Abstract
Rovalpituzumab tesirine (Rova-T) offers a targeted therapy for ~85% of SCLC patients whose tumors express DLL3, but clinical dosing is limited due to off-target toxicities. We hypothesized that a sub-efficacious dose of Rova-T combined with anti-PD1, which alone shows a clinical benefit to ~15% of SCLC patients, might elicit a novel mechanism of action and extend clinical utility. Using a pre-clinical murine SCLC tumor model that expresses Dll3 and has an intact murine immune system, we found that sub-efficacious doses of Rova-T with anti-PD1 resulted in enhanced anti-tumor activity, compared to either monotherapy. Multiplex immunohistochemistry (IHC) showed CD4 and CD8 T-cells primarily in normal tissue immediately adjacent to the tumor. Combination treatment, but not anti-PD1 alone, increased Ki67+/CD8 T-cells and Granzyme B+/CD8 in tumors by flow cytometry and IHC. Antibody depletion of T-cell populations showed CD8+ T-cells are required for in vivo anti-tumor efficacy. Whole transcriptome analysis as well as flow cytometry and IHC showed that Rova-T activates dendritic cells and increases Ccl5, Il-12, and Icam more than anti-PD1 alone. Increased tumor expression of PDL1 and MHC1 following Rova-T treatment also supports combination with anti-PD1. Mice previously treated with Rova-T + anti-PD1 withstood tumor re-challenge, demonstrating sustained anti-tumor immunity. Collectively our pre-clinical data support clinical combination of sub-efficacious Rova-T with anti-PD1 to extend the benefit of immune checkpoint inhibitors to more SCLC patients.
Keywords: Antibody drug conjugates, Immunogenic cell death, Rovalpituzumab tesirine, Checkpoint inhibitor, PD1
Highlights
-
•
Sub-efficacious Rova-T combined with anti-PD1 regresses murine SCLC tumors.
-
•
Anti-SCLC efficacy seen with Rova-T + anti-PD1 requires CD8 T-cells.
-
•
Rova-T + anti-PD1 recruits and activates T-cells and dendritic cells within the tumor.
-
•
Combination of Rova-T and anti-PD1 results in prolonged anti-tumor immunity.
Introduction
Small cell lung cancer (SCLC) is a highly aggressive and metastatic pulmonary neuroendocrine carcinoma that develops in current and former heavy cigarette smokers. SCLC tumors are characterized by near universal loss and inactivation of two tumor suppressor genes, TP53 and RB1 [1]. Additionally, SCLC tumors express neuroendocrine markers such as synaptophysin (SYP), chromogranin A (CHGA) and neural cell adhesion molecule 1 (NCAM1; CD56), while Notch signaling is repressed [2]. The transcription factor achaete-scute homologue 1 (ASCL1) is a lineage-specific oncogenic driver overexpressed in ~75% of SCLC. ASCL1 is the direct transcriptional regulator of delta-like protein 3 (DLL3), an atypical Notch ligand that could function in Notch pathway repression [3,4]. DLL3 expression on the surface of SCLC tumor cells has been therapeutically targeted with rovalpituzumab tesirine (Rova-T), an antibody-drug conjugate (ADC) consisting of a monoclonal antibody targeting DLL3, that delivers a pyrrolobenzodiazepine dimer (PBD) cytotoxin into tumor cells to elicit cell death [5].
Front-line standard of care for extensive stage SCLC is platinum chemotherapy (cisplatin or carboplatin) with etoposide, and while 44–78% of patients initially respond, patients relapse rapidly, and mean overall survival is 9–11 months [6]. Topotecan is currently the only approved second-line SCLC therapy and response can be predicted according to response to front-line therapy and time to progression [42]. Recently, several immunotherapies that activate or enhance a patient's immune system to recognize and kill tumor cells that target programmed cell death-1 (PD1) on immune cells or PD1 ligand (PDL1) on tumor cells [7] have been approved clinically for SCLC. Atezolizumab, an anti-PDL1 monoclonal antibody, increased median overall survival of SCLC patients to 12.3 months versus 10.3 months when used both with induction carboplatin/etoposide chemotherapy and in the frontline maintenance setting, leading to FDA approval [8]. Nivolumab and pembrolizumab, both anti-PD1 monoclonal antibodies, are approved in third line SCLC [9,10].
Only 18% of SCLC cases have PDL1 expression in tumor-infiltrating macrophages, and 48% showed PD1 positive lymphocytes with genomic amplification of PDL1 only seen in 2% of SCLC tumors [11,12]. PDL1 expression on tumors, a high level of tumor mutation burden, and high levels of tumor immune infiltrate correlate with patient response to immune checkpoint inhibitors, but these biomarkers alone do not predict tumor subtypes or patients that will respond [13]. While SCLC is characterized by high tumor mutation burden, it also shows high immunosuppression with low counts of tumor infiltrating lymphocytes and reduced antigen presentation [14]. Despite the high tumor mutation burden in SCLC, response rates in clinical trials suggest that SCLC patients with the highest mutation burden have a greater clinical benefit with nivolumab alone or in combination with ipilimumab, an anti-CTLA-4 immune checkpoint inhibitor [15,16]. Therefore, a subset of SCLC patients benefit from immune checkpoint inhibitors, and their use in combination with targeted therapies or cytotoxic agents might extend efficacy to more SCLC patients.
One approach to enhance the efficacy of immune checkpoint inhibitors is to combine them with cancer therapies that elicit immunogenic cell death (ICD), an apoptotic cell death process that results in the release of antigenic molecules that activate the adaptive immune response [[17], [18], [19]]. PBD based ADCs induce ICD and demonstrate synergistic antitumor responses with anti-PD1 and anti-PDL1 inhibitors in pre-clinical models [20]. Additionally, poly ADP-ribose polymerase (PARP) inhibitors and checkpoint kinase 1 (CHK1) inhibitors increase expression of PDL1 on tumor cells, activate the STING innate immune pathway, and show synergistic pre-clinical activity with anti-PDL1 in murine SCLC tumor models [13].
A phase II clinical trial evaluating Rova-T dosed twice at 0.3 mg/kg, six weeks apart, in recurrent SCLC with DLL3+ tumor cells, showed a 19% response rate and median survival of 5.7 months, with 40% of patients developing ≥ grade 3 toxicities including pleural effusions, edema and photosensitivity rash [21]. More recently, phase III trials evaluating Rova-T in the second line and frontline maintenance settings have not met clinical endpoints, due to the narrow therapeutic window for PBD-based ADCs [22]. These off-target treatment related side effects are seen across PBD containing ADCs [23]. Rova-T (0.3 mg/kg) and nivolumab (360 mg) in SCLC patients showed durable responses, but, given safety data, only strategies that enable lower doses of PBD based ADCs in combination with immunotherapy agents could provide a clinical path for SCLC [24].
To evaluate the combination of Rova-T + anti-PD1 pre-clinically, we used KP1, a SCLC genetically engineered mouse tumor model that lacks tumor suppressors TP53 and RB1 and endogenously expresses Dll3. Our first objective was to confirm that KP1 tumor bearing mice show a dose response to single agent Rova-T. Next, we tested combination of Rova-T + anti-PD1 to determine if sub-efficacious doses of Rova-T showed combination activity with anti-PD1. The mechanism behind the combination efficacy was explored by examining the immune infiltrates of the tumor model in response to therapy, through whole transcriptome, flow cytometry and immunofluorescence studies. Finally, dependency on specific immune cells was demonstrated through depletion studies, and long-term immune memory was confirmed in re-challenge studies. Collectively, our results demonstrate that sub-efficacious doses of Rova-T can elicit an antitumor response that increases the effectiveness of immunotherapies in a preclinical SCLC experimental model.
Results
Rova-T is efficacious in a mouse tumor model of SCLC
Rova-T is an ADC targeting DLL3 that elicits an anti-tumor response pre-clinically in patient derived xenograft models and clinically in patients with SCLC [5,25]. The antibody component of Rova-T, SC16.56, binds to a region of DLL3 with high homology between rat, mouse, and human (Supplementary Fig. 1A). Consistent with structural similarity, SC16.56 binds mouse Dll3 and human DLL3 to a similar extent as measured by flow cytometry in 293T overexpressing murine Dll3 and human DLL3 (Supplementary Fig. 1B) [5]. The KP1 cell line model was derived from a genetically defined model of SCLC [26] and recapitulates many molecular features of human SCLC [1]. KP1 cells express the immune-suppressive ligand PDL1 and are responsive to anti-PD1 therapies in vivo [27]. Flow staining confirmed that the KP1 cells express Dll3 on the cell surface, consistent with the high prevalence of Dll3 expression in human SCLC tumors (Supplementary Fig. 2A) [5]. The strong in vitro potency of Rova-T on KP1 cells indicates that the ADC induces murine Dll3 internalization and release of the cytotoxic PBD toxin (Supplementary Fig. 2B). Collectively these data show that KP1 endogenously express Dll3 on the cell surface and that Rova-T binds murine Dll3 and elicits in vitro cytotoxicity.
