Abstract
Healthcare-associated infections (HAIs) are the most frequent and severe complication acquired in healthcare settings with high impact in terms of morbidity, mortality and costs. Many bacteria could be implicated in these infections, but, expecially multidrug resistance bacteria could play an important role. Many microbial typing technologies have been developed until to the the bacterial whole-genome sequencing and the choice of a molecular typing method therefore will depend on the skill level and resources of the laboratory and the aim and scale of the investigation. In several studies the molecular investigation of pathogens involved in HAIs was performed with many microorganisms identified as causative agents such as Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Clostridium difficile, Acinetobacter spp., Enterobacter spp., Enterococcus spp., Staphylococcus aureus and several more minor species. Here, we will describe the most and least frequently reported clonal complex, sequence types and ribotypes with their worldwide geographic distribution for the most important species involved in HAIs. (www.actabiomedica.it)
Keywords: molecular epidemiology, healthcare associated infections
Background
Healthcare-associated infections (HAIs) are the most frequent and severe complication acquired in healthcare settings with high impact in terms of morbidity, mortality and costs. Many bacteria could be implicated in these infections, expecially multidrug resistance bacteria (1), which had the capability to efficiently spread from patient to patient and to easily acquire antibiotic resistance determinants (2).
Microbial typing is often employed to determine the source of infections with an important role of bacterial epidemiological typing (3). In fact, althought conventional microbial typing methods have been useful to describe the epidemiology of infectious disease, they are variable, poor reproducible and especially labour intensive and time-consuming (4). So, many microbial typing technologies have been developed until to the the bacterial whole-genome sequencing (WGS) (5).
The choice of a molecular typing method therefore will depend on the skill level and resources of the laboratory and the aim and scale of the investigation. (6, 7)
The more classical molecular methods most often used and cited for HAIs are ribotyping, PFGE and MLST (2). In several studies the molecular investigation of pathogens involved in HAIs was performed with many microorganisms identified as causative agents such as Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Clostridium difficile, Acinetobacter baumannii, Enterobacter spp., Enterococcus spp., Staphylococcus aureus and several more minor species.
The aim of this review is to describe the most and least frequently reported clonal complex, sequence types (STs) and ribotypes (RTs) with their worldwide geographic distribution for the most important species involved in HAIs.
Materials and methods
Search strategy and selection criteria
We carried out a systematic review to identify all study dealing with the identification of molecular epidemiology of HAIs of multi drug reisistance bacteria (MDR).
We searched the main scientific databases (PubMed, Sciverse Scopus, Web of knowledge) for the following search terms: “molecular”; “epidemiology”; “healthcare”; “associated”; “infections”, using the function “AND”. The bibliographies of all relevant articles, including reviews, were screened for further references. No language restrictions were imposed; papers in languages we were unable to read were translated using Google Translate. We developed the search terms in accordance with the Medical Subject Headings thesaurus, using a combination of test searches and via collaboration between independent researchers and knowledge users. After deleting duplicates, we further screened titles, abstracts, or entire articles using exclusion criteria. Screening was carried out independently by two authors (CG, SD). Any disagreement about eligibility between reviewers was resolved by a third author (RS and VLF). The first two authors extracted data from included papers using a data extraction form reviewed by the other co-authors (RS, VLF, AS, CA, GBC, FF). Every author contributes to the manuscript writing. These procedures comply with the PRISMA guidelines for reporting systematic reviews (8, 9).
Data extraction
Two independent reviewers (CG and SD) identified potentially relevant articles and collected the data.
Figure 1.
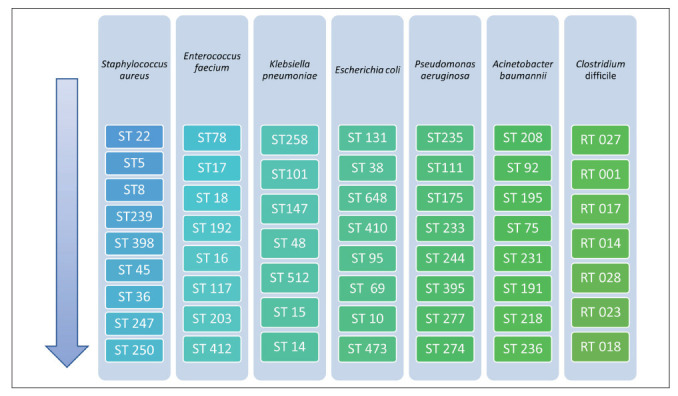
Representation by order of the most and least reported STs and RTs for important species involved in HAIs
Figure 2.
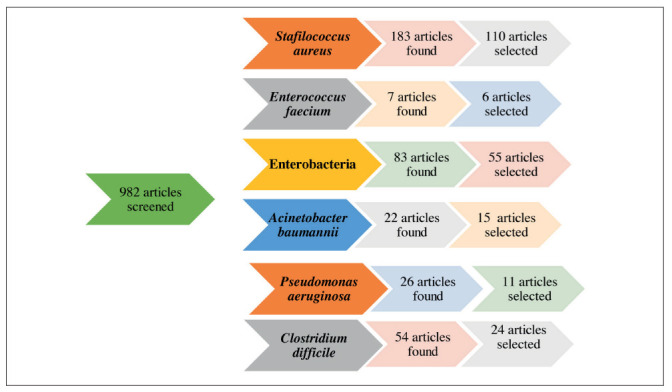
Flowchart of the evaluation and inclusion process for the review
Staphilococcus aures
Pulsed-field gel electrophoresis (PFGE) is the “gold standard” for epidemiological investigation of methicillin-resistant Staphylococcus aureus (MRSA), but several DNA sequence-based methods have been developed (10). As regard the clonal distribution in North America the multilocus sequence type (ST) 8-IV (USA300 clone) predominated while community-associated MRSA in Europe is characterised by clonal heterogeneity. The most common European strain is the European clone (ST80-IV), despite reports of USA300 are also present in literature. Several MRSA clones have arisen in Europe, i.e. the ST398-V pig-associated MRSA clone in the Netherlands and Denmark. Here below, give to the highest number of articles detected we will provide a distribution of the main findings by country.
In Asia two pandemic HA-MRSA clones, ST 239 and ST5, are disseminated internationally, whereas the molecular epidemiology of CA-MRSA is characterized by clonal heterogeneity, similar to that in Europe (11-19). Also, invasive S. aureus infections caused by PVL-positive strains are rare in Asia althought several PVL-positive MRSA clones, predominantly ST8-SCCmecIVa and ST30-SCCmecIVc, were circulating and causing sporadic cases of invasive infections in the community and hospital settings (20).
