Abstract
The neural control system underlying breathing is sexually dimorphic with males being more vulnerable to dysfunction. Microglia also display sex differences, and their role in the architecture of brainstem respiratory rhythm circuitry and modulation of cervical spinal cord respiratory plasticity is becoming better appreciated. To further understand the molecular underpinnings of these sex differences, we performed RNA sequencing of immunomagnetically isolated microglia from brainstem and cervical spinal cord of adult male and female rats. We used various bioinformatics tools (Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, Reactome, STRING, MAGICTRICKS) to functionally categorize identified gene sets, as well as to pinpoint common transcriptional gene drivers that may be responsible for the observed transcriptomic differences. We found few sex differences in the microglial transcriptomes derived from the brainstem, but several hundred genes were differentially expressed by sex in cervical spinal microglia. Comparing brainstem and spinal microglia within and between sexes, we found that the major factor guiding transcriptomic differences was central nervous system (CNS) location rather than sex. We further identified key transcriptional drivers that may be responsible for the transcriptomic differences observed between sexes and CNS regions; enhancer of zeste homolog 2 emerged as the predominant driver of the differentially downregulated genes. We suggest that functional gene alterations identified in metabolism, transcription, and intercellular communication underlie critical microglial heterogeneity and sex differences in CNS regions that contribute to respiratory disorders categorized by dysfunction in neural control. These data will also serve as an important resource data base to advance our understanding of innate immune cell contributions to sex differences and the field of respiratory neural control.
SIGNIFICANCE STATEMENT
The contributions of central nervous system (CNS) innate immune cells to sexually dimorphic differences in the neural circuitry controlling breathing are poorly understood. We identify key transcriptomic differences, and their transcriptional drivers, in microglia derived from the brainstem and the C3–C6 cervical spinal cord of healthy adult male and female rats. Gene alterations identified in metabolism, gene transcription, and intercellular communication likely underlie critical microglial heterogeneity and sex differences in these key CNS regions that contribute to the neural control of breathing.
Introduction
Breathing is a critical physiologic process that must be fully operational at birth and adapt to ever changing needs throughout the lifespan, until death. Regardless of its necessity in both sexes, the respiratory system displays significant sex differences including airway and lung anatomy, respiratory muscle oxygen consumption, and, importantly, the respiratory-related neuronal network (Becklake and Kauffmann, 1999; Behan and Wenninger, 2008; Dominelli et al., 2015; Dougherty et al., 2017; Iturri et al., 2017; Lozo et al., 2017; Rousseau et al., 2017; LoMauro and Aliverti, 2018). Despite these well known differences, comparatively little is known regarding sex-dependent dimorphisms in the respiratory control system, although reports suggest dimorphisms in response to ventilatory challenges (Jensen et al., 2005; Ahuja et al., 2007) and the capacity for respiratory plasticity (Zabka et al., 2006; Dougherty et al., 2017). There is also clear evidence of sex differences in some ventilatory control disorders, such as sudden infant death syndrome and central sleep apnea, which are more prevalent in males (Rousseau et al., 2017; Townsel et al., 2017). Disruptions in brainstem respiratory rhythm generation and pattern formation circuits contribute to these pathologies (Benarroch, 2018; Kinney and Haynes, 2019).
Little is known about the mechanistic underpinnings of sexual dimorphism in the neural circuits controlling breathing, including the role of intercellular communication between respiratory neurons and other cell types. Central nervous system (CNS) resident innate immune cells (microglia) are becoming better appreciated as important players both in the architecture of the medullary respiratory rhythm generator (Thoby Brisson et al., 2019) and as potentially contributing to neuroinflammation induced by various stimuli that is now well established to impair respiratory plasticity in adult animals and humans (Huxtable et al., 2013, 2015; Hocker et al., 2017; Lynch et al., 2017; Agosto-Marlin et al., 2018; Hocker and Huxtable, 2019; Sandhu et al., 2019). An adverse maternal and/or early life environment can also negatively impact the developing respiratory control system (Fournier et al., 2013, 2015; Gulemetova et al., 2013; Delhaes et al., 2014; Soliz et al., 2016; Baldy et al., 2018), an effect that may be due to inflammation (Johnson et al., 2018; Hocker et al., 2019; Kiernan et al., 2019; Knutson and Watters, 2020). Interestingly, these early life exposures appear to impact the male respiratory neural control system more severely than the female.
Based on the expression of select markers and morphology, microglia have long been believed to be functionally heterogeneous (Lawson et al., 1990; Schmid et al., 2002; Hanisch, 2013; De Biase et al., 2017; McCarthy, 2017) and sexually dimorphic cells, showing alterations in morphology and transcriptomes in various forebrain regions (Schwarz et al., 2012; Hanamsagar and Bilbo, 2016; Hanamsagar et al., 2017; Bordeleau et al., 2019; Kodama and Gan, 2019). Although the molecular and functional contributions of microglia to neural function have been widely studied in forebrain CNS regions (like the cortex and hippocampus), the same is not true of microglia from CNS regions in which the respiratory neural control circuitry is housed. Nevertheless, evidence is emerging to suggest that microglia also locally impact the neural control of breathing and physiologic adaptations to respiratory challenge (Stokes et al., 2017; Tenorio-Lopes et al., 2017; Baldy et al., 2018), although to our knowledge, no studies have investigated the microglial transcriptome in CNS regions responsible for respiratory rhythm and patterning, or sex- or region-specific differences in microglial transcriptomes in respiratory control regions.
Here, we investigated the transcriptomes of microglia derived from two regions important for respiratory control, the brainstem and the cervical spinal cord in adult male and female rats. The brainstem houses the neuronal circuitry responsible for generating and sculpting the neuronal signal to breathe (Ikeda et al., 2017; Del Negro et al., 2018), which is then transmitted to inspiratory motor neurons, the most important of which reside in the cervical spinal cord (Fogarty et al., 2018; Shinozaki et al., 2019). We report that although there were significant sexual dimorphisms in the genes expressed in microglia derived from the cervical spinal cord, few sex differences were noted in brainstem microglia. Moreover, the greatest factor driving transcriptomic differences between microglia from both sexes and regions appeared to be the CNS region from which they were derived. Interestingly, there were significant sex differences in the functional classes of genes expressed by microglia derived from the cervical spinal cord compared with those from the brainstem; however, most of the functional gene categories lacked overlap between sexes. Lastly, we identified unique transcriptional drivers responsible for the region- and sex-dependent differences in microglial gene signatures.
Materials and Methods
Animal Procedures.
All animal experimental procedures were performed according to the National Institutes of Health guidelines set forth in the Guide for the Care and Use of Laboratory Animals. All protocols were approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee. Rats were housed in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care on a 12:12 light:dark cycle, with food and water ad libitum. Because maternal inflammation can impact respiratory frequency and tidal volume in newborn pups (Ramirez et al., 2019) and maternal breathing dysfunction is thought to cause maternal inflammation that can cause fetal growth restriction, low birth weights, and premature birth in some human studies (reviewed in Izci Balserak (2015)), we used adult rats born from dams that were reared in controlled room air conditions during their pregnancy. Due to our interest in studying microglia from CNS regions involved in respiratory neural control, we housed timed pregnant Sprague-Dawley rats (arrived day E9; Charles River; Wilmington, MA) in environmentally controlled conditions from gestational day 10 to 21. The conditions consisted of computer-controlled room air delivery to maintain CO2 levels below 0.3%. Pups were birthed in filter-topped cages, and each litter was culled to eight (four males and four females whenever possible) by postnatal day 3 to control for potential differences arising due to maternal care. Microglia from ∼8-week-old young adult rats (one male and one female from each of five independent litters; n = 5 each sex) were immunomagnetically isolated and sent for RNA sequencing (Novogene, Sacramento, CA).
Data presented in this study represent a subanalysis of a larger study investigating the impact of maternal intermittent hypoxia on microglia from respiratory control regions in adult offspring. Here, we describe results from the control animals that were exposed to normoxia.
CD11b+ Cell Isolation.
Offspring were euthanized and perfused with cold PBS to remove circulating immune cells from the vasculature of the central nervous system. Whole brainstems were dissected between the pontomedullary junction and the obex. Spinal cervical C2–C6 vertebrae were removed, and dorsal/ventral C3–C6 cervical spinal segments were extracted based on identification of the spinal roots. Tissues were dissociated into single cell suspensions using papain enzymatic digestion. CD11b+ cells were immunomagnetically isolated as previously described (Crain and Watters, 2009; Crain et al., 2009, 2013; Nikodemova and Watters, 2012). Isolated CD11b+ cells will be hereafter referred to as “microglia.”
RNA Sequencing and Analysis.
