Abstract
Spermatogenesis is a complex process that establishes male fertility and involves proper communication between the germline (spermatozoa) and the somatic tissue (Sertoli cells). Many factors that are important for spermatozoa production are also required for Sertoli cell function. Recently, we showed that the transcriptional cofactor ubiquitously expressed transcript (UXT) encodes a protein that is essential in germ cells for spermatogenesis and fertility. However, the role of UXT within Sertoli cells and how it affects Sertoli cell function was still unclear. Here we describe a novel role for UXT in the Sertoli cell’s ability to support spermatogenesis. We find that the conditional deletion of Uxt in Sertoli cells results in smaller testis size and weight, which coincided with a loss of germ cells in a subset of seminiferous tubules. In addition, the deletion of Uxt has no impact on Sertoli cell abundance or maturity, as they express markers of mature Sertoli cells. Gene expression analysis reveals that the deletion of Uxt in Sertoli cells reduces the transcription of genes involved in the tight junctions of the blood–testis barrier (BTB). Furthermore, tracer experiments and electron microscopy reveal that the BTB is permeable in UXT KO animals. These findings broaden our understanding of UXT’s role in Sertoli cells and its contribution to the structural integrity of the BTB.
Keywords: Sertoli cells, blood–testis barrier
Summary Statement An intact BTB is key to spermatogenesis, and little is known about the proteins regulating its integrity. Here we demonstrate that a transcription cofactor, UXT regulates the integrity of the BTB through the modulation of a BTB tight junction protein CLDN11.
Introduction
The establishment and maintenance of germ cells are critical processes that ensure the propagation and survival of a species [1]. The male germ cell, spermatozoon, is created by the complex process of spermatogenesis, which involves the tight control of hormonal and other signaling pathways [2]. Many of these hormonal and signaling pathways are not just controlled in a cell intrinsic manner, but are mediated through the somatic tissue as well. Within the testis, the Sertoli cells are the only somatic cell to have direct contact with developing germ cells, and germ cells are dependent upon Sertoli cells for growth and survival [3]. Any disruptions to the signaling pathways between germ cells and Sertoli cells can interfere with sperm production, resulting in infertility.
Recently our laboratory has demonstrated that ubiquitously expressed transcript (UXT) (also known as ART-27, STAP1, and SKP2 Associated-Alpha PFD1, NM_013840.3) is critical in germ cells for their completion of spermatogenesis [4]. UXT is a transcriptional cofactor that is important in transcriptional regulation [4–9]. With no enzymatic activity and no DNA binding domain, UXT exerts its effects on transcription as part of a larger protein complex [5, 10, 11]. We have previously demonstrated that the deletion of Uxt in germ cells is not cell lethal but results in the loss of germ cells starting at 7 days postpartum (dpp). The deletion of Uxt eventually results in a Sertoli cell-only phenotype around 23dpp, and this ultimately leads to sterility. Due to the specific time frame of germ cell loss, it appears that Uxt is not simply a housekeeping gene required for germ cell viability. UXT negative germ cells are able to form and enter the first wave of spermatogenesis, but the process is never completed. Our gene expression analysis demonstrated that many genes involved in germ cell differentiation are downregulated, suggesting a role for UXT in germ cell differentiation. Although germ cells are highly reliant on Sertoli cells for their development, our previous studies did not address a putative role of UXT in Sertoli cells, the topic of this manuscript.
Sertoli cells are located within the seminiferous tubules of the mammalian testis. They have often been characterized as the nurse cell of the testes because of the role they play in nurturing and supporting developing germ cells [12]. Germ cells and Sertoli cells are the only two cell types within the seminiferous tubule. Sertoli cells are greatly outnumbered by germ cells in the seminiferous tubule as one Sertoli cell supports multiple germ cells. During spermatogenesis Sertoli cells have a secretory role, which contributes to spermatogonia stem cell (SSC) self-renewal and differentiation [13]. Sertoli cells also provide nutrients and growth factors for haploid germ cells [14–17]. Additionally, one unique Sertoli cell function is the ability to create a specialized environment that separates mitotic germ cells from meiotic germ cells [18]. The Sertoli cell does this by creating a barrier between neighboring Sertoli cells known as the blood–testis barrier (BTB) [19, 20]. By sequestering meiotic germ cells behind this barrier, the Sertoli cell is able to protect them from harmful toxins, control the regulatory factors and nutrients meiotic germ cells are exposed to, and create a separate environment for pre-meiotic and meiotic germ cells to develop [21]. Additionally, the BTB plays a part in sequestering haploid germ cells away from the immune system. This is important because germ cell maturation begins after immune competence is established. Therefore, the expression of novel surface antigens found on haploid germ cells is potentially recognized as foreign by the immune system which could lead to an autoimmune response. The BTB, along with local immune suppression from Sertoli cells, protects germ cells from the immune system. Altogether, these Sertoli cell functions are essential for spermatogenesis.
To characterize the role of UXT in Sertoli cells, we created a mouse model with a conditional deletion of Uxt in Sertoli cells. Our results demonstrate that Sertoli cells remain viable and express markers of mature Sertoli cells. However, while the deletion of Uxt is incomplete, the Sertoli cells are unable to support spermatogenesis as we observe a loss of germ cells in a subset of seminiferous tubules. Gene expression analysis on primary Sertoli cells demonstrated that the deletion of Uxt largely affects genes important in cell adhesion. Many of these cell adhesion genes are BTB tight junction proteins. Our follow-up studies confirmed that the deletion of Uxt results in the reduction of the BTB tight junction protein Claudin 11 (CLDN11). Electron microscopy and tracer experiments confirmed that the downregulation of cell adhesion molecules (CAMs) in Sertoli cells resulted in a permeable BTB. Overall, we show that the deletion of Uxt in Sertoli cells reduces the expression of BTB tight junction proteins, including CLDN11, thus compromising the BTB.
Results
Sertoli cell deletion of UXT impairs spermatogenesis
To understand the role of UXT in Sertoli cells, we created a mouse model harboring a conditional deletion of Uxt (Chromosome X: 20,941,722-20,962,017) in Sertoli cells. To induce Sertoli cell-specific deletion of Uxt, we mated UxtF/Y males [4] to Amh-Cre females. AMH is specifically expressed in Sertoli cells starting at 12 days post coitum (dpc), which results in an embryonic deletion of UXT [22]. At 2 months of age, testis size and weight were reduced in KO animals compared to littermate controls, suggesting a defect in spermatogenesis (Figure 1A and B).
Figure 1.
