Abstract
Pimozide is a dopamine receptor antagonist indicated for the treatment of Tourette syndrome. Prior in vitro studies characterized N-dealkylation of pimozide to 1,3-dihydro-1-(4-piperidinyl)-2H-benzimidazol-2-one (DHPBI) via CYP3A4 and, to a lesser extent, CYP1A2 as the only notable routes of pimozide biotransformation. However, drug-drug interactions between pimozide and CYP2D6 inhibitors and CYP2D6 genotype–dependent effects have since been observed. To reconcile these incongruities between the prior in vitro and in vivo studies, we characterized two novel pimozide metabolites: 5-hydroxypimozide and 6-hydroxypimozide. Notably, 5-hydroxypimozide was the major metabolite produced by recombinant CYP2D6 (Km ∼82 nM, Vmax ∼0.78 pmol/min per picomoles), and DHPBI was the major metabolite produced by recombinant CYP3A4 (apparent Km ∼1300 nM, Vmax ∼2.6 pmol/min per picomoles). Kinetics in pooled human liver microsomes (HLMs) for the 5-hydroxylation (Km ∼2200 nM, Vmax ∼59 pmol/min per milligram) and N-dealkylation (Km ∼3900 nM, Vmax ∼600 pmol/min per milligram) reactions were also determined. Collectively, formation of DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide accounted for 90% of pimozide depleted in incubations of NADPH-supplemented pooled HLMs. Studies conducted in HLMs isolated from individual donors with specific cytochrome P450 isoform protein abundances determined via mass spectrometry revealed that 5-hydroxypimozide (r2 = 0.94) and 6-hydroxypimozide (r2 = 0.86) formation rates were correlated with CYP2D6 abundance, whereas the DHPBI formation rate (r2 = 0.98) was correlated with CYP3A4 abundance. Furthermore, the HLMs differed with respect to their capacity to form 5-hydroxypimozide relative to DHPBI. Collectively, these data confirm a role for CYP2D6 in pimozide clearance via 5-hydroxylation and provide an explanation for a lack of involvement when only DHPBI formation was monitored in prior in vitro studies.
SIGNIFICANCE STATEMENT
Current CYP2D6 genotype–guided dosing information in the pimozide label is discordant with available knowledge regarding the primary biotransformation pathways. Herein, we characterize the CYP2D6-dependent biotransformation of pimozide to previously unidentified metabolites. In human liver microsomes, formation rates for the novel metabolites and a previously identified metabolite were determined to be a function of CYP2D6 and CYP3A4 content, respectively. These findings provide a mechanistic basis for observations of CYP2D6 genotype–dependent pimozide clearance in vivo.
Introduction
Tourette syndrome is a neuropsychiatric disorder that affects approximately 1% of children worldwide (Robertson, 2008). Two of the typical antipsychotics (pimozide and haloperidol) and a single atypical antipsychotic (aripiprazole) are the only drugs holding Food and Drug Administration–approved indications for the treatment of Tourette syndrome symptoms (Sallee et al., 2017). Although these drugs appear to be comparably effective, both pimozide and aripiprazole elicit fewer extrapyramidal adverse effects than haloperidol (Shapiro et al., 1989; Sallee et al., 1997; Yoo et al., 2011). However, in the case of pimozide, clinical use remains limited because of concerns for potassium channel–dependent QT interval prolongation (Haddad and Anderson, 2002; Gulisano et al., 2011; Beach et al., 2013).
Although evidence suggests that the association between pimozide administration and QT interval prolongation is concentration-dependent (Flockhart et al., 2000; Drolet et al., 2001), there are conflicting data regarding which clearance pathways meaningfully affect pimozide exposure. An early clinical study in a small set of subjects who were administered tritiated pimozide found that an N-dealkylation product of pimozide constituted the major source of radioactivity in the urine (Baro et al., 1972). Subsequent experiments in human liver microsomes (HLMs) further characterized 1,3-dihydro-1-(4-piperidinyl)-2H-benzimidazol-2-one (DHPBI), an N-dealkylation product of pimozide (Desta et al., 1998). Additional experiments applying cytochrome P450 isoform–specific inhibitors to HLMs and using recombinant cytochrome P450 expression systems demonstrated that the biotransformation of pimozide to DHPBI was principally mediated by CYP3A and, to a much lesser extent, by CYP1A2 (Desta et al., 1998). Interestingly, one study observed depletion of pimozide by recombinant CYP2D6 that was inhibitable by quinidine, a selective inhibitor of CYP2D6. However, DHPBI formation was not observed, and the product of the CYP2D6-mediated reaction was not reported (Flockhart et al., 1997). Subsequent experiments confirmed that quinidine coincubation did not reduce DHPBI formation in HLMs (Desta et al., 1998).
In the 1990s, case reports of life-threatening and fatal interactions between pimozide and other drugs, notably clarithromycin and certain selective serotonin reuptake inhibitors, emerged (Ahmed et al., 1993; Horrigan and Barnhill, 1994; Flockhart et al., 1996; McIntyre et al., 1997);. In response, clinical studies were conducted to better characterize the magnitude of the interactions with clarithromycin and the selective serotonin reuptake inhibitors sertraline and paroxetine (Desta et al., 1999; Alderman, 2005; http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/020031s05920710s023lbl.pdf). Although clarithromycin is known to be a potent inhibitor of CYP3A isoforms (Zhou et al., 2007), sertraline and paroxetine have traditionally been classified as inhibitors of CYP2D6, with a low potency for inhibiting CYP3A (Hemeryck and Belpaire, 2002). Nevertheless, all three drugs were observed to be capable of increasing pimozide exposure in vivo.
In 2007, a clinical study in adults demonstrated that the oral clearance of pimozide was 3-fold less in genotypic CYP2D6 poor metabolizers (PMs) compared with those genotyped as CYP2D6 normal metabolizers (Nucci and Gomeni, 2007). The identification of a potential role for CYP2D6 in the metabolism of pimozide provided a basis for rationalizing how sertraline and paroxetine, both established CYP2D6 inhibitors, could result in alterations in pimozide oral clearance. Although a proper explanation for the discrepancies between the in vitro and in vivo data remained absent, the Food and Drug Administration nevertheless requested changes to the pimozide product labeling to include a contraindication for the coadministration of strong CYP2D6 inhibitors (Rogers et al., 2012). Furthermore, multiple-dose simulations relying upon parameter estimates from the single-dose clinical study were used to establish CYP2D6 genotype–dependent dosing recommendations for pimozide in the Food and Drug Administration –approved labeling (Rogers et al., 2012).
In light of these changes in the pimozide labeling and the drug’s narrow therapeutic index, it is crucial to reconcile the contrasting results from the in vitro and in vivo investigations of pimozide metabolism. As such, we have revisited the characterization of pimozide metabolism in HLMs and recombinant cytochrome P450 expression systems. In the process, we have characterized two novel metabolites of pimozide that are preferentially produced by CYP2D6. Because the previous in vitro characterization of pimozide metabolism was conducted at exclusively supratherapeutic concentrations that might have underestimated the relative contribution of high-affinity, low-capacity enzymes to overall metabolism in vivo (Desta et al., 1998), we have expressly designed our kinetic experiments to include therapeutically relevant pimozide concentrations (McCreadie et al., 1984). Through the use of HLMs isolated from individual donors with known cytochrome P450 isoform abundances, we also evaluated the suitability of the formation of pimozide metabolites as potential probes of CYP2D6 and CYP3A4 function in vitro. The data suggest that CYP2D6-mediated biotransformation may represent a more dominant clearance pathway for pimozide than CYP3A4 in certain individuals.
