Abstract
The implementation of endotype-driven effective intervention strategies is now considered as an essential component for sepsis management. Rapid screening and frequent monitoring of immune responses are critical for evidence-based informed decisions in the early hours of patient arrival. Current technologies focus on pathogen identification that lack rapid testing of the patient immune response, impeding clinicians from providing appropriate sepsis treatment. Herein, we demonstrate a first-of-its-kind novel point-of-care device that uses a unique approach by directly monitoring a panel of five cytokine biomarkers (IL-6, IL-8, IL-10, TRAIL & IP-10), that is attributed as a sign of the body's host immune response to sepsis. The developed point-of-care device encompasses a disposable sensor cartridge attached to an electrochemical reader. High sensitivity is achieved owing to the unique sensor design with an array of nanofilm semiconducting/metal electrode interface, functionalized with specific capture probes to measure target biomarkers simultaneously using non-faradaic electrochemical impedance spectroscopy. The sensor has a detection limit of ~1 pg/mL and provides results in less than five minutes from a single drop of undiluted plasma sample. Furthermore, the sensor demonstrates an excellent correlation (Pearson's r > 0.90) with the reference method for a total n = 40 clinical samples, and the sensor's performance is ~30 times faster compared to the standard reference technique. We have demonstrated the sensor's effectiveness to enhance diagnosis with a mechanistic biomarker-guided approach that can help disease endotypying for effective clinical management of sepsis at the patient bedside.
Keywords: Sepsis endotyping, Multiplexed cytokine detection, Rapid point-of-care testing, Electrochemical sensing, IL-6IL-8 IL-10, TRAIL, IP-10, Clinical validation
Highlights
-
•
Developed biosensor simultaneously detects IL-6, IL-8, IL-10, TRAIL and IP-10 in undiluted plasma.
-
•
Robust, sensitive and specific detection using non-faradaic impedance spectroscopy.
-
•
Excellent correlation (Pearson's r > 0.90) with the reference methods for a total n = 40 clinical samples.
-
•
95–97% of original activity retained post five weeks of storage in 4 °C, indicating excellent operational stability.
-
•
Sample-to-detection time measured for all five biomarkers was ~5 min.
1. Introduction
Endotypes are biological subtypes characterized by distinct pathophysiological functions, described by specific biomarkers. The interpretation and verification of sepsis endotypes can save lives by encouraging early identification of patient groups for accurate therapy. Sepsis endotyping enables physicians to provide critical care and precision medicine as it showcases the patient's immune and treatment response to sepsis. Thus, there has been a promising transition from predicting the outcome to pathobiology driven understanding of host response heterogeneity to sepsis, leveraging innovative high-performance translational techniques and analytical methods to identify distinct biomarker subgroups of the host response. Medical communities have further acknowledged the value of biological markers as they continue to enhance sepsis diagnosis, which allows the classification of patients within the specific clinical category (Akdis, 2012; Burke, 2016). Sepsis is recognized as a global health crisis affecting more than 49 million people every year (Rudd et al., 2020). It is a life-threatening condition that represents the systemic immunological reaction of the body to an infectious incident that leads to death (Singer et al., 2016). It is well known that sepsis-related fatality is not directly caused by the invading pathogen rather, the clinical complexity is triggered by the dysregulated host immune response that leads to multiple organ dysfunction (Schouten et al., 2008). The pathogen or the causative agent triggering sepsis may differ the host's immune response being a key indicator in assessing fatality and the need for complex medical treatments. Ultimately, the combination of pathogen load, infection site, and host susceptibility leads to clinical presentation and course of the disease. Sepsis may be caused by any form of infection; the most common cause is a bacterial infection (pneumonia or urinary tract infection) that affects the body and triggers bacterial sepsis. Whereas viral sepsis is caused by a viral infection (flu), and more viral sepsis cases are triggered by COVID-19, which has caused a pandemic in 2020. Additionally, recent studies have reported mortality in COVID-19 patients triggered by sepsis, especially for elderly patients with pre-existing chronic illness (Alhazzani et al., 2020; Arentz et al., 2020; Bhatraju et al., 2020; Phua et al., 2020).
Currently, rapid diagnostics exist mainly for pathogen identification, such as those highlighted in supplementary (Table S1) while a traditional clinical workflow is illustrated in Fig. 1 A. Briefly, technologies such as SeptiFast (Roche) identifies multiple pathogens in 6 h, HYPLEX (BAG) relies on the PCR technique to recognize relevant pathogens within 3 h, and the Film Array device (BioFire) requires an hour for pathogen detection. These commercially available and those in development focus solely on detecting sepsis based on the pathogen. However, it is crucial to understand the nature of sepsis and its form of representation in the patient by tracking the host immune response. As sepsis initiates, both pro-inflammatory and anti-inflammatory mechanisms start promptly with a predominant initial hyper-inflammatory phase, as shown in Fig. 1B. In most cases, the innate immune response destroys the invading pathogen, but occasionally the pathogen prevails, and the host response may become unbalanced and destructive. The increased production of cytokines and chemokines is attributed to the severity and prevalence in sepsis, implying that a chain of unregulated inflammation has initiated displaying signs of both excessive inflammations as well as immune suppression, the severity of which varies from patient to patient. This perplexing phenomenon of the host immune response has been thought to be a race to death between the invading microbes and the host immune response, where the pathogens seek and benefit by suppressing various facets of host immunity according to Hotchkiss and coworkers (Hotchkiss et al., 2013). Therefore, monitoring cytokine biomarkers can assist in clinical decision-making and forecast sepsis-related outcomes to treatments (Dupuy et al., 2013). For instance, measuring the cytokine levels near-patient at different time points would help administer specific types of drugs, where, immunosuppressants may be prescribed early in the disease etiology addressing the hyperimmune state of the patient and immunomodulators at a later stage in conjunction with antimicrobial therapy. Hence, it is critical to additionally track immune response imbalance triggered by inflammatory & anti-inflammatory cytokine immune response to channelize appropriate treatment strategy. As a point-of-care treatment option, to date, no molecular host biomarker panel is available which makes an informed decision on the specific intervention based on the diagnosis of the immune response or the ability to detect improvements in the status of patients with sepsis (Albert-Vega et al., 2018; Gunsolus et al., 2019). This gives rise to a major diagnostic gap in near-patient testing capabilities. To address this technological gap, this work demonstrates first-of-a-kind near-patient testing ‘DETecT Sepsis’ (Direct Electrochemical Technique Targeting Sepsis) sensor, which directly measures a panel of five host immune biomarkers in <5 min to guide the physician with active feedback on patient immune status for better therapeutic administration. DETecT sepsis sensor enables a mechanistic approach for sepsis stratification by leveraging the use of endotypes as defined by specific biomarkers to classify based on pathophysiological process rather than the clinical representation of sepsis, which is a step towards precision medicine. Electrochemical sensing modality in conjunction with affinity-based capture probes specifically quantifies levels of pro- and anti-inflammatory biomarkers (IL-6, IL-8, IL-10, TRAIL, and IP-10) using minimally acquired blood plasma samples.