In Shenzhen, China in a study at nine sentinel hospitals ST72-SCCmec type IV is the predominant clone confirming previously studies (21). Also, in India a study shows that healthcare-associated MRSA strains may harbour community-acquired MRSA genetic markers (22).
Another possible application of molecular methods was screening and identification of MRSA carriage, useful for controlling MRSA dissemination in hospitals, such as described by Wu TH et al in Taiwan. In this study the leading ST were ST59 (44.4%), ST45 (30.6%), and ST239 (8.3%) for all isolates (31) while in another study in South Korea instead of classic MRSA clones responsible for HAIs, ST 72 accounted for 52.8% of the isolates during 2013-2014 (21,23). Other sequenced type identified by Chen et al were ST15 in CA and ST188 in HA (24,25). Despite ST239 was the predominant HA-MRSA clones in some studies was replaced by the continually growing ST5 and ST59, ST398, ST642 and ST107 (11, 26). The American strains was also be detected in some outbreaks (27). Finally some athpycal clones could be detected (28).
In Europe each country had a distinct epidemiology, with ST8-IVc (USA500) being most prevalent, especially in France and Spain. The main clonal complex is CC8/239 and other prevalent ones were CC5 and CC22 (29-38). Despite this some eceptions were described in literature;in a study conducted in UK WGS data from 20 historical outbreaks of MRSA were analysed and CC30 was the most common clonal complex followed by CC2 and CC8. CC30 was implicated in household infections while CC22 and CC8 in hospital settings, with the highest number of cases for CC8 (187 only for one outbreak) (38-40). Also, in Italy, similarly to other countries, prevalent strains were ST262 (34, 41-43) or the USA 300 one that in USA represented the dominant community-associated methicillin-resistant S. aureus lineage (44-52)). Although this, also USA100 clone, USA 300, USA500 and USA800 could be detected. Despite being relatively understudied, USA500 strains cause a significant burden of disease and were the third most common methicillin-resistant S.aureus (MRSA) strains identified in the U.S.A (52-55).
In Colombia, several HA-MRSA clones have been found, including the pediatric clone (CC5-ST5-SCC mec IV), the Brazilian clone (CC8-ST239-SCC mec III), and the Chilean/Cordobés clone (CC5-ST5-SCC mec I). Moreover, the CA-MRSA clone USA300 has been reported as causing hospital-acquired infections (56,57). ST 22 and ST 30 was identified in other studies (57-60). Also, ST 398 was detetcted in one sporadic case (61). While ST8-IV USA300 being the commonest clone in North America the ST30-IV Southwest Pacific clone established as the dominant clone in New Zealand for the past two decades, although recently unidentified PVL-negative ST5-IV spa t002 clone replaced it as the dominant CA-MRSA clone. Of particular concern was the finding of several successful and virulent MRSA clones from other geographic settings, including ST93-IV (Queensland CA-MRSA), ST8-IV (USA300) and ST772-V (Bengal Bay MRSA) (61).
On December 2017, in Australia two prevalent HA MRSA clones were detected, ST22-IV and ST239-III while CA ones were characterised as ST93-IV, ST5-IV, ST1-IV and ST45-VT. CA-MRSA, in particular the ST45-VT clone has acquired multiple antimicrobial resistance determinants (62,63). These results confirmed previously study that highligheted the presence in invasive isolates predominantly of ST93 (26.6 %) and pvl positive (54.3 %) while non-invasive isolates were rarely ST93 (1.9 %) or pvl positive (7.4 %) (64).
More recently, methicillin-susceptible S. aureus (MSSA) belonging to CC398 have been increasingly reported as a cause of invasive infections in patients without livestock contact leading to bloodstream infections associated with high mortality (65).
In Africa BenDarif et al. isolated PVL-negative CC5 isolates most frequently (38 %), more of which were similar to the HA USA100/800 strain type; CA-MRSA CC80 strains were the second most frequent (27 %) followed by CC22. The minor groups (<10 % each) were CC15, CC1, CC8/ST239, CC45, CC152, CC30 and CC88 (66, 67). In a recent study performed in South Africa 29.1% of cases were identified as MRSA infection (2.3% were considered CA-MRSA and 26.8% HA-MRSA). The most common sequence types were ST239 and ST612 of CC8 and a novel ST (ST4121) was obtained for one isolate (68). In a recent study HA MRSA (ST239 and ST22) and CA ones (ST80 and ST8) were found (69-70). In a public referral hospital in Kenya, in contrast to previous studies published, there was marked genetic diversity among clinical MRSA isolate and the predominant clonal complex was CC 5 (71,72). The European CA-MRSA clone in a study accounted for 14.1% of all HA MRSA infections (73). while the first description of the spread of the MRSA ST5-IVa clone was in 2014 (74). Finally, also USA 300 could be observed in some situations (75).
Other Staphylococcus spp
The gold standard for genotyping of S. epidermidis is pulsed-field gel electrophoresis (76), which could also be utilized to characterize other species of Staphilococcus. Genotyping methods used in studies on the molecular epidemiology of CoNS are mainly based on two different techniques: DNA banding pattern analysis and DNA sequencing and recently, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and Raman spectroscopy (77).
In a large study conducted in Iran the molecular epidemiology of Staphylococcus epidermidis and the comparation with a previously characterized collection of isolates origin from Northern Europe, Australia, and USA was perfomed. The study documented the dissemination of three MDRSE clones within and between hospitals in Iran and revealed an intercontinental spread of two clonal multidrug-resistant lineages (ST2 and ST5) in the hospital environment. Isolates of the predominant clones were significantly more frequently associated with multidrug-resistance and biofilm formation compared to nonclonal isolates. In particular three predominant PFGE clones were found. The PFGE patterns of the most common sequence type (PFGE type 040-ST2) showed 80% similarity to multidrug-resistant S. epidermidis (MDRSE) clinical isolates from eight hospitals in Northern Europe. The second most common (PFGE 024-ST22) showed a unique PFGE pattern, whereas the third most predominant genotype (PFGE 011-ST5) proved indistinguishable to the PFGE Co-ST5 identified in five hospitals in Northern Europe (78).