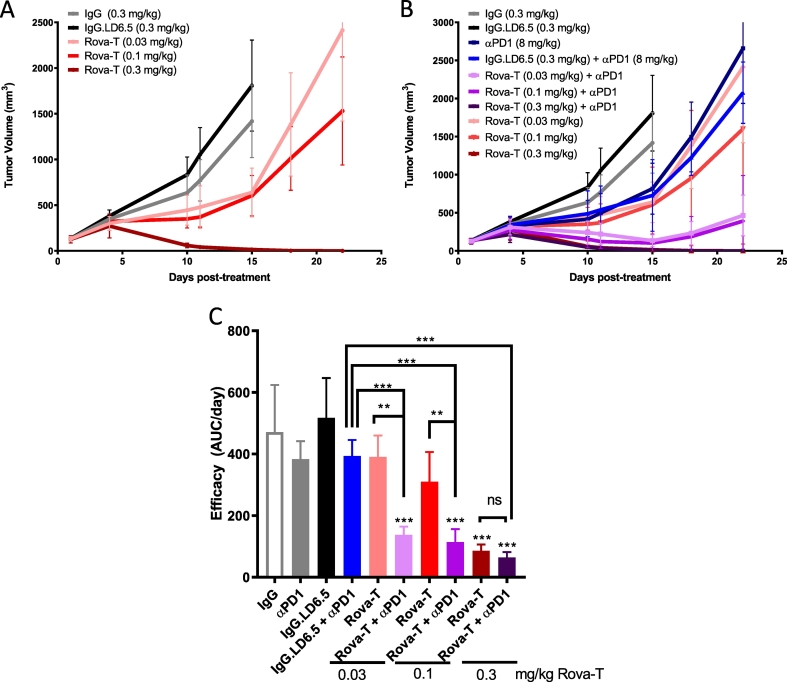
To determine whether the Dll3-positive KP1 tumor model is responsive to Rova-T therapy in an in vivo setting, B6129SF1 mice were inoculated with KP1 tumors, randomized into treatment cohorts with an average tumor volume of ~150 mm3, and given a single dose of 0.03, 0.1 or 0.3 mg/kg Rova-T. Non-targeting isotype ADC (IgG.LD6.5) dosed at 0.3 mg/kg had no significant effect on tumor growth as compared to the naked IgG control, demonstrating there is no target independent activity of the PBD conjugate (Fig. 1A). Monotherapy Rova-T resulted in dose-dependent reduction in tumor growth with 0.3 mg/kg showing a complete response with no measurable tumors for more than 80 days after treatment (Fig. 1A, data not shown). Thus, the murine SCLC tumor model KP1 is dose responsive to single agent Rova-T in vivo.
Fig. 1.
Evaluating single agent Rova-T activity with anti-PD1 in a murine model of SCLC. A) Mean KP1 tumor volumes in female B6129SF1 mice after treatment with a single dose of 0.3 mg/kg IgG, 0.3 mg/kg IgG.LD6.5 or varying doses (0.03, 0.1, or 0.3 mg/kg) of Rova-T. B) Mean tumor volumes (n = 5 mice per group) with 8 mg/kg Q4Dx4 anti-PD1 dosed alone or in combination with varying single dose levels of Rova-T as indicated. C) Summary of anti-tumor efficacy from panel B as quantified by area under tumor volume growth curve divided by days on study. One-way ANOVA analysis showed intergroup difference of P < 0.001. Statistical comparison (student's t-test) run for each treatment group as compared to IgG.LD6.5 as well as each combination group to each single agent therapy. (**P < 0.01, ***P < 0.001.) For all graphs, error bars represent SEM.
Combination of Rova-T + anti-PD1 therapy is more effective than either monotherapy
Clinically, the dose level of Rova-T is limited by target independent toxicities seen with other PBD-based ADCs including pleural effusions, peripheral edema and photosensitivity [25]. Therefore, combination treatments that enable lower doses of Rova-T could enhance the therapeutic index. We evaluated whether the magnitude of the anti-tumor response for Rova-T at lower doses was enhanced by co-administration of anti-PD1 therapy. Anti-PD1 alone or in combination with isotype ADC, IgG.LD6.5, resulted in a modest reduction in tumor growth, which was statistically significant 15 days after dosing (Fig. 1B). Monotherapy doses of 0.03 and 0.1 mg/kg Rova-T resulted in 24% and 40% reduction in efficacy, respectively (Fig. 1C). In combination with anti-PD1, doses of 0.03 and 0.1 mg/kg Rova-T achieved greater than 70% reduction in efficacy, resulting in robust combination activity (Fig. 1C).
Recent studies demonstrated a clinical benefit of Atezolizumab (anti-PDL1) in combination with carboplatin and etoposide as a frontline treatment in SCLC [8]. KP1 tumor bearing mice were was also dosed with Rova-T + anti-PDL1 combination. Consistent with the anti-PD1 study, anti-PDL1 in combination with 0.03 mg/kg Rova-T had a stronger percent tumor growth inhibition compared to either single agent (Supplementary Fig. 3).
Rova-T is immunomodulatory in mouse SCLC tumors
Previous reports indicate that cytotoxic agents, including PBDs, can induce immunogenic cell death, a process that results in enhanced dendritic cell (DC) activation and subsequent T cell infiltration and activation [20,28,29]. To evaluate potential immunomodulatory activity of Rova-T in SCLC tumors, the KP1 study was repeated using a single 0.03 mg/kg dose of Rova-T alone or in combination with anti-PD1. Tumors were harvested 3 and 8 days after treatment to assess changes to the immune microenvironment by flow cytometry, transcriptomics and immunofluorescence.
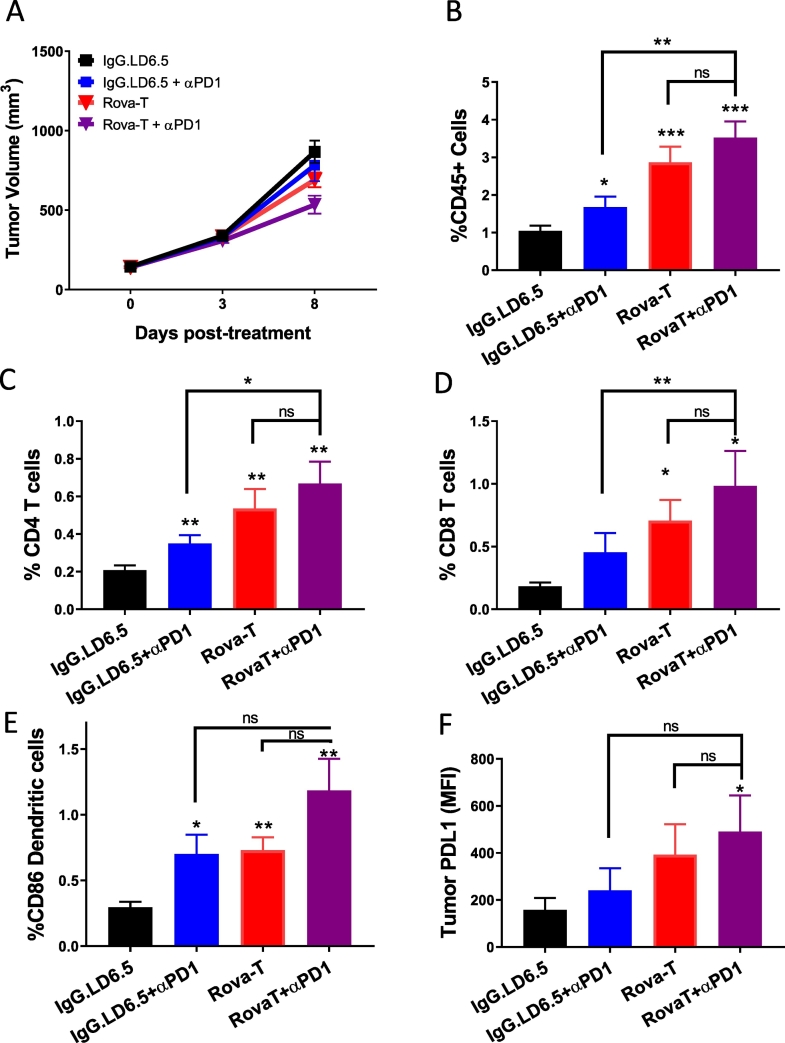
Eight days post-treatment, only minimal changes to tumor volume were observed, but a trend toward reduced tumor growth was seen for the Rova-T + anti-PD1 combination group, consistent with the previous efficacy study (Figs. 1B, 2A). Tumors were dissociated and subjected to flow cytometry analysis to evaluate infiltrating immune cells by gating on all live cells in the tumor. Anti-PD1 treatment had negligible impact on the proportion of CD45+ immune cells in the tumor; however, tumors from Rova-T dosed animals had more than 2-fold higher CD45+ immune cells than isotype ADC control-dosed animals (Fig. 2B). The combination of Rova-T + anti-PD1 had 3-fold higher CD45+ immune cells than IgGLD6.5 though this difference was not statistically different from the Rova-T single agent group, suggesting that anti-PD1 does not change immune cell infiltration triggered by Rova-T (Fig. 2B). Similar trends were observed 3 days post-treatment, when no measurable change to tumor volume was observed (Supplementary Fig. 4B).
