Abstract
Type 2 diabetes mellitus (T2DM) is a polygenic metabolic disease described by hyperglycemia, which is caused by insulin resistance or reduced insulin secretion. The interaction between various genetic variants and environmental factors triggers T2DM. The aim of this study was to find risk associated with genetic variants rs5210 and rs2237895 of KCNJ11 and KCNQ1 genes, respectively, in the development of T2DM in the Indian population. A total number of 300 cases of T2DM and 100 control samples were studied to find the polymorphism in KCNJ11 and KCNQ1 through PCR-RFLP. The genotype and allele frequencies in T2DM cases were significantly different compared to the control population. KCNJ11 rs5210 and KCNQ1 rs2237895 variants were found to be significantly associated with risk of T2DM in dominant (KCNJ11: OR, 2.07; 95% CI, 1.30–3.27; p − 0.001; KCNQ1: OR, 2.33; 95% CI, 1.46–3.70; p − 0.0003) and codominant models (KCNJ11: OR, 1.76; 95% CI, 1.09–2.84; p − 0.020; KCNQ1: OR, 1.85; 95% CI, 1.16–2.95; p − 0.009). We also compared clinicopathological characteristics between cases and control and observed a significant difference in all the parameters except HDL, gender, and family history. In this study, clinicopathological data with a carrier of a variant allele of both KCNJ11 and KCNQ1 genes were also analysed, and a significant association was found between the carrier of a variant allele with gender and PPG in KCNJ11 and with triglyceride in KCNQ1. We confirm the significant association of KCNJ11 (rs5210) and KCNQ1 (rs2237895) gene polymorphism with T2DM, indicating the role of these variants in developing risk for T2DM in Indian population.
1. Introduction
Type 2 Diabetes is a chronic metabolic disorder described by reduced insulin secretion and insulin action and hypergycemia. Among different types of diabetes, T2DM is the most common wherein multiple environmental and genetic factors are associated with the development of this disease [1, 2]. As per the International Diabetes Federation's (IDF) Diabetes Atlas, 8th edition, it has been estimated that the number of diabetes patients is expected to increase from 425 million in 2017 to 629 million by 2045. India is home to 74 million people with diabetes mellitus, with approximately 8.7% prevalence among the adult population, and considered as the “diabetes capital” of the world (IDF, 8th edition). Genome-wide association studies (GWAS) found various susceptibility genes for diabetes [3–5]. However, the mechanisms these genes follow for the development of diabetes are still not clear. Many gene polymorphism findings demonstrated the association of various SNPs in the pathogenesis of T2DM, and in different populations, each SNP may exhibit dissimilar association with T2DM. Therefore, there is a necessity to discover various genetic risk markers of T2DM for its prevention and treatment.
KCNJ11 and KCNQ1 genes have gained significant attention as a potential candidate for T2DM susceptibility because of their position and function in regulating glucose-stimulated insulin secretion. The KCNJ11 gene is located at 11p15.1 on the human chromosome and lacks intronic sequences; this gene is a member of the potassium channel gene family [6]. The KCNJ11 gene encodes Kir6.2, an inward-rectifier potassium ion channel, which forms a KATP channel with sulfonylurea receptor 1 (SUR1). This KATP channel, through glucose metabolism, controls insulin secretion and production [7]. The KCNQ1 gene is located at the 11th chromosome 11p15.5, approximately 404 kb, and consists of 17 exons [8]. The potassium voltage-gated channel KQT-like subfamily, member 1 (KCNQ1), encodes the pore-forming subunit of KvLQT1, voltage-gated potassium channel, and inhibition of this channel by KCNQ1 inhibitors 293B increased insulin secretion [9], indicating the important role of the KCNQ1 channel in regulating insulin secretion. KCNJ11 and KCNQ1 are ATP-sensitive K+ channels and play crucial role in regulating insulin-secreting β cells making them potential susceptibility genes for T2DM. Therefore, the present case-control study was conducted to assess the association of KCNJ11 (rs5210) and KCNQ1 (rs2237895) gene variants with T2DM risk in Indian population and its association with various clinicopathological characteristics to understand the implications of ethnic diversity on the onset of T2DM.
2. Materials and Methods
2.1. Ethical Approval and Study Subjects
Collection of clinical data in pretext Performa and sample collection of the diabetic patients and control/nondiabetic of North Indian origin from Ansari Health Center Jamia Millia Islamia New Delhi were performed with due consent, after getting Ethical clearance from the Institute Ethical Committee, Jamia Millia Islamia, New Delhi (Proposal No. 17/9/14/JMI/IEC/2015 dated 14/01/2016). The present cohort study was conducted in the Department of Biotechnology, Jamia Millia Islamia, New Delhi, and a total of 400 samples (300 cases and 100 controls) were collected, fulfilling all the relevant selection criteria.
All the study participants had undergone detailed clinical investigation, and written informed consent was collected from the patients before their participation. Various clinicopathological characteristics such as age, BMI, PPG, FPG, FPI, HbA1c, T-cholesterol, systolic BP, diastolic BP, triglycerides, LDL-C,HDL-C, gender, family history, smoking, and alcohol consumption were included in this study.
2.2. Single Nucleotide Polymorphism (SNP) Genotyping by PCR-RFLP
Genomic DNA was extracted from a fresh blood sample using the phenol chloroform method. The primers used for amplification were as follows: KCNJ11 (Forward: 5′- ATCCAGGGTGTTACAAGGCA-3′; reverse: 5′-TTTCAGGGACCAAGTAGAGCTG-3′) and KCNQ1 (Forward 5′-ATCCAGGGTGTTACAAGGCA-3'; reverse 5'- TTTCAGGGACCAAGTAGAGCT -3') in 20 ul of the reaction mixture volume using the Go TAQ Green Master Mix (Promega). For KCNJ11 rs5210, PCR conditions were as follows: an initial denaturation of 95°C for 5 minutes, followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, and final extension of 72°C for 5 min, and for KCNQ1 rs2237895, an initial denaturation of 94°C for 5 minutes, 35 cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 30 s), and final extension of 72°C for 10 m in a PeqSTAR 96 universal gradient thermocycler (Peqlab,VWR). Amplified products were, then, electrophoresed on 2% agarose gel, and images were captured by using the gel documentation system (Biorad).
KCNJ11 (6956A > G) and KCNQ1 (395974 A > C) polymorphism were detected by using a 6 ul PCR product with 2.5 units of restriction enzymes Hpy188III (NEB) and Ava I (NEB), respectively, incubated overnight at 37°C. Hpy188III digested the PCR product of the KCNJ11 gene; wild type allele (A/A) yielded 1 band of 316 bp; heterozygous (A/G) yielded 3 bands of 316 bp, 218 bp, and 98 bp, and risk allele (G/G) yielded two bands of 218 bp and 98 bp (Figure 1(a)), while Ava I digested the PCR product of the KCNQ1 gene; wild type allele (A/A) produced one band of 485 bp; heterozygous (A/C) produced three bands of 485 bp, 294 bp, and 191 bp, and risk allele (C/C) produced two bands of 294 bp and 191 bp (Figure 1(b)). The digested PCR products were subjected to electrophoresis in a 3% agarose gel, as shown in Figures 1(a) and 1(b)
Figure 1.
