Abstract
Ras mutations are present in only a subset of sporadic human cutaneous squamous cell carcinomas (cSCC) even though Ras is activated in most. This suggests that other mechanisms of Ras activation play a role in the disease. The aberrant expression of RasGRP1, a guanyl nucleotide exchange factor for Ras, is critical for mouse cSCC development through its ability to increase Ras activity. However, the role of RasGRP1 in human keratinocyte carcinogenesis remains unknown. Here we report that RasGRP1 is significantly elevated in human cSCC and that high RasGRP1 expression in human primary keratinocytes triggered activation of endogenous Ras and significant morphological changes including cytoplasmic vacuole formation and growth arrest. Moreover, RasGRP1-expressing cells were autophagic as indicated by LC3-II increase and the formation of LC3 punctae. In an in vitro organotypic skin model, wild type keratinocytes generated a well-stratified epithelium, while RasGRP1-expressing cells failed to do so. Finally, RasGRP1 induced transformation-like changes in skin cells from Li-Fraumeni patients with inactivating p53 mutations, demonstrating the oncogenic potential of this protein. These results support a role for RasGRP1 in human epidermal keratinocyte carcinogenesis and might serve as an important new therapeutic target.
Abbreviations: cSCC, human cutaneous squamous cell carcinomas; DAG, diacylglycerol; GEF, guanine nucleotide exchange factor; GFP, Green Fluorescent Protein; GST, Glutathione-S-Transferase; GTP, Guanosine - 5′- Tri-Phosphate; EGFR, Epidermal Growth Factor Receptor; EMT, epithelial to mesenchymal transition; ERK, Extracellular Signa-Regulated Kinase; FBS, Fetal Bovine Serum; HKn, newborn-derived primary Human Keratinocytes; LC3, microtubule-associated protein Light Chain 3; KEGG, Kyoto Encyclopedia of Genes and Genomes; LiF, Li-Fraumeni Syndrome; PARP, Poly ADP (Adenosine Diphosphate)-Ribose Polymerase; PBS, Phosphate Buffered Saline; Ras, Rat Sarcoma (GTPase); RasGRP1, Ras guanyl nucleotide-releasing protein 1
Introduction
The Ras proto-oncogene family can activate transformation of epidermal keratinocytes leading to cutaneous squamous cell carcinoma (cSCC) [1,2], a prevalent form of non-melanoma skin cancer [3]. The aberrant function of Ras and Ras signaling pathways is one of the most prevalent transformative events in human cancer with approximately 30% of human cancers having constitutively activating mutations in the Ras small GTPases themselves, in addition to frequent aberrations in other nodes of the Ras pathways [4]. Ras mutations have only been identified in a subset of sporadic human SCC samples, despite the fact that Ras is activated in the majority of human SCC [1,5], suggesting that other mechanisms of Ras activation play a role in the disease. In the skin, biochemical activation of wild type Ras by deregulated receptor tyrosine kinases, like EGFR [[6], [7], [8]] or increased secretion of growth factors [9,10] add to the oncogenic effects of mutant Ras, making it difficult to successfully block the Ras oncogenic signal. So far, no drugs targeting Ras activity itself are available for clinical use and therefore new targets in the Ras pathways may prove valuable alternatives [11].
The biochemical activation of Ras requires GTP loading catalyzed by guanine nucleotide exchange factors (GEFs) [12]. Therefore, identifying and targeting the GEFs that mediate aberrant activation of wild type Ras in different tumors can provide another molecular approach to control Ras downstream signaling and subsequent carcinogenesis [13,14]. The RasGRP1 Ras-GEF is abundantly expressed in lymphocytes, regulating their maturation and activation [[15], [16], [17], [18], [19]]. Analysis of transgenic mice overexpressing RasGRP1 in lymphocytes has shown that deregulated RasGRP1 expression leads to development of thymic lymphomas and T-cell leukemias [20]. Furthermore, leukemia models initiated by Ras hyperactivation depend upon RasGRP1 upregulation in the development of resistance to MEK inhibitor treatment [21]. RasGRP1 is also expressed in epidermal keratinocytes [17,22]. In the skin, RasGRP1 can increase the amount of active normal Ras by diacylglycerol analog-induced Ras activation and transgenic mice overexpressing RasGRP1 form spontaneous cutaneous squamous cell carcinoma (cSCC) [[23], [24], [25], [26]]. Moreover, RasGRP1 null-mutant mice, that do not show overt homeostatic changes in the skin, are significantly resistant to the development of Ras mutation-dependent skin tumors [27]. In contrast, EGFR-RasGRP1 signals have been shown to oppose EGFR-SOS proliferative signals in the intestinal epithelium and thus RasGRP1 can have a negative regulatory role in colorectal cancers [28]. Indeed, RasGRP1 mRNA levels were found to be low in colorectal cancer cell lines and patient samples. To understand the potential role of RasGRP1 in human skin carcinogenesis it is therefore essential to determine its effects in a human skin model.
In this study, we first assessed RasGRP1 over-expression in human cSCC by mining of genome-wide mRNA expression datasets in the public domain. Next, we analyzed the biological effects of RasGRP1 in human keratinocytes, utilizing primary cells as the experimental model. We found that in 3D keratinocyte culture, an in vitro skin model, newborn-derived primary keratinocytes (HKn) overexpressing RasGRP1 underwent growth arrest and autophagy and failed at generating a stratified epithelium. Additional mining yielded supporting evidence for a role for autophagy in human cSCC. Furthermore, enforced RasGRP1 expression induced cellular transformation-like changes in skin cells with p53 suppression, demonstrating the oncogenic potential of this protein. These results reveal a novel role for RasGRP1 in human keratinocyte transformation and justify further exploration of RasGRP1 involvement in Ras-induced transformation in human keratinocytes.
Materials and methods
Primary keratinocyte culture
Primary human epidermal keratinocytes were isolated from neonatal foreskins as previously described [29,30] with modifications. All work with human foreskin keratinocytes was reviewed and approved by the Institutional Review Board (IRB) at Hawaii Pacific Health (RP#06-030-1-HPH2NA). Only anonymous, discarded neonatal foreskins were used. Briefly, newborn foreskins were placed in defined keratinocyte serum-free medium (K-sfm) (Invitrogen, Grand Island, NY) with gentamycin (20 μg/ml) immediately following circumcision. Tissue was cleaned with 70% ethanol and subcutaneous fatty layers were removed. Skin was then cut into 3 mm2 pieces and submerged in dispase (BD Biosciences, San Jose, CA) at 4ᴏC overnight. The following day, the epidermis was mechanically separated from the dermis, minced with a sterile razor blade and placed in 0.05% Trypsin-EDTA at 37ᴏC for 12 min with agitation to dislodge the keratinocytes. Trypsin was neutralized with DMEM containing 10% FBS, and cells were collected by centrifugation, re-suspended in complete media and filtered to remove large debris. The keratinocytes were then plated on culture-treated flasks in K-sfm at 36 ᴏC in a humidified atmosphere of 6% CO2 in air. Validated human epidermal keratinocytes from adult skin (HKn) were purchased from Invitrogen (#C-005-5C) and cultured in Basal Medium 154 supplemented with HKGS (#S-001-K) according to the manufacturer's instructions. All keratinocytes were used only until passage 7 in these experiments.
LiF-Ep epidermal keratinocytes
LiF-Ep primary epidermal keratinocytes were purchased from the Rheinwald laboratory (Harvard Medical School, Boston, MA). These cells were isolated from the normal skin of a patient with Li-Fraumeni Syndrome and therefore harbor a heterozygous p53 (p53−/+) mutation that causes lack of function of p53. Cells were cultured as previously described [31]. Briefly, LiF-Ep cells were cultured in K-sfm (GIBCO, Invitrogen) plus the following reagents at final concentrations: bovine pituitary extract supplied with K-sfm (25 μg/ml), EGF (0.2 ng/ml), CaCl2 (0.4 mM). Cells were cultured at 36 °C in a humidified atmosphere of 6% CO2 in air.