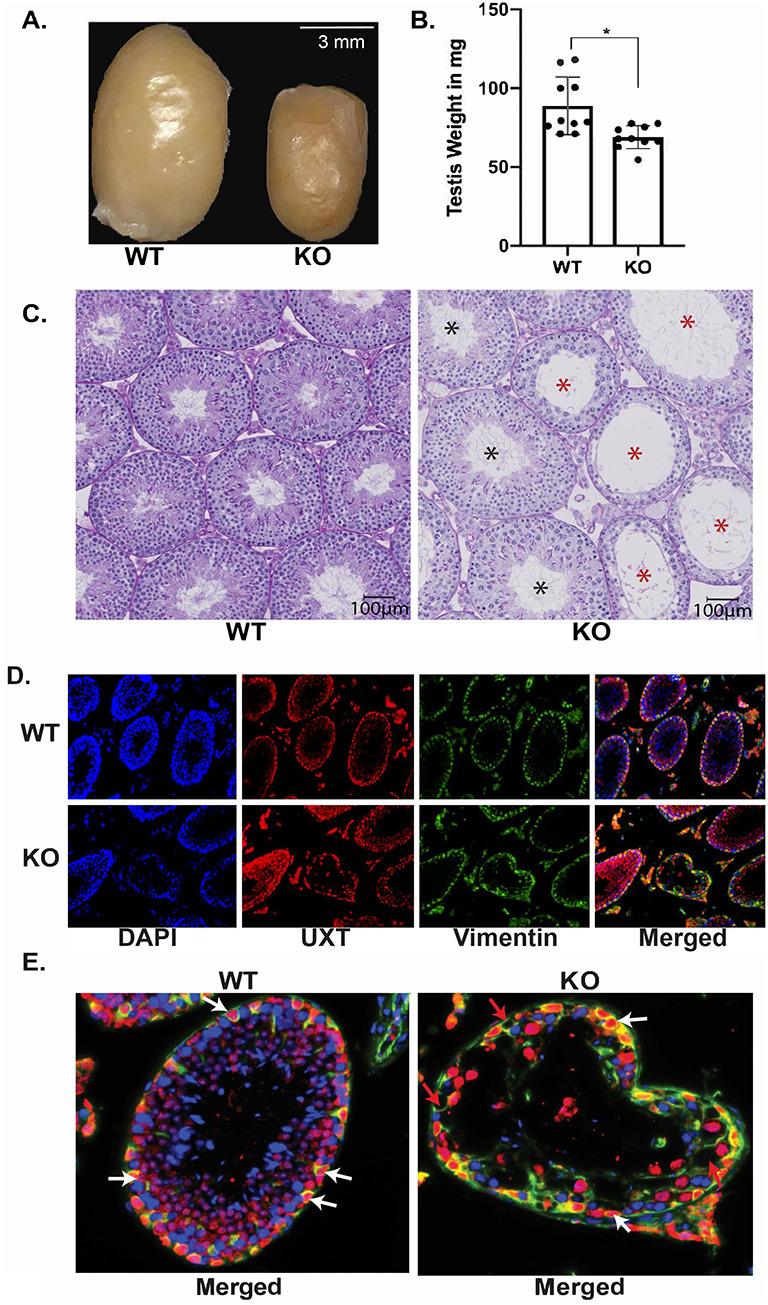
Ablation of UXT results in the loss of germ cells: (A) Gross anatomy of WT (left) and KO (right) testes. (B) Mass of UXT WT and KO testes in milligrams (mg). The graph demonstrates the mean mass of both WT and KO testes. Statistical significance was calculated using an unpaired student t-test. N = 10. (C) Periodic acid–Schiff (PAS) staining of UXT WT and KO testes. Scale bar is 100 μm. Black asterisk are tubules that appear unaffected. Red asterisk are tubules that show germ cell loss. (D) Immunofluorescence for UXT and Vimentin (Sertoli cell marker) in UXT WT and KO testes. (E) Higher magnification of merged Vimentin and UXT staining. White arrows show UXT positive Sertoli cells, and red arrows show UXT negative Sertoli cells.
Our previous study utilized a Vasa-Cre-driven deletion of Uxt in germ cells, which also resulted in reduced testis size and weight [4]. Further analysis demonstrated that the reduction in testis size and weight was due to the absence of germ cells in KO animals. To determine a putative function of UXT in Sertoli cells, we examined Amh-Cre-driven Uxt deletion in the testes from WT and KO animals. Interestingly, our histology results demonstrated that upon deletion of Uxt, there was a loss of germ cells in a subset of seminiferous tubules (red asterisks), while other seminiferous tubules appear to have no germ cell loss (black asterisks) (Figure 1C).
The heterogeneous loss of germ cells that is observed across different seminiferous tubules could be the result of inefficient deletion of UXT. It is possible that there is heterogeneous expression or inefficient activity of Amh-Cre. In order to evaluate the activity of Amh-Cre, we created a mouse model that utilizes tdTomato as a reporter. In both UXT WT and KO animals, tdTomato is under the control of the ubiquitously expressed Rosa26 promotor, but its expression is blocked by a stop codon that is flanked by loxP sites. Upon expression of Amh-Cre, the stop codon is specifically deleted, allowing for the expression of tdTomato. Our results showed that in both UXT WT and KO animals, tdTomato specifically marked all Sertoli cells (Figure 2A). This result suggests that CRE is present in every Sertoli cell, likely ruling out inefficient CRE expression as an explanation for the phenotypic heterogeneity. Although CRE is expressed homogenously, there are reports that CRE does not recombine all DNA loci at the same efficiency [23, 24]; the recombination of the Uxt loci appears to be inefficient. When we examined the expression of UXT in WT and KO animals, we observed that not every Sertoli cell in UXT KO animals is devoid of UXT (Figure 1D and E). Some Sertoli cells in phenotypically affected tubules (tubules that have germ cells loss) lack UXT, while others maintain UXT expression. The loss of germ cells indicate that Uxt plays a role in the Sertoli cell’s ability to support spermatogenesis.
Figure 2.
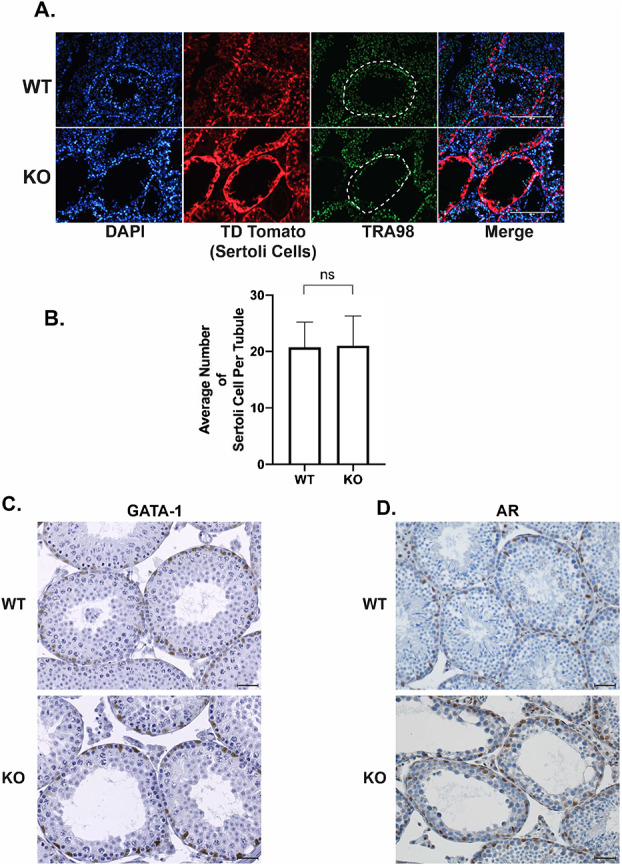
UXT negative Sertoli cells are viable and express mature markers: (A) Immunofluorescence for the germ cell marker TRA98 and tdTomato (genetic reporter for Sertoli cells) in UXT WT and KO testes. White dashed outline highlights the seminiferous tubule of focus. (B) Quantification of the average number of Sertoli cells per seminiferous tubule in WT and KO animals (WT (N = 3) and KO (N = 3)). (C) Immunohistochemistry (IHC) for GATA-1 in UXT WT and KO testes. (D) IHC for AR in UXT WT and KO testes. Scale bar is 100 μm.