Fig. 2.
Pharmacodynamic changes to immune cell infiltration in KP1 tumors dosed with Rova-T + anti-PD1. A) Mean KP1 tumor volume following a single dose of Rova-T or IgG.LD6.5 alone or in combination with 8 mg/kg anti-PD1 dosed every four days, with tumors harvested 3 and 8 days post treatment for flow cytometry, RNAseq, and immunofluorescence analysis. Percentage of live cells 8 days post treatment for B) CD45+ C) CD4+ T cells D) CD8+ T cells E) CD86+ DCs and F) mean fluorescence intensity of PDL1 staining among CD45 negative tumor cells. One-way ANOVA analysis showed intergroup difference of P < 0.01. Statistical comparison (student's t-test) run for each treatment group as compared to IgG.LD6.5 as well as each combination group to each single agent therapy. (*P < 0.05, **P < 0.01, ***P < 0.001.) For all graphs, error bars represent SEM.
Cytotoxic CD8+ T and helper CD4+ T cells activated by DCs drive the anti-tumor response to anti-PD1 therapy. Flow cytometry demonstrated increased percentage of both CD4+ and CD8+ T cells in the tumor 8 days after Rova-T monotherapy (Fig. 2C and D). While the percentage of CD4+ and CD8+ T-cells trended higher in the combination treatment compared to Rova-T alone, these differences were not statistically significant, suggesting that anti-PD1 does not change the DC-induced immune responses triggered by Rova-T (Fig. 2C and D). Three days post-treatment, the percentage of CD4+ and CD8+ T cells were already higher in the Rova-T and Rova-T + anti-PD1 dosed tumors (Supplementary Fig. 4C and D). These data indicate that Rova-T alone or in combination with anti-PD1 recruits CD4+ and CD8+ T cells to the tumor environment in this mouse model of SCLC.
Immunogenic cell death pathways are associated with enhanced activation of CD86-positive DCs which facilitate the priming and activation of T cells [30]. Flow cytometry on KP1 tumors from dosed mice showed an increase in CD86+ DC infiltration in the anti-PD1 and Rova-T monotherapy treatments 8 days after treatment (Fig. 2E) with some activation by Rova-T alone and the combination at 3 days (Supplementary Fig. 4E). The increase in CD86+ DC infiltration trended higher with combination treatment, but this change was not statistically different from Rova-T alone, suggesting that anti-PD1 does not change DC activation induced by Rova-T (Fig. 2E). These data indicate that Rova-T alone and in combination with anti-PD1 increases DC infiltration in the tumor microenvironment.
Cytotoxic therapies can result in the induction of PDL1, which can act to dampen the immune response against tumors [31]. PDL1 on tumor cells was measured by flow cytometry. Tumors harvested on both days 3 and 8 post dosing showed Rova-T alone elevated expression of PDL1 (Fig. 2F, Supplementary Fig. 4F), and the combination of Rova-T + anti-PD1 did not further induce PDL1 expression. The upregulation of this immune suppressive ligand in response to Rova-T further supports the use of an anti-PD(L)1 therapy as a combination to overcome immune evasion.
Combination of Rova-T + anti-PD1 alters transcripts associated with T cell activation
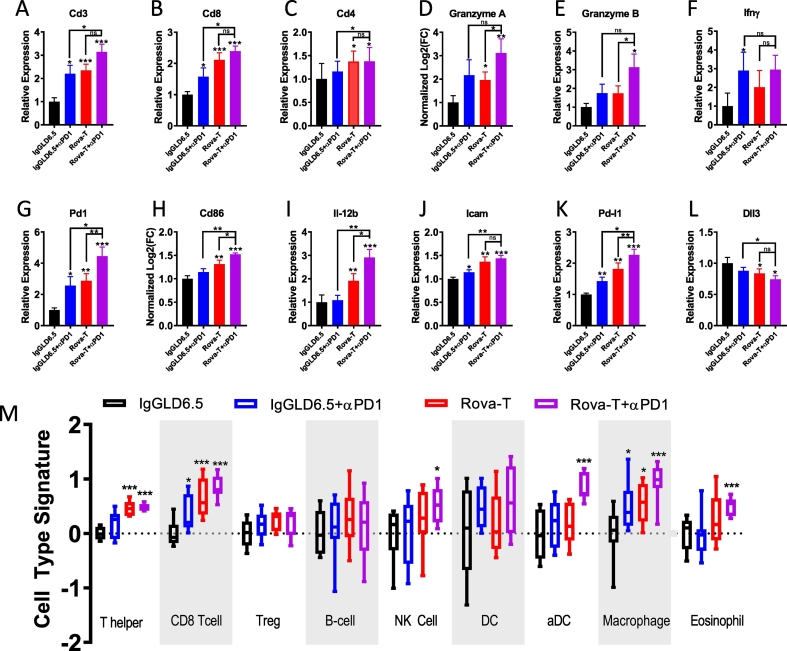
To better understand intratumoral changes in response to Rova-T, anti-PD1, and the combination, RNAseq analysis was run on RNA extracted from formaldehyde fixed paraffin embedded (FFPE) tumors collected 8 days post-treatment (Fig. 3). As seen with flow cytometry, transcripts for Cd45, Cd3, Cd8, and Cd4 were significantly upregulated with Rova-T monotherapy and combination dosing, consistent with enhanced infiltration of immune cells expressing these markers (Fig. 3A–C, Supplementary Fig. 5A). Tumors from combination dosed mice showed a trend toward higher CD3 expression, which was significantly higher than anti-PD1 alone, but did not reach significance relative to Rova-T monotherapy. Granzyme A and B, which are expressed upon activation of T cells [32], were upregulated particularly in the Rova-T + anti-PD1 combination group (Fig. 3D–E). In addition to cytotoxic proteins, activated T cells can express cytokines such as Ifnγ. Although expression was variable, all three treatment groups trended toward higher expression (Fig. 3F). When activated, effector T cells also express Pd1. Pd1 transcripts were highest in the tumors dosed with Rova-T + anti-PD1, consistent with enrichment of effector T cells in response to combination dosing (Fig. 3G).
Fig. 3.
Whole transcriptome analysis shows modulation of immune cells and markers of inflammation. A–L) Whole transcriptome analysis for immune-related transcripts in KP1 tumors 8 days after treatment for indicated transcripts. M) Nanostring PanCancer Immune Profiling analysis of RNAseq data in KP1 tumors 8 days after treatment. Immune signature score is indicated on y-axis and immune cell type is listed along the x-axis. Treatment groups are colored according to Fig. legend. For box and whisker plots, outer lines indicate range, box indicates boundary of upper and lower quartile, and central horizontal indicates median. For all groups with t-test analysis, P < 0.05 by one-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001 by student's t-test).
By flow cytometry, an increase in DC infiltration was observed after dosing (Fig. 2E). Similarly, Cd86 transcripts were upregulated in response to Rova-T alone and enhanced by the combination of Rova-T + anti-PD1 (Fig. 3H). Multiple DC subtypes can impact immune response in the tumor. By RNA, we found both CD11b and CD103 transcripts increased with combination treatment; however additional flow data would be needed to better understand changes to individual DC subtypes (Supplementary Fig. 5E, 5F). Additionally, cytokine secretion was upregulated in response to monotherapy agents but were significantly higher in the tumors dosed with Rova-T + anti-PD1 (Fig. 3I). One mechanism by which immune cells are recruited to tumors is through the binding and rolling of circulating T cells on activated endothelium [33]. Icam expression is a hallmark of activated endothelial and is expressed upon secretion of inflammatory cytokines. Icam expression was increased by Rova-T alone or in combination with anti-PD1, suggesting a possible mechanism for enhanced infiltration (Fig. 3J).