Three-dimensional organotypic cultures
Organotypic cultures as a 3D model for skin were prepared as previously described, with modifications [29,30]. Briefly, early passage human dermal fibroblasts were added to neutralized type I collagen (Organogenesis, Canton, MA) to a final concentration of 2.75 × 104 cells/ml. Three milliliter of this mixture were added to each well of a 6-well plate and incubated for 4 to 6 days while contraction of the cellular matrix occurred. 5 × 105 normal or adenoviral-transduced keratinocytes were then plated on the contracted collagen gel. Cultures were maintained submerged for 2 days in normal‑calcium (1.8 mM) epidermal growth medium that contained a 3:1 mixture of DMEM and Ham's F12 (VWR, Radnor, PA), 4 mM l-glutamine (Invitrogen), 5.4 μg/ml hydrocortisone (Sigma-Aldrich, St. Louis, MO), ITES (10 μg/ml insulin, 10 μg/ml transferrin, 10 mM ethanolamine, and 10 ng/ml selenium final concentration) (Lonza, Walkersville, MD), O-phosphorylethanolamine, CaCl2, triiodothyronine, progesterone, adenine (Sigma-Aldrich) and 0.1% chelated FBS (Invitrogen). After 2 days, cultures were maintained with epidermal growth medium consisting of a 3:1 mixture of DMEM and Ham's F12 containing all of the above supplements and 0.1% FBS for an additional 2 days. Cultures were raised to the air-liquid interface for 5 days. While grown at the interface, tissues were fed from below with a 1:1 mixture of DMEM and Ham's F12 containing 2% FBS with all the above supplements except progesterone. At the end of the protocol, skin tissue cultures were harvested, fixed with formalin for 6 h, transferred to 70% ethanol, and paraffin-embedded following standard histological techniques (Histology Core Facility, John A. Burns School of Medicine, University of Hawai'i at Mānoa, Honolulu, HI). Hemotoxylin and Eosin (H&E) stained slides were used for histopathological analysis.
Adenoviral vectors
Adenoviral expression approaches were utilized for enforced expression of RasGRP1, oncogenic H-RAS (RasQ61L) or the control GFP protein. Recombinant adenoviral vectors encoding rat RasGRP1 were generated with the Transpose-Ad system (Qbiogene, Irvine, CA) as described [22]. Similarly, Q61L active mutant H-Ras cDNA (Upstate Biotechnology, Lake Placid, NY) was used to create a recombinant adenovirus for the expression of oncogenic H-Ras. Adenovirus expressing the GFP protein (GFP) was obtained from Qbiogene. Adenoviral transduction was performed as previously described [22]. Briefly, adenoviruses were added to keratinocytes plated on 60-mm dishes at the appropriate multiplicity of infection in 1 ml medium. Following a 4 h incubation at 36 °C, 3 ml of warm K-sfm was added and the cells were cultured for an additional 24–94 h.
Immunoblot analysis
Primary human epidermal keratinocytes were harvested in lysis buffer 25 mM Tris-HCl (pH 7.4) containing 150 mM NaCl, 5 mM MgCl2, 1% IGEPAL, 5% glycerol, 1 mM Na2VO4, 25 mM NaF, Mini-Complete protease inhibitor and PhosSTOP phosphatase inhibitor (Roche Applied Science, Indianapolis, IN). For immunoblot analysis, 50 μg of whole-cell extract protein were loaded per lane. The primary antibodies used in this study were as follows: Pan-Ras clone RAS10 (#OP40) (Calbiochem, Darmstadt, Germany); LC3B (D11) (#3868), p44/p42 MAPK (ERK) (#9102), phospho-p44/42 MAPK Thr202/Tyr204 ERK1, Thr185/Tyr187 ERK2 (p-ERK) (#9101), Cleaved Caspase-3 (Asp175) (#9664), phospho-p53 (Ser15) (#9284), total p53 (#2527) (Cell Signaling Technology, Danvers, MA); RasGRP1 m199 (#sc-8430), (Santa Cruz Biotechnologies, Santa Cruz, CA); β-Actin (#A-2066), (Sigma-Aldrich, St. Louis, MO); hRasGRP1 (#AP5856c) (Abgent, San Diego, CA); and HRas (C-term) (#1521-1), (Epitomics, Burlingame, CA). β-actin was used as a loading control; the levels of protein expression were analyzed using UN-SCAN-IT (Silk Scientific, Orem, Utah).
Ras activation pull-down assay
Levels of active Ras (RasGTP) were measured by using the glutathione s-transferase-epitope tagged Ras binding domain (GST-RBD) of Raf-1 as a probe in a pull-down assay with 500 μg lysate protein as previously described [22]. Total lysate and pull-down samples were run in parallel and used for Western blotting with Pan-Ras (RAS10), H-Ras (C-term), p-ERK and total ERK antibodies.
Cell viability assay
HKn were plated in 96-well plates at 10,000 cells/well, and transduced with adenovirus containing GFP, RasGRP1 or H-RasQ61L. XTT Assay (Roche Applied Science) was performed 24 h, 48 h, 72 h and 96 h post transduction according to the manufacturer's instructions. Briefly, 50 μl of XTT labeling reagent containing electron coupling reagent was added to the wells and cells were incubated at 37 °C for 4 h. Absorbance was measured using the EnVision Multilabel Plate Reader (Perkin-Elmer, Waltham, MA) at 450 nm with a reference wavelength at 650 nm.
ApoTox-Glo triplex assay
HKn were plated in opaque 96-well plates at 10,000 cells/well, and transduced with adenovirus containing GFP, RasGRP1 or HRasQ61L. ApoTox-Glo Triplex Assay was performed 24 h, 48 h, 72 h and 96 h post transduction according to the Technical Manual (Promega, Madison, WI). Briefly, 20 μl of Viability/Cytotoxicity Reagent containing GF-AFC and bis-AAF-R110 Substrates was added to each well and cells were incubated for 1 h at 36 °C. Fluorescence was measured at 400 nm excitation/505 nm emission for viability and 485 nm excitation/520 nm emission for cytotoxicity. 100 μl of Caspase-Glo 3/7 Reagent was then added to each well and cells were incubated for 1 h at room temperature before luminescence was recorded. Fluorescence and luminescence were read using the EnVision Multilabel Plate Reader (Perkin-Elmer).
Propidium iodide staining for cell cycle analysis
HKn transduced with GFP, RasGRP1 or HRasQ61L were washed, fixed in 100% cold ethanol and stained with Propidium Iodide (PI) 72 or 96 h post transduction Cell cycle analysis was performed via flow cytometry (BD FACScalibur) and data was analyzed with FlowJo_V10 software.
Analysis of autophagic flux
HKn were transduced with respective adenoviruses, and 96 h later treated with 400 nM bafilomycin A1 (a vacuolar H+ ATPase inhibitor that prevents lysosomal degradation of autophagosome and inhibits late stage autophagy) (Tocris, Bristol, UK) for 4 h at 36 ᴏC. Cells were then harvested, and protein lysates were collected as previously described. Untreated controls for each group were run in parallel and immunoblot analysis for LC3-I/II expression was performed.
Immunofluorescence
For the immunofluorescence studies, HKn were seeded onto Permanox chamber slides and transduced 24 h later with the GFP, RasGRP1 or FLAG-tagged H-RasQ61L adenoviral constructs as described above. Immunofluorescence staining was performed as previously described [32]. Both fixed and live cells were viewed with an Olympus IX81 containing a QImaging CCD monochrome camera.
Transwell invasion assay
HKn transduced with GFP, RasGRP1 or HRasQ61L for 72 h were trypsinized and plated into the top of the transwell inserts (Greiner Bio One, Monroe, NC) coated with matrigel. Inserts were placed into a 24 well plate with the lower chamber containing K-sfm with EGF (0.3 ng/ml) for 24 h. Migrated cells were detached from the bottom part of the insert and stained with Calcein-AM (Sigma). Stained cells were then measured using a plate reader (Perkin-Elmer) and invasion was calculated as fold change between GFP transduced cells and RasGRP1 or HRasQ61L transduced cells.
Quantitative real-time polymerase chain reaction analysis of epithelial to mesenchymal transition
HKn transduced with GFP, RasGRP1 or HRasQ61L were harvested and total RNA was extracted using Trizol reagent (Invitrogen) according to manufacturer's protocol. The cDNA was reverse-transcribed from 2 μg of total RNA using ProtoScript II Reverse Transcriptase kit (New England Biolabs, Ipswich, MA). Quantitative real-time PCR was performed with SYBR green PCR master mix (Applied Biosystem, Foster City, CA). The C(t) values were normalized using GAPDH. Primers used were; Snail (FWD-AGACGAGGACAGTGGGAAAG, REV-AGATCCTTGGCCTCAGAGAG), E-cadherin (FWD-GTCAGGTGCCTGAGAACGAG, REV-GCCATCGTTGTTCACTGGAT), Vimentin (FWD-GAACTTTGCCGTTGAAGCTG, REV-TCTCAATGTCAAGGGCCATC), GAPDH (FWD-GTCTCCTCTGACTTCAACAGCG, REV-ACCACCCTGTTGCTGTAGCCAA).
Senescence assay
Cells in cultured in a 96 well plate were analyzed for senescence using the β-galactosidase assay according to manufacturer's protocol (Cell Signaling Technology, Danver, MA). Briefly, cells were fixed and incubated overnight with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in DMF at 37 °C. After PBS washing, cells were lysed (0.25% SDS, 10 mM Tris-HCl pH 7.6) and senescence determined by plate reader by relative enzyme activity absorbance at 650 nm wavelength for the X-Gal hydrolysis product.