Methicillin-resistant Staphylococcus lugdunensis (MRSL) is increasingly recognized in healthcare and community settings. A study perfomed in China highlighted the diversified structures of SCCmec elements among MRSL strains and their close relationship with SCCmec elements harboured by CA-MRSA; the sequence type was in descending order ST3-SCCmec V, ST27-SCCmec V, ST3-SCCmec IV and ST42-SCCmec V with CC2described such as dominant clonal complex in both community and hospital settings (79).
Enterococcus faecium
The main sequence type identified were ST78, ST17, ST18, ST192, ST16, ST117, ST203 and ST412 (2, 80-83).
Enterobacteriacee
The prevalence of carbapenemase-producing Enterobacteriaceae (CPE) is increasing worldwide. Regarding the resistance determinants, SHV, TEM, OXA-1-like and CTX-M-gp1 were predominant enzymatic variants, whereas CTX-M-gp9, CTX-M-gp2, KPC, VIM, GES, OXA-48-like, NDM and OXA-23-like were considered emerging enzymes (84) Also, some association between several risk factors and ESBL-KP infection such as length of hospitalization, use of cephalosporins, use of quinolones, presence of a nasogastric tube, presence of an intravenous catheter, mechanical ventilation and cerebrospinal fluid drainage were found (85-126).
Escherichia coli
Extra-intestinal pathogenic Escherichia coli are a significant cause of urinary tract infection and bacteraemia. Many studies were perfomed to investigate the prevalence and molecular epidemiology of ESBL-producing Escherichia coli causing HCAIs and CA infections. The prevalent clones were ST 131, ST 38, ST 648, ST 4120, ST 95, ST 69, ST10 and ST 473 (85-99). Also, such as the other bacteria some other clones could be described. In a study conducted in UK Trends in ExPEC serogroups were investigated: serogroups O25, O6, and O2 dominated and they were linked to the major ExPEC STs as follows: ST131-O25, ST73-O6, ST127-O6, and ST95-O2 (87), confirming other studies (100-101). In a Manchester hospital a large Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli outbreak was investigated: genomic analysis identified the spread of ST216 among patients and in the environment. Patient relocation and plumbing replacement were associated with control of outbreak; however, environmental contamination with CRE and patient CRE acquisitions recurred rapidly following this intervention (102).
Klebsiella pneumoniae
Klebsiella pneumoniae is an opportunistic pathogen and leading cause of HAIs infections. The main clones described were ST258, ST101, ST147, ST 48, ST 512, ST 15 and ST 14 (99,103-107). Some exceptions could be observed. In a multicentre prospective cohort study in Spain K. pneumoniae ST405 predominated (108,109). On November 2015, a KPC-producing Enterobacteriaceae outbreak occurred in a general hospital in South Korea due to a clonal spread of K. pneumoniae ST307 carrying a self-transferable IncX3-type plasmid harboring blaKPC-2. Sporadic emergence of K. pneumoniae ST697 and a ST11 isolate were observed (110).
K. pneumoniae were carbapenemase producers, expecially OXA-48-like (111-112). In an Italian study carbapenemase producers belonged to 10 different STs, with ST175 and ST621 being the most common lineages (113). The spread of carbapenemase-producing Enterobacteriaceae (CPE) is a great problem also in Russia, where in a study most of isolated strains under study were multi drug resistant; MDR mechanisms were based on carrying of epidemic extended-spectrum beta-lactamase bla CTX-M-15 gene, carbapenemase bla OXA-48-like gene and class 1 or 2 integrons (114). In a study performed in Colombia 85.7 % of K. pneumonia were positive for KPC carbapenemase, especially KPC-2 and 3 KPC-3, while for P. aeruginosa and E. cloacae, most isolates were non-carbapenemase producing (87.5 %). Molecular analisys revealed that most isolates belonged to ST14 for KP while ST170 and ST1804 were found in P. aeruginosa (108).
Apisarnthanarak et al detected that among 71 patients with HA infection due to an ESBL-producing strain of E. coli or K. pneumoniae, the gene for CTX-M, with or without other ESBL genes, was identified in all patients infected with an E. coli strain and in 90% patients infected with a K. pneumoniae one (107). In another study performed in Brunei ST 231 was the most isolated type, which may be representatives of a high-risk CRKP clone disseminating across Southeast Asia. Resistance of isolated strains was due to the production of OXA-232 and CTX-M-15 β-lactamases (109). Other clones could be associated with outbreak such as ST 307 or ST697 (115). In France, a study investigates epidemiological links between apparently unrelated cases of OXA-48-producing Klebsiella pneumoniae (Kp OXA-48) colonisation or infection. This study showed that environmental reservoirs should be considered as a source of CPE transmission (112), such as confirmed in other studies, i.e. in a Tunisian Hospital (ST167 and ST131) showing that strict control measures should be established to minimize this problem (113-116). K penumoniae could produce several carbapenamase: i.e. New Delhi metallo-β-lactamase-1 (NDM-1) is among the most recently discovered carbapenemases (117). In the last decade, hospital outbreaks involving KPC-producing K. pneumoniae have been predominantly attributed to isolates belonging to clonal group (CG) 258. However, results of recent epidemiological analysis indicate that ST 307, is emerging in different parts of the world and is a candidate to become a prevalent high-risk clone in the near future. A study showed that the ST307 genome encodes genetic features that may provide an advantage in adaptation to the hospital environment and the human host, in fact compared with the ST258 clone, capsulated ST307 isolates showed higher resistance to complement-mediated killing (118). In a study 31 patients were examined after returning to Poland from a trip to South and South-East Asia. The presence of New Delhi Metallo-β-lactamase-1 producing Escherichia coli and Klebsiella pneumoniae was confirmed in three patients (9.7%) returning to Poland from travels to India. All the positive patients were hospitalized during the trip in a New Delhi hospital (117).
Other Enterobacter ssp and miscellaneous
In UK a new E. cloacae complex isolate belonged to a novel ST (ST829) highliting the importance of phenotypic tests to detect carbapenemase activity when molecular assays are negative for the ‘big 5’ carbapenemase families to understand the possible circulation of rarer carbapenemases in clinical settings (119)
In a Spanich study a high number of OXA48KP isolates showed multidrug resistance (ST 15 and ST 29) and was associated with a high mortality and mainly hospital-acquired (120,121).
A hospital-wide point prevalence study and active surveillance were performed by Forde C. et al. to assess the prevalence of CRKP infection/colonization. During the surveillance period K. pneumoniae was the most frequently occurring species, followed by Enterobacter spp. All isolates involved in both outbreaks harboured the blaKPC gene, as demonstrated by PCR (122).
