Abstract
Introduction
The cell-surface ectonucleotidase CD39 is a key molecule of the immunosuppressive adenosine pathway within the tumor microenvironment. However, the relationship between CD39 and clear cell renal cell carcinoma (ccRCC) is rarely reported and still remains unclear.
Methods
CD39 expression was first analyzed using the Oncomine and the Tumor IMmune Estimation Resource (TIMER) databases, and then examined in ccRCC patients (n=367) who had undergone radical nephrectomy using immunohistochemistry (IHC) and real-time quantitative PCR analysis (qPCR). The prognosis value of CD39 in ccRCC was evaluated by Cox proportional hazards analysis. Functional and gene set enrichment analysis (GSEA) was performed using transcriptomic data of ccRCC from TCGA. Correlation analysis between CD39 and tumor-infiltrating lymphocytes (TILs) was performed using the TISIDB database. The impact of CD39 on immune checkpoint therapy (ICT) was evaluated by two public cohorts.
Results
CD39 mRNA and protein expression was upregulated in tumor tissues from ccRCC patients and aberrant expression of CD39 was associated with advanced tumor stage and poor prognosis in ccRCC patients. EMT, IL-2/STAT5, inflammatory response, interferon gamma and KRAS hallmark gene sets were identified as CD39-related signaling pathway. The expression level of CD39 was significantly and positively correlated with high abundance of the regulatory TILs including NK cells, macrophages, Th cells and Treg cells. CD39 was correlated with expression of several immune checkpoints and higher CD39 expression was associated with better OS of ccRCC patients who received ICT.
Conclusion
CD39 is a powerful prognostic marker of ccRCC patients. Increased tumor expression of CD39 mRNA is significantly correlated with infiltrating levels of TILs, and better efficacy of ICT to ccRCC. CD39 could be a novel therapeutic target for ccRCC.
Keywords: renal cell carcinoma, CD39, prognosis, TCGA, infiltrating immune cells, immune checkpoint therapy
Introduction
Renal cell carcinoma (RCC) is one of the most common genitourinary malignancies worldwide, contributing to about 3.8% of newly diagnosed cancers. In 2019, it was estimated that in the United States there were 73,750 new diagnoses of cancer of the kidney and renal pelvis, and 14,830 will die of the disease.1 An epidemiological study based on the Surveillance, Epidemiology, and End Results (SEER) database indicates that morbidity rates of RCC are rising on average 0.6% per year. Clear cell renal cell carcinoma (ccRCC) is the most common histological subtype of renal tumor with malignant potential, accounting for approximately 75% of cases, and is considered one of the most aggressive subtypes.2 Despite immune checkpoint inhibitor therapy, the prognosis of RCC is still unsatisfactory.3,4 There still exists undefined molecular regulatory networks that promote ccRCC to relapse and metastasize, as well as develop drug resistance. Therefore, it is of great significance to conduct more basic research to explore new molecular therapeutic targets for clinical application of ccRCC.
Tumor hypoxia, which is frequently observed in ccRCC, effectively regulates the tumorigenesis through a series of metabolic and immunological changes.5 Oxygen deprivation induces the accumulation of extracellular adenosine in tumors, which binds to the P1 purinergic receptors to restrain immune cell infiltration and activation.6 These immunosuppressive mechanisms may promote ccRCC to develop resistance to radiotherapy and immunotherapy, and inhibiting hypoxia and adenosine represents a potential target for anticancer therapy.7 The cell-surface ectonucleotidase CD39, also known as ectonucleoside triphosphate diphosphohydrolase-1 (ENTPD1), is a key molecule in the adenosine pathway. It specifically hydrolyses extracellular ATP to ADP and/or AMP, which is then dephosphorylated to adenosine in the presence of CD73 (another critical ectonucleotidase of the adenosine pathway).6 CD39 is expressed across a diverse range of cancer cell types, high expression of CD39 has been reported to be consistently correlated with poor prognosis in patients with non-small-cell lung cancers (NSCLCs), hepatocellular carcinoma (HCC), ovarian, gastric and breast cancers.8–12 Recently, the role of CD39 in regulating tumor immunity is emerging. Traditionally, CD39 was considered as a marker of Foxp3+ regulatory T cells in the tumor microenvironment, while more recently and in several studies, CD39 was identified as a marker for exhausted CD8+ T cells in cancer patients.13–16
Although the biological function of CD39 has been studied to some extent, the relationship between CD39 and ccRCC is rarely reported and still remains unclear. In this study, we have measured the expression of CD39 in surgical resected ccRCC tissue and evaluated the correlation of CD39 mRNA levels with clinicopathological factors. We have also investigated the association between tumor CD39 mRNA levels and prognosis of ccRCC patients to determine prognostic value. Moreover, we have identified the potential pathways where CD39 might be involved and analyzed the relativity between CD39 and tumor-infiltrating immune cells in ccRCC patients using public databases to comprehend the underlying mechanism.
Materials and Methods
Selection of Patients
A total of 367 patients receiving radical nephrectomy due to ccRCC (from Aug 2008 to Sept 2017) were recruited from the Department of Urology, Fudan University Shanghai Cancer Center (FUSCC) (Shanghai, China). The clinical and pathological information of each patient were recorded, including age at surgery, sex, depth of tumor invasion (T), lymph node metastasis (N), distant metastasis (M), International Society of Urological Pathology (ISUP) grading classification and pTNM stage (Table 1). Tissue samples, including paired tumor and para-carcinoma normal samples, were collected during surgery and preserved in the FUSCC tissue bank. Overall survival (OS) was defined as the time from radical nephrectomy to death or last follow-up. Patients’ progression-free survival (PFS) was calculated from the initiation of surgery until the first progression, including the start of a second-line treatment and death. This study was approved by the institutional ethics committee of FUSCC and written informed consent was obtained from all the patients preoperatively.