Materials and Methods
Materials and Reagents.
Pimozide was obtained from the United States Pharmacopeia (North Bethesda, MD). 5-Hydroxypimozide and 6-hydropxypimozide were custom synthesized by Dalton Research Molecules (Toronto, Canada). Pimozide-d5 and DHPBI-d5 were purchased from Toronto Research Chemicals (North York, Canada). EasyCYP Bactosome recombinant human cytochrome P450 isoforms with coexpressed cytochrome P450 reductase and human liver microsomes were obtained from Sekisui XenoTech (Kansas City, KS). Magnesium chloride, DHPBI, NADPH, EDTA, potassium monophosphate, and potassium diphosphate were obtained from Sigma-Aldrich (St. Louis, MO). Analytical-grade acetonitrile, methanol, and formic acid were purchased from Thermo Fisher Scientific (Waltham, MA).
Metabolite Identification.
Experiments to identify novel products of CYP2D6-mediated pimozide metabolism were conducted using recombinant human CYP2D6 with coexpressed cytochrome P450 reductase (product # CYP/EZ007) and HLMs from two individual donors (product/lot # H0033/0210015 and H0099/1110299) that were selected to represent the two extremes of CYP2D6 genotype–predicted phenotype: a PM (CYP2D6*4/*5) and an ultrarapid metabolizer (UM) (CYP2D6*1/*2x2). The CYP2D6 genotype for each of the HLM donors was provided by the supplier, Sekisui XenoTech. Each of these exploratory experiments was performed once in technical duplicate and deviated from the general incubation protocol that was subsequently optimized for the quantitative experiments. Recombinant CYP2D6 Bactosomes (10 pmol/ml, 250-μl incubation volume) with coexpressed cytochrome P450 reductase and HLMs (500 μg/ml, 250-μl incubation volume) were each preincubated for 5 minutes at 37°C in 100 mM potassium phosphate buffer (pH∼7.4) containing the following pimozide concentrations: 0, 25, 50, 100, 1000, and 10,000 nM. Reactions were initiated with 1 mM NADPH and quenched after 30 minutes with 125 μl of ice-cold methanol containing internal standards (100 nM pimozide-d5 and DHPBI-d5). Samples were centrifuged at 16.3g for 10 minutes. Supernatant was collected and applied to an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 100 mm) on an Acquity M-Class UPLC in tandem with a XEVO G2-XS quadrupole time-of-flight (Q-TOF) mass spectrometer (Waters Corporation, Milford, MA). The typical limit of detection for analytes on this instrument is 1–10 nM. The aqueous mobile phase consisted of 0.1% formic acid in water (solvent A), and the organic mobile phase consisted of 0.1% formic acid in acetonitrile (solvent B). The UPLC mobile gradient used is as follows: initial conditions of solvent were 1% solvent B held for 0.2 minutes; the percent of solvent B increased linearly to 99% at 6 minutes and was held until 7 minutes. Q-TOF data were assessed, in particular comparing the metabolite profile from the CYP2D6 PM and the UM incubated with 10,000 nM pimozide. Daughter fragmentation was assessed for pimozide and two putative metabolites with the same column and solvent gradient on a XEVO-TQD (Waters Corporation); further details for the assay conditions are provided in the Supplemental Information (Supplemental Table 1). SmartCYP software was then used to determine the most likely locations for CYP2D6-dependent oxidation of pimozide (Rydberg et al., 2010). Once standards for the suspected metabolites were synthesized, further incubations were conducted to identify the major site of metabolism by CYP2D6 and to confirm prior observations that DHPBI is the major metabolite of pimozide produced by CYP3A4. These follow-up incubations were conducted in singlet and used 10 nM (a nominally therapeutic concentration) of pimozide and 2 ml of 2 pmol/ml recombinant human CYP2D6 and CYP3A4. The incubations were quenched 8 minutes after initiation with NADPH and adhered to the general incubation conditions. These samples were analyzed according to the XEVO TQ-XS UPLC-MS/MS method described below, and a comparison of retention times and mass transitions was made to chromatograms from standards of DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide.
Quantitative UPLC-MS/MS Analysis.
Samples and calibration curve standards were centrifuged at 16.3g for 5 minutes to pellet protein. Subsequently, 100 μl of supernatant was transferred to glass vials containing 400 μl deionized water. Analytes were quantified via ultraperformance liquid chromatography (UPLC) and tandem mass spectrometry (MS/MS) on an Acquity UPLC I-Class coupled to a Xevo TQ-XS triple quadrupole mass spectrometer (Waters Corporation) equipped with an electrospray ionization source. Chromatographic separation of DHPBI, 5-hydroxypimozide, 6-hydroxypimozide, and pimozide was achieved on a Waters Acquity BEH C18 column (1.7 μm, 2.1 × 100 mm) heated to 60°C. Mobile phase A (aqueous) consisted of 0.1% formic acid in water, and mobile phase B (organic) consisted of 0.1% formic acid in acetonitrile. An initial flow rate of 0.4 ml/min of 95% phase A and 5% phase B was used. From 0.25 to 2 minutes, a linear gradient increasing the proportion of phase B to 25% was established, followed by a brief linear gradient to 30% phase B at 2.05 minutes. Proportion of phase B further increased along a linear gradient to 35% at 5 minutes and then along another linear gradient to 40% at 7 minutes. The column was then washed with 98% of phase B at 0.5 ml/min and equilibrated to initial conditions prior to the next sample injection. The mass spectrometer was operated in positive ion mode, and multiple reaction monitoring was employed. The mass transitions for the analytes were as follows: DHPBI was m/z 218.10→84.00, DHPBI-d5 was m/z 223.10→89.00, pimozide was m/z 462.30→328.10, pimozide-d5 was m/z 467.30→333.10, and 5-hydroxypimozide and 6-hydroxypimozide were m/z 478.30→328.10. Further details for the assay conditions are provided in the Supplemental Information (Supplemental Table 2). Standard curves were used in the estimation of analyte concentrations, and specific ranges are listed within respective methods sections. Peak areas were integrated, and sample concentrations were estimated in TargetLynx software version 4.2 (Waters Corporation).
General Incubation Conditions.
