Abstract
The cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway induces innate immunity by activating the production of inflammatory cytokines and type I interferons. Recently, studies revealed that self-DNA from by-products of chromosome instability and tumors could activate the cGAS-STING pathway, and subsequently promote or inhibit tumor development. However, the prognostic value and correlations with immune infiltrates of the cGAS-STING pathway in hepatocellular carcinoma (HCC) have not been clarified. In the present study, we used the Molecular Signatures Database, Oncomine, UALCAN, Human Protein Atlas, Kaplan–Meier plotter, LinkedOmics, and Tumor Immune Estimation Resource databases. Overexpression of XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and TMEM173 was markedly correlated with clinical stages and pathological grades in HCC. Moreover, higher mRNA expression of XRCC5, XRCC6, and PRKDC was significantly related with shorter overall survival. However, higher mRNA expression of IFI16, STAT6, NLRC3, and TMEM173 was associated with favorable overall survival. Our results suggested that the kinase targets of the cGAS-STING pathway included the SRC family of tyrosine kinases (LCK and LYN), phosphoinositide 3-kinase-related protein kinase (PIKK) family kinases (ATM and ATR), and mitogen-activated protein kinase 1 (MAPK1). Furthermore, we identified significant correlations among the expression of cGAS-STING pathway and infiltration of B cells, CD4+T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells in HCC. The expression of the cGAS-STING pathway also exhibited strong relationships with diverse immune marker sets in HCC. These findings suggest that cGAS-STING pathway members may be used as prognostic biomarkers and immunotherapeutic targets HCC patients.
Keywords: cGAS-STING, hepatocellular carcinoma, prognosis, tumor-infiltrating
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide [1]. The 5-year survival rate of patients with advanced liver cancer is poor due to high tumor recurrence, metastasis, and the lack of early diagnostic biomarkers with high sensitivity and specificity [2]. Owing to poor liver function correlated with cirrhosis and extrahepatic metastasis, most patients with HCC are resistant to common cytotoxic therapies [3]. Although doxorubicin was initially viewed as a first-choice drug for advanced HCC, a controlled trial showed that it was related with a low survival rate [4]. The overall life expectancy of patients with advanced HCC does not exceed 1 year even under treatment with sorafenib or regorafenib [5].Therefore, there is an urgent need to identify novel potential prognostic and therapeutic targets that are related with tumor formation and progression in patients with HCC.
Cyclic GMP-AMP synthase (cGAS) is activated to catalyze the synthesis of cyclic GMP-AMP (cGAMP) on binding to DNA. Moreover, cGAMP acts as a second messenger, which binds to and induces the stimulator of interferon genes (STING) [6,7]. The palmitoylation of STING mediates the recruitment and activation of TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) to produce cytokines, such as type I interferons (IFNs) [8,9]. STING also acts downstream of several other cytosolic DNA sensors, including DEAD-box helicase 41 (DDX41) and interferon gamma inducible protein 16 (IFI16) in the signaling pathway to produce type I IFNs. Knockdown of DDX41 expression inhibited the ability of myeloid dendritic cells (DCs) to mount cytokine and type I IFN responses to DNA, as well as DNA viruses. Lowering the expression of IFI16 by RNA-mediated interference blocked gene induction and activation of IRF3 [10,11]. The cGAS-STING pathway is an important DNA-sensing machinery in innate immunity and viral defense, and critically involved in tumor development [12,13]. Conditional deletion of TBK1 in lung epithelial cells could inhibit tumorigenesis of lung cancer in a mouse model. Besides promoting tumor growth, TBK1 was also involved in tumor-mediated immunosuppression [14]. Signal transducer and activator of transcription 6 (STAT6) is overexpressed in various human cancers, and is a regulator involved in multiple biological processes of cancers. Studies demonstrated that STAT6 silencing could induce apoptosis and growth inhibition in HCC-derived cells [15].
Recently, studies revealed that the prognosis of patients with tumors was correlated with the expression of cGAS-STING pathway members [16,17]. Cancer immunotherapy is an effective treatment against a number of cancers. Thomsen suggested that modulation of the cGAS-STING pathway could affect the tumor progression of HCC, and potentially be used as a treatment in patients with HCC [18]. Through overexpression and RNA interference, Wang et al. demonstrated that cGAS responded to exogenous dsDNA from the DNA damage response, and subsequently triggered the activation of STING/TBK1-mediated innate immunity in chicken liver cancer [19]. XRCC5 variants play a crucial role in determining susceptibility to HCC [20]. A previous study found that aberrant splicing of IRF3 could result in defects in IFN-mediated antiviral defenses in HCC [21]. Research demonstrated that DNA damage repair by XRCC6 could reverse TLR4-deficiency-worsened liver cancer development by recovering immunity to support autophagy and senescence [22]. An in vivo study showed that restored expression of IFI16 could effectively promote tumor regression, which could be partly abrogated by inhibition of the induced inflammasome in HCC [23]. These findings suggest that cGAS-STING pathway members are potential prognostic biomarkers and therapeutic targets and may be associated with immune infiltration in patients with HCC. However, the identification of suitable cGAS-STING pathway members for this purpose remains an important problem that requires urgent attention.
In our study, we searched for cGAS-STING pathway genes present in both the Molecular Signatures Database (MSigDB) and Oncomine database, and in relevant literature [7,13]. We selected 13 key genes of the cGAS-STING pathway members, namely XRCC5, IRF3, TRIM21, IFI16, STAT6, NLRC3, DDX41, TBK1, XRCC6, TREX1, PRKDC, cGAS, and STING (also termed TMEM173). The genes regulated by the cGAS and STING signaling axis include TBK1, IRF3, and STAT6. Moreover, the genes which regulate the pathway include TREX1, DDX41, IFI16, and NLRC3.These signature genes of the cGAS-STING pathway are linked to type I IFN response. First, we compared the expression of cGAS-STING pathway members in HCC tissues and normal tissues via Oncomine, UALCAN, and the Human Protein Atlas. We subsequently analyzed the correlations between these key genes and prognosis in patients with liver cancer using the Kaplan–Meier (K-M) plotter. In addition, we also investigated potential kinase targets of the cGAS-STING pathway members in patients with HCC using LinkedOmics. To further investigate potential immune therapeutic targets, we analyzed the correlations among the expression of cGAS-STING pathway members and immune cell infiltration and diverse immune marker sets in HCC microenvironments obtained from the Tumor Immune Estimation Resource (TIMER) database.
Materials and methods
MSigDB analysis
The comprehensive database MSigDB (www.gsea-msigdb.org/gsea/msigdb/) includes >10,000 gene sets and is widely used to perform gene set enrichment analysis [24]. In the present study, we searched this database to obtain cGAS-STING pathway genes.
Oncomine database analysis
Oncomine (www.oncomine.org) is a cancer microarray database, including 715 datasets and 86,733 samples for DNA or RNA sequence analysis [25]. Oncomine can be used to analyze differences in gene expression between tumor tissues and normal tissues. In the present study, we noted the transcriptional expression of cGAS-STING pathway genes between 20 different cancer samples and their normal adjacent tissues from the Oncomine database. The threshold was determined based on the following values: P=0.01, fold change = 1.5, gene rank = 10%, and data type for mRNA.
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu) is an interactive web server for the analysis of 31 types of cancer with transcriptome data from The Cancer Genome Atlas (TCGA). This tool is based on clinical data and level 3 RNA-sequence. UALCAN can be used to assess the association of transcriptional expression with relative clinicopathologic features [26]. In our study, UALCAN was utilized to analyze the mRNA expression of cGAS-STING pathway genes in HCC tissues, and estimate their association with clinicopathologic features. A P<0.05 denoted statistically significant difference.
Human protein atlas
The Human Protein Atlas (https://www.proteinatlas.org) is an online site containing immunohistochemistry-based expression data for approximately 20 common types of tumors. It is used to map the human proteins in organs, tissues, and cells through integration of different omics technologies, such as mass spectrometry-based proteomics and transcriptomics [27]. In the present study, the protein expression of different cGAS-STING pathway genes between human normal and HCC tissues was compared using immunohistochemistry images.