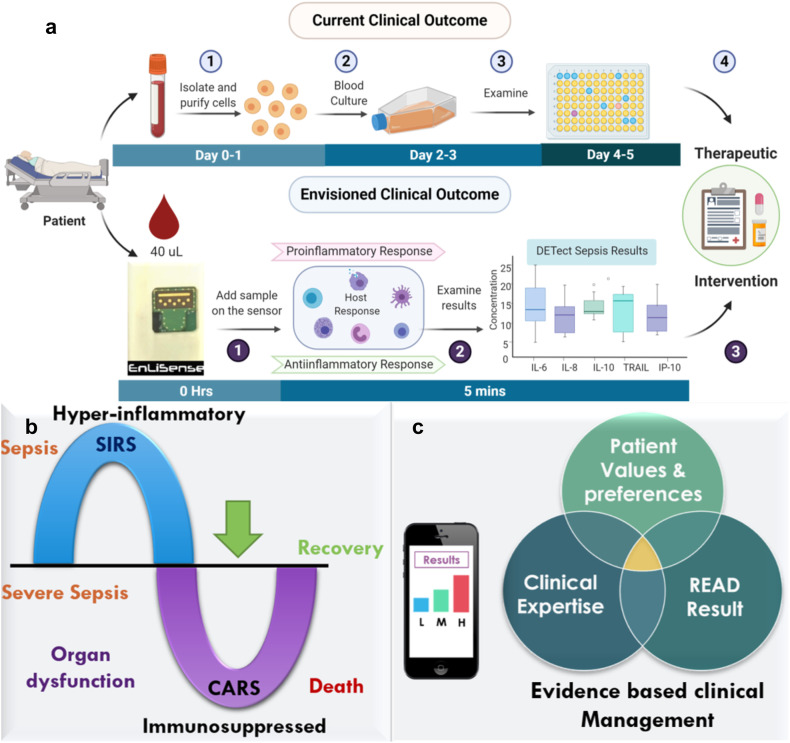
Fig. 1.
a) Illustration of the current clinical timeline vs. DETecT Sepsis approach as rapid near-patient testing for disease severity screening based on biomarker levels. Image is created with Biorender.com b) Hyper-inflammatory and Immunosuppressed phase of Sepsis hypothesis c) Proof of feasibility towards establishing evidence-based clinical management approach using n = 40 patient samples.
Fig. 1a illustrates the current clinical workflow, and Fig. 1b represents the opportunity the developed sensor offers for detecting sepsis and tracking the host immune response allowing evidence-based clinical management (Fig. 1c). Our point-of-treatment technology allows rapid detection of multiple host-immune response sepsis cytokine biomarkers with ease of sample handling coupled with low sample volume (~40 μL) to facilitate near-patient bedside monitoring towards enabling biomarker-guided patient stratification, endotyping, and improving treatment response within the critical golden hour post sepsis detection. The advantages of the DETecT Sepsis sensor over existing point-of-care tests are: (i) direct hassle-free measurement from a single drop of undiluted blood plasma; (ii) allows sepsis stratification based on the body's hyper and hypo immune response; (iii) specifically surface engineered sensor design facilitates high sensitivity and specificity; (iv) portable handheld format enables multi-measure capabilities at near-patient testing. Such a point-of-care-testing device would allow clinicians to make an evidence-based decision on immune-modulating treatments customized to the patient's inflammatory response within the “golden-hour” as illustrated in Fig. 1c.
2. Materials and methods
2.1. Experimental design
The objective of our study was to establish a robust sensing performance of the developed biosensing DETecT Sepsis sensor using EnLiSense's READ platform for the detection of sepsis. The sensor was designed to allow simultaneous detection of cytokine biomarker panel using minimal sample fluid (<40 μL) in human blood plasma. Sensor performance metrics (sensitivity, specificity, dynamic range, detection limit, precision, and accuracy) were tested for IL-6, IL-8, IL-10, TRAIL & IP-10 in plasma. DETect Sepsis sensor was further validated for clinical translation by testing 20 patient samples tested positive for sepsis at the time of admission compared to the 20 healthy cohorts. The performance of the developed sensor was compared with the Luminex standard as a reference method.
2.2. Reagents
Dithiobis (succinimidyl propionate) (DSP) and the Dimethyl sulphoxide (DMSO) solvent was purchased from Thermo fisher scientific (USA) along with the phosphate Buffer Saline (PBS) and SuperBlock. The antibodies and their specific antigens for IL-6, IL-8, IL-10, TRAIL, and IP-10 were purchased from Abcam. Pooled human plasma was obtained from Innovative Research, Inc. (USA) for sensor characteristic studies. Plasma from sepsis patients was obtained from the Austere-environments Consortium for Enhanced Sepsis Outcomes (ACESO), a consortium consisting of US Government, non-profit, academic, and industry partners. All the stock proteins and patient samples were stored at -20 °C or according to their storage conditions until further use. None of the proteins underwent more than 3 freeze-thaw cycles to avoid denaturing of the proteins. The antibodies were diluted in PBS to bring them to their optimized concentration, while their respective antigens were spiked in pooled human plasma in varying concentrations to perform calibrated response curves for each target biomarker.
2.3. Direct Electrochemical Technique Targeting Sepsis sensor on EnLiSense's READ platform
The DETecT (Direct Electrochemical Technique Targeting Sepsis) sensor uses EnLiSense's Rapid ElectroAnalytical Device (READ) platform that comprises of the following: (1) A disposable, single-use sensor cartridge with an array of sensing electrodes that are individually configured to detect multiple biomarkers simultaneously from the sample specimen in real-time. (2) A handheld, palm-sized form-factor electronic reader onto which the sensor is mounted, which transduces the electrical outputs resulting from the sample specimen to other electronic devices/data server through a software interface (configurable to support both wired and wireless communication). The subtle changes between antibody-antigen affinity interaction result in an electrochemical impedance signal response. Briefly, the detection mechanism is based on non-faradaic electrochemical impedance spectroscopy (EIS). Herein, a small input voltage (10 mV) is applied to the sensor over a frequency range and the resulting impedance response is measured by the portable electronic device. The functioning of the electronic reader has been previously demonstrated elsewhere by our group (Sankhala et al., 2018). EIS is a powerful technique that captures subtle interaction at the functionalized electrode surface. When a sample is introduced on the electrode surface and the electrode is polarized, the rearrangement of charges occurs at the electrode-solution interface. This results in a local built-up of excessive ions of opposite charge. The extent to which the exponential charge built-up decays, forms the electrical double layer (or double-layer capacitance). The target analyte binds to the specific capture probe antibody within this double layer leveraging the antibody-antigen affinity mechanism across each working electrode and impedance is measured (Jagannath et al., 2018; Tanak et al., 2019a, 2019b).