Incubations were conducted in 50 mM potassium phosphate buffer containing 5 mM magnesium chloride, 1 mM EDTA, and either purified recombinant cytochrome P450s (EasyCYP Bactosomes) or HLMs. Protein and substrate (pimozide) concentrations varied depending upon the specific experiment, but acetonitrile, the vehicle for pimozide, was limited to 0.5% v/v during all incubations. After a 5-minute preincubation step to equilibrate reactions to 37°C, experiments were initiated with the addition of NADPH (1 mM final concentration). Parallel incubations without NADPH were used as negative controls, with the exception of the recombinant cytochrome P450 panel that used Bactosomes prepared from a blank vector as a control. Duration of incubations varied across the sets of experiments. Reactions were quenched with 2-fold greater volume of ice-cold acetonitrile containing 40 nM DHPBI-d5 (internal standard for DHPBI) and 400 nM pimozide-d5 (internal standard for pimozide, 5-hydroxypimozide, and 6-hydroxypimozide). Any deviations from these general incubation conditions for any experiments are described in the relevant methods sections.
Molar Balance in Pooled HLMs.
Using the general incubation conditions, 200 μl of 200 μg/ml pooled HLMs (200 donors, 50% female, product/lot # H2620/1910096) was incubated with 50 nM pimozide for 20 minutes. These conditions were not linear with respect to time. The experiment was repeated in triplicate with technical duplicates. Concentrations of DHPBI, pimozide, 5-hydroxypimozide, and 6-hydroxypimozide were determined in the samples and compared with concentrations of pimozide in the incubation buffer at time 0 that were proportionally diluted with buffer absent of NADPH. Concentrations of analytes were determined via the quantitative UPLC-MS/MS assay using calibration curves ranging from 0.25 to 50 nM for 5-/6-hydroxypimozide and 1–50 nM DHPBI. To estimate substrate depletion, pimozide concentrations (calibration curve 5–500 nM) were also determined for this experiment. Analyte concentrations were converted to molar amounts, and the percentage of NADPH-dependent pimozide loss over the course of the incubation was calculated using eq. 1.
| (1) |
The amounts of DHPBI, 5-hydroxpimozide, 6-hydroxypimozide, and pimozide measured at 20 minutes are represented by the parameters DHPBIt=20, 5OHPIMt=20, 6OHPIMt=20, and PIMt=20, respectively. The starting amount of pimozide is represented by the PIMt=0 parameter.
Panel of Recombinant Cytochrome P450 Isoforms.
The capacity to catalyze the formation of DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide from pimozide (2500 nM) was evaluated in purified recombinant cytochrome P450 enzymes with coexpressed cytochrome P450 reductase, and some coexpressing cytochrome b5, for the following 14 known drug-metabolizing isoforms (EasyCYP Bactosome product numbers provided in parentheses): CYP1A1 (CYP/EZ018), CYP1A2 (CYP/EZ012), CYP2A6 (CYP/EZ011), CYP2B6 (CYP/EZ016), CYP2C8 (CYP/EZ047), CYP2C9 + b5 (CYP/EZ038), CYP2C18 (CYP/EZ022), CYP2C19 (CYP/EZ028), CYP2D6 (CYP/EZ013), CYP2E1 + b5 (CYP/EZ036), CYP2J2 (CYP/EZ034), CYP3A4 + b5 (CYP/EZ035), CYP3A5 + b5 (CYP/EZ045) and CYP3A7 + b5 (CYP/EZ060). The general incubation conditions were followed. All cytochrome P450 isoforms were evaluated at a protein concentration of 2 pmol/ml (final incubation volume = 200 μl), and Bactosomes prepared from a blank vector were used as a control. Incubations were quenched at 30 minutes after initiation with NADPH. These conditions were not confirmed to be linear with respect to time. The experiment was repeated in triplicate with technical singlets. Concentrations of analytes were determined via the quantitative UPLC-MS/MS assay using calibration curves with the following quantitative ranges: 1–250 nM for 5-/6-hydroxypimozide and 5–250 nM for DHPBI.
Enzyme Kinetics.
Under the general incubation conditions, EasyCYP Bactosomes for CYP2D6 (product # CYP/EZ013) and CYP3A4 (product # CYP/EZ035) were incubated at cytochrome P450 protein concentrations of 1 pmol/ml and 2 pmol/ml, respectively. Kinetic experiments using HLMs pooled from 200 donors (50% female, product/lot # H2620/1910096) were conducted at protein concentration of 50 μg/ml. A range of pimozide concentrations for CYP2D6 Bactosomes (10–2500 nM), CYP3A4 Bactosomes (10–5000 nM), and pooled HLMs (25–10,000 nM) were selected to span both nominally therapeutic and enzyme-saturating substrate concentrations. Final incubation volumes were 400 μl for recombinant expression systems and 200 μl for pooled HLMs. The durations of the incubations were 5 minutes for both pooled HLMs and CYP2D6 Bactosomes and 20 minutes for CYP3A4 after reaction initiation. These conditions were determined to be linear with respect to time. Parallel incubations without NADPH were used as negative controls. Samples were analyzed via UPLC-MS/MS using calibration curves that ranged from 0.25 to 500 nM for both 5-/6-hydroxypimozide and DHPBI. A single-enzyme Michaelis-Menten model was fit to the 5-hydroxypimozide and DHPBI formation rate data using GraphPad Prism version 8.1.2 (GraphPad Software, San Diego, CA). These experiments were conducted in triplicate with technical duplicates. Estimates for of Km and Vmax were determined for each replicate experiment, and the mean and S.D. of these point estimates were calculated.
Pimozide Metabolite Formation in HLMs from Individual Donors.
Protein expression of CYP2D6 and CYP3A4 in commercially acquired HLMs from seven separate donors was determined by Dr. Bhagwat Prasad (University of Washington, now Washington State University) via MS/MS according to a protocol established by his group (Vrana et al., 2017; Bhatt et al., 2018; Bhatt and Prasad, 2018). All donors were Caucasian and genotyped; none carried the CYP3A7*1C allele. Genotype information for CYP2D6 was provided by the HLM supplier (Sekisui XenoTech), and activity scores were assigned according to the Clinical Pharmacogenetics Implementation Consortium guidelines (Caudle et al., 2020). Tissue donor demographics, CYP2D6 genotypes, specific cytochrome P450 protein content, and relevant product/lot numbers of the HLMs are summarized in Table 1. Under the general incubation conditions, 200 μl of 200 μg/ml of HLMs from the separate donors was each incubated for 5 minutes with 5 μM pimozide. These incubation conditions were determined to be linear with respect to time. The experiment was repeated in triplicate with technical duplicates. Parallel incubations without NADPH were used as negative controls. Concentrations of metabolites were determined via UPLC-MS/MS using calibration curves ranging from 0.5 to 500 nM for 5-/6-hydroxypimozide and 1–500 nM for DHPBI. A simple linear regression model was fit to 5-/6-hydroxypimozide metabolite and DHPBI formation rate data and expression of CYP2D6 and CYP3A4 protein using GraphPad Prism software version 8.1.2 (San Diego, CA). As expected, the HLMs prepared from the single CYP2D6 PM (*4/*5 genotype) donor did not have detectable levels of CYP2D6 protein and were thus assigned an abundance of 0 pmol/mg microsomal protein. The formation rates for “gold-standard” phenotyping reactions for CYP2D6 (dextromethorphan O-demethylation) and CYP3A4 (midazolam 1′-hydroxylation and testosterone 6β-hydroxylation) were provided by the supplier (Sekisui XenoTech) for the HLMs from each of the donors. For purposes of comparison, a linear regression model was also fit to this vendor-supplied data. However, it should be noted that the exact protocol for the manufacturer-supplied phenotyping reactions is unknown and should be interpreted with caution.