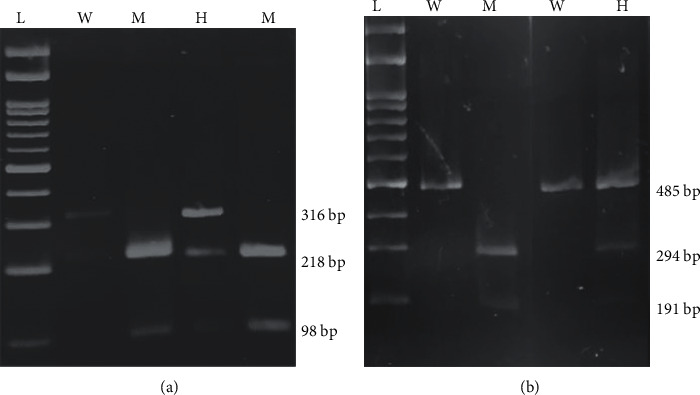
3% agarose gel electrophoresis for the digested PCR product with Hpy188III (NEB) and Ava I (NEB), respectively. (a) KCNJ11, L-100 bp DNA ladder; W- Homo Wild (A/A); M- Homo Mutant (G/G); H- Hetero (A/G) and (b) KCNQ1, L-100 bp DNA ladder; W- Homo Wild (A/A); M- Homo Mutant (C/C); H- Hetero (A/C).
2.3. Statistical Analysis
Statistical analysis was performed by SPSS Software (21.0 Version, IBM, United States). The chi-square test was applied for comparing genotype and allele frequencies for statistical significance between diabetic patients and controls. Data were presented as mean ± SD or as the number of cases. Comparative analysis of clinicopathological characteristics between cases and control was performed by Student's t-test, while the chi-square test was used for gender, family history, smoking, and alcohol consumption. The level of significance was set at 95% (i.e., p < 0.05).
3. Results
3.1. Clinicopathological Characteristics in T2DM Cases and Control
Various clinicopathological characteristics were compared between cases and control by using Student's t-test and the chi-square test. The average age of T2DM cases was (40.33 ± 9.76) years, whereas it was (35.29 ± 7.96) years in controls. Compared to control, T2DM cases had significantly higher BMI, PPG, FPG, FPI, HbA1c, T-cholesterol, systolic BP, diastolic BP, triglycerides, and LDL-C. A significant difference was found in the smoking and alcohol consumption between cases and control, while no significant difference was found in HDL, gender, and family history, as depicted in Table 1 and Figure 2.
Table 1.
Clinicopathological characteristics of T2DM patients and controls.
| Characteristic | T2DM (n = 300) | Control (n = 100) | p value |
|---|---|---|---|
| Age (years) | 40.33 ± 9.76 | 35.29 ± 7.96 | <0.0001∗ |
| BMI (kg/m2) | 28.8 ± 5.22 | 24.83 ± 2.33 | <0.0001∗ |
| PPG (mg/dl) | 208.5 ± 48.49 | 135 ± 13.02 | <0.0001∗ |
| FPG (mg/dl) | 140 ± 35.64 | 90.22 ± 7.11 | <0.0001∗ |
| FPI | 9.6 ± 1.35 | 8.66 ± 0.71 | <0.0001∗ |
| HbA1c | 7.12 ± 1.03 | 5.75 ± 0.54 | <0.0001∗ |
| T-cholesterol (mg/dl) | 245.58 ± 15.14 | 152.63 ± 18.82 | <0.0001∗ |
| Systolic BP (mmHg) | 146.79 ± 17.05 | 106.07 ± 10.39 | <0.0001∗ |
| Diastolic BP (mmHg) | 102.87 ± 16.19 | 75.85 ± 10.91 | <0.0001∗ |
| Triglycerides(mg/dl) | 356.32 ± 100.48 | 140.98 ± 5.52 | <0.0001∗ |
| HDL-C (mg/dl) | 45.89 ± 11.47 | 46.21 ± 8.7 | 0.7985 |
| LDL-C (mg/dl) | 194.60 ± 27.38 | 106.42 ± 19.92 | <0.0001∗ |
| Gender (male/female) | 177/123 | 68/32 | 0.11 |
| Family history (yes/no) | 131/169 | 39/61 | 0.414 |
| Smoking (yes/no) | 139/161 | 15/85 | <0.0001∗ |
| Alcohol consumption (yes/no) | 140/160 | 10/90 | <0.0001∗ |
Data are presented as mean ± SD except gender, family history, smoking, and alcohol consumption. p values were calculated by Student's t-test, while the Chi-square test was used for gender, family history, smoking, and alcohol consumption. ∗Significant at p < 0.05. BMI: body mass index; FPG: fasting plasma glucose; FPI: fasting plasma insulin; HbA1c: haemoglobin A1c HDL: high-density lipoprotein; LDL: low-density lipoprotein.
Figure 2.
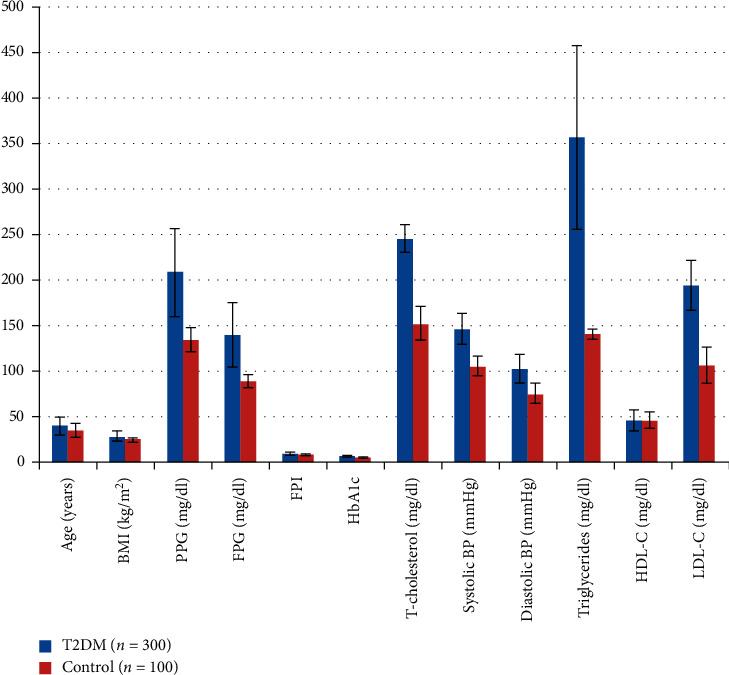
Graphical representation of clinicopathological characteristics of T2DM patients and controls.