Genetic ablation of Uxt does not impact Sertoli cell viability or maturity
Our studies demonstrate that the deletion of Uxt in Sertoli cells leads to a loss of germ cells in a subset of seminiferous tubules. Viable and mature Sertoli cells are critical to spermatogenesis [25]. As Sertoli cells mature and differentiate, they no longer proliferate, and they express markers such as GATA-1 and the androgen receptor (AR) [25]. Previously, our work highlighted the role of UXT in germ cell survival and development [4]. To test the possible role of UXT in Sertoli cell survival, we used the mouse model where tdTomato specifically marks Sertoli cells in UXT WT and UXT KO animals. We dissected the testes from these tdTomato UXT WT and KO animals and used immunofluorescence to visualize tdTomato (Figure 2A). Our results demonstrate that in WT and KO conditions, cells positive for tdTomato are still present, indicating that Sertoli cells are viable in the absence of Uxt (Figure 2A). To confirm the viability of Sertoli cells upon Uxt deletion, we quantified the number of Sertoli cells per tubule between WT and KO testes in 1-month-old animals (Figure 2B). Our results show that upon Uxt deletion, there is no difference in the number of Sertoli cells between WT and KO testes (Figure 2B). These results confirm that Sertoli cell viability is not affected by the deletion of Uxt. The equivalent number of Sertoli cells also suggests that they are not proliferating and have reached maturity.
Additionally, we also examined the expression of two markers of mature Sertoli cells, GATA-1 and AR. We conducted immunohistochemistry (IHC) for GATA-1 and AR in UXT WT and KO testes (Figure 2C and D). Our results demonstrate that KO Sertoli cells express both GATA-1 and AR (Figure 2C and D). These results indicate that Sertoli cells express markers of cell maturity in the absence of UXT.
While Sertoli cells were found to be viable and mature, the loss of germ cells led us to investigate whether there was an impact on fertility. Interestingly, we see that over time sperm concentration is reduced in UXT KO animals compared to WT animals (Supplementary Figure S1A). We also observed that the phenotype in the seminiferous tubules of UXT KO animals progressively become more severe. We quantified the number of tubules that showed a loss of germ cells (affected tubules) and saw that the number of affected tubules increased with age. The most severe phenotype occurred in 2-year-old KO animals (Supplementary Figure S1B). Two-year-old KO animals also exhibited a reduction in both the total number of litters and litter size when compared to 2-year-old WT animals (Supplementary Figure S1D and E). Overall this data indicates that the UXT deletion yields a phenotype that increases in severity over time. The increasing loss of germ cells may ultimately compromise fertility in older UXT KO animals.
Ablation of UXT leads to downregulation of cell adhesion molecules
Our previous studies of UXT in transcription regulation [5–7, 9] prompted us to conduct RNA-Seq analysis to uncover UXT regulated pathways that may regulate spermatogenesis. We hypothesized that the deletion of Uxt in Sertoli cells alters the Sertoli cell transcriptome, thereby impairing spermatogenesis. To test this hypothesis, we conducted RNA-Seq in primary WT and KO Sertoli cells to identify transcriptional changes that may explain germ cell loss.
We isolated and cultured primary Sertoli cells from WT and KO animals. Due to the heterogeneity of our phenotype, UXT was not deleted in all KO Sertoli cells (Figure 1D and E). To overcome this, we isolated primary Sertoli cells from UXT WT and KO animals and demonstrated the purity of our Sertoli cell culture via qPCR (Supplementary Figure S2A and B). We then infected the Sertoli cells with CRE recombinase to ensure the robust deletion of Uxt (Supplementary Figure S2A and C). While the addition of CRE reduced the levels of UXT in our KO Sertoli cells, it did not completely ablate UXT expression, further supporting the idea that CRE does not recombine all loci at the same efficiency (Supplementary Figure S2C). After selection with puromycin, we isolated RNA and protein from each sample (Supplementary Figure S2A).
Differential gene analysis demonstrated that upon Uxt KO, 773 genes were downregulated and 89 genes were upregulated (P ≤ 0.05; 1.5 log2 fold change) (Supplementary File S1). Figure 3A illustrates the top differentially regulated genes (P ≤ 0.05; 3.5 log2 fold change). The top downregulated genes according to Advaita Analysis iPathwayGuide revealed that cell adhesion molecules (CAMs) were the top downregulated pathway (Figure 3B) [26]. Our RNA-Seq results were validated via qPCR using RNA from primary Sertoli cells (Supplementary Figure S2D).
Figure 3.
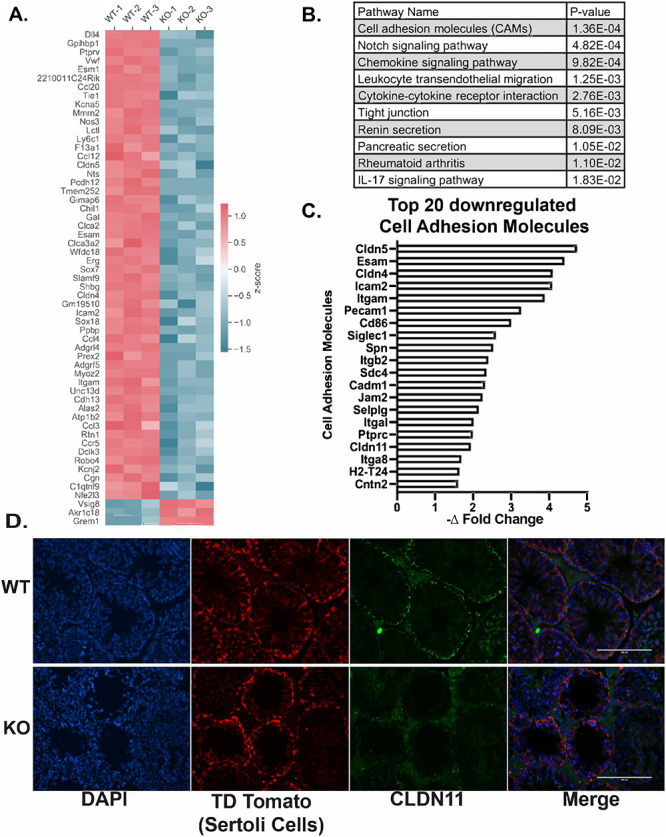
UXT deletion negatively impacts Sertoli cell adhesion: (A) Heatmap of the top differentially expressed genes (P ≤ 0.05; log2 fold change above or below 3.5). (B) Table of the top 10 downregulated pathways according to Advaita analysis iPathwayGuide (P ≤ 0.05; log2 fold change below 4). (C) Graphical representation of the top 20 downregulated cell adhesion molecules. (D) Immunofluorescence for CLDN11 and tdTomato (Sertoli cells) in WT and KO testes.
Cell adhesion is critical for Sertoli cell regulation of spermatogenesis. It allows Sertoli cells to form the blood–testis barrier (BTB) [21, 27]. The BTB is formed between adjacent Sertoli cells and is essential for providing a specialized environment for the development of spermatozoa. The BTB is composed of tight junctions (TJ), ectoplasmic specializations, desmosomes, and gap junctions [21]. Breach of the BTB leads to the destruction of this environment and the eventual loss of spermatozoa [21]. Therefore, we identified CAMs from our RNA-Seq data that have known roles in the structural integrity of the BTB.
