Abstract
Objective
The HBV HBx regulatory protein is required for transcription from the covalently closed circular DNA (cccDNA) minichromosome and affects the epigenetic control of both viral and host cellular chromatin.
Design
We explored, in relevant cellular models of HBV replication, the functional consequences of HBx interaction with DLEU2, a long non-coding RNA (lncRNA) expressed in the liver and increased in human hepatocellular carcinoma (HCC), in the regulation of host target genes and the HBV cccDNA.
Results
We show that HBx binds the promoter region, enhances the transcription and induces the accumulation of DLEU2 in infected hepatocytes. We found that nuclear DLEU2 directly binds HBx and the histone methyltransferase enhancer of zeste homolog 2 (EZH2), the catalytic active subunit of the polycomb repressor complex 2 (PRC2) complex. Computational modelling and biochemical evidence suggest that HBx and EZH2 share two preferential binding sites in DLEU2 intron 1. HBx and DLEU2 co-recruitment on the cccDNA displaces EZH2 from the viral chromatin to boost transcription and viral replication. DLEU2-HBx association with target host promoters relieves EZH2 repression and leads to the transcriptional activation of a subset of EZH2/PRC2 target genes in HBV-infected cells and HBV-related HCCs.
Conclusions
Our results highlight the ability of HBx to bind RNA to impact on the epigenetic control of both viral cccDNA and host genes and provide a new key to understand the role of DLEU2 and EZH2 overexpression in HBV-related HCCs and HBx contribution to hepatocytes transformation.
Keywords: hepatocellular carcinoma, hepatitis B, liver
Significance of this study.
What is already known on this subject?
HBx is recruited to and regulates the activity of a variety of coding genes and non-coding RNA promoters.
Long non-coding RNAs (lncRNAs) are frequently dysregulated in both HBV-related hepatocellular carcinoma (HCC) tissues and HBV-expressing/HBx-expressing cell lines.
LncRNAs can regulate multiple cellular processes by various mechanisms including chromatin remodelling.
A number of lncRNAs has been described to guide chromatin-modifying complexes, including enhancer of zeste homolog 2 (EZH2)/polycomb repressor complex 2 (PRC2).
What are the new findings?
HBx binds the promoter region of the DLEU2 lncRNA, enhances DLEU2 transcription and induces the accumulation of DLEU2 RNA.
HBx directly binds DLEU2 and functionally competes with EZH2 for partially overlapping sites on the DLEU2.
HBx and DLEU2 modulates transcription from the HBV covalently closed circular DNA minichromosome, hence boosting HBV replication, and from host genes promoters.
HBx cooperates with DLEU2 by activating the promoters of a subset of EZH2/PRC2 targets in HBV-replicating cells and in HBV-related HCC.
How might it impact on clinical practice in the foreseeable future?
-
These observations:
provide a new key to understand the role of EZH2 overexpression in HBV-related HCCs and the impact of HBV and HBx in hepatocyte transformation;
identify the interaction between HBx and DLEU2 as a new potential target to both inhibit HBV replication and hepatocyte transformation.
Introduction
Despite the availability of a preventive vaccine and antiviral treatments that arrest disease progression and reduce liver cancer risk, hepatitis B still remains a major global public health problem with about 257 million persons living with chronic HBV infection.1 2 HBV accounts for over 850 000 deaths/year and is responsible for >50% of the hepatocellular carcinomas (HCCs) worldwide.3 Currently, only 10% of chronically infected people are diagnosed and only 1% are treated.1 Hepatitis B surface antigen (HBsAg) loss is viewed as an important goal of therapy,4 but it is achieved only in a minority of patients treated with pegylated interferon alpha2a and nucleos(t)ide analogues (NUCs) and life-long treatments are recommended with NUCs.5 HBsAg loss is not accompanied by viral eradication4 and most patients never clear the HBV covalently closed circular DNA (cccDNA) and bear viral sequences integrated into the host DNA.6 The cccDNA minichromosome is the matrix for transcription of all viral mRNAs and the 3.5 kb pregenomic RNA (pgRNA) that serves as template for viral replication.7 The level of HBV replication is determined by the size of the cccDNA pool (ie, total number of cccDNA molecules) and its transcriptional productivity (ie, the amount of HBV RNAs transcribed from each cccDNA).7 cccDNA transcription is regulated by epigenetic modifications imposed to cccDNA-bound histones8 by viral and cellular proteins as well as inflammatory cytokines.7 HBx plays a key role through two independent mechanisms that cooperate to establish and maintain a transcriptionally active cccDNA minichromosome: (a) the degradation of the Smc5/6 restriction factors mediated by the activation of the DDB1-Cul4 E3 ligase early after infection9 and (b) the prevention of transcriptional repressor recruitment on the cccDNA.10 11 In addition to viral chromatin, HBx is recruited to and regulates a variety of coding genes and non-coding RNA promoters.12
Long non-coding RNAs (lncRNAs) are frequently dysregulated in HBV-related HCC and HBV-expressing/HBx-expressing cells.13 LncRNAs play a crucial role in the modulation of signalling pathways and gene expression.14 Viral infections may induce cellular lncRNAs with antiviral effect15 and viruses can use lncRNAs to regulate metabolic networks promoting viral survival.16 lncRNAs have been described to remodel chromatin14 by guiding chromatin-modifying complexes, including enhancer of zeste homolog 2 (EZH2)/polycomb repressor complex 2 (PRC2), towards their target loci.17 18 EZH2, the major cellular H3K27 trimethyltransferase, catalyses addition of methyl groups at lysine 27 of histone H3 (H3K27me3).19 EZH2, as a subunit of the canonical PRC2, along with EED, SUZ12 and RBBP4,19 is primarily involved in repression of transcription. Multiple DNA-binding proteins, including JARID220 and ATRX,21 contribute to PRC2 complex recruitment to target genes to silence gene expression. EZH2 is overexpressed in many cancers including HCC.22 23 Small molecule EZH2 inhibitors that target its catalytic SET domain are in clinical trials.24 However, EZH2 catalytic activity may not recapitulate all EZH2/PRC2 functions in cancer and EZH2/PRC2 recruitment can be uncoupled from H3K27me3 deposition/spreading.25 EZH2 also acts as a transcriptional co-activator of oestrogen-regulated, WNT-regulated and NOTCH-regulated proliferation pathways in breast cancer.26 27
Here we show, in relevant cellular models of HBV replication, that HBx activates the expression and directly binds the lncRNA DLEU2 to modulate transcription from the HBV cccDNA minichromosome, hence boosting HBV replication, and from host genes promoters. HBx-mediated increase in DLEU2 and HBx recruitment onto target gene regulatory sequences increases chromatin accessibility and activates a subset of EZH2/PRC2 targets in HBV-replicating cells and in HBV-related HCCs. These observations provide a new key to understand the role of DLEU2 and EZH2 overexpression in HBV-related HCCs and the impact of HBx in hepatocyte transformation.
