Abstract
Glioblastoma (GBM) is the most frequent and most aggressive form of glioma. It is characterized by marked genomic instability, which suggests that chromothripsis (CT) might be involved in GBM initiation. Recently, CT has emerged as an alternative mechanism of cancer development, involving massive chromosome rearrangements in a one-step catastrophic event. The aim of the study was to detect CT in GBM and identify novel gene fusions in CT regions. One hundred and seventy IDH-wild-type GBM were screened for CT patterns using whole-genome single nucleotide polymorphism (SNP) arrays. RNA sequencing was performed in 52 GBM with CT features to identify gene fusions within CT regions. Forty tumors (40/52, 77%) harbored at least one gene fusion within CT regions. We identified 120 candidate gene fusions, 30 of which with potential oncogenic activities. We validated 11 gene fusions, which involved the most recurrent fusion partners (EGFR, SEPT14, VOPP1 and CPM), by RT-PCR and Sanger sequencing. The occurrence of CT points to underlying gene fusions in IDH-wild-type GBM. CT provides exciting new research avenues in this highly aggressive cancer.
Abbreviations: BAF, B-allele frequency; chr, chromosome; CNA, copy number alteration; CNS, central nervous system; CT, chromothripsis; FPKM, fragments per kilobase of Exon per million fragments mapped; GBM, glioblastoma multiform; HD, homozygous deletion; LOH, loss of heterozygosity; RNA-Seq, RNA sequencing; RT-PCR, reverse transcriptase – polymerase chain reaction; SNP, single nucleotide polymorphism; WHO, World Health Organization
Introduction
Gliomas are the most frequent primary tumors of the central nervous system (CNS) [1]. More than half of gliomas are glioblastomas (GBM), which represent the most common and most aggressive form of glial tumors (WHO grade IV) [2]. Over 90% of GBM are primary (de novo) tumors, arising without a past history of lower-grade diffuse glioma, whereas secondary GBM results from the progression of a lower-grade diffuse glioma. Primary GBM develop rapidly, often in older patients (> 55 years-old), and are associated with a shorter survival compared to secondary GBM (median overall survival 15 months vs 2–3 years) [2]. The physiopathogenesis of GBM is still unknown. GBM harbor genomic instability with numerous copy number alterations (CNA); the most common chromosomal imbalances are gain of chromosome (chr) 7 and loss of chr 9p and 10. Primary GBM typically display EGFR amplification (and/or chr 7 gain), PTEN mutation or homozygous deletion, TERT promoter mutation, CDKN2A homozygous deletion, and chr 10 loss [[3], [4], [5], [6]]. Secondary GBM harbor, in most cases, isocitrate dehydrogenase 1 or 2 (IDH1/2) gene mutation, the earliest genetic event known in gliomagenesis and one of the most potent predictors of longer survival [7,8].
The genomic complexity observed in GBM has suggested CT as a potential mechanism of GBM initiation. Contrasting with the conventional (incremental, step-by-step) model of cancer development, CT is a single cataclysmic phenomenon (punctuated equilibrium) by which one or a few chr are shattered into tens to hundreds of pieces randomly reassembled by the DNA repair machinery [9,10] (Supplementary file 1). This one-step event may lead to the loss of tumor suppressor genes, the gain and/or amplification of oncogenes, and/or the formation of oncogenic gene fusions [9]. CT has been shown to occur in 8.7% of cancers, such as breast, ovarian, lung or colon adenocarcinomas, and in over a third (38.9%) of GBM [[11], [12], [13]]. This cataclysmic event might be involved in the pathogenesis of aggressive fast-growing tumors, such as IDH-wild-type GBM, which are additionally characterized by numerous chr aberrations. However, it is still unclear whether CT is a cause or a consequence of the dramatic chr instability observed in some tumors.
Supplementary file 1.
Chromothripsis is a one-step catastrophic event leading to massive chromosome rearrangements. A. Schematic example of CT, by which one chr is shattered into pieces randomly reassembled by the DNA repair machinery. The derivative chr might harbor fusion genes with potential oncogenic properties. Loss of DNA fragments may lead to the inactivation of tumor suppressor genes and (onco)gene amplification may lead to the formation of double minutes. B. Copy number profile from the same chr as shown in A. There is an oscillating pattern of copy number states with interspersed loss and retention of heterozygosity and focal amplification (fragment F). Chr: chromosome.
Gene fusions result from the juxtaposition of two genes during chr rearrangements that may lead to the expression of a chimeric protein. Since the Philadelphia chr (BCR-ABL1 gene fusion resulting from a translocation t(9;22)) has been identified as a key genetic alteration and therapeutic target (of imatinib mesylate) in chronic myeloid leukemia, detection of such potent driver gene fusions has been of great interest in cancer research [14]. Deep sequencing technologies have allowed the identification of gene fusions in hematological neoplasms, sarcomas, carcinomas, but also CNS tumors, including GBM [[15], [16], [17]]. The first oncogenic gene fusion reported in GBM was the FIG-ROS1 fusion in 2003 [18]. The EGFR-SEPT14 fusion has been observed in 4% of GBM [19]. The FGFR3-TACC3 fusion has been detected in 3% of GBM with promising therapeutic effects of FGFR inhibitors on these tumors [20,21]. In this context, CT provides novel insights into GBM pathogenesis and exciting new research avenues for the identification of targetable driver gene fusions.
Our study aimed to identify oncogenic gene fusions within CT regions in a cohort of 170 adult IDH-wild-type GBM. We identified CT patterns by whole-genome SNP arrays and performed RNA sequencing (RNA-seq) to detect novel gene fusions within CT regions. We selected gene fusions involving recurrent partners with potential oncogenic properties and validated the candidate fusions by RT-PCR followed by Sanger sequencing. Eleven potential oncogenic gene fusions were thus identified.
Material and methods
Patients and tumor samples
We selected 170 cases from the registries of the Pathology Department for which a diagnosis of GBM was made between 2005 and 2017. All cases met the following criteria: 1) histopathological diagnosis of IDH-wild-type GBM according to the 2016 WHO classification, 2) age at diagnosis >18 years-old, 3) available fresh frozen tissue containing at least 60% of tumor cells, and 4) written informed consent from each patient and approval of the research ethics committees of Angers University Hospital (Comité de Protection des Personnes, n° CP CB 2015/08). Frozen samples were retrieved from Angers University Hospital Biobank (CRB, biological resource center).
SNP array and copy number analyses
Tumor DNA of 170 frozen samples was extracted using the Nucleospin Tissue Kit (Macherey Nagel) and quantified using Qubit dsDNA BR Assay Kit (Life Technologies). Tumor DNA was hybridized with Infinium CytoSNP-850 K Illumina Beadchips (Illumina) according to the manufacturer's instructions. SNP arrays were scanned on an iScan (Illumina) and data were processed using the genotyping module in Genome Studio v2011.1 (Illumina) to calculate the B-allele frequencies (BAF) and logR ratios. The GAP method was used to call somatic CNA and assess the ploidy for each tumor [22,23].
CT identification
CT events were detected using segmented data (LogR ratio, BAF value) from SNP arrays, following the three major criteria described by Korbel and Campbell [24]:
-
1)
There were at least ten genomic rearrangements per chr arm with such rearrangements occurring in no more than four chr in a given sample,
-
2)
There was a clustering of breakpoints,
-
3)
There was interspersed loss and retention of heterozygosity with no more than two to three different copy number states (except for focal amplification or homozygous deletion (involving key cancer genes)).
After a manual screening, according to the criteria mentioned above, a validation analysis was performed on segmented data using CTLPScanner, a web server for the detection of CT patterns [25].
-
1.1.
RNA extraction and sequencing
Total RNA from tumor samples in which CT had been detected was extracted and purified using RNeasy Lipid Tissue Mini Kit (Qiagen) according to the supplier's recommendations. Evaluation of the total RNA for quantity and purity was performed using Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Library preparation and sequencing were performed by Integragen high-throughput sequencing platform. Briefly, the libraries were prepared with the TruSeq Stranded mRNA kit following the manufacturer's protocol and sequencing of the cDNA libraries was carried out using an Illumina HiSeq4000 with a 75-bp paired-end read length.