Transmission electron microscopy
Transduced cells were washed 2× with DPBS, tripsinized and pelleted at 2000 rpm for 5 min. Cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 and 2 mM CaCl2 for 1–2 h at room temperature. Following fixation, cell pellets were washed twice in 0.1 M cacodylate buffer for a total of 20–30 min, then post-fixed with 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h at room temperature. Specimens were dehydrated in a graded ethanol series (30%, 50%, 70%, 85%, 95%), 2–3 changes of 3–5 min for each dilution, followed by dehydration in 100% ethanol, with 3 changes of 10 min each. Cell pellets were then treated with propylene oxide, with 3 changes of 10 min each, and infiltrated with 1:1 propylene oxide:epoxy resin overnight. Samples were immersed in freshly prepared 100% epoxy resin for 2–8 h followed by a second immersion in freshly prepared 100% epoxy resin for 1–4 h. Immersed samples were placed in a mold with epoxy resin and allowed to polymerize at 60 °C for 2–3 days. The electron micrographs were taken with a Hitachi HT7700 all-digital 120 kV transmission electron microscopy with an AMT 2 k × 2 k CCD camera (Pacific Biosciences Research Center, University of Hawaii at Mānoa, Biological Electron Microscope Facility).
Public human cSCC mRNA expression dataset analysis
mRNA expression data from the public genome-wide datasets Nindl-15 (Affymetrix U133A array; GSE2503) and Riker-87 (Affymetrix U133P2 array; GSE7553) containing human cSCC and normal skin samples were retrieved from the NCBI Gene Expression Omnibus (GEO) site (http://www.ncbi.nlm.nih.gov/geo/). The R2 TranscriptView genomic analysis and visualization tool was used to check if Affymetrix probe-sets had a unique anti-sense position in an exon of the gene. R2 was developed in the Department of Oncogenomics at the Academic Medical Center – University of Amsterdam, the Netherlands and is accessible at http://r2.amc.nl. All probe-sets selected meet these criteria. The probe-set for RasGRP1 (205590_at) is directed against its 3’ UTR and thereby detects both RasGRP1 variants. All expression values and other details for the datasets used can be obtained thru their GSE number on the NCBI GEO website.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism V software. P values for the differential mRNA expression in the human cSCC datasets were calculated using the non-parametric Kruskal-Wallis test (for RasGRP1) or a 2log Student t-test (for all genes). P values for the in vitro experiments were calculated by Student t-test, One-Way, or Two-way ANOVA analysis as indicated. For all tests, P < 0.05 was considered statistically significant.
Results
RasGRP1 mRNA is overexpressed in human cSCC
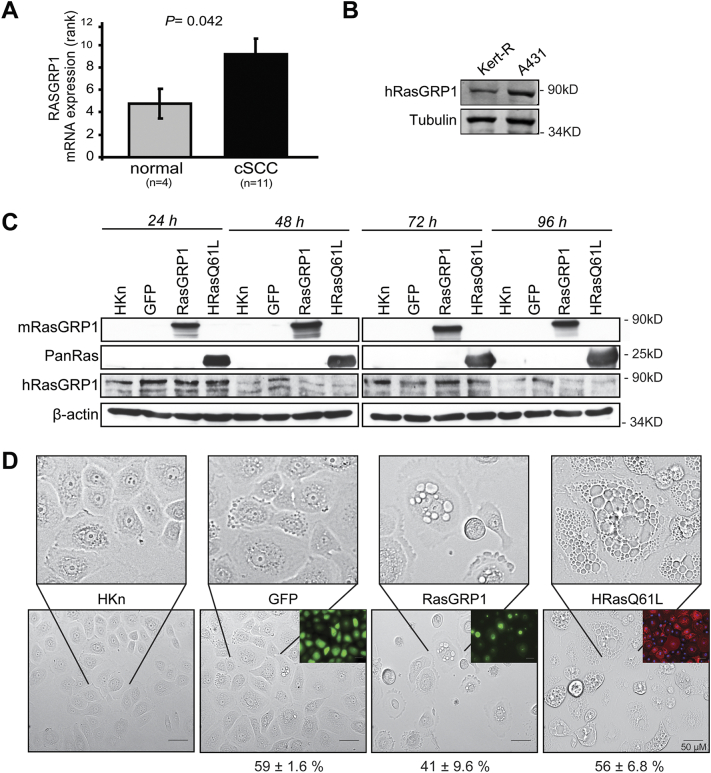
Because of the reported effects of RasGRP1 in skin carcinogenesis in the mouse, we examined its expression in human cutaneous squamous cell carcinoma (cSCC) samples by datamining of genome-wide mRNA expression datasets in the public domain (see Materials and Methods for details). Cutaneous SCC is under-represented among human cancer studies, in contrast to other skin cancer types such as melanoma. However, the Riker-87 skin cancer set [33] allowed a comparison of 11 cSCC versus 5 normal skin samples. RasGRP1 mRNA expression was significantly higher in cSCC than in normal skin (Fig. 1A), in support of a role for RasGRP1 in this human cancer. In additional comparisons in this dataset, we found that most Ras isoforms (i.e. HRas, KRas, NRas, and RRas2) were similarly significantly over-expressed in cSCC (Supplemental Table 1). This result, expected based on the literature [1,5], supports the validity of this datamining strategy. We further compared RasGRP1 protein expression in human keratinocytes to that in a human SCC cell line using immunoblotting. We find as in the mRNA analysis, RasGRP1 protein showed increased expression in human SCC (Fig. 1B).
Fig. 1.
RasGRP1 expression in human cSCC and normal skin and effects on human keratinocyte morphology. (A) Analysis of RasGRP1 mRNA expression in the Riker-87 dataset shows that RasGRP1 is significantly higher expressed in cSCC (11 samples) than in normal skin (4 samples). The results shown are for a rank-based Kruskal-Wallis test to prevent undue influence by very high or very low mRNA expression values. RasGRP1 also shows significant overexpression using 2log (3,21 ± 0,14 > 2,61 ± 0,21; P = 0,040) and normal t-tests (10,50 ± 0,84 > 6,95 ± 1,02; P = 0,039). (B) Immunoblot of endogenous RasGRP1 expression was determined for 1 keratinocyte and 1 human cSCC cell line. (C) Immunoblot analysis confirming the exogenous expression of mouse RasGRP1 (mRG1, using M199 antibody) and Ras in human keratinocytes (HKn) transduced with adenovirus at the indicated times post transduction. Expression of endogenous human RasGRP1 (hRG1) was also determined. (D) HKn were transduced with adenovirus encoding GFP, GFP-RasGRP1, or HRasQ61L. Images taken 72 h post transduction at 200× magnification (Olympus Microscope). Insets show transduction efficiency for GFP, RasGRP1, and HRasQ61L via fluorescence and staining. Scale bar = 50 μm. Percent of GFP positive transduced cells is indicated at the bottom.
High RasGRP1 induced changes in human keratinocyte morphology
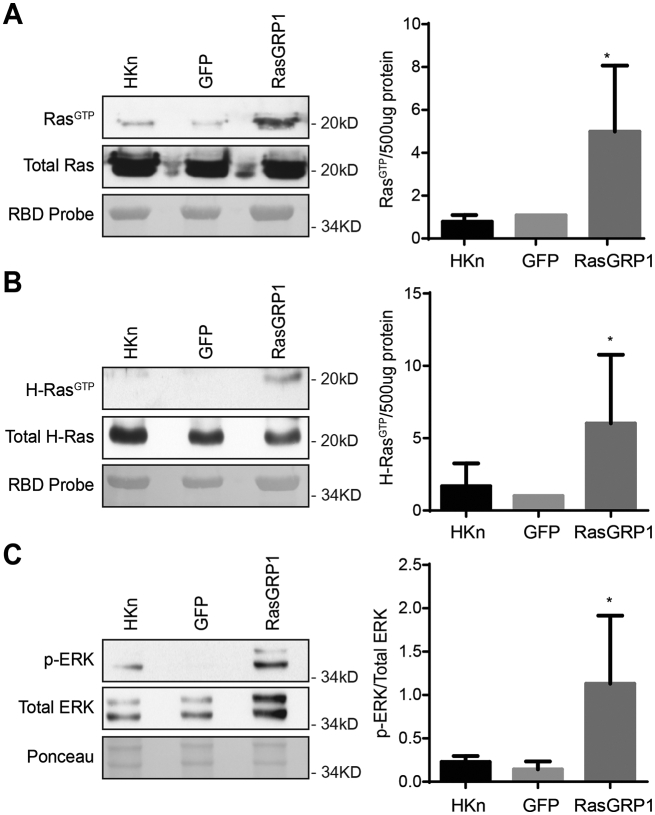
To study the role of RasGRP1 in human keratinocyte biology, we expressed GFP-tagged RasGRP1 using adenovirus as previously described [22]. Since RasGRP1 exerts its known effects through the activation of Ras, for comparison we also expressed oncogenic HRas (HRasQ61L) using adenovirus to determine if the effects of RasGRP1 mimicked those of active HRas as would be expected if RasGRP1 works via its GEF activity on endogenous Ras. To control for non-specific effects resulting from adenoviral transduction or GFP expression, adenovirus encoding GFP alone was transduced in parallel. Adenovirus transduction specifically and strongly induced exogenous expression of RasGRP1 in human keratinocytes (Fig. 1C). High expression of RasGRP1 and HRasQ61L caused significant changes in keratinocyte morphology, with induction of cytoplasmic vacuolization and cell flattening, as well as the appearance of apoptotic-like bodies (Fig. 1D). RasGRP1+ cells (HKn transduced with RasGRP1) cultured for longer periods of time (up to 11 days) showed more extensive flattening and vacuolization and some cell rounding. Activated Ras was previously reported to cause micropinocytosis vacuolization [34,35]. The expressed RasGRP1 was active and promoted Ras GTP loading and phosphorylation of ERK (Fig. 2A–C).