Results
DLEU2 lncRNA is a direct target of HBx upregulated in HBV infection and HBV-related HCC
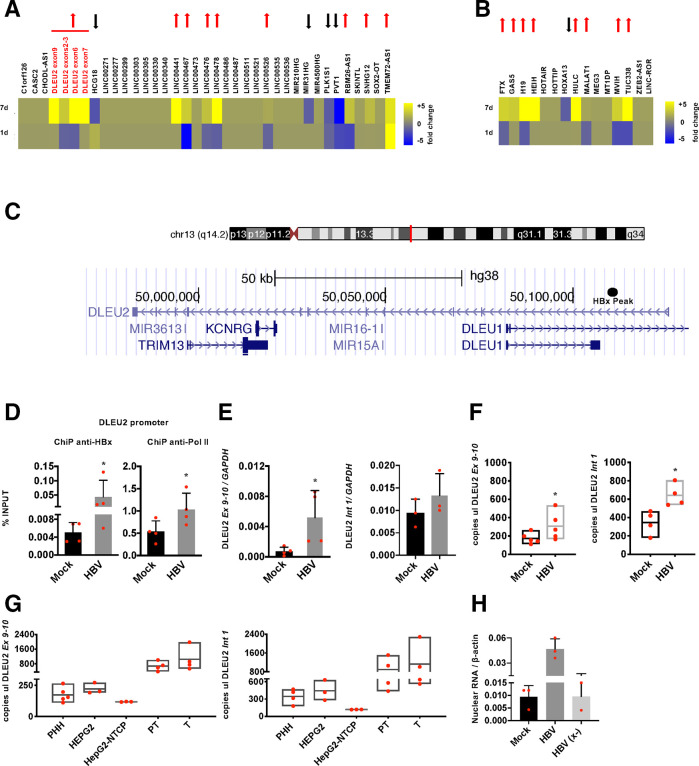
Several lncRNAs have been shown to be deregulated in HBV infection and in HBV-related HCCs.13 28 Recently, using a genome-wide analysis of HBx chromatin recruitment in HBV-replicating cells, we identified 16 lncRNA promoters and 26 lncRNA intragenic regions specifically bound by HBx12 (online supplementary table S1). The expression of the 34 unique lncRNAs that are directly targeted by HBx (figure 1A, online supplementary table S1) and of 15 additional lncRNAs that are known to be modulated in human HCCs (figure 1B) has been evaluated in mock-infected and HBV-infected primary human hepatocytes (PHHs) using a custom Nanostring-GE panel assay. As shown in figures 1A, 9 out of the 34 HBx-targeted lncRNAs were significantly upregulated and 4 were strongly downregulated by HBV at day 7 postinfection. Notably, 8 out of 15 HCC-modulated lncRNAs were strongly upregulated by HBV infection (figure 1B). Among the lncRNAs with the highest levels of induction in HBV-infected PHHs, DLEU2, LINC00441, LINC00478 are direct targets of HBx (ie, positive in the anti-HBx chromatin immunoprecipitation (ChIP)), whereas for HULC, HEIH, H19 and MALAT1 we do not have direct evidence of HBx promoter recruitment.12 We focused on DLEU2, which is differentially expressed during liver maturation29 and overexpressed in solid human cancers30 including HCCs28 31 (online supplementary figures S1a-b).
Figure 1.
HBx modulates DLEU2 expression. (A) Heatmap of the expression levels in HBV-infected primary human hepatocytes (PHHs) of 34 long non-coding RNAs (lncRNAs) that are direct transcriptional targets of HBx12 analysed by Nanostring. Blue and yellow colours in the heatmap indicate downregulated and upregulated lncRNAs, respectively. The red and black arrows identify lncRNAs upregulated >1.5-fold and downregulated <0.7-fold. LncRNA expression in HBV-infected samples is normalised to GAPDH and to mock condition. (B) Expression levels of 15 hepatocellular carcinoma (HCC)-associated lncRNAs, in HBV-infected PHHs. Nanostring analysis, data normalisation and heatmap representation as shown in figure 1A. (C) DLEU2 lncRNA genomic sequence, according to Genome Browser; the HBx peak, obtained by HBx chromatin immunoprecipitation sequencing (ChIP-Seq) searching, is indicated as black circle. (D) HBx and polymerase II (Pol II) occupancy on DLEU2 promoter region. Cross-linked chromatin from mock or HBV-infected PHHs (72 hours) was immunoprecipitated with anti-HBx or anti-Pol II antibodies, and then analysed by real-time quantitative PCR (qPCR) using specific DLEU2 promoter primer pairs. The detection of HBV covalently closed circular DNA (cccDNA) using specific primers in the ChIPed DNA from HBV-infected cells (cccDNA-ChIP) served as a technical positive control for the ChIP procedure. ChIP results are expressed as % of input. (E) Real-time qPCR of DLEU2 RNA (left panel: exons 9–10; right panel: intron 1) in mock-infected and HBV-infected PHHs (7 days). Results are expressed as relative values to endogenous human GAPDH mRNAs. (F) Droplet Digital PCR (ddPCR) quantification of DLEU2 RNA (left panel: exons 9–10; right panel: intron 1) from mock-infected and HBV-infected PHHs (7 days). Results are expressed as copies for 5 ng/μL of total RNA. (G) ddPCR quantification of DLEU2 RNA (left panel: exons 9–10; right panel: intron 1) from PHHs, HepG2 or HepG2-NTCP cells and from HBV-infected peritumour (PT) or tumour (T) tissue. Results are expressed as copies for 5 ng/μL of total RNA. (H) Real-time qPCR of DLEU2 RNA (intron 1) in nuclear lysates from of mock, HBV-infected and HBV (x-)-infected PHHs. Results are expressed as relative values to endogenous human β-actin mRNAs. (7 days). Data in panels (D), (E) and (F) represent means±SD from at least three independent experiments. In (D), (E) and (F), *p<0.05; Mann-Whitney U test.
gutjnl-2019-319637supp001.pdf (5.6MB, pdf)
DLEU2 gene extends for 142 990 bp in Ch13(q14.2) and overlaps with the miR-15a/miR-16-1 cluster, the TRIM13 gene and, partially, with the DLEU1 gene (figure 1C). The HBx peak detected by HBx chromatin immunoprecipitation sequencing (ChIP-Seq)12 (figure 1C, black circle) is located ~6500 bp downstream the DLEU2 promoter previously described.32 HBx binding on the HBx peak region was confirmed in independent ChIP assays performed in HBV-infected PHHs (figure 1D, left panel) and was accompanied by an increase in polymerase II (Pol II) binding (figure 1D, right panel) and DLEU2 steady state RNA levels (figure 1E–F). HBx recruitment on the DLEU2 promoter and increased DLEU2 expression were confirmed in other HBV infection/replication systems (online supplementary figure S2a-c). Figure 1G shows baseline levels of DLEU2 quantified by ddPCR in PHHs, HepG2 and HepG2-NTCP cells as well as in HBV-related HCCs and their paired HBV-infected non-tumorous liver tissues. The increase in DLEU2 RNA containing intron 1 sequences (figure 1E–G, right panels) and the accumulation of DLEU2 in the nuclear fraction of HBV-infected PHHs (figure 1H) as well as in HBV-replicating (online supplementary figures S2d) and HBx-HA-transfected HepG2 cells (online supplementary figure S2e), all supports the notion that DLEU2 induction by HBV occurs at the transcriptional level. The increase of DLEU2 levels after exogenous HBx expression in HepG2 cells (online supplementary figure S2e) and the loss of DLEU2 induction in PHHs infected with the HBx-deficient HBV (x-) virus (figure 1H) confirm the key role of HBx in increasing DLEU2 levels. Finally, HBx recruitment on the HBx peak region has no impact on the levels of the antisense DLEU1 lncRNA (online supplementary figure S2f).
