactions.
C. neoformans phenotypic variation affects patient disease outcomes
What phenotypes occur in vivo?
In response to the host environment, pathogens often alter their morphology through changes in gene expression. These changes are associated with adaptation and survival in the host and are classified as ‘virulence’ phenotypes or factors. The transition to the host environment elicits two types of virulence associated phenotypic changes in C. neoformans: cell surface changes and cell size changes (Figure 3). Alterations to the cell surface include production of a polysaccharide capsule, modifications to the composition of the cell wall, and melanin synthesis. More recently, cell size changes have been recognized as important virulence phenotypes in C. neoformans. Distinct cell size phenotypes include the typical yeast cells, the large titan cells, and the small micro and titanide cells. Both cell surface and size modifications impact the host–pathogen interaction, but how these traits are impacted by genetic polymorphisms has yet to be explored.
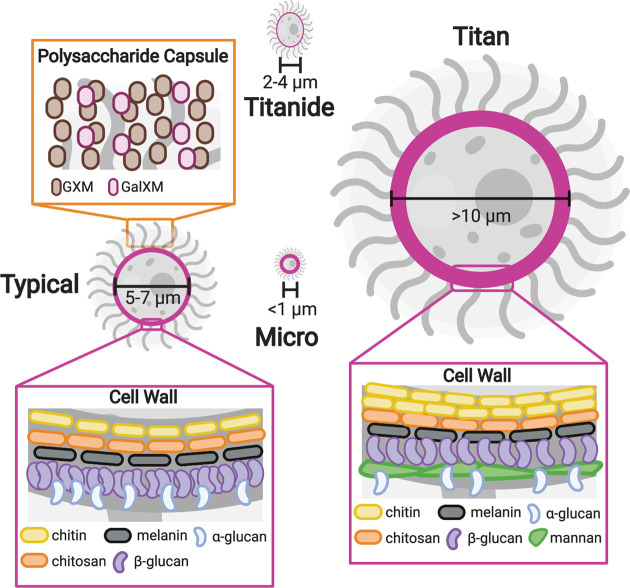
Figure 3. Cell surface alterations and cell size changes in C. neoformans.
Model illustrating the cell surface alterations and cell size phenotypes in C. neoformans. The outside of the C. neoformans cell consists of a cell wall (highlighted in pink) and polysaccharide capsule. Typical cells (5–7 μm in diameter) have a capsule predominantly composed of GXM and GalXM (orange box). The cell wall of typical cells contains chitin, chitosan, α-glucan, melanin, and β-glucan (left pink box). Titan cells (>10 μm in diameter) exhibit a thickened cell wall with an increase in chitin, decreased glucan, and a layer of mannan (right pink box) [59]. Micro cells (<1 μm in diameter) also display thickened cell walls [66]. Titanides are 2–4 μm in diameter and are oval in shape. The cell wall of titanides are thinner than typical cells [71]. Abbreviations: GXM, glucuronoxylomannan; GXMGal, glucuronoxylomannogalactin.
Cell surface changes have long been known to be major virulence determinants in C. neoformans. Because mammalian cells do not have cell walls, the immune system uses sensing of many of the components of the fungal cell wall, such as β-glucan and chitin, to trigger the immune response to fungal pathogens. In response to the host environment, C. neoformans produces a polysaccharide capsule that is anchored to the exterior surface of the cell wall and is primarily made up of glucuronoxylomannan (GXM) and glucuronoxylomannogalactin (GXMGal) (Figure 3). The capsule is thought to help prevent recognition of the cell wall by the immune system. It undergoes size and structural alterations in vivo that result in antigenic variability [56]. Capsule is also shed from the surface of the cell, and this shed capsule has independent immune modulatory functions [57,58]. Recent studies show the C. neoformans cell wall also undergoes dramatic remodeling in response to the host environment [59]. The cell wall consists primarily of α-glucan, β-glucan, chitin, and chitosan (Figure 3). In the host the amount of β-glucan in the cell wall is reduced, while chitin increases, possibly utilizing chitosan to facilitate this process [59,60]. Melanin is also deposited into the cell wall and plays a protective role during infection, likely through detoxification of free radicals generated by the immune response [61]. Cell wall integrity appears to be important throughout this process, likely for cell survival and proper capsule formation [62,63].
In addition to cell surface alterations, C. neoformans also undergoes cell size variation in vivo. When grown in vitro, C. neoformans is typically 5–7 µm in diameter. During a murine pulmonary infection, cells with diameters ranging from smaller than 1 µm to as large as 100 µm have been observed [64–66]. Cells that exhibit an enlarged phenotype, defined as a cell diameter greater than 10 µm, are referred to as titan cells [64,65]. The characterization of titan cells has largely been confined to mouse models, but titan cells have also been observed in human infections [67–69]. Cells that exhibit a decreased size were first described in a murine pulmonary infection and are known as micro cells. Micro cells are <1 µm in diameter and little is known about this population of cells [66,70]. Interestingly, Dambuza et al. recently described a second population of small cells, known as titanides [71]. Titanides are 2–4 µm in diameter and oval in morphology. Titanides are associated with in vitro titan inducing conditions and are yet to be observed in vivo [71]. In addition to their distinct cell sizes, all these cell populations also display cell surface alterations (Figure 3). Titan cells have a thickened cell wall with a mannan layer and a highly cross-linked capsule [64,65]. Micro cells also display a thickened cell wall, while titanides have a thinner cell wall [66,71]. Cell size appears to vary by site of infection. Cells collected from the lungs of mice exhibit larger capsules and larger cell body diameters, while cells isolated from the brain exhibit smaller capsules and cell body diameters [58,72,73]. This organ-dependent size variation has also been described in human infections. Histology sections of postmortem brain contained significantly smaller cryptococcal cells and capsules than histology sections of postmortem lung [74]. Cell size variation may be important during infection as different cell sizes may be beneficial at different stages of infection. For instance, smaller cells may promote dissemination and survival within macrophages, while larger cells may aid in pathogen survival during pulmonary infection. Consistent with this idea, titan cells are primarily found in the lungs and have been shown to promote pathogenesis through decreased rates of phagocytosis and increased stress tolerance [64,75,76].