Table 1.
Demographics and Clinical Characteristics of ccRCC Patients According to Their CD39 Expression
| Parameters | All Patients (n=367) | |||
|---|---|---|---|---|
| Low Expression (n=210) | High Expression (n=157) | p value | ||
| Age | 0.815 | |||
| <60 years | 243 (66.2) | 138 (65.7) | 105 (66.9) | |
| ≧60 years | 124 (33.8) | 72 (34.3) | 52 (33.1) | |
| Gender | 0.805 | |||
| Male | 2,48 (67.6) | 143 (68.1) | 105 (66.9) | |
| Female | 119 (32.4) | 67 (31.9) | 52 (33.1) | |
| Invasion deep | 0.003* | |||
| T1/T2 | 303 (82.6) | 184 (87.6) | 119 (75.8) | |
| T3/T4 | 64 (17.4) | 26 (12.4) | 38 (24.2) | |
| Lymph node metastasis | 0.127 | |||
| N0 | 322 (87.7) | 189 (90.0) | 133 (84.7) | |
| N1 | 45 (12.3) | 21 (10.0) | 24 (15.3) | |
| Distant metastasis | 0.002* | |||
| M0 | 308 (83.9) | 187 (89.0) | 121 (77.1) | |
| M1 | 59 (16.1) | 23 (11.0) | 36 (22.9) | |
| pTNM stage | 0.004* | |||
| I/II | 288 (78.5) | 176 (83.8) | 112 (71.3) | |
| III/IV | 79 (21.5) | 34 (16.2) | 45 (28.7) | |
| ISUP grade | 0.147 | |||
| 1/2 | 175 (47.7) | 107 (51.0) | 68 (43.3) | |
| ¾ | 192 (52.3) | 103 (49.0) | 89 (56.7) | |
Notes: Data expressed as count and percentage for categorical variables and were performed by Chi-square test. *p <0.05 between low-expression and high-expression groups.
Abbreviations: ccRCC, clear cell renal cell carcinoma; ISUP, International Society of Urological Pathology.
Immunohistochemistry
Six ccRCC and para-carcinoma normal tissues were collected from the overall patient cohort and fixed in 10% buffered formalin, embedded in paraffin. Immunohistochemistry was then performed to detect the expression of CD39 (recombinant rabbit monoclonal antibody, 1:50 dilution; ab223842, Abcam, USA) proteins, according to the manufacturer’s instructions. An HRP-conjugated secondary antibody and DAB kit (Dako, Agilent Technologists, CA, USA) were used to visualize antibody binding. Immunostaining reactivity was observed by two experienced pathologists independently.
Real-Time Quantitative PCR Analysis
Total RNA from harvested cells was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with attached protocols. RNA was reverse transcribed into cDNA using a PrimeScript RT reagent Kit (Thermo Fisher, USA). Primers were diluted and mixed in RNase free dH2O in SYBR® Green qPCR kit (Takara Biotechnology Co). β-actin RNA expression was measured for standardization. Primer sequences for human CD39 were as follows: forward 5ʹ- CTGATTCCTGGGAGCACATC-3ʹ and reverse 5ʹ- CTGGGATCATGTTGGTCAGG-3ʹ. Specific conditions of operating cycles for CD39 and β-action were performed according to SYBR® Green qPCR master mix (Applied Biosystems) manufacturer protocols. The relative CD39 mRNA expression was represented as ΔCt =Ct(CD39) – ΔCt(β-actin). Relative expression in ccRCC was represented using the ratio of CD39 expression in Tumor/Normal tissues (T/N). X-tile software was utilized to take the cut-off value of the T/N ratio of CD39 mRNA expression, where T/N ≤ 2.55 represented low expression and T/N > 2.55 represented high expression.
Bioinformatics Analysis
RNAseq gene expression data of ccRCC (“Level_3_RSEM_genes_normalized”) and patient clinical data were downloaded from the TCGA repository (https://portal.gdc.cancer.gov/). Bioinformatics analysis was performed using R software 3.5.2 (R Foundation for Statistical Computing, Vienna, Austria). The “limma” package was used to perform differential expression analysis (log FC>0.5, P<0.05).17 The “pheatmap” and the “ggplot2” package were then utilized to visualize differentially expressed genes (DEGs). GO function analysis and gene set enrichment analysis (GSEA) was performed using the “clusterprofiler” package based on hallmark gene sets.18,19 The DEGs gene-concept network was constructed based on the most significant hallmark gene clusters. We calculated the epithelial–mesenchymal transition (EMT)-related gene expression score via the arithmetic mean of the 200 EMT-related gene expression levels based on the work by Wang et al (log2-transformed).20
The Tumor IMmune Estimation Resource (TIMER) algorithm database (https://cistrome.shinyapps.io/timer/) and the Oncomine database (https://www.oncomine.org/resource/main.html) were used to estimate CD39 gene expression levels in various types of cancers.21,22 We then analyzed the relationship between CD39 expression and abundance of various types of tumor-infiltrating lymphocytes (TILs) in ccRCC patients via TISIDB (an integrated repository portal for tumor-immune system interactions) database (http://cis.hku.hk/TISIDB).23 The correlation analysis between the CD39 expression level and PD-1, PD-L1 and CTLA4 was also performed.
Survival Data of Immune Checkpoint Therapy Patient Cohorts
Miao et al performed whole exome sequencing of 35 metastatic ccRCC patients who received anti-programmed cell death-1 receptor (anti-PD-1) therapy (nivolumab).24 Sequencing and survival data of these metastatic ccRCC patients were downloaded for further analysis. The primary outcome to evaluate the response of patients to ICT was OS.