Total RNA was extracted from freshly isolated microglia with TriReagent according to the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO) as we have done before (Crain and Watters, 2009; Crain et al., 2009, 2013; Nikodemova and Watters, 2012). Total RNA was submitted to Novogene for library construction and paired-end (PE-150) sequencing with an Illumina NovaSeq. Index of the reference Rnor6.0 genome was built using Bowtie version 2.2.3. Reads were aligned using TopHat version 2.0.12. Gene counts were made using HTSeq version 0.6.1. Count files were imported to R and filtered such that only genes with a CPM >1 expressed in three samples were retained. Counts were normalized using the trimmed mean of M-values method and analyzed for differential expression using edgeR (Robinson et al., 2010). Differentially expressed genes were identified as statistically significant if the false discovery rate (FDR) was <5%. Results were uploaded to the National Center for Biotechnology Information Gene Expression Omnibus with reference number GSE142478 (https://www.ncbi.nlm.nih.gov/gds). The STRING data base (Szklarczyk et al., 2019) was used to visualize functional network associations and to perform gene ontology analyses for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (Kanehisa et al., 2019), Reactome pathway enrichment (Fabregat et al., 2018), and Gene Ontology (GO) class Biologic Process (The Gene Ontology Consortium, 2019).
MAGICTRICKS Analysis.
To determine if there were common transcriptional drivers of the observed gene expression differences between sexes and CNS regions, differentially expressed genes were evaluated for putative regulatory factors using MAGICTRICKS (Khan et al., 2019; A. Roopra, preprint, DOI: https://doi.org/10.1101/492744). Background lists of genes were created based on the criteria used for differential expression analyses (CPM >1 in three samples). The list of background genes and the list of upregulated or downregulated genes for each comparison were used as MAGICTRICKS input according to the software instructions. Cumulative distribution graphs and summary scores were generated by the MAGICTRICKS software, and the top 30 transcriptional drivers are shown.
Results
The Transcriptomes of Adult Brainstem–Derived Microglia are Similar Between Males and Females.
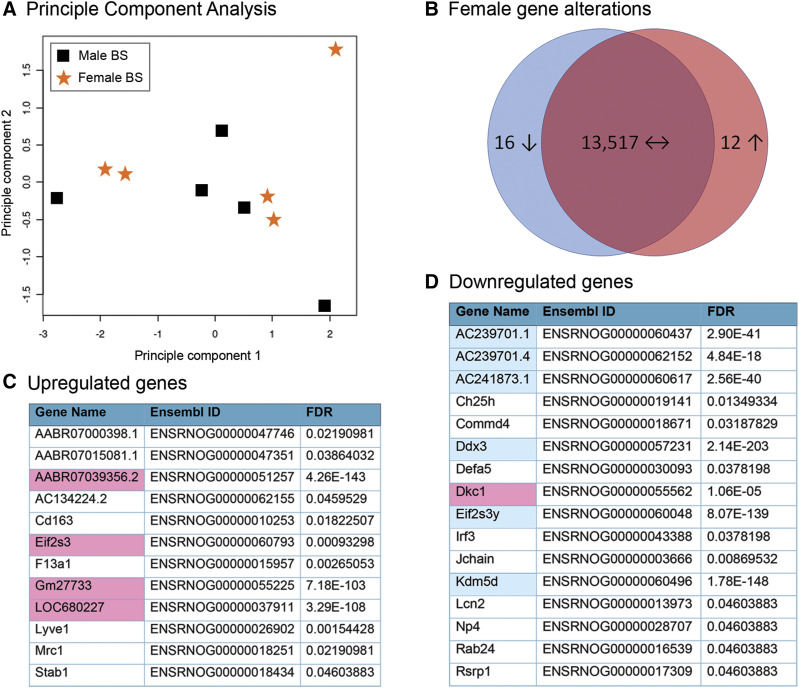
To examine microglia in regions of respiratory control, CD11b+ cells were immunomagnetically isolated from the brainstem (n = 5 independent adult male and female rats). We first compared microglial gene expression in the brainstem between males and females (Fig. 1). In brainstem-derived microglia, principle component analysis showed no major overall difference between male and female transcriptomes (Fig. 1A), indicating that both transcriptomes were nearly identical. Of the total genes expressed in males and females, we found that although >13,500 of them were shared, only 28 were differentially expressed by sex (FDR < 0.05; Fig. 1B). Of these differentially expressed genes, 12 were upregulated in females compared with males (Fig. 1C), and 16 were downregulated in females compared with males (Fig. 1D). Interestingly, several of the upregulated genes (Stab1, Mrc1, Lyve1, Cd163, and F13a1) were related to angiogenesis and lymphocyte homing and have been described to be expressed in different functional populations of macrophages derived from the lung (Poczobutt et al., 2016). Conversely, within the downregulated genes, several (Lcn2, Defa5, Np4, and Irf3) were involved in the immune response to bacterial and fungal infections (Takeuchi and Akira, 2010; Chakraborty et al., 2012; Zhao and Lu, 2014). As expected, approximately 40% of the differentially expressed genes were encoded on sex chromosomes. For example, of the 12 upregulated genes, four (Eif2s, AABR07039356.2, Gm27733, LOC680227) were X chromosome–linked; Eif2s is involved in initiation of protein translation, and this gene escapes X chromosome inactivation (Ehrmann et al., 1998). Of the 16 downregulated genes, only one gene [dyskerin 1, involved in maintaining telomere length (Chatterjee, 2017)] was encoded on the X chromosome (pink shading), and six (AC2392701.1, AC2392701.4, AC24187.1, Ddx3, Eif2s3y, Kdm5d) were Y chromosome–linked (blue shading). Ddx3 encodes an RNA helicase involved in viral innate immunity (Fullam and Schroder, 2013), Eif2s3y encodes a Y chromosome–specific initiation factor involved in protein translation (Armoskus et al., 2014), and Kdm5d encodes a Jumonji family histone demethylase that targets di- and trimethylated histone 3 lysine 4 to repress gene transcription (Wynder et al., 2010).
Fig. 1.
Female and male brainstem microglia share similar transcriptomes. (A) Principle component analysis of male and female brainstem (BS) microglia (n = 5/treatment). (B) Venn diagram showing the total numbers of shared and unique genes between males and females. There were 13,517 shared genes that did not significantly differ in either sex. There were 28 differentially expressed genes that significantly differed (FDR < 0.05) between females and males. Upregulated genes are shown in red, and downregulated genes are shown in blue. (C) Significantly upregulated genes (12) in female brainstem microglia vs. males. Genes highlighted in pink are X chromosome–linked. (D) Significantly downregulated genes (16) in female brainstem microglia vs. males. Genes highlighted in pink are X chromosome–linked, and those in blue are Y chromosome–linked.
The Transcriptomes of Adult Cervical Spinal Cord–Derived Microglia are Sexually Dimorphic.