3.2. Genotype Distribution and Allelic Frequencies of KCNJ11 and KCNQ1 Genes among T2DM Cases and Controls
The genotype and allele distribution of the KCNJ11 gene is shown in Table 2. The distribution of the genotypes was significantly different between T2DM cases and controls. When the AA genotype was set as the reference, both AG and GG genotypes were found to be associated with increased risk for T2DM (OR, 2.05; 95% CI, 1.25–3.37; p − 0.004 for AG; and OR, 2.12; 95% CI, 0.99–4.5; p − 0.046 for GG, respectively, Table 2). The statistical analysis of the observed genotypic frequencies for KCNJ11 (rs5210) showed a significant association (p − 0.007). The frequency of KCNJ11 rs5210 G allele was significantly higher in cases than in controls (37% vs. 26%, respectively). When the A allele of rs5210 was set as reference, the G allele of KCNJ11 contributed to increased risk of T2DM (OR, 1.67; 95% CI, 1.16–2.38; p − 0.004). We found a significant relationship between KCNJ11 rs5210 A ⟶ G gene polymorphism and T2DM risk under the dominant (OR, 2.07; 95% CI, 1.30–3.27; p − 0.001) and codominant models (OR, 1.76; 95% CI, 1.09–2.84; p − 0.020), whereas no significant relationship was found under the recessive model (OR, 1.54; 95% CI, 0.74–3.20; p − 0.239).
Table 2.
Genotypic and allelic distribution and association analysis of KCNJ11 rs5210 gene polymorphism and risk of T2DM under different genetic models.
| Genotype/allele | Cases (n = 300) | Control (n = 100) | Odds ratio (95% CI) | p value | |
|---|---|---|---|---|---|
| AA | 120 (40%) | 58 (58%) | Ref | Ref | |
| AG | 136 (45.3%) | 32 (32%) | 2.05 (1.25–3.37) | 0.004∗ | |
| GG | 44 (14.6%) | 10 (10%) | 2.12 (0.99–4.5) | 0.046∗ | |
| p value −0.007∗ | |||||
| Recessive model | GG | 44 | 10 | 1.54 (0.74–3.20) | 0.239 |
| AG + AA | 256 | 90 | |||
| Dominant model | AG + GG | 180 | 42 | 2.07 (1.30–3.27) | 0.001∗ |
| AA | 120 | 58 | |||
| Codominant model | AG | 136 | 32 | 1.76 (1.09–2.84) | 0.020∗ |
| AA + GG | 164 | 68 | |||
| Allele | |||||
| A | 378 (63%) | 148 (74%) | 1.67(1.16–2.38) | 0.004∗ | |
| G | 222 (37%) | 52 (26%) | |||
OR: odds ratio, CI: confidence interval, n: number in sample.∗Significant at p < 0.05.
The genotype and allele distribution of the KCNQ1 gene are shown in Table 3. The distribution of the genotypes was significantly different between T2DM cases and controls. When the AA genotype was set as the reference, both AC and CC genotypes were found to be associated with increased risk for T2DM (OR, 2.36; 95% CI, 1.43–3.89; p − 0.0007 for AC; and OR, 2.26; 95% CI, 1.14–4.46; p − 0.017 for CC). The statistical analysis of the observed genotypic frequencies for KCNQ1 showed a significant association (p − 0.001). The frequency of KCNQ1 rs2237895C allele was significantly higher in cases than in controls (44% vs. 32%, respectively). When the A allele of rs2237895 was set as reference, the C allele of KCNQ1 contributed to the increased risk of T2DM (OR, 1.66; 95% CI, 1.19–2.34; p − 0.003). We found a significant relationship between KCNQ1 rs2237895 A⟶C gene polymorphism and T2DM risk under the dominant model (OR, 2.33; 95% CI, 1.46–3.70; p − 0.0003) and codominant model (OR, 1.85; 95% CI, 1.16–2.95; p − 0.009), whereas no significant relationship was found under the recessive model (OR, 1.44; 95% CI, 0.76–2.71; p − 0.259).
Table 3.
Genotypic and allelic distribution and association analysis of KCNQ1 rs2237895 genes polymorphism and risk of T2DM under different genetic models.
| Genotype | Cases (n = 300) | Control (n = 100) | Odds ratio (95% CI) | p value | |
|---|---|---|---|---|---|
| AA | 90 (30%) | 50 (50%) | Ref | Ref | |
| AC | 153 (51%) | 36 (36%) | 2.36 (1.43–3.89) | 0.0007∗ | |
| CC | 57 (19%) | 14 (14%) | 2.26 (1.14–4.46) | 0.017∗ | |
| p value −0.001∗ | |||||
| Recessive model | CC | 57 | 14 | 1.44 (0.76–2.71) | 0.259 |
| AC + AA | 243 | 86 | |||
| Dominant model | AC + CC | 210 | 50 | 2.33 (1.46–3.70) | 0.0003∗ |
| AA | 90 | 50 | |||
| Codominant model | AC | 153 | 36 | 1.85 (1.16–2.95) | 0.009∗ |
| AA + CC | 147 | 64 | |||
| Allele | |||||
| A | 336 (56%) | 136 (68%) | 1.66 (1.19 to 2.34) | 0.003∗ | |
| C | 264 (44%) | 64 (32%) | |||
3.3. Observed and Expected Genotypes of KCNJ11 and KCNQ1 Polymorphism in Control and Cases
The observed and expected genotype frequencies of KCNJ11 and KCNQ1 gene polymorphism in controls and cases showed no deviation from the Hardy–Weinberg equilibrium. The chi-square test demonstrated that there was no significant deviation from the Hardy–Weinberg equilibrium for KCNJ11 and KCNQ1 SNP genotypes (p > 0.05), as shown in Table 4.
Table 4.