Statistical Analysis
SPSS Statistics software package, version 23.0 (Chicago, IL, USA) was used for the statistical analysis. Demographic characteristics were summarized by count and percentage for categorical variables, and chi-square was performed to compare the distribution of categorical data between different CD39 mRNA expression sets. Spearman correlation coefficient was utilized to evaluate the correlation of gene expression. Survival analysis was performed using the Kaplan–Meier method and P values were determined using Log rank test. Univariate and multivariate Cox logistic regression models were used to evaluate the hazard ratios of prognostic factors. All statistic assessments were evaluated at a two-sided P value of 0.05.
Results
Expression of CD39 in ccRCC
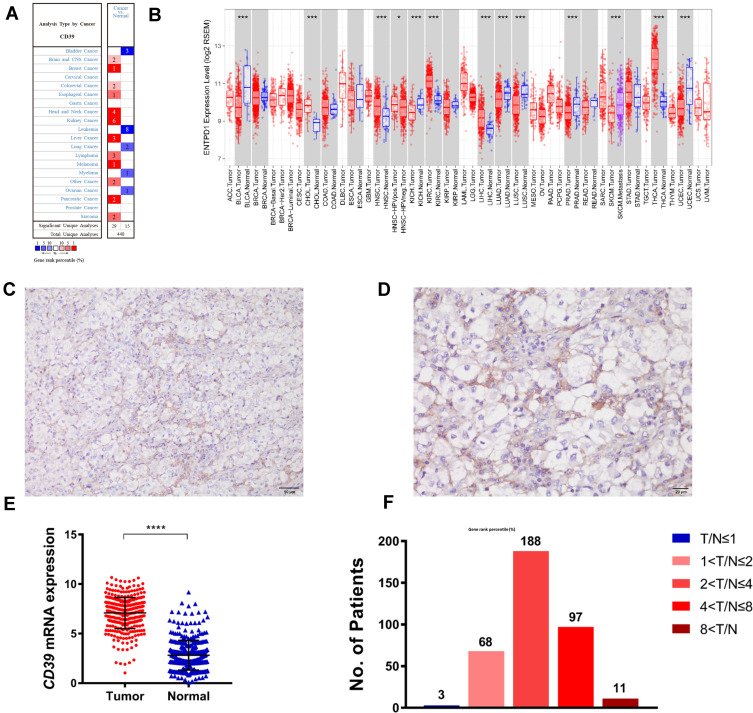
In this study, we first investigated the expression of CD39 in ccRCC tissues both on protein and transcription level to preliminarily estimate its role in ccRCC. Public database analysis using the TIMER algorithm database (using TCGA RNA-sequencing data) and the Oncomine database revealed that mRNA level of CD39 in ccRCC significantly increased compared with normal tissues (Figure 1A and B). To further validate CD39 expression in ccRCC, we performed IHC and real-time PCR on the specimens collected from ccRCC patients. According to immunohistochemistry, CD39 was mainly expressed in ccRCC tissues, and was primarily localized to the cell membrane. There was also positive CD39 expression in the stromal fibroblasts and lymphocytes, suggesting a complex function of CD39 in the tumor microenvironment (Figure 1C and D plus Supplementary Figure 1). The CD39 mRNA expression profile in ccRCC tumor tissue based on real-time PCR data revealed a much higher expression level of CD39 in tumor tissues compared with normal tissues, indicating that CD39 may be involved in the development and progression of ccRCC (Figure 1E). The overall ccRCC patient cohort was then stratified into low-CD39-expression group (210, 57.2%) and high-CD39-expression group (157, 42.8%) according to the T/N ratio. Figure 1F shows the dramatic difference in the distribution concerning the T/N ratio.
Figure 1.
Expression of CD39 in ccRCC. (A and B) CD39 expression level in different cancer types in the Oncomine database and TCGA database (analyzed by TIMER). (C and D) CD39 protein expression in ccRCC tissues (C, 200×magnification; D, 400×magnification). (E) CD39 mRNA expression in ccRCC and para-carcinoma normal tissue. (F) The percentage of different CD39 mRNA expression described by different T/N (ccRCC/normal tissue).
The Relationship Between CD39 Expression and Clinicopathological Characteristics in ccRCC Patients
We next investigated the relationship between CD39 level and various clinicopathological characteristics in ccRCC patients. Demographic and clinical characteristics of ccRCC patients are summarized in (Table 1 plus Supplementary Table 1). Patients with high CD39 mRNA expression significantly correlated with advanced T stage (p=0.003), M stage (p=0.002) and pTNM stage (p=0.004). However, the expression of CD39 was not associated with age, sex, positive lymph node metastasis (advanced N stage) and advanced ISUP grade (p>0.05).
Cox Regression Analyses and Survival Outcomes of the Cohorts
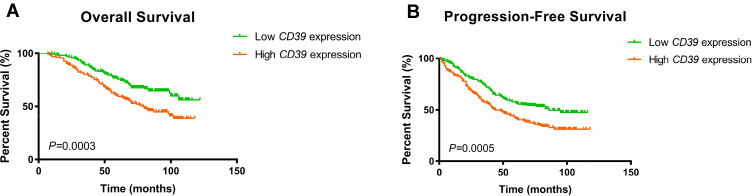
We further examined the associations between the CD39 mRNA expression and survival outcome in a ccRCC patients. The Kaplan–Meier survival curve and Log rank test analysis revealed that elevated CD39 expression was significantly associated with poorer PFS (p<0.001) and OS (p<0.001) (Figure 2A and B).
Figure 2.
Kaplan–Meier survival analyses of ccRCC patients stratified by the mRNA expression level of CD39. (A) Kaplan–Meier curves showing OS of patients. (B) Kaplan–Meier curves showing PFS of patients.