Bioinformatic analyses
The quality of the reads was evaluated for each sample using FastQC (V.0.11.4; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and RNA-SeQC [26]. Alignment was performed by STAR (V.2.5; https://github.com/alexdobin/STAR). To detect candidate gene fusions in RNA sequencing data, we used FusionCatcher (V.0.99.7c; Start with fastq files) and Star-Fusion (V.0.8.0; Start with alignment files). Then, in silico validation of predicted fusion transcripts was performed by FusionInspector (https://github.com/FusionInspector/FusionInspector). The candidate gene fusions were annotated according to 1) a list of known false positives (1000G, chimerdb2, gtex), 2) databases of known gene fusions found in healthy individuals, and 3) cancer databases (cosmic and 18cancers). EGFRvIII rearrangement (exon 2–7 skipping of EGFR gene) was identified by manual review of RNAseq data.
Potential oncogenic gene fusions were selected when any predicted in-frame fusion involved at least one partner with a potential or well-known role in cancer. For the selection of oncogenic fusion partners, we used Oncoscore (V.1.12.0, https://github.com/danro9685/OncoScore), a bioinformatic tool that measures the association of genes to cancer based on citation frequencies in biomedical literature (OncoScore cut-off threshold = 21.09 according to the developer's recommendations and published data) [27]. Any gene involved in a predicted in-frame gene fusion was evaluated by the Oncoscore algorithm. The exact role of the potentially cancer-related genes identified (Oncoscore >21.09) was thoroughly checked manually on PubMed.
Circos plot were created with Circa (http://omgenomics.com/circa). Fusion transcripts and putative derived chimeric proteins were visualized using chimeraviz (V1.8.5, https://github.com/stianlagstad/chimeraviz) and AGfusion (V1.251, https://github.com/murphycj/AGFusion) [28,29].
Expression levels of transcripts were measured with FPKM normalization method using Stringtie software (V.1.3.6, https://github.com/gpertea/stringtie).
Gene fusion validation
Briefly, 500 ng of total tumor RNA was retrotranscribed using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR kit (Thermo Fisher scientific) following the manufacturer's instructions. Synthetized single-stranded cDNA was amplified using forward and reverse primer combinations, which were designed within the margins of the paired-end read sequences detected by RNA-seq (Supplementary file 2). Direct Sanger sequencing of cDNA products was performed to confirm the DNA sequence and translation frame.
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 7.0 for MacOS, GraphPad Software (www.graphpad.com). Fisher's exact probability test was used to analyze the association of EGFR amplification, MDM2 and/or CDK4 amplification and CDKN2A homozygous deletion with CT (p-value: 0.12 (ns), 0.033 (*), 0.002 (**), <0.001 (***)). The statistical significance of FPKM differences observed between groups with and without CT was assessed by Mann-Whitney tests (p-value: 0.12 (ns), 0.033 (*), 0.002 (**), <0.001 (***)).
Results
Patient and tumor characteristics
The median age of the 170 patients at diagnosis was 61 years-old (range: 22–84). The male-to-female ratio was 1.39. Chr imbalances, gene amplifications, and homozygous deletions were investigated by SNP arrays (Fig. 1). The most common CNA were chr 10q loss (153/170, 90%), chr 10p loss (144/170, 84.7%), chr 7q gain (132/170, 77.6%), chr 7p gain (131/170, 77.0%), and chr 9p loss (85/170, 50.0%). CDKN2A homozygous deletion was the most frequent gene alteration (90/170, 52.9%), followed by EGFR amplification (70/170, 41.2%). MDM2 amplification and CDK4 amplification were found in 19 (19/170, 11.2%) and 24 GBM (24/170, 14.1%), respectively. MDM2 and CDK4 were co-amplified in 17 cases (17/170, 10.0%).
Fig. 1.
Landscape of copy number alterations in 170 IDH-wild-type GBM.
Whole chr losses are in dark green, partial losses in light green, chr gains in red (which were mostly whole chr gains) and copy neutral loss of heterozygosity (LOH) in light blue. Amplifications and homozygous deletions of key cancer genes in GBM are shown in orange and CT is shown in dark blue. Chr: chromosome; CT: chromothripsis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
CT is a common phenomenon in IDH-wild-type GBM
To evaluate the frequency of CT in IDH-wild-type GBM, we generated copy number profiles of 170 tumors by SNP arrays to detect CT patterns. Sixty-six cases (66/170, 38.8%) exhibited CT features (Fig. 1). Most glioblastoma cases with CT harbored only one rearranged chromosome (44/66, 66.7%). Glioblastomas harboring 2 chr (14/66, 21.2%), 3 chr (6/66, 9.1%) or 4 chr (2/66, 3.0%) with CT features were less frequent. CT mostly involved chr 7 (26/66, 39.4%), chr 9 (19/66, 28.8%), and chr 12 (12/66, 18.2%) and was associated with EGFR amplification (chr7p11), CDKN2A homozygous deletion (chr9p21) and CDK4/MDM2 co-amplification (chr12q14/12q15), respectively (p < 0.001, Fisher's exact probability test) (Fig. 2).
Fig. 2.
Chromothripsis occurrence across the genome in 170 IDH-wild-type GBM
A. CT mostly involved chr 7, 9 and 12.
B.EGFR amplification was more frequent in cases with CT on chr 7 compared to cases without CT on chr 7 (22/26, 84.6% vs 49/144, 34.0%, p < 0.001).
C.CDKN2A homozygous deletion was more frequent in cases with CT on chr 9 compared to cases without CT on chr 9 (18/19, 94.7% vs 79/151, 52.3%, p < 0.001).
D. Amplification of MDM2 and/or CDK4 were more frequent in cases with CT on chr 12 compared to cases without CT on chr 12 (12/12, 100.0% vs 14/158, 8.9%, p < 0.001).
Exact Fisher test; p-value: 0.12 (ns), 0.033 (*), 0.002 (**), <0.001 (***). CT: chromothripsis, chr: chromosome, HD: homozygous deletion.
CT regions harbor gene fusions in IDH-wild-type GBM
Out of 66 GBM with CT, 14 cases were excluded from RNA-seq because of insufficient tumor material or suboptimal nucleic acid quality or quantity. RNA-seq was performed in 52 GBM with CT features. The mean number of gene fusions was 1.7 per chromosome exhibiting CT features and 0.27 per chromosome without CT features (p < 0.001, Mann-Whitney test). 120 putative gene fusions were identified within CT regions (Supplementary file 2, Supplementary file 3). Gene fusions were mostly the results of intrachromosomal (109/120, 90.8%) rather than interchromosomal (11/120, 9.2%) rearrangements (Supplementary file 3). We detected at least one fusion transcript in 40 cases (40/52, 76.9%) and at least one in-frame gene fusion in 30 cases (30/52, 57.7%). The mean number of gene fusions and predicted in-frame fusion transcripts observed per tumor was 2.5 (range: 0–9) and 1.1 (range: 0–5), respectively. Further analysis allowed the identification of 30 in-frame fusions with potential oncogenic activities in 22 GBM (22/52, 42.3%) (Fig. 3, Fig. 4, Supplementary file 2). EGFR was the most recurrent partner involved in potential oncogenic fusions (5/52, 9.6%), and EGFR-SEPT14 and EGFR-VSTM2A were the most frequent fusions detected (two occurrences each). No other recurrent gene fusion was observed but, aside from EGFR, the most recurrent putative oncogenic partners were SEPT14 (4/52, 7.7%), VOPP1 (3/52, 5.8%) and CPM (3/52, 5.8%) (see below). Candidate gene fusions involving recurrent partners were validated by RT-PCR followed by Sanger sequencing (Table 1).
Supplementary file 3.
Detection of 120 gene fusions within CT regions in 52 IDH-wild type GBM. Putative in-frame oncogenic fusions are represented by arcs joining the two fusion partners. RNA-seq allowed the identification of 120 gene fusions, mostly resulting from intra-chromosomal rearrangements (109/120, 90.8%).
Fig. 3.
Selection of the fusion partners with potential oncogenic properties.