A recent study reported that targeting DNA damage response pathways enhances the efficacy of anti-PDL1 in part through enhanced T cell recruitment by Cxcl10 and Ccl5, transcriptional targets of the STING pathway [27]. Since Rova-T delivers a PBD warhead that induces DNA damage, we probed whether Cxcl10 or Ccl5 transcripts were significantly changed following dosing. Both Rova-T and anti-PD1 resulted in an increase of Ccl5 and Cxcl10 expression, and the combination treatment further increased expression, particularly for Ccl5 (Supplementary Fig. 5B, C) These data suggest that the induction of DNA damage by Rova-T can elicit a similar induction of STING pathway components as targets of DNA damage response inhibitors.
By flow cytometry, increased expression of PDL1 on tumor cells was observed (Fig. 2F). Consistent with those data, Pdl1 transcripts were markedly higher in response to Rova-T or the combination of Rova-T + anti-PD1 (Fig. 3K). In order to mediate cytotoxic killing, tumor cells must present reactive epitopes on their cell surface bound to MHC Class I (MHC1). We found that Rova-T alone or in combination with anti-PD1 enhanced MHC Class I expression as measured by anti-H-2Kb expression (Supplementary 5D). Together, these data suggest that while Rova-T enhanced MHC1 expression, the concomitant increase in Pdl1 may work to block cytotoxic killing. These data provide additional rationale for the combination of Rova-T with an inhibitor of PDL1 mediated immune suppression.
Given that Rova-T is an ADC that targets cells based on Dll3 expression, tumor cells with higher levels of Dll3 expression may be preferentially targeted by Rova-T. Rova-T monotherapy resulted in a small but significant reduction in Dll3 transcripts in the tumor (Fig. 3L). These transcriptional data were supported by flow cytometry analyses which revealed a reduction in the average Dll3 staining in tumor cells after Rova-T dosing (Supplementary Fig. 4A).
To look more globally at immune cells within the tumor, an algorithm was employed which makes use of gene signatures associated with immune cell subtypes [34]. Based on this transcriptional analysis, the biggest changes in the tumor in response to Rova-T monotherapy were associated with T helper cells, CD8+ T cells and macrophages (Fig. 3M). The changes in lymphocytes were consistent with the flow data presented in Fig. 2. The increase in macrophages could support an additional mechanism of tumor clearance and/or antigen presentation. Interestingly, while total DCs did not change significantly based on transcriptional analysis, the number of infiltrating DCs significantly increased, particularly in the combination group, consistent with CD86 staining by flow and cd86 transcript data (Figs. 3M, H, 2E). Similarly, NK cells and eosinophils showed a slight but significant increase in response specifically to the Rova-T + anti-PD1 combination, suggesting these cells could also play a role in anti-tumor response. Regulatory T and B cells did not significantly change with treatment (Fig. 3M). Additional flow studies would be needed to further understand the changes to myeloid cells and T cell subsets. Whole transcriptome analysis supports the recruitment of CD8+ and CD4+ T cells and the infiltration of DCs with Rova-T alone and in combination with anti-PD1, while the combination uniquely elicits a response through macrophages, eosinophils and NK cells.
A global transcriptional analysis was run for each dosing cohort relative to the IgG.LD6.5 control to better understand alteration of tumor microenvironment (Supplementary Fig. 6A, B). Setting a cutoff for significance of FDR < 1% and log2FC > 1.2, no genes were significantly altered in the anti-PD1 treatment while 95 genes were altered in the Rova-T monotherapy dosed tumors (Supplementary Fig. 6A, B). The vast majority of genes upregulated by Rova-T were further upregulated upon combination with anti-PD1 (Supplementary Fig. 6A). While gene upregulation did not meet significance for the anti-PD1 monotherapy group, about half of the genes upregulated by Rova-T also trended higher with the anti-PD1 group (Supplementary Fig. 6A).
To understand the biological mechanisms driving transcriptional changes in the tumor, differentially regulated Gene Ontology (GO) terms in each treatment group were identified using linear models. Across all GO ontology terms, most gene enrichment sets were associated with immune activation, including regulation of inflammatory response to antigenic stimulus, toll like receptor 9 signaling, response to interferon, T cell chemotaxis, T cell proliferation, and chemokine secretion (Supplementary Fig. 6C). Similar to observations at the single gene level, gene signatures tended to be modestly upregulated by either anti-PD1 or Rova-T alone, and highest with combination dosing (Supplementary Fig. 6C). These data support a substantial alteration of the tumor microenvironment characterized by signatures of immunogenic cell death spanning activation of pattern recognition receptors, chemokine secretion, lymphocyte chemotaxis and T cell proliferation in response to combination Rova-T + anti-PD1.
Rova-T + anti-PD1 alters activation and distribution of T cells within the tumor microenvironment
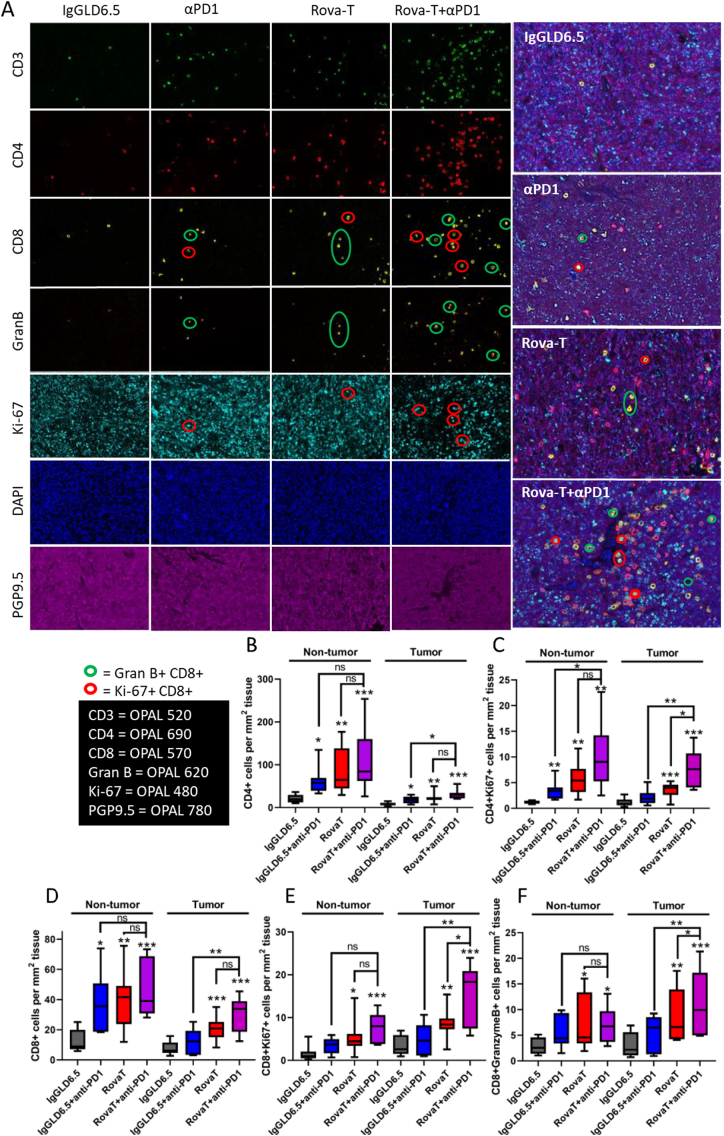
Based on our flow data, the percentage of CD4+ and CD8+ T cells in the tumor were similar between Rova-T and Rova-T + anti-PD1, suggesting that additional mechanisms beyond DC-induction must drive the combination anti-tumor effects observed with Rova-T + anti-PD1. To examine additional mechanisms that contribute to the combination activity, spatial distribution and activation status of immune cells was measured using a multiplex immunofluorescence assay that simultaneously measures CD3, CD4, and CD8 cells in FFPE KP1 tumors 8 days post-dosing. DAPI identified individual cells and PGP9.5, a neuronal marker, distinguished tumor cells from the microenvironment. Ki67 identified proliferating cells, and Granzyme B marked activated CD8+ T cells primed to induce tumor cell lysis. Multiplex staining with 20× magnification allowed for single cell resolution and spatial distribution of immune and tumor cells in a single section that spanned the entire tumor (Fig. 4).
Fig. 4.
Immunofluorescence analysis of immune cell subtypes, their activation and distribution in KP1 tumors.
(A) Multiplex IHC to evaluate the location, proliferation, and activation of T cell subsets within KP1 tumors. Representative single channel images are shown on the left with merged images on the right. Green circles highlight cells that are dual positive for Granzyme B and CD8. Red circles highlight cells that are dual positive for Ki67 and CD8. (B–F) Quantitative image analysis to enumerate immune cells in the tumor or tumor adjacent tissues. Tumor tissue was segmented using PGP9.5. All T cells were segmented using CD3, but CD4 (B, C) and CD8 (D–F) T cell subsets were evaluated separately. Ki67 was used as measure of proliferation and Granzyme B was used as a maker of CD8 T cell activation. For all groups with t-test analysis, P < 0.05 by one-way ANOVA. (*P < 0.05, **P < 0.01, ***P < 0.001 by student's t-test.) For box and whisker plots, outer lines indicate range, box indicates boundary of upper and lower quartile, and central horizontal indicates median.