In addition to factors associated with cell surface and size, additional virulence factors including growth at 37°C, urease and phospholipase production, sphingolipid utilization, among others have been identified as major virulence factors. Extensive efforts to identify in vitro phenotypes, genes important to their formation, and subsequent mutant analysis—primarily using murine models—has been undertaken. For more information on these studies of virulence factor phenotypes in C. neoformans, and how they promote pathogenesis, please refer to the following reviews [77–80]. The analysis of virulence phenotypes has largely been confined to in vitro studies and animal models, but changes in virulence phenotypes may be responsible for differences observed in patient outcomes between clinical isolates.
How does C. neoformans phenotype impact patient outcome?
Although there has been evidence of capsule enlargement, melanin synthesis, and titan cell formation during human infections, the extent of the phenotypic changes that occur in C. neoformans during human infection is largely unknown [61,67–69,81]. It is also unknown how these phenotypic changes may ultimately affect patient outcome and if they could be used as reliable indicators for patient outcome. However, within the last decade, progress has been made to better understand how C. neoformans phenotype relates to patient outcome in the context of human disease. Phenotypic analysis of clinical isolates isolated from the cerebrospinal fluid (CSF) of patients with significant disease paired with known clinical parameters has allowed for better understanding of the association of C. neoformans phenotype to patient outcome.
Recent studies have begun to investigate the correlation between human disease progression and C. neoformans cell surface changes using clinical data and virulence factor analysis of clinical isolates [81–88]. Several studies observed a significant correlation between capsule size and strain virulence [81–83]. Clancy et al. observed strains that produced smaller in vivo capsule size in mice were also more likely to have a higher lethal dose for 50% of mice (LD50) [82]. Interestingly, the correlation between LD50 and capsule size was not observed with in vitro induced capsule size [82]. Robertson et al. similarly showed that ex vivo capsule from human CSF samples correlated with several clinical parameters, including increased intracranial pressure and slower rate of fungal clearance [81]. This study also noted lack of correlation with ex vivo capsule and in vitro induced capsule formation [81]. Additionally, in the previously mentioned Wiesner et al. MLST study, strains associated with higher patient mortality displayed increased capsule shedding or melanin production [48]. Interestingly, the association with capsule shedding and patient mortality in the present study was not detected in vivo at time of patient diagnosis but rather associated with in vitro capsule shedding [48]. In contrast with these studies, Mukaremera et al. found no association between in vivo or in vitro capsule size and virulence in the mouse model [84]. The lack of agreement on the importance of the capsule to virulence throughout studies may be due to the different methodologies used to assess capsule size between studies. Typical in vitro capsule induction assays may not be sufficient to accurately evaluate the capsule formation of an isolate during human infection [81,82].
The phenotypic analysis of clinical isolates has largely focused on easy to measure virulence phenotypes such as in vitro capsule enlargement and/or shedding, melanin synthesis, growth at 37°C, urease activity, and phospholipase activity [77,85]. Several studies did not find a direct link between patient outcome and the virulence factors tested, especially with the use of in vitro assays [81–84,86–88]. Overall, these studies did highlight the high level of virulence factor heterogeneity between clinical isolates and the lack of association between a single virulence phenotype in determining disease outcome. Instead, a combination of multiple virulence phenotypes is likely to contribute to patient outcome.
Interestingly, two studies that incorporated analysis beyond the major cell surface virulence phenotypes showed the rate of in vitro phagocytosis was associated with patient mortality [86,88]. Alanio et al. showed isolates with high rates of in vitro phagocytosis and high intracellular proliferation displayed a higher risk of patient mortality [88]. A subsequent study by Sabiiti et al. similarly showed that increased rates of in vitro phagocytosis correlated with increased risk of mortality at 10 weeks post-diagnosis [86]. This study went on to show that isolates with increased rates of in vitro phagocytosis also displayed an increase in initial baseline fungal burden and increased laccase activity, indirectly suggesting a role for melanin in phagocytosis [86]. These studies show it may be necessary to go beyond the previously described virulence factors to better understand how the relationship between C. neoformans cell surface changes and host cell interactions impacts patient outcome.
In addition to phenotypic changes between isolates, it is important to note that C. neoformans also displays a wide range of phenotypes within a single population throughout the course of infection [66,70,89–92]. Phenotypic variation within an isolate may allow for rapid adaptation to the various host niches encountered during infection, suggesting the degree of phenotypic plasticity displayed by an isolate may also contribute to its virulence and the differences seen in disease outcome between isolates. A recent study investigating the size variability across clinical isolates from Botswana showed a correlation between size phenotype and clinical markers [70]. ‘Small’ phenotypes, which included micro cells and shed capsule, were found to be associated with clinical parameters important during later stages of infection, while ‘large’ phenotypes, which included increase in cell diameter and capsule size, were found to be associated with clinical parameters important during earlier stages of infection. Isolates typically exhibited either ‘large’ or ‘small’ phenotypes but rarely exhibited both. Interestingly, the four isolates that exhibited both ‘small’ and ‘large’ phenotypes all resulted in patient death [70]. Additionally, Thanh et al. showed clinical isolates able to infect immunocompetent patients displayed a significant increase in within-strain variation with regard to capsule and cell size [83]. These studies provide support to the idea that phenotypic heterogeneity within a population may contribute to host adaptation, which may subsequently impact patient survival. Similarly, Mukaremera et al. suggested that a single virulence factor would not dominate in C. neoformans, but rather the ability of an isolate to overcome a wider variety of stresses may result in increased virulence [84]. Thus, a strain’s ability to adapt to the host may be critical to disease outcome.
Microevolution
Does microevolution of C. neoformans occur during human infections?