Advantages of non-faradaic EIS over faradaic method includes (i) label-free technique that can directly measure the subtle binding interactions without a redox label for measuring impedance response, thus, making non-faradaic EIS considerably more compatible in point-of-care applications (ii) Non-faradaic impedance measurement eliminates the need for a DC potential; thus, it does not denature the biomolecules immobilized on the sensing electrode surface. The sensing layer was surface engineered through a standard sputter fabrication technique using RF magnetron to deposit a 200 nm thickness of semiconducting thin film on the gold electrodes. Before deposition, a solvent cleaning strategy was applied where the surface was thoroughly cleaned with isopropyl alcohol (IPA), acetone, and DI water to eliminate any impurities. Modulating the metal oxide layer's surface chemistry helps to improve the rearrangement of charges near the electrode-solution interface. Furthermore, Zinc oxide semiconductor has unique properties, including a large bandgap (3.367 eV), is non-toxic, and has high excitation binding energy (60 eV) that helps to increase overall sensitivity. Additionally, due to its high adsorption capability owing to its high isoelectric point (~9.5), chemical stability and good electrical conductivity enhance its use for sensitive electrochemical biosensing applications (Tanak et al., 2019a). Nanoscale dimensions of the semiconducting thin film allow size-based matching to the target analyte, which effectively increases surface area to volume ratio. Additionally, the structural morphology of the nanofilm offers selective biomolecular binding for the functionalized capture probes. With the increase in the surface-to-volume ratio, nanofilm's surface structures can significantly modulate the charge carrier densities within the material and increase band bending (Cho et al., 2020; Kim et al., 2013). Previously, our group has experimentally demonstrated a three-fold increase in sensitivity for the nanofilm as compared to planar nontextured microelectrodes (Shanmugam et al., 2016). Thus, leveraging the ZnO nanofilm surface engineered layer's unique properties, we have demonstrated sensitive electrochemical biosensing for the detection of multiplexed biomarkers.
2.4. Immunoassay development
The sensor surface was immobilized with 10 mM DSP dissolved in DMSO and incubated in the dark at room temperature. Specific capture antibodies (IL-6, IL-8, IL-10, TRAIL, and IP-10, 10 μg/mL each) were individually functionalized on each working electrode of the sensors. Superblock was used to hydrolyze unbound linker sites to avoid non-specific interaction. A calibrated response was established for each of the pentaplex biomarkers against varying dose concentrations spiked in pooled human plasma. Data were represented as a percentage change in impedance with respect to baseline (plasma blank without the target biomarkers), using Equation S1 (supplementary information). Cross-reactive study was tested by preparing a cocktail of non-specific biomolecules in low (10 pg/mL) and high concentrations (1000 pg/mL) with the absence of the target biomarker. Individual response for the cross-reacting molecules was measured for each sensor functionalized with the target capture antibody. To test the specificity of the sensor in the presence of interfering biomarkers, the target biomolecule was spiked to the previously prepared cocktail solution, and the response was measured for each of the pentaplex biomarkers. All the data represented is measured from n = 3 replicates. The limit of detection (LoD) described as the lowest measured concentration was calculated as 3 times SD of blank plasma.
2.5. Patient sample acquisition
Plasma samples were collected under written informed consent as part of an ongoing observational trial of sepsis in resource-limited settings conducted by ACESO(Schully et al., 2017; Schully and Clark, 2019). Briefly, patients presenting to the emergency department of a participating hospital with at least two SIRS features and a suspected infection (SEPSIS-2 criteria) were eligible for enrollment. For this work, plasma samples collected 24 h after enrollment from sepsis patients were used. Samples were provided stripped of all identifiers by ACESO to Biomedical Microdevices and Nanotechnology Laboratory, UT Dallas following the Material Transfer Agreement (MTA), approved by the Institutional Review Board (IRB# 19MRO151) at the University of Texas at Dallas. The samples were stored at -20 °C immediately on arrival until further use and did not undergo more than two freeze-thaw cycles.
2.6. Statistical analysis
Statistical analysis was performed using GraphPad Prism Software (GraphPad Software Inc., La Jolla, CA). ns: non-significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data represented as mean ± SEM for n = 3 replicates unless stated otherwise. One-way analysis of variance (ANOVA) was used for the comparison between three or more groups. T-test was used to compare significance for specific signal against non-specific interferons in the cross-reactive study. Differences between healthy and septic cohort were assessed using non-parametric unpaired Mann-Whitney tests.
3. Results
3.1. DETecT sepsis sensor evaluation for multiplexed quantification of immune biomarkers
There is an immense unmet need for a rapid diagnostic to enable patient stratification in sepsis towards effective disease management. Therefore, the point-of-care-tests should be highly sensitive with a wide dynamic range for a panel of host response biomarkers that can be leveraged for assessing the patient's immune state towards stratification and disease management. Based on the wide body of scientific literature and clinical evidence, our approach was to target a combination of pro and anti-inflammatory cytokines in conjunction with cell apoptosis monitoring protein. Therefore, we selected IL-6, IL-8, IP-10, IL-10, and TRAIL for this study as the levels of these biomarkers provide a composite snapshot into a patient's immune response state towards establishing disease severity and mortality risk in the patient. TRAIL and IP-10 were included for discriminating between the viral and bacterial loadings on the host (Chaudhry et al., 2013). DETecT sensor device was calibrated for each of the pentaplex biomarker panels in pooled human plasma (control set with no infections). Herein, varying dose concentrations were measured for specific analytes to establish a calibrated dose-response on the multiplexed affinity capture probe functionalized sensor array. Signal impedance response between antibody and the target analyte was captured using electrochemical impedance spectroscopy and represented as a percentage change in impedance with respect to the baseline (described in the methods section) in pooled plasma as shown in Fig. 2 a-e.