Consistent with the flow and transcriptomics data, the number of CD4+ and CD8+ immune cells in the tumor and the non-tumor microenvironment per unit area were increased after dosing with anti-PD1 or Rova-T + anti-PD1 combination (Fig. 4A, B, D). Within the tumor, the combination of anti-PD1 + Rova-T significantly increased the proliferation of T cells (Ki-67 positive) more than either single agent (Fig. 4A (red circles), 4C, 4E). In addition to increased proliferation, primed CD8+ cells express Granzyme B, and the combination of Rova-T + anti-PD1 significantly increased the number of Granzyme B+ T cells especially within the tumor (Fig. 4A (green circles), 4F). While there was weak staining of Granzyme B+ and Ki67+ CD8+ within the non-tumor microenvironment, the magnitude and significance were stronger within the tumor, supporting a direct role for these CD8+ cells in mediating the anti-tumor response observed with Rova-T + anti-PD1 (Fig. 4E, F).
Combination Rova-T + anti-PD1 efficacy requires cytotoxic CD8+ T cells
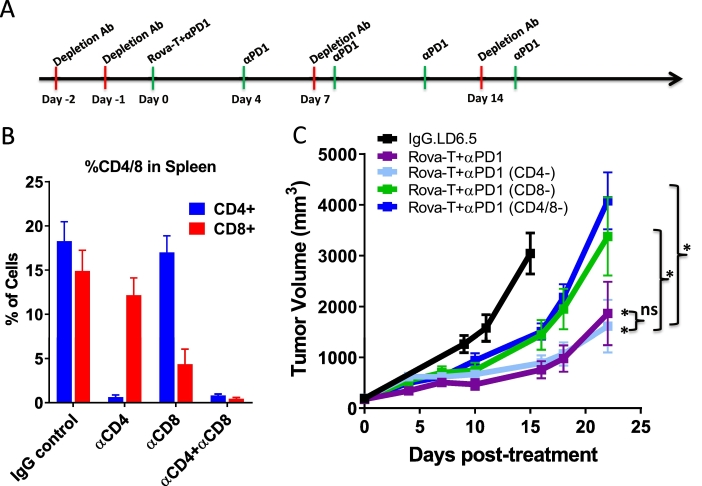
T cell infiltration and markers of T cell activation are associated with Rova-T + anti-PD1 combination treatment (Fig. 2, Fig. 3, Fig. 4). To determine whether T cells drive the anti-tumor activity, efficacy studies were repeated after depletion of CD4+, CD8+ or both CD8+ and CD4+ T cells. For two days prior to randomization, animals bearing KP1 tumors received an 8 mg/kg dose of an immunodepleting antibody against CD4 or CD8, alone or in combination, before the animals were dosed with Rova-T + anti-PD1 (Fig. 5A). At end of the study, spleens were harvested, dissociated, and subjected to flow cytometry to confirm T cell depletion. As compared to IgG control treatment, animals dosed with anti-CD4 or anti-CD8 showed marked reduction in the abundance of CD4+ and CD8+ T cells, respectively (Fig. 5B). Co-treatment of anti-CD4 and anti-CD8 significantly depleted both populations of T cells (Fig. 5B).
Fig. 5.
CD8+ T cells are required for anti-tumor activity of Rova-T + anti-PD1 combination. A) Schematic representation of dosing regimen for T cell depletion study. Treatment initiation beings on Day 0 with anti-PD1 (green hash) dosed at four day intervals and the depletion antibody (red hash) dosed at 7 day intervals with pre-loading for two days prior to dosing initiation. B) Percentage of CD4+ and CD8+ cells among all live cells in spleens harvested from animals at end of study from each of the depletion cohorts. C) Mean tumor volume of KP1 tumors in mice dosed with 0.1 mg/kg IgG.LD6.5 or 0.1 mg/kg Rova-T + 8 mg/kg anti-PD1 combination with 8 mg/kg of immunodepleting antibodies against CD4, CD8 or CD4 + CD8 T cells. Statistical comparison (student's t-test) run for each treatment group as compared to IgG.LD6.5 as well as the non-T cell-depleted combination group to each depletion cohort. For all groups with t-test analysis, P < 0.05 by one-way ANOVA. *P < 0.05 by student's t-test. For all graphs, error bars represent SEM.
The efficacy previously seen with Rova-T + anti-PD1 was not impacted by depletion of CD4+ T cells (Fig. 5C). In contrast, depletion of CD8+ T cells alone or in combination with CD4+ T cell depletion, significantly mitigated the anti-tumor activity of the combination, indicating a role for CD8+ T cells in the anti-tumor efficacy of Rova-T + anti-PD1 (Fig. 5C). These data demonstrate that CD8+ T cells are required for the anti-tumor efficacy of Rova-T + anti-PD1 combination therapy.
Rova-T induces prolonged anti-tumor immunity
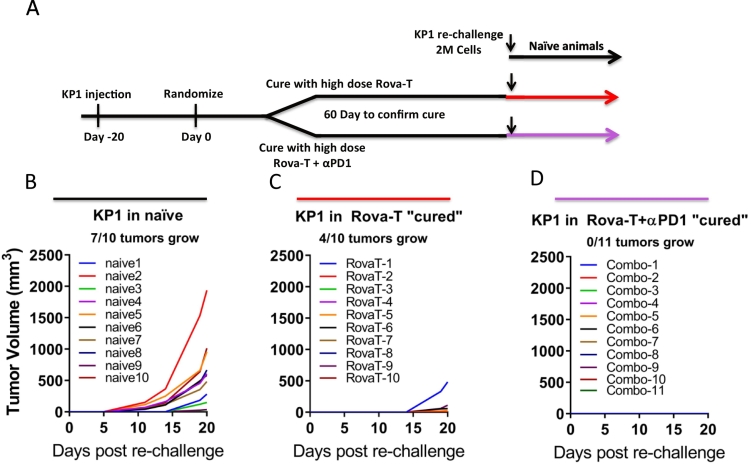
Anti-tumor immunity is a hallmark of immunogenic cell death and is an indicator of T cell memory against tumor antigens. To evaluate whether animals dosed with Rova-T or combination Rova-T + anti-PD1 developed anti-tumor immunity, animals harboring KP1 tumors were dosed with high dose Rova-T (0.3 mg/kg) alone or in combination with anti-PD1 to elicit durable efficacy (Fig. 6A). Mice were monitored for 60 days to confirm that there was no residual disease and tumor regrowth before re-challenge with injection of 2 million KP1 tumor cells on the opposite flank. Within 20 days post inoculation, 7 of the 10 naïve mice inoculated with KP1 had detectable tumor growth (Fig. 6B). In contrast, only 4 of the 10 animals exhibited tumor growth in animals that were in complete remission following previous Rova-T dosing, and tumor growth was significantly slower than in treatment naïve animals (Fig. 6C), indicating prolonged anti-tumor immunity. Remarkably, no tumors were detected in any of the 11 animals previously in complete remission after dosing with the Rova-T + anti-PD1, suggesting the addition of anti-PD1 further enhanced the development of immune memory (Fig. 6D). Collectively this data shows that Rova-T monotherapy elicits ICD that results in long term immune memory upon tumor re-challenge, which is further enhanced with Rova-T + anti-PD1.
Fig. 6.
Long term immune memory in animals cured with high dose Rova-T or Rova-T + anti-PD1. A) Schematic for KP1 re-challenge study design. At randomization, animals dosed with 0.3 mg/kg high dose Rova-T alone or in combination with anti-PD1 to achieve complete responses. B–D) Tumor volumes for KP1 reinjected into opposite flank of naïve mice or mice cured with Rova-T or Rova-T + ant-PD1 as indicated. Fraction of animals exhibiting measurable regrowth at Day 20 are indicated above graphs.
Discussion
Here we demonstrate that immunotherapy agents including anti-PD1 and anti-PDL1 combine with sub-efficacious doses of Rova-T for durable efficacy in a murine tumor model of SCLC. The combination of Rova-T + anti-PD1 increased the number of activated T cells in the KP1 SCLC tumors by flow cytometry, immunofluorescence and whole transcriptome analysis. DC activation is a hallmark of immunogenic cell death and one of the primary signals associated with T cell infiltration and activation [35]. Rova-T + anti-PD1 enhanced DC infiltration within the tumor as measured by flow cytometry and RNAseq (Figs. 2E and 3H). Consistent with increased DC activation, IL-12b and IFNg, pro-inflammatory cytokines associated with recruitment and activation of T cells, are transcriptionally upregulated (Fig. 3F, I). Additionally, transcriptional profiles of T cell and DC activation are among the most significant changes observed in the tumor microenvironment when an unbiased gene enrichment analysis was performed (Supplementary Fig. 6). While both CD4+ and CD8+ T cells are recruited into the tumor, immunodepletion studies confirmed CD8+ T cells drive the anti-tumor activity of the Rova-T + anti-PD1 combination (Fig. 5C). Long term immune memory is a hallmark of ICD. Mice bearing KP1 tumors, in long term remission following an efficacious dose of Rova-T monotherapy and with the Rova-T + anti-PD1 combination, effectively prevented tumor growth upon re-challenge, demonstrating immune memory. Together, these data illustrate that Rova-T induces ICD to enhance activity of anti-PD1 therapy in a CD8+ T cell dependent manner.