K-M plotter database analysis
The K-M plotter (www.kmplot.com) is used to estimate the effect of 54,000 genes for prognostic analysis in 21 common types of cancer. The database contains gene chip and RNA-sequence data-sources obtained from databases, such as the Gene Expression Omnibus [28,29]. We evaluated the prognostic value of the mRNA expression of cGAS-STING pathway genes in liver cancer via the K-M plotter database. We divided patients according to the median and accepted other default settings prior to the analysis. The hazard ratio (HR) with 95% confidence intervals and log-rank P-values was computed. A P<0.05 denoted statistically significant differences.
LinkedOmics
LinkedOmics (http://www.linkedomics.org/) is a publicly available portal containing all 32 types of cancer included in TCGA. It serves as a unique tool for biologists and clinicians to access, compare, and analyze cancer multi-omics data [30]. In this study, we determined the kinase target enrichment of cGAS-STING pathway genes using the “LinkInterpreter” module. We conducted analyses with a number of size of 3 and simulations of 500 in LIHC dataset. We considered 0.05 as the P-value cutoff and used the Spearman correlation test to analyze the results statistically.
TIMER database analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a reliable database for estimating immune cell infiltration using data from TCGA, including 10,897 samples from 32 types of cancer [31]. In the present study, we initially analyzed the expression of cGAS-STING pathway genes in HCC. Subsequently, we examined the correlation between the expression of cGAS-STING pathway members and immune cell infiltrates, including B cells, CD8+ T cells, CD4+ T cells, macrophages, DCs, and neutrophils with gene modules. Highly expressed genes in the tumor microenvironment tend to have negative correlations with tumor purity [32]. The generated scatterplots suggest statistical significance and provide the purity-corrected partial Spearman’s rho value. Further, correlations between the expression of cGAS-STING pathway members and gene markers of immune cells were further evaluated with correlation modules. The cGAS-STING pathway genes were represented on the X-axis, and related marker genes were used for the y-axis. Options for partial correlation under the condition of tumor purity were conducted.
Statistical analysis
The results generated from Oncomine were displayed with fold changes, P-values, and t-test results. Survival curves were generated via K-M plots. The results of KM plots were presented by P-values and HR obtained from a log-rank test. A P<0.05 denoted statistically significant differences.
Results
mRNA expression levels of cGAS-STING pathway members in HCC
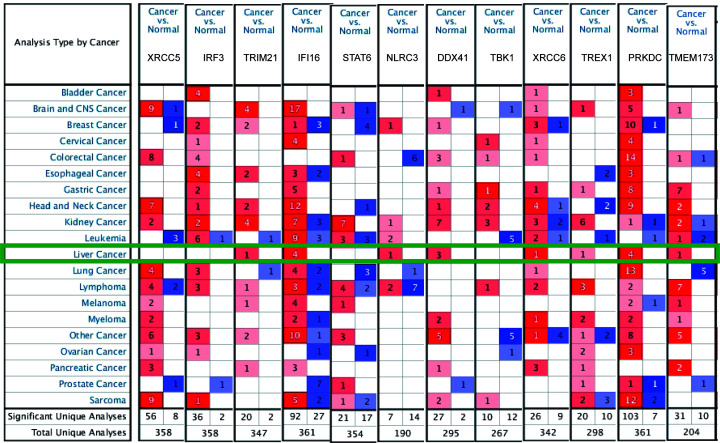
We analyzed the differences in the transcriptional levels of cGAS-STING pathway members in patients with HCC using the Oncomine database. The results revealed that the mRNA expression of TRIM21, IFI16, NLRC3, DDX41, XRCC6, TREX1, PRKDC, and TMEM173 was significantly higher in HCC tissues in multiple datasets (Figure 1 and Table 1). In the Wurmbach Liver dataset [33], higher expression of TRIM21 was revealed in HCC tissues compared with normal adjacent tissues (fold change = 1.607 and P = 4.78E-04). Up-regulation of IFI16 was also found in HCC tissues compared with normal adjacent tissues. The data from the Mas Liver dataset [34] showed increase of 3.474-fold (P = 4.23E-14) and 4.715-fold (P = 1.91E-16), respectively. Wurmbach [33] observed a 3.649-fold (P = 1.02E-06) and 2.250-fold (P = 4.06E-04) increase, respectively, in IFI16 mRNA expression in HCC samples. Wurmbach [33] also found a 2.197-fold (P = 1.35E-05) increase in NLRC3 mRNA expression in HCC tissues. Similarly, in the Roessler Liver dataset [35], 2.012-fold (P = 7.60E-8) and 1.701-fold (P = 2.95E-41) increases in DDX41 mRNA expression were found in HCC samples compared with normal tissues. Wurmbach [33] observed a 1.719-fold (P = 5.25E-7) increase in DDX41 mRNA expression in HCC samples. In the Roessler Liver 2 dataset [35], high expression of XRCC6 was observed in liver cancer tissues compared with normal adjacent tissues (fold change = 1.828 and P = 1.47E-70, respectively). In addition, TREX1 showed higher expression levels in HCC tissues versus normal tissues in Wurmbach Liver (fold change = 1.552 and P=0.002) [33]. Similarly, results from three datasets suggested that PRKDC expression was higher in HCC tissue versus normal adjacent tissue [33,35,36]. The mRNA expression of cGAS was not found on Oncomine.
Figure 1. Transcriptional expression of 12 cGAS-STING pathway members in 20 different types of cancer (Oncomine database).
Differences in transcriptional expression were compared using Student’s t-test. The cutoff criteria were as follows: P=0.01, fold change = 1.5, gene rank = 10%, and data type of mRNA.
Table 1. Significant changes in the transcription levels of cGAS-STING pathway members between HCC and normal liver tissues (Oncomine).
| Types of HCC versus liver | Fold change | P value | t-test | References | |
|---|---|---|---|---|---|
| TRIM21 | Hepatocellular carcinoma | 1.607 | 4.78E-04 | 3.847 | Wurmbach Liver [34] |
| IFI16 | Hepatocellular carcinoma | 3.474 | 4.23E-14 | 10.895 | Mas Liver [35] |
| Hepatocellular carcinoma | 4.715 | 1.91E-16 | 15.089 | Mas Liver [35] | |
| Hepatocellular carcinoma | 3.649 | 1.02E-06 | 7.012 | Wurmbach Liver [34] | |
| Hepatocellular carcinoma | 2.250 | 4.06E-04 | 3.861 | Wurmbach Liver [34] | |
| NLRC3 | Hepatocellular carcinoma | 2.197 | 1.35E-05 | 5.560 | Wurmbach Liver [34] |
| DDX41 | Hepatocellular carcinoma | 2.012 | 7.60E-8 | 6.619 | Roessler Liver [36] |
| Hepatocellular carcinoma | 1.719 | 5.25E-7 | 6.074 | Wurmbach Liver [34] | |
| Hepatocellular carcinoma | 1.701 | 2.95E-41 | 15.050 | Roessler Liver 2 [36] | |
| XRCC6 | Hepatocellular carcinoma | 1.828 | 1.47E-70 | 21.477 | Roessler Liver 2 [36] |
| TREX1 | Hepatocellular carcinoma | 1.552 | 0.002 | 3.172 | Wurmbach Liver [34] |
| PRKDC | Hepatocellular carcinoma | 2.792 | 4.91E-72 | 23.447 | Roessler Liver 2 [36] |
| Hepatocellular carcinoma | 3.011 | 9.67E-9 | 7.102 | Wurmbach Liver [34] | |
| Hepatocellular carcinoma | 1.651 | 8.50E-15 | 8.386 | Chen Liver [37] | |
| Hepatocellular carcinoma | 2.439 | 6.57E-8 | 7.031 | Roessler Liver [36] |
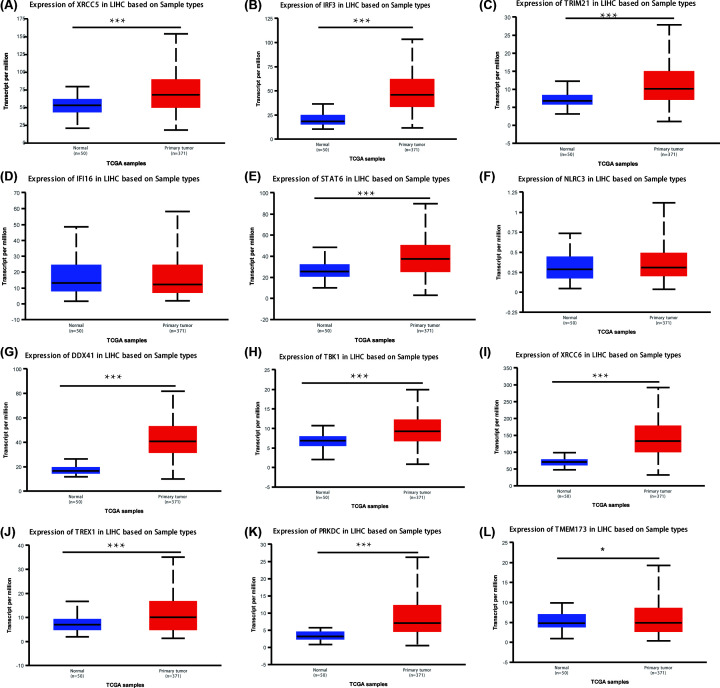
We also investigated the mRNA expression levels of cGAS-STING pathway members in HCC tissue with UALCAN. As shown in Figure 2, the transcriptional levels of XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC (all P<0.001) and TMEM173 (P<0.05) in HCC tissues were significantly upregulated (Figure 2A–C,E,G–L). However, the transcriptional levels of IFI16 (P = 7.04E-02) and NLRC3 (P = 9.95E-02) did not show significant differences (Figure 2D,F). The mRNA expression of cGAS was not found on UALCAN.