Fig. 2.
RasGRP1 expression led to Ras activation. Human Keratinocytes (HKn) were transduced with GFP or RasGRP1. RasGTP levels were measured by Ras activation pull-down assay. Immunoblot analysis for (A) PanRas (clone 10) and (B) H-Ras were performed to determine activation of specific Ras isoforms. Densitometry analysis of active protein normalized to 500 μg protein, relative to GFP. (C) Immunoblot analysis for p-ERK and total ERK further confirm activation of the MAPK pathway. Densitometry analysis of p-ERK protein normalized to total ERK. Results shown are from a representative of ≥5 independent experiments. Statistical analysis performed using unpaired t-test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, between GFP and RasGRP1.
High RasGRP1 induced growth arrest and reduced cell motility
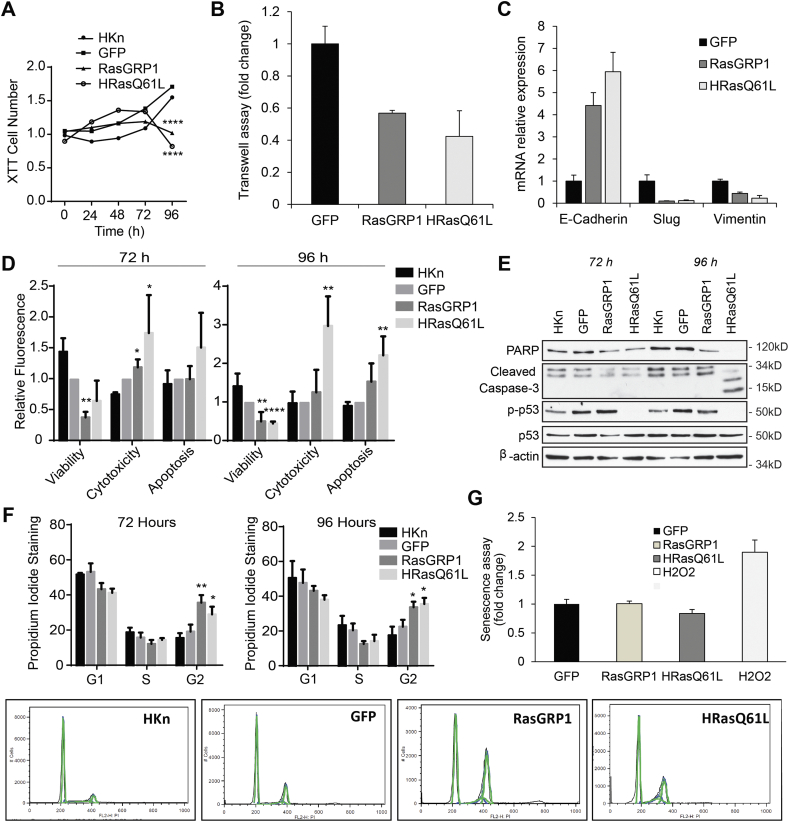
The morphological changes induced by RasGRP1 indicated a possible role in keratinocyte cell cycle arrest, cell death or cell motility. While wild type and GFP-expressing HKn cells proliferated normally, cell numbers significantly declined in RasGRP1+ and oncogenic Ras (HRasQ61L) keratinocytes at 96 h post-transduction (Fig. 3A). Cell motility significantly decreased in RasGRP1+ and HRasQ61L keratinocytes compared to GFP controls (Fig. 3B). In addition, mRNA expression of the EMT marker E-Cadherin was induced while Slug and Vimentin were decreased in RasGRP1 and HRasQ61L keratinocytes at 96 h post-transduction (Fig. 3C). We further assessed the viability, cytotoxicity as well as necrotic and apoptotic events in response to RasGRP1 overexpression in comparison to RasQ61L. Forced RasGRP1 expression led to a significant decrease in cell number by 72 and 96 h. No increase in cytotoxicity or apoptosis could be found in RasGRP1+ cells even at 96 h post-transduction whereas both were significantly higher in RasQ61L-expressing keratinocytes (Fig. 3D). In addition, no increased apoptosis was detected when RasGRP1-expressing cells were analyzed for Caspase-3 or PARP cleavage whereas HRasQ61L induced clear Caspase-3 and PARP cleavage at 96 h (Fig. 3E). Interestingly whereas RasGRP1 upregulated phosphorylation of p53 at ser15, oncogenic HRasQ61L downregulated phosphorylation at this site. These data suggested that the decrease in cell number for RasGRP1 may be the result of cell cycle arrest. When RasGRP1+ cells were analyzed for DNA content at 72 h post-transduction, they showed a modest decrease in cells in G1 and a significant increase in cells arrested in G2, in comparison to the controls (Fig. 3F). This suggested that RasGRP1 overexpression did not initially induce apoptosis, but instead prevented cells from exiting G2 and entering G1 for the next round of cell division. The G2-arrested cells might then undergo senescence or activate autophagy as a failsafe mechanism. We found no evidence of activation of senescence using a β galactosidase assay (Fig. 3G).
Fig. 3.
RasGRP1 expression led to cell cycle arrest in Human keratinocytes (HKn) whereas oncogenic Ras activated apoptosis. (A) HKn were transduced with GFP or RasGRP1, and cell numbers were measured by XTT staining (Roche) at the indicated times post transduction. Data represent the means of four technical replicates and are representative of four biological replicates. Statistical analysis was done using two-way ANOVA followed by a Tukey test. ****, P < 0.0001, between GFP and RasGRP1. (B) HKn cells were transfected as shown and the ability of the cells to invade through Matrigel across a membrane was determined by staining and counting cells that traversed the membrane using a Calcein-AM assay. (C) Epithelial to mesenchymal transition of HKn cells was assayed by measuring changes in mRNA expression of E-Cadherin, Slug, and Vimentin. (D) Viability, cytotoxicity and Caspase-3/7 activity were all measured using the ApoTox-Glo Triplex Assay (Promega) in HKn at 72 and 96 h post transduction. For each sample, the average background (no cell media control) was subtracted from each technical replicate (n = 4). Data shown are representative of ≥3 independent biological replicates. Cytotoxicity and apoptosis values were normalized to viability values for each sample. Statistical analysis was performed using an unpaired t-test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, between GFP and RasGRP1. (E) RasGRP1 overexpression did not trigger apoptosis. HKn were harvested at the indicated time points post transduction. Immunoblot analysis for PARP, Cleaved Caspase-3, and phospho-p53 (ser15) was performed. Results shown are from a representative experiment of at least three independent experiments. (F) Propidium iodide staining was performed at 72 and 96 h post transduction and samples were analyzed by flow cytometry. Raw flow cytometry data from one experiment is shown at bottom. Cell cycle phase gates were drawn as approximations of the Dean-Jett-Fox cell cycle modeling algorithm and data plotted according to phase as a bar graph. Statistical analysis was performed using an unpaired t-test (n = 3). ***, P ≤ 0.001, between GFP and RasGRP1. (G) β galactosidase assay showing no initiation of senescence by RasGRP1 or HRas. H2O2 is a positive control.