Our RNA-Seq data identified several CAM family proteins that form the tight junctions of the BTB (CLDN5, JAM2, and CLDN11) that were downregulated as a result of Uxt deletion (Figure 3C). We hypothesized that downregulation of genes that make up the various junctions of the BTB may result in a compromised BTB possibly explaining the loss of germ cells seen in UXT KO mice. To test this hypothesis, we conducted immunofluorescence for these tight junction proteins to see if the downregulation of their mRNA was indicative of their protein levels in vivo. Of the tight junction proteins (CLDN5, JAM2, and CLDN11) identified in our RNA-Seq analysis, Claudin 11 (Cldn11) was the only gene to have a noticeable reduction at the protein level in KO testes (Figure 3D). Upon deletion of Uxt, CLDN11 expression is noncontiguous at the BTB. The deletion of Cldn11 in Sertoli cells has been shown to result in infertility as a function of a permeable BTB [28]. This result indicates that UXT may, directly or indirectly, regulate the expression of CLDN11. Additionally, the loss of CLDN11 may provide an explanation for the germ cell loss caused by Uxt deletion in Sertoli cells.
Ablation of UXT impairs cell adhesion and blood–testis barrier integrity
Because the deletion of Uxt in Sertoli cells downregulates a vital component of the BTB tight junctions, CLDN11, we investigated the integrity of the BTB in UXT KO animals. We conducted electron microscopy to visualize the cell junctions formed by Sertoli cells (Figure 4A). The junctions formed by WT Sertoli cells were tightly sealed, with several electron rich areas, which indicate “kissing” points along the cell–cell junction. However, the junctions formed by KO Sertoli cells are discontinuous with regions where adjacent membranes are widely separated (Figure 4A). This phenotype was never seen in WT animals. This result suggests that there are defects in the cell junctions formed by KO Sertoli cells.
Figure 4.
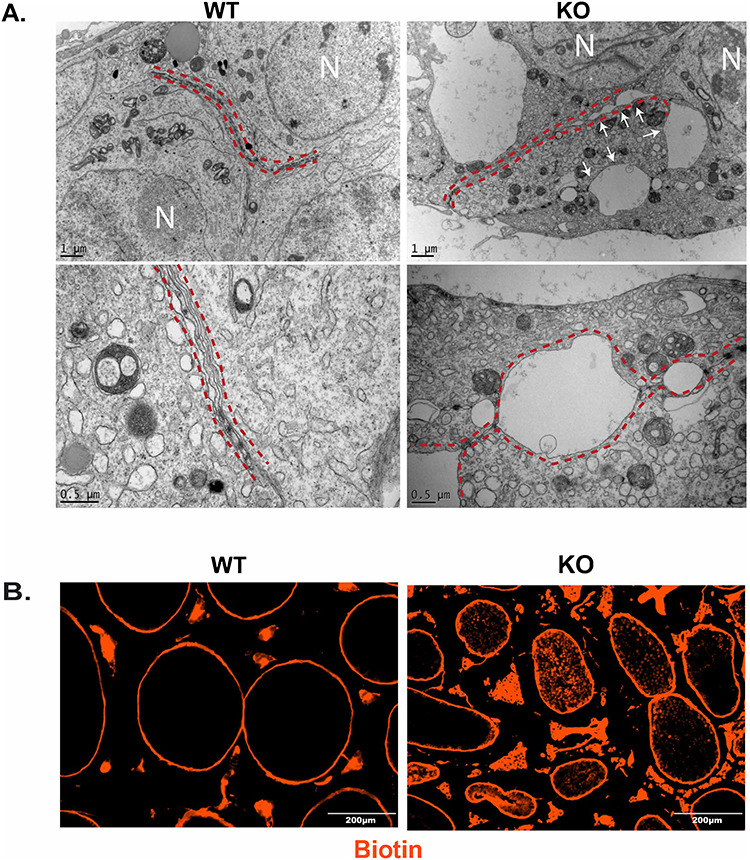
Ablation of UXT results in a permeable BTB: (A) Electron microscopy visualizing cell junctions in WT and KO testes WT (N = 3) and KO (N = 3). Red dashed lines outline cell junctions. White arrows indicate gaps in cell junction. Cell nucleus labeled by N. Bottom panels are higher magnification images. (B) BTB permeability was assayed using a biotin tracer (red). EVOS imaging system was used to visualize the exclusion of biotin in the lumen of WT seminiferous tubules and the presence of biotin in the lumen of KO seminiferous tubules.
To test for permeability of the BTB, we performed a functional assay to assess BTB integrity [28–31]. We injected a biotin tracer into the testes of WT and KO animals, letting the tracer diffuse throughout the tissue. We then fixed the tissue and analyzed the biotin diffusion into the seminiferous tubules beyond the BTB in WT and KO animals. In WT animals, the tracer was tightly restricted outside of the seminiferous tubules (Figure 4B). However, in KO animals, the tracer was present in the lumen of the seminiferous tubules (Figure 4B). Interestingly, in KO animals, we see that biotin infiltrates tubules that seemingly appear normal. It may be possible that these tubules will eventually lose germ cells due to the permeable BTB, but we would need to track these tubules over time to assess that possibility. Overall these results indicate that the BTB of KO animals is permeable.
We also investigated the possibility that the BTB is initially formed in UXT KO animals but becomes compromised or the deletion of UXT prevents or hinders the formation of the BTB. The BTB in mice is initially formed between 15 and 18 dpp [32]. To investigate KO Sertoli cells’ ability to form the BTB, we conducted IF on CLDN11 before the BTB is formed at 10 dpp and then after barrier formation at 18 dpp and 23 dpp. Our results demonstrate that while CLDN11 localizes at the tight junctions at the BTB in WT animals by 18 dpp, it is disorganized and discontinuous in UXT KO animals (Supplementary Figure S3). Overall, these results indicate that in KO animals, the reduction of CLDN11 expression results in an absent or permeable BTB.
Discussion
This is the first study examining the role of UXT in Sertoli cells. We find that the loss of UXT in a subset of Sertoli cells leads to a range of phenotypes. We did not observe a loss of germ cells in every seminiferous tubule upon the deletion of Uxt. Likewise, not every tubule showed a loss of CLDN11 or a compromised BTB. This is likely the result of UXT not being deleted in every Sertoli cell. While this was surprising, CRE activity can vary from gene to gene [23, 24]. Although incomplete deletion of Uxt reduced the severity of the observed phenotypes, loss of UXT produced significant effects on germ cells and the BTB. We believe that if the deletion of Uxt were complete, every seminiferous tubule would show a loss of germ cells, and the subsequent effects we observed would be much more severe.
One of the ways we tried to address the incomplete deletion of UXT in our RNA-Seq studies is through the addition of exogenous CRE. It is important to note that this could have biased our results as the levels of UXT were lower than the in vivo setting. However, Cldn11, which was identified in our RNA-Seq studies, was validated in our mouse model. Consistent with this finding, we were also able to confirm the defects in the BTB. Overall these findings validate our RNA-Seq analysis and provide an understanding of the transcriptional role of UXT in Sertoli cells.