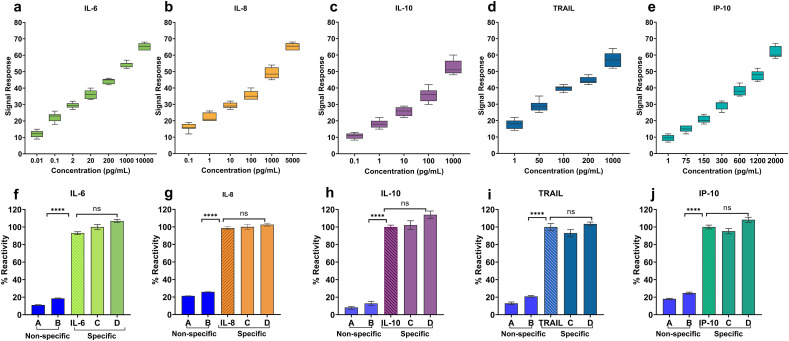
Fig. 2.
(a–e) Calibrated dose-response for IL-6, IL-8, IL-10, TRAIL, and IP-10 in pooled human blood plasma with a clinically relevant dynamic range. (f–j) The cross-reactive study demonstrating the specificity of DETecT Sepsis sensor for each of the target biomarker. A; Low concentration of non-specific biomarker mixture, B; High Concentration of non-specific biomarker mixture, C; Target marker along with a low concentration of non-specific biomarker mixture, D; Target marker along with a high concentration of non-specific biomarker mixture.
A dose-dependent increasing trend in impedance signal response was observed for all the biomarkers. The dynamic range for each biomarker was aimed to capture the healthy as well as a diseased state within the physiologically relevant range of clinical samples. IL-6 demonstrated a wide dynamic range of 0.01 pg/mL to 10 ng/mL with a limit of detection (LoD) of 0.1 pg/mL in spiked plasma samples (Fig. 2b). The box plots for each biomarker display no overlapping inter-quartile ranges with a minimum variation for each concentration, indicating good repeatability with the least variance. The signal impedance response is reflective of the affinity binding mechanism between the specific capture probe and the target analyte that indicates the biomarker concentration in pooled plasma. The unique multiplexed sensor design coupled with specific surface functionalization augments the signal response and has previously been described elsewhere (Tanak et al, 2019a, 2019b). IL-8 demonstrated a dynamic range of 0.1 pg/mL to 5 ng/mL with a detection limit of 0.1 pg/mL (Fig. 2c). IL-6 and IL-8 are known to be major mediators of an inflammatory response, and their levels elevate in patients with sepsis, which act as key indicators during the development of severe sepsis. Similarly, the dynamic range for IL-10 was observed to be from 0.1 pg/mL to 1 ng/mL (Fig. 2d) with an LoD of 1 pg/mL in pooled plasma. IL-10 belongs to the group of immunoregulatory molecules called anti-inflammatory cytokines that prevents the body from the adverse effects of excess inflammatory immune reactions. The key risk factor for sepsis severity and the fatal outcome is the chronic overproduction of IL-10, which indicates patients with sepsis are in a deep immunosuppression state (Gogos et al., 2000). Data in Fig. 2e shows the dynamic range of TRAIL from 1 pg/mL to 1 ng/mL with a detection limit of 1 pg/mL whereas IP-10 displayed a dynamic range of 1 pg/mL to 2 ng/mL (Fig. 2f) respectively. Additionally, each biomarker demonstrated a statically significant difference between concentrations as determined by one-way ANOVA with 95% confidence intervals (Table S2). The developed DETecT sepsis sensor demonstrated sensitive detection for biomarkers below normal threshold levels as well as the dynamic ranges extended beyond elevated levels predicted in disease states. The surface engineered semi-conducting nanofilm allows sensitive detection by potentially increasing the surface to volume ratio, allowing plenty of biomolecules to be immobilized onto the electrode surface.
After establishing a sensitive and robust calibration response, the selectivity and specificity of the DETecT Sepsis sensor on EnLiSense's READ platform were evaluated. Every analyte was tested with a series of non-specific markers, starting with the lowest concentration of the cross-reacting molecule, followed by the highest concentration as seen in Fig. 2(f-j). Selectivity of the biosensor is extremely important while testing actual clinical samples where the concentrations of the analyte can be much lower than that of the non-specific molecule. Thus, to mimic realistic scenarios, the sensor was additionally tested with a cocktail mixture of low and high non-specific molecules along with the target analyte spiked plasma sample to validate sensor device platform specificity. Non-specific biomarkers (represented as bar's A and B) in Fig. 2(f–j) showed less than 10% reactivity compared to the specific response of the target biomarker. Additionally, despite the presence of varying concentrations of cross-reacting molecules (represented as bar C and D in Fig. 2(f–j) along with the target biomarker, the developed sensor demonstrated similar results (100% reactivity) to that of the specific individual analyte. DETecT sepsis sensor's capability tobind to the functionalized antibody selectively is attributed to the surface-functionalized highly specific monoclonal antibody combined with the effect of blocking buffer. By blocking the active functional groups on the electrode surface, the blocking buffer (superblock) prevents non-specific binding and can help stabilize the biomolecule attached to the electrode surface, thereby increasing the specificity of the biosensor (Xiao and Isaacs, 2012). Specificity is a vital vital sensor metric to reduce false-positive results and provide accurate detection capability with increased resolution in distinguishing disease state. Overall, the data confirmed the developed DETEecT sepsis sensor demonstrated high sensitivity coupled with specific and selective response despite the presence of non-specific biomolecules for the multiplexed cytokine biomarker panel in plasma using EIS as the detection technique.
3.2. Evaluating DETecT sepsis sensor performance for repeatability, reproducibility stability, and accuracy in pooled human blood plasma
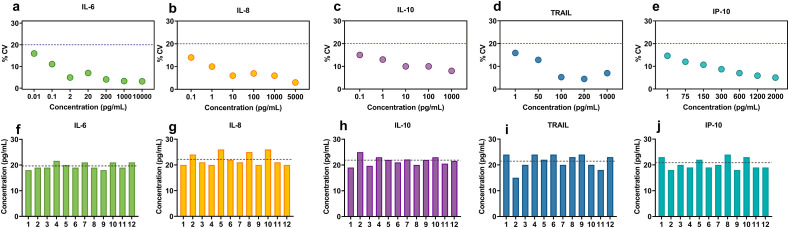
Repeatability, reproducibility, accuracy, and stability are the main considerations that need to be assessed while evaluating the sensing platform's effectiveness. The coefficient of variation (%CV) was calculated for all the study biomarkers as a measure to assess the dispersion within each reported concentration from n = 10 sensors, as shown in Fig. 3 (a–e). Generally, lower concentration tends to show higher variability and the developed sensor demonstrates the CV range between 3 and 16%. The results displayed in Fig. 3(a–e) exhibit a CV < 20% which is clinically accepted as per the guidelines set by the Clinical and Laboratory Standards Institute (CLSI), thereby demonstrating the repeatability of DETecT sepsis sensor within a wide dynamic range (CLSI, 2014). Fig. 3(f–j) demonstrates the reproducibility of the electrochemical response for 12 identical sensors with an average concentration indicated by the dotted line for each target analyte. The relative standard deviation (RSD) across all twelve sensors was ~ 10%. The value of RSD indicates good reproducibility and repeatability of the DETecT Sepsis sensor platform.