Observed and expected genotypes of KCNJ11 and KCNQ1 polymorphism in control and cases.
| Genotype | Observed genotype | Expected genotype | p value | Chi square (X2) |
|---|---|---|---|---|
| Control | ||||
|
| ||||
| KCNJ11 rs5210 | ||||
|
| ||||
| AA | 58 | 54.76 | 0.2422 | 2.836 |
| AG | 32 | 38.48 | ||
| GG | 10 | 6.76 | ||
|
| ||||
| KCNQ1 rs2237895 | ||||
|
| ||||
| AA | 50 | 46.24 | 0.2247 | 2.986 |
| AC | 36 | 43.52 | ||
| CC | 14 | 10.24 | ||
|
| ||||
| Cases | ||||
|
| ||||
| KCNJ11 rs5210 | ||||
| AA | 120 | 117.8133 | 0.8645 | 0.2912 |
| AG | 136 | 140.3733 | ||
| GG | 44 | 41.8133 | ||
| KCNQ1 rs2237895 | ||||
| AA | 90 | 92.4075 | 0.8535 | 0.3167 |
| AC | 153 | 148.185 | ||
| CC | 57 | 59.4075 | ||
3.4. Association of the Clinicopathological Characteristic of Type 2 Diabetes Patients with the Carrier of a Variant Allele of KCNJ11 and KCNQ1 Genes
We analysed the clinicopathological data with the carrier of a variant allele (AG + GG) of the KCNJ11 gene and found out that there was no statistically significant association between the carrier of a variant allele with age, family history, smoking, alcohol consumption, BMI, FPG, HB1Ac, systolic BP, diastolic BP, cholesterol, FPI, triglyceride, HDL, and LDL. We observed a significant association of gender and PPG with the carrier of a variant allele (AG + GG), as shown in Table 5 and Figure 3 (the result is significant at p > 0.05).
Table 5.
Clinicopathological characteristics of T2DM patients between wild and carrier of a variant allele of the KCNJ11 gene.
| Characteristic | Total | Wild (AA) | Carrier (AG + GG) | p value |
|---|---|---|---|---|
| Age | ||||
| ≤45 | 183 | 77 | 106 | 0.358 |
| >45 | 117 | 43 | 74 | |
|
| ||||
| Gender | ||||
| Male | 177 | 84 | 93 | 0.0015∗ |
| Female | 123 | 36 | 87 | |
|
| ||||
| Family history | ||||
| Yes | 131 | 49 | 82 | 0.419 |
| No | 169 | 71 | 98 | |
|
| ||||
| Smoking | ||||
| Yes | 139 | 59 | 80 | 0.421 |
| No | 161 | 68 | 100 | |
|
| ||||
| Alcohol consumption | ||||
| Yes | 140 | 62 | 78 | 0.156 |
| No | 160 | 5 | 102 | |
|
| ||||
| BMI | ||||
| ≤25 | 86 | 34 | 52 | 0.916 |
| >25 | 214 | 83 | 128 | |
|
| ||||
| FPG | ||||
| ≤140 | 214 | 87 | 127 | 0.715 |
| >140 | 86 | 3 | 53 | |
|
| ||||
| PPG | ||||
| ≤208 | 171 | 77 | 94 | 0.040∗ |
| >208 | 129 | 43 | 86 | |
|
| ||||
| HBA1C | ||||
| ≤7 | 192 | 75 | 117 | 0.658 |
| >7 | 108 | 45 | 63 | |
|
| ||||
| Systolic BP | ||||
| ≤146 | 128 | 46 | 82 | 0.215 |
| >146 | 172 | 74 | 98 | |
|
| ||||
| Diastolic BP | ||||
| ≤102 | 206 | 90 | 116 | 0.053 |
| >102 | 94 | 30 | 64 | |
|
| ||||
| Cholesterol | ||||
| ≤245 | 192 | 84 | 108 | 0.077 |
| >245 | 108 | 36 | 72 | |
|
| ||||
| FPI | ||||
| ≤9 | 135 | 52 | 83 | 0.635 |
| >9 | 165 | 68 | 97 | |
|
| ||||
| Triglyceride | ||||
| ≤355 | 156 | 66 | 90 | 0.395 |
| >355 | 144 | 54 | 90 | |
|
| ||||
| HDL-C | ||||
| ≤45 | 189 | 77 | 112 | 0.732 |
| >45 | 111 | 43 | 68 | |
|
| ||||
| LDL-C | ||||
| ≤194 | 93 | 39 | 54 | 0.646 |
| >194 | 207 | 81 | 126 | |
p values were determined by the chi-square test. ∗Significant association at p > 0.05.
Figure 3.
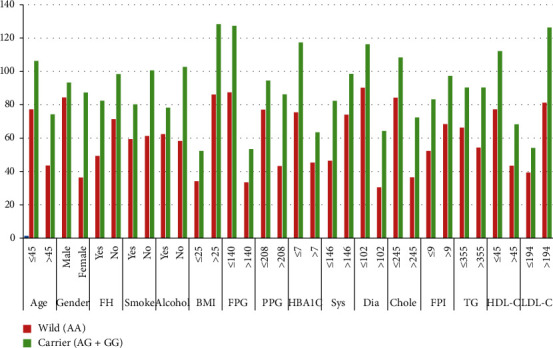
Graphical representation of clinicopathological characteristics of T2DM patients between wild and carrier of a variant allele of the KCNJ11 gene.
Similarly, clinicopathological data with the carrier of a variant allele (AC + CC) of the KCNQ1 gene were analysed, and we found out that there was no statistically significant association between the carrier of a variant allele with age, gender, family history, smoking, alcohol consumption, BMI, FPG, PPG, HB1Ac, systolic BP, diastolic BP, cholesterol, FPI, HDL-C, and LDL-C. We found a significant association of triglyceride with the carrier of a variant allele (AC + CC), as shown in Table 6 and Figure 4 (the result is significant at p > 0.05).
Table 6.
Clinicopathological characteristics of T2DM patients between wild and carrier of a variant allele of the KCNQ1 gene.
| Characteristic | Total | Wild (AA) | Carrier (AC + CC) | p value |
|---|---|---|---|---|
| Age | ||||
| ≤45 | 183 | 56 | 127 | 0.77 |
| >45 | 117 | 34 | 83 | |
|
| ||||
| Gender | ||||
| Male | 177 | 59 | 118 | 0.130 |
| Female | 123 | 31 | 92 | |
|
| ||||
| Family history | ||||
| Yes | 131 | 39 | 92 | 0.939 |
| No | 169 | 51 | 118 | |
|
| ||||
| Smoking | ||||
| Yes | 139 | 42 | 97 | 0.939 |
| No | 161 | 48 | 113 | |
|
| ||||
| Alcohol consumption | ||||
| Yes | 140 | 45 | 95 | 0.448 |
| No | 160 | 45 | 115 | |
|
| ||||
| BMI | ||||
| ≤25 | 86 | 27 | 59 | 0.738 |
| >25 | 214 | 63 | 151 | |
|
| ||||
| FPG | ||||
| ≤140 | 214 | 64 | 150 | 0.955 |
| >140 | 86 | 26 | 60 | |
|
| ||||
| PPG | ||||
| ≤208 | 171 | 50 | 121 | 0.740 |
| >208 | 129 | 40 | 89 | |
|
| ||||
| HBA1C | ||||
| ≤7 | 192 | 62 | 130 | 0.248 |
| >7 | 108 | 28 | 80 | |
|
| ||||
| Systolic BP | ||||
| ≤146 | 128 | 33 | 95 | 0.168 |
| >146 | 172 | 57 | 115 | |
|
| ||||
| Diastolic BP | ||||
| ≤102 | 206 | 62 | 144 | 0.956 |
| >102 | 94 | 28 | 66 | |
|
| ||||
| Cholesterol | ||||
| ≤245 | 192 | 63 | 129 | 0.156 |
| >245 | 108 | 27 | 81 | |
|
| ||||
| FPI | ||||
| ≤9 | 135 | 42 | 93 | 0.704 |
| >9 | 165 | 48 | 117 | |
|
| ||||
| Triglyceride | ||||
| ≤355 | 156 | 36 | 120 | 0.006∗ |
| >355 | 144 | 54 | 90 | |
|
| ||||
| HDL-C | ||||
| ≤45 | 189 | 57 | 132 | 0.937 |
| >45 | 111 | 33 | 78 | |
|
| ||||
| LDL-C | ||||
| ≤194 | 93 | 29 | 64 | 0.764 |
| >194 | 207 | 61 | 146 | |
p values were determined by the chi-square test. ∗Significant association at p > 0.05.