A multicenter study showed that in pediatric patients the ST 131 was the most prevalent sequence type among all resistant E. coli isolates (30%), and the CG 258 was the most prevalent allele among all resistant K. pneumoniae isolates (10%) (123).
Carbapenemase producing Citrobacter freundii infections are still uncommon in European countries, In a study were identified ST11, ST18, ST22 and ST64 and 6 new STs (ST89, ST90, ST91, ST92, ST92 and ST94). In this study of Villa J et al the dissemination of the blaVIM-1 gene among various clones suggests a successful horizontal transfer of integron carrying elements that play a dominant role in the development of multidrug resistance in Enterobacteriaceae (124).
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a leading cause of HAIs and often shows MDR phenotypes. It could be related also to a wide variety of clinical syndromes in neonatal intensive care unit patients, including sepsis, pneumonia, meningitis, diarrhea, conjunctivitis and skin infections. In these cases, molecular investigation and WGS provided detailed information without the need for further typing and could be useful to understand outbreak situations rapidly and with certainty (125-126). The most frequent isolates belong to ST235, ST111, ST175, ST 233, ST 244, ST 395, ST 277 and ST 274 (2, 125-126).
Also, Carbapenem-resistant isolates of Pseudomonas aeruginosa producing metallo-β-lactamases (MBLs) are increasingly reported worldwide and often belong to particular ‘high-risk clones’. In particular multidrug-resistant Pseudomonas aeruginosa expressing VIM-metallo-beta-lactamase is an emerging infection control problem. The source of many such infections is unclear, though there are reports of hospital outbreaks of P. aeruginosa related to environmental contamination, including tap water (127). In an Italian study overall, 5.1% isolates were positive for carbapenemase genes, including blaVIM, blaIMP and blaGES-5, while the remaining ceftolozane/tazobactam-resistant isolates tested negative for carbapenemase production. In a study, an outbreak in an intensive care burn unit was due especially to DLST 1-18; carbapenemase producers belonged to 10 different STs, with ST175 and ST621 being the most common lineages (128). In a UK study VIM-type MBLs predominated (91% of all MBLs found), but a few IMP- and NDM-type enzymes were also identified. Diverse VNTR types were seen, but 86% of isolates belonged to six major complexes. MLST of representative isolates from each complex showed that they corresponded to ST 111, 233, 235, 357, 654 and 773, respectively (129,130). Despite in several studies most isolates carried VIM-2, others carried IMP-1 or IMP-13, NDM-1, VIM-2 and IMP-18 or no metallo-beta-lactamase (MBL) gene were identified (194-195). In an Estonian study clinically relevant beta-lactamases (OXA-101, OXA-2 and GES-5) were found in 12% of strains, 27% of which were located in plasmids;whereas ST108 was associated with localized spread in one hospital and mostly carbapenem-resistant phenotype, ST260 strains occurred in all hospitals, mostly with multi-resistant phenotype and carried different resistanc genotype/machinery (131).
Acinetobacter baumannii
Acinetobacter baumannii is an important hospital-acquired pathogen in healthcare facilities that frequently causes bacteraemia and ventilator-associated pneumonia in intensive care units. Acinetobacter baumannii can be isolated from various sites in the hospital environment like medical equipment, bed linen, medical personnel and indwelling catheters. Multidrug resistance in the nosocomial pathogen Acinetobacter baumannii limits therapeutic options and impacts on clinical care. Resistance against carbapenems, a group of last-resort antimicrobials for treating multidrug-resistant (MDR) A. baumannii infections, is associated with the expression (and over-expression) of carbapenemases encoded by the blaOXA genes. The most common species isolated were ST 208, ST 92, ST 195, ST 75, ST 231, ST 191, ST 218 and ST 236 (132,133).
In a study performed in three hospitals in southern Vietnam from 2012 to 2014 antimicrobial resistance was common (74% of isolates were both MDR and XDR). High-level imipenem resistance for 91.6 % of the XDR imipenem-nonsusceptible organisms, of which the majority (86.7 %) harboured the blaOXA-51 and blaOXA-23 genes associated with an ISAba1 element (134).
The first description of NDM in A. baumannii in newborn units in Turkey was found in 2016 (135). In a study performed in South Africa all the A. baumannii isolates showed a high MDR (100 % resistance to ampicillin, amoxicillin, cefuroxime, cefuroximine axetil, cefoxitin, cefotaxime and nitrofurantoin; 7% were resistant to amikacin; 67 % to ceftazidime, cefepime, imipenem, meropenem, gentamicin, ciprofloxacin and trimethoprim/sulfalethoxazole). The most dominant ST among the collected isolates was ST758, member of the EUI group, but other ST identified were ST258, ST339, ST502 and ST848. The M-PCR assays showed that 99 % of the isolates contained the OXA-51 gene and 77 % contained the OXA-23 gene and was not restricted to a specific ST (136).
In a study in Brazil 91.9 % isolates were resistant to imipenem and 98.8 % were susceptible to colistin. The blaOXA-23 gene (78.2 %) and its upstream insertion ISAba1 (55.2 %) were predominant, followed by blaOXA-24 (55.2 %) and blaOXA-143 (28.7 %). The blaOXA-23 gene and ISAba1 were independently associated with resistance to imipenem (P<0.05). Different sequence types (STs) were detected among the 35 isolates: ST1 (25.7 %), ST162 (22.8 %) and ST730 (17.1 %) were the most common, and four new STs were identified. The isolates were grouped into five clonal complexes (CC1, CC15, CC79, CC108 and CC162) (137). In a study conducte in South America the phenotypic identification of isolated showed that the isolates belong mainly to A. calcoaceticus-A. baumannii complex. All of them were multi-resistant to almost the whole antibiotics except to tigecycline and sulperazon, and they were grouped into five (I to V) different antibiotypes, being the antibiotype I the most common (50.0%). The percent of beta-lactamases detected was: blaTEM (17.3%), blaCTX-M (9.6%), blaVIM (21.2%), blaIMP (7.7%), blaOXA-58 (21.2%), and blaOXA-51 (21.2%). The phylogenetic tree analysis showed that the isolates were clustering to A. baumannii (74.1%), A. nosocomialis (11.1%) and A. calcoaceticus (7.4 %). Besides, the integron class 1 and class 2 were detected in 23.1% and 17.3% respectively (138).