RasGRP1 activity triggered autophagy
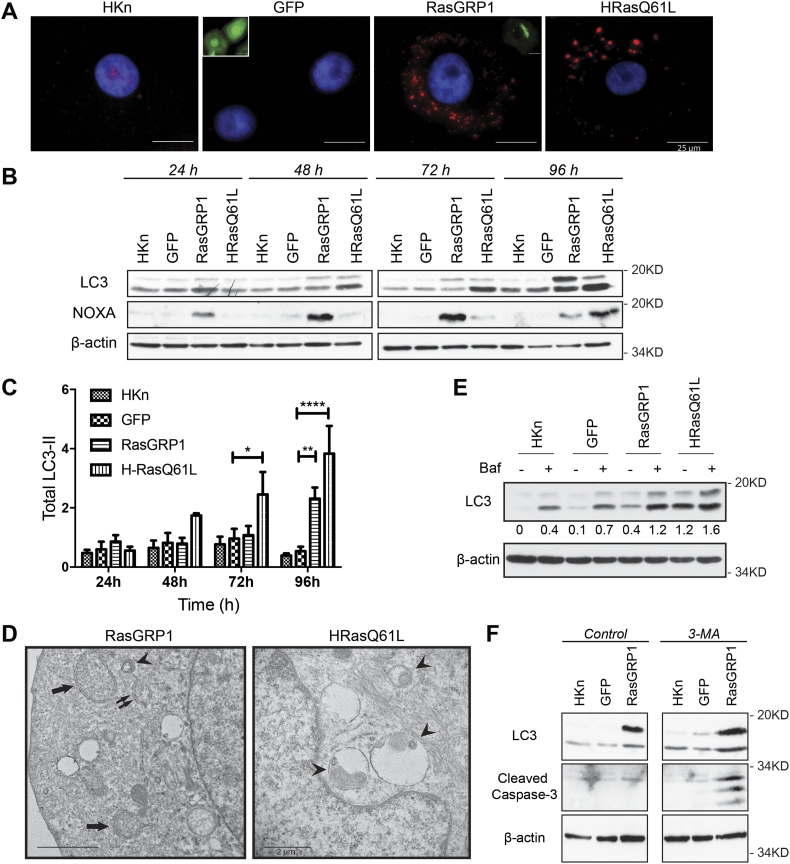
We next determined if RasGRP1 overexpression triggered autophagy in HKn cells. Ras pathways can activate autophagy in various human cell types [36,37]. One cellular hallmark of autophagy is a change in the lipidation and subsequent aggregation of the microtubule-associated protein light chain 3 (LC3). LC3 has two forms, a cytoplasmic soluble form (LC3-I) and a lipidated form that is associated with autophagosomes (LC3-II) [38]. In healthy cells, LC3 is expressed in a diffuse pattern. During autophagy, LC3-II levels increase, and show a distinctive LC3-II punctuate pattern due to localization in the autophagosomes [39]. We observed this pattern in the RasGRP1-expressing and HRasQ61L keratinocytes but not in the control cells (Fig. 4A). In addition, we investigated LC3-I to LC3-II conversion and mobility shift by immunoblot analysis. RasGRP1+ and HRasQ61L keratinocytes displayed a significant increase in LC3-II levels at 72 h and 96 h post transduction (Fig. 4B, C). Interestingly, this was accompanied by induction of NOXA, which has been previously shown to promote autophagy downstream of oncogenic Ras by displacement of Mcl-1 from Beclin [40]. Electron microscopy was used as a third method to confirm the presence of autophagic vacuoles that indicate cytosolic department degradation [39]. Indeed, upon RasGRP1 transduction, elongated isolation membranes (double arrows), double-membrane autophagosomes (arrows), as well as autophagosome-lysosome fusion vesicles (autolysosomes; arrowheads) were detected (Fig. 4D). Non-autophagic vacuoles of only a single membrane were also detected. The presence of autophagic vacuoles together with an induction of LC3-II expression indicated that enforced expression of RasGRP1 triggered autophagy-associated changes in HKn cells.
Fig. 4.
RasGRP1 expression triggered autophagy in Human keratinocytes (HKn). (A) HKn expressing GFP, RasGRP1, or oncogenic HRasQ61L were stained 96 h post transduction with an antibody against LC3. Red puncta represent LC3 aggregation in the cells. Microphotographs taken at 600× magnification. Insets show the transduction efficiencies for GFP and RasGRP1 via fluorescence microscopy. Scale bar = 25 μm. (B) Relative abundance of LC3-I/LC3-II was measured by immunoblot analysis. Data are representative of ≥3 independent experiments. (C) Total LC3-II quantitated by spot densitometry from 4 independent experiments and normalized to actin loading. (D) Electron microscopic analysis of HKn transduced with RasGRP1 for 96 h. Arrows indicate autophagosomes, arrowheads indicate autophagolysosomes and double arrows indicate isolation membranes (cup-shaped structures). Scale bar = 2 μm. (E) HKn were transduced with the indicated adenovirus and treated at 96 h post transduction with bafilomycin (400 nM) for 4 h. Untreated controls for each group were run in parallel. Immunoblot analysis of LC3 is shown. Results shown are representative of 3 independent experiments. (F) Inhibition of autophagy accelerated cell death by apoptosis in RasGRP1 overexpressing HKn. Primary human epidermal keratinocytes were treated with 5 mM 3-methyladenine (3-MA, an autophagy inhibitor that blocks type III PI-3K) prior to transduction with GFP or RasGRP1-GFP and through the duration of the experiment (72 h). Untreated controls for each group were run in parallel. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although increased LC3-II and NOXA expression suggests autophagy activation, LC3-II may also accumulate due to impairment of autophagosome-lysosome fusion or faulty protein degradation as LC3-II itself is localized to autophagosomes and is subjected to autophagic protein degradation [41]. To measure functional autophagic flux, induction of vacuole formation and lysosomal degradation of the autophagosomes, a late stage autophagy and lysosomal inhibitor, Bafilomycin A1 (Baf A1), was used. Indeed, Baf A1 treatment of RasGRP1+ cells led to increases in LC3-II expression compared to control cells, indicating active autophagic flux is induced in our model (Fig. 4E). Finally, pharmacologic inhibition of RasGRP1-induced autophagy resulted in apoptosis (Fig. 4F), suggesting that in this context, RasGRP1-induced autophagy is playing a cell survival role. Taken together, these data indicate that RasGRP1 induced autophagy in primary human keratinocytes.
Components of the autophagy pathway are overexpression in human cSCC
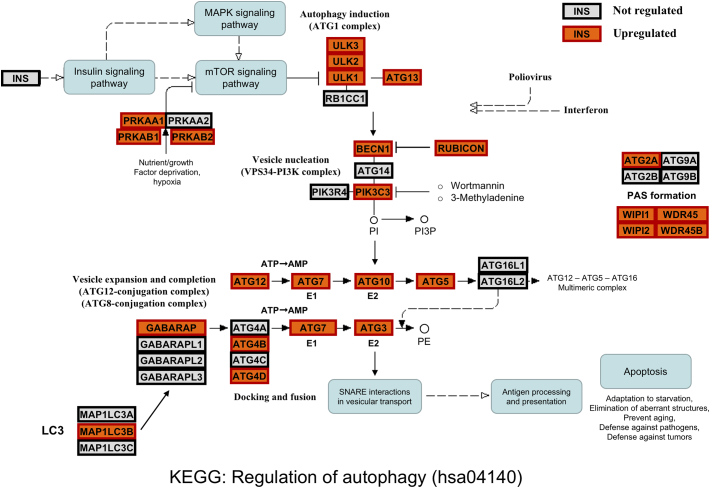
Our data indicate induction of autophagy by RasGRP1 overexpression in vitro. Mining of public datasets showed that RasGRP1 mRNA expression was significantly higher in human cSCC than in normal skin. We therefore investigated whether we could detect more activity of the autophagy pathway in human cSCC than in normal skin samples. mRNA expression analysis of autophagy pathway genes – as defined by KEGG - in human cSCC datasets showed that many of these genes are over-expressed in cSCC versus normal skin samples (Fig. 5, Supplemental Table 1). Although we realize that the autophagy is extensively modulated on the protein level, and these regulations cannot be detected using mRNA expression studies, we assume that the continuous, long-term autophagy we observe in our in vitro experiments requires sustained over-expression of key pathway genes also at the mRNA level. We would therefore propose that autophagy is upregulated in human cSCC in patients in cohort with RasGRP1.
Fig. 5.