DLEU2 lncRNA physically interacts with EZH2 and HBx
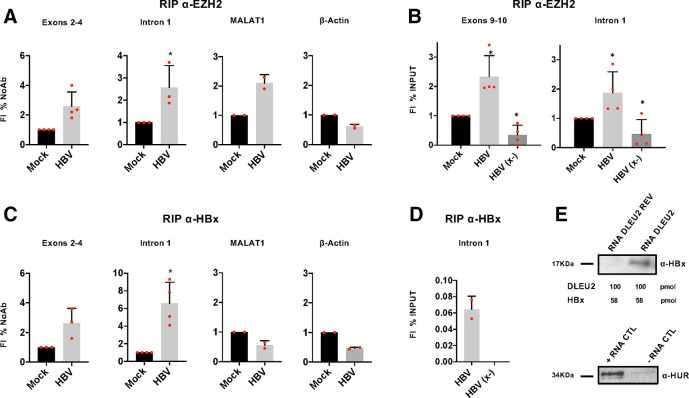
LncRNAs have been shown to guide chromatin-modifying complexes towards their chromatin targets and to regulate gene expression.14 33 Multiple lncRNAs interact in vivo with the EZH2/PRC2 complex in embryonic stem cells (ESCs).17 In EZH2 RIP-Seq performed in colon cancer cells,18 DLEU2 is highly enriched, together with the lncRNAs XIST, TSIX, MALAT1 and GAS5. Since HBx has been reported to modulate EZH2/PRC2 host target gene transcription,34 35 we sought to further investigate whether DLEU2 interacts with EZH2 and HBx in PHHs and HCC cell lines. As shown in figure 2A, we found that DLEU2 and EZH2 interact in PHHs. As compared with mock-infected cells, HBV-infected PHHs displayed a significant enrichment of DLEU2 in anti-EZH2 nuclear RIP (figure 2A). MALAT1, a lncRNA known to bind EZH2,14 served as positive control (figure 2A). The increased association of EZH2 with DLEU2 in HBV-infected PHHs and not in HBx-defective HBV (x-)-infected PHHs (figure 2B) is consistent with the HBx-dependent induction of DLEU2 in HBV-infected hepatocytes. Next, we performed anti-HBx RIPs in HBV-infected PHHs (figure 2C) and HepG2 cells transfected with wild-type (wt) monomeric HBV genomes (online supplementary figure S3a). We found that HBx binds DLEU2 in HBV-infected/HBV-replicating hepatocytes (figure 2C and online supplementary figure S3a). No enrichment of MALAT1 binding to HBx was observed (figure 2C and online supplementary figure S3a). As expected, no DLEU2 can be immunoprecipitated with anti-HBx antibodies in HBx-defective HBV (x-)-infected PHHs (figure 2D). These results indicate that HBx and DLEU2 interact in the nuclei of HBV-infected cells but do not exclude that this association might be mediated by additional partners. To ascertain whether HBx and DLEU2 directly interact, we performed RNA pull-down experiments using recombinant HBx and synthetic DLEU2 species designed in intron 1 sequence. Desthiobiotinylated DLEU2 60-mer oligonucleotides were able to pull down recombinant HBx in contrast to biotinylated reverse DLEU2 control oligonucleotides (figure 2E, upper panel). The specificity of the pulled down recombinant HBx band was confirmed by immunoblot (online supplementary figure S3b) and the same antibody immunoprecipitated a 17Kd HBx band that was recognised specifically by two anti-HBx antibodies in HBV-infected HepG2-NTCP cells (online supplementary figure S3c). Altogether, these results indicate that DLEU2 binds EZH2 in liver cells and, in HBV-infected cells, the viral protein HBx.
Figure 2.
DLEU2 long non-coding RNA (lncRNA) interacts with enhancer of zeste homolog 2 (EZH2) and directly binds HBx. (A) RNA binding protein immunoprecipitation (RIP) using anti-EZH2 antibody. Nuclear lysates of mock-infected and HBV-infected primary human hepatocytes (PHHs) (72 hours) were subjected to immunoprecipitation with anti-EZH2 antibody or control IgG. DLEU2 exons 2–4 or intron 1, MALAT1 (positive control) and β-actin (negative control) were detected by PCR and analysed by relative densitometry. (B) RIP using anti-EZH2 antibody. Nuclear lysates of mock, HBV-infected and HBV (x-)-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-EZH2 antibody or control IgG. DLEU2 exons 9–10 or intron 1 were detected by quantitative PCR (qPCR) assay. RIP results are expressed as fold induction (FI) of the % of input with respect to mock. (C) RIP using anti-HBx antibody. Nuclear lysate of mock-infected and HBV-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-HBx antibody or control IgG. DLEU2 exons 2–4 or intron 1, MALAT1 and β-actin (negative controls) were detected by PCR assay and analysed by densitometry relative to the mock. RIP data in (A) and (B) represent means±SD from at least three independent experiments. (D) RIP using anti-HBx antibody. Nuclear lysate of mock, HBV-infected and HBV (x-)-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-HBx antibody or control IgG. Intron 1 was detected by real-time qPCR assay. RIP results are expressed as % of input after subtraction of mock. (E) Immunoblot analysis of RNA pull-downs showing the interaction between the DLEU2 RNA (100 pmol of 3’-desthiobiotinylated RNA oligonucleotide) and recombinant HBx (1 µg), using a DLEU2 reverse sequence (DLEU2 REV) RNA as negative control (upper panel). Immunoblot analysis of positive and negative controls (provided by the manufacturer), which respectively contain RNA regions with or without binding sites for HUR, a RNA-binding proteins that regulates mRNA stability (lower panel). Data from one representative experiment out of three independent experiments are shown. In panels (A) and (B), *p<0.05; Mann-Whitney U test.
Modelling DLEU2-EZH2-HBx interaction in silico
Our experimental data do not allow to conclude whether HBx and EZH2 compete for DLEU2 binding or whether HBx, EZH2 and DLEU2 may form a tripartite complex. We modelled via structural bioinformatics tools the binary DLEU2-EZH2 and DLEU2-HBx as well as the ternary DLEU2-HBx-EZH2 protein-RNA complexes. The CatRapid sequence-based method36 and three-dimensional (3D) atomistic structures generated using the HEX and HADDOCK tools37 38 were used to model the physical interactions between proteins and DLEU2. Methodological details and challenges related to atomistic structure prediction are discussed in online supplementary materials.
In contrast to EZH2 for which a 3D structure is known,39 the 3D experimental structures of HBx and DLEU2 remain elusive. Using a homology modelling approach, we confirmed that HBx comprises disordered regions,40 essential for the interaction with protein and nucleic acid partners,41 as well as more structured domains (online supplementary figure S4). Using RNABindRplus,42 we found that the HBx residues predicted to bind RNA mainly comprise charged amino acids as arginines and lysines that are mostly located in the disordered region corresponding to the first half of the sequence (online supplementary figure S4a). DLEU2 analysis by CatRapid36 corroborated the interaction of intron 1 of DLEU2 with HBx and EZH2, whereas in line with the RIP experiments (figure 2C), our analysis does not favour a direct interaction between HBx and MALAT1.
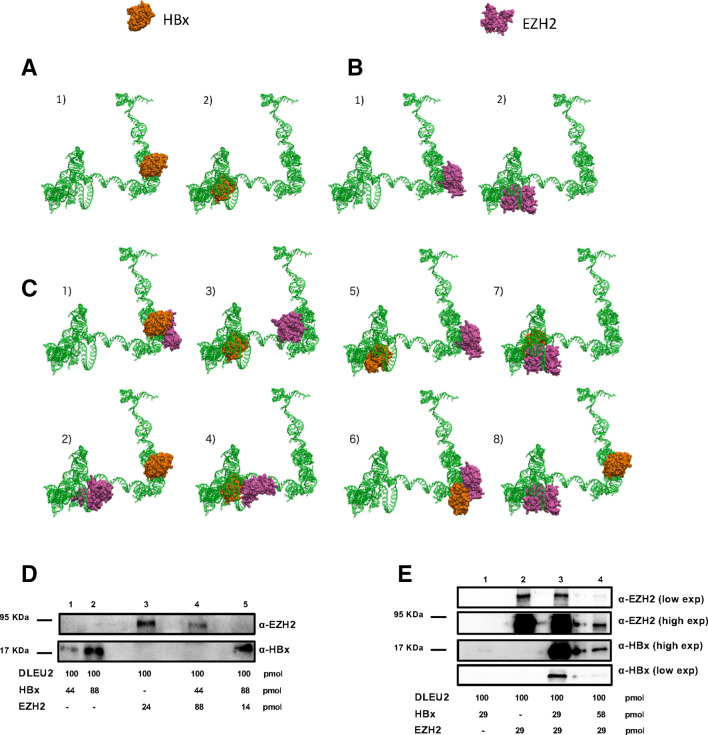
Next, we modelled the secondary (online supplementary figure S5) and 3D structure (figure 3) of the DLEU2 intron 1 region encompassing the sequences that interact with HBx in the RNA pull-down experiments (figure 2C) and exploited the HEX and HADDOCK biomolecular docking methods37 38 43 to model the binary and ternary interactions of DLEU2 with EZH2 and HBx. We found that two loci on intron 1 display a high probability to be occupied by HBx (figure 3A). Interestingly, HBx preferentially binds to internal loop motifs predicted in the secondary structure of DLEU2 intron 1 (online supplementary figure S5). Using the same biomolecular docking calculations, we confirmed the well-known interaction between HBx and DDB1 (PDB entry: 3I7H44) and the docking results for HBx-DDBI and HBx-DLEU2 point for similar interaction strength. We identified two loci on intron 1, close but not completely overlapping with the regions potentially targeted by HBx that display a nearly equal probability to be occupied by EZH2 (figure 3B). When EZH2 is already docked to DLEU2 intron 1, HBx is still predicted to bind DLEU2 intron 1 but it is docked either to the non-occupied EZH2 site (figure 3C, panels 5 and 8) or to a docking site that is partially overlapping with the one occupied by EZH2 (figure 3C, panels 6 and 7). Notably, in both cases the presence of EZH2 slightly alters HBx docking (compare HBx docking in figure 3C, panels 5 and 7, with HBx docking in figure 3A, panel 2; HBx docking in figure 3C, panels 6 and 8, with HBx docking in figure 3A, panel 1). Similar results were obtained when HBx is docked first on DLEU2 intron 1 (compare EZH2 docking in figure 3B vs figure 3C).