In a study performed in Brazil isolates were only susceptible to amikacin, gentamicin, tigecycline, and colistin, and contained the ISAba1 insertion sequence upstream ofblaOXA-23 and blaOXA-51 genes. Twenty-six OXA-23-producing A. baumannii strains belonged to the ST79 (CC79) clonal group, and patients infected or colonised by these isolates had a higher mortality rate (34.6%) (139).
In some studies, high mortality rate was detected for infections with some ST such as ST457 a clone that exclusively shared a few virulence factors with the hypervirulence strain LAC-4, including a capsule biosynthesis locus (KL49) that is supposed to be important for the hypervirulence in LAC-4 (140). Carbapenem resistance in Acinetobacter baumannii in China was mainly mediated by OXA-23-like carbapenemases, while OXA-24/40-like carbapenemases were rarely identified with OXA-72 as one variant of this latter (140). Also, in other countries the same carbapenemase was identified such as i.e. in Iran: in a study almost all A. baumannii isolates were extensively drug-resistant (98 %) and carried blaOXA-23-like (98 %) and class 1 integrons (48 %). PFGE and MLST analysis identified three major genotypes, all belonging to clonal complex 92 (CC92): ST848, ST451 and ST195. CC92 has previously been documented in the hospital setting in northern Iran, and ST195 has been reported in Arab States of the Persian Gulf (141). In African study ST391 was the predominant ST detected, 70.5% of which harbored blaOXA-23 alone, both blaOXA-23 and blaKPC in 11.8%. Carbapenem resistance due to blaOXA-23 carbapenemase was detected in 72% of isolated, followed by blaOXA-23 concomitant with blaKPC in 14%, while blaNDM with blaOXA-58 in 6% and blaNDM alone in 1 case (2%) (142). Other sequence types were identified such as ST1, ST162 and ST730 and ST 22 and ST26 in Hong Kong (140).
Clostridium
Clostridium difficile is an emerging cause of healthcare-associated infections with increasing frequency and severity attributed to highly virulent ribotypes such as 027. The changing epidemiology of Clostridum and the emergence of epidemic 027 necessitate continued surveillance to identify shifts in antibiotic susceptibility (143-145). Clostridium difficile can be characterized according to its ribotyping which is performed using the polymerase chain reaction. Several different ribotypes have been associated with Clostridum difficile infection (CDI). The ribotypes 001, 002, 014, 046, 078, 126, and 140 have been found to be prevalent in the Middle East. In Asia, ribotypes 001, 002, 014, 017, and 018 are more prevalent (2, 143-164).
| Predominant strains | Country |
| RT 001, 014, 020, 027, 078å | Europe and North America |
| 001, 002, 014, 046, 078, 126, 140 | Middle East |
| 002, 014, 017, 018 | Asia |
Ribotype 027 was found to have reduced susceptibility to metronidazole, rifampicin, moxifloxacin, clindamycin, imipenem, and chloramphenicol and also it leads to severe disease presentation, high morbidity and mortality rates, spread more easily within the hospital because they can resist the hospital environment, cleaning, and disinfectants (215-217). In fact, the cost of CDI is estimated to be about 3000 million euro/year in Europe and is expected to almost double over the next four decades (165). ClosER, currently the largest pan-European epidemiological study of C. difficile ribotype distribution and antibiotic susceptibility, aimed to undertake antimicrobial resistance surveillance pre- and post-introduction of fidaxomicin. In this study ribotypes 027, 014, 001, 078, 020, 002, 126, 015 and 005 were most frequently isolated, and emergent ribotypes 198 and 356 were identified in Hungary and Italy, respectively. All isolates were susceptible to fidaxomicin, with scarce resistance to metronidazole (0.2%, 6/2694), vancomycin (0.1%, 2/2694) and tigecycline (0%). Rifampicin, moxifloxacin and clindamycin resistance was evident in multiple ribotypes. Epidemic ribotypes (027/001) were associated with multiple antimicrobial resistance, and ribotypes 017, 018 and 356 with high-level resistance (146,149,150). This data was confirmed in other study both in Italy that in other countries (148-160). Also, in Portugal RT027 was the most frequent among healthcare facility-associated isolates (19.6%), while RT014 was the most common among community-associated isolates (12%) (158). In Asia the toxigenic ribotypes 043 and 017 were most common (both 14%) (159) and the latter was also found in a study in Germany (160). Althought approximately 30-40% of children <1 year of age are Clostridium difficile colonized; they could represent a reservoir for adult CDI. In New Zeland PCR ribotyping was performed on 32 C. difficile isolates cultured from the stool specimens of children with CDI founding that most belong to ribotype 014 (161). Similar findings were discovered in a Croatian University Hospital where excepet to the rybotipe 014 the 001 was the most prevalent one (162). In Australia were found RTs 014/020, 002, 056 and 070, similar to a previously study conducted in 2010. Proportions of RTs 014/020 and 002 remained similar respect to the past, while RTs 056, 015, 017 and 244 increased in prevalence (163). Also, other clones could be isolated in healthcare settings such as ribotype sa026 and 176 (164).
Conclusion
Undetected pathogen clusters can often be a source of spreading in-hospital infections. Unfortunately, detection of clusters can be problematic because epidemiological connection is not always easily established. Infection prevention and control (IPC) measures, however, are most effective when applied at the earliest possible stage.
Implementing daily routine molecular typing is effective for detecting and analyzing pathogen clusters. Falsely suspected outbreaks can be quickly resolved, whereas actual outbreaks can be identified faster, so that targeted IPC measures can be applied earlier; also the molecular epidemiology is importat to identify if the causative microrganism of the infection is really of environmental origin, in this case carrying out ad hoc sanitization procedures (165,166).
In fact, persistent contamination of hospital surfaces contributes to HAI transmission, and it is not always efficiently controlled by conventional cleaning, which does not prevent recontamination, has a high environmental impact and can favour selection of MDR strains (166-175).
Molecular epidemiology is an indispensable tool and should be part of a multidisciplinary approach in the proper management of HAI. In fact, althought these bacteria could lead to infections expecially in immunodeficient patients sometimes is possible to found case in immunocompetent ones, leading to high economic cost for the healthcare national system and also to unexpected death (176-179).
So, it is important to apply the best practices (such as vaccination, sanification, apply and improvement of guidelines, etc…) in time to reduce the prevalence of microorganism into healthcare area. In like of this scenario also the immunization of HCWs had a big role to prevent infection both of patients that of their collegues, depite the spread of vaccine hesitance (180-193).