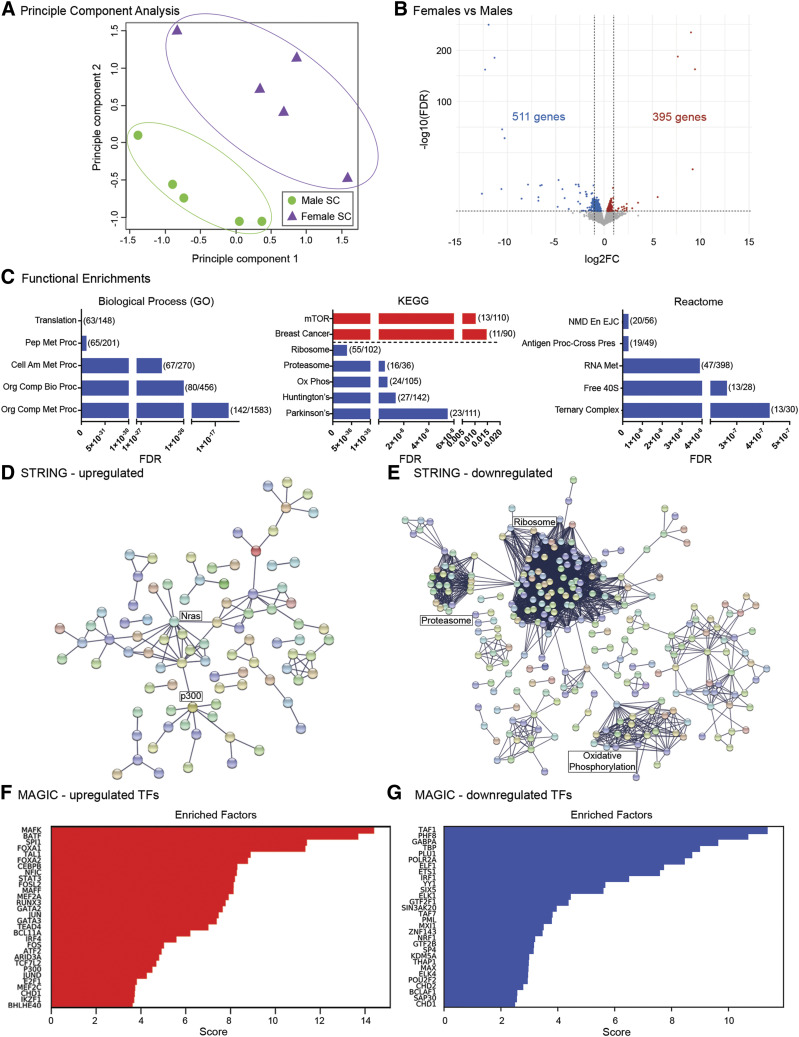
We next examined sex differences in the transcriptomes of microglia immunomagnetically isolated from a region of the cervical spinal cord containing the phrenic motor neurons that innervate the diaphragm, the major inspiratory muscle (Fig. 2). In contrast to the brainstem, principle component analysis of microglia from the male and female cervical spinal cord displayed no overlap (Fig. 2A), suggesting significant sex differences in their transcriptomes. Of the 12,517 total genes expressed in males and 12,534 genes expressed in females, a volcano plot of the RNA sequencing analyses revealed that 906 of these genes were differentially expressed between male and female microglia. Of these, 395 genes were upregulated compared with males, and 511 genes were downregulated compared with males (Fig. 2B; Supplemental Tables 1 and 2). We performed GO (Biologic Process category), KEGG, and Reactome analyses on the 395 genes that were upregulated in female spinal microglia to determine whether they segregated into specific functional gene categories. Neither GO nor Reactome analyses pinpointed functional gene categories, although KEGG analysis identified mammalian target of rapamycin and breast cancer signaling pathways as common functional categories into which several of the upregulated genes fell (Fig. 2C, red bars). Conversely, there were several functional categories identified for the 511 downregulated genes in females (Fig. 2C, blue bars). For example, processes involved in cellular and protein metabolism were identified by GO. Genes involved in ribosome and proteasome function, oxidative phosphorylation, and neurodegenerative diseases were identified by KEGG, and RNA splicing and metabolism as well as ribosome activity were pinpointed by Reactome analysis. We next performed STRING analyses to identify putative relationships between the proteins encoded by the differentially expressed genes. First, we examined the genes that were differentially upregulated in female microglia and identified a very small network of nodes, with proteins such as the monomeric G protein Nras, the microtubule-associated dynein complex, and the p300 histone acetyltransferase being at the hubs (Fig. 2D). On the other hand, several large gene network clusters appeared upon STRING analysis of the downregulated genes, including hubs involved in oxidative phosphorylation, ribosome, and proteasome function (Fig. 2E). The gene networks identified to be downregulated in female cervical spinal microglia suggest that male spinal microglia may have higher protein synthesis and posttranslational protein regulatory capacity, as well as higher oxidative metabolism than female microglia from the same region.
Fig. 2.
Cervical spinal cord microglial transcriptomes vary in females compared with males. (A) Principle component analysis of male and female cervical spinal cord microglia (SC; n = 5/treatment). (B) Volcano plot of differentially expressed genes in female vs. male cervical spinal cord microglia. Red dots represent significantly (FDR < 0.05) upregulated genes, whereas blue dots represent significantly downregulated genes. FC, fold change. (C) Functional enrichment analyses of differentially expressed genes from Biologic Process (GO; left), KEGG pathway (middle), and Reactome pathway (right). Shown are the top five most significant functional enrichments in upregulated genes (red) and in downregulated genes (blue). Enrichment categories are displayed on the y-axis and graphed by FDR, with lower FDR values indicating greater significance. Parentheses show the number of genes differentially expressed compared with the number of total background genes expressed in that category. Biologic Process (GO) categories: translation, peptide metabolic process (Pep Met Proc), cellular amide metabolic process (Cell Am Met Proc), organonitrogen compound biosynthetic process (Org Comp Bio Proc), organonitrogen compound metabolic process (Org Comp Met Proc). KEGG pathway categories: mammalian target of rapamycin signaling pathway (mTOR), breast cancer, ribosome, proteasome, oxidative phosphorylation (Ox Phos), Huntington’s disease (Huntington’s), Alzheimer’s disease (Alzheimer’s). Reactome pathway categories: nonsense mediated decay enhanced by the exon junction complex (NMD En EJC); antigen processing–cross presentation (Antigen Proc – Cross Pres); metabolism of RNA (RNA Met); formation of the ternary complex and, subsequently, the 43S complex (Ternary Complex). (D) STRING network analyses of the 395 upregulated genes. (E) STRING network analyses of the 511 downregulated genes. (F) MAGICTRICKS summary output for upregulated genes in female cervical spinal microglia (top 30 factors). (G) MAGICTRICKS summary output for downregulated genes in female cervical spinal microglia (top 30 factors). TF, transcription factor.
MAGIC analysis indicated that the topmost major transcriptional drivers (MAFK, BATF, SP1, FOXA1, TAL1) of the differentially upregulated genes (Fig. 2F) differed significantly from those (TAF1, PHF8, GABPA, TBP, PLU1) driving the expression of the differentially downregulated genes (Fig. 2G; Supplemental Table 9).
Microglia Derived from the Cervical Spinal Cord and Brainstem are Transcriptomically Distinct in Males.
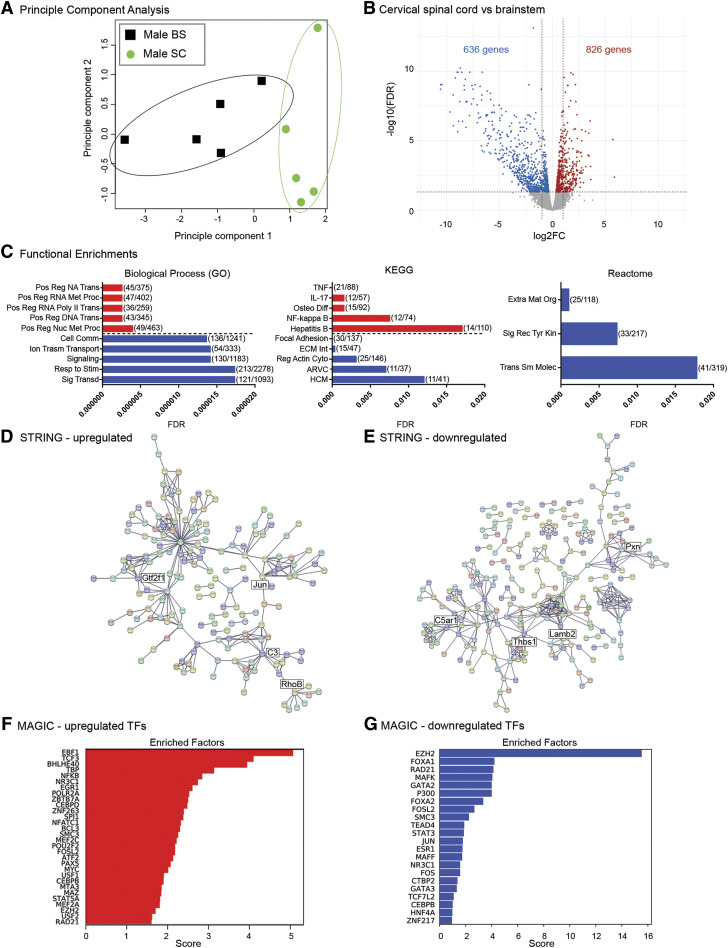
Given the region-specific sexual dimorphism in brainstem and cervical spinal cord microglia, we next interrogated whether within the same sex, there were general transcriptomic differences between brainstem and cervical spinal microglia (Fig. 3). Principle component analysis of male brainstem and cervical microglial transcriptomes indicated significant differences between both regions (Fig. 3A). We found that over 1400 genes were differentially expressed between the two CNS regions, 826 of which were higher in cervical spinal microglia, and 636 of which were lower, relative to expression in brainstem microglia (Fig. 3B; Supplemental Tables 3 and 4). GO, KEGG, and Reactome analyses were performed to determine whether the 826 differentially upregulated genes were segregated into specific functional gene categories (Fig. 3C, red bars). GO analysis indicated that the most upregulated functional gene categories included DNA and RNA synthesis, metabolism and transcription, whereas KEGG analysis identified proinflammatory cytokine and nuclear factor kappa B (NF-κB) signaling as common functional categories into which the upregulated genes fell. Reactome analysis identified no functional gene categories. Of the 636 differentially downregulated genes in male cervical spinal microglia, GO analysis pinpointed cell communication, signaling/signal transduction, and response to stimulus as functional gene categories (Fig. 3C, blue bars). KEGG analysis identified cytoskeleton and extracellular matrix functional gene categories, and Reactome analysis pinpointed extracellular matrix and receptor tyrosine kinase signaling. STRING analyses of the differentially upregulated genes showed clustering into distinct hubs at which complement C3, RhoB GTPase, and transcriptional regulators such as c-Jun and TFIIF were at the nodes (Fig. 3D). Conversely, analyses of the 636 genes that were lower in spinal microglia compared with brainstem microglia had multiple small, loosely grouped gene clusters, some of which had nodes revolving around cytoskeletal protein paxillin, thrombospondin 1, the extracellular matrix protein laminin subunit beta 2, and the complement component 5a receptor (Fig. 3E). Together, these data suggest that cervical microglia may be more metabolically and transcriptionally active than brainstem microglia and perhaps less mobile and interactive with other cells as well.