Figure 3.
HBx and enhancer of zeste homolog 2 (EZH2) directly bind DLEU2 and compete for partially overlapping sites. (A) Configurations obtained from docking of HBx (surface representation, orange) on three-dimensional (3D) structure of DLEU2 intron 1 (green). Panels 1–2 present the two most probable loci for HBx adsorption on DLEU2 intron 1. (B) Configurations obtained from docking of EZH2 (surface representation, mauve) on 3D structure of DLEU2 intron 1 (green). Panels 1–2 present the two most probable loci for EZH2 adsorption on DLEU2 intron 1. (C) Panels 1–2: two most probable configurations obtained from docking of EZH2 on the most probable HBx-DLEU2 intron 1 complex (panel (A)—1). Panels 3–4: two most probable configurations obtained from docking of EZH2 on the second most probable HBx-DLEU2 intron 1 complex (panel (A)—2). Panels 5–6: two most probable configurations obtained from docking of HBx on the most probable EZH2-DLEU2 intron 1 complex (panel (B)—1). Panels 7–8: two most probable configurations obtained from docking of HBx on the second most probable EZH2-DLEU2 intron 1 complex (panel (B)—2). (D) Immunoblot analysis of RNA pull-downs showing the interaction between DLEU2 RNA, HBx and EZH2. HBx (0.75 µg=44 pmol or 1.5 µg=88 pmol) or EZH2 (1.2 µg=14 pmol, 2 µg=24 pmol or 7.5 µg=88 pmol) recombinant proteins alone or in combination (EZH2:HBx ratios of 2:1 or 1:6) were added to a fixed amount of DLEU2 RNA (100 pmol). (E) Immunoblot analysis of RNA pull-downs showing the interaction between DLEU2 RNA (100 pmol), HBx and EZH2. Increasing concentrations of HBx (0.5 µg=29 pmol, 1 µg = 58 pmol) were added at a fixed concentration of EZH2 (2.5 µg=29 pmol) recombinant proteins corresponding to EZH2:HBx ratios of 1:1 and 1:2, respectively. Data from one representative experiment out of three independent experiments are shown.
Our in silico modelling is compatible with the formation of a ternary DLEU2-EZH2-HBx complex and, depending on the relative abundance of EZH2 and HBx, with a competition for partially overlapping or close by sites that may impact on EZH2/PRC2 complex functions. To verify this hypothesis, we performed DLEU2 RNA pull-down experiments with different stoichiometric ratios of HBx and EZH2.
As shown in figure 3D, both EZH2 (lane 3) and HBx (lanes 1–2) can individually bind directly DLEU2 and HBx binding is dose-dependent (lanes 1–2). At a higher EZH2:HBx ratio (2:1), EZH2 binds DLEU2 and prevents its interaction with HBx (lane 4). Conversely, at higher HBx:EZH2 ratios (6:1), HBx binding to DLEU2 precludes EZH2 binding (lane 5). In the presence of equimolar amounts of EZH2 and HBx, we observe a strong cooperative effect on HBx binding (figure 3E, lanes 1 and 3; online supplementary figure S3d), whereas EZH2 binding does not change (figure 3E, lanes 2–3). Thus, EZH2 seems to favour HBx binding to DLEU2, whereas the binding of EZH2 does not appear to be influenced by HBx. These findings can be explained if EZH2 facilitates HBx binding to DLEU2 sites that are not bound when HBx is alone. Notably, when the amount of HBx increases, EZH2 binding to DLEU2 decreases (compare figure 3E, lane 4 (HBx:EZH2 ratio 2:1) with lane 3 (equimolar amounts of HBx and EZH2) or lane 2 (EZH2 alone)). The binding of HBx in figure 3E, lane 4 (58 pmol) is higher than in lane 1 (29 pmol) but lower than in lane 3 (HBx 29 pmol and EZH2 29 pmol). The lower binding of HBx to DLEU2 observed in lane 4 as compared with lane 3 is likely the result of the displacement of EZH2 and the consequent loss of its ability to facilitate HBx binding on alternative binding sites. Altogether, the in silico modelling and the results of the RNA pull-down experiments reveal a highly dynamic interaction between EZH2 and HBx for DLEU2 binding that leads, depending on their relative abundance, to a functional competition between HBx and EZH2 for partially overlapping sites on DLEU2.
HBx-DLEU2 interaction regulates cccDNA transcription and HBV replication
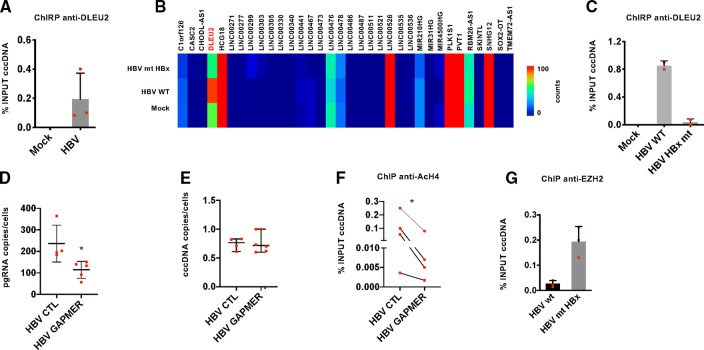
HBx is required for the transcripion of HBV mRNAs from the cccDNA, including the HBV pgRNA that serves as template for reverse transcription and HBV replication.9 10 45 Since DLEU2 interacts with HBx and EZH2, we next sought to assess whether DLEU2, and its interaction with HBx, contributes to cccDNA transcription. We first performed cccDNA chromatin isolation by RNA purification (ChIRP) experiments in HBV-replicating cells, using three independent antisense probe sets that specifically recover DLEU2 intronic sequences, to ascertain the binding of DLEU2 to the cccDNA (figure 4A). Notably, in HBV HBx mt-transfected HepG2 cells DLEU2 expression is reduced (figure 4B) and DLEU2 binding to the cccDNA is abrogated (figure 4C). DLEU2 depletion by specific locked nucleic acid (LNA) longRNA Gapmers (online supplementary figure S6a) resulted in a reduction of both HBV pgRNA levels (figure 4D) and total HBV DNA (data not shown) in HBV-infected PHHs, whereas cccDNA levels are not affected (figure 4E). According to these results, DLEU2 interaction with HBx and its recruitment on the viral minichromosome affect cccDNA transcription and viral replication but do not interfere with the establishment of the cccDNA pool. Similar results were obtained in both HBV-transfected HepG2 cells and HBV-infected HepG2-NTCP cells (online supplementary figure S6b). Since EZH2 is recruited onto the cccDNA in HBV-infected HepG2-NTCP cells,46 we sought to investigate whether the ability of DLEU2 to interact with EZH2 (figure 2A) could be relevant in the regulation of cccDNA transcription. Although in the absence of HBx, the K9 methyltransferase SETDB111 and the arginine protein methyltransferase PRMT547 seem to play a predominant role in cccDNA transcriptional repression, EZH2-mediated K27 methylation of cccDNA-bound H3 also increases11 and EZH2 silencing results in increased of HBsAg production.47 Notably, in HBV HBx mt-transfected HepG2 cells the recruitment of EZH2 onto the cccDNA is significantly increased (figure 4F). These results and the reduction of cccDNA-bound H4 histone acetylation in the presence of DLEU2-targeting Gapmers (figure 4G) implicate DLEU2 in the epigenetic control of cccDNA transcription and in HBV replication.