Figure 4.
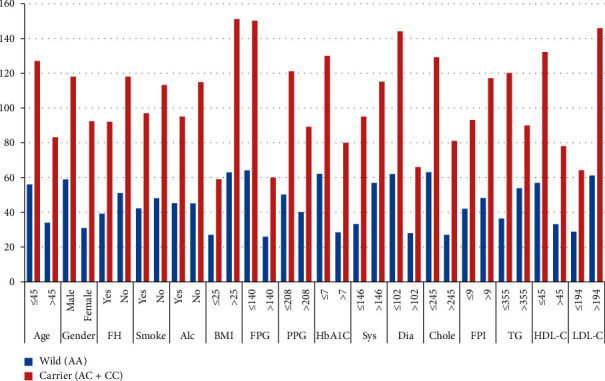
Graphical representation of clinicopathological characteristics of T2DM patients between wild and carrier of a variant allele of the KCNQ1 gene.
3.5. Combined Effect of KCNJ11 + KCNQ1 Genotypes on Clinicopathological Parameters
The effect of KCNJ11 + KCNQ1 genotypes together had a significant impact on the regulation of PPG, FPG, cholesterol, LDL, and diastolic BP (Table 7).
Table 7.
Combined effect of KCNJ11 + KCNQ1 genotypes on clinicopathological parameters.
| Parameters | Genotype variables KCNJ11 + KCNQ1 | Mean ± SD | p value |
|
| |||
| BMI | (AA + AA) | 27.79 ± 4.47 | 0.069 |
| (AG + AC) | 29.24 ± 5.07 | ||
| (GG + CC) | 25.99 ± 3.37 | ||
| (AG + CC) | 27.88 ± 5.49 | ||
| (AG + AA) | 27.45 ± 4.8 | ||
| (GG + AA) | 27.47 ± 5.00 | ||
|
| |||
| PPG | (AA + AA) | 177.41 ± 53.14 | 0.003∗ |
| (AG + AC) | 193.88 ± 39.85 | ||
| (GG + CC) | 177.59 ± 47.63 | ||
| (AG + CC) | 210.09 ± 68.61 | ||
| (AG + AA) | 184.07 ± 46.72 | ||
| (GG + AA) | 196.50 ± 62.67 | ||
|
| |||
| FPG | (AA + AA) | 121.12 ± 40.06 | 0.030∗ |
| (AG + AC) | 132.55 ± 38.45 | ||
| (GG + CC) | 120.45 ± 26.15 | ||
| (AG + CC) | 132.07 ± 43.79 | ||
| (AG + AA) | 122.18 ± 30.65 | ||
| (GG + AA) | 134.38 ± 42.72 | ||
|
| |||
| FPI | (AA + AA) | 9.04 ± 1.20 | 0.29 |
| (AG + AC) | 9.54 ± 1.41 | ||
| (GG + CC) | 8.98 ± 1.14 | ||
| (AG + CC) | 9.42 ± 1.24 | ||
| (AG + AA) | 9.39 ± 1.23 | ||
| (GG + AA) | 9.42 ± 1.40 | ||
|
| |||
| HbA1C | (AA + AA) | 6.57 ± 1.10 | 0.238 |
| (AG + AC) | 6.94 ± 1.12 | ||
| (GG + CC) | 6.59 ± 0.93 | ||
| (AG + CC) | 6.79 ± 1.08 | ||
| (AG + AA) | 6.74 ± 1.08 | ||
| (GG + AA) | 6.92 ± 1.24 | ||
|
| |||
| Cholesterol | (AA + AA) | 211.97 ± 46.99 | 0.009∗ |
| (AG + AC) | 230.80 ± 38.24 | ||
| (GG + CC) | 218.63 ± 52.46 | ||
| (AG + CC) | 234.34 ± 36.31 | ||
| (AG + AA) | 216.69 ± 44.70 | ||
| (GG + AA) | 223.84 ± 45.10 | ||
|
| |||
| HDL | (AA + AA) | 46.69 ± 9.18 | 0.117 |
| (AG + AC) | 45.03 ± 10.48 | ||
| (GG + CC) | 47.72 ± 9.52 | ||
| (AG + CC) | 45.39 ± 12.97 | ||
| (AG + AA) | 46.80 ± 10.20 | ||
| (GG + AA) | 44.23 ± 12.98 | ||
|
| |||
| LDL | (AA + AA) | 159.88 ± 48.49 | 0.005∗ |
| (AG + AC) | 182.14 ± 41.31 | ||
| (GG + CC) | 170.90 ± 53.74 | ||
| (AG + CC) | 183.42 ± 41.27 | ||
| (AG + AA) | 166.94 ± 47.61 | ||
| (GG + AA) | 176.92 ± 45.11 | ||
|
| |||
| Triglycerides | (AA + AA) | 277.84 ± 133.61 | 0.45 |
| (AG + AC) | 317.14 ± 125.36 | ||
| (GG + CC) | 274.09 ± 109.46 | ||
| (AG + CC) | 336.88 ± 123.27 | ||
| (AG + AA) | 289.20 ± 128.33 | ||
| (GG + AA) | 313.30 ± 120.98 | ||
|
| |||
| Systolic BP | (AA + AA) | 132.08 ± 24.79 | 0.28 |
| (AG + AC) | 138.63 ± 21.56 | ||
| (GG + CC) | 133.36 ± 25.96 | ||
| (AG + CC) | 141.96 ± 22.52 | ||
| (AG + AA) | 134.87 ± 24.31 | ||
| (GG + AA) | 138.53 ± 23.17 | ||
|
| |||
| Diastolic BP | (AA + AA) | 93.84 ± 22.38 | 0.010∗ |
| (AG + AC) | 100.10 ± 17.54 | ||
| (GG + CC) | 95.45 ± 21.38 | ||
| (AG + CC) | 100.39 ± 16.99 | ||
| (AG + AA) | 93.22 ± 19.21 | ||
| (GG + AA) | 94.87 ± 15.87 | ||
∗Significant at p > 0.05.