At a molecular level, we see that the deletion of UXT results in an altered transcriptional program that leaves Sertoli cells unable to support spermatogenesis. While our results suggest that many of these effects could be driven through altered cell adhesion in Sertoli cells, other molecular pathways could contribute to the observed phenotypes. Upon deletion of UXT, our gene expression analysis indicates that over 800 genes were differential expressed. Some of the top downregulated pathways such as Notch, chemokine, and cytokine signaling could impact Sertoli cell function. Notch signaling, in Sertoli cells, has been demonstrated to play a role in germ cell homeostasis, while chemokine signaling is critical in establishing and maintaining the spermatogonia stem cell niche [33–35]. While aberrant signaling of these two pathways could alter spermatogenesis, the established role for cytokines in BTB dynamics could further explain the phenotypes that we observed in our model [36]. Cytokines are important for the restructuring of the BTB and apical ectoplasmic specialization (ES) that allow Sertoli cell–Sertoli cell interactions and Sertoli cell–germ cell interactions, respectively. It is possible that the deletion of UXT alters cytokine signaling, and this results in decreased cell adhesion in Sertoli cells. Cytokines have been reported to effect CLDN11 [37]; however, studies are needed to investigate the interplay between UXT and cytokine signaling.
CLDN11 is an integral membrane protein that belongs to the claudin family. Of the 24 claudins, CLDN3, CLDN5, and CLDN11 have been implicated in the structural integrity of the BTB [28, 31, 38]. Both Cldn5 and Cldn11 were downregulated in our RNA-Seq results upon the deletion of Uxt. However, upon examining the protein levels of CLDN5 and CLDN11, we only observed a decrease in CLDN11 protein expression. CLDN11 has been studied in Sertoli cells and is known to be a major contributor to the tight junctions of the BTB [28]. The constitutive and conditional deletion of Cldn11 in Sertoli cells results in sterility and a permeable BTB [28, 39]. In line with these findings, we show that the permeable BTB in UXT KO mice could be an effect of reduced CLDN11 expression, resulting in the loss of germ cells in UXT KO testes.
The idea that the sole function of the BTB is to create an immune-privileged environment was questioned in mouse models where Cldn11 was deleted. While the deletion of Cldn11 resulted in a permeable BTB and germ cell loss, there was no sign of an autoimmune response in these animals [39]. This result is consistent with our findings, which demonstrate that upon deletion of Uxt, there is a permeable BTB and germ cell loss with no histological evidence of an immune response in UXT KO animals. In addition to its role as an immunological barrier, the BTB has other functions in the seminiferous tubule, such as conferring polarity, germ cell differentiation, and protecting germ cells from harmful toxins [18]. This suggests that the permeability of the BTB in UXT KO animals may result in a disruption of the specialized microenvironment needed for spermatogenesis to occur.
Given that UXT has no DNA binding domain, we do not know precisely how it regulates Cldn11 gene transcription. We do know that UXT exists in a larger protein complex known as the unconventional prefoldin-like complex (UPC) [10]. Our lab, as well as others, has identified UXT, along with its binding partners: unconventional prefoldin RPB5 interacting protein (URI), prefoldin 2 (PFDN2), prefoldin subunit 6 (PFDN6), P53, and DNA damage regulated 1 (PDRG1), to interact with proteins involved in transcription, including the polymerase subunit RPB5/POLR2E and the ATP-dependent DNA helicases TIP48 and TIP49 [10, 11]. We conducted an IP/mass spectrometry analysis of UXT interactors in mouse Sertoli cells and identified several components of the UPC: complex URI, PFND2, PFDN6, PDRG1, and RPB5, suggesting that the UPC complex also exists in Sertoli cells. Our studies on the prostate have shown that the loss of UXT destabilizes the UPC [6, 7, 10]. If this is true in Sertoli cells, it would be consistent with our RNA-Seq results, which show that the majority of genes are downregulated upon Uxt deletion. One possibility is that in the UXT WT mouse, the UPC interacts with RNA polymerase, mediating normal transcription. Upon deletion of Uxt, the UPC becomes destabilized, losing its interaction with RNA polymerase, leading to an altered transcriptional program. Further studies are needed to understand the role of the UPC in transcriptional regulation within Sertoli cells.
We have shown a previously undescribed role of UXT as a transcriptional regulator in Sertoli cells. Upon the deletion of Uxt, CAMs, including CLDN11, are downregulated at the mRNA and protein level. Ultimately, we find that the deletion of UXT results in a permeable BTB and loss of germ cells. While these findings implicate UXT in the Sertoli cells’ ability to form tight junctions, what remains unclear is the impact UXT has on the apical attachments formed between Sertoli cells and spermatids. It is possible that the cell adhesion molecules allowing Sertoli cells to attach to germ cells are also negatively impacted. This phenomenon could lead to an early release of spermatids and negatively impact fertility. Further studies will be needed in order to understand the impact of UXT on the apical attachments. However, this may be difficult to evaluate in UXT KO animals considering a permeable BTB impacts the presence of spermatids. Taken together, this work describes an important role for UXT in Sertoli cell adhesion and a potential regulator of CLDN11.
Materials and methods
Ethic’s statement
This study protocol was approved by the Institutional Animal Care and Use Committee of NYU School of Medicine (Protocol 170102). Mice were euthanized with carbon dioxide delivered at a regulated flow rate followed by cervical dislocation. Mice were maintained and observed for signs of distress in accordance with the Guidelines for the Care and Use of Laboratory Animals at NYU School of Medicine.
Generating UxtF/Y; AMH-Cre mice
Uxt F/Y animals were previously generated in our laboratory, as described in [4]. Amh-Cre (129S.FVB-TG (AMH-cre) 8815REB/) females were purchased from Jackson Laboratory. Mixed background UxtF/Y males were mated to Amh-Cre females to generate a Sertoli cell-specific deletion of Uxt (UXT KO).
Additionally, we mated Uxt F/+; Amh-Cre females to Rosa26; Lox-stop-Lox tdTomato (B6.Cg-Gt (ROSA) 26Sortm14 (CAG-tdTomato) Hze/J) males from Jackson Laboratory. The resulting cross generated UxtF/Y; Amh-Cre; Lox-stop-Lox tdTomato (KO) males and Uxt X/Y; Amh-Cre; Lox-stop-Lox tdTomato (WT) males. These animals allowed us to specifically mark and visualize the Sertoli cells in WT and KO animals with tdTomato. This study protocol was approved by the Institutional Animal Care and Use Committee of NYU School of Medicine (Protocol 170102). Mice were euthanized with carbon dioxide delivered at a regulated flow rate followed by cervical dislocation. Mice were maintained and observed for signs of distress in accordance with the Guidelines for the Care and Use of Laboratory Animals at NYU School of Medicine.
Histology
The testes were dissected and cut in half. They were fixed in 4% buffered formalin for 24 h at 4 °C then dehydrated in two changes each of 50, 70, 95, and 100% ethanol. The testes were then cleared in xylenes and infiltrated with paraffin overnight at 60 °C.
The testes that were processed for periodic–acid Schiff (PAS) staining were dissected and halved. They were fixed overnight in Bouin solution at 4 °C and then dehydrated in 50 and 70% ethanol for two 20-min changes. Next, the testes were placed into 70% ethanol saturated with lithium chloride until the tissue turned white.
Immunofluorescence
The testes that were used for immunofluorescence were either processed as mentioned above or dissected and immediately snap frozen. They were then placed into optimal cutting temperature (OCT) to create frozen blocks. The blocks were then sectioned using a cryostat to produce 3 μm sections, and paraffin blocks were sectioned using a standard microtome to obtain 5 μm sections. Frozen slides were immediately placed into ice-cold acetone for 10 min for fixation.