Figure 4.
DLEU2 controls HBV replication. (A) DLEU2-qChIRP (chromatin isolation by RNA purification) for covalently closed circular DNA (cccDNA) using a DLEU2-specific antisense biotinylated DNA probe (100 pmol) in mock-replicating and HBV-replicating HepG2 cells (48 hours). Affinity-purified material was analysed by real-time quantitative PCR (qPCR) with DLEU2 RNA-specific primer pairs and the results are expressed as % of input. (B) Heatmap of the expression levels of 34 long non-coding RNAs (lncRNAs) that are direct transcriptional targets of HBx analysed by Nanostring in HepG2 cells replicating wild-type HBV (HBV wt) or HBV with mutant HBx (HBV mt HBx) (48 hours). Colours in the heatmap indicate log2 counts normalised to GAPDH. (C) DLEU2-qChIRP for cccDNA using a DLEU2-specific antisense biotinylated DNA probe (100 pmol) in HBV or HBV HBx mt replicating HepG2 cells (48 hours). Affinity-purified material was analysed as in 4A. Results are expressed as % of input. (D) ddPCR quantification of HBV pregenomic RNA (pgRNA) (copies/cell) from HBV-infected primary human hepatocytes (PHHs) (4 days) transfected with scrambled Gapmer (CTL) or DLEU2-specific Gapmer pools. Results are expressed as copies for 10 ng of total RNA after normalisation to endogenous human β-globin. (E) ddPCR quantification of HBV cccDNA (copies/cell) from HBV-infected PHHs (4 days) transfected with scrambled Gapmer (CTL) or DLEU2-specific Gapmer pools. Results are expressed as copies for 10 ng of total RNA after normalisation to endogenous human β-globin. (F) cccDNA-bound histone H4 acetylation. Cross-linked chromatin from HBV-replicating HepG2 cells transfected with scrambled Gapmer (CTL) or DLEU2-specific Gapmer pools (48 hours) was subjected to immunoprecipitation with anti-AcH4 antibody or IgG controls, and then analysed by real-time qPCR using cccDNA-specific primer pairs. Chromatin immunoprecipitation (ChIP) results are expressed as % of input. (G) Enhancer of zeste homolog 2 (EZH2) occupancy on cccDNA in HBV wt-replicating or HBV mt HBx-replicating HepG2 cells (48 hours). ChIP results are expressed as % of input. Data in panels (A), (B), (C), (D) and (F) represent means±SD from at least three independent experiments. In (D), and (F), *p<0.05; Mann-Whitney U test.
HBx and DLEU2 coregulation of host genes
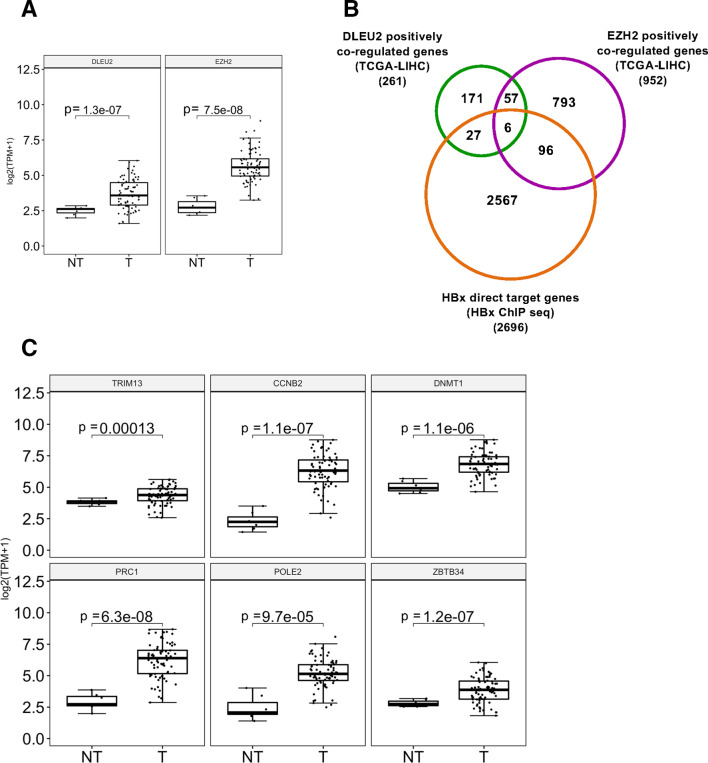
In view of the ability of HBx to complex with DLEU2 and EZH2, which are both overexpressed in tumour liver tissues of patients with HBV-related HCC (figure 5A), we sought to determine whether DLEU2 and EZH2 might regulate a shared set of genes and to evaluate the involvement of HBx in this process. To this aim, we extracted the genes whose expression is positively correlated with DLEU2 or EZH2 in 76 HBV-related HCCs from the transcriptomic TCGA_LIHC dataset and intersected them with the genes identified as HBx direct targets by Chip-Seq.12 As shown in figures 5B, 261 genes were coregulated with DLEU2 in HBV-related HCCs (online supplementary table S2), with a significant enrichment in pathways associated with Gene expression and cell cycle, DNA damage response and mRNA splicing (online supplementary table S3). The intersection of DLEU2 coregulated genes and HBx ChIP data identified as HBx direct targets 33 genes (figure 5B) that enrich the same pathways with a significant propensity for splicing (online supplementary table S4). In the same patients, among the 952 genes that were coregulated with EZH2, 102 genes are direct HBx targets and 63 genes were coregulated with DLEU2. Notably, the 63 genes coregulated by both EZH2 and DLEU2 and the 102 HBx target genes that are coregulated with EZH2 enrich several pathways associated with DNA replication, cell cycle and mitosis (online supplementary tables S5-S6). Six genes, namely TRIM13, CCNB2, DNMT1, PRC1, POLE2 and ZBTB34 (online supplementary table S7), are coregulated by HBx, DLEU2 and EZH2 (figure 5B). Notably, they are all upregulated in tumour liver tissues of patients with HBV-related HCC (figure 5C) and, with the exception of TRIM13, already reported to act as tumour suppressors in breast cancer cell lines and to be downregulated in aggressive breast cancer,48 49 associated with a reduced overall survival (online supplementary figure S7).
Figure 5.
DLEU2 and enhancer of zeste homolog 2 (EZH2) coregulated HBx direct target gene expression in human HBV-related hepatocellular carcinoma (HCC). (A) DLEU2 and EZH2 expression data from TCGA in non-tumour (NT) and in tumour human liver tissues (T) from 76 HBV-related patients with HCC. (B) Venn diagram showing the intersection of genes whose expression is positively correlated with DLEU2 or EZH2 in 76 HBV-related HCCs from the transcriptomic TCGA-LIHC dataset accessed via the R2 platform (https://r2.amc.nl) with the genes identified as HBx direct targets by chromatin immunoprecipitation sequencing (ChIP-Seq). (C) TRIM13, CCNB2, DNMT1, PRC1, POLE2 and ZBTB34 mRNA levels in NT and T human liver tissues from HBV-related patients with HCC (transcriptomic data from TCGA, HBV-related HCCs=76).