Figure 2. mRNA expression of different cGAS-STING pathway members in HCC samples and adjacent normal liver samples (UALCAN).
The mRNA expression of XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and TMEM173 was found to be up-regulated in HCC tissues versus normal tissues (A–C,E,G–L). The transcriptional levels of IFI16 and NLRC3 did not show significant differences (D and F); ***P<0.001.
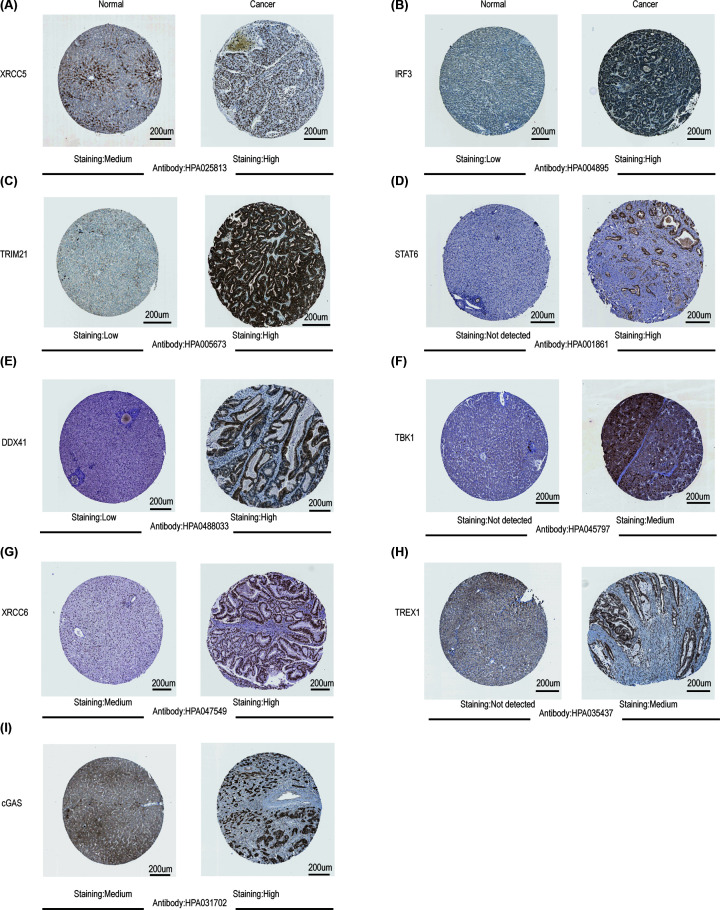
After confirming the up-regulation of mRNA expression levels of cGAS-STING pathway members in HCC tissues, we subsequently assessed their protein expression levels based on the Human Protein Atlas website. As shown in Figure 3, STAT6, TBK1, and TREX1 proteins were not expressed in normal liver tissues, whereas they exhibited medium or high expressions in HCC samples (Figure 3D,F,H). In addition, low or medium protein expression of XRCC5, IRF3, TRIM21, DDX41, XRCC6, and cGAS was observed in normal liver tissues, while high protein expression was detected in HCC tissues (Figure 3A–C,E,G,I). However, the immunohistochemical images of IFI16, NLRC3, PRKDC, and TMEM173 expression were not found in this website. In general, these results revealed that the transcription and protein levels of most cGAS-STING pathway members were upregulated in patients with HCC.
Figure 3. Representative immunohistochemistry images of different cGAS-STING pathway members in HCC tissues and normal liver tissues (Human Protein Atlas).
STAT6, TBK1, and TREX1 proteins were not expressed in normal liver tissues, whereas they exhibited medium or high expression in HCC samples (D,F,H). In addition, low or medium protein expression of XRCC5, IRF3, TRIM21, DDX41, XRCC6, and cGAS was observed in normal liver tissues; high expression of these proteins was noted in HCC tissues (A–C,E,G,I).
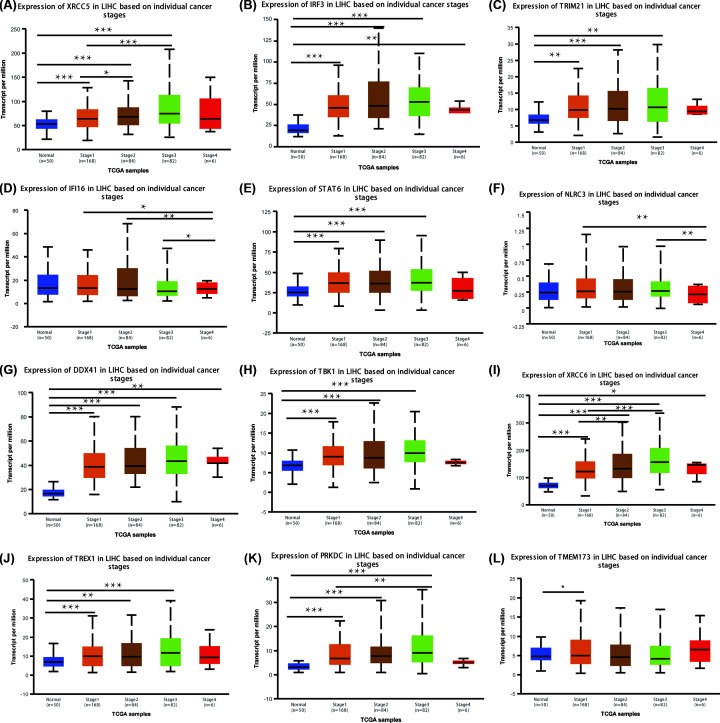
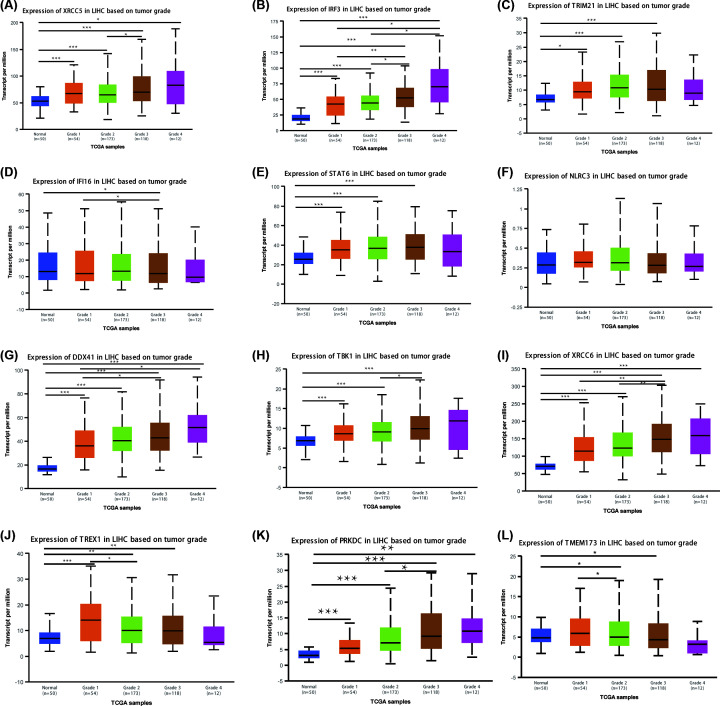
We next analyzed the correlation between the mRNA expression of cGAS-STING pathway members and the pathological stage of patients with liver cancer according to UALCAN. The results are presented in Figures 4 and 5. The mRNA expression of cGAS-STING pathway members, except for IFI16 and NLRC3, was significantly related with individual cancer stages and tumor grades in HCC. Furthermore, patients in advanced stages and tumor grades of HCC had higher mRNA expression levels of cGAS-STING pathway members compared with those in early stages of the disease. In Figure 4, the highest mRNA expression of DDX41 was found in stage 4 (Figure 4G), while the highest mRNA expression of XRCC5, IRF3, TRIM21, STAT6, TBK1, XRCC6, TREX1, and PRKDC was found in stage 3 (Figure 4A–C,E,H–K), which may be attributed to the small sample numbers (only six patients with stage 4 HCC). Figure 4L shows that the highest mRNA expression of TMEM173 was found in stage 1. Similarly, the highest mRNA expression of XRCC5, IRF3, DDX41, TBK1, XRCC6, and PRKDC was found in grade 4 tumors (Figure 5A,B,G–I,K), while the highest mRNA expression of TRIM21, STAT6, and TMEM173 was found in grade 3 (Figure 5C,E,L). However, the highest mRNA expression of IFI16 and TREX1 was found in grade 1 or 2; as the tumor grade increased, their mRNA expression tended to be lower (Figure 5D,J). These results suggested that XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and TMEM173 were strongly correlated with tumorigenesis and tumor progression in patients with HCC.