Fig. 3.
(a–e) Coefficient of variation plot examining the precision of DETecT Sepsis sensor for IL-6, IL-8, IL-10, TRAIL and IP-10 in pooled plasma. The dotted line at 20% represents the acceptable limit according to CLSI guidelines (f–j) Repeatability and reproducibility of DETecT sepsis sensor across 12 sensors for IL-6, IL-8, IL-10, TRAIL and IP-10 in pooled plasma respectively. The dotted line represents the average concentration across 12 replicates.
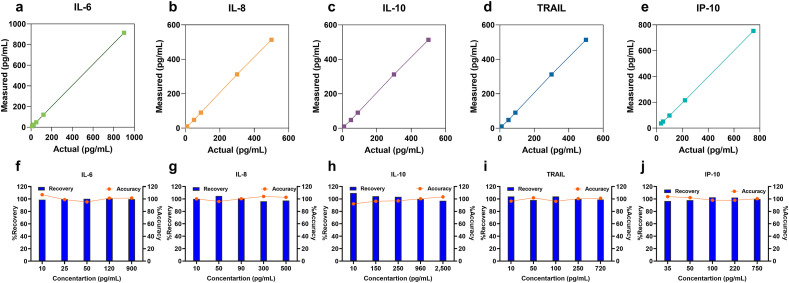
Next, DETecT Sepsis sensor performance was tested for its accuracy by the spike and recovery study. Known concentrations (actual) spiked in triplicate correlated with an R2 value of 0.99 with the measured concentration calculated based on the previously established calibration curve. Fig. 4 (a–e) demonstrates reliable detection of actual spiked concentration across all five target biomarkers. The recovery percentage was then calculated along with the accuracy of the sensor as represented in Fig. 4(f–j). As observed from the results, the percent recovery was between ~89 and 110% for IL-6, IL-8, IL-10 TRAIL, and IP-10, which lies well within the acceptable range for assay validation according to CLSI standards (Andreasson et al., 2015). Once the accuracy was established, the operational stability of DETecT Sepsis sensor was tested for up to five weeks (35 days) with the sensors stored in 4 °C. 1 pg/mL concentration for TRAIL and IP-10 was measured thrice (n = 3). No significant change in response was observed for TRAIL with only a 3% loss of signal at the end of 5 weeks as seen in supplementary Fig S1. Similarly, a loss of only 5% of the signal response was seen at the end of five weeks specific to IP-10 as seen in supplementary Fig S2. In summary, both TRAIL and IP-10 retained 95–97% of its original activity post five weeks of storage in 4 °C, indicating excellent operational stability. To our knowledge, this is the first demonstration of a pentaplex biomarker sensor that allows simultaneous quantification of pro-and anti-inflammatory biomarkers with a single drop of plasma sample with reliable sensor performance metrics.
Fig. 4.
Repeatability, Reproducibility, and stability of DETecT Sepsis. (a–e) Actual and measured concentration in pooled plasma represented as correlation plots using the DETecT sepsis sensor. (f–j) Repeatable and reliable sensing capability highlighting the high recovery rate and accuracy of the developed sensing platform.
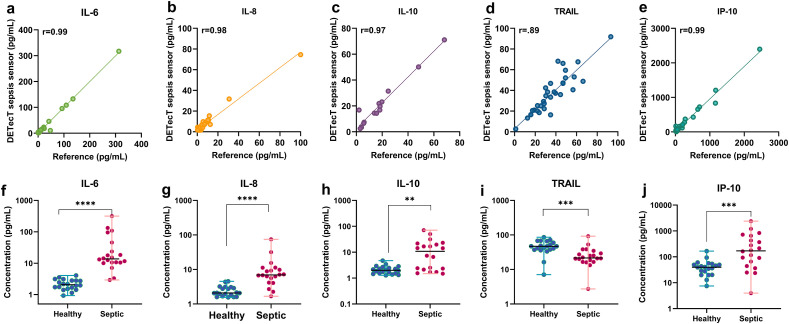
3.3. Validating DETecT sepsis sensor with clinical patient samples
Clinical translation for the multiplexed POCT technologies requires validation with patient samples. For this study, 20 septic patient plasma samples along with 20 control (Non-septic) healthy plasma samples (Table S3) were evaluated using the DETecT Sepsis sensor and Luminex as the reference standard. The onset of sepsis was confirmed at 24 h time point from the hospital. All the 40 samples were tested for IL-6, IL-8, IL-10, TRAIL, and IP-10 using the developed sensor device. Concentrations measured using the sensor correlated well with a Pearson's r ≥ 0.90 for all the five test biomarkers as seen in Fig. 5 a-e. Fig. 5(f–j) represents the DETecT Sepsis sensor's capability to distinguish healthy vs. sepsis patient cohorts for IL-6, IL-8 IL-10, TRAIL, and IP-10. We observed that levels of IL-6, IL-8, IL-10, TRAIL, and IP-10 of the healthy cohort were significantly different from the Septic cohort as confirmed using Mann-Whitney U statistical analysis test. Mean plasma IL-6 levels for the septic cohort were 44.05 ± 74.32 pg/mL as compared to 2.2 ± 0.83 pg/mL of the healthy cohort. IL-8 levels for the healthy cohort were all below 2.44 ± 0.88 pg/mL while mean levels in the septic cohort were around 11.65 ± 16.16 pg/mL. Similarly, for anti-inflammatory IL-10 biomarkers, the mean level established for the healthy cohort was 2.17 ± 0.84 pg/mL and septic patients' mean levels were 15.47 ± 18.18 pg/mL. As seen in Fig. 5d, TRAIL levels were significantly lower in septic patients with a mean value of 27.62 ± 18.62 pg/mL as compared to the healthy cohort mean levels of 47.69 ± 18.74 pg/mL. Studies have shown to correlate lower levels of TRAIL to poor patient outcomes, thus indicating the overall severity of illness (Schenck et al., 2019; Tian et al., 2013). Mean healthy concentrations for IP-10 were 45.27 ± 34.57 pg/mL whereas septic sample mean levels were measured to be 397.3 ± 572.5 pg/mL. The DETecT Sepsis method was also able to distinguish all the five biomarkers in healthy and septic patient samples with a good statistical significance of p < 0.001 (Table S4).