The association of genes to cancer was estimated with Oncoscore, a bioinformatic tool that ranks cancer-related genes based on citation frequencies in the literature (OncoScore cut-off threshold = 21.09 (horizontal dashed line), according to the developer's recommendations and publications). The relevance of the putative oncogenes (triangle) and tumor suppressor genes (square) were manually checked from the available Pubmed literature. IF: in-frame.
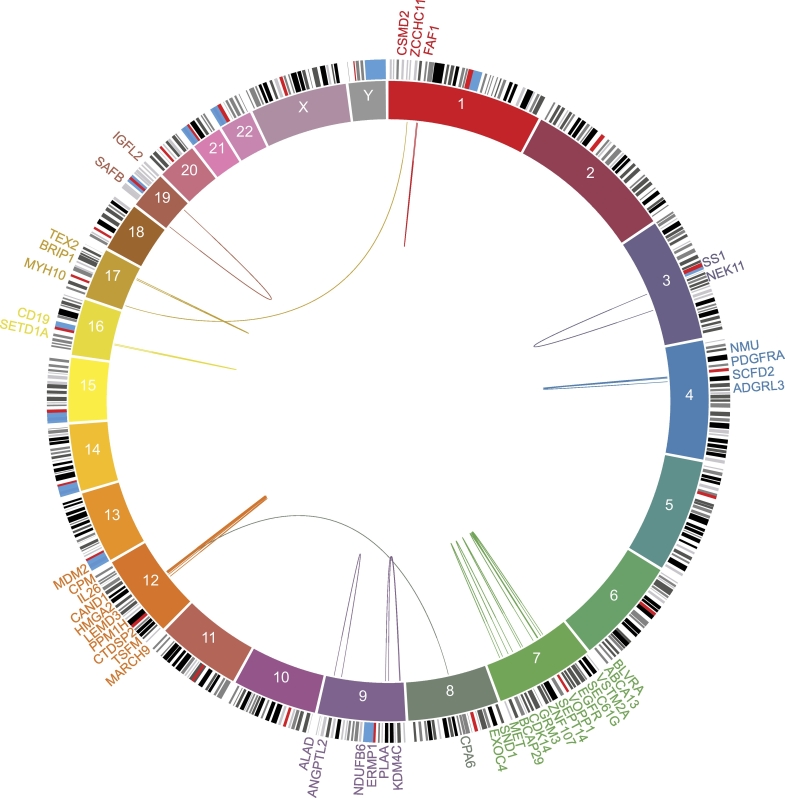
Fig. 4.
Identification of 30 putative in-frame oncogenic gene fusions within CT regions in 22 IDH-wild-type GBM.
Chromosomes are represented in blocks in the inner ring. The outer ring indicates the names of the genes. Putative in-frame oncogenic fusions are represented by arcs joining the two fusion partners. Extensive inter- or intra-chromosomal rearrangements may lead to the formation of oncogenic fusions. Putative in-frame gene fusions within CT regions that involved at least one partner with potential or well-known oncogenic properties were selected.
Table 1.
Potential oncogenic fusions involving recurrent partners in CT regions.
| Sample | Gene fusion | Gene 1 |
Gene 2 |
||||
|---|---|---|---|---|---|---|---|
| Name | Chr | Position | Name | Chr | Position | ||
| 28 | EGFR-VSTM2A | EGFR | 7 | 55,087,058 | VSTM2A | 7 | 54,612,315 |
| 56 | EGFR-VSTM2A | EGFR | 7 | 55,087,058 | VSTM2A | 7 | 54,544,622 |
| 73 | EGFR-SEPT14 | EGFR | 7 | 55,268,106 | SEPT14 | 7 | 55,796,092 |
| 123 | EGFR-SEPT14 | EGFR | 7 | 55,268,106 | SEPT14 | 7 | 55,863,785 |
| 15 | EGFR-VOPP1 | EGFR | 7 | 55,268,106 | VOPP1 | 7 | 55,521,130 |
| 95 | VOPP1-SEPT14 | VOPP1 | 7 | 55,639,964 | SEPT14 | 7 | 55,886,916 |
| 105 | VOPP1-ABCA13 | VOPP1 | 7 | 55,639,964 | ABCA13 | 7 | 48,266,859 |
| 109 | BLVRA-SEPT14 | BLVRA | 7 | 43,840,171 | SEPT14 | 7 | 55,914,330 |
| 144 | LEMD3-CPM | LEMD3 | 12 | 65,171,118 | CPM | 12 | 68,885,889 |
| 38 | CPM-MDM2 | CPM | 12 | 69,326,458 | MDM2 | 12 | 69,202,988 |
| 59 | CPA6-CPM | CPA6 | 8 | 68,396,003 | CPM | 12 | 69,279,669 |
Eleven gene fusions involving recurrent oncogenic partners (EGFR, SEPT14, VOPP1 and CPM) were selected and validated by RT-PCR followed by Sanger sequencing. EGFR was the most recurrent partner involved in potential oncogenic fusions (5/52, 9.6%), followed by SEPT14 (4/52, 7.7%), VOPP1 (3/52, 5.8%), and CPM (3/52, 5.8%).
IDH-wild-type GBM with CT harbor recurrent EGFR fusions
The EGFR fusions (n = 5) involved three different gene partners: SEPT14 (2/52, 3.8%), VSTM2A (2/52, 3.8%) and VOPP1 (1/52, 1.9%). All five cases also harbored an EGFR gene amplification but lacked the EGFRvIII rearrangement (Supplementary file 4). We evaluated whether an EGFR fusion had a direct influence on EGFR FPKM values, which schematically reflect the expression levels of EGFR transcripts from RNA-seq analysis. EGFR FPKM values were not significantly different in EGFR-amplified tumors whether or not there was an additional EGFR fusion (Mann-Whitney test; p-value: 0.69).
Supplementary file 4.
Characterization of EGFR status in 52 IDH-wild-type GBM with CT. A.EGFR amplification was detected in 51.9% (27/52) of IDH-wild-type GBM with CT (irrespective of the chr involved). All five cases harboring an EGFR gene fusion also showed an amplification of EGFR (detected by SNP arrays) but consistently lacked the EGFRvIII rearrangement (identified by RNA-seq). EGFRvIII rearrangement was observed in 18.5% (5/27) of EGFR-amplified GBM.B. FPKM values reflect expression levels of mRNA. When EGFR gene was amplified, EGFR transcript levels were not significantly different whether there was an additional EGFR gene fusion or an additional EGFRvIII rearrangement.
Further analysis of the EGFR fusion transcripts showed that the EGFR-SEPT14 gene fusions (n = 2) and the EGFR-VOPP1 gene fusion (n = 1) shared the same breakpoint within EGFR; they both coded for the N-terminal portion of EGFR (982 residues), including the tyrosine kinase domain, fused respectively with a coiled-coil domain from SEPT14 and a transmembrane helical domain from VOPP1 (Supplementary file 5). The EGFR-VSTM2A gene fusions (n = 2) shared the same breakpoint within EGFR; they involved the N-terminal portion of EGFR, including only 29 residues that would not allow the potential chimeric protein to have EGFR signaling activities. The N-terminal portion of EGFR was fused with the C-terminal portion (210 residues) of the non-oncogenic VSTM2A protein (Supplementary file 5).
Supplementary file 5.
Examples of EGFR gene fusions identified within CT regions in IDH-wild-type GBM. Each figure shows 1) chr ideograms (top), with a red line indicating the location of the two partner genes within each chr, 2) transcript portions (greyed-out) of each partner gene whose fusion is represented by a connecting red line (middle), and 3) predicted chimeric protein (bottom) with the protein domain annotations. A.EGFR-SEPT14 gene fusion (sample 123). B.EGFR-VOPP1 gene fusion (sample 15). c.EGFR-VSTM2A gene fusion (sample 51). Chr: chromosome; Rec_L_domain: Receptor L domain; TMhelix: Transmembrane Helix; I-set V-Set: Immunoglobulin V-set domain.
CPM, VOPP1 and SEPT14 are recurrent fusion partners within CT regions in IDH-wild-type GBM
Three patients had a gene fusion involving carboxypeptidase M (CPM) gene and either MDM2, CPA6, or LEMD3 gene (3/52, 5.8%). They partially involved the peptidase domain of CPM (Fig. 5). The CPM-MDM2 transcript fusion comprised the C-terminal portion of MDM2 (492 residues) that includes the p53 interaction domain (Fig. 5).