TABLE 1.
Donor demographics, CYP2D6 genotype, and cytochrome P450 protein content in single-donor HLMs
All single-donor HLMs obtained for Sekisui Xenotech. Activity scores were assigned according to the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines (Caudle et al., 2020). Protein abundances for CYP2D6 and CYP3A4 were determined via MS/MS analysis.
| Donor Product no. | Lot no. | Sex | Age (y) | CYP2D6 Genotype | CYP2D6 Activity Score | CYP2D6 (pmol/mg HLM protein) | CYP3A4 (pmol/mg HLM protein) |
|---|---|---|---|---|---|---|---|
| H0289 | 0710505 | F | 60 | 1/*2x2 | >2.25a | 11.93 | 71.71 |
| H0439 | 0710037 | F | 78 | *1/*4 | 1.0b | 3.31 | 4.97 |
| H0446 | 0710044 | M | 21 | *4/*41 | 0.5b | 0.80 | 48.85 |
| H0455 | 0710053 | F | 37 | *1/*4 | 1.0b | 4.25 | 35.53 |
| H0485 | 0710139 | M | 10 | *1/*2 | 2.0c | 4.20 | 2.41 |
| H0486 | 0710140 | F | 49 | *4/*5 | 0.0d | <LOQ | 44.44 |
| H0493 | 0710147 | F | 64 | *1/*1 | 2.0c | 7.49 | 34.94 |
F, female; LOQ, limit of quantification; M, male; No., number.
Ultrarapid metabolizer status predicted.
Intermediate metabolizer status predicted.
Normal metabolizer status predicted.
Poor metabolizer status predicted.
Results
Prediction and Identification of the Product of CYP2D6-Mediated Metabolism of Pimozide.
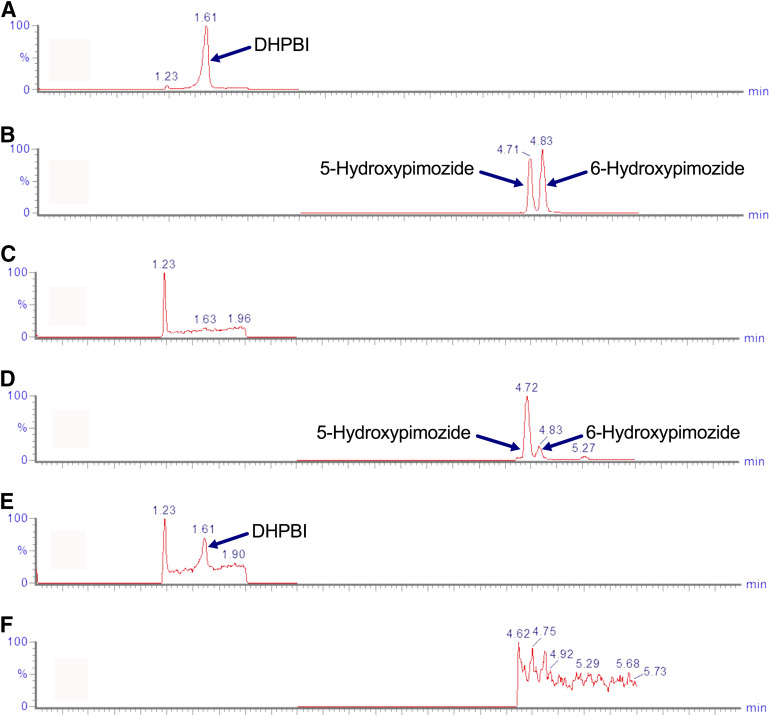
The Q-TOF analysis of metabolite profiles from the exploratory incubations of pimozide in recombinant CYP2D6 and HLMs from a CYP2D6 UM revealed a metabolite that was consistent with hydroxylation (i.e., +16 Da). This peak was not present in the incubation using HLMs isolated from a CYP2D6 PM. The fragmentation pattern of the +16-Da metabolite produced by incubating 10 μM pimozide in CYP2D6 UM HLM incubations was analyzed on a XEVO-TQD mass spectrometer and suggested that oxidation occurred on the benzimidazolone moiety of the pimozide molecule; the major fragment (daughter ion m/z 328) from both pimozide and hydroxypimozide molecules occurred between the piperidine ring and benzimidazolone moiety (Supplemental Fig. 1). SmartCYP software was used to determine the most likely sites for CYP2D6-mediated oxidation of pimozide. It assigned the greatest likelihood to the five position on the aromatic ring of the benzimidazolone moiety; the adjacent six position was determined to be the second most likely site of CYP2D6-mediated metabolism (Supplemental Fig. 2). When recombinant CYP2D6 was incubated with a therapeutically relevant concentration of pimozide (10 nM) and the incubation product was analyzed via UPLC-MS/MS, peaks with the same m/z transition and corresponding elution times to NMR-validated standards for 5- and 6-hydroxypimozide were observed (Fig. 1). The peak height and area were larger for 5-hydroxypimozide than 6-hydroxypimozide. It is important to note that 4- or 7-hydroxypimozide could theoretically coelute with 5-hydroxypimozide under the chromatographic conditions used. Therefore, the absence of authentic standards for the products of the less likely 4- and 7-hydroxylation reactions preclude definitive confirmation that 5-hydroxypimozide is the major metabolite produced by CYP2D6. When recombinant CYP3A4 was incubated with pimozide under otherwise identical incubation conditions, a peak corresponding to the same m/z transition and elution time as a commercially available authentic standard for DHPBI was observed (Fig. 1). It is worth noting that CYP3A4 did not produce detectable amounts of either of the hydroxylated pimozide metabolites under these therapeutically relevant conditions. However, when recombinant CYP3A4 was incubated with substantially greater pimozide concentrations as part of subsequent kinetic studies (described in the Kinetics of Pimozide Metabolism methods section), detectable formation of both hydroxylated metabolites was observed. Relative to CYP2D6, recombinant CYP3A4 was less selective for the production of one hydroxypimozide isomer over the other (Supplemental Fig. 3).
Fig. 1.
Elution of hydroxypimozide isomers and DHPBI with products of CYP2D6- and CYP3A4-mediated metabolism. Chromatograms for combined standards of 50 nM DHPBI (m/z 218.10 → 84.00) (A) and 5-/6-hydroxypimozide metabolites (m/z 478.30 → 328.10) (B) are provided for reference. Chromatograms from incubations of therapeutically relevant concentrations (10 nM) of pimozide in 2 pmol/ml recombinant CYP2D6, m/z 218.10 → 84.00 (C) and m/z 478.30 → 328.10 (D), and CYP3A4, m/z 218.10 → 84.00 (E) and m/z 478.30 → 328.10 (F), are also displayed. Retention times are listed in blue above the respective peaks.
Molar Balance of Pimozide Metabolism in Pooled HLMs.