Fig. 5.
Clinical validation of DETecT Sepsis sensor with Sepsis Patient samples. (a–e) Correlation between the Reference method (Luminex) and DETecT sepsis sensor platform obtained by analyzing n = 40 patient blood plasma samples with a Pearson's r ≥ 0.90 for IL-6, IL-8, IL-10, TRAIL, and IP-10 biomarkers. (f–j) DETecT Sepsis sensors capability to significantly distinguish healthy vs. sepsis patient cohorts for IL-6, IL-8 IL-10, TRAIL and IP-10. Note: **p < 0.01, ***P < 0.001, ****P < 0.0001.
4. Discussion
Patients with sepsis can be stratified based on evaluating specific immunological response patterns by a promising approach of cytokine profiling (Marshall, 2006; Ulloa et al., n.d.). Research has been proposed that a combination of biomarkers may yield better results, as no single biomarker exhibits an accuracy of 100% to predict a reliable outcome. Accordingly, in this work, we developed a unique strategy targeting a combination of five pro and anti-inflammatory cytokine biomarkers to rapidly detect sepsis using the DETecT Sepsis sensor to assess the patient's host response useful in a clinical setting. The empirical research approach adopted for this work was based on the evolving knowledge of host response to infection during sepsis. It is well-identified that sepsis fatality is not specifically induced by infectious microorganisms or pathogens; instead, the subsequent pathological outcome is triggered by dysregulation of the host immune response with a combination of pro and anti-inflammatory processes, contributing to multiple organ failure. DETecT Sepsis sensor provides a descriptive understanding of the host immune response with the pentaplex biomarker strategy enabling patient stratification to predict timely evidence on the arc of sepsis. The sensitive and specific aspect combined with a wide dynamic range of the DETecT sensor device platform allows sepsis stratification to differentiate the patient state enabling appropriate therapeutic interventions.
The combination of pro and anti-inflammatory markers (IL-6, IL-8, and IL-10) reveals host immune response during the early stages of sepsis whereas TRAIL and IP-10 provide information to differentiate between bacterial or viral sources of infection. Each biomarker provides key information based on the specific pathophysiology. Timing of cytokines release and the symbiosis between pro- and anti-inflammatory agents determines the degree of infection, and their excessive production can be associated with deleterious effects. A clinical investigation detected a high concentration of cytokines in plasma of critically ill patients affected with COVID-19, suggesting that cytokine storm was associated with the severity of the disease as well (Huang et al., 2020). Cytokine mediators are elevated in both pediatric and adult patients and are responsible for illicit symptoms including fever, hypotension, and production of acute-phase proteins (Banyer et al., 2000; Nedeva et al., 2019). They are likely to be prognostic as primary regulators during the early stage of sepsis (Damas et al., 1992; Viallon et al., 2000). Additionally, elevated IL-6 and IL-8 levels are linked with early 48 h and 28-day mortality in sepsis patients (Bozza et al., 2007). Moreover, IL-8 is a known mediator of the inflammatory response which plays a major role in neutrophil activation. Our results are in agreement with previous research which illustrated the increase of IL-6 and IL-8 during the first 24 h of hospital admission as compared to the healthy cohort (Bozza et al., 2007; Hou et al., 2015; Thao et al., 2018), thus, demonstrating the role of cytokines biomarkers in identifying sepsis prognosis at an early stage. The role of anti-inflammatory biomarker IL-10 during sepsis is complicated as it depends on the time of intervention of either being protective or destructive to the host. Overproduction of IL-10 in septic patients is an indicator of severity and fatal outcome as the body spirals in a state of immunoparalysis. Therefore, IL-10 in the DETecT pentaplex panel acts as an indicator of immune suppression, and thereby reflects the severity of the patient's condition for sepsis-induced immunosuppression. Since the patient samples tested were collected at a 24-h time point, the IL-10 levels were not as significant compared to the healthy cohort, thereby indicating an early stage of sepsis. Our results demonstrating the down-regulated TRAIL profile support the hypothesis that TRAIL participates in sepsis by controlling inflammatory cell apoptosis and promotes inflammation resolution (Renshaw et al., 2003). The presence of TRAIL and IP-10 has been independently used to differentiate viral from bacterial infection. (Oved et al., 2015; van der Does et al, 2016, 2018; van Houten et al., 2017). Moreover, the hypothesis that TRAIL participates in sepsis by controlling inflammatory cell apoptosis and promotes inflammation resolution (Renshaw et al., 2003). Broadly, each selected biomarker demonstrates key elements in activating immune response during sepsis. The approach of multiplexed cytokine profiling would enable rapid sepsis endotyping based on biomarker levels using the DETecT Sepsis sensing platform. Physicians can actively monitor patient status for better prognosis and provide enhanced therapeutic interventions, with multiple measures at patient bedside.
Clinical management of sepsis can be divided into three phases: (i) patient screening; (ii) patient stratification based on evidence-based clinical management and response to treatment therapies; and (iii) prognostic monitoring. As discussed earlier, much effort has been utilized in the first and third phases, while very little progress has been reported with patient stratification methods. Existing methodology lacks the potential to deliver rapid POC results deprived of demanding post-processing to facilitate adequate diagnosis strategies for septic patients. To date, very little work has been done on enabling multiplexed biomarker detection at point-of-care-testing allowing timely treatment. Moreover, transitioning these findings into a clinically feasible test requires a rapid, convenient method that can resolve the lengthy testing process (1–8 h) while providing accurate results. The developed self-integrated sensing device improvements over existing techniques include (i) direct patient sample measurement without the need for sample preparation or dilution (ii) low sample volume utilization (~40 μL) of blood plasma in a portable hand-held format, that could be used to collect data over multiple time points with (iii) rapid results achieved within ~5 min (iv) simultaneous multiplexed detection of cytokine panel biomarkers, for classifying patients depending on the levels reflecting severity of illness (v) Sensitive, selective, specific and stable biosensing response enhances the reliability of the detecting mechanism.
Many researchers have leveraged the use of IL-6, IL-8, IL-10, TRAIL, and IP-10 individually, but to our knowledge, this work is the first demonstration of a simultaneous pentaplex biomarker panel for early sepsis diagnosis and monitoring using low sample volume (<40 μL) achievable rapidly. DETecT Sepsis sensor can be used effectively in the emergency department for early sepsis screening, or it can be used to monitor sepsis prognosis for patients as a bedside monitoring device. Biomarkers linked with sepsis are attributed to the complex immune response pathways, therefore rapid multiplexed detection would enable early therapeutic intervention and improve patient outcomes. Additionally, the sample-to-detection time, measured for all five biomarkers from time of sampling until sensor readout, was ~5 min, which is > 30 times faster than the standard reference method (~5 h). The repeatable and reproducible results demonstrated by the DETecT Sepsis sensor shows evidence of an accurate and reliable electrochemical biosensing mechanism. When tested against common interferants, the specific sensing capability for the developed pentaplex sensor was not affected. Moreover, the sensor displayed a stable response for over five weeks. To our knowledge, no multiplex point-of-treatment device is available for sepsis detection for near-patient testing without sample dilution with rapid response time. Our data shows the first demonstration of a truly novel multiplexed platform capable of monitoring host response using pentaplex biomarkers for sepsis detection that would enable patient stratification and sepsis endotypying. This rapid sample-to-result detection capability demonstrated by the DETecT Sepsis sensor establishes it as a value-added point-of-care testing approach in hospitals and emergency departments towards risk stratification of sepsis severity and responses to treatment.
5. Conclusions
In summary, DETecT Sepsis device platform provides the first proof of concept for rapid diagnostic screening of sepsis leveraging a host immune response biomarker pane. The developed sensor showed comparable results to the reference standard as shown with Pearson's r > 0.97 for all the five biomarkers. However, the developed DETecT sepsis sensor achieved quicker response time with lesser completed assay procedure, leveraging a combination of unique surface engineered sensing strategy coupled with an affinity biosensing principle. The results of this research demonstrate a robust, sensitive, specific, and stable performance by the DETecT Sepsis sensor which is highly expected from a point-of-care device. Moreover, it possesses three significant advantages over current detection methodology. Firstly, a specific affinity-based transduction mechanism allows the simultaneous detection of IL-6, IL-8, IL-10, TRAIL, and IP-10. Multiplexed detection capability will help provide a precise molecular fingerprint of every patient encouraging initiatives towards precision medicine. Secondly, rapid response time offered by the DETecT sensor enables faster decision-making for physicians to operate within the “golden hour” and initiate required treatment thus, avoiding the dynamic sequence of irreversible organ failure and subsequent death caused due to delayed response time. Finally, small form factor and ease in handling allow flexibility in using the device in a versatile environment (in an emergency department, or for bed-side monitoring) while ultra-low sample volume (<40 μL) encourages physicians to collect multiple measurements within a day to monitor patients host immune response and assess the severity of sepsis. The study was limited to a small sample size of septic patients. However, these observations have several implications for research to integrate different host immune response biomarkers with self-integrated point-of-care devices for a better patient outcome. We are currently in the process of expanding our multiplexed capabilities and investigating sepsis pathophysiology via a larger patient cohort. In conclusion, this work has pioneered a potential solution to the existing sepsis dilemma, by providing host response strategy to address complexity, shifting the paradigm of the on-going sepsis diagnostic approach.
CRediT authorship contribution statement
Ambalika S. Tanak: Conceptualization, Resources, Methodology, Validation, Data curation, Investigation, Writing - original draft, Visualization. Sriram Muthukumar: Conceptualization, Resources, Writing - review & editing. Subramaniam Krishnan: Resources, Writing - review & editing. Kevin L. Schully: Resources, Writing - review & editing. Danielle V. Clark: Resources, Writing - review & editing. Shalini Prasad: Conceptualization, Resources, Writing - review & editing.
Declaration of competing interest
Drs. Shalini Prasad and Sriram Muthukumar have a significant interest in EnLiSense LLC, a company that may have a commercial interest in the results of this research and technology. The potential individual conflict of interest has been reviewed and managed by The University of Texas at Dallas, and played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report, or in the decision to submit the report for publication.
Drs. Danielle V Clark and Subramaniam Krishnan report no conflict of interest. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
Dr. Kevin L Schully is an employee of the US government. This work was prepared as a part of his official duties. Title 17 U.S.C. 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of a person's official duties. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Defense Threat Reduction Agency, Department of Defense, nor the U.S. Government.
Acknowledgments
We would like to acknowledge Devang Sankhala for helping with the electronic reader. We would like to thank Yashaswee Tamrakar, Jad Moumen, Thien Nguyen, Fayha Khan, Raqeeb Ali and Bayan Hamed, for their support in performing experiments. We would also like to thank Badrinath Jagannath for providing feedback on the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112726.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Akdis C.A. Nat. Med. 2012 doi: 10.1038/nm.2754. [DOI] [PubMed] [Google Scholar]
- Albert-Vega C., Tawfik D.M., Trouillet-Assant S., Vachot L., Mallet F., Textoris J. 2018. [DOI] [PMC free article] [PubMed]
- Alhazzani W., Møller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., Oczkowski S., Levy M.M., Derde L., Dzierba A., Du B., Aboodi M., Wunsch H., Cecconi M., Koh Y., Chertow D.S., Maitland K., Alshamsi F., Belley-Cote E., Greco M., Laundy M., Morgan J.S., Kesecioglu J., McGeer A., Mermel L., Mammen M.J., Alexander P.E., Arrington A., Centofanti J.E., Citerio G., Baw B., Memish Z.A., Hammond N., Hayden F.G., Evans L., Rhodes A. Intensive Care Med. 2020;46:854–887. doi: 10.1007/s00134-020-06022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson U., Perret-Liaudet A., van Waalwijk van Doorn L.J.C., Blennow K., Chiasserini D., Engelborghs S., Fladby T., Genc S., Kruse N., Kuiperij H.B., Kulic L., Lewczuk P., Mollenhauer B., Mroczko B., Parnetti L., Vanmechelen E., Verbeek M.M., Winblad B., Zetterberg H., Koel-Simmelink M., Teunissen C.E. Front. Neurol. 2015;6 doi: 10.3389/fneur.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., Lee M. JAMA, J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyer J.L., Hamilton N.H.R., Ramshaw I.A., Ramsay A.J. Rev. Immunogenet. 2000 [PubMed] [Google Scholar]
- Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., Greninger A.L., Pipavath S., Wurfel M.M., Evans L., Kritek P.A., West T.E., Luks A., Gerbino A., Dale C.R., Goldman J.D., O'Mahony S., Mikacenic C. N. Engl. J. Med. 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza F.A., Salluh J.I., Japiassu A.M., Soares M., Assis E.F., Gomes R.N., Bozza M.T., Castro-Faria-Neto H.C., Bozza P.T. Crit. Care. 2007;11:R49. doi: 10.1186/cc5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke H.B. 2016. BIC; p. S33380. [DOI] [Google Scholar]
- Chaudhry H., Zhou J., Zhong Y., Ali M.M., Mcguire F., Nagarkatti P.S., Nagarkatti M. 2013. In vivo (Brooklyn) [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Le T.A., Lee H. 2020. [DOI]
- CLSI . approved guideline—third ed. CLSI; Wayne (PA): 2014. Evaluation of Precision of Quantitative Measurement Procedures. [Google Scholar]
- Damas P., Ledoux D., Nys M., Vrindts Y., De Groote D., Franchimont P., Lamy M. Ann. Surg. 1992;215:356–362. doi: 10.1097/00000658-199204000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy A.M., Philippart F., Péan Y., Lasocki S., Charles P.E., Chalumeau M., Claessens Y.E., Quenot J.P., Guen C.G. Le, Ruiz S., Luyt C.E., Roche N., Stahl J.P., Bedos J.P., Pugin J., Gauzit R., Misset B., Brun-Buisson C. 2013. Ann. Intensive Care. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos C.A., Drosou E., Bassaris H.P., Skoutelis A. Pro‐ versus anti‐inflammatory. J. Infect. Dis. 2000;181:176–180. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- Gunsolus I.L., Sweeney T.E., Liesenfeld O., Ledeboer N.A. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.00425-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss R.S., Monneret G., Payen D. Lancet Infect. Dis. 2013 doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Huang D., Zeng R., Ye Z., Zhang Y. J. Clin. Exp. Med. 2015;8:15238–15245. [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath B., Muthukumar S., Prasad S. Anal. Chim. Acta. 2018;1016:29–39. doi: 10.1016/j.aca.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Kim D.K., Fafarman A.T., Diroll B.T., Chan S.H., Gordon T.R., Murray C.B., Kagan C.R. ACS Nano. 2013;7:8760–8770. doi: 10.1021/nn403132x. [DOI] [PubMed] [Google Scholar]
- Marshall J.C. Curr. Infect. Dis. Rep. 2006 doi: 10.1007/s11908-006-0045-1. [DOI] [Google Scholar]
- Nedeva C., Menassa J., Puthalakath H. Front. Cell Dev. Biol. 2019 doi: 10.3389/fcell.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved K., Cohen A., Boico O., Navon R., Friedman T., Etshtein L., Kriger O., Bamberger E., Fonar Y., Yacobov R., Wolchinsky R., Denkberg G., Dotan Y., Hochberg A., Reiter Y., Grupper M., Srugo I., Feigin P., Gorfine M., Chistyakov I., Dagan R., Klein A., Potasman I., Eden E. 2015. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua J., Weng L., Ling L., Egi M., Lim C.M., Divatia J.V., Shrestha B.R., Arabi Y.M., Ng J., Gomersall C.D., Nishimura M., Koh Y., Du B. 2020. Lancet Respir. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw S.A., Parmar J.S., Singleton V., Rowe S.J., Dockrell D.H., Dower S.K., Bingle C.D., Chilvers E.R., Whyte M.K.B. J. Immunol. 2003;170:1027–1033. doi: 10.4049/jimmunol.170.2.1027. [DOI] [PubMed] [Google Scholar]
- Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., Fleischmann-Struzek C., Machado F.R., Reinhart K.K., Rowan K., Seymour C.W., Watson R.S., West T.E., Marinho F., Hay S.I., Lozano R., Lopez A.D., Angus D.C., Murray C.J.L., Naghavi M. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankhala D., Muthukumar S., Prasad S. SLAS Technol. 2018;23:529–539. doi: 10.1177/2472630318759257. [DOI] [PubMed] [Google Scholar]
- Schenck E.J., Ma K.C., Price D.R., Nicholson T., Oromendia C., Gentzler E.R., Sanchez E., Baron R.M., Fredenburgh L.E., Huh J.W., Siempos I.I., Choi A.M.K. 2019. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten M., Wiersinga W.J., Levi M., van der Poll T. J. Leukoc. Biol. 2008;83:536–545. doi: 10.1189/jlb.0607373. [DOI] [PubMed] [Google Scholar]
- Schully K.L., Berjohn C.M., Prouty A.M., Fitkariwala A., Som T., Sieng D., Gregory M.J., Vaughn A., Kheng S., Te V., Duplessis C.A., Lawler J.V., Clark D.V. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schully K.L., Clark D.V. Elsevier; 2019. pp. 105–115. [DOI] [Google Scholar]
- Shanmugam N.R., Muthukumar S., Prasad S. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep33423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Deutschman C.S., Seymour C., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., Poll T. Der, Vincent J.L., Angus D.C. JAMA, J. Am. Med. Assoc. 2016 doi: 10.1001/jama.2016.0287. [DOI] [Google Scholar]
- Tanak A.S., Jagannath B., Tamrakar Y., Muthukumar S., Prasad S. Anal. Chim. Acta X. 2019;3:100029. doi: 10.1016/j.acax.2019.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanak A.S., Muthukumar S., Hashim I.A., Prasad S. Bioelectron. Med. 2019;2:13–27. doi: 10.2217/bem-2019-0011. [DOI] [Google Scholar]
- Thao P.T.N., Tra T.T., Son N.T., Wada K. BMC Emerg. Med. 2018;18 doi: 10.1186/s12873-018-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Tao T., Zhu J., Zou Y., Wang J., Li J., Bo L., Deng X. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa, L., medicine, K.T.-T. in Molecular, 2005, Undefined, n.d., Elsevier.
- van der Does Y., Rood P.P.M., Ramakers C., Schuit S.C.E., Patka P., van Gorp E.C.M., Limper M. Clin. Microbiol. Infect. 2018;24:1297–1304. doi: 10.1016/j.cmi.2018.09.007. [DOI] [PubMed] [Google Scholar]
- van der Does Y., Tjikhoeri A., Ramakers C., Rood P.P.M., van Gorp E.C.M., Limper M. J. Infect. 2016 doi: 10.1016/j.jinf.2016.03.004. [DOI] [PubMed] [Google Scholar]
- van Houten C.B., de Groot J.A.H., Klein A., Srugo I., Chistyakov I., de Waal W., Meijssen C.B., Avis W., Wolfs T.F.W., Shachor-Meyouhas Y., Stein M., Sanders E.A.M., Bont L.J. Lancet Infect. Dis. 2017;17:431–440. doi: 10.1016/S1473-3099(16)30519-9. [DOI] [PubMed] [Google Scholar]
- Viallon A., Zeni F., Pouzet V., Lambert C., Quenet S., Aubert G., Guyomarch S., Tardy B., Bertrand J.C. Intensive Care Med. 2000;26:1082–1088. doi: 10.1007/s001340051321. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Isaacs S.N. J. Immunol. Methods. 2012;384:148–151. doi: 10.1016/j.jim.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.