Fig. 3.
Transcriptomic divergence between male cervical spinal and brainstem microglia. (A) Principle component analysis of male cervical spinal cord (SC) and brainstem (BS) microglia (n = 5/treatment). (B) Volcano plot of differentially expressed genes in male cervical spinal cord microglia. Red dots represent significantly (FDR < 0.05) upregulated genes, blue dots represent significantly downregulated genes. FC, fold change. (C) Functional enrichment analyses of differentially expressed genes from Biologic Process (GO; left), KEGG pathway (middle), and Reactome pathway (right). Shown are the top five most significant functional enrichments in upregulated genes (red) and in downregulated genes (blue). Enrichment categories are displayed on the y-axis and graphed by FDR, with lower FDR values indicating greater significance. Parentheses show the number of genes differentially expressed in that category compared with the number of total background genes expressed in the category. Biologic Process (GO) categories: positive regulation of nucleic acid–templated transcription (Pos Reg NA Trans); positive regulation of RNA metabolic process (Pos Reg RNA Met Proc); positive regulation of transcription by RNA polymerase II (Pos Reg RNA Poly II Trans); positive regulation of transcription, DNA templated (Pos Reg DNA Trans); positive regulation of nucleobase-containing metabolic compound (Pos Reg Nuc Met Proc); cell communication (Cell Comm); ion transmembrane transport (Ion Transm Transport); signaling; response to stimulus (Resp to Stim); signal transduction (Sig Transd). KEGG pathway categories: tumor necrosis factor signaling pathway (TNF), interleukin-17 signaling pathway (IL-17), osteoclast differentiation (Osteo Diff), NF-κB signaling pathway, hepatitis B, focal adhesion, extracellular matrix–receptor interaction (ECM Int), regulation of actin cytoskeleton (Reg Act Cyto), arrhythmogenic right ventricular cardiomyopathy (ARVC), hypertrophic cardiomyopathy (HCM). Reactome pathway categories: extracellular matrix organization (Extra Mat Org), signaling by receptor tyrosine kinases (Sig Rec Tyr Kin), transport of small molecules (Trans Sm Molec). (D) STRING network analyses of the 826 upregulated genes. (E) STRING network analyses of the 636 downregulated genes. (F) MAGICTRICKS summary output for upregulated genes in male cervical spinal microglia (top 30 factors). (G) MAGICTRICKS summary output for downregulated genes in male cervical spinal microglia (all 22 factors). TF, transcription factor.
Interestingly, MAGIC analysis identified that the differentially upregulated genes were driven by multiple transcriptional regulators, including early B cell factor 1 (EBF1), NF-κB, the glucocorticoid receptor, and early growth response factor 1 (Fig. 3F; Supplemental Table 10), whereas the most significant transcriptional driver of the differentially downregulated genes in cervical spinal microglia was identified as the repressive histone H3 lysine 27 histone methylase, enhancer of zeste homolog 2 (EZH2) (Nutt et al., 2020) (Fig. 3G; Supplemental Table 10).
Microglia Derived from the Cervical Spinal Cord and Brainstem Are Transcriptomically Distinct in Females.
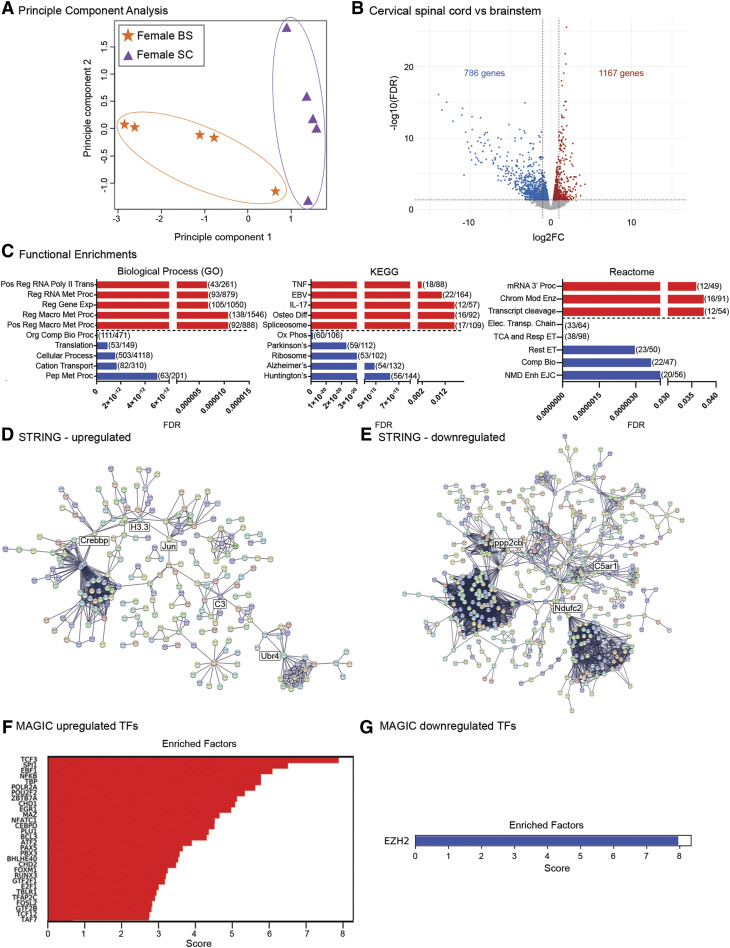
We next evaluated microglial transcriptomic differences between respiratory control regions in females (Fig. 4). Principle component analysis of female brainstem and cervical microglial transcriptomes indicated significant differences between both regions (Fig. 4A). We found that over 1900 genes were differentially expressed, 1167 of which were differentially upregulated and 786 which were downregulated in female cervical spinal microglia compared with brainstem microglia (Fig. 4B; Supplemental Tables 5 and 6). GO, KEGG, and Reactome analyses were performed to determine whether the 1167 differentially upregulated genes segregated into specific functional gene categories (Fig. 4C, red bars). GO analysis indicated that the most upregulated functional gene categories included regulation of RNA transcription and gene expression, together with macromolecule metabolism, whereas KEGG analysis identified proinflammatory cytokine viral infection and spliceosome as common functional categories into which the upregulated genes fell. Reactome analyses identified mRNA processing and termination as well as chromatin modifying enzymes as key functional categories that were upregulated in female spinal microglia. Of the 786 differentially downregulated genes in female cervical spinal microglia, GO analysis pinpointed nitrogen compound biosynthesis, protein translation, and peptide metabolism as functional gene categories. KEGG analysis identified oxidative phosphorylation, ribosome, and neurodegenerative disease categories, whereas Reactome analysis pinpointed mitochondrial electron transport and RNA splicing as functional gene categories into which the differentially expressed genes fell (Fig. 4C, blue bars). STRING analyses of the upregulated genes revealed several clusters with ubiquitin E3 ligase, complement C3, c-Jun, Crebbp, and histone H3.3 at their hubs (Fig. 4D), whereas STRING analysis of the downregulated genes demonstrated multiple, larger gene network clusters with protein phosphatase 2A, ndufc2 mitochondrial respiratory chain protein, and the complement C5a receptor at their hubs (Fig. 4E). These data imply that in females, cervical spinal microglia are more transcriptionally active and may have higher resting levels of proinflammatory cytokines than brainstem microglia but that they are less metabolically active.
Fig. 4.