The NADPH-dependent conversion of pimozide to DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide was evaluated in pooled HLMs. The most abundant metabolite formed was DHPBI, followed by 5-hydroxypimozide and then 6-hydroxypimozide (Table 2). According to eq. 1, 90% of the pimozide that was depleted over the course of the experiment could collectively be accounted for by these three pimozide metabolites. The conditions used in this experiment are nonlinear with respect to time, and caution is advised regarding any kinetic interpretation of the data.
TABLE 2.
Molar balance of pimozide metabolism in pooled HLMs
Mean (S.D.) amounts (picomoles) of pimozide molecules depleted and pimozide metabolites formed by pooled (n = 200 donors) human liver microsomes incubated with pimozide (10 pmol starting amount, 50 nM starting concentration) for 20 min. Percentage standardization of metabolites formed relative to pimozide molecules depleted is also provided.
| 5-Hydroxypimozide | 6-Hydroxypimozide | DHPBI | Sum of Metabolitesa | Pimozide Depletedb | |
|---|---|---|---|---|---|
| Amount (pmol) | 0.27 (0.040) | 0.11 (0.035) | 2.4 (0.20) | 2.8 (0.17) | 3.1 (0.80) |
| % Depleted Accountedc | 8.7 (1.3) | 3.5 (1.1) | 78 (6.5) | 90 (5.5) | N/A |
N/A, not applicable.
Denotes sum of amounts of DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide formed during each incubation.
Denotes the difference between pimozide amounts measured at time 0 and 20 min in each incubation.
Sum of metabolite amounts divided by amount of pimozide depleted over the course of the incubation, expressed as a percentage (see eq. 1).
Recombinant Cytochrome P450 Screen.
CYP2D6 was the predominate isoform to produce 5- and 6-hydroxypimozide and favored the production of 5-hydroxypimozide over 6-hydroxypimozide (Table 3). However, other cytochrome P450 isoforms, including CYP1A1, CYP1A2, CYP2C18, CYP2J2, CYP3A4, CYP3A5, and CYP3A7, also catalyzed the formation of lesser amounts of the hydroxypimozide isomers. Only CYP3A isoforms formed detectable amounts of DHPBI; CYP3A4 was the most active of the CYP3A isoforms.
TABLE 3.
Comparison of pimozide metabolite concentrations produced in incubations of recombinant cytochrome P450 isoforms
Recombinant human cytochrome P450 isoforms (2 pmol/ml) were incubated with 2500 nM pimozide for 30 min. Numbers represent the mean (S.D.) from triplicate experiments for the concentrations (nanomolars) of the specific metabolites observed after quenching of incubations. A “—” denotes a cytochrome P450 isoform that was not observed to produce detectable amounts of the listed metabolites. The lower LOQs for this experiment were 1 nM for 5-/6-hydroxypimozide and 5 nM for DHPBI. Experimental conditions were not confirmed to be linear with respect to time.
| Cytochrome P450 Isoform | 1A1 | 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C18 | 2C19 | 2D6 | 2E1 | 2J2 | 3A4 | 3A5 | 3A7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DHPBI | — | — | — | — | — | — | — | — | — | — | — | 78 (6.5) | <LOQ | 14 (1.5) |
| 5-OH-Pimozide | <LOQ | <LOQ | — | — | — | — | <LOQ | — | 35 (11) | — | <LOQ | 2.4 (0.24) | 2.5 (0.22) | <LOQ |
| 6-OH-Pimozide | <LOQ | <LOQ | — | — | — | — | <LOQ | — | 5.8 (1.2) | — | <LOQ | 1.7 (0.33) | 1.4 (0.16) | <LOQ |
LOQ, limit of quantification. 5-OH, 5-hydroxy. 6-OH, 6-hydroxy.
Kinetics of Pimozide Metabolism.
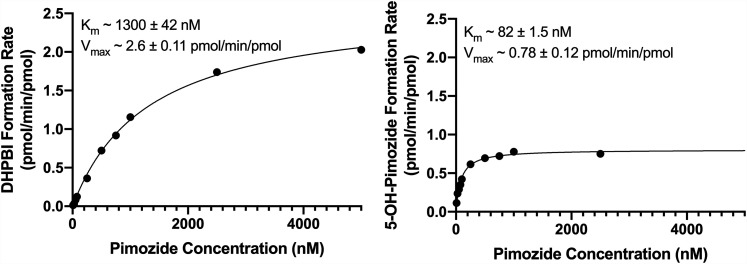
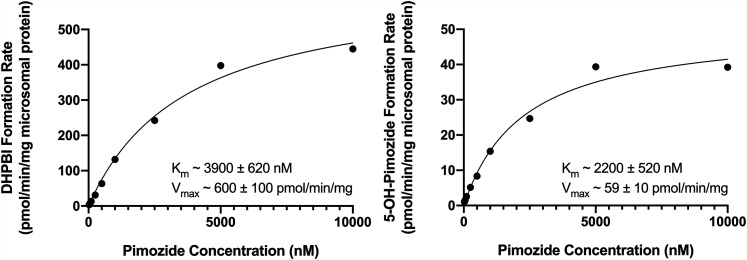
Experiments evaluating the kinetics of pimozide biotransformation via CYP2D6 and CYP3A4 to their principle metabolite products, 5-hydroxypimozide and DHPBI, respectively, were conducted in recombinant human cytochrome P450s and pooled HLMs. Formation rates were plotted against substrate (i.e., pimozide) concentrations, and a simple single-enzyme Michaelis-Menten model was fit to the data. A representative Michaelis-Menten curve and estimates for Km and Vmax for the recombinant enzymes and pooled HLMs are presented in Figs. 2 and 3, respectively. The Km and Vmax for DHPBI formation in CYP3A4 Bactosomes were, respectively, 15- and 3-fold greater than 5-hydroxypimozide formation in CYP2D6 Bactosomes. However, when subsequent incubations were conducted in pooled HLMs, the differences in affinity for the principally CYP3A4-mediated N-dealkylation reaction (Km ∼3900 nM) and the principally CYP2D6-mediated 5-hydroxylation reaction (Km ∼2200 nM) were less pronounced. The Vmax for the N-dealkylation reaction (∼600 pmol/min per milligram protein) was greater than that of the 5-hydroxylation reaction (∼59 pmol/min per milligram protein) in pooled HLMs.
Fig. 2.
Kinetics of pimozide biotransformation to DHPBI and 5-hydroxypimozide (5-OH-Pimozide) by recombinant CYP3A4 (left) and CYP2D6 (right), respectively. The displayed results are from a representative experiment. Individual data points represent the means of technical duplicates at a given pimozide concentration from the same experimental replicate, and the solid line reflects the fit of a single-enzyme Michaelis-Menten model to the data. Mean ± S.D. for parameter (Km and Vmax) estimates from n = 3 experiments using each isoform appear as text at the top of the respective graphs.
Fig. 3.
Kinetics of pimozide biotransformation to DHPBI (left) and 5-hydroxypimozide (5-OH-Pimozide) (right) in pooled human liver microsomes. The displayed results are from a representative experiment. Individual data points represent the means of technical duplicates at a given pimozide concentration from the same experimental replicate, and the solid line reflects the fit of a single-enzyme Michaelis-Menten model to the data. Mean ± S.D. for parameter (Km and Vmax) estimates from n = 3 experiments appear as text at the bottom of the respective graphs for each reaction.