We also utilized univariate and multivariate Cox regression to estimate survival outcomes in ccRCC patient cohort. Univariate Cox regression showed that high CD39 expression was independently associated with poor PFS (HR: 1.627; p=0.001) and OS (HR: 1.782; p<0.001) for ccRCC patients, as were age at surgery (PFS: p=0.025, OS: p=0.017) and advanced ISUP grade (PFS: p<0.001, OS: p<0.001), while sex was not assessed as prognostic indicators of PFS (p=0.076) and OS (0.906) in our study (Tables 2 and 3). Multivariate Cox regression included factors that proved to be prognostic indicators in univariate analysis. Notably, high CD39 expression remained a significant independent prognostic factor of PFS (HR: 1.394; p=0.023) and OS (HR: 1424; p=0.037) in ccRCC patients in multivariate analysis, as were lymph node metastases (PFS: p<0.001, OS: p=0.003), distant metastases (PFS: p=0.024, OS: p=0.017), pTNM stage (PFS: p<0.001, OS: p<0.001) and ISUP grade (PFS: p<0.001, OS: p=0.004). Patients with positive lymph node metastasis, positive distant metastasis, advanced stage tumor and ISUP grade had worse PFS and OS. While age at surgery (PFS: p=0.330, OS: p=0.140) and tumor invasion depth (PFS: p=0.368, OS: p=0.351) were not considered as independent prognostic factors of ccRCC patients (Tables 2 and 3).
Table 2.
Univariate and Multivariate Cox Regression Analyses of PFS in 367 Enrolled ccRCC Patients
| Covariates | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age at surgery | 1.014 (1.002–1.026) | 0.025* | 1.006 (0.994–1.019) | 0.330 |
| Sex (male vs female) | 0.756 (0.556–1.030) | 0.076 | ||
| Invasion deep (T1/T2 vs T3/T4) | 9.811 (6.946–13.858) | <0.001* | 1.313 (0.726–2.374) | 0.368 |
| Lymph node metastasis (N0 vs N1) | 12.235 (8.318–17.997) | <0.001* | 2.389 (1.468–3.888) | <0.001* |
| Distant metastasis (M0 vs M1) | 8.781 (6.293–12.253) | <0.001* | 1.735 (1.076–2.799) | 0.024* |
| pTNM stage (Ⅰ/Ⅱ vs III/Ⅳ) | 12.538 (8.885–17.694) | <0.001* | 4.148 (2.046–8.409) | <0.001* |
| ISUP grade (1/2 vs 3/4) | 2.760 (2.054–3.709) | <0.001* | 1.769 (1.285–2.436) | <0.001* |
| CD39 expression (low vs high) | 1.627 (1.234–2.146) | 0.001* | 1.394 (1.048–1.855) | 0.023* |
Note: *p <0.05.
Abbreviations: PFS, progression-free survival; ccRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval; ISUP, International Society of Urological Pathology.
Table 3.
Univariate and Multivariate Cox Regression Analyses of OS in 367 Enrolled ccRCC Patients
| Covariates | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age at surgery | 1.016 (1.003–1.030) | 0.017* | 1.010 (0.997–1.025) | 0.140 |
| Sex (male vs female) | 0.980 (0.698–1.376) | 0.906 | ||
| Invasion deep (T1/T2 vs T3/T4) | 11.455 (7.979–16.444) | <0.001* | 1.332 (0.729–2.433) | 0.351 |
| Lymph node metastasis (N0 vs N1) | 11.027 (7.438–16.346) | <0.001* | 2.055 (1.270–3.325) | 0.003* |
| Distant metastasis (M0 vs M1) | 10.347 (7.244–14.779) | <0.001* | 1.856 (1.118–3.082) | 0.017* |
| pTNM stage (Ⅰ/Ⅱ vs III/Ⅳ) | 14.517 (10.026–21.019) | <0.001* | 4.839 (2.338–10.015) | <0.001* |
| ISUP grade (1/2 vs 3/4) | 3.387 (2.366–4.849) | <0.001* | 1.798 (1.206–2.681) | 0.004* |
| CD39 expression (low vs high) | 1.782 (1.295–2.450) | <0.001* | 1.424 (1.022–1.984) | 0.037* |
Note: *p <0.05.
Abbreviations: OS, overall survival; ccRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval; ISUP, International Society of Urological Pathology.
Taking these data together, elevated CD39 mRNA expression was an independent predictor of poor prognosis for ccRCC patients suggesting CD39 can be a potential prognostic biomarker for ccRCC patients.