Microevolution refers to rapid changes over a short period of time. During microevolution, selection of genetic and phenotypic alterations that allow for increased fitness occurs. There is evidence of microevolution in C. neoformans during passage in the lab, murine infection, and human infection [91,93–103]. The study of C. neoformans microevolution has primarily focused on recurrent infections in humans. Changes in karyotypes between recurrent isolates from humans result in altered virulence in a murine model [102–104]. The use of WGS coupled with phenotypic analyses has allowed for the identification of more specific genetic differences to be observed between initial and recurrent isolates and the phenotypic changes underlying these genetic changes to be assessed [99,101]. The genetic changes in recurrent isolates correlated with increased fluconazole resistance, and a smaller fraction correlated with other virulence phenotypes [101]. Exploring the genetic and phenotypic changes in recurrent human isolates collected from the CSF allows for better insight into the adaptations specific to the central nervous system. The changes in C. neoformans important for adaptation earlier in infection, such as establishment and progression of pulmonary infection, are yet to be explored in human samples. In a murine model, microevolution is thought to be organ dependent, and distinct subpopulations arise during a pulmonary infection, highlighting the heterogeneous initial response to the host environment [97,105]. The role each of these subpopulations plays during infection is unknown. Of particular interest is the population thought to be dormant cells [105]. Dormant cells may be important in host adaptation and reactivation of latent infections. There is epidemiological evidence of reactivation of dormant cells in human infections, but the characterization of dormant cells and their role in microevolution are yet to be assessed in clinical isolates [106].
What are the possible mechanisms underlying microevolution in C. neoformans?
Genetic and phenotypic analyses of serial isolates from recurrent infections has provided insight into the genetic changes and subsequent phenotypic changes that occur during human infections that may promote host adaptation [99,101]. Although we now have a better understanding of the genetic and phenotypic changes C. neoformans undergoes during human infections, the mechanisms underlying these changes are still largely unexplored. In this section, we discuss the possible mechanisms that may promote microevolution in the human host, focusing on those that produce genetic alterations. The possible mechanisms discussed in this section include transposable elements (TEs), mutators, and titan cells (Figure 4).
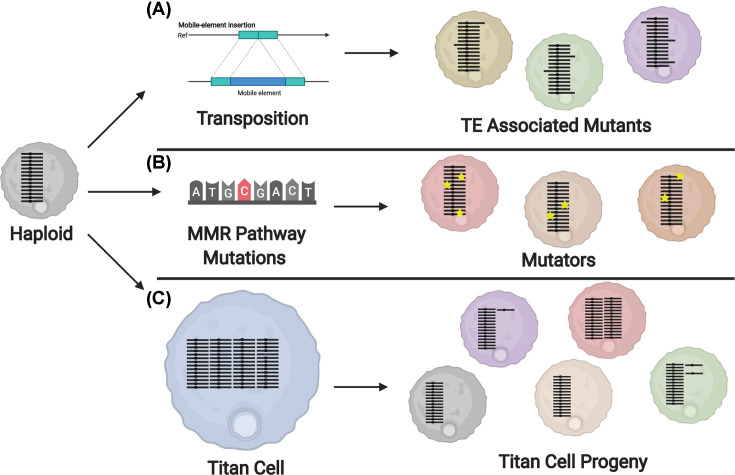
Figure 4. Mechanisms of in vivo microevolution.
Model displaying possible mechanisms underlying the genetic and subsequent phenotypic changes seen during infection. Individual chromosomes are shown in black, and phenotype changes are represented by change in cell color. (A) During infection, TEs move throughout the genome. Transposon insertion into genes that may result in a range of phenotypes [111]. (B) Mutations in the MMR pathway result in cells with a higher rate of mutations (yellow stars) that may display various phenotypes [114,116]. (C) Polyploid titan cells undergo a reductive division to produce progeny with reduced genome sizes and genomic alterations, including cells with various whole chromosome aneuploidies (purple and green cells). Phenotype changes have also been observed in titan progeny that have no identified genomic alterations (tan cell) [76]. Abbreviation: MMR, mismatch repair.
TEs, or transposons, are segments of DNA that can move throughout the genome of an organism. TEs can independently proliferate, either by encoding their own enzymes for transposition or though reverse transcription (retrotransposons). TEs are common in eukaryotes and make up 5% of the C. neoformans genome, where they preferentially cluster on each chromosome into 40–100 kb regions near the centromere [95]. TEs can create or reverse mutations and cause changes in gene expression or regulation. As TEs can negatively disrupt the genome, most eukaryotes have evolved methods to defend against TE-induced mutagenesis. C. neoformans uses endogenously produced small RNAs (endo-siRNA) to silence TEs [107–110]. While limiting transposon mobilization to prevent detrimental mutagenesis is important under normal conditions, when adapting to a new, high-stress environment, transposons may prove beneficial. Since TEs are found in the C. neoformans genome and do contribute to mutagenesis, it is a logical hypothesis that TEs are involved in C. neoformans adaptation to the host through genetic and subsequent phenotypic alterations (Figure 4A). A recent paper analyzing transposons in C. deneoformans found an increase in TE activation when cells were grown at 37°C [111]. This same study hinted that a similar phenomenon exists in C. neoformans, where transposon activation during infection may act as a mechanism for microevolution (Figure 4A).
Mutators, or isolates that mutate rapidly under stress, have been identified in many species of fungi. Mutation phenotypes in Saccharomyces cerevisiae, C. gattii, and C. neoformans have been found to occur due to mutations in the mismatch repair (MMR) pathway, particularly in the MMR gene MSH2 [112–114]. Mutations in genes involved in the MMR pathway of C. neoformans—MSH2, MLH1, and PMS1—result in isolates that showed a change in lung proliferation, suggesting a role in virulence [115]. Naturally arising mutations in these same three genes cause a mutator phenotype [114,116]. A 2017 study closely investigated the role of the MMR pathway in C. neoformans by deleting MSH2, MLH1, and PMS1 and making double mutants of each strain [114]. The study observed that deletions in the MMR pathway led to rapid microevolution and rapid resistance to antifungal agents. Interestingly, the deletion mutants for the MMR pathway also had a different profile of mutations compared with wild type, showing an increase in both transition mutations and single nucleotide insertions and deletions occurring in homopolymeric tracts. In contrast with previous work, deletions in MLH1 and MSH2 did not cause any changes in virulence compared with wild type [114,115]. Mutations in the MMR pathway cause an increase in 200-fold in rate of mutations compared with wild type, suggesting a mechanism for rapid microevolution in C. neoformans [114]. WGS of relapse isolates have revealed SNPs in MSH2, perhaps due to the increased drug resistance caused by that mutation [94]. A higher rate of microevolution has also been observed in C. deneoformans strains that have a mutation in POL3, a gene that encodes for the DNA polymerase Δ subunit [117,118]. Thus far, two independent pathways have been identified in the C. neoformans species complex for the development of mutator phenotypes. These mutators are able to rapidly adapt resistance to antifungal drugs, in a way that is less harmful to fungal cell fitness than aneuploidy. Mutators are also able to quickly develop new phenotypes (Figure 4B) [114]. Though they do not seem to have improved virulence compared with wild type, future studies may reveal that C. neoformans isolates with mutator phenotypes are able to more quickly adapt to the host environment.