3.6. Regression Analysis: Dependent Variable Cases and Control
Regression analysis was performed to check the involvement of other variables with the occurrence of T2DM. It was observed that the variables such as age, BMI, PPG, FPG,FPI, HbA1C, cholesterol, LDL, systolic BP, diastolic BP, and triglycerides showed a significant association with the study subject and R2 showed an association with T2DM (Table 8), while no association was observed with gender and HDL.
Table 8.
Regression analysis: dependent variable cases and control.
| Variables | R 2 | p value |
|---|---|---|
| Age | 0.05 | <0.0001 |
| Gender | 0.006 | 0.11 |
| BMI | 0.119 | <0.0001 |
| PPG | 0.360 | <0.0001 |
| FPG | 0.317 | <0.0001 |
| FPI | 0.099 | <0.0001 |
| HbA1C | 0.290 | <0.0001 |
| Cholesterol | 0.862 | <0.0001 |
| HDL | 0.00 | 0.80 |
| LDL | 0.689 | <0.0001 |
| Systolic BP | 0.560 | <0.0001 |
| Diastolic BP | 0.378 | <0.0001 |
| Triglycerides | 0.535 | <0.0001 |
| Cases and controls as dependent variables | ||
∗Significant at p > 0.05.
4. Discussion
In the present case-control study, we investigated the association of SNPs (rs5210) and (rs2237895) within the KCNJ11 and KCNQ1 genes, respectively, with the susceptibility to T2DM in Indian population. The KCNJ11 polymorphism association with T2DM risk has been extensively studied among European population; however, their relationship in the Indian subcontinent is yet to be validated. Various GWAS also demonstrated that several KCNQ1 variants (i.e., rs2237892, rs2237895, and rs2237897) are associated with T2DM risk and reduced insulin secretion in Europeans, Asians, and American Indians [10–12]. However, studies in different races and regions have demonstrated different findings. It has also been stated that Indians might have different genetic predisposition to diabetes than Europeans [13]. In this study, we compared various clinicopathological characteristics between cases and control and observed a significant difference in all the parameters except HDL, gender, and family history. Dorman and Bunker. suggested that people with a T2DM family history are three times more susceptible to this disease [14]. However, in our study, we have not observed any association of family history with T2DM risk between cases and control. However, family history is considered as a strong and independent risk factor for diabetes [15–19].
In humans, the significance of KCNJ11 in insulin secretion was suggested by its function in permanent neonatal diabetes [20] and familial persistent hyperinsulinemic hypoglycaemia of infancy [21]. KCNJ11 is considered as a promising candidate susceptibility gene for T2DM due to the protein encoded by KCNJ11, which is crucial for pancreatic beta-cell function. In the present study, we observed a significant difference in the distribution of KCNJ11 genotypes among T2DM cases and healthy controls and higher risk allele distribution was observed among cases as compared to healthy controls. Also, the KCNJ11 rs5210 variant demonstrated a significant association between T2DM cases and controls under dominant and codominant models. Our findings indicated an association of the KCNJ11 rs5210 polymorphism with T2DM in Indian patients. Our results were found to be similar to other studies which confirmed the association of KCNJ11 variant 3p + 215 (rs5210) with T2DM [22, 23]. Similarly, meta-analysis in the East Asian population and genotypic and allelic contrast also suggested a significant association of KCNJ11 and T2DM for rs5210 [24]. Furthermore, the rs5210 variant of the KCNJ11 gene was indicated to be related with T2DM risk in meta-analysis of 5 studies, and it was significantly heterogeneous (p=0.02) [25]. Similar results were also observed among the Mexican, Finnish, and Korean populations [26–28]. Our results were found to be inconsistent with other studies in which no association was found between KCNJ11 E23K polymorphism and T2DM in Iranian [29], Czech [30], Moroccan [31], and Mongolian population [32].
KCNQ1 comprises of 676 amino acids with 1 ion-selective P loop and 6 transmembrane regions. The P loop is composed of 4 identical α subunits, which is an ion-filter duct. It consists of a porous structure which is highly conservative leading to higher selectivity to potassium. The KCNQ1 knockout mouse showed prominent increase in insulin sensitivity, suggesting that KCNQ1 may act as a novel element affecting insulin sensitivity through glucose metabolism [33]. In our study, the genotype and allele distribution of KCNQ1 rs2237895 showed a significant difference between T2DM cases and controls and higher risk allele distribution was observed among cases as compared to healthy controls. Also, the KCNQ1 rs2237895 variant showed a significant association between T2DM cases and controls under dominant and codominant models. Our findings indicated an association of the KCNQ1 rs2237895 polymorphism with T2DM in Indian patients. Our findings were found to be consistent with the previous study in which KCNQ1 gene polymorphism (rs2074196, rs2237892, rs2237895, rs2283228, and rs2237897) was linked with T2DM risk among Japanese populations in 2 independent genome-wide association studies [3, 10]. Furthermore, in European population, according to GWA scans and replication studies, SNPs (rs151290, rs2237892, and rs2237895) of the KCNQ1 were indicated to be a genetic risk factor for T2DM [3, 10, 34–36]. In addition, many previous studies proposed that various variants of KCNQ1 rs2237895, rs2237892, rs2237897, and rs151290 showed an association with T2DM in China [37], Germany [35], Pakistan, and the Netherlands [38, 39].
In East Asian origin patients, the KCNQ1 gene has been found to play a significant role in contributing to T2D susceptibility [3, 10, 40]. Some studies were contradictory to our finding which did not find any association of KCNQ1 variants (rs2237892 and rs2273895) and T2DM risk [41, 42]. A study on the South Indian population from Hyderabad also showed a significant association of KCNJ11 (rs5210) and KCNQ1 (rs2283228) with T2DM risk, which corroborates with our results [43]. However, we did not come across any such study investigating the association of KCNJ11 (rs5210) and KCNQ1 (rs2237895) gene polymorphism with T2DM risk in North Indian origin patients.
Additionally, we analysed clinicopathological data with the carrier of a variant allele of KCNJ11 and KCNQ1. We found an association between the carrier of a variant allele with gender and PPG in KCNJ11 and with triglyceride in KCNQ1.Chen et al. also suggested that KCNQ1 was associated with triglyceride levels in the Chinese Han population [44].This study will help in understanding the genetic background of T2DM, which is very essential for the identification of high-risk individuals and to include prevention plan in these high-risk individuals and general population.