Identifying the CD39-Related Signaling Pathway Based on TCGA ccRCC Dataset
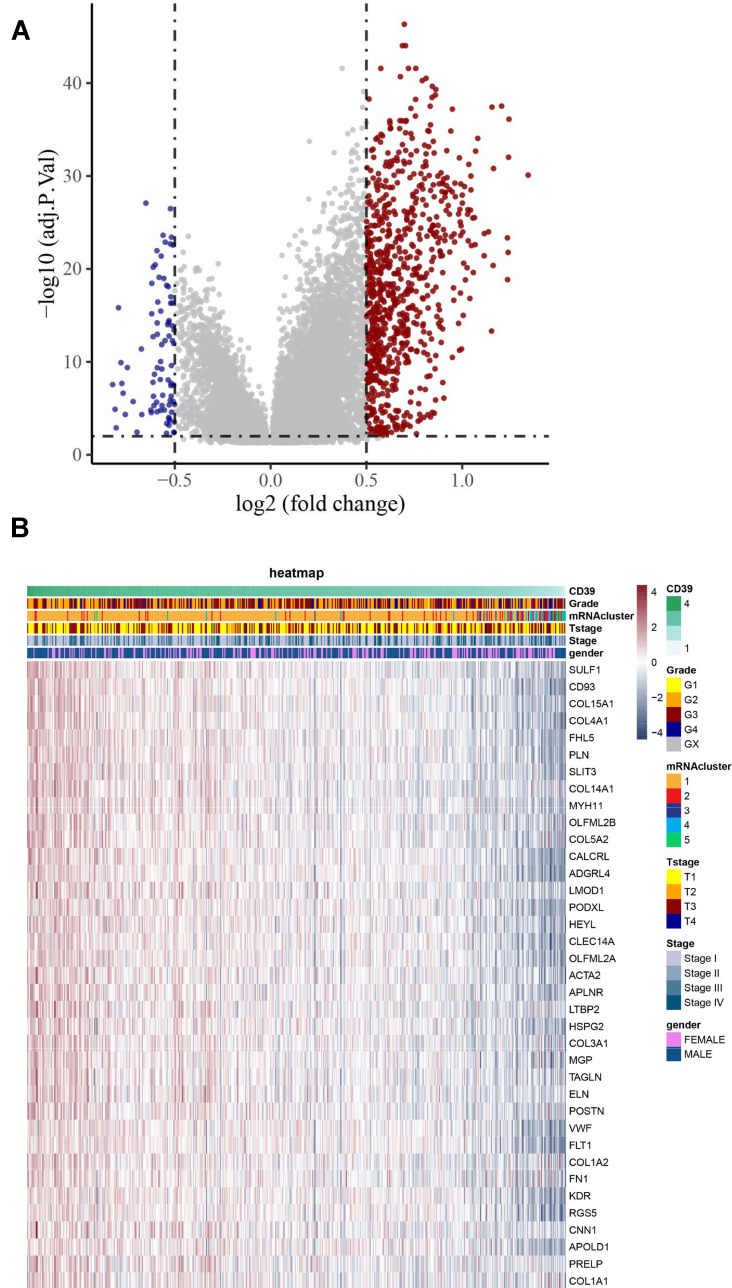
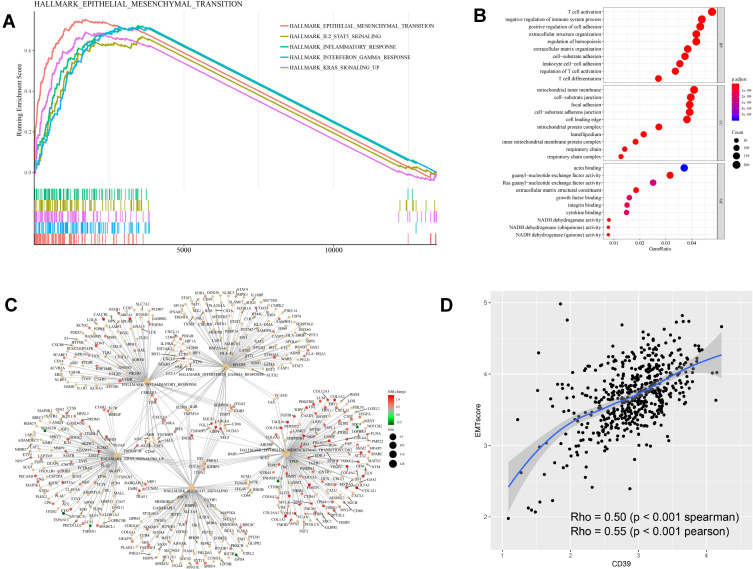
To investigate the underlying mechanism of CD39 in promoting tumor progression, we conducted differential expression analysis based on the RNAseq data of ccRCC from TCGA and utilized a volcano plot to visualize the result (Figure 3A). We also utilized a heatmap to show the correlation between the DEGs and clinicopathological characteristics of the TCGA ccRCC cohort (Figure 3B). We then conducted GSEA of the DEGs in ccRCC based on hallmark gene sets to identify CD39-associated signaling pathways. The results revealed that differentially expressed genes of CD39 were enriched in the following gene set: HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION, HALLMARK_IL2_STAT5_SIGNALING, HALLMARK_INFLAMMATORY_RESPONSE, HALLMARK_INTERFERON_GAMMA_RESPONSE, HALLMARK_KRAS_SIGNALING_UP (Figure 4A). A gene-concept network was constructed to display the most significant enriched hallmark gene clusters and the genes involved with these significant terms (Figure 4C). GO functional analysis revealed that the DEGs were mainly enriched by T cell activation and negative regulation of the immune system process in the biological process (BP) group. The mitochondrial inner membrane and cell-substrate junction were the most enriched terms in the cellular component (CC) group. For the molecular function (MF) group, the DEGs were mainly enriched in actin-binding and guanyl-nucleotide exchange factor activity (Figure 4B). Moreover, we further demonstrated that CD39 was significantly correlated with a high EMT-related gene expression score (Spearman’s rho=0.5, p<0.001) (Figure 4D).
Figure 3.
Differential expression analysis based on the RNAseq data of ccRCC from TCGA. (A) Volcano plot showing DEGs based on CD39 mRNA expression. The red dots represent up-regulated genes and the blue dots represent down-regulated genes. (B) Expression profiles of DEGs across TCGA ccRCC samples.
Figure 4.
Functional and gene set enrichment analysis of CD39 based on TCGA ccRCC samples. (A) Pathway enrichment analysis of DEGs based on the hall mark gene sets. (B) GO function analysis of DEGs. The top group represents BP group, the middle group represents CC group and the bottom group represents MF group. (C) DEGs gene-concept network based on the most significant hallmark gene clusters. (D) Plot of the correlation between CD39 and EMT-related gene in TCGA ccRCC samples.