As mentioned earlier, titan cells are enlarged cells that arise during the initial pulmonary infection. In the lungs, 20–30% of C. neoformans cells convert into titan cells [64]. While C. neoformans is primarily a haploid yeast consisting of one set of 14 chromosomes, titan cells are polyploid [64]. Despite their increased size and ploidy, titan cells are able to bud and undergo cytokinesis to produce typical-sized daughter cells with reduced ploidy [76]. Gerstein et al. showed that titans are capable of generating haploid and aneuploid daughter cell populations in vitro that exhibit increased resistance to fluconazole, oxidative, and nitrosative stress (Figure 4C) [76]. Aneuploid formation underlying fluconazole resistance is well documented in C. neoformans and was recently shown to occur during fluconazole treatment in human infections [119,120]. Aneuploidy is often thought to be detrimental to an organism’s fitness, but under certain conditions, especially during times of stress, aneuploidy may be beneficial [121]. The finding that titan cells are able to produce aneuploid populations suggests they may act as a source of rapid adaptation during infection (Figure 4C). Further analysis of the genomic changes produced by titan cells in vivo is necessary to better understand the contribution of titan cells to host adaptation and provide further insight into the genomic changes that are important for survival in the host.
In this section, we summarized three potential mechanisms underlying microevolution in C. neoformans that result in DNA alterations. It is important to note that changes in the regulation of genes important to survival and host adaptation are not limited to genetic alterations. Epigenetic modifications can also produce rapid changes in gene expression. These modifications and subsequent transcriptome changes likely also facilitate host adaptation in C. neoformans. Previous studies have shown that chromatin remodeling controlled by histone deacetylases results in changes to virulence phenotypes [122,123] and the post-transcriptional process of mRNA decay also plays a critical role in stress response and thermotolerance [124].
Concluding remarks
Historically, studies on the biology and genetics of the Cryptococcus sp. have largely been performed independently from clinical studies. Yet, recent translational studies have highlighted the importance of the genotype and phenotype of C. neoformans strains to clinical outcome in patients. These studies have uncovered critical gaps in our understanding of how C. neoformans and the other species within the Cryptococcus species complex adapt to and cause disease within the host. Future research needs to focus on better understanding of the host–pathogen interaction, development of useful models that accurately recapitulate human disease phenotypes, and a better understanding of the path from genetic variation to phenotypic variation and ultimately disease characteristics of the Cryptococcus species.
Acknowledgements
The figures were created with BioRender.com.
Abbreviations
- CSF
cerebrospinal fluid
- GWAS
genome-wide association study
- INDEL
insertion/deletion
- LD50
lethal dose for 50% of mice
- MLST
multilocus sequence typing
- MMR
mismatch repair
- SNP
single nucleotide polymorphism
- ST
sequence type
- TE
transposable element
- WGS
whole-genome sequencing
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Institutes of Health [grant numbers R01AI134636, R01NS118538, R21NS108715 (to K.N.), K12GM119955 (to S.A.), F31AI148047 (to K.M.J.)].
Author Contribution
S.A.: initial concept and outline. Figures 1 and 2 were created by K.M.J. Figure 4 was created by K.M.J. and S.A., and Figure 3 was created by S.A. K.M.J., K.N., and S.A.: writing, editing, and approval of final manuscript.
References
- 1.Hagen F., Khayhan K., Theelen B., Kolecka A., Polacheck I., Sionov E. et al. (2015) Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78, 16–48 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 2.Kwon-Chung K.J., Bennett J.E., Wickes B.L., Meyer W., Cuomo C.A., Wollenburg K.R. et al. (2017) The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2, e00357–16, 10.1128/mSphere.00357-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorrell T.C., Chen S.C.-A., Phillips P. and Marr K.A. (2011) Clinical perspectives on Cryptococcus neoformans and Cryptococcus gattii: implications for diagnosis and management. In Cryptococcus: From Human Pathogen to Model Yeast(Heitman J., Kozel T.R., Kwon-Chung K.J., Perfect J.R. and Casadevall A., eds), pp. 595–606, ASM Press, Washington DC, U.S.A. [Google Scholar]
- 4.Maziarz E.K. and Perfect J.R. (2016) Cryptococcosis. Infect. Dis. Clin. North Am. 30, 179–206 10.1016/j.idc.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans E.E. (1950) The antigenic composition of Cryptococcus neoformans. I. A serologic classification by means of the capsular and agglutination reactions. J. Immunol. 64, 423–430 [PubMed] [Google Scholar]
- 6.Wilson D.E., Bennett J.E. and Bailey J.W. (1968) Serologic grouping of Cryptococcus neoformans. Proc. Soc. Exp. Biol. Med. 127, 820–823 10.3181/00379727-127-32812 [DOI] [PubMed] [Google Scholar]
- 7.Kwon-Chung K.J. (1975) A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67, 1197–1200 10.1080/00275514.1975.12019866 [DOI] [PubMed] [Google Scholar]
- 8.Kwon-Chung K.J. (1976) A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68, 943–946 10.1080/00275514.1976.12019972 [DOI] [PubMed] [Google Scholar]
- 9.Kwon-Chung K.J. (1976) Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68, 821–833 10.1080/00275514.1976.12019959 [DOI] [PubMed] [Google Scholar]
- 10.Kwon-Chung K.J. and Bennett J.E. (1978) Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108, 337–340 10.1093/oxfordjournals.aje.a112628 [DOI] [PubMed] [Google Scholar]
- 11.Ellis D.H. and Pfeiffer T.J. (1990) Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336, 923–925 10.1016/0140-6736(90)92283-N [DOI] [PubMed] [Google Scholar]
- 12.Bennett J.E., Kwon-Chung K.J. and Theodore T.S. (1978) Biochemical differences between serotypes of Cryptococcus neoformans. Sabouraudia 16, 167–174 10.1080/00362177885380231 [DOI] [PubMed] [Google Scholar]