Autophagy pathway gene expression in human cSCC. Graphical overview of the KEGG “Regulation of autophagy” pathway (hsa04140) showing constituent genes with a significant over-expression in cSCC versus normal skin samples in the Riker-87 dataset. The KEGG pathway graph is overlayed with boxes representing genes with significant cSCC over-expression (gene name on an orange background with thick red border), or with no differential expression (grey background with thick black border). There were no pathway genes with significant lower expression in cSCC than in normal skin. The overview shows widespread over-expression in cSCC, strongly suggesting that the autophagy pathway is activated in cSCC cells. Full expression data are in Supplemental Table 1. KEGG can be accessed at http://www.genome.jp/kegg/. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Autophagy pathway gene expression in human cSCC. Graphical overview of the KEGG “Regulation of autophagy” pathway (hsa04140) showing constituent genes with a significant over-expression in cSCC versus normal skin samples in the Riker-87 dataset. The KEGG pathway graph is overlayed with boxes representing genes with significant cSCC over-expression (gene name on an orange background with thick red border), or with no differential expression (grey background with thick black border). There were no pathway genes with significant lower expression in cSCC than in normal skin. The overview shows widespread over-expression in cSCC, strongly suggesting that the autophagy pathway is activated in cSCC cells. Full expression data are in Supplemental Table 1. KEGG can be accessed at http://www.genome.jp/kegg/. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
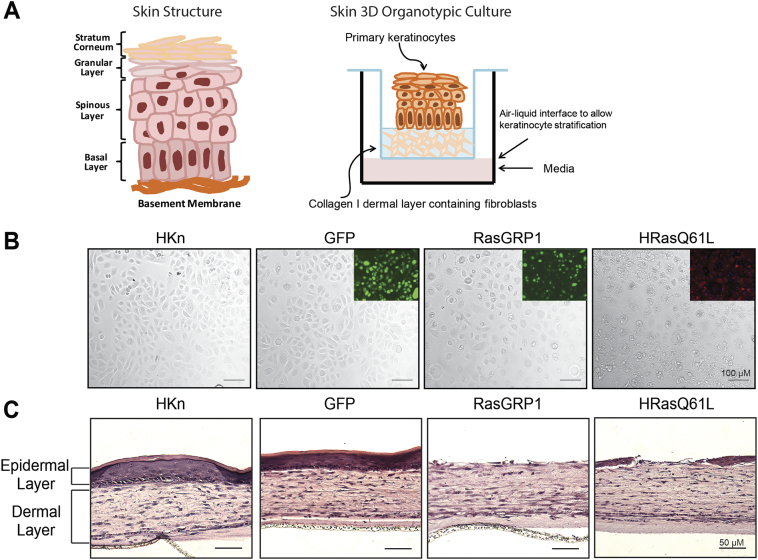
High RasGRP1 led to defective stratification in skin tissue reconstructs
To better understand how RasGRP1-expressing cells behave in a model approximating physiological architecture, and as an extension of the datamining results, we utilized an in vitro 3D skin tissue reconstruction model. This model allows for cell interaction with the microenvironment and mimics skin tissue polarity and differentiation patterns (Fig. 6A). Micrographs of transduced keratinocytes used for the construction of the 3D skin model were taken prior to seeding onto the collagen dermal layer (Fig. 6B). Insets for GFP+, RasGRP1+, and HRasQ61L+ show immunofluorescence staining demonstrating >90% transduction efficiency. In this skin model, wild type and GFP-transduced HKn formed well-organized stratified epidermal layers, while exogenous HRasQ61L expression completely blunted this phenotype (Fig. 6C) as previously described [42,43]. Importantly, keratinocytes overexpressing RasGRP1 also failed to form a normal stratified epithelium, mimicking enforced oncogenic HRas expression (Fig. 6C). 8 out of 9 independent experiments with RasGRP1+ cells, and 9 of 9 experiments with oncogenic HRas+ cells resulted in defective epithelium formation in 3D culture, confirming the reproducibility of these findings.
Fig. 6.
RasGRP1 expression led to defective stratification in skin tissue reconstructs. (A) Left: Diagram illustrating the architecture of normal human skin including basal and suprabasal layers. Right: Diagram of the skin reconstruct procedure for generation of a 3D organotypic culture. Keratinocytes transduced with adenoviral vectors encoding GFP, RasGRP1 or HRasQ61L were seeded onto the dermal layer of a 3D skin reconstruction culture. (B) Micrographs of HKn 48 h post transduction prior to harvesting for preparation of the 3D cultures. Insets shown are fluorescence images for GFP, RasGRP1 or HRasQ61L-expressing HKn. (C) HKn were seeded onto the dermal layer of a 3D skin reconstruction culture 48 h post transduction. Tissue reconstructs were harvested after 10 days and processed by H&E staining for histological assessment.
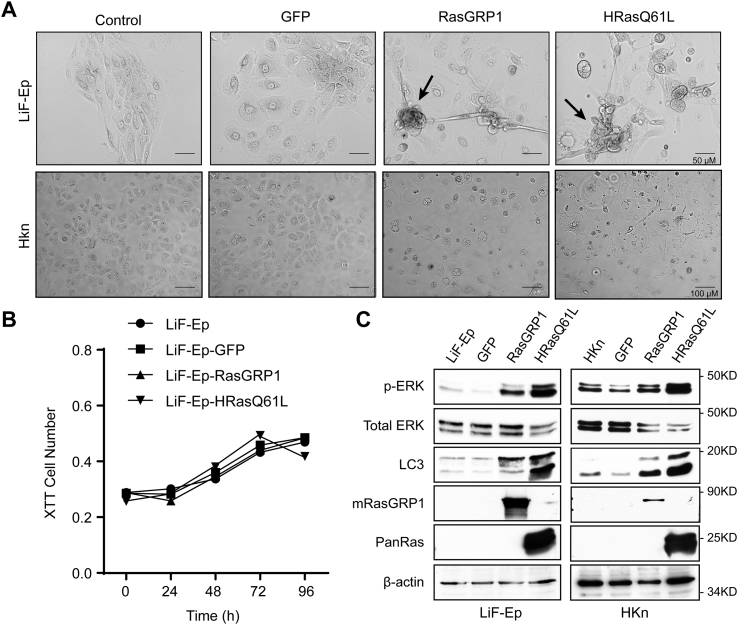
High RasGRP1 expression led to partial transformation in skin cells with a heterozygous p53 mutation
In human SCC, Ras pathway activations frequently occur on the background of p53 mutations that predispose to SCC and BCC formation [44]. We therefore considered that a p53 aberration as a second genetic change might cooperate with RasGRP activity and enable oncogenic transformation of skin cells. As a model, we utilized primary human epidermal keratinocytes from normal skin of patients with Li-Fraumeni Syndrome [31]. These “LiF-Ep” cells have a heterozygous p53 mutation (p53+/−) that causes lack of function of p53, but otherwise show normal culture growth. Li-Fraumeni patients therefore are predisposed to a variety of tumors including non-melanoma skin cancers. LiF-Ep cells were transduced with GFP, RasGRP1, or HRasQ61L as described above for HKn cells. At 7 days post transduction, RasGRP1+ and HRasQ61L+ cells showed morphological changes including spindle-like cell elongation and focus formation, suggestive of transformation, while control, non-infected and GFP+ cells and transduced HKn cells both maintained a smooth, cobblestone-like morphology (Fig. 7A). XTT assays with transduced LiF-Ep cells showed no changes in cell number compared to the untreated and GFP transduced cells at 96 h, further suggesting rescue of the RasGRP1-induced growth arrest by the p53 copy loss (Fig. 7B). Immunoblot analysis confirmed the overexpression of RasGRP1 and HRasQ61L and showed that this led to increased levels of p-ERK and LC3, confirming activation of the MAPK pathway and an increase in autophagy, respectively (Fig. 7C). Taken together these experiments suggested that RasGRP1 expression led to partial transformation in skin cells with decreased p53 levels, supporting the hypothesis that RasGRP1 can act as a tumor-promoting gene in human skin cancer in the absence of Ras oncogenes.
Fig. 7.
RasGRP1 overexpression led to partial transformation in Li-Fraumeni patient-derived skin cells with p53 mutations. Li-Fraumeni skin cells (LiF-Ep; p53+/−) keratinocytes were transduced with GFP, RasGRP1 or HRasQ61L. (A) Representative micrographs of the Lif-Ep cultures or HKn cells at 7 days post transduction. Images taken at 200 x magnification (Olympus Microscope). Scale bar = 50 μm. (B) LiF-Ep were transduced, and cell numbers were determined by XTT staining at the indicated times post transduction. Data are mean of 3 biological replicates. Statistical analysis was performed using two-way ANOVA followed by a Turkey test. (C) Immunoblots analysis of RasGRP1 (M199), PanRas, LC3 and p-ERK/total ERK in LiF-Ep and HKn cells 72 h post transduction. Data are representative of 3 independent experiments.