Paraffin blocks were deparaffinized at 60 °C for 1 h. Slides were then dewaxed in xylenes for two 10-min changes and rehydrated in 100, 95, and 75% ethanol for two 5-min changes. Slides were immediately placed into antigen unmasking solution (Vector H-3300) and heated in a microwave for 20 min for antigen retrieval. All slides were washed in PBS and blocked in 20% normal goat serum (Vector Labs). Next slides were incubated with primary antibodies (Table 1) that were diluted in 2% BSA and incubated overnight at 4 °C. Primary antibodies were removed, and slides were washed with PBS. Secondary antibodies (Table 1) were diluted in 2% BSA and incubated with slides for 30 min at room temperature. Slides were then washed and incubated with DAPI for 5 min. Slides were then washed and coverslipped using Fluoromount-G (SouthernBiotech). Slides were imaged using the EVOS imaging system (Invitrogen EVOS FL Auto Digital Inverted Fluorescence System).
Table 1.
Antibodies.
| Antibody | Vendor/catalog number | Dilution |
|---|---|---|
| UXT (2361) | 1:00–1:1000 | |
| Vimentin | Abcam/Ab194719 | 1:250 |
| Streptavidin, Alexa Fluor 555 conjugate | Invitrogen/S21381 | 1:250 |
| Anti-Oligodendrocyte Specific Protein antibody (CLDN11) | Abcam/Ab53041 | 1:00–1:500 |
| TRA98 | Abcam/Ab82527 | 1:50 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen/A-21247 | 1:250 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen/A-21244 | 1:250 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen/A-21428 | 1:250 |
| ERK 1 Antibody (K-23) | Santa Cruz Biotechnology/sc-94 | 1:800 |
| ECL Rabbit IgG, HRP-linked whole Ab | Millipore Sigma/NA934 | 1:5000 |
| GATA-1 | Cell Signaling Technology/3535 | 1:100 |
| AR (N-20) | Santa Cruz Biotechnology/sc-5093 | 1:50 |
Breeding study
Three Uxt WT and three KO animals were aged for 2 years. At 2 years of age, these males were placed into separate housing for 3 days. After 3 days, two 6-week-old WT (C57BL/6J, Jackson Laboratory) females were placed with each Uxt WT and KO animal. Females were monitored to observe the presence of vaginal plugs. Each breeding cage was monitored for the number of litters and the size of each litter. After each litter, the two females were rotated to be housed with a different male mouse. This study was carried out over 6 months. At the end of 6 months, the testes from Uxt WT and KO animals were dissected and prepared for histological examination.
Quantification of histology
The testes from the Uxt WT and KO animals used in the breeding study were dissected and prepared for histological examination and PAS staining, as mentioned above. Testes blocks were cut to obtain 5 μm sections. Sections were subjected to PAS staining. Over 350 seminiferous tubules were counted per animal. All seminiferous tubules were evaluated for the presence or absence of germ cells. Tubules that had a loss of germ cells were counted and labeled as an affected tubule. An unpaired t-test was conducted to determine statistical significance.
RNA isolation and sequencing
The testes were dissected and subjected to two enzymatic digestions. The first enzymatic digestion (1 mg/ml collagenase and 4 mg/ml DNase dissolved in 10 mL of Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12)) was carried out at 37 °C in a shaking water bath at 150 RPM for 4 min. The first enzymatic digestion was removed and replaced with the second enzymatic digestion (1 mg/ml collagenase, 0.5 mg/ml trypsin, 1.5 mg/ml hyaluronidase, and 4 mg/ml DNase dissolved in 10 mL of DMEM/F12). This digestion was carried out at 37 °C in a shaking water bath at 150 RPM for 8 min. Cells were resuspended, on ice, for 3 min using a transfer pipette. The cells were then filtered through a 70 μm filter and pelleted at 300 × G for 8 min. The cell pellet was then resuspended in DMEM/F12 media containing 5% horse serum and 2.5% fetal bovine serum. Ten percent of the cell pellet was used to collect RNA (pre-population) and the remainder plated for Sertoli cell enrichment.
Cells were cultured for 24 h and washed twice with PBS, and the media was replaced with serum-free DMEM/F12 for 24 h to remove Sertoli cells (differential plating). After 24 h, the cells were washed twice with PBS and subjected to lentiviral infection. Enriched Sertoli cells that were obtained from UxtF/Y (WT) animals were transduced with an empty vector containing a puromycin selectable marker. Enriched Sertoli cells that were obtained from UxtF/Y; AMH-Cre (KO) animals were transduced with a vector containing Cre and puromycin selectable marker (Addgene plasmid # 17408). WT and KO Sertoli cells were subjected to two consecutive infections and then selected in puromycin for 1 week.
After puromycin selection, the cells were lysed to collect RNA and protein. RNA was obtained by using the RNeasy Plus Mini kit (Qiagen), and gDNA was removed via the gDNA removal column according to the manufacturer’s protocol. Sertoli cell purity was assessed by conducting qPCR for Sox9 (Sertoli cell marker), Cyp17a1 (Leydig cell marker), and Sycp3 (germ cell marker), on RNA obtained before and after differential plating. This same procedure that was used above to obtain RNA for sequencing was utilized to validate the RNA-Seq findings by qPCR (primers Table 2). The remaining RNA was submitted for library preparation and sequencing.
Table 2.
qPCR primers.
| Primer name | Sequence |
|---|---|
| UXT F1 | ACAAGGTATATGAGCAGCTGTCCGT |
| UXT R1 | TCTGGGACCACTGTGTCAACGA |
| GATA4 F1 | CACCCCAATCTCGATATGTTTGA |
| GATA4 R1 | GCACAGGTAGTGTCCCGTC |
| CLDN11 F1 | GTGGTGGGTTTCGTCACGAG |
| CLDN11 R1 | CGTCCATTTTTCGGCAGGTG |
| CLDN4 F1 | GTCCTGGGAATCTCCTTGGC |
| CLDN4 R1 | TCTGTGCCGTGACGATGTTG |
| CLDN5 F1 | GCAAGGTGTATGAATCTGTGCT |
| CLDN5 R1 | GTCAAGGTAACAAAGAGTGCCA |
| ESAM F1 | TTGCTGCGGGTTTTGTTCCT |
| ESAM R1 | TCTACCGCTTCCAATTTGTTGAG |
| ICAM2 F1 | TGGTCCGAGAAGCAGATAGTAG |
| ICAM2 R1 | GAGGCTGGTACACCCTGATG |
| GAPDH F1 | AATGGATTTGGACGCATTGGT |
| GAPDH R1 | TTTGCACTGGTACGTGTTGAT |
RNA quality was assessed via Agilent bioanalyzer. RNA-Seq library were prepared using the Illumina TruSeq Stranded mRNA LT kit (Cat #RS- RS-122-2101 or RS-122-2102), on a Beckman Biomek FX instrument, using 250 ng of total RNA as input, amplified by 12 cycles of PCR, and run on an Illumina 2500 (v4 chemistry), as single 50-base pair reads.