Figure 4. Relationship between the mRNA expression of different cGAS-STING pathway members and individual cancer stages in patients with HCC.
The mRNA expression of 10 cGAS-STING pathway members was strongly related with the cancer stages of individual patients. Patients who were in more advanced stages tended to express higher mRNA levels of cGAS-STING pathway members. The highest mRNA expression of DDX41 was found in stage 4 (G), while the highest mRNA expression of XRCC5, IRF3, TRIM21, STAT6, TBK1, XRCC6, TREX1, and PRKDC was found in stage 3 (A–C,E,H–K). The highest mRNA expression of TMEM173 was found in stage 1 (L). However, the mRNA expression of IFI16 and NLRC3 did not show a correlation with tumor grades in patients with liver cancer (D,F); *P<0.05, **P<0.01, ***P<0.001.
Figure 5. Association of mRNA expression of different cGAS-STING pathway members with tumor grades in patients with HCC.
The mRNA expression of 10 cGAS-STING pathway members was significantly related with tumor grades; as the tumor grade increased, the mRNA expression of cGAS-STING pathway members tended to increase in parallel. The highest mRNA expression of XRCC5, IRF3, DDX41, TBK1, XRCC6, and PRKDC was found in grade 4 tumors (A,B,G–I,K), while the highest mRNA expression of TRIM21, STAT6, and TMEM173 was found in grade 3 tumors (C,E,L). However, the highest mRNA expression of IFI16 and TREX1 was found in grade 1 or 2; as the tumor grade increased, the mRNA expression of IFI16 and TREX1 tended to decrease (D,J). NLRC3 mRNA expression did not show a correlation with tumor grades in patients with liver cancer (F); *P<0.05, **P<0.01, ***P<0.001.
Prognostic value of mRNA expression of cGAS-STING pathway members among patients with HCC
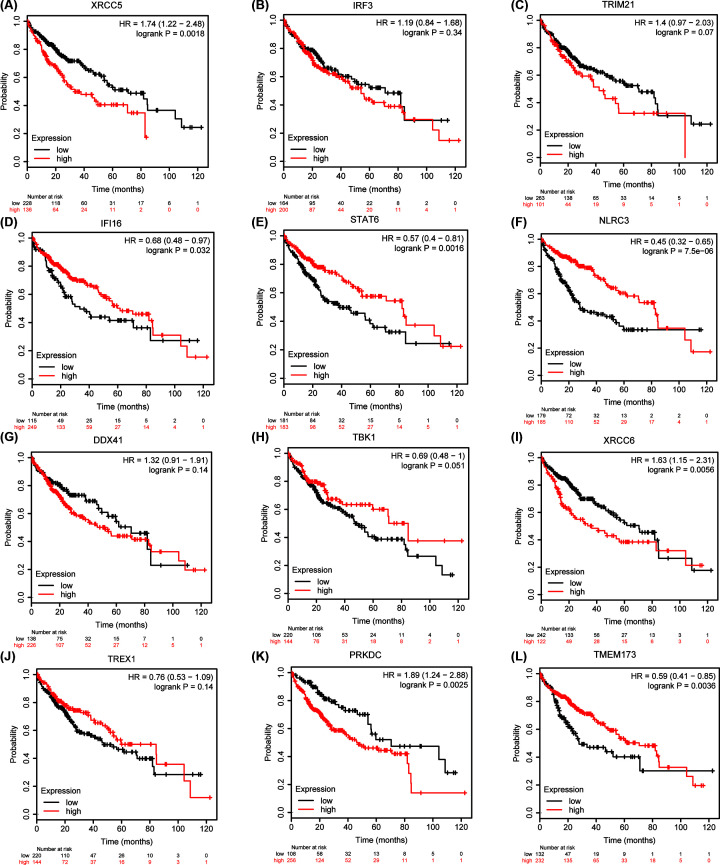
We further analyzed the prognostic potential of mRNA expression of cGAS-STING pathway members in patients with HCC using the K-M plotter. Overall survival (OS) curves are illustrated in Figure 6. Higher mRNA expression of XRCC5 (HR: 1.74, 95% confidence interval [CI]: 1.22-2.48, P=0.0018), XRCC6 (HR: 1.63, 95% CI: 1.15–2.31, P=0.0056), and PRKDC (HR: 1.89, 95% CI: 1.24–2.88, P=0.0025) were related with shorter OS (Figure 6A,I,K). In contrast, higher mRNA expression of IFI16 (HR: 0.68, 95% CI: 0.48–0.97, P=0.032), STAT6 (HR: 0.57, 95% CI: 0.4–0.81, P=0.0016), NLRC3 (HR: 0.45, 95% CI: 0.32–0.65, P = 7.5e-06), and TMEM173 (HR: 0.59, 95% CI: 0.41–0.85, P=0.0036) was correlated with favorable OS (Figure 6D–F,L). However, the mRNA expression of IRF3 (P=0.34), TRIM21 (P=0.07), DDX41 (P=0.14), TBK1(P=0.051), and TREX1 (P=0.14) did not show a correlation with prognosis in patients with HCC (Figure 6B,C,G,H,J). These results suggested that the mRNA expression of XRCC5, XRCC6, PRKDC, IFI16, STAT6, NLRC3, and TMEM173 was associated with the prognosis of patients with HCC. The prognostic data of cGAS were not found in the K-M plotter.
Figure 6. Prognostic value of mRNA expression of different cGAS-STING pathway members in patients with HCC (Kaplan–Meier plotter).
Higher mRNA expression of XRCC5, XRCC6, and PRKDC was significantly associated with shorter OS in patients with liver cancer (A,I,K). In contrast, higher mRNA expression of IFI16, STAT6, NLRC3, and TMEM173 was significantly related with favorable OS in patients with liver cancer (D–F,L). However, IRF3, TRIM21, DDX41, TBK1, and TREX1 mRNA expression did not show a correlation with prognosis in patients with HCC (B,C,G,H,J).