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. doi: 10.1111/j.1469-0691.2011.03570. x. [DOI] [PubMed] [Google Scholar]
- 2.Mirande C, Bizine I, Giannetti A, Picot N, van Belkum A. Epidemiological aspects of healthcare-associated infections and microbial genomics. Eur J Clin Microbiol Infect Dis. 2018;37(5):823–31. doi: 10.1007/s10096-017-3170-x. doi: 10.1007/s10096-017-3170-x. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich GD, Post JC. The Time Is Now for Gene- and Genome-Based Bacterial Diagnostics: “You Say You Want a Revolution”. JAMA Intern Med. 2013 Aug 12;173(15):1405–6. doi: 10.1001/jamainternmed.2013.7042. doi: 10.1001/jamainternmed. 2013.7042. [DOI] [PubMed] [Google Scholar]
- 4.Franco-Duarte R, Černáková L, Kadam S, et al. Advances in Chemical and Biological Methods to Identify Microorganisms-From Past to Present. Microorganisms. 2019 May 13;7(5) doi: 10.3390/microorganisms7050130. pii: E130. doi: 10.3390/microorganisms7050130. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quainoo S, Coolen JPM, van Hijum SAFT, et al. Whole-Genome Sequencing of Bacterial Pathogens: The Future of Nosocomial Outbreak Analysis. Clin Microbiol Rev. 2017 Oct;30(4):1015–1063. doi: 10.1128/CMR.00016-17. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol. 2014 Jan;37(1):1–15. Epub 2014 Jan 15. Review. [PubMed] [Google Scholar]
- 7.Signorelli C. Molecular epidemiology in healthcare-associated infections: guidelines of the Italian Society of Hygiene, Preventive Medicine and Public Health (SItI) Ig Sanita Pubbl. 2015 May-Jun;71(3):241–3. Italian. [PubMed] [Google Scholar]
- 8.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009 Oct;62(10):e1–34. doi: 10.1016/j.jclinepi.2009.06.006. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. Available on: www.cochrane-handbook.org . [Google Scholar]
- 10.Huang CC, Ho CM, Chen HC, et al. Evaluation of double locus (clfB and spa) sequence typing for studying molecular epidemiology of methicillin-resistant Staphylococcus aureus in Taiwan. J Microbiol Immunol Infect. 2017 Oct;50(5):604–612. doi: 10.1016/j.jmii.2015.10.002. doi: 10.1016/j.jmii.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Chen YJ, Liu KL, Chen CJ, Huang YC. Comparative Molecular Characteristics of Community-Associated and Healthcare-Associated Methicillin-Resistant Staphylococcus aureus Isolates From Adult Patients in Northern Taiwan. Medicine (Baltimore) 2015 Dec;94(49):e1961. doi: 10.1097/MD.0000000000001961. doi: 10.1097/MD.0000000000001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu D, Wang Z, Wang H, et al. Predominance of ST5-II-t311 clone among healthcare-associated methicillin-resistant Staphylococcus aureus isolates recovered from Zhejiang, China. Int J Infect Dis. 2018 Jun;71:107–112. doi: 10.1016/j.ijid.2018.04.798. doi: 10.1016/j.ijid.2018.04.798. [DOI] [PubMed] [Google Scholar]
- 13.Cho OH, Park KH, Song JY, et al. Prevalence and Microbiological Characteristics of qacA/B-Positive Methicillin-Resistant Staphylococcus aureus Isolates in a Surgical Intensive Care Unit. Microb Drug Resist. 2018 Apr;24(3):283–289. doi: 10.1089/mdr.2017.0072. doi: 10.1089/mdr.2017.0072. [DOI] [PubMed] [Google Scholar]
- 14.Sonnevend Á, Blair I, Alkaabi M, et al. Change in meticillin-resistant Staphylococcus aureus clones at a tertiary care hospital in the United Arab Emirates over a 5-year period. J Clin Pathol. 2012 Feb;65(2):178–82. doi: 10.1136/jclinpath-2011-200436. doi: 10.1136/jclinpath-2011-200436. [DOI] [PubMed] [Google Scholar]
- 15.Nakaminami H, Noguchi N, Ito A, et al. Characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals in Tokyo, Japan. J Infect Chemother. 2014;20(8):512–5. doi: 10.1016/j.jiac.2014.03.006. doi: 10.1016/j.jiac.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Wang WK, Han LZ, et al. Epidemiological and genetic diversity of Staphylococcus aureus causing bloodstream infection in Shanghai, 2009-2011. PLoS One. 2013 Sep 9;8(9):e72811. doi: 10.1371/journal.pone.0072811. doi: 10.1371/journal.pone.0072811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu FF, Han LZ, Chen X, et al. Molecular characterization of Staphylococcus aureus from surgical site infections in orthopedic patients in an orthopedic trauma clinical medical center in Shanghai. Surg Infect (Larchmt) 2015 Feb;16(1):97–104. doi: 10.1089/sur.2014.027. doi: 10.1089/sur.2014.027. [DOI] [PubMed] [Google Scholar]
- 18.Wu HS, Kuo SC, Chen LY, et al. Comparison between patients under hemodialysis with community-onset bacteremia caused by community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus strains. J Microbiol Immunol Infect. 2013 Apr;46(2):96–103. doi: 10.1016/j.jmii.2012.02.004. Epub 2012 Apr 19. [Google Scholar]
- 19.Huang YC, Ho CF, Chen CJ, Su LH, Lin TY. Comparative molecular analysis of community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus isolates from children in northern Taiwan. Clin Microbiol Infect. 2008 Dec;14(12):1167–72. doi: 10.1111/j.1469-0691.2008.02115.x. doi: 10.1111/j.1469-0691.2008.02115.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu TH, Lee CY, Yang HJ, et al. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus among nasal carriage strains isolated from emergency department patients and healthcare workers in central Taiwan. J Microbiol Immunol Infect. 2019 Apr;52(2):248–254. doi: 10.1016/j.jmii.2018.08.015. doi: 10.1016/j.jmii.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Kang S, Lee J, Kim M. Medicine The association between Staphylococcus aureus nasal colonization and symptomatic infection in children in Korea where ST72 is the major genotype: A prospective observational study. Medicine (Baltimore) 2017 Aug;96(34):e7838. doi: 10.1097/MD.0000000000007838. doi: 10.1097/MD.0000000000007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A, Prasad KN, Rahman M, et al. High frequency of SCCmec type V and agr type I among heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) in north India. J Glob Antimicrob Resist. 2017 Mar;8:110–14. doi: 10.1016/j.jgar.2016.11.006. doi: 10.1016/j.jgar.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Wu HS, Kuo SC, Chen LY, et al. Comparison between patients under hemodialysis with community-onset bacteremia caused by community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus strains. J Microbiol Immunol Infect. 2013 Apr;46(2):96–103. doi: 10.1016/j.jmii.2012.02.004. [Google Scholar]