Pimozide Metabolite Formation Correlates with Protein Abundance of CYP2D6 and CYP3A4 in HLMs.
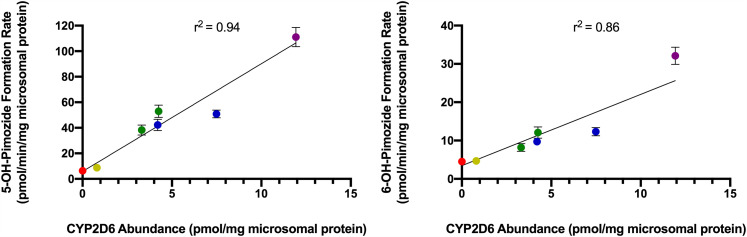
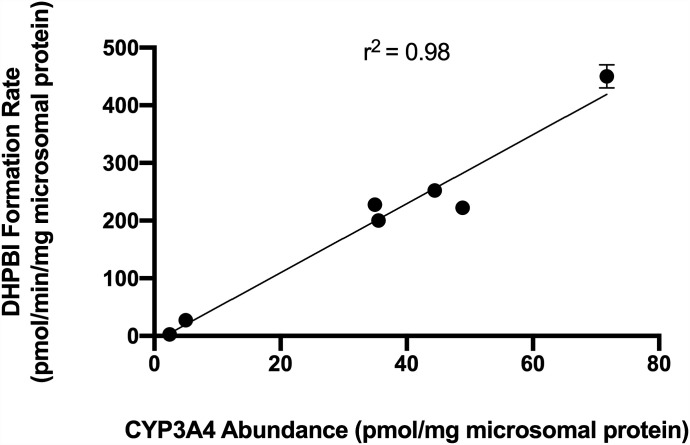
Both pimozide 5-hydroxylation (r2 = 0.94) and 6-hydroxylation (r2 = 0.86) rates were positively linearly correlated with abundance of CYP2D6 protein (Fig. 4). Notably, the correlation between the vendor-supplied rate of dextromethorphan O-demethylation to dextrorphan and CYP2D6 protein abundance (r2 = 0.93) was comparable to that of 5-hydroxypimozide (Supplemental Fig. 4). There was poor linear correlation between either pimozide 5-hydroxylation (r2 = 0.08) or 6-hydroxylation (r2 = 0.26) rates and CYP3A4 abundance (Supplemental Fig. 5). Expression levels of CYP2D6 protein and formation rates for 5-hydroxypimozide and 6-hydroxypimozide were observed generally to be greater in individuals with a higher activity score assignment. However, there was overlap between the 1.0 (green) and 2.0 (blue) CYP2D6 activity score groups (Fig. 4). The DHPBI formation rate was positively correlated with abundance of CYP3A4 protein (r2 = 0.98) (Fig. 5). For comparison, a simple linear regression model fit the vendor-supplied rates for either testosterone 6β-hydroxylation or midazolam 1′-hydroxylation, and CYP3A4 protein abundance yielded r2 values of 0.77 and 0.39, respectively (Supplemental Fig. 6). There was poor linear correlation (r2 = 0.12) between DHPBI formation rate and CYP2D6 abundance (Supplemental Fig. 7).
Fig. 4.
Correlation of CYP2D6 protein abundance and the rate of biotransformation of pimozide to 5-hydroxypimozide (5-OH-Pimozide) (left) and 6-hydroxypimozide (6-OH-Pimozide) (right) in HLMs isolated from individual donors (n = 7). Data points represent the means of separate experiments (n = 3), and error bars reflect the S.D.; error bars are not visible for many points because of low interexperimental variability. The r2 corresponding to the fit of a simple linear regression model for each metabolite is listed at the top of each respective graph. Expression of CYP2D6 protein was determined via MS/MS analysis. Data points were color coded to reflect activity score assignments for CYP2D6 genotypes: 0 (red), 0.5 (yellow), 1.0 (green), 2.0 (blue), and >2.25 (purple) (Caudle et al., 2020).
Fig. 5.
Correlation of CYP3A4 protein abundance and the rate of pimozide N-dealkylation to DHPBI in HLMs isolated from individual donors (n = 7). Data points represent the means of separate experiments (n = 3), and error bars reflect the S.D.; error bars are not visible for most points because of low interexperimental variability. The r2 corresponding to the fit of a simple linear regression model for each metabolite is listed at the top of each respective graph. Expression of CYP3A4 protein was determined via MS/MS analysis.
Discussion
Initial clinical studies observed that pimozide was primarily excreted in the form of N-dealkylation products in the urine (Baro et al., 1972; Pinder et al., 1976). Ensuing in vitro investigations focused solely on N-dealkylation to DHPBI (Desta et al., 1998; Desta et al., 2002), establishing DHPBI formation as the primary route of pimozide metabolism and identifying CYP3A4 (and to a much lesser extent CYP1A2) as the mediating enzymes (Desta et al., 1998). However, subsequent in vivo studies demonstrated that CYP2D6 genotype and the coadministration of CYP2D6 inhibitors have an effect on pimozide exposure (Alderman, 2005; Nucci and Gomeni, 2007; http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/020031s05920710s023lbl.pdf). The findings described herein present a plausible case for reconciling the discordance between the previously published in vitro and in vivo studies. We have identified two novel metabolites of pimozide, 5- and 6-hydroxypimozide (Fig. 1), and determined that both metabolites, but particularly 5-hydroxypimozide, are formed via CYP2D6 (Table 3). This finding, in conjunction with confirmation of previous observations of negligible DHPBI formation by CYP2D6 (Desta et al., 1998), suggests that assessment of N-dealkylation rate is not the most appropriate means to evaluate the contribution of CYP2D6 to pimozide biotransformation. Furthermore, our experiments comparing the kinetics of pimozide metabolism to 5-hydroxypimozide and DHPBI by CYP2D6 and CYP3A4, respectively, suggest that CYP2D6 represents a moderately higher affinity (i.e., lower Km) pathway (Figs. 2 and 3).
Collectively, the three metabolites monitored in our experiments (DHPBI, 5-hydroxypimozide, and 6-hydroxypimozide) account for 90% of the products of NADPH-dependent biotransformation in pooled HLMs, strongly suggesting that we are capturing a near-complete picture of the extent of NADPH-dependent pimozide metabolism occurring in HLMs (Table 2). Although DHPBI represented the major metabolite formed by pooled HLMs (n = 200 donors) (Fig. 3), we observed that the relative rates of production for the three metabolites varied across HLMs sourced from individual donors and were dependent upon the underlying abundance of CYP2D6 (Fig. 4) and CYP3A4 (Fig. 5) protein in the microsomes. Interestingly, in HLMs from two of the seven individual donors evaluated, the pimozide 5-hydroxylation rate exceeded the N-dealkylation rate, suggesting that CYP2D6 may be the major route of metabolism in those individuals. Notably, CYP2D6 and CYP3A4 protein expression did not correlate with each other (Supplemental Fig. 8). One limitation of these experiments was that none of the HLMs expressed CYP3A7 (no neonatal/fetal donors or adults possessing the CYP3A7*1C functional adult expresser allele). Despite observations of pimozide N-dealkylation by recombinant CYP3A7 (Table 3), we were unable to evaluate the contribution of hepatic CYP3A7 abundance on pimozide N-dealkylation.