Discussion
RasGRP1 plays a critical role in mediating diacylglycerol signaling, TPA-induced Ras activation, and keratinocyte transformation leading to cSCC in mice. Transgenic mice with RasGRP1 overexpression in the basal epidermis (K5.RasGRP1) developed larger and more aggressive skin tumors when subjected to the DMBA/TPA protocol [23,25]. In contrast, RasGRP1 null-mutant mice were significantly resistant to DMBA/TPA-induced carcinogenesis [27]. However, due to differences in mouse and human skin that could affect responses to oncogenic insults, these results require validation in a human model before clinical translation can be explored. We show here that human epidermal keratinocytes express RasGRP1 and that aberrant high expression of this exchange factor results in growth arrest and induction of autophagy in cells with normal p53 function. High expression of RasGRP1 mRNA and of the majority of autophagy pathway genes was verified in human cSCC samples. Furthermore, high RasGRP1 expression led to partial transformation of human skin cells with p53 mutations, suggesting a role for RasGRP1 in human skin carcinogenesis.
The Ras proto-oncogene family consists of major contributors to the transformation of epidermal keratinocytes and is frequently found activated in human cSCC either by oncogenic mutation [45,46] or EGFR-enhanced signaling [6,47]. However, in primary cells aberrant Ras activation does not lead to cell transformation unless accompanied by a cooperating mutation. Normal primary cells have powerful failsafe mechanisms to balance potentially oncogenic growth signals and prevent clonal expansion and subsequent neoplastic progression. These include cell cycle arrest, cell senescence and apoptosis, depending on the cell type and strength of oncogenic signaling [48,49]. Our data show failsafe mechanism-related phenotypes in human epidermal keratinocytes overexpressing RasGRP1 that include cell cycle arrest and induction of autophagy.
Autophagy is a cellular process in which cytoplasmic components such as the mitochondria of a cell are degraded, for example in times of nutrient deprivation, and reused to support survival of the cell. Autophagy can also lead to cell death in some contexts. Oncogenic Ras can activate autophagy and this can play a role in tumor formation and progression depending on the stage and signaling context [37]. Under our experimental conditions, RasGRP1-induced autophagy was not associated with apoptosis as the overexpression of RasGRP1 failed to induce PARP cleavage, Caspase-3 activation or DNA damage (Fig. 3B). In contrast, oncogenic HRasQ61L activated autophagy in these human keratinocytes accompanied by apoptosis as reflected in PARP cleavage, caspase 3 activation (Fig. 3B). Importantly oncogenic Ras lead to a reduction in p53 phosphorylation whereas RasGRP1 initially led to an increase as expected [50] downstream of activated ERK (Fig. 3B). This change in p53 may in part be responsible for the difference in RasGRP1 and HRasQ61L effects. Phosphorylation at ser 15 stabilizes p53 by blocking MDM2 binding and promotes p53 dependent transcription which can activate genes encoding proteins involved in cell cycle arrest, DNA repair, and/or apoptosis, and also represses transcriptional activation of growth-promoting genes. Furthermore, pharmacologic inhibition of RasGRP1-induced autophagy resulted in apoptosis (Fig. 4F), suggesting that in this context, RasGRP1-induced autophagy is playing a cell survival role. We have also observed similar results for RasGRP1 LC3 in LiF-Ep (p53+/−) cells. RasGRP1 overexpression in LiF-Ep cells led to continued cell proliferation and focus formation suggestive of transformation in monolayer, indicating that the heterozygous p53 mutation may cooperate with activated RasGRP1 signaling to maintain proliferation and transform cells.
A 3D organotypic model that mimics many aspects of human skin architecture was used to study the transformative potential of RasGRP1 in human keratinocytes. We generated organized and well-stratified epithelia using normal primary keratinocytes (HKn) and fibroblasts isolated from neonatal tissue. When these HKn cells were manipulated to overexpress RasGRP1, the ability of cells to grow and stratify in a 3D culture was severely impaired. These data are consistent with the in vitro data from 2D culture that RasGRP1 induced the failsafe mechanism-related phenotype of cell cycle arrest in a normal p53 background. Expression of oncogenic Ras in primary keratinocytes leads to cell death and/or senescence, which impairs their ability to form normal organotypic cultures [42,43]. Therefore, RasGRP1-induced keratinocyte stratification failure might be partially due to Ras hyper-activation. In fact, the overexpression of RasGRP1 led to a 5-fold increase in levels of active Ras, with H-Ras being the predominant isoform being activated (Fig. 2). In this context, HRasQ61L-expressing cells also failed to form a normal organotypic culture, validating the previously described phenomenon under our experimental conditions.
Our data show that RasGRP1 can activate failsafe mechanisms when expressed at high levels in keratinocytes with normal p53 and Ras and that it can activate transformation processes in cells mutant for p53 (but with normal wild type Ras). This strongly supports a potential role for RasGRP1 in promoting cSCC in patients with high RasGRP1. It remains to be determined what signals upstream of RasGRP1 such as diacylglycerol or oncogenic growth factors may contribute. Further it is possible that RasGRP1 may work in tangent with Ras mutations to amplify their transforming ability. Determining how these upstream and downstream signals affect RasGRP1 function in cSCC will be an important next step in determining the value of RasGRP1 as a diagnostic and therapeutic target in cSCC.
In summary, our results reveal a novel transformation-related role of RasGRP1 in human keratinocytes. This includes a previously unknown ability to activate autophagy. Our results, together with previous findings of the oncogenic role of RasGRP1 in mouse keratinocytes, warrant further investigation into the participation of RasGRP1 in keratinocyte transformation, and the potential contribution of RasGRP1 in the development of cutaneous squamous cell carcinoma. The results further support exploring targeting RasGRP1 signaling as a potential new approach in treating cSCC.
The following is the supplementary data related to this article.
KEGG Autophagy pathway gene expression in human cSCCs.
Funding
This work was supported by the National Institute of Health [R01 CA096841 to JJ/JWR] and funds provided by the University of Hawaii Cancer Center.
Authors' contributions
LF and WSY carried out the molecular and cellular studies and helped draft the manuscript. DG carried out the bioinformatics studies and helped draft and revise the manuscript. JWR, JJ and PSL conceived of the study. JWR, JT, and JJ participated in its design and coordination and helped to draft and revise the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We dedicate this work to the memory of Patricia S. Lorenzo. Dr. Lorenzo will be remembered for her strength, courage and above all, as an exceptional Scientist and mother. This work was originally conceived and begun by Dr. Lorenzo, but she passed away before its completion. The authors thank Miyoko Bellinger of the Histopathology Core at the John A. Burns School of Medicine for her help with histological processing of skin tissue reconstructs and Tina Carvalho of the Biological Electron Microscope Facility at the Pacific Bioscience Research Center for her assistance with the transmission electron microscopy studies. We also thank Yuka Endo for assistance with cell biological experiments.
Contributor Information
Junfang Ji, Email: junfangji@zju.edu.cn.
Joe W. Ramos, Email: joeramos@hawaii.edu.