Kinase targets of cGAS-STING pathway members in HCC
Based on the significantly different expression of cGAS-STING pathway members in HCC tissues versus normal tissues, we estimated possible kinase targets of the differentially expressed cGAS-STING pathway members. In the present study, we investigated the top two kinase targets of cGAS-STING pathway members via the LinkedOmics database. As shown in Table 2, PLK1 and ATR were the top two kinase targets of XRCC5. DYRK1A and EGFR were noted as the targets for the IRF3 kinase-target network. LYN and LCK were mainly correlated with TRIM21. Constituents of the IFI16 kinase-target network were primarily correlated with SYK and LCK. ATR and PLK1, and LCK and SYK were the top two kinase targets of STAT6 and NLRC3, respectively. Mitogen-activated protein kinase 1 (MAPK1) and DYRK1A were regarded as the kinase targets of DDX41. ATM and DAPK1 were mainly related with TBK1. AURKB and PLK1 were shown as kinase targets of XRCC6. ATM and CDK2, as well as ATM and CDK1, were the top two targets of TREX1 and PRKDC, respectively. BUB1 and HCK were regarded as the kinase targets of TMEM173.The kinase targets of cGAS were not found in the LinkedOmics database.
Table 2. The kinase target networks of cGAS-STING pathway members in HCC (LinkedOmics).
| cGAS-STING pathway | Enriched kinase target | Description | Leading EdgeNum | P value |
|---|---|---|---|---|
| XRCC5 | Kinase_PLK1 | Polo-like kinase 1 | 51 | 0 |
| Kinase_ATR | ATR serine/threonine kinase | 32 | 0 | |
| IRF3 | Kinase_DYRK1A | Dual specificity tyrosine phosphorylation regulated kinase 1A | 7 | 0.004 |
| Kinase_EGFR | Epidermal growth factor receptor | 17 | 0.008 | |
| TRIM21 | Kinase_LYN | LYN proto-oncogene, Src family tyrosine kinase | 22 | 0 |
| Kinase_LCK | LCK proto-oncogene, Src family tyrosine kinase | 31 | 0 | |
| IFI16 | Kinase_SYK | Spleen associated tyrosine kinase | 21 | 0 |
| Kinase_LCK | LCK proto-oncogene, Src family tyrosine kinase | 25 | 0 | |
| STAT6 | Kinase_ATR | ATR serine/threonine kinase | 31 | 0 |
| Kinase_PLK1 | Polo-like kinase 1 | 32 | 0 | |
| NLRC3 | Kinase_LCK | LCK proto-oncogene, Src family tyrosine kinase | 24 | 0 |
| Kinase_SYK | Spleen associated tyrosine kinase | 20 | 0 | |
| DDX41 | Kinase_MAPK1 | Mitogen-activated protein kinase 1 | 60 | 0 |
| Kinase_DYRK1A | Dual specificity tyrosine phosphorylation regulated kinase 1A | 6 | 0.011 | |
| TBK1 | Kinase_ATM | ATM serine/threonine kinase | 56 | 0 |
| Kinase_DAPK1 | Death-associated protein kinase 1 | 6 | 0 | |
| XRCC6 | Kinase_AURKB | Aurora kinase B | 41 | 0 |
| Kinase_PLK1 | Polo-like kinase 1 | 34 | 0 | |
| TREX1 | Kinase_ATM | ATM serine/threonine kinase | 69 | 0 |
| Kinase_CDK2 | Cyclin-dependent kinase 2 | 124 | 0 | |
| PRKDC | Kinase_ATM | ATM serine/threonine kinase | 63 | 0 |
| Kinase_CDK1 | Cyclin-dependent kinase 1 | 125 | 0 | |
| TMEM173 | Kinase_BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | 4 | 0.029 |
| Kinase_HCK | HCK proto-oncogene, Src family tyrosine kinase | 10 | 0.028 |
Immune cell infiltration of cGAS-STING pathway members in HCC
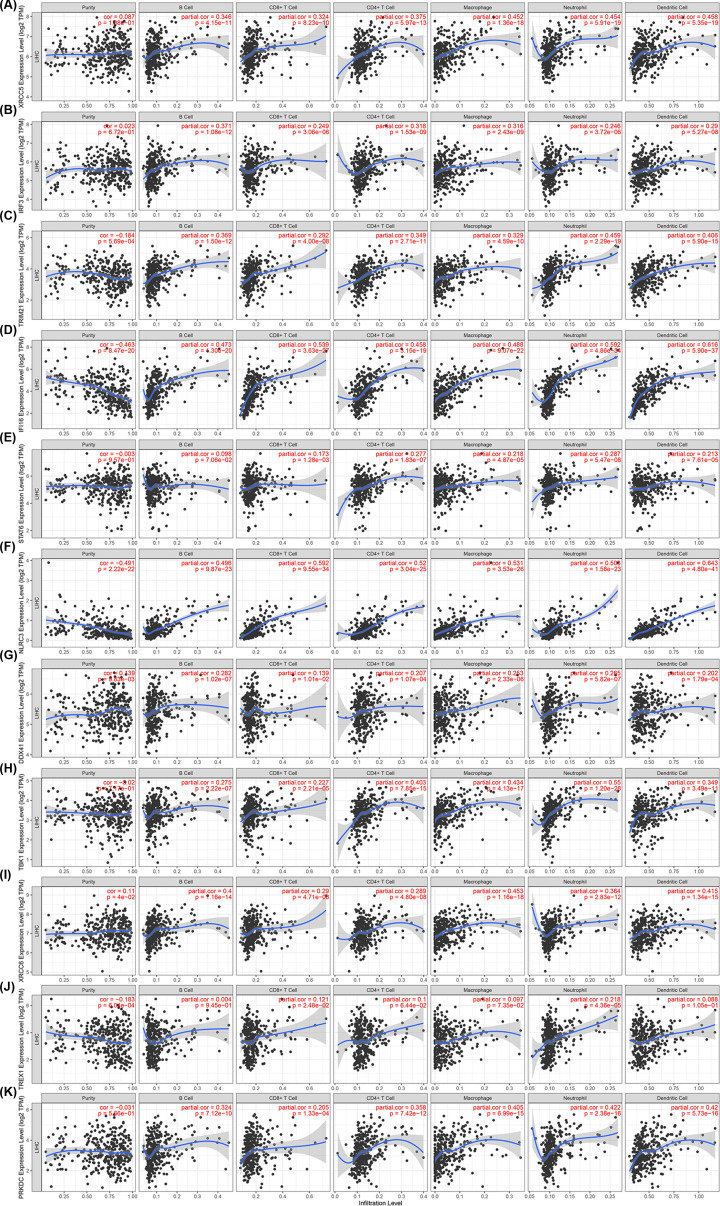
Tumor-infiltrating lymphocytes can be used as independent predictors for sentinel lymph node status and survival in cancer [37]. Therefore, we investigated the correlations between differentially expressed cGAS-STING pathway members and the infiltration of immune cells using the TIMER database. As shown in Figure 7, there was a positive relationship between the expression of XRCC5, IRF3, TRIM21, IFI16, NLRC3, DDX41, TBK1, XRCC6, and PRKDC, and the infiltration of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and DCs (Figure 7A–D,F–I,K). STAT6 expression (Figure 7E) was positively associated with the infiltration of CD8+ T cells (correlation: 0.173, P = 1.28e−03), macrophages (correlation: 0.218, P = 4.87e−05), neutrophils (correlation: 0.287, P = 5.47e−08), CD4+ T cells (correlation: 0.277, P = 1.83e−07), and DCs (correlation: 0.213, P = 7.61e−05). Similarly, the expression of TREX1 (Figure 7J) was correlated with the infiltration of CD8+ T cells (correlation: 0.121, P = 2.48e−02), and neutrophils (correlation: 0.218, P = 4.36e−05). These results strongly suggested that, in patients with liver cancer, the cGAS-STING pathway may act a specific role for immune cell infiltration, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs.
Figure 7. Correlation between differentially expressed cGAS-STING pathway members and immune cell infiltration (TIMER).