RNA-Seq analysis
Data was analyzed by Rosalind (https://rosalind.onramp.bio/), with a HyperScale architecture developed by OnRamp BioInformatics, Inc. (San Diego, CA). Reads were trimmed using Cutadapt [40]. Quality scores were assessed using FastQC [41]. Reads were aligned to the Mus musculus genome build mm10 using STAR [42]. Individual sample reads were quantified using HTseq [43] and normalized via relative log expression (RLE) using DESeq2 R library [44]. Read distribution percentages, violin plots, identity heatmaps, and sample MDS plots were generated as part of the QC step using RSeQC [45]. DEseq2 was also used to calculate fold changes and P values. Clustering of genes for the final heatmap of differentially expressed genes was done using the PAM (partitioning around medoids) method using the fpc R library [46]. Functional enrichment analysis of pathways, gene ontology, domain structure, and other ontologies was performed using HOMER [47]. Several database sources were referenced for enrichment analysis, including Interpro [48], NCBI [49], MSigDB [50, 51] REACTOME [52], and WikiPathways [53]. Enrichment was calculated relative to a set of background genes relevant for the experiment. Additional gene enrichment is available from the following partner institutions: Advaita (http://www.advaitabio.com/ipathwayguide) [54, 55].
Transmission electron microscopy
Mice were anesthetized and perfused with 4% paraformaldehyde in 0.1 M PBS (pH 7.4). The perfused testes were dissected and placed in a fixative solution containing 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h and post-fixed with 2% osmium tetroxide for 1 h and then block stained in 1% aqueous uranyl acetate, processed in a standard manner and embedded in EMbed 812 (Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections (70 nm) were cut, mounted on copper grids, and stained with uranyl acetate and lead citrate. Stained grids were examined under Philips CM-12 electron microscope and photographed with a Gatan (4k × 2.7k) digital camera.
Biotin tracer studies
Two-month-old Uxt WT and Uxt KO animals were used to assess the integrity of the BTB. The experiment was performed as described in [38] using a biotin tracer EZ-Link Sulfo-NHS-LC-Biotin (molecular weight, 557 Da) (21335; Thermo Scientific, www.piercenet.com). The localization of the tracer was visualized with the EVOS imaging system (Invitrogen EVOS FL Auto Digital Inverted Fluorescence System).
Quantification of Sertoli cells
Tissue sections were obtained from three 1-month-old Uxt WT and three Uxt KO animals. IHC was conducted for GATA1 on each tissue section to mark Sertoli cells. Slides were then evaluated for tubules that contained GATA1-positive Sertoli cells (WT = 249 tubules KO = 230 tubules). Oblong tubules that contained GATA1-positive Sertoli cells were excluded from analysis. Tubules were then manually counted using the Photoshop counting tool for the number of GATA 1-positive Sertoli cells. The number of GATA1-positive Sertoli cells were then averaged together for WT and KO animals.
Data availability
RNA-Seq data files are available on NCBI’s Gene Expression Omnibus accession number GSE155412.
Supplementary Material
Acknowledgments
We would like to acknowledge the shared resources of the NYUMC Genome Technology Center, High Throughput Biology Core (HTB), Histopathology Core, which are all partially supported by the Cancer Center Support Grant NIH/NCI P30CA016087 at the Laura and Isaac Perlmutter Cancer Center. We would also like to thank the NYU Langone Health DART Microscopy Lab, Alice Liang, Chris Petzold, and Kristen Dancel-Manning, for their assistance with TEM work. This core lab is partially funded by NYU Cancer Center Support Grant NIH/NCI P30CA016087. The HTB is also supported by NYSTEM Contract C026719. We also thank Dr. Margarita Vigodner (Stern College, Yeshiva University) and Dr. Myles Wilkinson (UC San Diego School of Medicine) for valuable advice. Much of this work was also supported by other members of the lab including Russell Ledet and Jeffery Schneider. Lastly, we thank Michael Garabedian for all of his invaluable advice and suggestions regarding this work.
Footnotes
† Grant Support: This research was supported by the National Institutes of Health (R01CA112226) (to SKL). This work was also partially funded by the Howard Hughes Medical Institute Gilliam Fellowship, the Ford Foundation Fellowship, and the National Institutes of Health Research Supplement to Promote Diversity in Health-Related Research (to PAT).
Contributor Information
Phillip A Thomas, Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, NY, USA.
Eric D Schafler, Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, NY, USA.
Sophie E Ruff, Department of Urology, New York University School of Medicine, New York, NY, USA; Department of Microbiology, New York University School of Medicine, New York, NY, USA.
Maud Voisin, Department of Microbiology, New York University School of Medicine, New York, NY, USA.
Susan Ha, Department of Urology, New York University School of Medicine, New York, NY, USA.
Susan K Logan, Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, NY, USA; Department of Urology, New York University School of Medicine, New York, NY, USA.
Conflict of interest
The authors declare no competing or financial interests.
References
- 1. Neto FT, Bach PV, Li, PS and Goldstein, M. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol 2016; 59:10–26. [DOI] [PubMed] [Google Scholar]
- 2. O'Shaughnessy PJ. Hormonal control of germ cell development and spermatogenesis. Semin Cell Dev Biol 2014; 29:55–65. [DOI] [PubMed] [Google Scholar]
- 3. Griswold MD. The central role of Sertoli cells in spermatogenesis . Semin Cell Dev Biol 1998; 9(4):411–6. [DOI] [PubMed] [Google Scholar]
- 4. Schafler ED, Thomas PA, Ha S, Wang Y, Bermudez-Hernandez K, Tang Z, Fenyö D, Vigodner, Logan SK. UXT is required for spermatogenesis in mice . PLoS One 2018; 13(4):e0195747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Markus SM, Taneja SS, Logan SK, Li W, Ha S, Hittelman AB, Rogatsky I, Garabedian MJ. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus . Mol Biol Cell 2002; 13(2):670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nwachukwu JC, Li W, Pineda-Torra I, Huang HY, Ruoff R, Shapiro E, Taneja SS, Logan SK, Garabedian MJ. Transcriptional regulation of the androgen receptor cofactor androgen receptor trapped clone-27 . Mol Endocrinol 2007; 21(12):2864–76. [DOI] [PubMed] [Google Scholar]
- 7. Nwachukwu JC, Mita P, Ruoff R, Ha S, Wang Q, Huang J, Taneja SS, Brown M, Gerald WL, Garabedian MJ, Logan SK. Genome-wide impact of androgen receptor trapped clone-27 loss on androgen-regulated transcription in prostate cancer cells. Cancer Res 2009; 69(7):3140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Su S, Li CY, Lei PJ, Wang X, Zhao QY, Cai Y, Wang Z, Li L, Wu M. The EZH1-SUZ12 complex positively regulates the transcription of NF-kappaB target genes through interaction with UXT . J Cell Sci 2016; 129(12):2343–53. [DOI] [PubMed] [Google Scholar]
- 9. Taneja SS, Ha S, Swenson NK, Pineda Torra I, Rome S, Walden PD, Huang HY, Shapiro E, Garabedian MJ, Logan SK. ART-27, an androgen receptor coactivator regulated in prostate development and cancer . J Biol Chem 2004; 279(14):13944–52. [DOI] [PubMed] [Google Scholar]
- 10. Mita P, Savas JN, Ha S, Djouder N, Yates 3rd JR, Logan SK. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex . PLoS One 2013; 8(5):e63879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI . Science 2003; 302(5648):1208–12. [DOI] [PubMed] [Google Scholar]