Transcriptomic divergence between female cervical spinal and brainstem microglia. (A) Principle component analysis of female cervical spinal cord (CSC) and brainstem (BS) microglia (n = 5/treatment). (B) Volcano plot of differential gene expression changes in female C3–C6 cervical spinal cord vs. brainstem microglia. Red dots represent significantly (FDR < 0.05) upregulated genes, and blue dots represent significantly downregulated genes. FC, fold change. (C) Functional enrichment analyses of differentially expressed genes from Biologic Process (GO; left), KEGG pathway (middle), and Reactome pathway (right). Shown are the top five most significant functional enrichments in upregulated genes (red) and in downregulated genes (blue). Enrichment categories are displayed on the y-axis and graphed by FDR, with lower FDR values indicating greater significance. Parentheses show the number of genes differentially expressed in that category compared with the number of total background genes expressed in the category. Biologic Process (GO) categories: positive regulation of transcription by RNA polymerase II (Pos Reg RNA Poly II Trans), regulation of RNA metabolic process (Reg RNA Met Proc), regulation of gene expression (Reg Gene Exp), regulation of macromolecule metabolic process (Reg Macro Met Proc), positive regulation of macromolecule metabolic process (Pos Reg Macro Met Proc), organonitrogen compound biosynthetic process (Org Comp Bio Proc), translation, cellular process, cation transport, peptide metabolic process (Pep Met Proc). KEGG pathway categories: tumor necrosis factor signaling pathway (TNF), Epstein-Barr virus signaling pathway (EBV), interleukin-17 signaling pathway (IL-17), osteoclast differentiation (Osteo Diff), spliceosome, oxidative phosphorylation (Ox Phos), Parkinson’s disease (Parkinson’s), ribosome, Alzheimer’s disease (Alzheimer’s), Huntington’s disease (Huntington’s). Reactome pathway categories: mRNA 3′-end processing (mRNA 3′ Proc); chromatin modifying enzymes (Chrom Mod Enz); cleavage of the growing transcript in the termination region (Transcript cleavage); respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins (Elect. Transp. Chain); the citric acid cycle and respiratory electron transport (TCA and Resp ET); respiratory electron transport (Resp ET); complex I biogenesis (Comp Bio); nonsense mediated decay enhanced by the exon junction complex (NMD Enh EJC). (D) STRING network analyses of the 1167 upregulated genes. (E) STRING network analyses of the 786 downregulated genes. (F) MAGICTRICKS summary output for cervical spinal cord upregulated genes (top 30 factors). (G) EZH2 is the sole factor identified from MAGICTRICKS summary output for downregulated genes in cervical spinal microglia. TF, transcription factor.
MAGIC analysis of the upregulated genes in females showed that they were driven by multiple transcriptional drivers, some of which were similar to those found in males. The top driver identified was T-cell factor 3 (TCF3), and others included SP1 and other interacting factors such as ZBTB7a, EBF1, and NF-κB (Fig. 4F; Supplemental Table 11). Interestingly, for the differentially downregulated genes, MAGIC identified only a single transcriptional driver, EZH2, similar to what was observed in male cervical spinal microglia (Fig. 4G).
Gene Ontology, KEGG, and Reactome Functional Gene Categories Poorly Overlap in Males and Females.
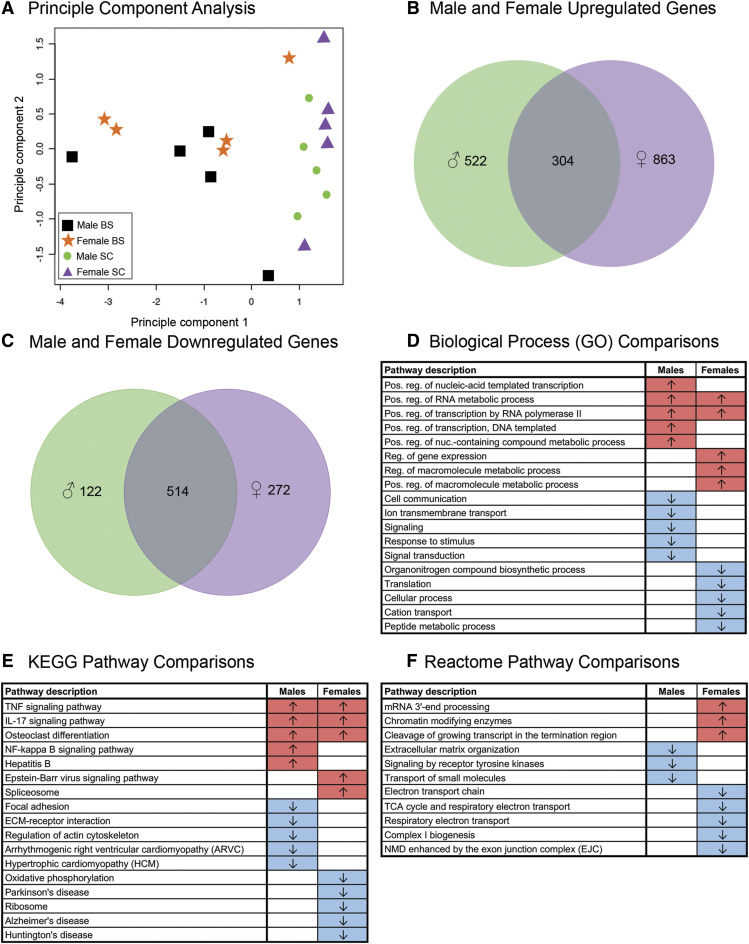
An initial principle component analysis of both sexes and CNS regions revealed that CNS region was a stronger driver of differential gene expression than was sex (Fig. 5A). We found that 13,120 genes were expressed both in male and female spinal microglia. Male spinal microglia expressed 826 differentially upregulated genes compared with male brainstem microglia, and females 1167 genes (Figs. 3 and 4). Of these upregulated genes, 304 were shared between male (green) and female (purple) spinal microglia (Fig. 5B; Supplemental Table 7). Male spinal microglia expressed 636 differentially downregulated genes compared with male brainstem microglia, and females 786 genes (Figs. 3 and 4). Of these downregulated genes, 514 were shared between male (green) and female (purple) spinal microglia (Fig. 5C; Supplemental Table 8). However, to determine whether the gene categories into which the most differentially expressed genes fell were similar between the sexes, we compared the top five up- and downregulated GO (Fig. 5D), KEGG (Fig. 5E), and Reactome pathway (Fig. 5F) categories in spinal microglia (compared with expression in brainstem microglia) derived from data in Fig. 3 (males) and Fig. 4 (females). Upregulated (red) and downregulated (blue) gene categories are shown in males (left column) and females (right column). We noted in all cases, that functional gene categories into which the differentially expressed genes fell differed between males and females, with the exception of two overlapping categories in GO Biologic Process terms and three terms in the KEGG pathway analysis. The very slight overlap in functional gene categories between male and female microglia suggests that there is significant functional divergence between male and female cervical microglia, perhaps underscoring potential functional differences between the sexes.
Fig. 5.
Functional gene categories have little overlap between males and females. (A) Principle component analyses of microglia from male and female brainstem (BS) and cervical spinal cord (SC) (n = 5/treatment) show that CNS region, not sex, is the stronger driver of differential gene expression. (B) Venn diagram showing the total number of differentially upregulated genes in male and female spinal microglia compared with brainstem microglia. Of the differentially upregulated genes in spinal compared with brainstem microglia, 304 were shared by both sexes, 522 genes were unique to males, and 863 were unique to females. Genes expressed in males are shown in green, and genes expressed in females are shown in purple. (C) Venn diagram showing the total number of differentially upregulated genes in male and female spinal microglia compared with brainstem microglia. Of the differentially upregulated genes in spinal compared with brainstem microglia, 514 were shared by both sexes, 122 genes were unique to males, and 272 were unique to females. Genes expressed in males are shown in green, and genes expressed in females are shown in purple. Tables Biologic Process (GO class) (D), KEGG pathway (E), and Reactome pathway (F) summarize the top five functional gene categories into which the differentially expressed genes in spinal microglia fall, among both both sexes. Categories that are comparatively upregulated (↑ red shading) and downregulated (↓ blue shading) in male and female spinal microglia (relative to the expression of those genes in male and female brainstem microglia) are shown separately and demonstrate that very few functional gene categories overlap between the sexes. ECM, extracellular matrix; IL-17, interleukin-17; NMD, nonsense mediated decay; TCA, tricarboxylic acid; TNF, tumor necrosis factor.