- 13.Polacheck I. and Kwon-Chung K.J. (1980) Creatinine metabolism in Cryptococcus neoformans and Cryptococcus bacillisporus. J. Bacteriol. 142, 15–20 10.1128/JB.142.1.15-20.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polacheck I. and Kwon-Chung K.J. (1986) Canavanine resistance in Cryptococcus neoformans. Antimicrob. Agents Chemother. 29, 468–473 10.1128/AAC.29.3.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min K.H. and Kwon-Chung K.J. (1986) The biochemical basis for the distinction between the two Cryptococcus neoformans varieties with CGB medium. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261, 471–480 [DOI] [PubMed] [Google Scholar]
- 16.Kwon-Chung K.J., Wickes B.L., Booth J.L., Vishniac H.S. and Bennett J.E. (1987) Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect. Immun. 55, 1751–1754 10.1128/IAI.55.8.1751-1754.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickes B.L., Moore T.D. and Kwon-Chung K.J. (1994) Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140, 543–550 10.1099/00221287-140-3-543 [DOI] [PubMed] [Google Scholar]
- 18.Franzot S.P., Salkin I.F. and Casadevall A. (1999) Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 37, 838–840 10.1128/JCM.37.3.838-840.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon-Chung K.J., Boekhout T., Fell J.W. and Diaz M. (2002) (1557) Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51, 804–806 10.2307/1555045 [DOI] [Google Scholar]
- 20.Hagen F., Lumbsch H.T., Arsic Arsenijevic V., Badali H., Bertout S., Billmyre R.B. et al. (2017) Importance of resolving fungal nomenclature: the case of multiple pathogenic species in the Cryptococcus genus. mSphere 2, e00238–17, 10.1128/mSphere.00238-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litvintseva A.P., Thakur R., Vilgalys R. and Mitchell T.G. (2006) Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172, 2223–2238 10.1534/genetics.105.046672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khayhan K., Hagen F., Pan W., Simwami S., Fisher M.C., Wahyuningsih R. et al. (2013) Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE 8, e72222 10.1371/journal.pone.0072222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira-Paim K., Andrade-Silva L., Fonseca F.M., Ferreira T.B., Mora D.J., Andrade-Silva J. et al. (2017) MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in southeastern Brazil. PLoS Negl. Trop. Dis. 11, e0005223 10.1371/journal.pntd.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrade-Silva L.E., Ferreira-Paim K., Ferreira T.B., Vilas-Boas A., Mora D.J., Manzato V.M. et al. (2018) Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS ONE 13, e0193237 10.1371/journal.pone.0193237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer W., Aanensen D.M., Boekhout T., Cogliati M., Diaz M.R., Esposto M.C. et al. (2009) Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47, 561–570 10.1080/13693780902953886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day J.N., Qihui S., Thanh L.T., Trieu P.H., Van A.D., Thu N.H. et al. (2017) Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl. Trop. Dis. 11, e0005628 10.1371/journal.pntd.0005628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang W., Fa Z. and Liao W. (2015) Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genet. Biol. 78, 7–15 10.1016/j.fgb.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 28.Kiertiburanakul S., Wirojtananugoon S., Pracharktam R. and Sungkanuparph S. (2006) Cryptococcosis in human immunodeficiency virus-negative patients. Int. J. Infect. Dis. 10, 72–78 10.1016/j.ijid.2004.12.004 [DOI] [PubMed] [Google Scholar]
- 29.Pappas P.G., Perfect J.R., Cloud G.A., Larsen R.A., Pankey G.A., Lancaster D.J. et al. (2001) Cryptococcosis in human immunodeficiency virus-negative patients in the era of effective azole therapy. Clin. Infect. Dis. 33, 690–699 10.1086/322597 [DOI] [PubMed] [Google Scholar]
- 30.Chan M., Lye D., Win M.K., Chow A. and Barkham T. (2014) Clinical and microbiological characteristics of cryptococcosis in Singapore: predominance of Cryptococcus neoformans compared with Cryptococcus gattii. Int. J. Infect. Dis. 26, 110–115 10.1016/j.ijid.2014.05.019 [DOI] [PubMed] [Google Scholar]
- 31.Chen S., Sorrell T., Nimmo G., Speed B., Currie B., Ellis D. et al. (2000) Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin. Infect. Dis. 31, 499–508 10.1086/313992 [DOI] [PubMed] [Google Scholar]
- 32.Dromer F., Varma A., Ronin O., Mathoulin S. and Dupont B. (1994) Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J. Clin. Microbiol. 32, 2364–2371 10.1128/JCM.32.10.2364-2371.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S.C., Meyer W. and Sorrell T.C. (2014) Cryptococcus gattii infections. Clin. Microbiol. Rev. 27, 980–1024 10.1128/CMR.00126-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz J.H. (2020) The disease ecology, epidemiology, clinical manifestations, and management of emerging Cryptococcus gattii complex infections. Wilderness Environ. Med. 31, 101–109 10.1016/j.wem.2019.10.004 [DOI] [PubMed] [Google Scholar]
- 35.Hoang L.M.N., Maguire J.A., Doyle P., Fyfe M. and Roscoe D.L. (2004) Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre: epidemiology, microbiology and histopathology. J. Med. Microbiol. 53, 935–940 10.1099/jmm.0.05427-0 [DOI] [PubMed] [Google Scholar]
- 36.Harris J.R., Lockhart S.R., Debess E., Marsden-Haug N., Goldoft M., Wohrle R. et al. (2011) Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin. Infect. Dis. 53, 1188–1195 10.1093/cid/cir723 [DOI] [PubMed] [Google Scholar]
- 37.Fyfe M., MacDougall L., Romney M., Starr M., Pearce M., Mak S. et al. (2008) Cryptococcus gattii infections on Vancouver Island, British Columbia, Canada: emergence of a tropical fungus in a temperate environment. Can. Commun. Dis. Rep. 34, 1–12 [PubMed] [Google Scholar]
- 38.Engelthaler D.M., Hicks N.D., Gillece J.D., Roe C.C., Schupp J.M., Driebe E.M. et al. (2014) Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5, e01464–14 10.1128/mBio.01464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang Y.C., Stins M.F., McCaffery M.J., Miller G.F., Pare D.R., Dam T. et al. (2004) Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect. Immun. 72, 4985–4995 10.1128/IAI.72.9.4985-4995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barchiesi F., Cogliati M., Esposto M.C., Spreghini E., Schimizzi A.M., Wickes B.L. et al. (2005) Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J. Infect. 51, 10–16 10.1016/j.jinf.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 41.Dromer F., Mathoulin S., Dupont B., Letenneur L. and Ronin O. (1996) Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. French Cryptococcosis Study Group. Clin. Infect. Dis. 23, 91–96 10.1093/clinids/23.1.91 [DOI] [PubMed] [Google Scholar]