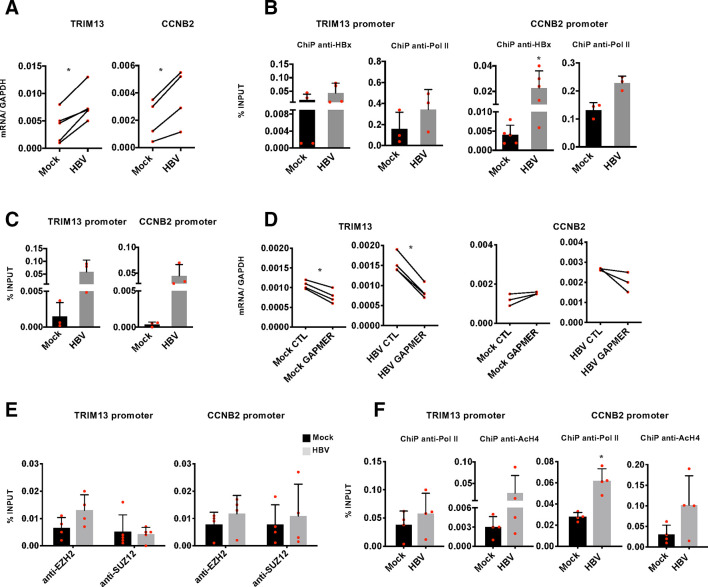
LncRNAs can regulate the expression of both neighbouring genes in cis and distantly located genes in trans.14 To gain mechanistic insights on how DLEU2 and HBx cooperate in the transcriptional regulation of host genes, and on the role of EZH2 in this process, we focused on TRIM13 and CCNB2, as examples of genes coregulated by EZH2 and DLEU2 and targeted by DLEU2 in cis and in trans, respectively. TRIM13 and CCNB2 expression is increased in HBV-infected PHHs (figure 6A) and other cell-based HBV infection/replication models (online supplementary figure S8a). The recruitment of HBx on the TRIM13 and CCNB2 promoters (figure 6B) in HBV-infected PHHs is accompanied by an increase in promoter-bound Pol II (figure 6B) and H4 acetylation (online supplementary figure S8b). Next, we investigated DLEU2 involvement in the regulation of TRIM13 and CCNB2 genes. ChIRP experiments showed that DLEU2 is recruited onto the TRIM13 and CCNB2 promoter regions in HBV-negative cells (figure 6C) and, consistent with the increased DLEU2 expression in HBV-infected cells (figure 1E–F and online supplementary figure S2b), its binding is further enriched in HBV-replicating cells (figure 6C). Selective degradation of DLEU2 by Gapmers (online supplementary figure S6a) resulted in decreased TRIM13 and CCNB2 expression in mock-infected and HBV-infected PHHs (figure 6D) and HepG2-NTCP cells (online supplementary figure S8c) as well as in wt HBV-replicating HepG2 cells (online supplementary figure S8d). Notably, TRIM13 transcription is not induced in HBx mt HBV-replicating HepG2 cells and is not affected by DLEU2-specific Gapmers (online supplementary figure S8d, left panel). In HBV-replicating HepG2 cells co-transfected with HBV monomers and DLEU2-specific Gapmers, where the degradation of DLEU2 is more efficient (online supplementary figure S6a), we also showed a sharp decrease of H4 acetylation at the TRIM13 promoter (online supplementary figure S8e). Collectively, these results indicate that DLEU2 and HBx cooperate to increase TRIM13 and CCNB2 expression. Next, we investigated whether the increased recruitment of DLEU2 on the TRIM2 and CCN2 promoters (figure 6C) and their transcriptional activation is accompanied by changes in EZH2/PRC2 occupancy. As shown in figure 6E, both EZH2 and SUZ12 bind TRIM13 and CCNB2 promoters in uninfected cells and their chromatin occupancy on both promoters remains stable or even increases in HBV-replicating HepG2-NTCP cells (figure 6E) and is accompanied by an increase of Pol II occupancy and H4 acetylation (figure 6F). The protein levels of PRC2 complex components in uninfected and HBV-infected PHHs and HepG2-NTCP cells are shown in online supplementary figure S9. Finally, in HBV-replicating HepG2 cells both EZH2 chromatin occupancy and the repressive mark H3me3K27 decrease at the TRIM13 promoter, whereas in the absence of HBx both EZH2 binding and H3me3K27 increase (online supplementary figure S8f).
Figure 6.
DLEU2 and HBx cooperate to activate TRIM13 and CCNB2 transcription. (A) Real-time quantitative PCR (qPCR) of TRIM13 and CCNB2 mRNA in mock-infected and HBV-infected primary human hepatocytes (PHHs) (7 days). Results are expressed as relative values to endogenous human GAPDH mRNAs. (B) HBx occupancy and modulation of polymerase II (Pol II) binding on TRIM13 and CCNB2 promoters in mock-infected and HBV-infected PHHs (72 hours) by chromatin immunoprecipitation (ChIP) assay. The detection of HBV covalently closed circular DNA (cccDNA) using specific primers in the ChIPed DNA from HBV-infected cells (cccDNA-ChIP) served as a technical positive control for the ChIP procedure. ChIP results are expressed as % of input. (C) DLEU2-ChIRP for TRIM13 promoter or CCNB2 promoter using DLEU2-specific antisense DNA probe in mock-replicating and HBV-replicating HepG2 cells (48 hours). (D) Real-time qPCR of TRIM13 and CCNB2 cDNA in mock-infected or HBV-infected PHHs transfected (4 days) with scrambled Gapmer (CTL) or with DLEU2 Gapmer (GAPMER). Results are expressed as relative values to endogenous human GAPDH mRNAs. (E) Enhancer of zeste homolog 2 (EZH2) and SUZ12 occupancy on TRIM13 and CCNB2 promoter regions by ChIP assay in mock-infected and HBV-infected HepG2-NTCP cells (7 days). Anti-CTCF ChIPs were included as a technical positive control. Results are expressed as % of input. (F) Modulation of H4 acetylation and Pol II binding on the TRIM13 and CCNB2 promoters in HBV-infected HepG2-NTCP cells (7 days). Results are expressed as % of input. Data in panels (A) to (F) represent means±SD from at least three independent experiments. In (A), (B), (D) and (F), *p<0.05, Mann-Whitney U test.
Altogether, these results suggest that HBx and DLEU2 cooperate to activate transcription from a subset of cellular genes, including genes that are negatively regulated by EZH2/PRC2 in non-infected cells and activated in HBV-infected cells as well as HBV-related HCCs.