Discussion
Microglia, resident CNS macrophages, play a role in both respiratory rhythm generation and the neuroinflammation that can impair respiratory motor neuronal control. Although microglia make up 5%–10% of all CNS cells (diminishing in the rostral-caudal direction), they have the ability to alter brain architecture in all brain regions, including in the vicinity of brainstem respiratory neurons (Thoby Brisson et al., 2019). Although microglial number and morphology has been studied in brainstem regions involved in autonomic regulation of cardiovascular function (Kapoor et al., 2016a,b; Cohen et al., 2019), here, we analyze the transcriptomes of microglia isolated from the brainstem and cervical spinal cord and report significant sexual dimorphism in cervical spinal microglial gene signatures. These observations are consistent with the profound differences in morphology and transcriptome in male and female forebrain microglia (Schwarz et al., 2012; Hanamsagar and Bilbo, 2016; Hanamsagar et al., 2017; Bordeleau et al., 2019; Kodama and Gan, 2019). Sex differences in respiratory anatomy and function are well known, including the tendency for the female diaphragm to be shorter (Bellemare et al., 2003) and a higher ratio of large to small airways in females, enabling higher flow rates than in males (Becklake and Kauffmann, 1999). Males also have larger nasal cavities and longer nasal floors than females of comparable size (García-Martínez et al., 2016).
Despite these significant and well studied anatomic differences in breathing between the sexes, comparatively less is understood about sexual dimorphisms in respiratory neural control, including responses to ventilatory challenges (Jensen et al., 2005; Ahuja et al., 2007) and the capacity for respiratory plasticity (Zabka et al., 2006; Dougherty et al., 2017). The transcriptomic differences identified here may begin to shed light on cellular mechanisms underlying these differences. Indeed, the impact of stress and neuroinflammatory challenge on the developing respiratory control system also differs by sex; early life stress more strongly impacts respiratory control in male offspring (Fournier et al., 2013, 2015; Rousseau et al., 2017), effects that were associated with alterations in microglial density and morphology in the medulla of both sexes (Baldy et al., 2018). Although sex differences in brainstem microglial responses to early life stress were not detected in this model (Baldy et al., 2018), male and female microglia from the ventrolateral periaqueductal gray of adult rats (another brainstem region involved in respiratory neural control) do behave dimorphically after endotoxin challenge (Doyle et al., 2017). Profound sex-specific differences have also been reported in microglial inflammatory responses from non–respiratory-associated spinal regions (Sorge et al., 2011; Nacka-Aleksić et al., 2015; Mapplebeck et al., 2016; Taves et al., 2016; Fernandez-Zafra et al., 2019), suggesting that sex differences may also exist in spinal microglia from breathing-associated regions. It is not yet clear how microglia impact normal respiratory function or neuroplasticity; studies are underway to evaluate this.
Here we report that the microglial transcriptome exhibits both sex- and region-specific differences in CNS regions important for respiratory rhythm generation and patterning. Because male-prevalent breathing disorders, such as sudden infant death syndrome and central sleep apnea among others, may result from irregularity or interruption in the drive to breathe, often attributed to brainstem dysfunction (Benarroch, 2018; Kinney and Haynes, 2019), we initially predicted considerable sex differences in brainstem microglial gene signatures. Surprisingly, we found that brainstem microglia exhibited few gene expression differences based on sex (only 28 genes were differentially expressed between males and females). Also, unexpectedly, unlike brainstem microglia, we found marked sex differences in the transcriptomes of microglia isolated from the cervical spinal cord. Principal component analyses indicated a greater impact of CNS region than sex on the microglial transcriptome. We surmise that transcriptomic conservation in brainstem microglia between the sexes may subserve the importance of maintaining universal cardiorespiratory control in both sexes. One caveat to our study is that due to technical limitations, microglia were harvested from the entirety of the brainstem and cervical spinal cord and, as such, do not necessarily represent exclusively “respiratory” regions. A more detailed analysis of microglia isolated solely from the vicinity of respiratory neurons would be of considerable interest in future studies.
The functional gene categories into which the differentially expressed genes in male and female cervical spinal microglia segregated suggest that male spinal microglia may have greater capacities for oxidative metabolism, protein synthesis, and posttranslational protein regulation than female spinal microglia, potentially indicating that they may be more robust than female microglia. When comparing cervical spinal to brainstem microglia in males, brainstem microglia appeared to be less metabolically and transcriptionally active than cervical microglia and potentially more mobile and interactive with other CNS cell types, indicating that brainstem microglia may be more readily impacted by disruptions in brainstem function. This was similar in females as well, where brainstem microglia were less transcriptionally, but more metabolically, active. However, when compared with spinal microglia, female brainstem microglia appeared to have lower basal transcript levels of proinflammatory cytokines, possibly indicating that brainstem microglia may be more sensitive to the impact of inflammation than spinal microglia. Nonetheless, the near absence of overlap in functional gene categories between the differentially expressed genes in male and female cervical spinal microglia suggests that microglial activities in the cervical spinal cord in each sex likely differ. Consistent with this notion is the observation that phrenic nerve neuroplasticity appears to be more labile in males than females. For example, phrenic long-term facilitation diminishes with age in males (Zabka et al., 2001a), whereas it increases with age in females (Zabka et al., 2001b). Since proinflammatory microglia impair phrenic long-term facilitation in males (Huxtable et al., 2011, 2013, 2015), future studies should address a specific role for microglia in their sexually dimorphic contributions to spinal respiratory plasticity.
We performed bioinformatic analyses to identify whether common transcriptional drivers of the differentially expressed genes in microglia from both sexes and CNS regions. To our surprise, although there were significant sex differences in cervical spinal microglial transcriptomes, we found that the primary transcriptional modulator driving repression of the differentially downregulated genes in spinal microglia in both sexes was the histone methylase EZH2, an enzymatic subunit of the polycomb repressive complex 2 that catalyzes the trimethylation of histone H3 lysine 27 involved in gene silencing. EZH2 plays an important role in microglial plasticity and the rapid changes in gene expression that support their phenotype adaptations in different environmental situations (Cheray and Joseph, 2018). Conversely, the differentially upregulated genes in spinal microglia of both sexes were driven by combinations of essentially the same transcriptional regulators; in males EBF1 and TCF3 were the first and second most influential gene drivers, whereas in females, the top two drivers were TCF3 and SP1. Multiple other factors within the top several transcriptional regulators overlapped in both sexes. EBF1 is well known to regulate diversity in immune and inflammatory gene expression in microglia (Natoli et al., 2011; Griffin et al., 2013; Grabert et al., 2016). TCF3 often functions as a repressor that interacts with the Wnt signaling pathway to enhance the T helper cell type 2 response (Cadigan and Waterman, 2012; Marchetti and Pluchino, 2013). SP1 is a pleiotropic repressive transcription factor that controls microglial development and expression of multiple genes including immune genes (Delpech et al., 2016). It is interesting to note that our MAGIC analyses indicated that some of the major transcriptional factors driving differentially upregulated genes in both sexes often appear to function as transcriptional repressors; perhaps their binding is relieved to enable upregulation of these genes, or their binding removes another repressive factor to enable transcription. Additional studies are necessary to further distinguish these mechanisms.
One caveat important to mention here is that although the brainstem and cervical spinal cord house circuitry integral to breathing, these regions are also involved in other important physiologic systems, such as cardiovascular function, neuromodulation, and forelimb control. Thus, the differences in microglia that we report here will likely also impact several other critical physiologic systems in addition to breathing. Although females were included in all analyses in the present studies, we did not control for estrous cycle stage, which may contribute to the biologic variability observed in the principle component analyses of female transcriptomes in both CNS regions. This may be especially relevant to respiratory neuroplasticity given that it varies across the estrous cycle (Dougherty et al., 2017), as does the microglial transcriptome in cortical brain regions (Duclot and Kabbaj, 2015).
In summary, we provide an important transcriptomic resource data base to the field of respiratory neural control. The significant variations in microglial transcriptomes between male and female brainstem and cervical spinal cord, and differences in their functional gene categories, may underlie important, sexually dimorphic physiologic divergences in the control of breathing. We also identify novel transcriptional drivers that may be important for regulating these sex- and CNS region–specific differences in microglial transcriptomes. This information will be useful for future studies aimed at harnessing advantageous sex and regional differences in microglial gene expression to treat breathing disorders.
Acknowledgments
The authors wish to thank the University of Wisconsin-Madison Biotechnology Center Bioinformatics Research Core for their expert help and guidance with our bioinformatics analyses. We also wish to acknowledge Novogene (Sacramento, CA), which performed RNA sequencing on a fee-for-service basis.
Abbreviations
- CNS
central nervous system
- EBF1
early B cell factor 1
- EZH2
enhancer of zeste homolog 2
- FDR
false discovery rate
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NF-κB
nuclear factor kappa B
- TCF3
T-cell factor 3
Authorship Contributions
Participated in research design: Ewald, Kiernan, Baker, Watters.
Conducted experiments: Ewald, Timko.
Contributed new reagents or analytic tools: Roopra.
Performed data analysis: Ewald, Kiernan, Roopra, Radcliff.
Wrote or contributed to the writing of the manuscript: Ewald, Kiernan, Radcliff, Baker, Watters.