- 12. França LR, Hess RA, Dufour JM, Hofmann MC, Griswold MD. The Sertoli cell: one hundred fifty years of beauty and plasticity . Andrology 2016; 4(2):189–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015; 149(4):R159–67. [DOI] [PubMed] [Google Scholar]
- 14. Dorrington JH, Fritz IB, Armstrong DT. Control of testicular estrogen synthesis . Biol Reprod 1978; 18(1):55–64. [DOI] [PubMed] [Google Scholar]
- 15. Robinson R, Fritz IB. Metabolism of glucose by Sertoli cells in culture. Biol Reprod 1981; 24(5): 1032–41. [DOI] [PubMed] [Google Scholar]
- 16. Lacroix M, Smith FE, Fritz IB. Secretion of plasminogen activator by Sertoli cell enriched cultures . Mol Cell Endocrinol 1977; 9(2):227–36. [DOI] [PubMed] [Google Scholar]
- 17. Skinner MK, Griswold MD. Sertoli cells synthesize and secrete transferrin-like protein. J Biol Chem 1980; 255(20):9523–5. [PubMed] [Google Scholar]
- 18. Cheng CY, Mruk DD. The blood-testis barrier and its implications for male contraception . Pharmacol Rev 2012; 64(1):16–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiquoine AD. Observations on the early events of cadmium necrosis of the testis. Anat Rec 1964; 149:23–35. [DOI] [PubMed] [Google Scholar]
- 20. Kormano M. Dye permeability and alkaline phosphatase activity of testicular capillaries in the postnatal rat. Histochemie 1967; 9(4):327–38. [DOI] [PubMed] [Google Scholar]
- 21. Mruk DD, Cheng CY. The mammalian blood-testis barrier: its biology and regulation . Endocr Rev 2015; 36(5):564–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beau C, Vivian N, Münsterberg A, Dresser DW, Lovell-Badge R, Guerrier D. In vivo analysis of the regulation of the anti-Mullerian hormone, as a marker of Sertoli cell differentiation during testicular development, reveals a multi-step process . Mol Reprod Dev 2001; 59(3):256–64. [DOI] [PubMed] [Google Scholar]
- 23. Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2001; 2(4):292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coppoolse ER, de Vroomen MJ, van Gennip F, Hersmus BJM, van Haaren MJJ. Size does matter: Cre-mediated somatic deletion efficiency depends on the distance between the target lox-sites . Plant Mol Biol 2005; 58(5):687–98. [DOI] [PubMed] [Google Scholar]
- 25. Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood . Reproduction 2003; 125(6):769–84. [DOI] [PubMed] [Google Scholar]
- 26. Ahsan S, Draghici S. Identifying significantly impacted pathways and putative mechanisms with iPathwayGuide. Curr Protoc Bioinformatics 2017; 57:7–15, 1–7 15 30. [DOI] [PubMed] [Google Scholar]
- 27. Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KAL, Sharpe RM, Saunders PTK, Swinnen JV, Verhoeven G. The effect of a sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice . Mol Endocrinol 2006; 20(2):321–34. [DOI] [PubMed] [Google Scholar]
- 28. Mazaud-Guittot S, Meugnier E, Pesenti S, Wu X, Vidal H, Gow A, Le Magueresse-Battistoni B. Claudin 11 deficiency in mice results in loss of the Sertoli cell epithelial phenotype in the testis . Biol Reprod 2010; 82(1):202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Islam R, Yoon H, Kim BS, Bae HS, Shin HR, Kim WJ, Yoon WJ, Lee YS, Woo KM, Baek JH, Ryoo HM. Blood-testis barrier integrity depends on Pin1 expression in Sertoli cells . Sci Rep 2017; 7(1):6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCabe MJ, Foo CF, Dinger ME, Smooker PM, Stanton PG. Claudin-11 and occludin are major contributors to Sertoli cell tight junction function, in vitro . Asian J Androl 2016; 18(4):620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morrow CM, Tyagi G, Simon L, Carnes K, Murphy KM, Cooke PS, Hofmann MC, Hess RA. Claudin 5 expression in mouse seminiferous epithelium is dependent upon the transcription factor ets variant 5 and contributes to blood-testis barrier function . Biol Reprod, Cambridge, MA, USA, 2009; 81(5): 871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hogarth C. Retinoic acid metabolism, signaling, and function in the adult testis In: Sertoli Cell Biology. Elsevier: Waltham, MA; 2015: 247–272. [Google Scholar]
- 33. Garcia TX, Parekh P, Gandhi P, Sinha K, Hofmann MC. The NOTCH ligand JAG1 regulates GDNF expression in Sertoli cells . Stem Cells Dev 2017; 26(8):585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen SR, Tang JX, Cheng JM, Li J, Jin C, Li XY, Deng SL, Zhang Y, Wang XX, Liu YX. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 2015; 6(35):37012–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simon L, Ekman GC, Garcia T, Carnes K, Zhang Z, Murphy T, Murphy KM, Hess RA, Cooke PS, Hofmanna MC. ETV5 regulates sertoli cell chemokines involved in mouse stem/progenitor spermatogonia maintenance . Stem Cells 2010; 28(10):1882–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu S, Yan M, Ge R, Cheng CY. Crosstalk between Sertoli and germ cells in male fertility . Trends Mol Med 2020; 26(2):215–231. [DOI] [PubMed] [Google Scholar]
- 37. Oh YS, Jo NM, Park JK, Gye MC. Changes in inflammatory cytokines accompany deregulation of Claudin-11, resulting in inter-Sertoli tight junctions in Varicocele rat testes. J Urol 2016; 196(4):1303–12. [DOI] [PubMed] [Google Scholar]
- 38. Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier . Proc Natl Acad Sci USA 2005; 102(46):16696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice . Cell 1999; 99(6):649–59. [DOI] [PubMed] [Google Scholar]
- 40. Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads . EMBnet.journal, 2011; 17(1):3. [Google Scholar]
- 41. Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge, UK: Babraham Bioinformatics, Babraham Institute; 2010. [Google Scholar]
- 42. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner . Bioinformatics 2013; 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data . Bioinformatics 2015; 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 . Genome biology 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012; 28(16):2184–2185. [DOI] [PubMed] [Google Scholar]
- 46. Vomhof-DeKrey EE, Lee J, Lansing J, Brown C, Darland D, Basson MD. Schlafen 3 knockout mice display gender-specific differences in weight gain, food efficiency, and expression of markers of intestinal epithelial differentiation, metabolism, and immune cell function. PloS One 2019; 14(7):e0219267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities . Molecular cell 2010; 38(4):576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations . Nucleic Acids Res 2018; 47(D1):D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI biosystems database . Nucleic Acids Res 2009; 38(suppl_1):D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert SL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles . Proc Natl Acad Sci 2005; 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0 . Bioinformatics 2011; 27(12):1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Duenas Roca C, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2017; 46(D1):D649–-D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, Mélius J, Cirillo E, Coort SL, Digles D, Ehrhart F, Giesbertz P, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research . Nucleic Acids Res 2017; 46(D1): D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Donato M, Xu Z, Tomoiaga A, Granneman JG, Mackenzie RG, Bao R, Than NG, Westfall PH, Romero R, Draghici S. Analysis and correction of crosstalk effects in pathway analysis. Genome Res 2013; 23(11):1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis . Genome Res 2007; 17(10):1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data files are available on NCBI’s Gene Expression Omnibus accession number GSE155412.