5. Conclusions
In the present study, we identify a significant association of KCNJ11 (rs5210) and KCNQ11 (rs2237895) gene polymorphism with T2DM risk, suggesting the role of these variants in the increased risk of developing T2DM in the Indian population. We also found that the G allele of rs5210 and C allele of rs2237895 polymorphism of KCNJ11 and KCNQ1, respectively, are the major risk factors that confer susceptibility to T2DM. Furthermore, these genetic variants will enable us to find the risk prediction of T2DM development and its underline pathogenesis, which will be applicable for individualized treatment of T2DM.
Acknowledgments
The authors would like to acknowledge all the study participants who were included in this study. VK would like to acknowledge DBT for providing fellowship, under the DBT JRF/SRF scheme.
Abbreviations
- IDF:
International Diabetes Federation
- KCNJ11:
Potassium inwardly rectifying channel, subfamily J, member 11
- KCNQ1:
Potassium voltage-gated channel KQT-like subfamily, member 1
- GWAS:
Genome-wide association studies
- PCR-RFLP:
Polymerase chain reaction-restriction fragment length polymorphism
- SUR1:
Sulfonylurea receptor 1
- KATP:
ATP- sensitive K channel
- OR:
Odds ratio
- CI:
Confidence interval
- HWE:
Hardy–Weinberg equilibrium
- SNP:
Single nucleotide polymorphism.
Data Availability
We confirm that the data used during the research will not be shared with anybody or broadcasted in any public domain, since it is impermissible as per the policy instructions of JMI. The metadata, supporting the study outcomes, can be accessed from JMI through proper consent; however, privileged data with restricted open accessibility, cedes institutional authorization, and discretion and other related information and data can also be retrieved from the authors with permission of JMI.
Additional Points
Highlights. (i) The genotype and allele frequencies in type 2 diabetes cases were significantly different compared to the control population. (ii) Clinicopathological parameters were analysed, and a significant association was found between the carrier of a variant allele with gender and PPG in KCNJ11 and with triglyceride in KCNQ1. (iii) KCNJ11 (rs5210) and KCNQ1 (rs2237895) gene variants are significantly associated with type 2 diabetes in the Indian population.
Ethical Approval
Institutional Ethics Committee of Jamia Millia Islamia (Proposal No. 17/9/14/JMI/IEC/2015 dated 14/01/2016) approved this study.
Consent
Written informed consent was obtained before inclusion in the study.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Amit Kumar Verma, Vasiuddin Khan, and Deepti Bhatt equally contributed to this work. Conceptualization was carried out by KD, AKV and MYS; VK, AKV, and SK were responsible for the methodology; DB, AKV, and AHR were responsible for the software; validation was performed by AKV, YG, RH, and MAA; formal Analysis was performed by VK, SK, and AKV; data Curation was performed by VK, AKV, and DB; VK, DB, and AKV wrote the original draft; review and editing were performed by YG, SR, AA, BM, AKV, and KD; visualization was performed by BM, AKV, and KD; and supervision was performed by KD.
References
- 1.Cho Y. S., Lee J.-Y., Park K. S., Nho C. W. Genetics of type 2 diabetes in East Asian populations. Current Diabetes Reports. 2012;12(6):686–696. doi: 10.1007/s11892-012-0326-z. [DOI] [PubMed] [Google Scholar]
- 2.Ali O. Genetics of type 2 diabetes. World Journal of Diabetes. 2013;4(4):p. 114. doi: 10.4239/wjd.v4.i4.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasuda K., Miyake K., Horikawa Y., et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nature Genetics. 2008;40(9):1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 4.Voight B. F., Scott L. J., Steinthorsdottir V., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi T., Hara K., Maeda S., et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nature Genetics. 2010;42(10):864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 6.Haghvirdizadeh P., Mohamed Z., Abdullah N. A., Haghvirdizadeh P., Haerian M. S., Haerian B. S. KCNJ11: genetic polymorphisms and risk of diabetes mellitus. Journal of Diabetes Research. 2015;2015:9. doi: 10.1155/2015/908152.908152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McTaggart J. S., Clark R. H., Ashcroft F. M. Symposium review: the role of the KATPchannel in glucose homeostasis in health and disease: more than meets the islet. The Journal of Physiology. 2010;588(17):3201–3209. doi: 10.1113/jphysiol.2010.191767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y.-H., Kang E. S., Kim S. H., et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. Journal of Human Genetics. 2008;53(11-12):991–998. doi: 10.1007/s10038-008-0341-8. [DOI] [PubMed] [Google Scholar]
- 9.Ullrich S., Su J., Ranta F., et al. Effects of IKs channel inhibitors in insulin-secreting INS-1 cells. Pflügers Archiv–European Journal of Physiology. 2005;451(3):428–436. doi: 10.1007/s00424-005-1479-2. [DOI] [PubMed] [Google Scholar]
- 10.Unoki H., Takahashi A., Kawaguchi T., et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nature Genetics. 2008;40(9):1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 11.Hanson R. L., Guo T., Muller Y. L., et al. Strong parent-of-origin effects in the association ofKCNQ1Variants with type 2 diabetes in American Indians. Diabetes. 2013;62(8):2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y.-y., Wang X.-m., Lu X.-z. KCNQ1rs2237892 C⟶T gene polymorphism and type 2 diabetes mellitus in the Asian population: a meta-analysis of 15,736 patients. Journal of Cellular and Molecular Medicine. 2014;18(2):274–282. doi: 10.1111/jcmm.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chauhan G., Spurgeon C. J., Tabassum R., et al. Impact of common variants ofPPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, andCDKAL1on the risk of type 2 diabetes in 5,164 Indians. Diabetes. 2010;59(8):2068–2074. doi: 10.2337/db09-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman J. S., Bunker C. H. HLA-DQ locus of the human leukocyte antigen complex and type 1 diabetes mellitus: a huge review. Epidemiologic Reviews. 2000;22(2):218–227. doi: 10.1093/oxfordjournals.epirev.a018034. [DOI] [PubMed] [Google Scholar]
- 15.Harrison T. A., Hindorff L. A., Kim H., et al. Family history of diabetes as a potential public health tool. American Journal of Preventive Medicine. 2003;24(2):152–159. doi: 10.1016/s0749-3797(02)00588-3. [DOI] [PubMed] [Google Scholar]
- 16.O’Rahilly S., Barroso I., Wareham N. J. Genetic factors in type 2 diabetes: the end of the beginning? Science. 2005;307(5708):370–373. doi: 10.1126/science.1104346. [DOI] [PubMed] [Google Scholar]
- 17.Newell A. M. Genetics for targeting disease prevention: diabetes. Primary Care: Clinics in Office Practice. 2004;31(3):743–766. doi: 10.1016/j.pop.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Arslanian S. A., Bacha F., Saad R., Gungor N. Family history of type 2 diabetes is associated with decreased insulin sensitivity and an impaired balance between insulin sensitivity and insulin secretion in white youth. Diabetes Care. 2005;28(1):115–119. doi: 10.2337/diacare.28.1.115. [DOI] [PubMed] [Google Scholar]