- 24.Chen YJ, Chen PA, Chen CJ, Huang YC. Molecular characteristics and clinical features of pediatric methicillin-susceptible Staphylococcus aureus infection in a medical center in northern Taiwan. BMC Infect Dis. 2019;19(1):402. doi: 10.1186/s12879-019-4033-0. doi: 10.1186/s12879-019-4033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PY, Chuang YC, Wang JT, Chang SC. Impact of prior healthcare-associated exposure on clinical and molecular characterization of methicillin-susceptible Staphylococcus aureus bacteremia: results from a retrospective cohort study. Medicine (Baltimore) 2015 Feb;94(5):e474. doi: 10.1097/MD.0000000000000474. doi: 10.1097/MD.0000000000000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain S, Chowdhury R, Datta M, Chowdhury G, Mukhopadhyay AK. Characterization of the clonal profile of methicillin resistant Staphylococcus aureus isolated from patients with early post-operative orthopedic implant-based infections. Ann Clin Microbiol Antimicrob. 2019 Feb;18(1):8. doi: 10.1186/s12941-019-0307-z. doi: 10.1186/s12941-019-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uehara Y, Mori M, Tauchi M, et al. First report on USA300 outbreak in a neonatal intensive care unit detected by polymerase chain reaction-based open reading frame typing in Japan. J Infect Chemother. 2019 May;25(5):400–3. doi: 10.1016/j.jiac.2018.12.002. doi: 10.1016/j.jiac.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Hon PY, Koh TH, Tan TY, et al. Changing molecular epidemiology and high rates of mupirocin resistance among meticillin-resistant Staphylococcus aureus in Singaporean hospitals. J Glob Antimicrob Resist. 2014 Mar;2(1):53–55. doi: 10.1016/j.jgar.2013.10.002. doi: 10.1016/j.jgar.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Hetem DJ, Derde LP, Empel J, et al. Molecular epidemiology of MRSA in 13 ICUs from eight European countries. J Antimicrob Chemother. 2016 Jan;71(1):45–52. doi: 10.1093/jac/dkv298. doi: 10.1093/jac/dkv298. [DOI] [PubMed] [Google Scholar]
- 30.Mkrtchyan HV, Xu Z, Yacoub M, et al. Detection of diverse genotypes of Methicillin-resistant Staphylococcus aureus from hospital personnel and the environment in Armenia. Antimicrob Resist Infect Control. 2017;6:19. doi: 10.1186/s13756-017-0169-0. [Google Scholar]
- 31.Mammina C, Calà C, Bonura C, et al. Polyclonal non multiresistant methicillin resistant Staphylococcus aureus isolates from clinical cases of infection occurring in Palermo, Italy, during a one-year surveillance period. Ann Clin Microbiol Antimicrob. 2012 Jun 19;11:17. doi: 10.1186/1476-0711-11-17. doi: 10.1186/1476-0711-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espadinha D, Faria NA, Miragaia M, et al. Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS One. 2013 Apr 3;8(4):e59960. doi: 10.1371/journal.pone.0059960. doi: 10.1371/journal.pone.0059960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hetem DJ, Westh H, Boye K, Jarløv JO, Bonten MJ, Bootsma MC. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish Hospitals. J Antimicrob Chemother. 2012 Jun;67(7):1775–80. doi: 10.1093/jac/dks125. doi: 10.1093/jac/dks125. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez-Baño J, Angeles Domínguez M, Blas Millán A, et al. Clinical and molecular epidemiology of community-acquired, healthcare-associated and nosocomial methicillin-resistant Staphylococus aureus in Spain. Clin Microbiol Infect. 2009 Dec;15(12):1111–8. doi: 10.1111/j.1469-0691.2009.02717.x. doi: 10.1111/j.1469-0691.2009.02717.x. [DOI] [PubMed] [Google Scholar]
- 35.González-Domínguez M, Seral C, Potel C, et al. Antimicrobial resistance, virulence factors and genetic lineages of hospital-onset methicillin-resistant Staphylococcus aureus isolates detected in a hospital in Zaragoza. Enferm Infecc Microbiol Clin. 2015 Nov;33(9):590–6. doi: 10.1016/j.eimc.2015.01.015. doi: 10.1016/j.eimc.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Szymanek-Majchrzak K, Mlynarczyk A, Mlynarczyk G. Characteristics of glycopeptide-resistant Staphylococcus aureus strains isolated from inpatients of three teaching hospitals in Warsaw, Poland. Antimicrob Resist Infect Control. 2018 Aug 29;7:105. doi: 10.1186/s13756-018-0397-y. doi: 10.1186/s13756-018-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich PJ, Boyle MG, Hogan PG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains in the neonatal intensive care unit: an infection prevention and patient safety challenge. Clin Microbiol Infect. 2016 Jul;22(7):645.e1–8. doi: 10.1016/j.cmi.2016.04.013. doi: 10.1016/j.cmi.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drougka E, Foka A, Liakopoulos A, et al. A 12-year survey of methicillin-resistant Staphylococcus aureus infections in Greece: ST80-IV epidemic. Clin Microbiol Infect. 2014 Nov;20(11):796–803. doi: 10.1111/1469-0691.12624. doi: 10.1111/1469-0691.12624. [DOI] [PubMed] [Google Scholar]
- 39.Vandendriessche S, Hallin M, Catry B, et al. Previous healthcare exposure is the main antecedent for methicillin-resistant Staphylococcus aureus carriage on hospital admission in Belgium. Eur J Clin Microbiol Infect Dis. 2012 Sep;31(9):2283–92. doi: 10.1007/s10096-012-1567-0. doi: 10.1007/s10096-012-1567-0. [DOI] [PubMed] [Google Scholar]
- 40.Ellington MJ, Yearwood L, Ganner M, East C, Kearns AM. Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J Antimicrob Chemother. 2008 Jan;61(1):73–7. doi: 10.1093/jac/dkm422. [DOI] [PubMed] [Google Scholar]