Relationship between the abundance of immune cells and expression of (A) XRCC5, (B) IRF3, (C) TRIM21, (D) IFI16, (E) STAT6, (F) NLRC3, (G) DDX41, (H) TBK1, (I) XRCC6, (J) TREX1, and (K) PRKDC in HCC.
Correlation analysis between the expression of the cGAS-STING pathway and immune marker sets
We investigated the relationships between cGAS-STING pathway members and different marker genes of immune cells of HCC using the TIMER database. The immune marker sets of various immune cells, such as monocytes, M1 and M2 macrophages, tumor-associated macrophages (TAMs), and DCs were analyzed. In addition, we investigated various functional T cells, such as T helper 1 (Th1), Th2, and regulatory T (Treg) cells. The results are presented in Supplementary Tables S1–3. After correlation adjustment by purity, we found that the expression levels of cGAS-STING pathway members were significantly related with most immune marker sets of different immune cells and various functional T cells in liver cancer.
Importantly, the expression of immune marker sets of TAMs, monocytes, M1 and M2 macrophages was associated with the expression levels of most cGAS-STING pathway members, including XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC (Supplementary Tables S1–3).
Furthermore, the results also suggested that the expression of most marker sets of DCs, such as HLA-DPB1, CD1C, NRP1, and ITGAX had strong correlations with XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC expression in HCC (Supplementary Tables S1–3). These results suggested that there was a strong correlation between cGAS-STING pathway members and DC infiltration. In addition, there was a significant connection between FOXP3 and TGFβ1 for Treg cells and cGAS-STING pathway members in HCC.
Significant correlations between cGAS-STING pathway members, (e.g., XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC) and marker gene sets of Treg and T cell exhaustion (e.g., FOXP3, STAT5B, CCR8, PDCD1, TGFβ1, LAG3, CTLA4, and HAVCR2) are also presented in Supplementary Tables S1–3. The cGAS-STING pathway members exhibited significant correlations with immune infiltrating cells in HCC, and may play an important role in immune escape in the liver cancer microenvironment.
Discussion
The cGAS-STING pathway has emerged as a potential mechanism to induce inflammation-mediated tumorigenesis. Actually, persistent activation of this pathway and its downstream effectors, such as TBK1, has been connected with chronic inflammation and cancer progression [38,39]. The development of HCC is a multistep process that involves continuous inflammatory damage, such as hepatocyte necrosis [40]. Some studies have observed correlations between cGAS-STING pathway members, the tumor microenvironment, and cancer immunotherapy. However, the prognostic value and potential therapeutic targets of the cGAS-STING pathway in HCC are poorly characterized. In the present study, we investigated the expression, prognostic values, and correlations with immune infiltrates of different cGAS-STING pathway members in HCC.
We first investigated the expression of cGAS-STING pathway members and its correlations with the pathological stage in HCC. Based on the Oncomine database, the mRNA expression of TRIM21, IFI16, NLRC3, DDX41, XRCC6, TREX1, PRKDC, and TMEM173 was significantly higher in HCC tissues in multiple datasets. Moreover, in patients with HCC, high expression of mRNA and protein was found in cGAS-STING pathway member genes, including XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and TMEM173. Furthermore, the mRNA expression of these 10 genes was strongly associated with cancer stages and tumor grades in patients with HCC. These data demonstrate that differentially expressed cGAS-STING pathway members may play a significant role in HCC. Recently, high expression of XRCC5 has been found in breast and gastric cancer [17,41]. A strong correlation was found between the variable number tandem repeat polymorphism in the XRCC5 gene and the risk of breast cancer [17]. Overexpression of XRCC5 was also detected in gastric cancer, in which XRCC5 regulated the overexpression of chloride channel 3 (CLC3) [41]. Additionally, Liu [42] found that higher expression of XRCC5 was related with metastasis through the Wnt/β-catenin signaling pathway in patients with HCC, which was consistent with our results. In conclusion, XRCC5 may participate in the tumorigenesis of HCC.
Recently, Shi found that pharmacological targeting or knockdown of IRF3 using amlexanox. This drug is used for anti-inflammatory treatment, and can inhibit gastric tumor growth in a Yes-associated protein-dependent manner. The expression of IRF3 is up-regulated and prognosticates patient survival in gastric cancer [43]. A study showed that the expression of TBK1 was increased in mesenchymal small cell lung cancer cell lines [44]. Research found that TRIM21 modulated epithelial–mesenchymal transition (EMT) by regulating the stability of Snail in breast cancer [45]. To our knowledge, this was the first study to investigate the correlation among IRF3, TBK1, and TRIM21 with HCC. Our results suggested that all three may participate in the tumorigenesis of HCC.
The potential prognostic value of the mRNA expression levels of cGAS-STING pathway members in patients with HCC was subsequently investigated. The findings revealed that higher mRNA expression of XRCC5, XRCC6, and PRKDC was significantly associated with shorter OS. In contrast, higher mRNA expression of IFI16, STAT6, and NLRC3 was significantly related with better OS. Liu [42] observed that the high mRNA expression of XRCC5 predicted poor prognosis in patients with HCC, which was in accordance with our results. Studies have shown that NLRC3 was a negative regulator in innate immune signaling activated by STING. NLRC3 could inhibit STING–TBK1 interaction and the production of downstream type I IFN [46]. IFI16 enhanced STING activation by interacting with STING via the PYRIN domain. Moreover, the IFI16-induced inflammasome was found to inhibit HCC growth and metastasis, and was a tumor suppressor during the development [23]. In our study, we found that NLRC3 and IFI16 were independent prognostic factors for favorable OS in patients with liver cancer.
We also investigated the kinase targets of the cGAS-STING pathway members. We discovered that the SRC family of tyrosine kinases (LCK and LYN), phosphoinositide 3-kinase-related protein kinase (PIKK) family kinases (ATM and ATR), and MAPK1 were potential kinase targets of the cGAS-STING pathway members. These kinase targets affect the progression of cell cycle, DNA damage, and EMT [47–51]. Furthermore, they are involved in tumor progression by mediating tumor cell invasion, apoptosis, and migration [51–53]. As a result, differentially expressed cGAS-STING pathway members may modulate DNA repair, the EMT, and cell cycle progression by regulating these kinases in patients with HCC.
Another important finding in the present study is that the expression of cGAS-STING pathway members is associated with various immune cell infiltration levels in liver cancer. The cGAS-STING pathway can mediate protective immune defense toward infection with a great number of DNA-containing pathogens, as well as generate intrinsic antitumor immunity [54]. The accumulation of tumor DNA could activate STING-IRF3-induced IFN signaling to enforce tumor-antigen presentation on DCs and cross-prime CD8+ T cells for antitumor immunity [55]. There is an increasing body of evidence supporting that immune cell infiltration could affect cancer recurrence and progression, and also play an important role in clinical outcome and response to immunotherapy [56,57]. CD4+ T cells may be involved in the recognition of tumor antigens, and the activation of M1 macrophages may mediate the inhibition of tumor growth [58]. Our results demonstrate that there is a significant correlation between the expression of cGAS-STING pathway members and the infiltration of immune cells, such as T cells, macrophages, and DCs. This implies that cGAS-STING pathway members may act as potential prognostic indicators, as well as reflect the immune status. STING activation in hepatic macrophages could induce the production of proinflammatory cytokines, leading to nonalcoholic steatohepatitis that is characterized by hepatic steatosis [59].
Moreover, the correlations between the expression of cGAS-STING pathway members and the marker sets of immune cells suggest the role of cGAS-STING pathway members in regulating tumor immunology in HCC. Firstly, the M1 macrophage markers (e.g., IRF5 and PTGS2) and the marker genes of M2 macrophages (e.g., VSIG4, and MS4A4A) were significantly associated with the expression of XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC. Collectively, the results suggest the regulatory role of these cGAS-STING pathway genes in the polarization of TAMs. Moreover, our results reveal a strong relationship between the expression of XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC and DC infiltration. These genes also play a potential role in activating Treg cells and inducing T-cell exhaustion. DCs promote tumor metastasis by reducing the cytotoxicity of CD8+ T cells and increasing Treg cells [60]. Further studies are warranted to investigate whether these genes are important factors in inducing the DCs and tumor metastasis.