- 19.Annis A. M., Caulder M. S., Cook M. L., Duquette D. Family History, diabetes, and other demographic and risk factors among participants of the national health and nutrition examination survey 1999–2002. Preventing Chronic Disease. 2005;2(2)1327713 [PMC free article] [PubMed] [Google Scholar]
- 20.Gloyn A. L., Pearson E. R., Antcliff J. F., et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. New England Journal of Medicine. 2004;350(18):1838–1849. doi: 10.1056/nejmoa032922. [DOI] [PubMed] [Google Scholar]
- 21.Aguilar-Bryan L., Bryan J. Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocrine Reviews. 1999;20(2):101–135. doi: 10.1210/er.20.2.101. [DOI] [PubMed] [Google Scholar]
- 22.Florez J. C., Burtt N., De Bakker P. I. W., et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53(5):1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 23.Barroso I., Luan J., Middelberg R. P., et al. Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS Biology. 2003;1(1) doi: 10.1371/journal.pbio.0000020.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L., Zhou X., Luo Y., et al. Association between KCNJ11 gene polymorphisms and risk of type 2 diabetes mellitus in East Asian populations: a meta-analysis in 42,573 individuals. Molecular Biology Reports. 2012;39(1):645–659. doi: 10.1007/s11033-011-0782-6. [DOI] [PubMed] [Google Scholar]
- 25.Qin L. J., Lv Y., Huang Q. Y. Meta-analysis of association of common variants in the KCNJ11-ABCC8 region with type 2 diabetes. Genetics and Molecular Research. 2013;12(3):2990–3002. doi: 10.4238/2013.august.20.1. [DOI] [PubMed] [Google Scholar]
- 26.Koo B. K., Cho Y. M., Park B. L., et al. Polymorphisms ofKCNJ11(Kir6.2 gene) are associated with Type 2 diabetes and hypertension in the Korean population. Diabetic Medicine. 2007;24(2):178–186. doi: 10.1111/j.1464-5491.2006.02050.x. [DOI] [PubMed] [Google Scholar]
- 27.Willer C. J., Bonnycastle L. L., Conneely K. N., et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes. 2007;56(1):256–264. doi: 10.2337/db06-0461. [DOI] [PubMed] [Google Scholar]
- 28.Cruz M., Valladares-Salgado A., Garcia-Mena J., et al. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes/metabolism Research and Reviews. 2010;26(4):261–270. doi: 10.1002/dmrr.1082. [DOI] [PubMed] [Google Scholar]
- 29.Keshavarz P., Habibipour R., Ghasemi M., Kazemnezhad E., Alizadeh M., Omami M. H. H. Lack of genetic susceptibility of KCNJ11 E23K polymorphism with risk of type 2 diabetes in an Iranian population. Endocrine Research. 2014;39(3):120–125. doi: 10.3109/07435800.2013.860607. [DOI] [PubMed] [Google Scholar]
- 30.Čejková P., Novota P., Černá M., et al. KCNJ11 E23K polymorphism and diabetes mellitus with adult onset in Czech patients. Folia Biologica. 2007;53:173–175. doi: 10.14712/fb2007053050173. [DOI] [PubMed] [Google Scholar]
- 31.Benrahma H., Charoute H., Lasram K., et al. Association analysis of IGF2BP2, KCNJ11, and CDKAL1 polymorphisms with type 2 diabetes mellitus in a Moroccan population: a case–control study and meta-analysis. Biochemical Genetics. 2014;52(9-10):430–442. doi: 10.1007/s10528-014-9658-5. [DOI] [PubMed] [Google Scholar]
- 32.Odgerel Z., Lee H. S., Erdenebileg N., et al. Genetic variants in potassium channels are associated with type 2 diabetes in a Mongolian population. Journal of Diabetes. 2012;4(3):238–242. doi: 10.1111/j.1753-0407.2011.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boini K. M., Graf D., Hennige A. M., et al. Enhanced insulin sensitivity of gene-targeted mice lacking functional KCNQ1. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2009;296(6):R1695–R1701. doi: 10.1152/ajpregu.90839.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmkvist J., Banasik K., Andersen G., et al. The type 2 diabetes associated minor allele of rs2237895 KCNQ1 associates with reduced insulin release following an oral glucose load. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0005872.e5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müssig K., Staiger H., Machicao F., et al. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes. 2009;58(7):1715–1720. doi: 10.2337/db08-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grallert H., Herder C., Marzi C., et al. Association of genetic variation in KCNQ1 with type 2 diabetes in the KORA surveys. Hormone and Metabolic Research. 2010;42(02):149–151. doi: 10.1055/s-0029-1241170. [DOI] [PubMed] [Google Scholar]
- 37.Lu S., Xie Y., Lin K., et al. Genome-wide association studies-derived susceptibility loci in type 2 diabetes: confirmation in a Chinese population. Clinical and Investigative Medicine. 2012;35(5):E327–E333. doi: 10.25011/cim.v35i5.18706. [DOI] [PubMed] [Google Scholar]
- 38.Rees S. D., Hydrie M. Z. I., Shera A. S., et al. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia. 2011;54(6):1368–1374. doi: 10.1007/s00125-011-2063-2. [DOI] [PubMed] [Google Scholar]
- 39.Van Vliet-Ostaptchouk J. V., Van Haeften T. W., Landman G. W., et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0032148.e32148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura M., Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [review] Endocrine Journal. 2011;58(9):723–739. doi: 10.1507/endocrj.ej11-0113. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z., Zhang X., Ma G., Qian Q., Yao Y. Association study of four variants in KCNQ1 with type 2 diabetes mellitus and premature coronary artery disease in a Chinese population. Molecular Biology Reports. 2010;37(1):207–212. doi: 10.1007/s11033-009-9597-0. [DOI] [PubMed] [Google Scholar]
- 42.Been L. F., Ralhan S., Wander G. S., et al. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Medical Genetics. 2011;12(1):p. 18. doi: 10.1186/1471-2350-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan I. A., Vattam K. K., Jahan P., Mukkavali K. K., Hasan Q., Rao P. Correlation between KCNQ1 and KCNJ11 gene polymorphisms and type 2 and post-transplant diabetes mellitus in the Asian Indian population. Genes & Diseases. 2015;2(3):276–282. doi: 10.1016/j.gendis.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X.-d, Yang Y.-j, Li S.-y, et al. Several polymorphisms of KCNQ1 gene are associated with plasma lipid levels in general Chinese populations. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0034229.e34229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We confirm that the data used during the research will not be shared with anybody or broadcasted in any public domain, since it is impermissible as per the policy instructions of JMI. The metadata, supporting the study outcomes, can be accessed from JMI through proper consent; however, privileged data with restricted open accessibility, cedes institutional authorization, and discretion and other related information and data can also be retrieved from the authors with permission of JMI.