Fig. 5.
Candidate gene fusions identified within CT regions of IDH-wild type GBM.
Each figure shows 1) chr ideograms (top), with a vertical line indicating the location of the partner gene within each chr, 2) transcript portions (greyed-out) of each partner gene whose fusion is represented by a connecting line (middle), and 3) predicted chimeric protein (bottom) with the protein domain annotations. A.CPM-MDM2 gene fusion (case 38). B.CPA6-CPM gene fusion (case 59). C.LEMD3-CPM gene fusion (case 144). D.VOPP1-SEPT14 gene fusion (case 95). E.BLVRA-SEPT14 gene fusion (case 109). F.VOPP1-ABCA13 gene fusion (case 105). Chr: chromosome; TMhelix: transmembrane helix; LEM: LAP2, emerin, MAN1.
Three gene fusions involved VOPP1 gene and either EGFR, SEPT14, or ABCA13 gene. The EGFR-VOPP1 gene fusion has been discussed above. We identified a VOPP1-SEPT14 fusion transcript, coding for the first 18 residues of VOPP1 fused with the C-terminal portion of SEPT14 that includes part of its GTP-binding septin domain (Fig. 5). We detected a VOPP1-ABCA13 fusion transcript retaining the same N-terminal portion of VOPP1 fused with a large C-terminal portion of the non-oncogenic ABCA13 protein (Fig. 5).
SEPT14 gene fusions were identified in 4 GBM (4/52, 7.7%). The EGFR-SEPT14 (2/52, 3.8%) and VOPP1-SEPT14 fusions have been discussed above. The BLVRA-SEPT14 fusion transcript we detected coded for the N-terminal portion of the non-oncogenic BLVRA protein fused with the C-terminal portion of SEPT14 (514 residues) that includes its whole septin domain (Fig. 5).
Discussion
In most cancers, genomic rearrangements are thought to occur in a stepwise manner during tumor development [10]. Recent findings suggest CT as an alternative mechanism, involving massive chr rearrangements in a one-step catastrophic event [9]. By generating copy number profiles from 170 primary IDH-wild-type GBM by SNP arrays, we showed that up to 38.8% of these tumors exhibited CT features, which is consistent with previously published data [11,12]. Because CT may lead to gene fusions, we analyzed RNA-seq data from primary GBM harboring CT patterns and successfully detected potential oncogenic fusions within CT regions. Overall, RNA-seq performed in 52 GBM led to the identification of 30 putative oncogenic gene fusions, 11 of which were validated by RT-PCR followed by Sanger sequencing. Most cases with CT harbored only one rearranged chromosome which implies that intra-chromosomal rearrangements are more frequent compared to inter-chromosomal rearrangements. Intra-chromosomal rearrangements may be facilitated by spatial proximity of the fusion partners. Interestingly, the candidate gene fusions we identified within a given CT region were not observed in glioblastomas without CT in that region. The identification of driver gene fusions may help understand tumor pathogenesis and open new therapeutic avenues. Singh et al. first reported FGFR3-TACC3 fusions in 3% of GBM [20]; such fusions confer sensitivity to FGFR inhibitors with promising preliminary results in the clinic [30,31]. Herein, we selected any predicted in-frame fusion involving at least one partner with a potential or well-known role in cancer. The gene fusions that we have identified have never been reported previously, except for the EGFR-SEPT14 gene fusion.
EGFR amplification is the most common gene alteration in IDH-wild-type GBM, detected in 40% of the cases [32]. EGFR was the most recurrent fusion partner in our series (5/52, 9.6%) and all EGFR fusions co-occurred with CT at 7p11. This is consistent with the work of Frattini et al. who reported EGFR fusions in 7.6% of GBM (but CT was not studied) [19]. Moreover, we observed an EGFR-SEPT14 fusion in 3.8% of GBM. Frattini et al. reported this fusion in 3.2% (6/185) of GBM and additionally demonstrated that such fusions confer sensitivity to EGFR inhibitors [19]. We showed that EGFR-SEPT14 and EGFR-VOPP1 gene fusions shared the same breakpoint within EGFR that had previously been described in the literature [19,33]. Our results suggest that EGFR-SEPT14 and EGFR-VOPP1 may have similar biological activities. Herein, we demonstrate that fusions involving EGFR may be the results of CT in GBM.
CPM fusions, detected in 5.8% of GBM (3/52), might be potential driver events. CPM gene is located at the multi-aberration 12q13–15 locus [34]. Although few data are available, CPM is an extracellular membrane-bound peptidase that cleaves the C-terminal arginine of epidermal growth factor (EGF), resulting in des-Arg-EGF which binds EGFR with equal or higher affinity than native EGF [35]. It is not known whether CPM modulates EGFR signaling but CPM was recently identified as a recurrently amplified gene in liposarcoma and a potential oncogene involved in EGFR signaling in vitro and in vivo (murine xenograft model) [36]. Kanojia et al. showed that CPM knockdown in liposarcoma xenograft significantly decreased tumor growth in vivo [36]. Since our results suggest that CPM fusion and EGFR amplification might be mutually exclusive events (3 cases with CPM fusion that constantly lacked EGFR amplification), we hypothesize that CPM fusions might constitute an alternative mechanism of EGFR activation in a subset of IDH-wild-type GBM. Nonetheless, it is important to mention that CPM gene fusions partially retained the peptidase domain of CPM and that MDM2 and/or CDK4 amplification consistently co-existed with CPM fusions. CPM fusions may be bystander events in GBM. Further investigations are needed to define the exact role of CPM fusions in gliomagenesis, independently of MDM2 and/or CDK4 amplification.
SEPT14, a recurrent fusion partner in our series (4/52, 7.7%), belongs to the septin family, comprised of Ras-like GTPases known to be involved in cancer [37]. Although septins seem to play a role in mechanisms such as tumor proliferation, resistance to apoptosis, cell migration and invasion, a direct relationship between septins and tumorigenesis has yet to be established [37]. Herein, BLVRA-SEPT14 was the only gene fusion that entirely retained the SEPT14 GTPase domain and its C-terminal coiled-coil domain, potentially leading to a chimeric protein with the putative oncogenic functions of SEPT14.
VOPP1 was identified as a potential oncogenic fusion partner. It is a frequently EGFR-coamplified gene (amplified in at least one-third of EGFR-amplified GBM) that has been shown to be a key regulator of NF-kb signaling and to contribute to resistance to apoptosis [38]. However, in our series, gene fusions involving VOPP1 did not preserve the functional domains of VOPP1 and thus, might not be considered as oncogenic.
First described by Stephens et al. in 2011, CT is a cataclysmic phenomenon whose detection, among complex chr rearrangements, is still challenging [9]. The criteria of CT, as defined by Korbel and Campbell, were historically based on whole-genome paired-end DNA sequencing data. They include 1) clustering of breakpoints, 2) regularity of oscillating copy-number states, 3) prevalence of regions with interspersed loss and retention, 4) prevalence of rearrangements affecting a single haplotype, 5) randomness of DNA fragment joins, 6) randomness of DNA fragment order, and 7) ability to walk the derivative chr [9,24]. Herein, CT patterns were investigated by SNP arrays, which allow the detection of the major (first three) features of CT according to this definition [9,24]. As suggested by Korbel and Campbell, the diagnostic criteria must be considered as part of an evolving definition. For instance, for Stephens et al., at least 50 breakpoints per chr should be present but many publications used less stringent criteria, with a threshold of 5 to 20 breakpoints per chr [15,[39], [40], [41]]. Herein, we chose a threshold of 10 breakpoints per chr arm, as most authors did [15,39,[42], [43], [44], [45]]. Using statistical simulation, Kinsella et al. pointed out how challenging CT identification and its distinction from “CT-like events” (which may result from sequential rearrangements) might be [46]. This emphasizes the need for further investigations to refine the diagnostic criteria for CT [46]. Nonetheless, in our study, a subset of GBM harbored chr that had undergone complex genomic rearrangements highly suggestive of CT [9,24]. Studies on larger cohorts are needed in order to confirm our results and potentially identify a recurrent gene fusion (apart from the already known EGFR-SEPT14 fusion) rather than a recurrent partner. At this point, it is, however, difficult to state if the lack of recurrence is due to the relatively small size of our cohort or the random dimension of CT, which by definition reassembles shattered chr haphazardly. Last but not least, we selected potential oncogenic fusion genes based on Oncoscore (measurement of the association of genes to cancer based on citation frequencies in the literature). The relevance of the gene fusions identified needs to be assessed through functional studies. Lentiviral transduction of human (glial or glioma) cell lines would allow assessment of proliferation and migration of cells expressing the fusion gene. Investigating the downstream signaling pathways and the potential sensitivity to specific inhibitors would provide answers as to the role of the gene fusions in GBM.