In addition, the increase in the expression of XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC was significantly related with the expression of Treg and T-cell exhaustion gene markers, such as FOXP3, STAT5B, TGFβ1, PDCD1, CTLA4, HAVCR2, and LAG3 in patients with HCC. HAVCR2 is an important surface protein from exhausted T cells [61]. In human squamous cell carcinomas, STING signaling abrogated tumor immunogenicity by recruiting Treg cells [62]. This is highly correlated with the expression of XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC in HCC. Furthermore, significant associations were also found between the expression of XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC and the regulation of some marker genes of T helper cells (e.g., Th1, Th2, and Th17). These associations suggest the potential mechanism through which these cGAS-STING pathway members may regulate T-cell functions in HCC. Collectively, the results suggest that cGAS-STING pathway members (e.g., XRCC5, TRIM21, IFI16, STAT6, NLRC3, TBK1, XRCC6, and PRKDC) may play a crucial role in the regulation and recruitment of immune infiltrating cells in HCC.
The present study had some limitations. First, all the data in the present study were obtained from online databases; hence, further studies involving larger sample sizes, and in vitro and in vivo experiments are warranted to confirm our results. Second, analysis based on the transcriptional levels may indicate a few aspects of the immune status; however, its ability to detect global changes is limited. Finally, we did not analyze the potential mechanisms of cGAS-STING pathway members in HCC. Future studies should be performed to investigate the detailed mechanism among cGAS-STING pathway members and HCC.
Conclusions
XRCC5, IRF3, TRIM21, STAT6, DDX41, TBK1, XRCC6, TREX1, PRKDC, and TMEM173 were found to be significantly positive correlated with clinical cancer stages and tumor grades in patients with liver cancer. In addition, high mRNA expression of XRCC5, XRCC6, and PRKDC was significantly related with poor OS. In contrast, high mRNA expression of IFI16, STAT6, NLRC3, and TMEM173 was strongly correlated with favorable OS in HCC. Increased expression levels of cGAS-STING pathway members are correlated with increased infiltration levels of immune cells, including B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages, and DCs in HCC. Therefore, cGAS-STING pathway members, especially XRCC5, IFI16, STAT6, NLRC3, XRCC6, and PRKDC, are likely involved in immune infiltration and can be used as potential prognostic biomarkers for patients with HCC. We hope our results provide a new perspective for the design of new immunotherapeutic drugs against HCC.
Supplementary Material
Abbreviations
- cGAMP
cyclic GMP-AMP
- cGAS
cyclic GMP-AMP synthase
- DC
dendritic cell
- DDX41
DEAD-box helicase 41
- HCC
hepatocellular carcinoma
- IFI16
interferon gamma inducible protein 16
- IRF3
interferon regulatory factor 3
- MAPK1
mitogen-activated protein kinase 1
- PIKK
phosphoinositide 3-kinase-related protein kinase
- STAT6
signal transducer and activator of transcription-6
- STING
stimulator of interferon response cGAMP interactor
- TBK1
TANK-binding kinase 1
- TMEM173
transmembrane protein 173
Contributor Information
Bingtian Bi, Email: bibt@sysucc.org.cn.
Jingdun Xie, Email: xiejd6@mail.sysu.edu.cn.
Data Availability
The data that support the findings of our study are openly available from Molecular Signatures Database (MSigDB), Oncomine database, UALCAN, Human Protein Atlas, Kaplan-Meier plotter, LinkedOmics, and Tumor Immune Estimation Resource (TIMER) database (https://www.gsea-msigdb.org/gsea/msigdb/, www.oncomine.org, http://ualcan.path.uab.edu, https://www.proteinatlas.org, www.kmplot.com, http://www.linkedomics.org/, https://cistrome.shinyapps.io/timer/).
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China [grant number 2018YFC2001901]; National Natural Science Foundation of China [grant number 81701074]; and Research Foundation of Guangdong Medical Science and Technology [grant number A2018236].
Author Contribution
Zhenhua Qi designed the project. Zhenhua Qi and Fang Yan wrote the original manuscript and conducted the formal analysis. Dongtai Chen and Wei Xing contributed to the bioinformatics analysis. Qiang Li and Weian Zeng reviewed the manuscript. Bingtian Bi and Jingdun Xie contributed to data curation and acquisition of funding.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Greten T.F., Wang X.W. and Korangy F. (2015) Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut 64, 842–848 10.1136/gutjnl-2014-307990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assenat E., Pageaux G.-P., Thézenas S., Peron J.-M., Bécouarn Y., Seitz J.-F. et al. (2019) Sorafenib alone vs. sorafenib plus GEMOX as 1-line treatment for advanced HCC: the phase II randomised PRODIGE 10 trial. Br. J. Cancer 120, 896–902 10.1038/s41416-019-0443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gish R.G., Porta C., Lazar L., Ruff P., Feld R., Croitoru A. et al. (2007) Phase III randomized controlled trial comparing the survival of patients with unresectable hepatocellular carcinoma treated with nolatrexed or doxorubicin. J. Clin. Oncol. 25, 3069–3075 10.1200/JCO.2006.08.4046 [DOI] [PubMed] [Google Scholar]
- 5.Bruix J., Qin S., Merle P., Granito A., Huang Y.-H., Bodoky G. et al. (2017) Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 389, 56–66 10.1016/S0140-6736(16)32453-9 [DOI] [PubMed] [Google Scholar]
- 6.Wu J., Sun L., Chen X., Du F., Shi H., Chen C. et al. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motwani M., Pesiridis S. and Fitzgerald K.A. (2019) DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 10.1038/s41576-019-0151-1 [DOI] [PubMed] [Google Scholar]
- 8.Zhao B., Du F., Xu P., Shu C., Sankaran B., Bell S.L. et al. (2019) A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722 10.1038/s41586-019-1228-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T. et al. (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Yuan B., Bao M., Lu N., Kim T. and Liu Y.-J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S. et al. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Z., Jia S., Shao C. and Shi Y. (2020) Irradiation induces cancer lung metastasis through activation of the cGAS-STING-CCL5 pathway in mesenchymal stromal cells. Cell Death Dis. 11, 326 10.1038/s41419-020-2546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon J. and Bakhoum S.F. (2020) The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 10, 26–39 10.1158/2159-8290.CD-19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu L., Li Y., Xie X., Zhou X., Gu M., Jie Z. et al. (2019) TBKBP1 and TBK1 form a growth factor signalling axis mediating immunosuppression and tumourigenesis. Nat. Cell Biol. 21, 1604–1614 10.1038/s41556-019-0429-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qing T., Yamin Z., Guijie W., Yan J. and Zhongyang S. (2017) STAT6 silencing induces hepatocellular carcinoma-derived cell apoptosis and growth inhibition by decreasing the RANKL expression. Biomed. Pharmacother. 92, 1–6 10.1016/j.biopha.2017.05.029 [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Wang H., Ren J., Chen Q. and Chen Z.J. (2017) cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 114, E4612–E4620 10.1073/pnas.1705499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Z., Li Y., Yang X., Yu M., Chen Z., Zhao C. et al. (2018) Overexpression of CLC-3 is regulated by XRCC5 and is a poor prognostic biomarker for gastric cancer. J. Hematol. Oncol. 11, 115 10.1186/s13045-018-0660-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomsen M.K., Skouboe M.K., Boularan C., Vernejoul F., Lioux T., Leknes S.L. et al. (2020) The cGAS-STING pathway is a therapeutic target in a preclinical model of hepatocellular carcinoma. Oncogene 39, 1652–1664 10.1038/s41388-019-1108-8 [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Ba G., Han Y.-Q., Ming S.-L., Wang M.-D., Fu P.-F. et al. (2020) Cyclic GMP-AMP synthase is essential for cytosolic double-stranded DNA and fowl adenovirus serotype 4 triggered innate immune responses in chickens. Int. J. Biol. Macromol. 146, 497–507 10.1016/j.ijbiomac.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 20.Li R., Yang Y., An Y., Zhou Y., Liu Y., Yu Q. et al. (2011) Genetic polymorphisms in DNA double-strand break repair genes XRCC5, XRCC6 and susceptibility to hepatocellular carcinoma. Carcinogenesis 32, 530–536 10.1093/carcin/bgr018 [DOI] [PubMed] [Google Scholar]