Footnotes
This work was supported by National Institute of Health National Heart, Lung and Blood Institute [R01 HL142752] (to J.J.W. and T.L.B.) and National Institute of Neurologic Disorders and Stroke [F31NS100229] (to E.A.K.).
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Agosto-Marlin IM, Nichols NL, Mitchell GS. (2018) Systemic inflammation inhibits serotonin receptor 2-induced phrenic motor facilitation upstream from BDNF/TrkB signaling. J Neurophysiol 119:2176–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja D, Mateika JH, Diamond MP, Badr MS. (2007) Ventilatory sensitivity to carbon dioxide before and after episodic hypoxia in women treated with testosterone. J Appl Physiol (1985) 102:1832–1838. [DOI] [PubMed] [Google Scholar]
- Armoskus C, Moreira D, Bollinger K, Jimenez O, Taniguchi S, Tsai H-W. (2014) Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res 1562:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldy C, Fournier S, Boisjoly-Villeneuve S, Tremblay ME, Kinkead R. (2018) The influence of sex and neonatal stress on medullary microglia in rat pups. Exp Physiol 103:1192–1199. [DOI] [PubMed] [Google Scholar]
- Becklake MR, Kauffmann F. (1999) Gender differences in airway behaviour over the human life span. Thorax 54:1119–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan M, Wenninger JM. (2008) Sex steroidal hormones and respiratory control. Respir Physiol Neurobiol 164:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemare F, Jeanneret A, Couture J. (2003) Sex differences in thoracic dimensions and configuration. Am J Respir Crit Care Med 168:305–312. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. (2018) Brainstem integration of arousal, sleep, cardiovascular, and respiratory control. Neurology 91:958–966. [DOI] [PubMed] [Google Scholar]
- Bordeleau M, Carrier M, Luheshi GN, Tremblay ME. (2019) Microglia along sex lines: from brain colonization, maturation and function, to implication in neurodevelopmental disorders. Semin Cell Dev Biol 94:152–163. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Waterman ML. (2012) TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4:a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Kaur S, Guha S, Batra SK. (2012) The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochim Biophys Acta 1826:129–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. (2017) Telomeres in health and disease. J Oral Maxillofac Pathol 21:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheray M, Joseph B. (2018) Epigenetics control microglia plasticity. Front Cell Neurosci 12:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen EM, Mohammed S, Kavurma M, Nedoboy PE, Cartland S, Farnham MMJ, Pilowsky PM. (2019) Microglia in the RVLM of SHR have reduced P2Y12R and CX3CR1 expression, shorter processes, and lower cell density. Auton Neurosci 216:9–16. [DOI] [PubMed] [Google Scholar]
- Crain JM, Nikodemova M, Watters JJ. (2009) Expression of P2 nucleotide receptors varies with age and sex in murine brain microglia. J Neuroinflammation 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain JM, Nikodemova M, Watters JJ. (2013) Microglia express distinct M1 and M2 phenotypic markers in the postnatal and adult central nervous system in male and female mice. J Neurosci Res 91:1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain JM, Watters JJ. (2009) Cytokine and BDNF expression vary with age and sex in mouse microglia. J Neurochem 108:138. [Google Scholar]
- De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, et al. (2017) Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95:341–356.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaes F, Fournier S, Tolsa JF, Peyter AC, Bairam A, Kinkead R. (2014) Consequences of gestational stress on GABAergic modulation of respiratory activity in developing newborn pups. Respir Physiol Neurobiol 200:72–79. [DOI] [PubMed] [Google Scholar]
- Del Negro CA, Funk GD, Feldman JL. (2018) Breathing matters. Nat Rev Neurosci 19:351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech JC, Wei L, Hao J, Yu X, Madore C, Butovsky O, Kaffman A. (2016) Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain Behav Immun 57:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominelli PB, Render JN, Molgat-Seon Y, Foster GE, Romer LM, Sheel AW. (2015) Oxygen cost of exercise hyperpnoea is greater in women compared with men. J Physiol 593:1965–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty BJ, Kopp ES, Watters JJ. (2017) Nongenomic actions of 17-β estradiol restore respiratory neuroplasticity in young ovariectomized female rats. J Neurosci 37:6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle HH, Eidson LN, Sinkiewicz DM, Murphy AZ. (2017) Sex differences in microglia activity within the periaqueductal gray of the rat: a potential mechanism driving the dimorphic effects of morphine. J Neurosci 37:3202–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclot F, Kabbaj M. (2015) The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol 16:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrmann IE, Ellis PS, Mazeyrat S, Duthie S, Brockdorff N, Mattei MG, Gavin MA, Affara NA, Brown GM, Simpson E, et al. (1998) Characterization of genes encoding translation initiation factor eIF-2γ in mouse and human: sex chromosome localization, escape from X-inactivation and evolution. Hum Mol Genet 7:1725–1737. [DOI] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. (2018) The reactome pathway knowledgebase. Nucleic Acids Res 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Zafra T, Gao T, Jurczak A, Sandor K, Hore Z, Agalave NM, Su J, Estelius J, Lampa J, Hokfelt T, et al. (2019) Exploring the transcriptome of resident spinal microglia after collagen antibody-induced arthritis. Pain 160:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty MJ, Mantilla CB, Sieck GC. (2018) Breathing: motor control of diaphragm muscle. Physiology (Bethesda) 33:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier S, Gulemetova R, Baldy C, Joseph V, Kinkead R. (2015) Neonatal stress affects the aging trajectory of female rats on the endocrine, temperature, and ventilatory responses to hypoxia. Am J Physiol Regul Integr Comp Physiol 308:R659–R667. [DOI] [PubMed] [Google Scholar]
- Fournier S, Steele S, Julien C, Fournier S, Gulemetova R, Caravagna C, Soliz J, Bairam A, Kinkead R. (2013) Gestational stress promotes pathological apneas and sex-specific disruption of respiratory control development in newborn rat. J Neurosci 33:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullam A, Schröder M. (2013) DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta 1829:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez D, Torres-Tamayo N, Torres-Sanchez I, García-Río F, Bastir M. (2016) Morphological and functional implications of sexual dimorphism in the human skeletal thorax. Am J Phys Anthropol 161:467–477. [DOI] [PubMed] [Google Scholar]
- Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, McColl BW. (2016) Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED. (2013) Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem 288:35925–35939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulemetova R, Drolet G, Kinkead R. (2013) Neonatal stress augments the hypoxic chemoreflex of adult male rats by increasing AMPA receptor-mediated modulation. Exp Physiol 98:1312–1324. [DOI] [PubMed] [Google Scholar]
- Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, Bilbo SD. (2017) Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65:1504–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Bilbo SD. (2016) Sex differences in neurodevelopmental and neurodegenerative disorders: focus on microglial function and neuroinflammation during development. J Steroid Biochem Mol Biol 160:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch UK. (2013) Functional diversity of microglia - how heterogeneous are they to begin with? Front Cell Neurosci 7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocker AD, Beyeler SA, Gardner AN, Johnson SM, Watters JJ, Huxtable AG. (2019) One bout of neonatal inflammation impairs adult respiratory motor plasticity in male and female rats. eLife 8:e45399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocker AD, Huxtable AG. (2019) Viral mimetic-induced inflammation abolishes Q-pathway, but not S-pathway, respiratory motor plasticity in adult rats. Front Physiol 10:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocker AD, Stokes JA, Powell FL, Huxtable AG. (2017) The impact of inflammation on respiratory plasticity. Exp Neurol 287:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxtable AG, Smith SM, Peterson TJ, Watters JJ, Mitchell GS. (2015) Intermittent hypoxia-induced spinal inflammation impairs respiratory motor plasticity by a spinal p38 MAP kinase-dependent mechanism. J Neurosci 35:6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxtable AG, Smith SMC, Vinit S, Watters JJ, Mitchell GS. (2013) Systemic LPS induces spinal inflammatory gene expression and impairs phrenic long-term facilitation following acute intermittent hypoxia. J Appl Physiol (1985) 114:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxtable AG, Vinit S, Windelborn JA, Crader SM, Guenther CH, Watters JJ, Mitchell GS. (2011) Systemic inflammation impairs respiratory chemoreflexes and plasticity. Respir Physiol Neurobiol 178:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kawakami K, Onimaru H, Okada Y, Yokota S, Koshiya N, Oku Y, Iizuka M, Koizumi H. (2017) The respiratory control mechanisms in the brainstem and spinal cord: integrative views of the neuroanatomy and neurophysiology. J Physiol Sci 67:45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturri P, Bairam A, Soliz J. (2017) Efficient breathing at neonatal ages: a sex and Epo-dependent issue. Respir Physiol Neurobiol 245:89–97. [DOI] [PubMed] [Google Scholar]