The exact mechanisms of CT are yet to discover. The most common hypothesis relies on the formation of a micronucleus, which is a consequence of chr segregation errors during mitosis (especially in cells deficient in the p53 checkpoint). Mis-segregated chr become encapsulated into micronuclei with fragmentation of the trapped chr during the following mitosis [47]. Chr pulverization and subsequent non-homologous end-joining (NHEJ)-mediated repair lead to inter- and intra-chromosomal rearrangements and genomic instability [48,49]. Rausch et al. demonstrated an association between TP53 mutation and CT occurrence in Sonic Hedgehog medulloblastoma and acute myeloid leukemia [39]. Herein, our results do not suggest such an association since only 17.3% (9/52) of GBM with CT harbored a TP53 mutation.
In conclusion, CT points to underlying gene fusions in IDH-wild-type GBM. CT detection should incite searching for gene fusions in GBM patients. The functional relevance of CT-related fusions has yet to be demonstrated. Introduction of the fusion genes into cell lines using lentiviral vectors may be a way to assess the potential oncogenic functions of the fusions. Showing enhanced cell proliferation or migration would get scientists and clinicians one step closer to understanding the role of the gene fusions in GBM.
The following are the supplementary data related to this article.
List of the 120 gene fusions identified within CT regions in 52 IDH-wild-type GBM. RNA-seq was performed in 52 GBM detecting 120 gene fusions within CT regions. Thirty gene fusions involving at least one partner with oncogenic properties were identified in 22 IDH-wild-type GBM with CT. List of primers used for validation by RT-PCR followed by Sanger sequencing. Chr: chromosome.
Funding
This work was supported by a grant from La Ligue Contre le Cancer 44, 45 and 49.
CRediT authorship contribution statement
Franck Ah-Pine: Conceptualization, Formal analysis, Investigation, Writing – Original Draft, Visualization
Déborah Casas : Conceptualization, Investigation, Validation, Writing - Review & Editing
Philippe Menei : Resources
Blandine Boisselier: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Writing - Review & Editing
Emmanuel Garcion: Conceptualization, Methodology, Supervision, Project administration, Writing- Review & Editing
Audrey Rousseau: Conceptualization, Methodology, Supervision, Project administration, Funding acquisition, Writing - Review & Editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Déborah Casas, Email: deborah.casas@etud.univ-angers.fr.
Philippe Menei, Email: phmenei@chu-angers.fr.
Emmanuel Garcion, Email: emmanuel.garcion@univ-angers.fr.
Audrey Rousseau, Email: AuRousseau@chu-angers.fr.
References
- 1.Ostrom Q.T., Gittleman H., Liao P., Vecchione-Koval T., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncology. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D.N., Ohgaki H., Wiestler O., Cavenee W. WHO Classification of Tumours of the Central Nervous System. 2016. http://publications.iarc.fr/Book-And-Report-Series/Who-Iarc-Classification-Of-Tumours/Who-Classification-Of-Tumours-Of-The-Central-Nervous-System-2016 International Agency For Research On Cancer. [DOI] [PMC free article] [PubMed]
- 3.Fujisawa H., Reis R.M., Nakamura M., Colella S., Yonekawa Y., Kleihues P., Ohgaki H. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab. Investig. 2000;80:65–72. doi: 10.1038/labinvest.3780009. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.R. McLendon, A. Friedman, D. Bigner, E.G. Van Meir, D.J. Brat, G. M. Mastrogianakis, J.J. Olson, T. Mikkelsen, N. Lehman, K. Aldape, W.K. Alfred Yung, O. Bogler, S. VandenBerg, M. Berger, M. Prados, D. Muzny, M. Morgan, S. Scherer, A. Sabo, L. Nazareth, L. Lewis, O. Hall, Y. Zhu, Y. Ren, O. Alvi, J. Yao, A. Hawes, S. Jhangiani, G. Fowler, A. San Lucas, C. Kovar, A. Cree, H. Dinh, J. Santibanez, V. Joshi, M.L. Gonzalez-Garay, C.A. Miller, A. Milosavljevic, L. Donehower, D.A. Wheeler, R.A. Gibbs, K. Cibulskis, C. Sougnez, T. Fennell, S. Mahan, J. Wilkinson, L. Ziaugra, R. Onofrio, T. Bloom, R. Nicol, K. Ardlie, J. Baldwin, S. Gabriel, E.S. Lander, L. Ding, R.S. Fulton, M.D. McLellan, J. Wallis, D.E. Larson, X. Shi, R. Abbott, L. Fulton, K. Chen, D.C. Koboldt, M.C. Wendl, R. Meyer, Y. Tang, L. Lin, J.R. Osborne, B.H. Dunford-Shore, T.L. Miner, K. Delehaunty, C. Markovic, G. Swift, W. Courtney, C. Pohl, S. Abbott, A. Hawkins, S. Leong, C. Haipek, H. Schmidt, M. Wiechert, T. Vickery, S. Scott, D.J. Dooling, A. Chinwalla, G.M. Weinstock, E.R. Mardis, R.K. Wilson, G. Getz, W. Winckler, R.G.W. Verhaak, M.S. Lawrence, M. O'Kelly, J. Robinson, G. Alexe, R. Beroukhim, S. Carter, D. Chiang, J. Gould, S. Gupta, J. Korn, C. Mermel, J. Mesirov, S. Monti, H. Nguyen, M. Parkin, M. Reich, N. Stransky, B.A. Weir, L. Garraway, T. Golub, M. Meyerson, L. Chin, A. Protopopov, J. Zhang, I. Perna, S. Aronson, N. Sathiamoorthy, G. Ren, J. Yao, W.R. Wiedemeyer, H. Kim, S. Won Kong, Y. Xiao, I.S. Kohane, J. Seidman, P.J. Park, R. Kucherlapati, P.W. Laird, L. Cope, J.G. Herman, D.J. Weisenberger, F. Pan, D. Van Den Berg, L. Van Neste, J. Mi Yi, K.E. Schuebel, S.B. Baylin, D.M. Absher, J.Z. Li, A. Southwick, S. Brady, A. Aggarwal, T. Chung, G. Sherlock, J.D. Brooks, R.M. Myers, P.T. Spellman, E. Purdom, L.R. Jakkula, A. V. Lapuk, H. Marr, S. Dorton, Y. Gi Choi, J. Han, A. Ray, V. Wang, S. Durinck, M. Robinson, N.J. Wang, K. Vranizan, V. Peng, E. Van Name, G. V. Fontenay, J. Ngai, J.G. Conboy, B. Parvin, H.S. Feiler, T.P. Speed, J.W. Gray, C. Brennan, N.D. Socci, A. Olshen, B.S. Taylor, A. Lash, N. Schultz, B. Reva, Y. Antipin, A. Stukalov, B. Gross, E. Cerami, W. Qing Wang, L.-X. Qin, V.E. Seshan, L. Villafania, M. Cavatore, L. Borsu, A. Viale, W. Gerald, C. Sander, M. Ladanyi, C.M. Perou, D. Neil Hayes, M.D. Topal, K.A. Hoadley, Y. Qi, S. Balu, Y. Shi, J. Wu, R. Penny, M. Bittner, T. Shelton, E. Lenkiewicz, S. Morris, D. Beasley, S. Sanders, A. Kahn, R. Sfeir, J. Chen, D. Nassau, L. Feng, E. Hickey, J. Zhang, J.N. Weinstein, A. Barker, D.S. Gerhard, J. Vockley, C. Compton, J. Vaught, P. Fielding, M.L. Ferguson, C. Schaefer, S. Madhavan, K.H. Buetow, F. Collins, P. Good, M. Guyer, B. Ozenberger, J. Peterson, E. Thomson, Comprehensive genomic characterization defines human glioblastoma genes and core pathways, Nature. 455 (2008) 1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed]