The ability of Rova-T to induce signatures of immunogenic cell death has significant implications for the clinical development of Rova-T and checkpoint inhibitors. First, atezolizumab is currently approved as front-line therapy for SCLC in combination with carboplatin [8]. While platinum-based therapies induce immunogenic cell death, they may also induce toxicity of proliferating immune cells, including CD4+ and CD8+ T cells [36,37]. Since Rova-T specifically targets DLL3-positive tumor cells, while inducing ICD and demonstrating combined efficacy with anti-PD(L)1 (Fig. 1), this approach may spare proliferating immune cells. Consistent with this hypothesis, T cell proliferation profiles were upregulated in GSEA analysis of Rova-T + anti-PD1 (Supplementary Fig. 6), suggesting that Rova-T may provide a benefit over systemic chemotherapies as an adjuvant for checkpoint inhibitors. Secondly, Rova-T demonstrates monotherapy activity in 29% of second line SCLC patients at 0.3 mg/kg but is dose limiting due to off-target toxicities including pleural effusions, peripheral edema and photosensitivity [25]. The pre-clinical dosing of a murine SCLC tumor model demonstrates that in combination with anti-PD(L)1, Rova-T can be dosed 10×-fold lower than required for monotherapy efficacy. Indeed, in a Phase 1/2 study, the combination of Rova-T with nivolumab resulted in durable responses; however, Rova-T was dosed at 0.3 mg/kg, resulting in adverse events [24]. Low dose Rova-T + anti-PD1 combination may achieve clinical benefits at tolerated dose levels.
While atezolizumab in combination with chemotherapy is now standard of care frontline therapy in patients with SCLC, only a small percentage of patients showed increased response compared to chemotherapy alone [8]. Our pre-clinical data shows that Rova-T increases expression of PDL1 on tumor cells and enhances the infiltration of immune cells in the tumor, suggesting that Rova-T may shift tumors into a more checkpoint-responsive state. Multiple biomarkers are associated with responsiveness to checkpoint inhibition, including PDL1 expression on tumors, high level of tumor mutation burden, and higher levels of immune infiltrate in the tumor [13]. Loss of MHC1 expression is associated with resistance to anti-PD(L)1 therapies [38], and the significant upregulation of MHC1 by Rova-T may counteract this. While KP1 tumors recapitulate some molecular aspects of SCLC, like loss of both TP53 and RB1, they lack the high tumor mutation burden observed in SCLC patients with a history of heavy cigarette smoking [1]. Clinically, SCLC patients with the highest mutation burden benefit most from immune checkpoint inhibition [15,16]. Therefore, the pre-clinical combination activity observed between Rova-T + anti-PD(L)1 in the KP1 model may underestimate the magnitude of response in SCLC patients with higher tumor mutation burden.
Mechanistically, our data suggest that while the ICD triggered by Rova-T is sufficient to drive T cell infiltration, the combination with anti-PD1 is required to fully activate infiltrating immune cells. Although the Rova-T + anti-PD1 combination trended toward higher infiltration of T cells, these changes were not statistically different from Rova-T alone (Fig. 2C, D). Icam expression, which facilitates immune cell rolling on blood vessels proximal to inflamed tissue, was also similar between Rova-T and the combination group, suggesting that additional mechanisms beyond DC-induction and T-cell infiltration must drive the combination effect (Fig. 3J). Unlike T cell infiltration, however, markers of T cell activation by multiple metrics suggest the combination outperformed either single agent. Transcriptional upregulation of Granzyme B, for example, was substantially increased in combination dosed tumors compared to either single agent (Fig. 3E). By immunofluorescence, the proliferation and activation of T cells was markedly changed in tumors dosed with the combination as compared to either single agent even though the absolute number of T cells was not statistically different in the combination compared to Rova-T alone (Fig. 4). Upregulation of DC activation markers, such as CD86 and IL-12b, suggest that Rova-T enhances T cell activation through priming of these antigen presenting cells.
Rova-T offers a targeted therapeutic strategy for the majority of SCLC patients whose tumor cells express DLL3, but off-target toxicities associated with the PBD warhead limit clinical dosing. Despite the approval of atezolizumab in combination with front-line chemotherapy, a minority of SCLC patients show a clinical benefit. Our pre-clinical data support the combination of anti-PD1 with a sub-efficacious dose of Rova-T, 0.03 mg/kg, which is 10×-fold lower than the dose used in single agent clinical trials (0.3 mg/kg), as well as in the combination trial of Rova-T + nivolumab. Combination therapy of Rova-T + anti-PD(L)1 will benefit patients through synergistic activation of multiple pathways that can enhance the effects of targeting the immune system while directing a cytotoxic agent directly to SCLC tumor cells expressing DLL3.
Methods
Antibodies and compounds
Rova-T was prepared as previously described [5]. Anti–PD1 antibody (Clone 17D2 [muIgG2a/k]) and anti-PDL1 (YW243.55.S70 [muIgG2a/k]) was formulated in phosphate buffered saline (PBS). For flow cytometric analyses, all antibody concentrations and catalog numbers can be found in Supplementary Table 1. For IHC antibodies, catalog numbers and concentrations are summarized in Supplementary Table 2.
Cell lines
KP1, a murine SCLC tumor cell line derived from genetically engineered mice (Rb−/−; p53−/−) [39], were grown in suspension in RPMI1640 medium (Gibco) with 10% fetal bovine serum (Thermo Fisher 16000-044) and pen/strep (Invitrogen), cultured at 37 °C in a humidified chamber with 5% CO2. KP1 cells were confirmed to be negative for pathogens including Mycoplasma (IDEXX BioAnalytics).
In vitro cytotoxicity
Cells were cultured in a T75 flask to ~80% confluency and harvested with trypsin into a single cell suspension. Five hundred (500) KP1cells per well were seeded in tissue culture plates at 50 μL/well culture media and incubated at 37 °C for 18–24 h. SC16 or HuIgG conjugated to LD6.5 were serially diluted and applied to cells for 96 h. CellTiter Glo reagent was prepared per manufacturer's instructions and added at 100 μL/well to the cultures (Promega G7570). Luminescence for untreated wells set to 100% viability.
Immune profiling with multicolor flow cytometry
KP1 murine SCLC xenograft tumors were disaggregated to single-cell suspensions by mincing with razor blades and passing through 40-micron nylon filters. Suspension cells were plated in a round-bottom 96-well plate at 1 × 105 cells per well and centrifuged at 900 ×g, 4 °C for 5 min. Cell pellets were washed in FACS buffer (PBS + 2% FBS, 1 mmol/L EDTA) and again centrifuged. Cells were blocked with Mouse BD Fc Block reagent (Becton Dickinson) and surface stained with 2.5 μg/mL of each antibodies in FACS buffer for 30 min on ice. The resultant single cells were then washed 3 times in FACS buffer and resuspended in FACS buffer containing 2 mg/mL DAPI. Data acquisition was performed with a FACSCanto II (BD Biosciences) cytometer. Viable cells were selected and analyzed for fluorescence intensity. Rainbow beads (BD Biosciences) were used as calibrators to transform mean fluorescence intensities. Data was analyzed using FlowJo (FlowJo). To confirm T cell depletion, spleens were harvested from KP1 bearing mice, dissociated and subject to flow cytometric analysis as described previously.
In vivo efficacy studies
All in vivo studies were conducted in accordance to Stemcentrx Institutional Animal Care and Use Committee (protocols SCAR-3-2008 and SCAR-5-2008) and performed in accordance with the American Association for Laboratory Animal Science and American Veterinary Medical Association guidelines. Animal health was monitored daily, and mouse weights and tumor volumes were measured at least weekly.