- 21.Marozin S., Altomonte J., Stadler F., Thasler W.E., Schmid R.M. and Ebert O. (2008) Inhibition of the IFN-β Response in Hepatocellular Carcinoma by Alternative Spliced Isoform of IFN Regulatory Factor-3. Mol. Ther. 16, 1789–1797 10.1038/mt.2008.201 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Lin H., Hua F. and Hu Z-w (2013) Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy 9, 925–927 10.4161/auto.24229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W., Zhao Z., Ni Z., Zhao Y., Du W. and Chen S. (2017) IFI16 restoration in hepatocellular carcinoma induces tumour inhibition via activation of p53 signals and inflammasome. Cell Prolif. 50, 10.1111/cpr.12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi BVSK et al. (2017) UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A. et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 28.Szász A.M., Lánczky A., Nagy Á, Förster S., Hark K., Green J.E. et al. (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy Á, Lánczky A., Menyhárt O. and Győrffy B. (2018) Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasaikar S.V., Straub P., Wang J. and Zhang B. (2018) LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S. et al. (2017) TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77, e108–e110 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., Severson E., Pignon J.-C., Zhao H., Li T., Novak J. et al. (2016) Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17, 174 10.1186/s13059-016-1028-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wurmbach E., Chen Y.-b., Khitrov G., Zhang W., Roayaie S., Schwartz M. et al. (2007) Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45, 938–947 10.1002/hep.21622 [DOI] [PubMed] [Google Scholar]
- 34.Mas V.R., Maluf D.G., Archer K.J., Yanek K., Kong X., Kulik L. et al. (2009) Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol. Med. 15, 85–94 10.2119/molmed.2008.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roessler S., Jia H.-L., Budhu A., Forgues M., Ye Q.-H., Lee J.-S. et al. (2010) A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 10.1158/0008-5472.CAN-10-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Cheung S.T., So S., Fan S.T., Barry C., Higgins J. et al. (2002) Gene expression patterns in human liver cancers. Mol. Biol. Cell 13, 1929–1939 10.1091/mbc.02-02-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azimi F., Scolyer R.A., Rumcheva P., Moncrieff M., Murali R., McCarthy S.W. et al. (2012) Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 30, 2678–2683 10.1200/JCO.2011.37.8539 [DOI] [PubMed] [Google Scholar]
- 38.Ahn J., Xia T., Konno H., Konno K., Ruiz P. and Barber G.N. (2014) Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 10.1038/ncomms6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F. et al. (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forner A., Reig M. and Bruix J. (2018) Hepatocellular carcinoma. Lancet 391, 1301–1314 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 41.Al-Eitan L.N., Rababa'h D.M., Alghamdi M.A. and Khasawneh R.H. (2019) Genetic association of gene polymorphisms with breast cancer among Jordanian women. Onco Targets Ther 12, 7923–7928 10.2147/OTT.S220226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z.H., Wang N., Wang F.Q., Dong Q. and Ding J. (2019) High expression of XRCC5 is associated with metastasis through Wnt signaling pathway and predicts poor prognosis in patients with hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 23, 7835–7847 [DOI] [PubMed] [Google Scholar]
- 43.Jiao S., Guan J., Chen M., Wang W., Li C., Wang Y. et al. (2018) Targeting IRF3 as a YAP agonist therapy against gastric cancer. J. Exp. Med. 215, 699–718 10.1084/jem.20171116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cañadas I., Thummalapalli R., Kim J.W., Kitajima S., Jenkins R.W., Christensen C.L. et al. (2018) Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 24, 1143–1150 10.1038/s41591-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazzari E., Korczeniewska J., Ní Gabhann J., Smith S., Barnes B.J. and Jefferies C.A. (2014) TRIpartite motif 21 (TRIM21) differentially regulates the stability of interferon regulatory factor 5 (IRF5) isoforms. PLoS ONE 9, e103609 10.1371/journal.pone.0103609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., Mo J., Swanson K.V., Wen H., Petrucelli A., Gregory S.M. et al. (2014) NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40, 329–341 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engen J.R., Wales T.E., Hochrein J.M., Meyn M.A., Banu Ozkan S., Bahar I. et al. (2008) Structure and dynamic regulation of Src-family kinases. Cell. Mol. Life Sci.: CMLS 65, 3058–3073 10.1007/s00018-008-8122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukumoto Y., Morii M., Miura T., Kubota S., Ishibashi K., Honda T. et al. (2014) Src family kinases promote silencing of ATR-Chk1 signaling in termination of DNA damage checkpoint. J. Biol. Chem. 289, 12313–12329 10.1074/jbc.M113.533752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.al-Ramadi B.K., Zhang H. and Bothwell A.L. (1998) Cell-cycle arrest and apoptosis hypersusceptibility as a consequence of Lck deficiency in nontransformed T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 95, 12498–12503 10.1073/pnas.95.21.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavin M.F., Scott S., Gueven N., Kozlov S., Peng C. and Chen P. (2004) Functional consequences of sequence alterations in the ATM gene. DNA Repair (Amst.) 3, 1197–1205 10.1016/j.dnarep.2004.03.011 [DOI] [PubMed] [Google Scholar]
- 51.Kouhkan F., Mobarra N., Soufi-Zomorrod M., Keramati F., Hosseini Rad S.M.A., Fathi-Roudsari M. et al. (2016) MicroRNA-129-1 acts as tumour suppressor and induces cell cycle arrest of GBM cancer cells through targeting IGF2BP3 and MAPK1. J. Med. Genet. 53, 24–33 10.1136/jmedgenet-2015-103225 [DOI] [PubMed] [Google Scholar]
- 52.Montero J.C., Seoane S., Ocaña A. and Pandiella A. (2011) Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clin. Cancer Res. 17, 5546–5552 10.1158/1078-0432.CCR-10-2616 [DOI] [PubMed] [Google Scholar]
- 53.Pizarro J.G., Folch J., de la Torre A.V., Junyent F., Verdaguer E., Jordan J. et al. (2010) ATM is involved in cell-cycle control through the regulation of retinoblastoma protein phosphorylation. J. Cell. Biochem. 110, 210–218 [DOI] [PubMed] [Google Scholar]
- 54.Chen Q., Sun L. and Chen Z.J. (2016) Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- 55.Woo S.-R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y.K. et al. (2014) STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C. et al. (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 57.Liu X., Wu S., Yang Y., Zhao M., Zhu G. and Hou Z. (2017) The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed. Pharmacother. 95, 55–61 10.1016/j.biopha.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 58.Lin P., Guo Y.-N., Shi L., Li X.-J., Yang H., He Y. et al. (2019) Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 11, 480–500 10.18632/aging.101754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo X., Li H., Ma L., Zhou J., Guo X., Woo S.-L. et al. (2018) Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 155, 1971–1984 10.1053/j.gastro.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawant A., Hensel J.A., Chanda D., Harris B.A., Siegal G.P., Maheshwari A. et al. (2012) Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J. Immunol. 189, 4258–4265 10.4049/jimmunol.1101855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y.-H., Zhu C., Kondo Y., Anderson A.C., Gandhi A., Russell A. et al. (2016) Corrigendum: CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 536, 359 10.1038/nature17421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang D., Xiao-Feng H., Guan-Jun D., Er-Ling H., Sheng C., Ting-Ting W. et al. (2015) Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim. Biophys. Acta 1852, 2494–2503 10.1016/j.bbadis.2015.08.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of our study are openly available from Molecular Signatures Database (MSigDB), Oncomine database, UALCAN, Human Protein Atlas, Kaplan-Meier plotter, LinkedOmics, and Tumor Immune Estimation Resource (TIMER) database (https://www.gsea-msigdb.org/gsea/msigdb/, www.oncomine.org, http://ualcan.path.uab.edu, https://www.proteinatlas.org, www.kmplot.com, http://www.linkedomics.org/, https://cistrome.shinyapps.io/timer/).