- 6.Labussière M., Boisselier B., Mokhtari K., Di Stefano A.-L., Rahimian A., Rossetto M., Ciccarino P., Saulnier O., Paterra R., Marie Y., Finocchiaro G., Sanson M. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83:1200–1206. doi: 10.1212/WNL.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 7.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K.W., Velculescu V.E., Vogelstein B., Bigner D.D. IDH1 and IDH2 mutations in Gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobusawa S., Watanabe T., Kleihues P., Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary Glioblastomas. Clin. Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 9.P.J. Stephens, C.D. Greenman, B. Fu, F. Yang, G.R. Bignell, L.J. Mudie, E.D. Pleasance, K.W. Lau, D. Beare, L.A. Stebbings, S. McLaren, M.-L. Lin, D.J. McBride, I. Varela, S. Nik-Zainal, C. Leroy, M. Jia, A. Menzies, A.P. Butler, J.W. Teague, M.A. Quail, J. Burton, H. Swerdlow, N.P. Carter, L.A. Morsberger, C. Iacobuzio-Donahue, G.A. Follows, A.R. Green, A.M. Flanagan, M.R. Stratton, P.A. Futreal, P.J. Campbell, Massive genomic rearrangement acquired in a single catastrophic event during cancer development, Cell. 144 (2011) 27–40. doi: 10.1016/J.CELL.2010.11.055. [DOI] [PMC free article] [PubMed]
- 10.Jones S., Chen W.-D., Parmigiani G., Diehl F., Beerenwinkel N., Antal T., Traulsen A., Nowak M.A., Siegel C., Velculescu V.E., Kinzler K.W., Vogelstein B., Willis J., Markowitz S.D. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra A., Lindberg M., Faust G.G., Leibowitz M.L., Clark R.A., Layer R.M., Quinlan A.R., Hall I.M. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J.M. Furgason, R.F. Koncar, S.K. Michelhaugh, F.H. Sarkar, S. Mittal, A.E. Sloan, J.S. Barnholtz-Sloan, E.M. Bahassi, Whole genome sequence analysis links chromothripsis to EGFR, MDM2, MDM4, and CDK4 amplification in glioblastoma, Oncoscience. 2 (2015) 618–28. doi:10.18632/oncoscience.178. [DOI] [PMC free article] [PubMed]
- 13.B. Boisselier, F. Dugay, M.-A. Belaud-Rotureau, A. Coutolleau, E. Garcion, P. Menei, P. Guardiola, A. Rousseau, Whole genome duplication is an early event leading to aneuploidy in IDH-wild type glioblastoma, Oncotarget. 9 (2018) 36017–28. doi:10.18632/oncotarget.26330. [DOI] [PMC free article] [PubMed]
- 14.S.G. O'Brien, F. Guilhot, R.A. Larson, I. Gathmann, M. Baccarani, F. Cervantes, J.J. Cornelissen, T. Fischer, A. Hochhaus, T. Hughes, K. Lechner, J.L. Nielsen, P. Rousselot, J. Reiffers, G. Saglio, J. Shepherd, B. Simonsson, A. Gratwohl, J.M. Goldman, H. Kantarjian, K. Taylor, G. Verhoef, A.E. Bolton, R. Capdeville, B.J. Druker, Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia, N. Engl. J. Med. 348 (2003) 994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed]
- 15.P.A. Northcott, D.J.H. Shih, J. Peacock, L. Garzia, A.S. Morrissy, T. Zichner, A.M. Stütz, A. Korshunov, J. Reimand, S.E. Schumacher, R. Beroukhim, D.W. Ellison, C.R. Marshall, A.C. Lionel, S. Mack, A. Dubuc, Y. Yao, V. Ramaswamy, B. Luu, A. Rolider, F.M.G. Cavalli, X. Wang, M. Remke, X. Wu, R.Y.B. Chiu, A. Chu, E. Chuah, R.D. Corbett, G.R. Hoad, S.D. Jackman, Y. Li, A. Lo, K.L. Mungall, K.M. Nip, J.Q. Qian, A.G.J. Raymond, N.T. Thiessen, R.J. Varhol, I. Birol, R.A. Moore, A.J. Mungall, R. Holt, D. Kawauchi, M.F. Roussel, M. Kool, D.T.W. Jones, H. Witt, A. Fernandez-L, A.M. Kenney, R.J. Wechsler-Reya, P. Dirks, T. Aviv, W.A. Grajkowska, M. Perek-Polnik, C.C. Haberler, O. Delattre, S.S. Reynaud, F.F. Doz, S.S. Pernet-Fattet, B.-K. Cho, S.-K. Kim, K.-C. Wang, W. Scheurlen, C.G. Eberhart, M. Fèvre-Montange, A. Jouvet, I.F. Pollack, X. Fan, K.M. Muraszko, G.Y. Gillespie, C. Di Rocco, L. Massimi, E.M.C. Michiels, N.K. Kloosterhof, P.J. French, J.M. Kros, J.M. Olson, R.G. Ellenbogen, K. Zitterbart, L. Kren, R.C. Thompson, M.K. Cooper, B. Lach, R.E. McLendon, D.D. Bigner, A. Fontebasso, S. Albrecht, N. Jabado, J.C. Lindsey, S. Bailey, N. Gupta, W.A. Weiss, L. Bognár, A. Klekner, T.E. Van Meter, T. Kumabe, T. Tominaga, S.K. Elbabaa, J.R. Leonard, J.B. Rubin, L.M. Liau, E.G. Van Meir, M. Fouladi, H. Nakamura, G. Cinalli, M. Garami, P. Hauser, A.G. Saad, A. Iolascon, S. Jung, C.G. Carlotti, R. Vibhakar, Y.S. Ra, S. Robinson, M. Zollo, C.C. Faria, J.A. Chan, M.L. Levy, P.H.B. Sorensen, M. Meyerson, S.L. Pomeroy, Y.-J. Cho, G.D. Bader, U. Tabori, C.E. Hawkins, E. Bouffet, S.W. Scherer, J.T. Rutka, D. Malkin, S.C. Clifford, S.J.M. Jones, J.O. Korbel, S.M. Pfister, M.A. Marra, M.D. Taylor, Subgroup-specific structural variation across 1,000 medulloblastoma genomes., Nature. 488 (2012) 49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed]
- 16.Iwakawa R., Takenaka M., Kohno T., Shimada Y., Totoki Y., Shibata T., Tsuta K., Nishikawa R., Noguchi M., Sato-Otsubo A., Ogawa S., Yokota J. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosom. Cancer. 2013;52:802–816. doi: 10.1002/gcc.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R.D. Morin, K. Mungall, E. Pleasance, A.J. Mungall, R. Goya, R.D. Huff, D.W. Scott, J. Ding, A. Roth, R. Chiu, R.D. Corbett, F.C. Chan, M. Mendez-Lago, D.L. Trinh, M. Bolger-Munro, G. Taylor, A. Hadj Khodabakhshi, S. Ben-Neriah, J. Pon, B. Meissner, B. Woolcock, N. Farnoud, S. Rogic, E.L. Lim, N.A. Johnson, S. Shah, S. Jones, C. Steidl, R. Holt, I. Birol, R. Moore, J.M. Connors, R.D. Gascoyne, M.A. Marra, Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing., Blood. 122 (2013) 1256–65. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed]
- 18.Charest A., Lane K., McMahon K., Park J., Preisinger E., Conroy H., Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21), genes. Chromosom. Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 19.Frattini V., Trifonov V., Chan J.M., Castano A., Lia M., Abate F., Keir S.T., Ji A.X., Zoppoli P., Niola F., Danussi C., Dolgalev I., Porrati P., Pellegatta S., Heguy A., Gupta G., Pisapia D.J., Canoll P., Bruce J.N., McLendon R.E., Yan H., Aldape K., Finocchiaro G., Mikkelsen T., Privé G.G., Bigner D.D., Lasorella A., Rabadan R., Iavarone A. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013;45:1141. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. Singh, J.M. Chan, P. Zoppoli, F. Niola, R. Sullivan, A. Castano, E.M. Liu, J. Reichel, P. Porrati, S. Pellegatta, K. Qiu, Z. Gao, M. Ceccarelli, R. Riccardi, D.J. Brat, A. Guha, K. Aldape, J.G. Golfinos, D. Zagzag, T. Mikkelsen, G. Finocchiaro, A. Lasorella, R. Rabadan, A. Iavarone, Transforming Fusions of FGFR and TACC Genes in Human Glioblastoma, Science (80-. ). 337 (2012) 1231–35. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed]