- Izci Balserak B. (2015) Sleep disordered breathing in pregnancy. Breathe (Sheff) 11:268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D, Wolfe LA, O’Donnell DE, Davies GAL. (2005) Chemoreflex control of breathing during wakefulness in healthy men and women. J Appl Physiol (1985) 98:822–828. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Randhawa KS, Epstein JJ, Gustafson E, Hocker AD, Huxtable AG, Baker TL, Watters JJ. (2018) Gestational intermittent hypoxia increases susceptibility to neuroinflammation and alters respiratory motor control in neonatal rats. Respir Physiol Neurobiol 256:128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor K, Bhandare AM, Farnham MM, Pilowsky PM. (2016a) Alerted microglia and the sympathetic nervous system: a novel form of microglia in the development of hypertension. Respir Physiol Neurobiol 226:51–62. [DOI] [PubMed] [Google Scholar]
- Kapoor K, Bhandare AM, Nedoboy PE, Mohammed S, Farnham MM, Pilowsky PM. (2016b) Dynamic changes in the relationship of microglia to cardiovascular neurons in response to increases and decreases in blood pressure. Neuroscience 329:12–29. [DOI] [PubMed] [Google Scholar]
- Khan N, Schoenike B, Basu T, Grabenstatter H, Rodriguez G, Sindic C, Johnson M, Wallace E, Maganti R, Dingledine R, et al. (2019) A systems approach identifies Enhancer of Zeste Homolog 2 (EZH2) as a protective factor in epilepsy. PLoS One 14:e0226733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan EA, Wang T, Vanderplow AM, Cherukuri S, Cahill ME, Watters JJ. (2019) Neonatal intermittent hypoxia induces lasting sex-specific augmentation of rat microglial cytokine expression. Front Immunol 10:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney HC, Haynes RL. (2019) The serotonin brainstem hypothesis for the sudden infant death syndrome. J Neuropathol Exp Neurol 78:765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson AO, Watters JJ. (2020) All roads lead to inflammation: is maternal immune activation a common culprit behind environmental factors impacting offspring neural control of breathing? Respir Physiol Neurobiol 274:103361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama L, Gan L. (2019) Do microglial sex differences contribute to sex differences in neurodegenerative diseases? Trends Mol Med 25:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, Gordon S. (1990) Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39:151–170. [DOI] [PubMed] [Google Scholar]
- LoMauro A, Aliverti A. (2018) Sex differences in respiratory function. Breathe (Sheff) 14:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozo T, Komnenov D, Badr MS, Mateika JH. (2017) Sex differences in sleep disordered breathing in adults. Respir Physiol Neurobiol 245:65–75. [DOI] [PubMed] [Google Scholar]
- Lynch M, Duffell L, Sandhu M, Srivatsan S, Deatsch K, Kessler A, Mitchell GS, Jayaraman A, Rymer WZ. (2017) Effect of acute intermittent hypoxia on motor function in individuals with chronic spinal cord injury following ibuprofen pretreatment: a pilot study. J Spinal Cord Med 40:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapplebeck JC, Beggs S, Salter MW. (2016) Sex differences in pain: a tale of two immune cells. Pain 157 (Suppl 1):S2–S6. [DOI] [PubMed] [Google Scholar]
- Marchetti B, Pluchino S. (2013) Wnt your brain be inflamed? Yes, it Wnt! Trends Mol Med 19:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM. (2017) Location, location, location: microglia are where they live. Neuron 95:233–235. [DOI] [PubMed] [Google Scholar]
- Nacka-Aleksić M, Djikić J, Pilipović I, Stojić-Vukanić Z, Kosec D, Bufan B, Arsenović-Ranin N, Dimitrijević M, Leposavić G. (2015) Male rats develop more severe experimental autoimmune encephalomyelitis than female rats: sexual dimorphism and diergism at the spinal cord level. Brain Behav Immun 49:101–118. [DOI] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, Barozzi I. (2011) The genomic landscapes of inflammation. Genes Dev 25:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodemova M, Watters JJ. (2012) Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J Neuroinflammation 9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Keenan C, Chopin M, Allan RS. (2020) EZH2 function in immune cell development. Biol Chem Jul 28;401(8):933-943. doi: 10.1515/hsz-2019-0436. [DOI] [PubMed]
- Poczobutt JM, De S, Yadav VK, Nguyen TT, Li H, Sippel TR, Weiser-Evans MCM, Nemenoff RA. (2016) Expression profiling of macrophages reveals multiple populations with distinct biological roles in an immunocompetent orthotopic model of lung cancer. J Immunol 196:2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez SC, Koschnitzky JE, Youngquist TM, Baertsch NA, Smith CV, Ramirez JM. (2019) Perinatal breathing patterns and survival in mice born prematurely and at term. Front Physiol 10:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau JP, Tenorio-Lopes L, Baldy C, Janes TA, Fournier S, Kinkead R. (2017) On the origins of sex-based differences in respiratory disorders: lessons and hypotheses from stress neuroendocrinology in developing rats. Respir Physiol Neurobiol 245:105–121. [DOI] [PubMed] [Google Scholar]
- Sandhu MS, Gray E, Kocherginsky M, Jayaraman A, Mitchell GS, Rymer WZ. (2019) Prednisolone pretreatment enhances intermittent hypoxia-induced plasticity in persons with chronic incomplete spinal cord injury. Neurorehabil Neural Repair 33:911–921. [DOI] [PubMed] [Google Scholar]
- Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS, Sutcliffe JG, Carson MJ. (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J Neurochem 83:1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Sholar PW, Bilbo SD. (2012) Sex differences in microglial colonization of the developing rat brain. J Neurochem 120:948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Yokota S, Miwakeichi F, Pokorski M, Aoyama R, Fukuda K, Yoshida H, Toyama Y, Nakamura M, Okada Y. (2019) Structural and functional identification of two distinct inspiratory neuronal populations at the level of the phrenic nucleus in the rat cervical spinal cord. Brain Struct Funct 224:57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliz J, Tam R, Kinkead R. (2016) Neonatal maternal separation augments carotid body response to hypoxia in adult males but not female rats. Front Physiol 7:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge RE, LaCroix-Fralish ML, Tuttle AH, Sotocinal SG, Austin JS, Ritchie J, Chanda ML, Graham AC, Topham L, Beggs S, et al. (2011) Spinal cord Toll-like receptor 4 mediates inflammatory and neuropathic hypersensitivity in male but not female mice. J Neurosci 31:15450–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes JA, Arbogast TE, Moya EA, Fu Z, Powell FL. (2017) Minocycline blocks glial cell activation and ventilatory acclimatization to hypoxia. J Neurophysiol 117:1625–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140:805–820. [DOI] [PubMed] [Google Scholar]
- Taves S, Berta T, Liu DL, Gan S, Chen G, Kim YH, Van de Ven T, Laufer S, Ji RR. (2016) Spinal inhibition of p38 MAP kinase reduces inflammatory and neuropathic pain in male but not female mice: sex-dependent microglial signaling in the spinal cord. Brain Behav Immun 55:70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio-Lopes L, Baldy C, Jochmans-Lemoine A, Mercier O, Pothier-Piccinin O, Seaborn T, Joseph V, Marc I, Kinkead R. (2017) Consequences of maternal omega-3 polyunsaturated fatty acid supplementation on respiratory function in rat pups. J Physiol 595:1637–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoby Brisson M, Cardoit L, Mayeur RM-E, Pascual O. (2019) Microglia shape the embryonic development of mammalian respiratory networks, in Proceedings of the Annual Meeting of the Society for Neuroscience; 2019 October 19–23; Chicago, IL. [Google Scholar]
- Townsel CD, Emmer SF, Campbell WA, Hussain N. (2017) Gender differences in respiratory morbidity and mortality of preterm neonates. Front Pediatr 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynder C, Stalker L, Doughty ML. (2010) Role of H3K4 demethylases in complex neurodevelopmental diseases. Epigenomics 2:407–418. [DOI] [PubMed] [Google Scholar]
- Zabka AG, Behan M, Mitchell GS. (2001a) Long term facilitation of respiratory motor output decreases with age in male rats. J Physiol 531:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabka AG, Behan M, Mitchell GS. (2001b) Selected contribution: time-dependent hypoxic respiratory responses in female rats are influenced by age and by the estrus cycle. J Appl Physiol (1985) 91:2831–2838. [DOI] [PubMed] [Google Scholar]
- Zabka AG, Mitchell GS, Behan M. (2006) Conversion from testosterone to oestradiol is required to modulate respiratory long-term facilitation in male rats. J Physiol 576:903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Lu W. (2014) Defensins in innate immunity. Curr Opin Hematol 21:37–42. [DOI] [PubMed] [Google Scholar]