KP1 cells were cultured and passaged by pipetting to single cells and split 1 to 3. 1 × 106 cells were implanted subcutaneously in the flank of 6- to 8-week-old B6129SF1/J female mice (Jackson lab, stock no. 101043). Tumor bearing mice were randomized into groups of five to eight animals with average tumor volumes of 140 to 200 mm3 per cohort, typically 2 to 3 weeks after implantation. Mice were dosed with a single dose of control IgG (saline, intraperitoneally, day 1), control IgG1LD6.5 (0.3 to 1 mg/kg, intraperitoneally), SC16.LD6.5 (0.3 to 1 mg/kg, intraperitoneally), anti-PD1 (8 mg/kg, intraperitoneally, Q4D) or anti-PDL1 (10 mg/kg, intraperitoneally, Q4D).
Tumors were measured with digital calipers in two dimensions, long and short axis (in millimeters), and tumor volume (mm3) was calculated as the volume of a prolate ellipsoid: 0.5 × long axis × short axis2. Data collection was stopped, and the mice were euthanized if they exhibited ≥20% weight loss, inactivity, or poor body condition.
For the rechallenge study, 2 × 106 cells were implanted subcutaneously in the opposite flank of tumor cured mice. Naïve (never tumor-inoculated) mice were also inoculated with 2 × 106 KP1 cells for comparison.
For T cell depletion studies, CD4 (clone GK1.5; BioXCell), CD8 (clone 53-6.72; BioXCell), or both CD4 and CD8 depleting antibodies was administered (8 mg/kg) on days −2, −1 and then on day 7 and day 14, before and after drug treatment.
Transcriptome sequencing
RNA was isolated from 10μm FFPE curls using FFPE RNA & DNA Extraction Kit (Norgen, Cat. No. 54300). RNA-Seq libraries were made using KAPA Hyper with RiboErase kit (Roche, Cat. No. KK8560) from 500 ng of input total RNA. All libraries were then sequenced using the Illumina HiSeq sequencing system to generate 150 bp paired-end reads. Quality assessment on alignment results was performed using MultiQC [40]. Sequencing reads were mapped to mouse genome GRCm38 annotations and TPM values from SALMON [41] were used for downstream analyses.
Tumor immune profiling from whole genome RNAseq was performed based on the nCounter NanoString PanCancer Immune Profiling Panel where 109 immune related markers are used to collectively represent 24 immune cell types. Cell type specific expression scores for individual samples were calculated by averaging the expression on log2TPM unit (with 0.01 as offset) across genes within a specific cell type. The cell type specific scores were then centered to 0 across all samples.
For differential expression analysis on a per gene level, we fitted a linear regression model with expression as response and treatment group as variable, where IgG groups was coded as reference. Empirical Bayesian moderated F-statistics similar to ANOVA test for assessing overall significance for each gene estimates associated with treatment arms were generated through R package limma (v3.38.2) [43] for downstream inference. Multiple tests were then corrected using Benjamini & Hochberg method [44] for control false positive rate (FDR). Differentially expression genes were identified at FDR < 1% and with absolute log2 fold changes larger than 1.2.
For pathway analysis, a pathway score was computed for individual samples using single sample GSEA (ssGSEA) [45] against all Gene Ontology (GO) terms in MSigDB (v6.2) C5 collection. Similar to the previous analysis on gene level, differentially expressed pathways were then inferred using linear regression model (R package limma v3.38.2) by regressing pathway scores on treatment groups. Briefly, a linear regression model was fitted per pathway with pathway score as response and treatment group as variable, where IgG groups were coded as reference. Empirical Bayesian moderated F-statistics and the associated P value were generated using limma (v3.38.2) [43], followed by multiple tests correction using Benjamini & Hochberg method [44]. Differentially expressed pathways were identified at FDR < 1%.
Mouse to human annotation mapping was carried out by Ensembl by biomaRt R interface (v2.38.0). All analysis was carried out in R statistical environment [46]. Multiple testing was corrected using Benjamini & Hochberg method [44].
Immunofluorescent and multiplex staining
Immunofluorescent multiplex staining was accomplished by utilizing Akoya Biosciences' OPAL technology (Akoya Biosciences, Hopkinton, MA). Murine derived KP1 tumors once harvested were fixed in 10% neutral buffered formalin for 24 h at RT and subsequently processed for FFPE embedding. Tumors were sectioned at a thickness of 4 μm and placed on charged slides. Sections were then baked for 20 min at 65C and then placed on the automated Ventana DISCOVERY Ultra platform (Roche Diagnostics, Tucson, AZ) where all subsequent steps of the staining procedure were performed using Universal Software v19 (Roche Diagnostics).
Once slides are placed on the DISCOVERY Ultra platform, sections are depariffinized using EZ Prep Solution (Cat # 950-102; Roche Diagnostics) and pretreated by incubating slides in Benchmark Ultra CC1 buffer (Cat # 950-224; Roche Diagnostics) for 40 min at 97C. Endogenous peroxidase was then blocked by incubating sections in DISCOVERY Inhibitor (Cat # 760-4840; Roche Diagnostics) for 8 min at RT. A detailed description of the antibodies and the sequence they were applied are depicted in Supplementary Table 2. The horse radish peroxidase (HRP) conjugated secondary reagent utilized to detect all six primary antibodies was the OMNIMAP anti-rabbit-HRP multimer (Roche Diagnostics; Cat # 760-4311). In order to avoid cross reactivity between the six rabbit derived primary antibodies, sections are incubated in Benchmark Ultra CC2 buffer (Cat # 950-223; Roche Diagnostics) for 8 min at 100 °C after each and every OPAL Dye incubation as a means of “stripping” off the previous antibody complex. No stripping occurs after incubation in OPAL 780 reagent. Nuclei were visualized by incubating slides in Spectral DAPI (Cat # FP1490; Akoya Biosciences) for 16 min at RT and cover slipped with ProLong Gold Diamond Anti-fade mounting media (Cat # P36970; Thermo Fisher Scientific, Waltham, MA).
The spectral information from a multiplexed panel of targets is captured through the Vectra 3.0 Automated Quantitative Pathology Imaging System. In order for the spectral information to be reliably unmixed and quantitated, correct examples of each fluorophore emission spectra, as well as a representative autofluorescence spectra from an unstained sample, in the context to be used, must be registered in a multispectral library. Each of the individually stained sections (CD8a-OPAL 570, CD4-OPAL 690, CD3-OPAL 520, Granzyme B-OPAL 620, KI-67-OPAL 480, PGP9.5-OPAL 780 and DAPI) was used to establish the spectral library of fluorophores required for multispectral analysis. This spectral library forms the reference of target quantitation, as the intensity of each fluorescent target is extracted from the multispectral data using linear unmixing. Immunostained sections were scanned using the Vectra 3.0 Automated Quantitative Pathology Imaging system at 20 nm wavelength intervals from 420 to 720 nm and combined these captures to create a single stack image which retained the unique spectral signature of all IF markers. Once captured, images are unmix and prepared using Inform 2.4.3 software and then imported into HALO version 2.2.1870.31 software (Indica Labs, Albuquerque, NM) where the High Plex FL v2.0 module within HALO is then used for image capture and data analysis.
Statistics
Flow cytometry statistical analyses were performed with GraphPad Prism 8.1 software. For each experiment, we used a one-way ANOVA to assess if there were differences between groups. If the ANOVA P-value was less than 0.05, we a student's t-test to identify differences between individual treatment groups (P < 0.05).
CRediT authorship contribution statement
Philip Vitorino (Formal analysis, Visualization, Writing)
Chen-Hua Chuang (Methodology, Visualization, Investigation)
Alexandre Iannello (Conceptualization, Investigation)
Xi Zhao (Software, Visualization)
Wade Anderson (Investigation)
Ronald Ferrando (Investigation)
Zhaomei Zhang (Investigation)
Shravanthi Madhavan (Resources, Investigation)
Holger Karsunky (Supervision, Conceptualization)
Laura R. Saunders (Project administration, Supervision, Writing).
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors were employed at AbbVie, Inc. at time of study.
Acknowledgements
The authors thank Dr. Julien Sage (Stanford University) for sharing the KP1 SCLC murine model and for valuable input on the study plans (no funding to disclose).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100883.