- 42.Desnos-Ollivier M., Patel S., Raoux-Barbot D., Heitman J., Dromer F. and Group FCS (2015) Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. mBio 6, e00311–15 10.1128/mBio.00311-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon-Chung K.J. and Bennett J.E. (1984) Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120, 123–130 10.1093/oxfordjournals.aje.a113861 [DOI] [PubMed] [Google Scholar]
- 44.Neuville S., Dromer F., Morin O., Dupont B., Ronin O., Lortholary O. et al. (2003) Primary cutaneous cryptococcosis: a distinct clinical entity. Clin. Infect. Dis. 36, 337–347 10.1086/345956 [DOI] [PubMed] [Google Scholar]
- 45.Ngamskulrungroj P., Chang Y., Sionov E. and Kwon-Chung K.J. (2012) The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3, e00103–12, 10.1128/mBio.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajasingham R., Smith R.M., Park B.J., Jarvis J.N., Govender N.P., Chiller T.M. et al. (2017) Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881 10.1016/S1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bicanic T. and Harrison T.S. (2005) Cryptococcal meningitis. Br. Med. Bull. 72, 99–118, 10.1093/bmb/ldh043 [DOI] [PubMed] [Google Scholar]
- 48.Wiesner D.L., Moskalenko O., Corcoran J.M., McDonald T., Rolfes M.A., Meya D.B. et al. (2012) Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3, e00196–12, 10.1128/mBio.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beale M.A., Sabiiti W., Robertson E.J., Fuentes-Cabrejo K.M., O’Hanlon S.J., Jarvis J.N. et al. (2015) Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across Southern Africa. PLoS Negl. Trop. Dis. 9, e0003847 10.1371/journal.pntd.0003847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ponzio V., Chen Y., Rodrigues A.M., Tenor J.L., Toffaletti D.L., Medina-Pestana J.O. et al. (2019) Genotypic diversity and clinical outcome of cryptococcosis in renal transplant recipients in Brazil. Emerg. Microbes Infect. 8, 119–129 10.1080/22221751.2018.1562849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., Varma A., Diaz M.R., Litvintseva A.P., Wollenberg K.K. and Kwon-Chung K.J. (2008) Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 14, 755–762 10.3201/eid1405.071312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chau T.T., Mai N.H., Phu N.H., Nghia H.D., Chuong L.V., Sinh D.X. et al. (2010) A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam - high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect. Dis. 10, 199 10.1186/1471-2334-10-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Day J.N., Hoang T.N., Duong A.V., Hong C.T., Diep P.T., Campbell J.I. et al. (2011) Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VN1. J. Clin. Microbiol. 49, 658–664 10.1128/JCM.01985-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashton P.M., Thanh L.T., Trieu P.H., Van Anh D., Trinh N.M., Beardsley J. et al. (2019) Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat. Commun. 10, 2035 10.1038/s41467-019-10092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerstein A.C., Jackson K.M., McDonald T.R., Wang Y., Lueck B.D., Bohjanen S. et al. (2019) Identification of pathogen genomic differences that impact human immune response and disease during Cryptococcus neoformans infection. mBio 10, e01440–19 10.1128/mBio.01440-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McFadden D.C., Fries B.C., Wang F. and Casadevall A. (2007) Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot. Cell 6, 1464–1473 10.1128/EC.00162-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frases S., Nimrichter L., Viana N.B., Nakouzi A. and Casadevall A. (2008) Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot. Cell 7, 319–327 10.1128/EC.00378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denham S.T., Verma S., Reynolds R.C., Worne C.L., Daugherty J.M., Lane T.E. et al. (2018) Regulated release of cryptococcal polysaccharide drives virulence and suppresses immune cell infiltration into the central nervous system. Infect. Immun. 86, e00662–17 10.1128/IAI.00662-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukaremera L., Lee K.K., Wagener J., Wiesner D.L., Gow N.A.R. and Nielsen K. (2018) Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf. 1, 15–24 10.1016/j.tcsw.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker L.G., Specht C.A. and Lodge J.K. (2011) Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell 10, 1264–1268 10.1128/EC.05138-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nosanchuk J.D., Rosas A.L., Lee S.C. and Casadevall A. (2000) Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355, 2049–2050 10.1016/S0140-6736(00)02356-4 [DOI] [PubMed] [Google Scholar]
- 62.Reese A.J. and Doering T.L. (2003) Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50, 1401–1409 10.1046/j.1365-2958.2003.03780.x [DOI] [PubMed] [Google Scholar]
- 63.Reese A.J., Yoneda A., Breger J.A., Beauvais A., Liu H., Griffith C.L. et al. (2007) Loss of cell wall alpha(1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63, 1385–1398 10.1111/j.1365-2958.2006.05551.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okagaki L.H., Strain A.K., Nielsen J.N., Charlier C., Baltes N.J., Chrétien F. et al. (2010) Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6, e1000953 10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaragoza O., García-Rodas R., Nosanchuk J.D., Cuenca-Estrella M., Rodríguez-Tudela J.L. and Casadevall A. (2010) Fungal cell gigantism during mammalian infection. PLoS Pathog. 6, e1000945 10.1371/journal.ppat.1000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feldmesser M., Kress Y. and Casadevall A. (2001) Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147, 2355–2365 10.1099/00221287-147-8-2355 [DOI] [PubMed] [Google Scholar]
- 67.Cruickshank J.G., Cavill R. and Jelbert M. (1973) Cryptococcus neoformans of unusual morphology. Appl. Microbiol. 25, 309–312 10.1128/AEM.25.2.309-312.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love G.L., Boyd G.D. and Greer D.L. (1985) Large Cryptococcus neoformans isolated from brain abscess. J. Clin. Microbiol. 22, 1068–1070 10.1128/JCM.22.6.1068-1070.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J.M., Zhou Q., Cai H.R., Zhuang Y., Zhang Y.F., Xin X.Y. et al. (2014) Clinicopathological features of pulmonary cryptococcosis with cryptococcal titan cells: a comparative analysis of 27 cases. Int. J. Clin. Exp. Pathol. 7, 4837–4846 [PMC free article] [PubMed] [Google Scholar]