CD39 Expression is Associated with Several Types of Infiltrating Immune Cells and Better Response of ccRCC Patients to ICT
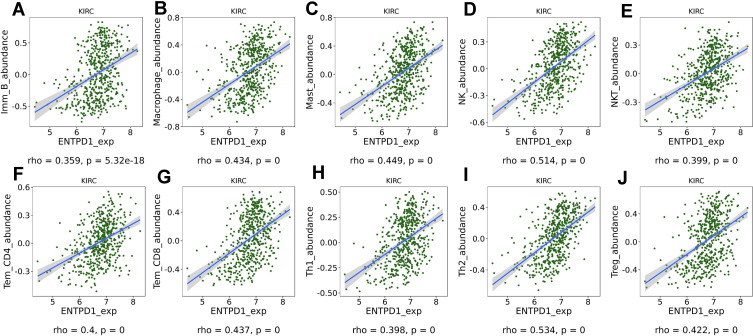
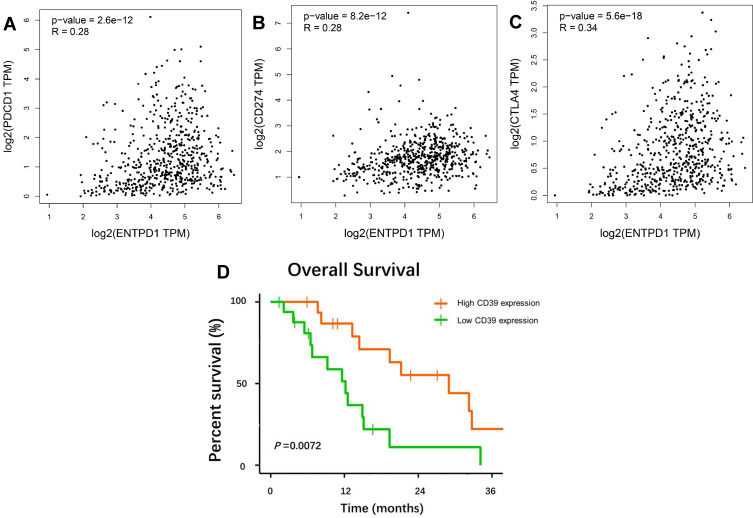
Finally, in order to investigate the role of CD39 in the immune microenvironment in ccRCC, we analyzed the correlation between CD39 expression and tumor-infiltrating immune cells via TISIDB. The expression level of CD39 was significantly and positively correlated with infiltrating levels of immature B cells (r=0.359, p=5.32e-18), macrophages (r=0.434, p=0), mast cells (r=0.449, p=0), natural killer (NK) cells (r=0.514, p=0), natural killer T (NKT) cells (r=0.339, p=0), effector memory CD4+ T cells (r=4, p=0), effector memory CD8+ T cells (r=0.437, p=0), type 1 T helper (Th1) cells (r=0.398, p=0), type 2 T helper (Th2) cells (r=0.534, p=0) and regulatory T (Treg) cell (r=0.422, p=0) in ccRCC (Figure 5). In addition, based on the TCGA database, we found that CD39 was correlated with PD-1 (Spearman’s rho=0.28, p<0.001), PD-L1 (Spearman’s rho=0.28, p<0.001) and CTLA4 (Spearman’s rho=0.34, p<0.001) expression levels (Figure 6A–C). We then used sequencing and survival data of ccRCC patient cohorts uploaded by Miao et al to query the impact of CD39 expression on OS of patients treated with ICT. The Kaplan–Meier survival curve and Log rank test analysis revealed that elevated CD39 expression was significantly associated with better OS both in ccRCC (p=0.0072) ICT-treated patients (Figure 6D). Together, these findings suggested that CD39 participated in the regulation of the tumor-immune microenvironment and could be a specific prognosticator of antitumor immune response of ICT in ccRCC patients.
Figure 5.
Correlation analysis of CD39 expression with infiltrating immune cells via the TIMER database. CD39 expression was significantly and positively correlated with infiltrating levels of immature B cells (A), macrophages (B), mast cells (C), natural killer (NK) cells (D), natural killer T (NKT) cells (E), effector memory CD4 T cells (F), effector memory CD8 T cells (G), type 1 T helper cells (H), type 2 T helper cells (I) and regulatory T (Treg) cells (J).
Figure 6.
Plot of the correlation between CD39 and PD-1 (A), PD-L1 (B), CTLA4 (C) in TCGA ccRCC samples. High-expression level of CD39 is associated with better overall survival in cohorts of ccRCC patients (D).
Discussion
As the rate-limiting enzyme in the hydrolysis of extracellular ATP/ADP, CD39 participates in the generation of adenosine, a major immunosuppressive molecule that promotes tumor growth and progression.6,25 CD39 was first identified as a dominant cell-surface ectonucleotidase in the purinergic regulation of inflammation during chronic inflammatory diseases.26,27 During the last decade, the role of CD39 in the context of tumorigenesis has been highlighted. Activation of the adenosinergic pathway occurs within hypoxic tumors including renal cell carcinoma, and elevated CD39 expression has been shown to be an independent poor prognostic factor in various types of cancer.10,11,28 ccRCC is characterized by a clear cytoplasm, which is due to glycogen and lipid accumulation, indicating that genetic mutations are involved in cellular metabolism.29 Complex metabolic reprogramming is frequently observed in this malignancy, including impairments of glycolysis, lipid and nucleotide metabolism, as well as mitochondrial bioenergetics.30,31 Abnormal activation of the adenosine pathway is an essential metabolic alteration involved in the progression of ccRCC. However, the role of CD39 in the development and progression of ccRCC is rarely reported and still largely unknown until now.
In the present study, we enrolled 367 patients who had undergone radical nephrectomy in our center to investigate CD39 protein and mRNA expression in ccRCC tissues. We found that CD39 was upregulated in tumor tissues of ccRCC both in protein and mRNA levels. Moreover, aberrant expression of CD39 was associated with advanced tumor stage and poor prognosis in ccRCC patients. Analysis based on Oncomine and TIMER datasets also came to the same conclusion. Our study thus demonstrated that aberrant CD39 expression was a potential prognostic biomarker for ccRCC patients.