Appendix A. Supplementary data
Supplementary figures
References
- 1.George J., Lim J.S., Jang S.J., Cun Y., Ozretic L., Kong G., Leenders F., Lu X., Fernandez-Cuesta L., Bosco G. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sriuranpong V., Borges M.W., Ravi R.K., Arnold D.R., Nelkin B.D., Baylin S.B., Ball D.W. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–3205. [PubMed] [Google Scholar]
- 3.Augustyn A., Borromeo M., Wang T., Fujimoto J., Shao C., Dospoy P.D., Lee V., Tan C., Sullivan J.P., Larsen J.E. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14788–14793. doi: 10.1073/pnas.1410419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang T., Collins B.J., Jin N., Watkins D.N., Brock M.V., Matsui W., Nelkin B.D., Ball D.W. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69:845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders L.R., Bankovich A.J., Anderson W.C., Aujay M.A., Bheddah S., Black K., Desai R., Escarpe P.A., Hampl J., Laysang A. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farago A.F., Keane F.K. Current standards for clinical management of small cell lung cancer. Transl. Lung Cancer Res. 2018;7:69–79. doi: 10.21037/tlcr.2018.01.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 8.Horn L., Gettinger S.N., Gordon M.S., Herbst R.S., Gandhi L., Felip E., Sequist L.V., Spigel D.R., Antonia S.J., Balmanoukian A. Safety and clinical activity of atezolizumab monotherapy in metastatic non-small-cell lung cancer: final results from a phase I study. Eur. J. Cancer. 2018;101:201–209. doi: 10.1016/j.ejca.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Antonia S.J., Lopez-Martin J.A., Bendell J., Ott P.A., Taylor M., Eder J.P., Jager D., Pietanza M.C., Le D.T., de Braud F. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–895. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 10.Chung H.C., Ros W., Delord J.P., Perets R., Italiano A., Shapira-Frommer R., Manzuk L., Piha-Paul S.A., Xu L., Zeigenfuss S. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019;37:1470–1478. doi: 10.1200/JCO.18.01265. [DOI] [PubMed] [Google Scholar]
- 11.George J., Saito M., Tsuta K., Iwakawa R., Shiraishi K., Scheel A.H., Uchida S., Watanabe S.I., Nishikawa R., Noguchi M. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin. Cancer Res. 2017;23:1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultheis A.M., Scheel A.H., Ozretic L., George J., Thomas R.K., Hagemann T., Zander T., Wolf J., Buettner R. PD-L1 expression in small cell neuroendocrine carcinomas. Eur. J. Cancer. 2015;51:421–426. doi: 10.1016/j.ejca.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Keenan T.E., Burke K.P., Van Allen E.M. Genomic correlates of response to immune checkpoint blockade. Nat. Med. 2019;25:389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazdar A.F., Minna J.D. Small cell lung cancers made from scratch. J. Exp. Med. 2019;216:476–478. doi: 10.1084/jem.20182216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellmann M.D., Callahan M.K., Awad M.M., Calvo E., Ascierto P.A., Atmaca A., Rizvi N.A., Hirsch F.R., Selvaggi G., Szustakowski J.D. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33(853–861) doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellmann M.D., Nathanson T., Rizvi H., Creelan B.C., Sanchez-Vega F., Ahuja A., Ni A., Novik J.B., Mangarin L.M.B., Abu-Akeel M. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(843–852) doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casares N., Pequignot M.O., Tesniere A., Ghiringhelli F., Roux S., Chaput N., Schmitt E., Hamai A., Hervas-Stubbs S., Obeid M. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J. Exp. Med. 2005;202:1691–1701. doi: 10.1084/jem.20050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Gerber H.P., Sapra P., Loganzo F., May C. Combining antibody-drug conjugates and immune-mediated cancer therapy: what to expect? Biochem. Pharmacol. 2016;102:1–6. doi: 10.1016/j.bcp.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Rios-Doria J., Harper J., Rothstein R., Wetzel L., Chesebrough J., Marrero A., Chen C., Strout P., Mulgrew K., McGlinchey K. Antibody-drug conjugates bearing pyrrolobenzodiazepine or tubulysin payloads are immunomodulatory and synergize with multiple immunotherapies. Cancer Res. 2017;77:2686–2698. doi: 10.1158/0008-5472.CAN-16-2854. [DOI] [PubMed] [Google Scholar]
- 21.Carbone D.P., Morgensztern D., Le Moulec S., Santana-Davila R., Ready N., Hann C.L. Efficacy and safety of rovalpituzumab tesirine in patients with DLL3-expressing, >= 3rd line small cell lung cancer: results from the phase 2 TRINITY study. J. Clin. Oncol. 2018;36 doi: 10.1158/1078-0432.CCR-19-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Release P. 2018. Phase 3 Trial of Rova-T as Second-line Therapy for Advanced Small-cell Lung Cancer (TAHOE Study) Halted (AbbVie) [Google Scholar]
- 23.Jackson P.J.M., Kay S., Pysz I., Thurston D.E. Use of pyrrolobenzodiazepines and related covalent-binding DNA-interactive molecules as ADC payloads: Is mechanism related to systemic toxicity? Drug Discov. Today Technol. 2018;30:71–83. doi: 10.1016/j.ddtec.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Malhotra J., Nikolinakos P., Leal T., Lehman J., Morgensztern D., Patel J.D., Wrangle J.M., Curigliano G., Dansin E., Greillier L. Ph1/2 study of Rova-T in combination with nivolumab (Nivo) +/− ipilimumab (Ipi) for patients (pts) with 2L+extensive-stage (ED) SCLC. J Clin Oncol. 2019;37 doi: 10.1016/j.jtho.2021.02.022. [DOI] [PubMed] [Google Scholar]
- 25.Rudin C.M., Pietanza M.C., Bauer T.M., Ready N., Morgensztern D., Glisson B.S., Byers L.A., Johnson M.L., Burris H.A., 3rd, Robert F. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park K.S., Liang M.C., Raiser D.M., Zamponi R., Roach R.R., Curtis S.J., Walton Z., Schaffer B.E., Roake C.M., Zmoos A.F. Characterization of the cell of origin for small cell lung cancer. Cell Cycle. 2011;10:2806–2815. doi: 10.4161/cc.10.16.17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen T., Rodriguez B.L., Chen L., Corte C.M.D., Morikawa N., Fujimoto J., Cristea S., Nguyen T., Diao L., Li L. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9:646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galluzzi L., Buque A., Kepp O., Zitvogel L., Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 29.Iwata T.N., Ishii C., Ishida S., Ogitani Y., Wada T., Agatsuma T. A HER2-targeting antibody-drug conjugate, trastuzumab deruxtecan (DS-8201a), enhances antitumor immunity in a mouse model. Mol. Cancer Ther. 2018;17:1494–1503. doi: 10.1158/1535-7163.MCT-17-0749. [DOI] [PubMed] [Google Scholar]
- 30.Konermann A., Deschner J., Allam J.P., Novak N., Winter J., Baader S.L., Jepsen S., Jager A. Antigen-presenting cell marker expression and phagocytotic activity in periodontal ligament cells. J. Oral Pathol. Med. 2012;41:340–347. doi: 10.1111/j.1600-0714.2011.01086.x. [DOI] [PubMed] [Google Scholar]
- 31.Lacour M., Hiltbrunner S., Lee S.Y., Soltermann A., Rushing E.J., Soldini D., Weder W., Curioni-Fontecedro A. Adjuvant chemotherapy increases programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer recurrence. Clin. Lung Cancer. 2019;20:391–396. doi: 10.1016/j.cllc.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Cullen S.P., Martin S.J. Mechanisms of granule-dependent killing. Cell Death Differ. 2008;15:251–262. doi: 10.1038/sj.cdd.4402244. [DOI] [PubMed] [Google Scholar]
- 33.McEver R.P. Selectins: initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc. Res. 2015;107:331–339. doi: 10.1093/cvr/cvv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cesano A. nCounter((R)) PanCancer Immune Profiling Panel (NanoString Technologies, Inc., Seattle, WA) J Immunother Cancer. 2015;3:42. doi: 10.1186/s40425-015-0088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J., Wang G., Chen Y., Wang H., Hua Y., Cai Z. Immunogenic cell death in cancer therapy: present and emerging inducers. J. Cell. Mol. Med. 2019;23:4854–4865. doi: 10.1111/jcmm.14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hato S.V., Khong A., de Vries I.J., Lesterhuis W.J. Molecular pathways: the immunogenic effects of platinum-based chemotherapeutics. Clin. Cancer Res. 2014;20:2831–2837. doi: 10.1158/1078-0432.CCR-13-3141. [DOI] [PubMed] [Google Scholar]
- 37.McCoy M.J., Lake R.A., van der Most R.G., Dick I.M., Nowak A.K. Post-chemotherapy T-cell recovery is a marker of improved survival in patients with advanced thoracic malignancies. Br. J. Cancer. 2012;107:1107–1115. doi: 10.1038/bjc.2012.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park K.S., Martelotto L.G., Peifer M., Sos M.L., Karnezis A.N., Mahjoub M.R., Bernard K., Conklin J.F., Szczepny A., Yuan J. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat. Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baize N., Monnet I., Greillier L., Quere G., Kerjouan M., Janicot H., Vergnenegre A., Auliac J.B., Chouaid C. Second-line treatments of small-cell lung cancers. Expert. Rev. Anticancer. Ther. 2017;17(11):1033–1043. doi: 10.1080/14737140.2017.1372198. [DOI] [PubMed] [Google Scholar]
- 43.Ritchie M.E., Phipson B., Wu D.I., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 45.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Core Team . 2017. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures