Abstract
Objective
Genome-wide association studies (GWAS) in rheumatoid arthritis (RA) have discovered over 100 RA loci, explaining patient-relevant RA pathogenesis but showing a large fraction of missing heritability. As a continuous effort, we conducted GWAS in a large Korean RA case–control population.
Methods
We newly generated genome-wide variant data in two independent Korean cohorts comprising 4068 RA cases and 36 487 controls, followed by a whole-genome imputation and a meta-analysis of the disease association results in the two cohorts. By integrating publicly available omics data with the GWAS results, a series of bioinformatic analyses were conducted to prioritise the RA-risk genes in RA loci and to dissect biological mechanisms underlying disease associations.
Results
We identified six new RA-risk loci (SLAMF6, CXCL13, SWAP70, NFKBIA, ZFP36L1 and LINC00158) with pmeta<5×10−8 and consistent disease effect sizes in the two cohorts. A total of 122 genes were prioritised from the 6 novel and 13 replicated RA loci based on physical distance, regulatory variants and chromatin interaction. Bioinformatics analyses highlighted potentially RA-relevant tissues (including immune tissues, lung and small intestine) with tissue-specific expression of RA-associated genes and suggested the immune-related gene sets (such as CD40 pathway, IL-21-mediated pathway and citrullination) and the risk-allele sharing with other diseases.
Conclusion
This study identified six new RA-associated loci that contributed to better understanding of the genetic aetiology and biology in RA.
Keywords: arthritis, rheumatoid; polymorphism, genetic; autoimmune diseases
Key messages.
What is already known about this subject?
Genome-wide association studies (GWAS) have identified >100 susceptibility loci for rheumatoid arthritis (RA).
Although the heritability of RA was estimated at 50%–65% in twin studies, previously reported loci were able to explain only about 15% of the total phenotypic variance for RA.
What does this study add?
We identified six new RA-risk loci (SLAMF6, CXCL13, SWAP70, NFKBIA, ZFP36L1 and LINC00158) that reached the genome-wide significance threshold (p=5.0×10−8) through a meta-analysis of newly generated GWAS results for 4068 RA cases and 36 487 healthy controls in a Korean population.
A series of bioinformatic analyses using the GWAS results identified 122 RA-relevant gene candidates from novel and known RA loci and suggested important roles of several tissues and gene sets in RA development.
How might this impact on clinical practice or future developments?
Our finding about novel RA loci refines the known genetic architecture of RA and provides genetic biomarkers for RA.
Introduction
Rheumatoid arthritis (RA) is a complex autoimmune disorder characterised by chronically inflamed joints and autoantibody production. A combination between genetic background and environmental triggers confers an increased risk for RA. The overall heritability of RA has been estimated to be about 50%–65% in twin studies.1 2 Genome-wide association studies (GWAS) have discovered over 100 RA-associated genetic variants in multiple ancestries.3 4 However, these loci collectively explained only a small portion (about 15%) of the phenotypic variance for RA.4 5 Thus, continuous effort in identifying additional RA variants is necessary to better understand the disease aetiology. Despite the substantial missing fraction of heritability, the reported RA loci have provided insights into the pathogenesis of RA by the so-called post-GWAS analysis that integrates GWAS data with multiple biological resources.4
Here, we performed GWAS to identify novel loci exceeding the genome-wide significance threshold (p=5×10−8) in Korean cohorts comprising 4068 RA cases and 36 487 controls, followed by subsequent post-GWAS analyses for prioritising RA-relevant genes and identifying variant-highlighted biology.
Methods
GWAS participants
A total of 4068 RA cases and 36 487 healthy controls from two independent case–control cohorts were analysed (3177 RA cases and 32 820 controls in cohort #1; 891 RA cases and 3667 controls in cohort #2). All the cases were recruited from eight participating university hospitals in Korea and diagnosed through the 1987 revised American College of Rheumatology RA classification criteria.6 Anti-citrullinated protein antibodies (ACPAs) were positive in 83.2%, negative in 14.6% and not examined in 2.2% of participants. The controls were recruited through the KoGES and Hanyang University Hospital for Rheumatic Diseases. The genomic DNAs from the KoGES samples were stored in the National Biobank of Korea. All participants provided written informed consent for the study and the Institutional Review Board of Hanyang University approved this study.
Genotyping and whole genome imputation
Cohort #1 was newly genotyped with a customised genotyping array, Korea Biobank Array (KoreanChip).7 Genotyping of the RA cases of cohort #2 was performed on an Illumina HumanOmni2.5Exome-8 BeadChip, while the control genotype data were produced using Illumina Human Omni1-Quad BeadChip. The overlapping variants between cases and controls in cohort #2 were merged and used in the downstream analyses. The genotyping data for each cohort were filtered based on the general criteria to retain good-quality genetic data (~465 K variants in set #1 of 3177 cases and 32 820 controls; ~559 K variants in set #2 of 891 cases and 3667 controls) that showed a high call rate per individual and variant (≥0.99), Hardy-Weinberg equilibrium (pHWE ≥5×10−6), no excessive heterozygosity, no difference in call rates per variant between cases and controls (p≥5×10−4), no cryptic first-degree relatedness among individuals, homogeneous genetic background among individuals and minor allele frequency (MAF) ≥0.005.
Imputation for autosomal variants was performed by Eagle28 and IMPUTE49 using the reference panel constructed from the 1000 Genomes Project (1KGP) phase 310 reference panel and whole genome sequencing data for 397 Koreans (Korean Personal Genome Project).11 Each imputation chunk included 10 000 variants with a 2 Mb buffer region. Imputation for the non-pseudo-autosomal region of the X chromosome was performed by Shapeit212 and Minimac313 with the 1KGP phase 310 reference panel, making imputation chunks of 5 Mb regions with 1 Mb buffer. The numbers of the imputed variants with good quality (showing imputation quality index ≥0.3, MAF ≥0.5% and pHWE in control ≥1×10−6) were 10 934 705 in cohort #1 and 10 868 813 in cohort #2. These variants were analysed in the subsequent analyses.
Association analysis
The genetic association between RA and each autosomal variant was tested by logistic regression adjusting for gender and the top four principal components using EPACTS.14 The association analysis for the X chromosome variants was performed separately for females and males by the same model excluding the gender variable. An inverse-variance-weighted fixed-effects meta-analysis for the genomic-control association results in the two independent cohorts were conducted using METAL.15 To investigate statistically independent association signals within each RA locus, we performed a conditional analysis by GCTA-COJO16 using the association summary statistics based on the reference linkage disequilibrium (LD) calculated in cohort #1.
Gene set and tissue-specific expression analysis
Gene-level association p values for 19 840 protein-coding genes were calculated by MAGMA17 using a variant-wide mean model based on variant-level association summary statistics within and around genes. Statistical significance for the association of the gene-level Z scores (converted from gene-level p values) with MSigDB18 gene sets including curated gene sets and gene ontology terms and tissue-specific gene expression in 54 tissues in Genotype-Tissue Expression (GTEx) v8 RNA Sequencing (RNA-Seq) data19 were tested according to the MAGMA17 regression models.
Enrichment analysis for tissue-specific histone modifications
GREGOR20 was deployed to calculate the enrichment estimates of RA variants within four tissue-specific histone marks (H3K4me1, H3K4me3, H3K27ac and H3K27me3) from Roadmap Epigenomics Project.21
Gene prioritisation in RA loci
For each RA locus, we identified a lead variant with the lowest pmeta value within a physically (±300 kb) or genetically (r2>0.1) defined region. FUMA22 was then employed to identify the most likely RA-relevant genes based on the lead RA-risk variants and the meta-analysis association summary statistics according to the following three mapping strategies—(1) Positional mapping: this strategy found genes within and 10 kb around the region containing a lead variant and its proxy variants in each RA locus. (2) Expression quantitative trait locus (eQTL) mapping: this mapping collected the genes regulated by known eQTL in LD with lead RA-risk variants in blood, spleen, lung and small intestine. The known eQTL was retrieved from GTEx,19 single-cell RNA-seq data in peripheral blood mononuclear cells,23 and DICE immune-cell data.24 (3) Chromatin interaction mapping: this approach found genes making promoter-involved or enhancer-involved chromatin interactions with the region with RA-risk variants in blood, spleen, lung and small intestine based on the GSE87112 (Hi-C) data25 and FANTOM526 annotations.
Variant-based heritability
An LD score regression method27 was performed to estimate the genome-wide and partitioned heritability28 using East Asian LD scores from the 1KGP phase 310 and the association summary statistics, assuming an RA prevalence of 0.01.
Ancestral genetic correlation of RA associations between Koreans and Europeans
A correlation of RA GWAS between Korean and European5 populations was examined by the Popcorn29 programme accounting for the ancestry-specific LD scores and cross-covariance scores in two ancestries in the 1KGP.
Correlation analysis of genetic architectures among diseases
We assessed the correlation of genome-wide disease associations between RA and other diseases (n=28; from Biobank Japan Project30 31 and Bentham et al 32) using the LD score regression method.27 The association summary statistics in 27 diseases excluding systemic lupus erythematosus were estimated from Japanese populations, whose LD structure is highly consistent with Korean ancestry.33 The data for systemic lupus erythematosus were generated from European case–control populations. Each set of genome-wide association summary statistics was generated using >1000 cases and displayed at least one disease locus with p<5×10−8.
Results
Identification of six novel RA loci
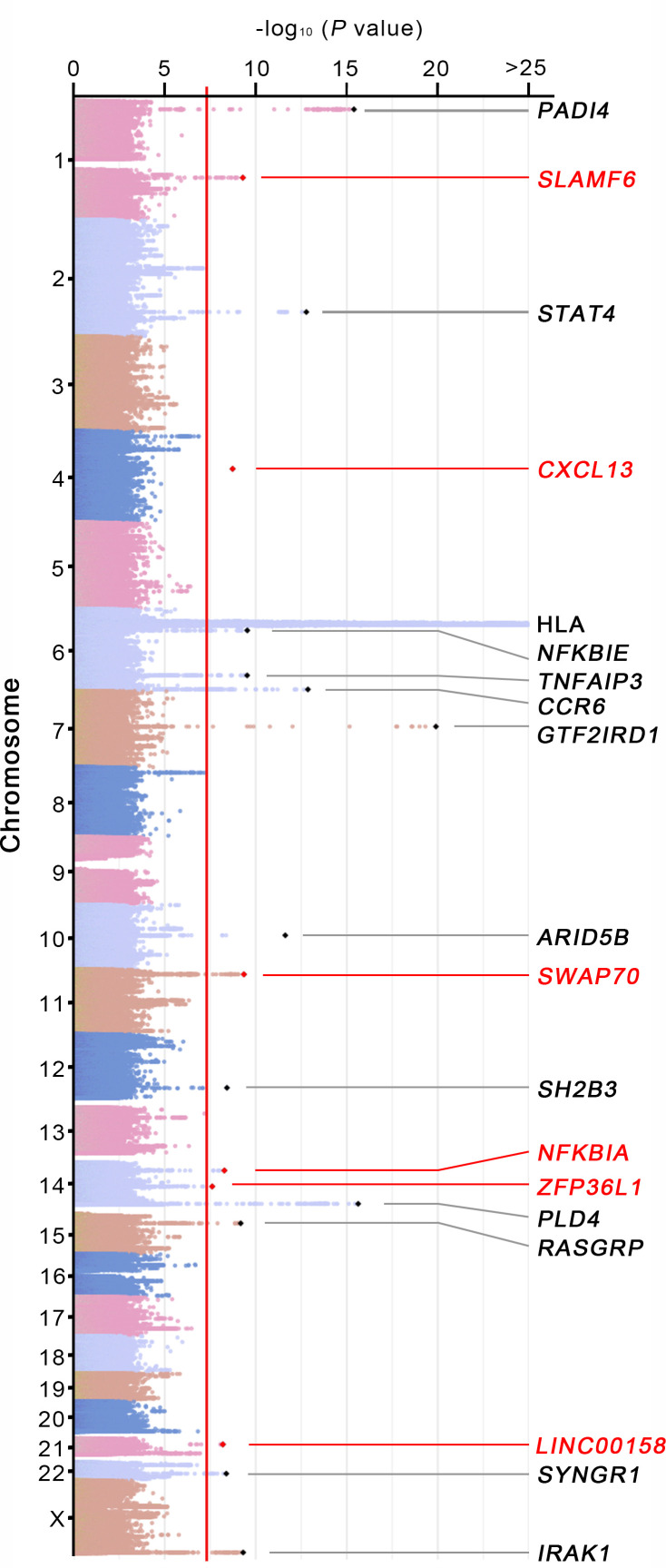
To identify new RA risk loci in a Korean population, we performed a fixed-effects meta-analysis of the RA association statistics of the newly generated, imputed variants in two independent Korean cohorts comprising 4068 RA cases and 36 487 controls. There were 10 404 202 intersecting variants between the two cohorts. The genomic control inflation factors (λGC) of the meta-analysis results including and excluding the major histocompatibility complex (MHC) region were estimated to be 1.035 and 1.020, respectively, showing no substantial population stratification in a quantile–quantile plot (online supplementary figure 1).
annrheumdis-2020-217663supp001.pdf (572.6KB, pdf)
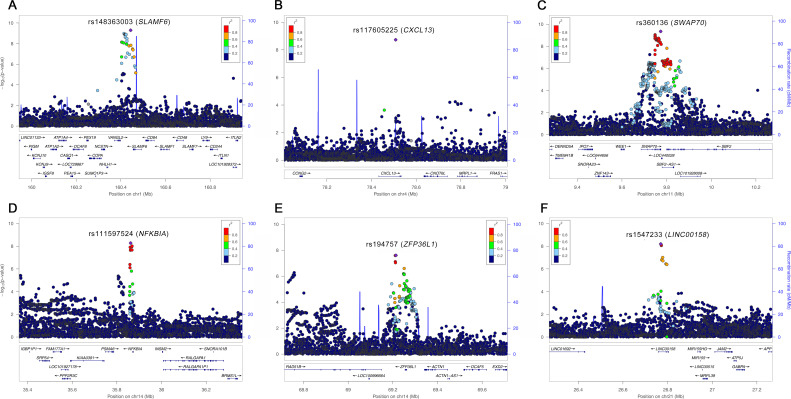
We identified 19 loci surpassing the genome-wide significance threshold (figure 1 and table 1). Of these loci, six (SLAMF6, CXCL13, SWAP70, NFKBIA, ZFP36L1 and LINC00158; figure 2) were novel loci that showed highly consistent effect sizes on the risk of RA in the two cohorts (table 1). The other 13 loci including the MHC region with the most significant association in the human genome had been reported in other RA genetic studies3 4 (figure 1 and table 1). No secondary association signals in novel RA loci were detected in conditional analyses.
Figure 1.
Genome-wide association analysis results for RA in a Korean population. A meta-analysis of genome-wide association results in two independent Korean cohorts identified 19 RA loci including 6 novel and 13 known loci. The minus log10-transformed association p values for the variants were plotted against their chromosomal positions. Novel and known RA-associated loci exceeding the genome-wide significance threshold (indicated by a red vertical line) are displayed with locus names in red and black, respectively. RA, rheumatoid arthritis.
Table 1.
RA associations of 19 loci in a meta-analysis
| Variant | Chr:Pos* | Gene† | Cohort #1 | Cohort #2 | Meta-analysis | ||||
| OR (95% CI)‡ | P value | OR (95% CI)‡ | P value | OR (95% CI)‡ | pmeta | pHet§ | |||
| rs11464953 | 1:17 645 185 | PADI4 | 1.23 (1.16–1.29) | 1.5×10−13 | 1.25 (1.12–1.40) | 8.9×10−5 | 1.23 (1.17–1.29) | 7.0×10−16 | 0.73 |
| rs148363003 | 1:160 443 973 | SLAMF6 | 1.63 (1.39–1.92) | 3.5×10−9 | 1.59 (1.12–2.27) | 1.0×10−2 | 1.62 (1.39–1.89) | 5.1×10−10 | 0.91 |
| rs11889341 | 2:191 943 742 | STAT4 | 1.21 (1.14–1.28) | 6.3×10−11 | 1.26 (1.12–1.41) | 1.3×10−4 | 1.22 (1.15–1.28) | 2.9×10−13 | 0.57 |
| rs117605225 | 4:78 508 197 | CXCL13 | 0.66 (0.57–0.76) | 1.6×10−8 | 0.59 (0.40–0.87) | 7.8×10−3 | 0.65 (0.56–0.75) | 1.9×10−9 | 0.63 |
| rs112062732 | 6:32 453 936 | HLA-DRB1 | 2.38 (2.25–2.52) | 1.1×10−193 | 2.79 (2.45–3.19) | 1.6×10−52 | 2.44 (2.31–2.58) | 5.5×10−227 | 0.034 |
| rs1044690 | 6:44 245 241 | NFKBIE | 1.19 (1.12–1.27) | 1.4×10−8 | 1.20 (1.06–1.37) | 4.6×10−3 | 1.20 (1.13–1.27) | 9.0×10−10 | 0.92 |
| rs9494893 | 6:138 223 490 | TNFAIP3 | 1.40 (1.27–1.55) | 4.7×10−11 | 1.15 (0.92–1.45) | 0.23 | 1.36 (1.23–1.49) | 3.5×10−10 | 0.13 |
| rs3093019 | 6:167 539 655 | CCR6 | 0.83 (0.78–0.87) | 2.3×10−12 | 0.84 (0.75–0.94) | 2.0×10−3 | 0.83 (0.79–0.87) | 1.4×10−13 | 0.85 |
| rs113066392 | 7:74 026 152 | GTF2IRD1 | 1.46 (1.35–1.57) | 6.3×10−22 | 1.22 (0.95–1.57) | 0.12 | 1.43 (1.33–1.55) | 1.3×10−20 | 0.20 |
| rs71508903 | 10:63 779 871 | ARID5B | 1.27 (1.19–1.36) | 2.9×10−13 | 1.12 (0.98–1.28) | 0.11 | 1.24 (1.17–1.32) | 2.4×10−12 | 0.092 |
| rs360136 | 11:9 773 517 | SWAP70 | 1.14 (1.08–1.20) | 1.4×10−6 | 1.31 (1.18–1.46) | 1.2×10−6 | 1.17 (1.11–1.23) | 4.5×10−10 | 0.026 |
| rs78894077 | 12:111 856 673 | SH2B3 | 1.30 (1.19–1.43) | 3.6×10−8 | 1.42 (1.10–1.83) | 7.2×10−3 | 1.32 (1.20–1.45) | 3.9×10−9 | 0.56 |
| rs111597524 | 14:35 861 800 | NFKBIA | 1.18 (1.12–1.26) | 1.3×10−8 | 1.15 (1.01–1.31) | 3.5×10−2 | 1.18 (1.12–1.24) | 5.3×10−9 | 0.68 |
| rs194757 | 14:69 213 851 | ZFP36L1 | 0.87 (0.83–0.92) | 2.9×10−6 | 0.81 (0.72–0.91) | 3.4×10−4 | 0.86 (0.82–0.91) | 2.5×10−8 | 0.24 |
| rs2841269 | 14:105 386 149 | PLD4 | 0.80 (0.76–0.85) | 5.9×10−15 | 0.82 (0.72–0.92) | 7.5×10−4 | 0.80 (0.76–0.85) | 2.4×10−16 | 0.77 |
| rs8032939 | 15:38 834 033 | RASGRP1 | 0.84 (0.80–0.89) | 1.6×10−10 | 0.93 (0.83–1.04) | 0.18 | 0.86 (0.81–0.90) | 9.5×10−10 | 0.13 |
| rs1547233 | 21:26 769 507 | LINC00158 | 1.17 (1.10–1.24) | 8.4×10−8 | 1.18 (1.05–1.33) | 6.3×10−3 | 1.17 (1.11–1.24) | 6.4×10−9 | 0.92 |
| rs2049985 | 22:39 756 650 | SYNGR1 | 0.78 (0.72–0.85) | 1.2×10−9 | 0.89 (0.75–1.05) | 0.17 | 0.80 (0.74–0.86) | 4.1×10−9 | 0.20 |
| rs1059702 | X:153 284 192 | IRAK1 | 0.86 (0.81–0.91) | 8.2×10−7 | 0.74 (0.65–0.84) | 3.3×10−6 | 0.84 (0.79–0.89) | 4.9×10−10 | 0.038 |
*Based on hg19.
†Novel RA loci are indicated in bold.
‡Frequencies, ORs and CIs for EAs.
§ P values for cross-cohort association heterogeneity (pHet) were calculated by Cochrane’s Q tests.
Chr, chromosome; EAs, effect alleles; Freq, frequency; NEA, non-effect allele; Pos, Position; RA, rheumatoid arthritis.
Figure 2.
Regional association plots for novel RA-associated loci. Regional association plots for the six new RA loci: (A) SLAMF6, (B) CXCL13, (C) SWAP70, (D) NFKBIA, (E) ZFP36L1 and (F) LINC00158, represent the minus log10-transformed p values for variants in each locus according to their genomic positions. The lead variants are indicated by purple points and the other variants are coloured according to r2 values with the lead variant in each locus. The names of lead variants and their closest genes are given on the top of the plots. RA, rheumatoid arthritis.
The overall variant-based heritability for RA on the liability scale was estimated to be 43.04%. The contribution of the non-MHC regions to heritability was 18.60%. When partitioned by the known non-MHC risk loci and the six novel loci, the heritability for RA was 6.65% from the known loci and 0.65% from the novel loci. The genetic correlation of RA associations between Korean and European5 populations was strong in a genome-wide level (correlation coefficient r=0.76; p=1.3×10−9) and in 100 known and six novel RA variants (r=0.76; online supplementary table 3 and figure 4).
Asian-specific RA-risk missense variant in the SH2B3 loci
Among the replicated known loci in this study, the RA association of the SH2B3 locus was previously explained by the intergenic variant rs10774624 in European populations5 that were neither common (MAF=0.56%) nor associated with RA (pmeta=0.64) in Korean populations (online supplementary table 3). However, we discovered that the SH2B3 missense variant (rs78894077, c.724C>T of exon 2; p.Phe242Ser of the PH domain) was associated with RA in the same locus (pmeta=3.89×10−9). Interestingly, the RA-risk allele T (p.242Ser) of rs78894077 was frequent in Korean populations (MAF=7.43%) but absent in other non-Asian ancestries according to the 1KGP phase 3 data. The RA-risk allele of the same variant in SH2B3 was also reported to associate with the increased risk of myeloproliferative neoplasms in East Asian populations.34 35
Tissues and gene sets implicated by genome-wide association results
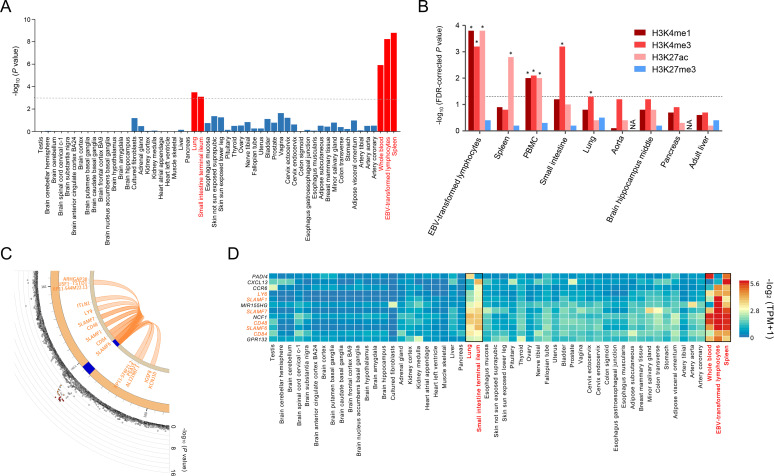
We calculated gene-level disease association p values from the variant-level association summary statistics using MAGMA17 and tested for the association between gene-level Z scores (converted from the gene-level p values) and tissue-specific expression level to provide deeper insights into the underlying disease-driving tissues in RA development. The analysis revealed five significant tissues with tissue-specific expression of RA-associated genes, including spleen (p=1.50×10−9), Epstein-Barr virus (EBV)-transformed lymphocytes (p=5.32×10−9), whole blood (p=1.12×10−6), lung (p=2.99×10−4) and small intestine (p=7.36×10−4), among 54 different tissues based on GTEx v8 RNA-Seq data19 (figure 3A and online supplementary table 1). Consistent with the association between the tissue specificity of gene expression and gene-level associations, we identified a significant enrichment of the identified lead and proxy variants within three transcription-activating histone marks (H3K4me1, H3K4me3 and H3K27ac) in the same tissues in a GREGOR20 enrichment analysis using Roadmap Epigenomic data21 at a false discovery rate (FDR) threshold of 5% (figure 3B). These results are supported by the well-documented knowledge about the aetiological roles of immune tissues and non-immune tissues in RA (eg, the increased risk of RA by EBV infection to B cells,36 the early production of RA autoantibody in lung37 and the enhanced inflammation in small intestine in RA38).
Figure 3.
Tissue-specific gene expression in RA loci including the SLAMF6 locus. (A) Tissue-specific gene expression levels in 54 GTEx tissues were analysed by MAGMA to identify tissue-specific expression of RA-associated genes. Five significant tissues (spleen, EBV-transformed lymphocytes, whole blood, lung and small intestine) are highlighted in red bars. The dashed line represents the Bonferroni-corrected significance threshold (=0.05/54). (B) Tissue-specific enrichment of RA variants within histone marks was analysed by GREGOR using Roadmap Epigenomics Project data. FDR-corrected p values for the overlap between RA-risk variants in 19 RA loci and 4 tissue-specific histone marks in 9 tissues were plotted with asterisks indicating significant enrichments with three transcription-activating histone marks in 5 tissues at the FDR threshold of 5% (represented by a dashed line). (C) The Circos plot for the SLAMF6 locus summarises the physical location (blue) with RA-risk variants and chromatin interaction (orange loops) involving both the RA-risk variant and the tissue-specific regulatory regions (including promoters and enhancers) in the five identified tissues. The outer layer in the Circos plot represents the minus log10-transformed RA association p values of variants according to the chromosomal position. The SLAMF6 variant (rs148363003) had chromatin interactions with 12 other coding genes (ARHGAP30, CD48, CD84, IGSF8, ITLN1, KCNJ10, LY9, PEA15, SLAMF1, SLAMF7, TSTD1 and USF1) on chromosome 1. (D) The heat map of gene expression levels represents the predominant expression levels of 11 prioritised RA-relevant RA genes in the 5 identified tissues (indicated by red tissue names and black box lines) among 54 tissues. The 11 RA genes that clustered in a gene expression analysis include known RA-associated genes and SLAMF genes (SLAMF6, LY9, SLAMF1, SLAMF7, CD48 and CD84). EBV, Epstein-Barr virus; RA, rheumatoid arthritis.
In addition, a MAGMA17 gene set analysis was performed using the gene-level Z scores and the predefined gene sets in MSigDB.18 The results suggested important roles of several immune-related gene sets such as the CD40 pathway (p=1.62×10-14), citrullination (p=8.60×10−13), IL-21-mediated signalling pathway (p=3.66×10−9) and lymphocyte activation (p=4.53×10−8) in developing RA (online supplementary table 2).
Prioritisation of RA-relevant genes in RA loci
Using the genome-wide association results, we further identified 122 potentially RA-relevant genes within 18 non-MHC RA loci by mapping the genes positioned on, cis-regulated by, and chromatin-interacted with RA-risk variants in blood, spleen, lung and small intestine (online supplementary table 3). Of the novel loci, the RA-risk variant in SLAMF6 (rs148363003) showed multiple chromatin interactions with several genes including five signalling lymphocytic activation molecule family (SLAMF) genes: SLAMF1, CD48 (also known as SLAMF2), CD84 (also known as SLAMF5), LY9 (also known as SLAMF3) and SLAMF7 (figure 3C). Interestingly, the expression levels of SLAMF6 and other interacting SLAMF genes were tissue-specifically overexpressed mostly in spleen, EBV-transformed B lymphocytes, whole blood, lung and small intestine terminal ileum. Similar tissue-specific expression patterns were observed in other well-known RA genes including PADI4, CCR6 and NCF1 (figure 3D).
Genetic correlation with other diseases
To assess the shared genetic risk between RA and other complex diseases using GWAS summary statics, we calculated the genetic correlation of genome-wide disease associations between RA (in our Korean cohorts) and other diseases (27 diseases in Japanese populations30 31 and systemic lupus erythematosus in a European population32). The genetic architecture of RA was positively correlated with that of RA (in Japanese populations), Graves' disease, systemic lupus erythematosus and atrial fibrillation, while a negative correlation was found with chronic hepatitis B infection and gastric cancer (online supplementary figure 2). The degrees of genetic correlations with RA were high in autoimmune diseases such as Graves’ disease (r=0.39) and systemic lupus erythematosus (r=0.33) (online supplementary figure 2).
Consistently, the prioritised RA-relevant genes in non-MHC RA loci in this study were significantly enriched in the susceptibility loci of inflammatory diseases such as systemic lupus erythematosus and inflammatory bowel disease at an FDR threshold of 0.05 (p≤2.43×10−7 in hypergeometric tests), according to GWAS catalog39 (online supplementary figure 3).
Discussion
In this study, we employed a new genome-wide array, KoreanChip, for the majority of the study subjects. The customised array KoreanChip has an extensive genomic coverage and powerful imputation performance for low-frequency variants in Korean ancestry compared with other commercial GWAS arrays with a similar number of probes. A high-density variant association result and a meta-analysis with another independent cohort helped us perform a powerful analysis to assess the genomic boundaries, the reliability (or cross-cohort heterogeneity) of disease association signals and functional annotations of RA loci.
The meta-analysis of GWAS data in Korean cohorts identified 19 susceptibility loci for RA, of which 6 loci have not been reported before, although there was considerably large missing heritability40 and a lack of functional studies. Of these new loci for RA, previous functional studies on the products of SLAMF6, CXCL13 and SWAP70 have been associated with RA or other inflammatory diseases. SLAMF6 is a member of superfamily immunoglobulin (Ig) domain-containing molecules that are involved in haemophilic interactions between naive B and T cells.41 SLAMF6 acts as a costimulatory molecule on the surface of T lymphocytes to increase T-cell adhesiveness by activating the small GTPase Rap1, promoting the TCR downstream signal transduction and the differentiation to Th17 cells expressing IL-17A.42 SLAMF members including SLAMF6 have been proposed as potential therapeutic targets for inflammatory and autoimmune diseases.43 In this study, RA-risk variants in SLAMF6 including rs148363003 had chromatin interaction with five SLAMF genes and the expression level of these six SLAMF genes in the locus was high in most of the potentially RA-relevant tissues. In other studies on rheumatic diseases, a SLAMF6 mutation (p.Phe238Cys) was found in T cells in RA44 and the increase in disease activity index in patients with systemic lupus erythematosus was associated with the enhanced levels of SLAMF3 and SLAMF6 on the surface of T lymphocytes and the high IL-17 level in serum.45
C–X–C motif chemokine ligand 13 (CXCL13), a B-cell chemoattractant, binds to its receptor CXCR5 to promote B-cell migration.46 The RA-risk variant (rs117605225) was located in a highly conserved region in the intron of CXCL13 according to two conservation scores (called GERP47 and SiPhy48). The RA-risk CXCL13 variant showed consistent effect sizes in the two independent cohorts and had no proxy variants based on the LD indices in the Korean cohorts and the 1KGP populations. Circulating CXCL13 levels had positive correlations with joint destruction rates and disease activity scores.49 Similarly, a transcriptome analysis displayed the enriched CXCL13 transcript levels in T cells in synovial fluid of patients with RA.50
SWAP70 is a Rho GTPase-regulatory protein that controls both cytoskeletal dynamics and the induction of interferon regulatory factor 4 (IRF4).51 52 SWAP70 is necessary for normal B-cell migration and immobilises F-actin filaments on phagosomes in human dendritic cells.51 53 Although a further investigation would be required to pinpoint and confirm SWAP70 as an RA-driving gene in the SWAP70 locus, an experimental murine model showed that genetic defects of SWAP70 resulted in lupus-like symptoms and the accumulation of Treg cells.53 54
This study emphasised the crucial role of the immune and non-immune tissues (lung and small intestine) for the development of RA by analysing the tissue-specific expression of RA-relevant genes, especially SLAMF member genes in the SLAMF6 locus and several known RA-associated genes including PADI4, CCR6 and NCF1. Among the RA-relevant tissues, lung has long been suggested as an early site of inflammation and ACPA in developing RA in an interaction of genetic factors (eg, HLA-DRB1,55 PADI4 56 57) with cigarette smoking58 and air pollution.59 Our previous bioinformatic study60 also identified the significant enrichment of RA-risk variants within lung-specific enhancer regions, indicating that non-coding RA-variants may regulate gene expression in allele-specific manner in lung.
Small intestine has large surfaces that are exposed to the gut environment and affects the human immune response with intestinal immune cells. As IgA-producing plasma cells of gut-associated lymphoid tissue are rich in lamina propria, the immune response by ACPA and rheumatoid factor of IgA isotype can be initiated in the intestinal mucosal tissue in RA.61 Interestingly, IL-21, highlighted by our gene set analysis, is primarily produced in the small intestine and is important in IgA responses to gut atypical commensals62 and T helper cell responses in intestinal inflammation in lamina propria in mice.63 In gut microbiota-induced arthritis in a murine model, T follicular helper cells were induced in lamina propria of gut before the onset of experimental arthritis and migrated to systemic lymphoid tissues, resulting in autoantibody production and arthritis.64
EBV infection has been associated with an increased risk of RA and other inflammatory diseases.36 65 An in silico study by Harley et al 66 analysed the binding sites of EBV-encoded EBNA2 proteins on the human genome in EBV-transformed B cells. The authors identified the significant enrichment of RA variants within the EBNA2-binding elements, which allowed the expression of RA-relevant genes to be highly allele-specific.
Functional studies on novel RA variants are needed in the future. Our bioinformatics approaches provide a list of 122 potentially RA-relevant genes in RA loci that may facilitate follow-up functional studies of candidate genes, patient stratification and drug target validation. The gene set analysis highlighted several immune-related pathways such as the CD40 signalling and IL-21-mediated pathways that had showed altered activities in patients with RA.67 68
In summary, this study discovered six novel RA loci in Korean populations and demonstrated variant-driven pathological features, evoking the importance of the identification of new RA loci and the integration of disease association results with other omics data.
Acknowledgments
We are grateful to all participants in this study and Jung-Ah Kim for the assistance in sample preparation.
Footnotes
Handling editor: Josef S Smolen
Y-CK, JL and S-YB contributed equally.
Contributors: KK and S-CB designed the study. Y-CK, JLim, EH, MYH, KY, KK and S-CB analysed the data and interpreted the results. S-YB, J-YC, D-HY, S-SL, JLee, WTC, T-HK, Y-KS, S-CS, C-BC, J-BJ, YMK, J-MS, Y-KL, S-KC, B-JK, H-SL and S-CB recruited and characterised patients with RA and controls. KK, S-YB, B-JK and S-CB generated genomic data. YCK, JLim, KK and S-CB wrote the manuscript. All authors reviewed and approved the manuscript.
Funding: This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (2017R1E1A1A01076388), the Korea Healthcare Technology R&D Project funded by the Ministry for Health and Welfare (HI15C3182) and Hanyang University Institute for Rheumatology Research. A part of genotype data was produced using the KoreanChip available through the K-CHIP consortium. KoreanChip was designed by Center for Genome Science, Korea National Institute of Health, Korea (4845–301, 3000–3031).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Ethics approval: The study was approved by the Institutional Review Board of Hanyang University.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. MacGregor AJ, Snieder H, Rigby AS, et al. . Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 2000;43:30–7. [DOI] [PubMed] [Google Scholar]
- 2. Terao C, Ikari K, Nakayamada S, et al. . A twin study of rheumatoid arthritis in the Japanese population. Mod Rheumatol 2016;26:685–9. 10.3109/14397595.2015.1135856 [DOI] [PubMed] [Google Scholar]
- 3. Okada Y, Eyre S, Suzuki A, et al. . Genetics of rheumatoid arthritis: 2018 status. Ann Rheum Dis 2019;78:446–53. 10.1136/annrheumdis-2018-213678 [DOI] [PubMed] [Google Scholar]
- 4. Kim K, Bang S-Y, Lee H-S, et al. . Update on the genetic architecture of rheumatoid arthritis. Nat Rev Rheumatol 2017;13:13–24. 10.1038/nrrheum.2016.176 [DOI] [PubMed] [Google Scholar]
- 5. Okada Y, Wu D, Trynka G, et al. . Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506:376–81. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnett FC, Edworthy SM, Bloch DA, et al. . The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. 10.1002/art.1780310302 [DOI] [PubMed] [Google Scholar]
- 7. Moon S, Kim YJ, Han S, et al. . The Korea Biobank array: design and identification of coding variants associated with blood biochemical traits. Sci Rep 2019;9:1382. 10.1038/s41598-018-37832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loh P-R, Danecek P, Palamara PF, et al. . Reference-based phasing using the haplotype reference Consortium panel. Nat Genet 2016;48:1443–8. 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bycroft C, Freeman C, Petkova D, et al. . The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, et al. . A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim Y-J, Kim B-J, Han B-H. The Korean Reference Genome project: construction of the reference panel for imputation analysis [Abstract]. The 61th Annual Meeting of The American Society of Human Genetics, 2015:P3163F. [Google Scholar]
- 12. O'Connell J, Gurdasani D, Delaneau O, et al. . A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet 2014;10:e1004234. 10.1371/journal.pgen.1004234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Howie B, Fuchsberger C, Stephens M, et al. . Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012;44:955–9. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang HM. Efficient and parallelizable association container toolbox (EPACTS). Available: http://genome.sph.umich.edu/wiki/EPACTS
- 15. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J, Ferreira T, Morris AP, et al. . Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–75. S1-3. 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Leeuw CA, Mooij JM, Heskes T, et al. . MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liberzon A, Subramanian A, Pinchback R, et al. . Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. GTEx Consortium Human genomics. The Genotype-Tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidt EM, Zhang J, Zhou W, et al. . GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 2015;31:2601–6. 10.1093/bioinformatics/btv201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stunnenberg HG, Hirst M, International Human Epigenome Consortium . the International human epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 2016;167:1145–9. 10.1016/j.cell.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 22. Watanabe K, Taskesen E, van Bochoven A, et al. . Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Wijst MGP, Brugge H, de Vries DH, et al. . Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet 2018;50:493–7. 10.1038/s41588-018-0089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmiedel BJ, Singh D, Madrigal A, et al. . Impact of genetic polymorphisms on human immune cell gene expression. Cell 2018;175:e16:1701–15. 10.1016/j.cell.2018.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmitt AD, Hu M, Jung I, et al. . A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 2016;17:2042–59. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lizio M, Abugessaisa I, Noguchi S, et al. . Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res 2019;47:D752–8. 10.1093/nar/gky1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finucane HK, Bulik-Sullivan B, Gusev A, et al. . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–35. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown BC, Ye CJ, Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, et al. . Transethnic Genetic-Correlation estimates from summary statistics. Am J Hum Genet 2016;99:76–88. 10.1016/j.ajhg.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Low S-K, Takahashi A, Ebana Y, et al. . Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet 2017;49:953–8. 10.1038/ng.3842 [DOI] [PubMed] [Google Scholar]
- 31. Ishigaki K, Akiyama M, Kanai M, et al. . Large scale genome-wide association study in a Japanese population identified 45 novel susceptibility loci for 22 diseases. bioRxiv 2019:795948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bentham J, Morris DL, Graham DSC, et al. . Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015;47:1457–64. 10.1038/ng.3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Lu D, Chung Y-J, et al. . Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas 2018;155:19. 10.1186/s41065-018-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y, Fang F, Hu Y, et al. . The polymorphisms in LNK gene correlated to the clinical type of myeloproliferative neoplasms. PLoS One 2016;11:e0154183. 10.1371/journal.pone.0154183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ha J-S, Jeon D-S. Possible new LNK mutations in myeloproliferative neoplasms. Am J Hematol 2011;86:866–8. 10.1002/ajh.22107 [DOI] [PubMed] [Google Scholar]
- 36. Costenbader KH, Karlson EW. Epstein-Barr virus and rheumatoid arthritis: is there a link? Arthritis Res Ther 2006;8:204. 10.1186/ar1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reynisdottir G, Karimi R, Joshua V, et al. . Structural changes and antibody enrichment in the lungs are early features of anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol 2014;66:31–9. 10.1002/art.38201 [DOI] [PubMed] [Google Scholar]