References
- 1.Pierceall W.E., Goldberg L.H., Tainsky M.A., Mukhopadhyay T., Ananthaswamy H.N. Ras gene mutation and amplification in human nonmelanoma skin cancers. Mol. Carcinog. 1991;4:196–202. doi: 10.1002/mc.2940040306. [DOI] [PubMed] [Google Scholar]
- 2.Madan V., Lear J.T., Szeimies R.-M. Non-melanoma skin cancer. Lancet. 2010;375:673–685. doi: 10.1016/S0140-6736(09)61196-X. [DOI] [PubMed] [Google Scholar]
- 3.Alam M., Ratner D. Cutaneous squamous-cell carcinoma. N. Engl. J. Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 4.Bos J.L. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 5.Spencer J.M., Kahn S.M., Jiang W., DeLeo V.A., Weinstein I.B. Activated ras genes occur in human actinic keratoses, premalignant precursors to squamous cell carcinomas. Arch. Dermatol. 1995;131:796–800. [PubMed] [Google Scholar]
- 6.Shimizu T., Izumi H., Oga A., Furumoto H., Murakami T., Ofuji R., Muto M., Sasaki K. Epidermal growth factor receptor overexpression and genetic aberrations in metastatic squamous-cell carcinoma of the skin. Dermatology. 2001;202:203–206. doi: 10.1159/000051637. [DOI] [PubMed] [Google Scholar]
- 7.Toll A., Salgado R., Yebenes M., Martin-Ezquerra G., Gilaberte M., Baro T., Sole F., Alameda F., Espinet B., Pujol R.M. Epidermal growth factor receptor gene numerical aberrations are frequent events in actinic keratoses and invasive cutaneous squamous cell carcinomas. Exp. Dermatol. 2010;19:151–153. doi: 10.1111/j.1600-0625.2009.01028.x. [DOI] [PubMed] [Google Scholar]
- 8.Maubec E., Duvillard P., Velasco V., Crickx B., Avril M.F. Immunohistochemical analysis of EGFR and HER-2 in patients with metastatic squamous cell carcinoma of the skin. Anticancer Res. 2005;25:1205–1210. [PubMed] [Google Scholar]
- 9.Kiguchi K., Beltran L., Rupp T., DiGiovanni J. Altered expression of epidermal growth factor receptor ligands in tumor promoter-treated mouse epidermis and in primary mouse skin tumors induced by an initiation-promotion protocol. Mol. Carcinog. 1998;22:73–83. [PubMed] [Google Scholar]
- 10.Kiguchi K., Beltran L.M., You J., Rho O., DiGiovanni J. Elevation of transforming growth factor-alpha mRNA and protein expression by diverse tumor promoters in SENCAR mouse epidermis. Mol. Carcinog. 1995;12:225–235. doi: 10.1002/mc.2940120407. [DOI] [PubMed] [Google Scholar]
- 11.Cox A.D., Der C.J. Ras family signaling: therapeutic targeting. Cancer Biol Ther. 2002;1:599–606. doi: 10.4161/cbt.306. [DOI] [PubMed] [Google Scholar]
- 12.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Aronheim A., Engelberg D., Li N., Al-Alawi N., Schlessinger J., Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 14.Jeng H.H., Taylor L.J., Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat. Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim R., Trubetskoy A., Suzuki T., Jenkins N.A., Copeland N.G., Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J. Virol. 2003;77:2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T., Shen H., Akagi K., Morse H.C., Malley J.D., Naiman D.Q., Jenkins N.A., Copeland N.G. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 17.Ebinu J.O., Bottorff D.A., Chan E.Y., Stang S.L., Dunn R.J., Stone J.C. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 18.Tognon C.E., Kirk H.E., Passmore L.A., Whitehead I.P., Der C.J., Kay R.J. Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol. Cell. Biol. 1998;18:6995–7008. doi: 10.1128/mcb.18.12.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebinu J.O., Stang S.L., Teixeira C., Bottorff D.A., Hooton J., Blumberg P.M., Barry M., Bleakley R.C., Ostergaard H.L., Stone J.C. RasGRP links T-cell receptor signaling to Ras. Blood. 2000;95:3199–3203. [PubMed] [Google Scholar]
- 20.Klinger M.B., Guilbault B., Goulding R.E., Kay R.J. Deregulated expression of RasGRP1 initiates thymic lymphomagenesis independently of T-cell receptors. Oncogene. 2005;24:2695–2704. doi: 10.1038/sj.onc.1208334. [DOI] [PubMed] [Google Scholar]
- 21.Lauchle J.O., Kim D., Le D.T., Akagi K., Crone M., Krisman K., Warner K., Bonifas J.M., Li Q., Coakley K.M., Diaz-Flores E., Gorman M., Przybranowski S., Tran M., Kogan S.C., Roose J.P., Copeland N.G., Jenkins N.A., Parada L., Wolff L., Sebolt-Leopold J., Shannon K. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambaratsingh R.A., Stone J.C., Blumberg P.M., Lorenzo P.S. RasGRP1 represents a novel non-protein kinase C phorbol ester signaling pathway in mouse epidermal keratinocytes. J. Biol. Chem. 2003;278:52792–52801. doi: 10.1074/jbc.M308240200. [DOI] [PubMed] [Google Scholar]
- 23.Oki-Idouchi C.E., Lorenzo P.S. Transgenic overexpression of RasGRP1 in mouse epidermis results in spontaneous tumors of the skin. Cancer Res. 2007;67:276–280. doi: 10.1158/0008-5472.CAN-06-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diez F.R., Garrido A.A., Sharma A., Luke C.T., Stone J.C., Dower N.A., Cline J.M., Lorenzo P.S. RasGRP1 transgenic mice develop cutaneous squamous cell carcinomas in response to skin wounding: potential role of granulocyte colony-stimulating factor. Am. J. Pathol. 2009;175:392–399. doi: 10.2353/ajpath.2009.090036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luke C.T., Oki-Idouchi C.E., Cline J.M., Lorenzo P.S. RasGRP1 overexpression in the epidermis of transgenic mice contributes to tumor progression during multistage skin carcinogenesis. Cancer Res. 2007;67:10190–10197. doi: 10.1158/0008-5472.CAN-07-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A., Luke C.T., Dower N.A., Stone J.C., Lorenzo P.S. RasGRP1 is essential for ras activation by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate in epidermal keratinocytes. J. Biol. Chem. 2010;285:15724–15730. doi: 10.1074/jbc.M109.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A., Fonseca L.L., Rajani C., Yanagida J.K., Endo Y., Cline J.M., Stone J.C., Ji J., Ramos J.W., Lorenzo P.S. Targeted deletion of RasGRP1 impairs skin tumorigenesis. Carcinogenesis. 2014;35:1084–1091. doi: 10.1093/carcin/bgu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Depeille P., Henricks L.M., van de Ven R.A., Lemmens E., Wang C.Y., Matli M., Werb Z., Haigis K.M., Donner D., Warren R., Roose J.P. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat. Cell Biol. 2015;17:804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangatirkar P., Paquet-Fifield S., Li A., Rossi R., Kaur P. Establishment of 3D organotypic cultures using human neonatal epidermal cells. Nat. Protoc. 2007;2:178–186. doi: 10.1038/nprot.2006.448. [DOI] [PubMed] [Google Scholar]
- 30.Turksen K. 2009. Epidermal Cells: Methods and Protocols., Springer Protocols. [Google Scholar]
- 31.Dickson M.A., Hahn W.C., Ino Y., Ronfard V., Wu J.Y., Weinberg R.A., Louis D.N., Li F.P., Rheinwald J.G. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamura S.M., Oki-Idouchi C.E., Lorenzo P.S. The exchange factor and diacylglycerol receptor RasGRP3 interacts with dynein light chain 1 through its C-terminal domain. J. Biol. Chem. 2006;281:36132–36139. doi: 10.1074/jbc.M605093200. [DOI] [PubMed] [Google Scholar]
- 33.Riker A.I., Enkemann S.A., Fodstad O., Liu S., Ren S., Morris C., Xi Y., Howell P., Metge B., Samant R.S., Shevde L.A., Li W., Eschrich S., Daud A., Ju J., Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genet. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., Rabinowitz J.D., Metallo C.M., Vander Heiden M.G., Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amyere M., Payrastre B., Krause U., Van Der Smissen P., Veithen A., Courtoy P.J. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-kinase and phospholipase C. Mol. Biol. Cell. 2000;11:3453–3467. doi: 10.1091/mbc.11.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byun J.Y., Yoon C.H., An S., Park I.C., Kang C.M., Kim M.J., Lee S.J. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis. 2009;30:1880–1888. doi: 10.1093/carcin/bgp235. [DOI] [PubMed] [Google Scholar]
- 37.Schmukler E., Kloog Y., Pinkas-Kramarski R. Ras and autophagy in cancer development and therapy. Oncotarget. 2014;5:577–586. doi: 10.18632/oncotarget.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elgendy M., Sheridan C., Brumatti G., Martin S.J. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Klionsky D.J., Cuervo A.M., Seglen P.O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 42.Dajee M., Lazarov M., Zhang J.Y., Cai T., Green C.L., Russell A.J., Marinkovich M.P., Tao S., Lin Q., Kubo Y., Khavari P.A. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida S., Kajitani N., Satsuka A., Nakamura H., Sakai H. Ras modifies proliferation and invasiveness of cells expressing human papillomavirus oncoproteins. J. Virol. 2008;82:8820–8827. doi: 10.1128/JVI.02363-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamin C.L., Ananthaswamy H.N. p53 and the pathogenesis of skin cancer. Toxicol. Appl. Pharmacol. 2007;224:241–248. doi: 10.1016/j.taap.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roop D.R., Lowy D.R., Tambourin P.E., Strickland J., Harper J.R., Balaschak M., Spangler E.F., Yuspa S.H. An activated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature. 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 46.Brown K., Quintanilla M., Ramsden M., Kerr I.B., Young S., Balmain A. v-ras genes from Harvey and BALB murine sarcoma viruses can act as initiators of two-stage mouse skin carcinogenesis. Cell. 1986;46:447–456. doi: 10.1016/0092-8674(86)90665-3. [DOI] [PubMed] [Google Scholar]
- 47.Ch'ng S., Low I., Ng D., Brasch H., Sullivan M., Davis P., Tan S.T. Epidermal growth factor receptor: a novel biomarker for aggressive head and neck cutaneous squamous cell carcinoma. Hum. Pathol. 2008;39:344–349. doi: 10.1016/j.humpath.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 49.Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 50.Melnikova V.O., Santamaria A.B., Bolshakov S.V., Ananthaswamy H.N. Mutant p53 is constitutively phosphorylated at Serine 15 in UV-induced mouse skin tumors: involvement of ERK1/2 MAP kinase. Oncogene. 2003;22:5958–5966. doi: 10.1038/sj.onc.1206595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KEGG Autophagy pathway gene expression in human cSCCs.