Conclusions
HBx, whose primary role in the HBV life cycle is to bind the nuclear cccDNA minichromosome in order to initiate and maintain viral transcription and drive HBV replication,10 11 45 also binds a large number of target sequences on the cellular genome.12 HBx interacts with multiple transcription factors, coregulators and chromatin modifying complexes12 50 in order to modulate the transcription of host genes. Here, we show that HBx binds the promoter region of DLEU2, enhances DLEU2 transcription and induces the accumulation of DLEU2 RNA species in infected hepatocytes. We showed by RIP experiments that DLEU2 binds both EZH2, the catalytic subunit of the PRC2 chromatin complex, in liver cells and HBx in HBV-infected cells. These results highlight a novel capacity of HBx to bind ncRNAs and prompted us to investigate the role of these interactions in the transcriptional regulation of cccDNA and HBx host target genes. cccDNA ChIRP experiments showed that DLEU2 binds to the cccDNA and functional experiments with DLEU2-specific Gapmers supported the role of DLEU2 recruitment in the regulation of cccDNA transcription. HBx interaction with DLEU2 on the cccDNA may represent a novel therapeutic target to silence cccDNA transcription and foster a functional cure of HBV infection.51
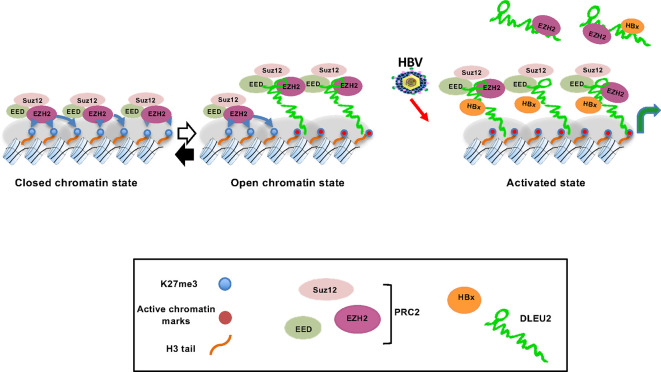
To gain mechanistic insights on how DLEU2 and HBx cooperate in the transcriptional regulation of host genes, and the role of the EZH2/PRC2 complex in this process, we focused on TRIM13 and CCNB2 as examples of genes targeted by DLEU2 in cis and in trans, respectively. Notably, TRIM13 and CCNB2 are part of a subset of six genes coregulated with DLEU2 or EZH2 in HBV-related patients with HCC and identified as direct HBx targets in ChIP-Seq experiments.12 The contribution of PRC2-mediated deposition of the H3K27me3 repressive chromatin mark to the epigenetic silencing during embryonic development and differentiation19 and the impact of its dysregulation in cancer22 are well established, although EZH2-PRC2-independent and PRC2-dependent transcriptional activation have been described.26 27 35 52 53 PRC2 complex binding to coding and non-coding RNAs is also well documented,17 18 but there is no unified view on how PRC2 interacts with chromatin-bound RNAs and the functional consequences of this interaction. Current models, which are not necessarily mutually exclusive, of how PRC2 binding to chromatin-bound RNAs impact on transcription include: a) PRC2 recruitment to its target sites by lncRNAs17 18; b) RNAs serving as decoys to prevent PRC2 binding to active genes or to evict PRC214 54–56; c) RNA binding to an allosteric regulatory site on EZH2 to inhibit the enzymatic activity of PRC2.56 Notably, PRC2 binding to RNA and association with chromatin appear to be antagonistic.54 55 57 By combining ChIRP and ChIP experiments, we showed that DLEU2 and HBx cooperate to increase TRIM13 and CNNB2 expression without changing EZH2 and SUZ12 occupancy on their promoters. Thus, HBx by increasing DLEU2 levels and by binding to DLEU2 interferes with EZH2/PRC2 repressive functions and activates transcription. Altogether, our results are consistent with a model in which HBx interaction with DLEU2 either evicts EZH2, a modality prevalent on cccDNA, or displaces the PRC2/EZH2 complex from close contact with chromatin in the case of host genes54–57 (figure 7).
Figure 7.
A model for DLEU2-PRC2/EZH2-HBx interaction. Left panel: PRC2-mediated epigenetic silencing of target genes; middle panel: DLEU2 recruitment displaces PRC2/EZH2 from close contact with target chromatin, resulting in a reduction of the methyl groups at lysine 27 of histone H3 (H3K27me3) repressive mark; right panel: in the presence of HBx (HBV-infected hepatocytes and HBV-related HCCs expressing HBx), the increase in DLEU2 levels and DLEU2 interaction with PRC2/EZH2 lead to a progressive increase of active chromatin marks associated with active transcription. HBx is part of the DLEU2-PRC2/EZH2 complex and recruited at the chromatin level together with transcription factors and co-activators to contribute to transcriptional activation (not shown in the cartoon). EZH2, enhancer of zeste homolog 2; PRC2, polycomb repressor complex 2.
The activation of DLEU2 by HBx and the impact of its direct interaction with DLEU2 on host target genes may have important consequences on viral pathogenesis and HCC development. It is worth underlining that the genes positively coregulated with DLEU2 or EZH2 in HBV-related HCCs are enriched for pathways associated to cell cycle, DNA damage response and mRNA splicing. The intersection of the transcriptomic data with HBx ChIP-data12 identified six genes coregulated by HBx, EZH2 and DLEU2, all overexpressed in HBV-related HCCs and, with one exception, associated with a reduced overall survival. Collectively, our data and these observations suggest that DLEU2 and HBx may coregulate a subset of target genes in HBV-infected cells and HCC. The ability of HBx to bind DLEU2 impacts on the epigenetic control of host genes and provide a new key to understand the role of HBx, and the overexpression of DLEU2 and EZH2 in HBV-related HCCs.
Experimental procedures
Materials and methods are described in the online supplementary information. Primer pairs and probes used throughout the manuscript are listed in online supplementary table 8.
Acknowledgments
The authors would like to thank Professor J Faivre, Dr M Ballarino and Professor F Zoulim for fruitful discussions. The authors would like to thank M. Rivoire for providing liver samples. For experimental assistance, the authors would like to thank Dr C. Caron de Fromentel, Dr L Calvo (Nanostring), Dr A D’Alfonso (RNA extraction).
Footnotes
Contributors: DS and LC share co-first authorship. FG and ML conceived and designed the study. FG supervised the study. DS, VA, OF, AP, M-LP, MZ, SJ, LB and FG performed experiments and analysed data. LC and GC conducted computational analysis. M-LP performed bioinformatic analysis. LC, ML and FG wrote the manuscript.
Funding: This work was supported by grants from Agence Nationale pour la Recherche sur le SIDA et les hepatites virales (ANRS) to ML (n° ECTZ8323; n. ECTZ27696; n. ECTZ66014); from the IDEX Package PALSE (Programme Avenir Lyon-St Etienne); from the Agence Nationale de la Recherche (ANR@TRACTION) to ML; from the EU project 667273 HEP-CAR to ML.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The transcriptomic data used to generate the Venn diagram in Figure 5B ('genes whose expression is positively correlated with DLEU2 or EZH2') can be accessed on the TGCA-LIHC dataset and extracted using the R2 platform (https://r2.amc.nl) with the function 'Find correlated Genes with a Single Gene'. The ChIP-Seq dataset used in Figure 5B ('genes identified as HBx direct genes'; listed in online supplementary table 8) can be accessed at http://www.ebi.ac.uk/ena/data/view/PRJEB13227. All other raw data, materials and reagents are available on request from the corresponding authors (FG and ML).