- 38. Nissinen R, Leirisalo-Repo M, Nieminen AM, et al. . Immune activation in the small intestine in patients with rheumatoid arthritis. Ann Rheum Dis 2004;63:1327–30. 10.1136/ard.2003.011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacArthur J, Bowler E, Cerezo M, et al. . The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res 2017;45:D896–901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Génin E. Missing heritability of complex diseases: case solved? Hum Genet 2020;139:103–13. 10.1007/s00439-019-02034-4 [DOI] [PubMed] [Google Scholar]
- 41. Radomir L, Cohen S, Kramer MP, et al. . T cells regulate peripheral naive mature B cell survival by cell-cell contact mediated through SLAMF6 and SAP. J Immunol 2017;199:2745–57. 10.4049/jimmunol.1700557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dragovich MA, Adam K, Strazza M, et al. . SLAMF6 clustering is required to augment T cell activation. PLoS One 2019;14:e0218109. 10.1371/journal.pone.0218109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dragovich MA, Mor A. The SLAM family receptors: potential therapeutic targets for inflammatory and autoimmune diseases. Autoimmun Rev 2018;17:674–82. 10.1016/j.autrev.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savola P, Kelkka T, Rajala HL, et al. . Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat Commun 2017;8:15869. 10.1038/ncomms15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chatterjee M, Rauen T, Kis-Toth K, et al. . Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol 2012;188:1206–12. 10.4049/jimmunol.1102773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi K, Hayashida K, Kaneko M, et al. . Lymphoid chemokine B cell-attracting chemokine-1 (CXCL13) is expressed in germinal center of ectopic lymphoid follicles within the synovium of chronic arthritis patients. J Immunol 2001;166:650–5. 10.4049/jimmunol.166.1.650 [DOI] [PubMed] [Google Scholar]
- 47. Cooper GM, Stone EA, Asimenos G, et al. . Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005;15:901–13. 10.1101/gr.3577405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garber M, Guttman M, Clamp M, et al. . Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 2009;25:i54–62. 10.1093/bioinformatics/btp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meeuwisse CM, van der Linden MP, Rullmann TA, et al. . Identification of CXCL13 as a marker for rheumatoid arthritis outcome using an in silico model of the rheumatic joint. Arthritis Rheum 2011;63:1265–73. 10.1002/art.30273 [DOI] [PubMed] [Google Scholar]
- 50. Takeshita M, Suzuki K, Kondo Y, et al. . Multi-dimensional analysis identified rheumatoid arthritis-driving pathway in human T cell. Ann Rheum Dis 2019;78:1346–56. 10.1136/annrheumdis-2018-214885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baranov MV, Revelo NH, Dingjan I, et al. . SWAP70 organizes the actin cytoskeleton and is essential for phagocytosis. Cell Rep 2016;17:1518–31. 10.1016/j.celrep.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manni M, Gupta S, Ricker E, et al. . Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat Immunol 2018;19:407–19. 10.1038/s41590-018-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quemeneur L, Angeli V, Chopin M, et al. . SWAP-70 deficiency causes high-affinity plasma cell generation despite impaired germinal center formation. Blood 2008;111:2714–24. 10.1182/blood-2007-07-102822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chandrasekaran U, Yi W, Gupta S, et al. . Regulation of effector Treg cells in murine lupus. Arthritis Rheumatol 2016;68:1454–66. 10.1002/art.39599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim K, Jiang X, Cui J, et al. . Interactions between amino acid-defined major histocompatibility complex class II variants and smoking in seropositive rheumatoid arthritis. Arthritis Rheumatol 2015;67:2611–23. 10.1002/art.39228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kochi Y, Thabet MM, Suzuki A, et al. . PADI4 polymorphism predisposes male smokers to rheumatoid arthritis. Ann Rheum Dis 2011;70:512–5. 10.1136/ard.2010.130526 [DOI] [PubMed] [Google Scholar]
- 57. Kilsgård O, Andersson P, Malmsten M, et al. . Peptidylarginine deiminases present in the airways during tobacco smoking and inflammation can citrullinate the host defense peptide LL-37, resulting in altered activities. Am J Respir Cell Mol Biol 2012;46:240–8. 10.1165/rcmb.2010-0500OC [DOI] [PubMed] [Google Scholar]
- 58. Di Giuseppe D, Discacciati A, Orsini N, et al. . Cigarette smoking and risk of rheumatoid arthritis: a dose-response meta-analysis. Arthritis Res Ther 2014;16:R61. 10.1186/ar4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Demoruelle MK, Weisman MH, Simonian PL, et al. . Brief report: airways abnormalities and rheumatoid arthritis-related autoantibodies in subjects without arthritis: early injury or initiating site of autoimmunity? Arthritis Rheum 2012;64:1756–61. 10.1002/art.34344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lim J, Kim K. Genetic variants differentially associated with rheumatoid arthritis and systemic lupus erythematosus reveal the disease-specific biology. Sci Rep 2019;9:2739. 10.1038/s41598-019-39132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kobayashi N, Takahashi D, Takano S, et al. . The roles of Peyer's patches and Microfold cells in the gut immune system: relevance to autoimmune diseases. Front Immunol 2019;10:2345. 10.3389/fimmu.2019.02345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cho H, Jaime H, de Oliveira RP, et al. . Defective IgA response to atypical intestinal commensals in IL-21 receptor deficiency reshapes immune cell homeostasis and mucosal immunity. Mucosal Immunol 2019;12:85–96. 10.1038/s41385-018-0056-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y, Jiang X, Zhu J, et al. . IL-21/IL-21R signaling suppresses intestinal inflammation induced by DSS through regulation of Th responses in lamina propria in mice. Sci Rep 2016;6:31881. 10.1038/srep31881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Teng F, Klinger CN, Felix KM, et al. . Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer's patch T follicular helper cells. Immunity 2016;44:875–88. 10.1016/j.immuni.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Draborg AH, Duus K, Houen G. Epstein-Barr virus in systemic autoimmune diseases. Clin Dev Immunol 2013;2013:535738:1–9. 10.1155/2013/535738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Harley JB, Chen X, Pujato M, et al. . Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet 2018;50:699–707. 10.1038/s41588-018-0102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Román-Fernández IV, García-Chagollán M, Cerpa-Cruz S, et al. . Assessment of CD40 and CD40L expression in rheumatoid arthritis patients, association with clinical features and DAS28. Clin Exp Med 2019;19:427–37. 10.1007/s10238-019-00568-5 [DOI] [PubMed] [Google Scholar]
- 68. Liu R, Wu Q, Su D, et al. . A regulatory effect of IL-21 on T follicular helper-like cell and B cell in rheumatoid arthritis. Arthritis Res Ther 2012;14:R255. 10.1186/ar4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2020-217663supp001.pdf (572.6KB, pdf)