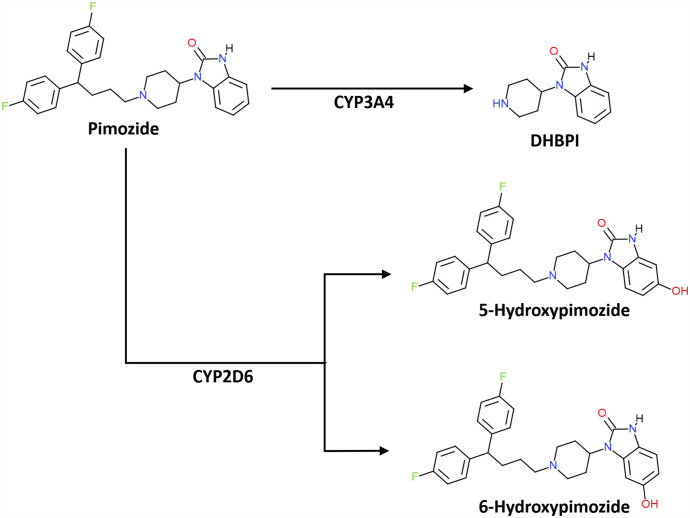
Collectively, these observations warrant a revised pathway for pimozide biotransformation (Fig. 6), with the understanding that the major pathway of systemic metabolism will be dependent on the relative degree of hepatic CYP2D6 and CYP3A4 activity in a given individual. Individual expression levels of CYP2D6 and CYP3A4 would also be anticipated to impact the magnitude of pharmacokinetic drug-drug interactions (DDIs) between pimozide and inhibitors of these enzymes (i.e., DDI sensitivity); the effects could range from negligible to profound depending on the relative fractional clearance via each of these enzymatic pathways. In light of this, the utilization of pathway-specific metabolites may provide a means for predicting individualized dosing and DDI sensitivity. The strong correlations we observed between the formation of the different pimozide metabolites and the expression of the respective enzymes that mediate their production is encouraging. In the future, DHPBI and 5-hydroxypimozide levels in the blood or urine may have utility in the prediction of the relative sensitivity for CYP2D6- versus CYP3A4-dependent DDIs with pimozide for a given individual. Additional studies will need to be conducted to determine the extent to which 5-hydroxypimozide is produced in vivo and present in the blood and excreta. Although the pharmacologic activity of 5-hydroxypimozide is unknown, it may also be necessary to characterize its activity profile if the metabolite is found to circulate in vivo. Interestingly, human clinical trials evaluating the effectiveness of 5-chloropimozide, an analog of pimozide that was ultimately never marketed, demonstrated the investigational drug to be a “highly potent” and “long acting” dopamine receptor antagonist (Janssen et al., 1975), thus suggesting that 5-hydroxypimozide may retain pharmacologic activity.
Fig. 6.
Revised pathway of pimozide biotransformation. The principle products of CYP2D6-mediated pimozide metabolism are 5- and 6-hydroxypimozide, and the major product of CYP3A4-mediated metabolism of pimozide is DHPBI. Pimozide and metabolite structures were made using ChemDoodle Web Components 2D Sketcher (iChemLabs, Chesterfield, VA).
In conclusion, our discovery of CYP2D6-mediated metabolism of pimozide to 5- and 6-hydroxypimozide illuminates a longstanding knowledge deficit in our understanding of the drug’s in vivo disposition. Our findings provide a mechanistic basis for prior clinical observations of reduced pimozide oral clearance in individuals who are CYP2D6 PMs or who are concomitantly taking certain CYP2D6 inhibitors (e.g., paroxetine and sertraline) (Alderman, 2005; Nucci and Gomeni, 2007; http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/020031s05920710s023lbl.pdf). Application of the approach we have used may prove useful in the reassessment of other drugs. As with pimozide, identification of novel metabolites could be required, and modern in silico tools such as SmartCYP may provide useful guidance in the process.
Acknowledgments
We would like to thank Dr. Leon Van Haandel, Dr. Annemarie Rompca, and Dr. Robin Pearce for their technical assistance. We would also like to thank Kim Gibson and Dr. Whitney Nolte for their assistance with the maintenance and operation of analytical instrumentation.
Abbreviations
- DDI
drug-drug interaction
- DHPBI
1,3-dihydro-1-(4-piperidinyl)-2H-benzimidazol-2-one
- HLM
human liver microsome
- MS/MS
tandem mass spectrometry
- PM
poor metabolizer
- Q-TOF
quadrupole time-of-flight
- UM
ultrarapid metabolizer
- UPLC
ultraperformance liquid chromatography
Authorship Contributions
Participated in research design: Chapron, Dinh, Leeder.
Conducted experiments: Chapron, Dinh, Toren, Gaedigk.
Contributed new reagents or analytic tools: Gaedigk, Leeder.
Performed data analysis: Chapron, Dinh, Toren, Gaedigk.
Wrote or contributed to the writing of the manuscript: Chapron, Dinh, Gaedigk, Leeder.
Footnotes
This work was funded by National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant 1 U54-HD090258] “Genomic and Ontogeny-Linked Dose Individualization and cLinical Optimization for KidS.” B.D.C. is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant T32-HD069038] “Research Fellowship Program in Pediatric Clinical/Developmental Pharmacology.”