References
- 1. World Health Organization Global hepatitis report 2017, 2017. Available: http://www.who.int/iris/handle/10665/255016
- 2. Schweitzer A, Horn J, Mikolajczyk RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015;386:1546–55. 10.1016/S0140-6736(15)61412-X [DOI] [PubMed] [Google Scholar]
- 3. Stanaway JD, Flaxman AD, Naghavi M, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the global burden of disease study 2013. Lancet 2016;388:1081–8. 10.1016/S0140-6736(16)30579-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lok AS, Zoulim F, Dusheiko G, et al. Hepatitis B cure: from discovery to regulatory approval. Hepatology 2017;66:1296–313. 10.1002/hep.29323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. European Association for the Study of the Liver EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370–98. 10.1016/j.jhep.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 6. Mason WS, Gill US, Litwin S, et al. Hbv DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016;151:986–98. 10.1053/j.gastro.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Testoni B, Levrero M, Zoulim F. Challenges to a cure for HBV infection. Semin Liver Dis 2017;37:231–42. 10.1055/s-0037-1606212 [DOI] [PubMed] [Google Scholar]
- 8. Pollicino T, Belloni L, Raffa G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 2006;130:823–37. 10.1053/j.gastro.2006.01.001 [DOI] [PubMed] [Google Scholar]
- 9. Decorsière A, Mueller H, van Breugel PC, et al. Hepatitis B virus X protein identifies the SMC5/6 complex as a host restriction factor. Nature 2016;531:386–9. 10.1038/nature17170 [DOI] [PubMed] [Google Scholar]
- 10. Belloni L, Pollicino T, De Nicola F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 2009;106:19975–9. 10.1073/pnas.0908365106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rivière L, Gerossier L, Ducroux A, et al. Hbx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol 2015;63:1093–102. 10.1016/j.jhep.2015.06.023 [DOI] [PubMed] [Google Scholar]
- 12. Guerrieri F, Belloni L, D'Andrea D, et al. Genome-Wide identification of direct HBx genomic targets. BMC Genomics 2017;18:184 10.1186/s12864-017-3561-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moyo B, Nicholson SA, Arbuthnot PB. The role of long non-coding RNAs in hepatitis B virus-related hepatocellular carcinoma. Virus Res 2016;212:103–13. 10.1016/j.virusres.2015.07.025 [DOI] [PubMed] [Google Scholar]
- 14. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–14. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu W, Ding C. Roles of lncRNAs in viral infections. Front Cell Infect Microbiol 2017;7:205 10.3389/fcimb.2017.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang P, Xu J, Wang Y, et al. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017;358:1051–5. 10.1126/science.aao0409 [DOI] [PubMed] [Google Scholar]
- 17. Zhao J, Ohsumi TK, Kung JT, et al. Genome-Wide identification of Polycomb-Associated RNAs by RIP-seq. Mol Cell 2010;40:939–53. 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guil S, Soler M, Portela A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 2012;19:664–70. 10.1038/nsmb.2315 [DOI] [PubMed] [Google Scholar]
- 19. Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature 2011;469:343–9. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li G, Margueron R, Ku M, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 2010;24:368–80. 10.1101/gad.1886410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sarma K, Cifuentes-Rojas C, Ergun A, et al. Atrx directs binding of PRC2 to Xist RNA and polycomb targets. Cell 2014;159:869–83. 10.1016/j.cell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wassef M, Margueron R. The multiple facets of PRC2 alterations in cancers. J Mol Biol 2017;429:1978–93. 10.1016/j.jmb.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 23. Cai M-Y, Tong Z-T, Zheng F, et al. Ezh2 protein: a promising immunomarker for the detection of hepatocellular carcinomas in liver needle biopsies. Gut 2011;60:967–76. 10.1136/gut.2010.231993 [DOI] [PubMed] [Google Scholar]
- 24. Kim W, Bird GH, Neff T, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol 2013;9:643–50. 10.1038/nchembio.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun 2019;10:1679 10.1038/s41467-019-09624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez ME, Moore HM, Li X, et al. Ezh2 expands breast stem cells through activation of Notch1 signaling. Proc Natl Acad Sci U S A 2014;111:3098–103. 10.1073/pnas.1308953111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Xi Y, Li W, et al. Trim28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene 2017;36:2991–3001. 10.1038/onc.2016.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421 10.1038/ncomms14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng L, Paulson A, Li H, et al. Developmental programming of long non-coding RNAs during postnatal liver maturation in mice. PLoS One 2014;9:e114917 10.1371/journal.pone.0114917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giulietti M, Righetti A, Principato G, et al. Lncrna co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis 2018;39:1016–25. 10.1093/carcin/bgy069 [DOI] [PubMed] [Google Scholar]
- 31. Villa E, Critelli R, Lei B, et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. results from a prospective study. Gut 2016;65:861–9. 10.1136/gutjnl-2014-308483 [DOI] [PubMed] [Google Scholar]
- 32. Lerner M, Harada M, Lovén J, et al. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res 2009;315:2941–52. 10.1016/j.yexcr.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 33. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu J-J, Song W, Zhang S-D, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep 2016;6:23521 10.1038/srep23521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan H, Zhang H, Pascuzzi PE, et al. Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene 2016;35:715–26. 10.1038/onc.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cirillo D, Blanco M, Armaos A, et al. Quantitative predictions of protein interactions with long noncoding RNAs. Nat Methods 2017;14:5–6. 10.1038/nmeth.4100 [DOI] [PubMed] [Google Scholar]
- 37. Ritchie DW, Kozakov D, Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics 2008;24:1865–73. 10.1093/bioinformatics/btn334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Zundert GCP, Rodrigues JPGLM, Trellet M, et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 2016;428:720–5. 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 39. Antonysamy S, Condon B, Druzina Z, et al. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain. PLoS One 2013;8:e84147 10.1371/journal.pone.0084147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minor MM, Slagle BL. Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system. Viruses 2014;6:4683–702. 10.3390/v6114683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mészáros B, Tompa P, Simon I, et al. Molecular principles of the interactions of disordered proteins. J Mol Biol 2007;372:549–61. 10.1016/j.jmb.2007.07.004 [DOI] [PubMed] [Google Scholar]
- 42. Walia RR, Xue LC, Wilkins K, et al. RNABindRPlus: a predictor that combines machine learning and sequence homology-based methods to improve the reliability of predicted RNA-binding residues in proteins. PLoS One 2014;9:e97725 10.1371/journal.pone.0097725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wass MN, Fuentes G, Pons C, et al. Towards the prediction of protein interaction partners using physical docking. Mol Syst Biol 2011;7:469 10.1038/msb.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li T, Robert EI, van Breugel PC, et al. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the Cul4-Ddb1 ubiquitin ligase machinery. Nat Struct Mol Biol 2010;17:105–11. 10.1038/nsmb.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lucifora J, Arzberger S, Durantel D, et al. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 2011;55:996–1003. 10.1016/j.jhep.2011.02.015 [DOI] [PubMed] [Google Scholar]
- 46. Zhang H, Xing Z, Mani SKK, et al. Rna helicase DEAD box protein 5 regulates polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 2016;64:1033–48. 10.1002/hep.28698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang W, Chen J, Wu M, et al. Prmt5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology 2017;66:398–415. 10.1002/hep.29133 [DOI] [PubMed] [Google Scholar]
- 48. Tomar D, Singh R. TRIM13 regulates ubiquitination and turnover of NEMO to suppress TNF induced NF-κB activation. Cell Signal 2014;26:2606–13. 10.1016/j.cellsig.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 49. Chen W-xian, Cheng L, Xu L-yun, et al. Bioinformatics analysis of prognostic value of TRIM13 gene in breast cancer. Biosci Rep 2019;39 10.1042/BSR20190285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slagle BL, Bouchard MJ. Hepatitis B virus X and regulation of viral gene expression. Cold Spring Harb Perspect Med 2016;6:a021402 10.1101/cshperspect.a021402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lok AS, Zoulim F, Dusheiko G, et al. Hepatitis B cure: from discovery to regulatory approval. J Hepatol 2017;67:847–61. 10.1016/j.jhep.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 52. Studach LL, Menne S, Cairo S, et al. Subset of Suz12/PRC2 target genes is activated during hepatitis B virus replication and liver carcinogenesis associated with HBV X protein. Hepatology 2012;56:1240–51. 10.1002/hep.25781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu J, Shao Z, Li D, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 2015;57:304–16. 10.1016/j.molcel.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beltran M, Yates CM, Skalska L, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 2016;26:896–907. 10.1101/gr.197632.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davidovich C, Wang X, Cifuentes-Rojas C, et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 2015;57:552–8. 10.1016/j.molcel.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Q, McKenzie NJ, Warneford-Thomson R, et al. Rna exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat Struct Mol Biol 2019;26:237–47. 10.1038/s41594-019-0197-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X, Goodrich KJ, Gooding AR, et al. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol Cell 2017;65:1056–67. 10.1016/j.molcel.2017.02.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319637supp001.pdf (5.6MB, pdf)