- 70.Fernandes K.E., Brockway A., Haverkamp M., Cuomo C.A., van Ogtrop F., Perfect J.R. et al. (2018) Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from Botswanan HIV/AIDs patients. mBio 9, e02016–18 10.1128/mBio.02016-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dambuza I.M., Drake T., Chapuis A., Zhou X., Correia J., Taylor-Smith L. et al. (2018) The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 14, e1006978 10.1371/journal.ppat.1006978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charlier C., Chrétien F., Baudrimont M., Mordelet E., Lortholary O. and Dromer F. (2005) Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 166, 421–432 10.1016/S0002-9440(10)62265-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivera J., Feldmesser M., Cammer M. and Casadevall A. (1998) Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 66, 5027–5030 10.1128/IAI.66.10.5027-5030.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie S., Sao R., Braun A. and Bottone E.J. (2012) Difference in Cryptococcus neoformans cellular and capsule size in sequential pulmonary and meningeal infection: a postmortem study. Diagn. Microbiol. Infect. Dis. 73, 49–52 10.1016/j.diagmicrobio.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 75.Okagaki L.H. and Nielsen K. (2012) Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot. Cell 11, 820–826 10.1128/EC.00121-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gerstein A.C., Fu M.S., Mukaremera L., Li Z., Ormerod K.L., Fraser J.A. et al. (2015) Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6, e01340–15 10.1128/mBio.01340-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kozel T.R. (1995) Virulence factors of Cryptococcus neoformans. Trends Microbiol. 3, 295–299 10.1016/S0966-842X(00)88957-X [DOI] [PubMed] [Google Scholar]
- 78.O’Meara T.R. and Alspaugh J.A. (2012) The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408 10.1128/CMR.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaragoza O. and Nielsen K. (2013) Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr. Opin. Microbiol. 16, 409–413 10.1016/j.mib.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaragoza O. (2019) Basic principles of the virulence of Cryptococcus. Virulence 10, 490–501 10.1080/21505594.2019.1614383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robertson E.J., Najjuka G., Rolfes M.A., Akampurira A., Jain N., Anantharanjit J. et al. (2014) Cryptococcus neoformans ex vivo capsule size is associated with intracranial pressure and host immune response in HIV-associated cryptococcal meningitis. J. Infect. Dis. 209, 74–82 10.1093/infdis/jit435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clancy C.J., Nguyen M.H., Alandoerffer R., Cheng S., Iczkowski K., Richardson M. et al. (2006) Cryptococcus neoformans var. grubii isolates recovered from persons with AIDS demonstrate a wide range of virulence during murine meningoencephalitis that correlates with the expression of certain virulence factors. Microbiology 152, 2247–2255 10.1099/mic.0.28798-0 [DOI] [PubMed] [Google Scholar]
- 83.Thanh L.T., Toffaletti D.L., Tenor J.L., Giamberardino C., Sempowski G.D., Asfaw Y. et al. (2020) Assessing the virulence of Cryptococcus neoformans causing meningitis in HIV infected and uninfected patients in Vietnam. Med. Mycol., 1–13 10.1093/mmy/myaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukaremera L., McDonald T.R., Nielsen J.N., Molenaar C.J., Akampurira A., Schutz C. et al. (2019) The mouse inhalation model of Cryptococcus neoformans infection recapitulates strain virulence in humans and shows that closely related strains can possess differential virulence. Infect. Immun. 87, e00046–19, 10.1128/IAI.00046-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alspaugh J.A. (2015) Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 78, 55–58 10.1016/j.fgb.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabiiti W., Robertson E., Beale M.A., Johnston S.A., Brouwer A.E., Loyse A. et al. (2014) Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J. Clin. Invest. 124, 2000–2008 10.1172/JCI72950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liaw S.J., Wu H.C. and Hsueh P.R. (2010) Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin. Microbiol. Infect. 16, 696–703 10.1111/j.1469-0691.2009.02930.x [DOI] [PubMed] [Google Scholar]
- 88.Alanio A., Desnos-Ollivier M. and Dromer F. (2011) Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio 2, e00158–11, 10.1128/mBio.00158-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lopes W., Cruz G.N.F., Rodrigues M.L., Vainstein M.H., Kmetzsch L., Staats C.C. et al. (2020) Scanning electron microscopy and machine learning reveal heterogeneity in capsular morphotypes of the human pathogen Cryptococcus spp. Sci. Rep. 10, 2362 10.1038/s41598-020-59276-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goldman D.L., Fries B.C., Franzot S.P., Montella L. and Casadevall A. (1998) Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc. Natl. Acad. Sci. U.S.A. 95, 14967–14972 10.1073/pnas.95.25.14967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fries B.C., Taborda C.P., Serfass E. and Casadevall A. (2001) Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Invest. 108, 1639–1648 10.1172/JCI13407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jain N., Cook E., Xess I., Hasan F., Fries D. and Fries B.C. (2009) Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryot. Cell 8, 858–866 10.1128/EC.00017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Franzot S.P., Mukherjee J., Cherniak R., Chen L.C., Hamdan J.S. and Casadevall A. (1998) Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 66, 89–97 10.1128/IAI.66.1.89-97.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arras S.D.M., Ormerod K.L., Erpf P.E., Espinosa M.I., Carpenter A.C., Blundell R.D. et al. (2017) Convergent microevolution of Cryptococcus neoformans hypervirulence in the laboratory and the clinic. Sci. Rep. 7, 17918 10.1038/s41598-017-18106-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Janbon G., Ormerod K.L., Paulet D., Byrnes E.J., Yadav V., Chatterjee G. et al. (2014) Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10, e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu G., Chen S.H., Qiu J., Bennett J.E., Myers T.G. and Williamson P.R. (2014) Microevolution during serial mouse passage demonstrates FRE3 as a virulence adaptation gene in Cryptococcus neoformans. mBio 5, e00941–14 10.1128/mBio.00941-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia-Hermoso D., Dromer F. and Janbon G. (2004) Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect. Immun. 