- 21.Parker B.C., Annala M.J., Cogdell D.E., Granberg K.J., Sun Y., Ji P., Li X., Gumin J., Zheng H., Hu L., Yli-Harja O., Haapasalo H., Visakorpi T., Liu X., Liu C., Sawaya R., Fuller G.N., Chen K., Lang F.F., Nykter M., Zhang W. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J. Clin. Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardina P.J., Lo K.C., Lee W., Cowell J.K., Turpaz Y. Ploidy status and copy number aberrations in primary glioblastomas defined by integrated analysis of allelic ratios, signal ratios and loss of heterozygosity using 500K SNP mapping arrays. BMC Genomics. 2008;9:489. doi: 10.1186/1471-2164-9-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova T., Manié E., Stoppa-Lyonnet D., Rigaill G., Barillot E., Stern M.H. Genome alteration print (GAP): A tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korbel J.O., Campbell P.J. Criteria for inference of Chromothripsis in Cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/J.CELL.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Liu J., Ouyang L., Chen Y., Liu B., Cai H. CTLPScanner: a web server for chromothripsis-like pattern detection. Nucleic Acids Res. 2016;44:W252–W258. doi: 10.1093/nar/gkw434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.-D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piazza R., Ramazzotti D., Spinelli R., Pirola A., De Sano L., Ferrari P., Magistroni V., Cordani N., Sharma N., Gambacorti-Passerini C. OncoScore: a novel, internet-based tool to assess the oncogenic potential of genes. Sci. Rep. 2017;7:46290. doi: 10.1038/srep46290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Lågstad, S. Zhao, A.M. Hoff, B. Johannessen, O.C. Lingjærde, R.I. Skotheim, chimeraviz: a tool for visualizing chimeric RNA, Bioinformatics. 33 (2017) 2954–2956. doi: 10.1093/bioinformatics/btx329. [DOI] [PMC free article] [PubMed]
- 29.Murphy C., Elemento O. AGFusion: annotate and visualize gene fusions. BioRxiv. 2016;080903 doi: 10.1101/080903. [DOI] [Google Scholar]
- 30.A.L. Di Stefano, A. Fucci, V. Frattini, M. Labussiere, K. Mokhtari, P. Zoppoli, Y. Marie, A. Bruno, B. Boisselier, M. Giry, J. Savatovsky, M. Touat, H. Belaid, A. Kamoun, A. Idbaih, C. Houillier, F.R. Luo, J.-C. Soria, J. Tabernero, M. Eoli, R. Paterra, S. Yip, K. Petrecca, J.A. Chan, G. Finocchiaro, A. Lasorella, M. Sanson, A. Iavarone, Detection, characterization, and inhibition of FGFR-TACC fusions in IDH Wild-type glioma., Clin. Cancer Res. 21 (2015) 3307–17. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed]
- 31.Lasorella A., Sanson M., Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro-Oncology. 2017;19:475–483. doi: 10.1093/neuonc/now240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C.W. Brennan, R.G.W. Verhaak, A. McKenna, B. Campos, H. Noushmehr, S.R. Salama, S. Zheng, D. Chakravarty, J.Z. Sanborn, S.H. Berman, R. Beroukhim, B. Bernard, C.-J. Wu, G. Genovese, I. Shmulevich, J. Barnholtz-Sloan, L. Zou, R. Vegesna, S.A. Shukla, G. Ciriello, W.K. Yung, W. Zhang, C. Sougnez, T. Mikkelsen, K. Aldape, D.D. Bigner, E.G. Van Meir, M. Prados, A. Sloan, K.L. Black, J. Eschbacher, G. Finocchiaro, W. Friedman, D.W. Andrews, A. Guha, M. Iacocca, B.P. O'Neill, G. Foltz, J. Myers, D.J. Weisenberger, R. Penny, R. Kucherlapati, C.M. Perou, D.N. Hayes, R. Gibbs, M. Marra, G.B. Mills, E. Lander, P. Spellman, R. Wilson, C. Sander, J. Weinstein, M. Meyerson, S. Gabriel, P.W. Laird, D. Haussler, G. Getz, L. Chin, C. TCGA Research Network, J. Barnholtz-Sloan, W. Barrett, Q. Ostrom, Y. Wolinsky, K.L. Black, B. Bose, P.T. Boulos, M. Boulos, J. Brown, C. Czerinski, M. Eppley, M. Iacocca, T. Kempista, T. Kitko, Y. Koyfman, B. Rabeno, P. Rastogi, M. Sugarman, P. Swanson, K. Yalamanchii, I.P. Otey, Y.S. Liu, Y. Xiao, J.T. Auman, P.-C. Chen, A. Hadjipanayis, E. Lee, S. Lee, P.J. Park, J. Seidman, L. Yang, R. Kucherlapati, S. Kalkanis, T. Mikkelsen, L.M. Poisson, A. Raghunathan, L. Scarpace, B. Bernard, R. Bressler, A. Eakin, L. Iype, R.B. Kreisberg, K. Leinonen, S. Reynolds, H. Rovira, V. Thorsson, I. Shmulevich, M.J. Annala, R. Penny, J. Paulauskis, E. Curley, M. Hatfield, D. Mallery, S. Morris, T. Shelton, C. Shelton, M. Sherman, P. Yena, L. Cuppini, F. DiMeco, M. Eoli, G. Finocchiaro, E. Maderna, B. Pollo, M. Saini, S. Balu, K.A. Hoadley, L. Li, C.R. Miller, Y. Shi, M.D. Topal, J. Wu, G. Dunn, C. Giannini, B.P. O'Neill, B.A. Aksoy, Y. Antipin, L. Borsu, S.H. Berman, C.W. Brennan, E. Cerami, D. Chakravarty, G. Ciriello, J. Gao, B. Gross, A. Jacobsen, M. Ladanyi, A. Lash, Y. Liang, B. Reva, C. Sander, N. Schultz, R. Shen, N.D. Socci, A. Viale, M.L. Ferguson, Q.-R. Chen, J.A. Demchok, L.A.L. Dillon, K.R.M. Shaw, M. Sheth, R. Tarnuzzer, Z. Wang, L. Yang, T. Davidsen, M.S. Guyer, B.A. Ozenberger, H.J. Sofia, J. Bergsten, J. Eckman, J. Harr, J. Myers, C. Smith, K. Tucker, C. Winemiller, L.A. Zach, J.Y. Ljubimova, G. Eley, B. Ayala, M.A. Jensen, A. Kahn, T.D. Pihl, D.A. Pot, Y. Wan, J. Eschbacher, G. Foltz, N. Hansen, P. Hothi, B. Lin, N. Shah, J. Yoon, C. Lau, M. Berens, K. Ardlie, R. Beroukhim, S.L. Carter, A.D. Cherniack, M. Noble, J. Cho, K. Cibulskis, D. DiCara, S. Frazer, S.B. Gabriel, N. Gehlenborg, J. Gentry, D. Heiman, J. Kim, R. Jing, E.S. Lander, M. Lawrence, P. Lin, W. Mallard, M. Meyerson, R.C. Onofrio, G. Saksena, S. Schumacher, C. Sougnez, P. Stojanov, B. Tabak, D. Voet, H. Zhang, L. Zou, G. Getz, N.N. Dees, L. Ding, L.L. Fulton, R.S. Fulton, K.-L. Kanchi, E.R. Mardis, R.K. Wilson, S.B. Baylin, D.W. Andrews, L. Harshyne, M.L. Cohen, K. Devine, A.E. Sloan, S.R. VandenBerg, M.S. Berger, M. Prados, D. Carlin, B. Craft, K. Ellrott, M. Goldman, T. Goldstein, M. Grifford, D. Haussler, S. Ma, S. Ng, S.R. Salama, J.Z. Sanborn, J. Stuart, T. Swatloski, P. Waltman, J. Zhu, R. Foss, B. Frentzen, W. Friedman, R. McTiernan, A. Yachnis, D.N. Hayes, C.M. Perou, S. Zheng, R. Vegesna, Y. Mao, R. Akbani, K. Aldape, O. Bogler, G.N. Fuller, W. Liu, Y. Liu, Y. Lu, G. Mills, A. Protopopov, X. Ren, Y. Sun, C.-J. Wu, W.K.A. Yung, W. Zhang, J. Zhang, K. Chen, J.N. Weinstein, L. Chin, R.G.W. Verhaak, H. Noushmehr, D.J. Weisenberger, M.S. Bootwalla, P.H. Lai, T.J. Triche, D.J. Van Den Berg, P.W. Laird, D.H. Gutmann, N.L. Lehman, E.G. VanMeir, D. Brat, J.J. Olson, G.M. Mastrogianakis, N.S. Devi, Z. Zhang, D. Bigner, E. Lipp, R. McLendon, The somatic genomic landscape of glioblastoma., Cell. 155 (2013) 462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed]