Hallmark gene sets are a collection of refined gene sets derived from multiple MSigDB sets, representing a specific biological process and providing more refined inputs for pathway enrichment analysis.18 After identifying the DEGs of CD39, we further demonstrated that these genes were primarily enriched in EMT. The correlation analysis between CD39 and EMT-related gene expression also proved this result. Moreover, GSEA also identified IL-2/STAT5, inflammatory response, interferon gamma and KRAS as CD39-related signaling pathways. EMT is a biological process that improved the invasion and metastasis capacity of cancers.32 KRAS is also an important tumor-promoting process involved in the tumor microenvironment.33 IL-2 and interferon gamma are key cytokines in the regulation of the anti-tumor immune system. Recently, several studies found that IL-2 and interferon gamma also participate in the anti-tumor process of ICT.31,34 Although CD39 was primarily recognized as a significant molecule in the adenosinergic pathway and participated in the tumor immunosuppression.6,27 However, the results of our research indicated that the role of CD39 plays in the tumor microenvironment was much more complicated and the mechanism of CD39 in the progression of ccRCC involved various activated signaling pathways.
ICT with anti-PD-1 and anti-PD-L1 therapies have revolutionized the treatment of various advanced cancer including ccRCC.3,4 Although immune checkpoint inhibitors (ICIs) can significantly improve prognosis of ccRCC patients, there still exist a considerable proportion of patients who remain unresponsive or resistant to ICT.35 ccRCC is characterized as one of the most immune-infiltrated tumors.36 Signals in the immune microenvironment including the accumulation of oncometabolites, or T-cell dysfunction may heavily affect the responses of patients with ccRCC to ICT.37,38 Previous studies have demonstrated that CD39 is broadly expressed across various types of immune cells, and recently the specific role of CD39 in these cells has been clarified gradually.6 Zhang et al proposed that CD39 suppressed NK cell-mediated immunity and promoted tumor metastases.39 Takenaka et al identified that CD39 driven by the aryl hydrocarbon receptor (AHR) participates in the regulation of tumor-associated macrophages and T cells in glioblastoma.40 Canale et al demonstrated that CD39 could define cell exhaustion in tumor-associated CD8+ T Cells.41 Simoni et al proposed CD39 as a molecule that could be used to address the presence of bystander T cells in cancer patients.15 Qi et al demonstrated that a high frequency of CD39+CD8+ T cells was independently associated with a poor prognosis in ccRCC.42 In this study, we also demonstrated that the DEGs of CD39 were remarkably enriched in T cell activation and negative regulation of the immune system process through GO function analysis. Moreover, we further validated and investigated the role of CD39 in the immune microenvironment of ccRCC patients. Based on public database analyses, we found that high CD39 expression always accompanied with a high abundance of regulatory TILs including NK cells, macrophages, Th cells and Treg cells. While association between CD39 and effector or activated cells was weak. Combining these results and the special role that CD39 plays in the adenosine pathway, we proposed that CD39 may participate in the regulation of several immune cells to create an immunosuppressive tumor microenvironment. Moreover, we observed correlations between CD39 and several immune checkpoints. Thus, we further utilized ccRCC ICT data from the study of Miao et al to investigate the impact of CD39 expression on the efficacy of ICT. The results revealed that higher CD39 expression was associated with better OS in the ccRCC patients. However, the molecular mechanism of the correlation between immunosuppressive CD39 and the patient response to ICT remains unclear. A possible explanation is that high CD39 expression always accompanied with high abundance of regulatory TILs, while most of these TILs exhibited a dysfunctional phenotype and lost immune control of tumors. Somehow immune checkpoint inhibitor may reactive the exhausted immune cells and restore tumor immunity. However, both in vivo and in vitro experiments should be conducted to explore the molecular mechanism of CD39 facilitating the formation of immunosuppressive environment.
In conclusion, our study demonstrated that the CD39 mRNA level was a powerful prognostic marker of ccRCC patients. Moreover, we observed a significant correlation between CD39 and EMT, IL-2/STAT5, inflammatory response, interferon gamma, KRAS and infiltrating levels of TILs. High tumor CD39 expression was also associated with better OS of ccRCC patients who received ICT, indicating that CD39 could be a novel therapeutic target for improving treatment strategies for ccRCC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Project 81772706). The paper was edited by International Science Editing (Supplementary material).
Data Sharing Statement
The authors elect to not share data.