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahmed I, Dagincourt PG, Miller LG, Shader RI. (1993) Possible interaction between fluoxetine and pimozide causing sinus bradycardia. Can J Psychiatry 38:62–63. [DOI] [PubMed] [Google Scholar]
- Alderman J. (2005) Coadministration of sertraline with cisapride or pimozide: an open-label, nonrandomized examination of pharmacokinetics and corrected QT intervals in healthy adult volunteers. Clin Ther 27:1050–1063. [DOI] [PubMed] [Google Scholar]
- Baro F, Brugmans J, Heykants J. (1972) [Absorption, metabolism and excretion of pimozide in man]. Clin Ter 63:239–249. [PubMed] [Google Scholar]
- Beach SR, Celano CM, Noseworthy PA, Januzzi JL, Huffman JC. (2013) QTc prolongation, torsades de pointes, and psychotropic medications. Psychosomatics 54:1–13. [DOI] [PubMed] [Google Scholar]
- Bhatt DK, Basit A, Zhang H, Gaedigk A, Lee SB, Claw KG, Mehrotra A, Chaudhry AS, Pearce RE, Gaedigk R, et al. (2018) Hepatic abundance and activity of androgen- and drug-metabolizing enzyme UGT2B17 are associated with genotype, age, and sex. Drug Metab Dispos 46:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DK, Prasad B. (2018) Critical issues and optimized practices in quantification of protein abundance level to determine interindividual variability in DMET proteins by LC-MS/MS proteomics. Clin Pharmacol Ther 103:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE, Gammal RS, Relling MV, Scott SA, Hertz DL, et al. (2020) Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the clinical pharmacogenetics implementation consortium and Dutch pharmacogenetics working group. Clin Transl Sci 13:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z, Kerbusch T, Flockhart DA. (1999) Effect of clarithromycin on the pharmacokinetics and pharmacodynamics of pimozide in healthy poor and extensive metabolizers of cytochrome P450 2D6 (CYP2D6). Clin Pharmacol Ther 65:10–20. [DOI] [PubMed] [Google Scholar]
- Desta Z, Kerbusch T, Soukhova N, Richard E, Ko JW, Flockhart DA. (1998) Identification and characterization of human cytochrome P450 isoforms interacting with pimozide. J Pharmacol Exp Ther 285:428–437. [PubMed] [Google Scholar]
- Desta Z, Soukhova N, Flockhart DA. (2002) In vitro inhibition of pimozide N-dealkylation by selective serotonin reuptake inhibitors and azithromycin. J Clin Psychopharmacol 22:162–168. [DOI] [PubMed] [Google Scholar]
- Drolet B, Rousseau G, Daleau P, Cardinal R, Simard C, Turgeon J. (2001) Pimozide (Orap) prolongs cardiac repolarization by blocking the rapid component of the delayed rectifier potassium current in native cardiac myocytes. J Cardiovasc Pharmacol Ther 6:255–260. [DOI] [PubMed] [Google Scholar]
- Flockhart DA, Drici MD, Kerbusch T, Soukhova N, Richard E, Pearle PL, Mahal SK, Babb VJ. (2000) Studies on the mechanism of a fatal clarithromycin-pimozide interaction in a patient with Tourette syndrome. J Clin Psychopharmacol 20:317–324. [DOI] [PubMed] [Google Scholar]
- Flockhart DA, Richard E, Soukova N, Kerbuscb T. (1997) Metabolism of pimozide by CYF3A and CYP2D6 in human liver microsomes. Clin Pharmacol Ther 61:232. [Google Scholar]
- Flockhart DA, Richard E, Woosley RL, Pearle PL, Drici MD. (1996) A metabolic interaction between clarithromycin and pimozide may result in cardiac toxicity. Clin Pharmacol Ther 59:189. [Google Scholar]
- Gulisano M, Cali PV, Cavanna AE, Eddy C, Rickards H, Rizzo R. (2011) Cardiovascular safety of aripiprazole and pimozide in young patients with Tourette syndrome. Neurol Sci 32:1213–1217. [DOI] [PubMed] [Google Scholar]
- Haddad PM, Anderson IM. (2002) Antipsychotic-related QTc prolongation, torsade de pointes and sudden death. Drugs 62:1649–1671. [DOI] [PubMed] [Google Scholar]
- Hemeryck A, Belpaire FM. (2002) Selective serotonin reuptake inhibitors and cytochrome P-450 mediated drug-drug interactions: an update. Curr Drug Metab 3:13–37. [DOI] [PubMed] [Google Scholar]
- Horrigan JP, Barnhill LJ. (1994) Paroxetine-pimozide drug interaction. J Am Acad Child Adolesc Psychiatry 33:1060–1061. [DOI] [PubMed] [Google Scholar]
- Janssen PA, Niemegeers CJ, Schellekens KH, Lenaerts FM, Wauquier A. (1975) Clopimozide (R 29 764), a new highly potent and orally long-acting neuroleptic of the diphenylbutylpiperidine series. Arzneimittelforschung 25:1287–1294. [PubMed] [Google Scholar]
- McCreadie RG, Mackie M, Wiles DH, Jorgensen A, Hansen V, Menzies C. (1984) Within-individual variation in steady state plasma levels of different neuroleptics and prolactin. Br J Psychiatry 144:625–629. [DOI] [PubMed] [Google Scholar]
- McIntyre IM, King CV, Staikos V, Gall J, Drummer OH. (1997) A fatality involving moclobemide, sertraline, and pimozide. J Forensic Sci 42:951–953. [PubMed] [Google Scholar]
- Nucci GMK, Gomeni R. (2007) Population pharmacokinetic modelling of pimozide and its relation to CYP2D6 genotype, in Annual Meeting of Population Approach Group in Europe, Copenhagen, Denmark.
- Pinder RM, Brogden RN, Swayer R, Speight TM, Spencer R, Avery GS. (1976) Pimozide: a review of its pharmacological properties and therapeutic uses in psychiatry. Drugs 12:1–40. [DOI] [PubMed] [Google Scholar]
- Robertson MM. (2008) The prevalence and epidemiology of Gilles de la Tourette syndrome. Part 1: the epidemiological and prevalence studies. J Psychosom Res 65:461–472. [DOI] [PubMed] [Google Scholar]
- Rogers HL, Bhattaram A, Zineh I, Gobburu J, Mathis M, Laughren TP, Pacanowski M. (2012) CYP2D6 genotype information to guide pimozide treatment in adult and pediatric patients: basis for the U.S. Food and Drug Administration’s new dosing recommendations. J Clin Psychiatry 73:1187–1190. [DOI] [PubMed] [Google Scholar]
- Rydberg P, Gloriam DE, Olsen L. (2010) The SMARTCyp cytochrome P450 metabolism prediction server. Bioinformatics 26:2988–2989. [DOI] [PubMed] [Google Scholar]
- Sallee F, Kohegyi E, Zhao J, McQuade R, Cox K, Sanchez R, van Beek A, Nyilas M, Carson W, Kurlan R. (2017) Randomized, double-blind, placebo-controlled trial demonstrates the efficacy and safety of oral aripiprazole for the treatment of tourette’s disorder in children and adolescents. J Child Adolesc Psychopharmacol 27:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee FR, Nesbitt L, Jackson C, Sine L, Sethuraman G. (1997) Relative efficacy of haloperidol and pimozide in children and adolescents with Tourette’s disorder. Am J Psychiatry 154:1057–1062. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Shapiro AK, Fulop G, Hubbard M, Mandeli J, Nordlie J, Phillips RA. (1989) Controlled study of haloperidol, pimozide and placebo for the treatment of Gilles de la Tourette’s syndrome. Arch Gen Psychiatry 46:722–730. [DOI] [PubMed] [Google Scholar]
- Vrana M, Whittington D, Nautiyal V, Prasad B. (2017) Database of optimized proteomic quantitative methods for human drug disposition-related proteins for applications in physiologically based pharmacokinetic modeling. CPT Pharmacometrics Syst Pharmacol 6:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HK, Lee JS, Paik KW, Choi SH, Yoon SJ, Kim JE, Hong JP. (2011) Open-label study comparing the efficacy and tolerability of aripiprazole and haloperidol in the treatment of pediatric tic disorders. Eur Child Adolesc Psychiatry 20:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF, Xue CC, Yu XQ, Li C, Wang G. (2007) Clinically important drug interactions potentially involving mechanism-based inhibition of cytochrome P450 3A4 and the role of therapeutic drug monitoring. Ther Drug Monit 29:687–710. [DOI] [PubMed] [Google Scholar]