72, 3359–3365 10.1128/IAI.72.6.3359-3365.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sullivan D., Haynes K., Moran G., Shanley D. and Coleman D. (1996) Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J. Clin. Microbiol. 34, 1739–1744 10.1128/JCM.34.7.1739-1744.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ormerod K.L., Morrow C.A., Chow E.W., Lee I.R., Arras S.D., Schirra H.J. et al. (2013) Comparative genomics of serial isolates of Cryptococcus neoformans reveals gene associated with carbon utilization and virulence. G3 (Bethesda) 3, 675–686 10.1534/g3.113.005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Y., Toffaletti D.L., Tenor J.L., Litvintseva A.P., Fang C., Mitchell T.G. et al. (2014) The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 5, e01087–13 10.1128/mBio.01087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y., Farrer R.A., Giamberardino C., Sakthikumar S., Jones A., Yang T. et al. (2017) Microevolution of serial clinical isolates of Cryptococcus neoformans var. grubii and C. gattii. mBio 8, e00166–17 10.1128/mBio.00166-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fries B.C., Chen F., Currie B.P. and Casadevall A. (1996) Karyotype instability in Cryptococcus neoformans infection. J. Clin. Microbiol. 34, 1531–1534 10.1128/JCM.34.6.1531-1534.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brandt M.E., Pfaller M.A., Hajjeh R.A., Graviss E.A., Rees J., Spitzer E.D. et al. (1996) Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated cryptococcosis. Cryptococcal Disease Active Surveillance Group. J. Infect. Dis. 174, 812–820 10.1093/infdis/174.4.812 [DOI] [PubMed] [Google Scholar]
- 104.Fries B.C. and Casadevall A. (1998) Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 178, 1761–1766 10.1086/314521 [DOI] [PubMed] [Google Scholar]
- 105.Alanio A., Vernel-Pauillac F., Sturny-Leclère A. and Dromer F. (2015) Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. mBio 6, e02580–14, 10.1128/mBio.02580-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garcia-Hermoso D., Janbon G. and Dromer F. (1999) Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37, 3204–3209 10.1128/JCM.37.10.3204-3209.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang X., Hsueh Y.P., Li W., Floyd A., Skalsky R. and Heitman J. (2010) Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 24, 2566–2582 10.1101/gad.1970910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Janbon G., Maeng S., Yang D.H., Ko Y.J., Jung K.W., Moyrand F. et al. (2010) Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet. Biol. 47, 1070–1080 10.1016/j.fgb.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dumesic P.A., Natarajan P., Chen C., Drinnenberg I.A., Schiller B.J., Thompson J. et al. (2013) Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 152, 957–968 10.1016/j.cell.2013.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burke J.E., Longhurst A.D., Natarajan P., Rao B., Liu J., Sales-Lee J. et al. (2019) A non-dicer RNase III and four other novel factors required for RNAi-mediated transposon suppression in the human pathogenic yeast Cryptococcus neoformans. G3 (Bethesda) 9, 2235–2244 10.1534/g3.119.400330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gusa A., Williams J.D., Cho J.E., Averette A.F., Sun S., Shouse E.M. et al. (2020) Transposon mobilization in the human fungal pathogen Cryptococcus is mutagenic during infection and promotes drug resistance in vitro. Proc. Natl. Acad. Sci. U.S.A. 117, 9973–9980 10.1073/pnas.2001451117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strand M., Prolla T.A., Liskay R.M. and Petes T.D. (1993) Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365, 274–276 10.1038/365274a0 [DOI] [PubMed] [Google Scholar]
- 113.Billmyre R.B., Croll D., Li W., Mieczkowski P., Carter D.A., Cuomo C.A. et al. (2014) Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5, e01494–14, 10.1128/mBio.01494-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boyce K.J., Wang Y., Verma S., Shakya V.P.S., Xue C. and Idnurm A. (2017) Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptoccous neoformans. mBio 8, e00595–17, 10.1128/mBio.00595-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu O.W., Chun C.D., Chow E.D., Chen C., Madhani H.D. and Noble S.M. (2008) Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135, 174–188 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rhodes J., Beale M.A., Vanhove M., Jarvis J.N., Kannambath S., Simpson J.A. et al. (2017) A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3 (Bethesda) 7, 1165–1176 10.1534/g3.116.037499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Magditch D.A., Liu T.B., Xue C. and Idnurm A. (2012) DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog. 8, e1002936 10.1371/journal.ppat.1002936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boyce K.J., Cao C., Xue C. and Idnurm A. (2020) A spontaneous mutation in DNA polymerase POL3 during in vitro passaging causes a hypermutator phenotype in Cryptococcus species. DNA Repair (Amst.) 86, 102751 10.1016/j.dnarep.2019.102751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sionov E., Lee H., Chang Y.C. and Kwon-Chung K.J. (2010) Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6, e1000848 10.1371/journal.ppat.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stone N.R., Rhodes J., Fisher M.C., Mfinanga S., Kivuyo S., Rugemalila J. et al. (2019) Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Invest. 129, 999–1014 10.1172/JCI124516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsai H.J. and Nelliat A. (2019) A double-edged sword: aneuploidy is a prevalent strategy in fungal adaptation. Genes (Basel) 10, 787, 10.3390/genes10100787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brandão F.A., Derengowski L.S., Albuquerque P., Nicola A.M., Silva-Pereira I. and Poças-Fonseca M.J. (2015) Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence 6, 618–630 10.1080/21505594.2015.1038014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brandão F., Esher S.K., Ost K.S., Pianalto K., Nichols C.B., Fernandes L. et al. (2018) HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci. Rep. 8, 5209 10.1038/s41598-018-21965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bloom A.L., Solomons J.T., Havel V.E. and Panepinto J.C. (2013) Uncoupling of mRNA synthesis and degradation impairs adaptation to host temperature in Cryptococcus neoformans. Mol. Microbiol. 89, 65–83 10.1111/mmi.12258 [DOI] [PMC free article] [PubMed] [Google Scholar]