Disclosure
The authors declare no conflicts of interest for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part a: renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029 [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Powles T, Atkins MB, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, Phase 3, randomised controlled trial. Lancet (London, England). 2019;393(10189):2404–2415. doi: 10.1016/S0140-6736(19)30723-8 [DOI] [PubMed] [Google Scholar]
- 4.Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380(12):1116–1127. doi: 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- 5.Alsaab HO, Sau S, Alzhrani RM, et al. Tumor hypoxia directed multimodal nanotherapy for overcoming drug resistance in renal cell carcinoma and reprogramming macrophages. Biomaterials. 2018;183:280–294. doi: 10.1016/j.biomaterials.2018.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayan D, Young A, Teng MWL, Smyth MJ. Targeting immunosuppressive adenosine in cancer. Nat Rev Cancer. 2017;17(12):765. doi: 10.1038/nrc.2017.110 [DOI] [PubMed] [Google Scholar]
- 7.Hatfield SM, Kjaergaard J, Lukashev D, et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7(277):277ra230. doi: 10.1126/scitranslmed.aaa1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastid J, Regairaz A, Bonnefoy N, et al. Inhibition of CD39 enzymatic function at the surface of tumor cells alleviates their immunosuppressive activity. Cancer Immunol Res. 2015;3(3):254–265. doi: 10.1158/2326-6066.CIR-14-0018 [DOI] [PubMed] [Google Scholar]
- 9.Turcotte M, Spring K, Pommey S, et al. CD73 is associated with poor prognosis in high-grade serous ovarian cancer. Cancer Res. 2015;75(21):4494–4503. doi: 10.1158/0008-5472.CAN-14-3569 [DOI] [PubMed] [Google Scholar]
- 10.Cai XY, Ni XC, Yi Y, et al. Overexpression of CD39 in hepatocellular carcinoma is an independent indicator of poor outcome after radical resection. Medicine (Baltimore). 2016;95(40):e4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai XY, Wang XF, Li J, et al. Overexpression of CD39 and high tumoral CD39(+)/CD8(+) ratio are associated with adverse prognosis in resectable gastric cancer. Int J Clin Exp Pathol. 2015;8(11):14757–14764. [PMC free article] [PubMed] [Google Scholar]
- 12.Abousamra NK, Salah El-Din M, Hamza Elzahaf E, Esmael ME. Ectonucleoside triphosphate diphosphohydrolase-1 (E-NTPDase1/CD39) as a new prognostic marker in chronic lymphocytic leukemia. Leuk Lymphoma. 2015;56(1):113–119. doi: 10.3109/10428194.2014.907893 [DOI] [PubMed] [Google Scholar]
- 13.Canale FP, Ramello MC, Montes CL. CD39 as a marker of pathogenic CD8+ T cells in cancer and other chronic inflammatory diseases. Oncoscience. 2018;5(3–4):65–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta PK, Godec J, Wolski D, et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 2015;11(10):e1005177. doi: 10.1371/journal.ppat.1005177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simoni Y, Becht E, Fehlings M, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579. doi: 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- 16.Duhen T, Duhen R, Montler R, et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun. 2018;9(1):2724. doi: 10.1038/s41467-018-05072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Saci A, Szabo PM, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9(1):3503. doi: 10.1038/s41467-018-05992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- 24.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allard B, Longhi MS, Robson SC, Stagg J. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol Rev. 2017;276(1):121–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takenaka MC, Robson S, Quintana FJ. Regulation of the T cell response by CD39. Trends Immunol. 2016;37(7):427–439. doi: 10.1016/j.it.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastid J, Cottalorda-Regairaz A, Alberici G, Bonnefoy N, Eliaou JF, Bensussan A. ENTPD1/CD39 is a promising therapeutic target in oncology. Oncogene. 2013;32(14):1743–1751. doi: 10.1038/onc.2012.269 [DOI] [PubMed] [Google Scholar]
- 28.Mandapathil M, Boduc M, Roessler M, Guldner C, Walliczek-Dworschak U, Mandic R. Ectonucleotidase CD39 expression in regional metastases in head and neck cancer. Acta Otolaryngol. 2018;138(4):428–432. doi: 10.1080/00016489.2017.1405278 [DOI] [PubMed] [Google Scholar]
- 29.Lucarelli G, Loizzo D, Franzin R, et al. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagn. 2019;19(5):397–407. doi: 10.1080/14737159.2019.1607729 [DOI] [PubMed] [Google Scholar]
- 30.Bianchi C, Meregalli C, Bombelli S, et al. The glucose and lipid metabolism reprogramming is grade-dependent in clear cell renal cell carcinoma primary cultures and is targetable to modulate cell viability and proliferation. Oncotarget. 2017;8(69):113502–113515. doi: 10.18632/oncotarget.23056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucarelli G, Rutigliano M, Sallustio F, et al. Integrated multi-omics characterization reveals a distinctive metabolic signature and the role of NDUFA4L2 in promoting angiogenesis, chemoresistance, and mitochondrial dysfunction in clear cell renal cell carcinoma. Aging (Albany NY). 2018;10(12):3957–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. [DOI] [PubMed] [Google Scholar]
- 33.Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016;2(6):805–812. doi: 10.1001/jamaoncol.2016.0405 [DOI] [PubMed] [Google Scholar]
- 34.Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology. 2016;5(6):e1163462. doi: 10.1080/2162402X.2016.1163462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roviello G, Corona SP, Nesi G, Mini E. Results from a meta-analysis of immune checkpoint inhibitors in first-line renal cancer patients: does PD-L1 matter? Ther Adv Med Oncol. 2019;11:1758835919861905. doi: 10.1177/1758835919861905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamma R, Rutigliano M, Lucarelli G, et al. Microvascular density, macrophages, and mast cells in human clear cell renal carcinoma with and without bevacizumab treatment. Urol Oncol. 2019;37(6):355 e311–355 e319. doi: 10.1016/j.urolonc.2019.01.025 [DOI] [PubMed] [Google Scholar]
- 37.Lucarelli G, Rutigliano M, Ferro M, et al. Activation of the kynurenine pathway predicts poor outcome in patients with clear cell renal cell carcinoma. Urol Oncol. 2017;35(7):461 e415–461 e427. doi: 10.1016/j.urolonc.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 38.Gigante M, Pontrelli P, Herr W, et al. miR-29b and miR-198 overexpression in CD8+ T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J Transl Med. 2016;14:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Vijayan D, Li XY, et al. The role of NK cells and CD39 in the immunological control of tumor metastases. Oncoimmunology. 2019;8(6):e1593809. doi: 10.1080/2162402X.2019.1593809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takenaka MC, Gabriely G, Rothhammer V, et al. Author correction: control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci. 2019;22(9):1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canale FP, Ramello MC, Nunez N, et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8(+) T cells. Cancer Res. 2018;78(1):115–128. doi: 10.1158/0008-5472.CAN-16-2684 [DOI] [PubMed] [Google Scholar]
- 42.Qi Y, Xia Y, Lin Z, et al. Tumor-infiltrating CD39(+)CD8(+) T cells determine poor prognosis and immune evasion in clear cell renal cell carcinoma patients. Cancer Immunol Immunother. 2020;69(8):1565–1576. doi: 10.1007/s00262-020-02563-2 [DOI] [PMC free article] [PubMed] [Google Scholar]