- 33.L. Erdem-Eraslan, M.J. van den Bent, Y. Hoogstrate, H. Naz-Khan, A. Stubbs, P. van der Spek, R. Böttcher, Y. Gao, M. de Wit, W. Taal, H.M. Oosterkamp, A. Walenkamp, L. V Beerepoot, M.C.J. Hanse, J. Buter, A.H. Honkoop, B. van der Holt, R.M. Vernhout, P.A.E.S. Smitt, J.M. Kros, P.J. French, Identification of patients with recurrent glioblastoma who may benefit from combined bevacizumab and CCNU therapy: a report from the BELOB trial, Cancer Res. 76 (2016) 525. doi: 10.1158/0008-5472.CAN-15-0776. [DOI] [PubMed]
- 34.Wanschura S., Kazmierczak B., Schoenmakers E., Meyen E., Bartnitzke S., Van de Ven W., Bullerdiek J., Schloot W. Regional fine mapping of the multiple-aberration region involved in uterine leiomyoma, lipoma, and pleomorphic adenoma of the salivary gland to 12q15. Genes Chromosom. Cancer. 1995;14:68–70. doi: 10.1002/gcc.2870140112. [DOI] [PubMed] [Google Scholar]
- 35.McGwire G.B., Skidgel R.A. Extracellular conversion of epidermal growth factor (EGF) to des-Arg53-EGF by carboxypeptidase M. J. Biol. Chem. 1995;270:17154–17158. doi: 10.1074/JBC.270.29.17154. [DOI] [PubMed] [Google Scholar]
- 36.D. Kanojia, Y. Nagata, M. Garg, D.H. Lee, A. Sato, K. Yoshida, Y. Sato, M. Sanada, A. Mayakonda, C. Bartenhagen, H.-U. Klein, N.B. Doan, J.W. Said, S. Mohith, S. Gunasekar, Y. Shiraishi, K. Chiba, H. Tanaka, S. Miyano, O. Myklebost, H. Yang, M. Dugas, L.A. Meza-Zepeda, A.W. Silberman, C. Forscher, J.W. Tyner, S. Ogawa, H.P. Koeffler, Genomic landscape of liposarcoma, Oncotarget. 6 (2015) 42429–44. doi:10.18632/oncotarget.6464. [DOI] [PMC free article] [PubMed]
- 37.Angelis D., Spiliotis E.T. Septin mutations in human cancers. Front. Cell Dev. Biol. 2016;4:122. doi: 10.3389/fcell.2016.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S., James C.D. ECop (EGFR-Coamplified and overexpressed protein), a novel protein, regulates NF-κB transcriptional activity and associated apoptotic response in an IκBα-dependent manner. Oncogene. 2005;24:2495–2502. doi: 10.1038/sj.onc.1208496. [DOI] [PubMed] [Google Scholar]
- 39.T. Rausch, D.T.W. Jones, M. Zapatka, A.M. Stütz, T. Zichner, J. Weischenfeldt, N. Jäger, M. Remke, D. Shih, P.A. Northcott, E. Pfaff, J. Tica, Q. Wang, L. Massimi, H. Witt, S. Bender, S. Pleier, H. Cin, C. Hawkins, C. Beck, A. von Deimling, V. Hans, B. Brors, R. Eils, W. Scheurlen, J. Blake, V. Benes, A.E. Kulozik, O. Witt, D. Martin, C. Zhang, R. Porat, 0o, Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations., Cell. 148 (2012) 59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed]
- 40.Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I., Hamdi M., van Nes J., Westerman B.A., van Arkel J., Ebus M.E., Haneveld F., Lakeman A., Schild L., Molenaar P., Stroeken P., van Noesel M.M., Ora I., Santo E.E., Caron H.N., Westerhout E.M., Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 41.C. Chiang, J.C. Jacobsen, C. Ernst, C. Hanscom, A. Heilbut, I. Blumenthal, R.E. Mills, A. Kirby, A.M. Lindgren, S.R. Rudiger, C.J. McLaughlan, C.S. Bawden, S.J. Reid, R.L.M. Faull, R.G. Snell, I.M. Hall, Y. Shen, T.K. Ohsumi, M.L. Borowsky, M.J. Daly, C. Lee, C.C. Morton, M.E. MacDonald, J.F. Gusella, M.E. Talkowski, Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration., Nat. Genet. 44 (2012) 390–7. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed]
- 42.T.-M. Kim, S.-H. Jung, C.H. An, S.H. Lee, I.-P. Baek, M.S. Kim, S.-W. Park, J.-K. Rhee, S.-H. Lee, Y.-J. Chung, Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity., Clin. Cancer Res. 21 (2015) 4461–72. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed]
- 43.Kim T.-M., Xi R., Luquette L.J., Park R.W., Johnson M.D., Park P.J. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res. 2013;23:217–227. doi: 10.1101/gr.140301.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Poppelen N.M., Yavuzyigitoglu S., Smit K.N., Vaarwater J., Eussen B., Brands T., Paridaens D., Kiliç E., de Klein A. Chromosomal rearrangements in uveal melanoma: Chromothripsis. Genes Chromosom. Cancer. 2018;57:452–458. doi: 10.1002/gcc.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abáigar M., Robledo C., Benito R., Ramos F., Díez-Campelo M., Hermosín L., Sánchez-Del-Real J., Alonso J.M., Cuello R., Megido M., Rodríguez J.N., Martín-Núñez G., Aguilar C., Vargas M., Martín A.A., García J.L., Kohlmann A., Del Cañizo M.C., Hernández-Rivas J.M. Chromothripsis is a recurrent genomic abnormality in high-risk myelodysplastic syndromes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinsella M., Patel A., Bafna V. The elusive evidence for chromothripsis. Nucleic Acids Res. 2014;42:8231–8242. doi: 10.1093/nar/gku525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ly P., Brunner S.F., Shoshani O., Kim D.H., Lan W., Pyntikova T., Flanagan A.M., Behjati S., Page D.C., Campbell P.J., Cleveland D.W. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019;51:705–715. doi: 10.1038/s41588-019-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.K. Crasta, N.J. Ganem, R. Dagher, A.B. Lantermann, E. V Ivanova, Y. Pan, L. Nezi, A. Protopopov, D. Chowdhury, D. Pellman, DNA breaks and chromosome pulverization from errors in mitosis, Nature. 482 (2012) 53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed]
- 49.Ly P., Cleveland D.W. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 2017;27:917–930. doi: 10.1016/j.tcb.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the 120 gene fusions identified within CT regions in 52 IDH-wild-type GBM. RNA-seq was performed in 52 GBM detecting 120 gene fusions within CT regions. Thirty gene fusions involving at least one partner with oncogenic properties were identified in 22 IDH-wild-type GBM with CT. List of primers used for validation by RT-PCR followed by Sanger sequencing. Chr: chromosome.