- 41.Ugolotti E, Di Marco E, Bandettini R, Biassoni R. Genomic characterization of a paediatric MRSA outbreak by next-generation sequencing. J Hosp Infect. 2018;98(2):155–60. doi: 10.1016/j.jhin.2017.08.009. doi: 10.1016/j.jhin.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Velasco C, López-Cortés LE, Caballero FJ, et al. Clinical and molecular epidemiology of meticillin-resistant Staphylococcus aureus causing bacteraemia in Southern Spain. J Hosp Infect. 2012 Aug;81(4):257–63. doi: 10.1016/j.jhin.2012.05.007. doi: 10.1016/j.jhin.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Miller R, Walker AS, Knox K, et al. ‘Feral’ and ‘wild’-type methicillin-resistant Staphylococcus aureus in the United Kingdom. Epidemiol Infect. 2010 May;138(5):655–65. doi: 10.1017/S0950268809991294. doi: 10.1017/S0950268809991294. [DOI] [PubMed] [Google Scholar]
- 44.Hultén KG, Kaplan SL, Lamberth LB, et al. Hospital-acquired Staphylococcus aureus infections at Texas Children’s Hospital, 2001-2007. Infect Control Hosp Epidemiol. 2010 Feb;31(2):183–90. doi: 10.1086/649793. doi: 10.1086/649793. [DOI] [PubMed] [Google Scholar]
- 45.Hudson LO, Murphy CR, Spratt BG, et al. Diversity of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from inpatients of 30 hospitals in Orange County, California. PLoS One. 2013;8(4):e62117. doi: 10.1371/journal.pone.0062117. doi: 10.1371/journal.pone.0062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diekema DJ, Richter SS, Heilmann KP, et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol. 2014;35(3):285–92. doi: 10.1086/675283. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 47.Tattevin P, Schwartz BS, Graber CJ, et al. Concurrent epidemics of skin and soft tissue infection and bloodstream infection due to community-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2012;55(6):781–8. doi: 10.1093/cid/cis527. doi: 10.1093/cid/cis527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Márquez-Ortiz RA, Álvarez-Olmos MI, Escobar Pérez JA, et al. USA300-related methicillin-resistant Staphylococcus aureus clone is the predominant cause of community and hospital MRSA infections in Colombian children. Int J Infect Dis. 2014;25:88–93. doi: 10.1016/j.ijid.2014.01.008. doi: 10.1016/j.ijid.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Jamrozy DM, Harris SR, Mohamed N, et al. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb Genom. 2016;2(5):e000058. doi: 10.1099/mgen.0.000058. doi: 10.1099/mgen.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenkins TC, McCollister BD, Sharma R, et al. Epidemiology of healthcare-associated bloodstream infection caused by USA300 strains of methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Infect Control Hosp Epidemiol. 2009;30(3):233–41. doi: 10.1086/595963. doi: 10.1086/595963. [DOI] [PubMed] [Google Scholar]
- 51.Carrillo-Marquez MA, Hulten KG, Mason EO, Kaplan SL. Clinical and molecular epidemiology of Staphylococcus aureus catheter-related bacteremia in children. Pediatr Infect Dis J. 2010;29(5):410–4. doi: 10.1097/INF.0b013e3181c767b6. doi: 10.1097/INF.0b013e3181c767b6. [DOI] [PubMed] [Google Scholar]
- 52.Bush K, Leal J, Fathima S, et al. The molecular epidemiology of incident methicillin-resistant Staphylococcus aureus cases among hospitalized patients in Alberta, Canada: a retrospective cohort study. Antimicrob Resist Infect Control. 2015;4:35. doi: 10.1186/s13756-015-0076-1. doi: 10.1186/s13756-015-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eko KE, Forshey BM, Carrel M, Schweizer ML, Perencevich EN, Smith TC. Molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization and infection isolates in a Veterans Affairs hospital. Antimicrob Resist Infect Control. 2015;4:10. doi: 10.1186/s13756-015-0048-5. doi: 10.1186/s13756-015-0048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frisch MB, Castillo-Ramírez S, Petit RA, 3rd, et al. Invasive Methicillin-Resistant Staphylococcus aureus USA500 Strains from the U.S. Emerging Infections Program Constitute Three Geographically Distinct Lineages. mSphere. 2018;3(3) doi: 10.1128/mSphere.00571-17. pii: e00571-17. doi: 10.1128/mSphere.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nair N, Kourbatova E, Poole K, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) among patients admitted to adult intensive care units: the STAR*ICU trial. Infect Control Hosp Epidemiol. 2011;32(11):1057–63. doi: 10.1086/662178. doi: 10.1086/662178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ocampo AM, Vélez LA, Robledo J, Jiménez JN. Changes over time in the distribution of dominant clonal complexes of methicillin-resistant Staphylococcus aureus in Medellín, Colombia. Biomedica. 2014;34(Suppl 1):34–40. doi: 10.1590/S0120-41572014000500005. doi: 10.1590/S0120-41572014000500005. [DOI] [PubMed] [Google Scholar]
- 57.Medina G, Egea AL, Otth C, et al. Molecular epidemiology of hospital-onset methicillin-resistant Staphylococcus aureus infections in Southern Chile. Eur J Clin Microbiol Infect Dis. 2013 Dec;32(12):1533–40. doi: 10.1007/s10096-013-1907-8. doi: 10.1007/s10096-013-1907-8. [DOI] [PubMed] [Google Scholar]
- 58.Lévesque S, Bourgault AM, Galarneau LA, Moisan D, Doualla-Bell F, Tremblay C. Molecular epidemiology and antimicrobial susceptibility profiles of methicillin-resistant Staphylococcus aureus blood culture isolates: results of the Quebec Provincial Surveillance Programme. Epidemiol Infect. 2015;143(7):1511–8. doi: 10.1017/S095026881400209X. doi: 10.1017/S095026881400209X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson AE, Davis MF, Julian KG, et al. Molecular and phenotypic characteristics of healthcare- and community-associated methicillin-resistant Staphylococcus aureus at a rural hospital. PLoS One. 2012;7(6):e38354. doi: 10.1371/journal.pone.0038354. doi: 10.1371/journal.pone.0038354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlemann AC, Hafer C, Miko BA, et al. Clin Emergence of sequence type 398 as a community- and healthcare-associated methicillin-susceptible Staphylococcus aureus in northern Manhattan. Infect Dis. 2013;57(5):700–3. doi: 10.1093/cid/cit375. doi: 10.1093/cid/cit375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson DA, Roberts SA, Ritchie SR, Coombs GW, Fraser JD, Heffernan H. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in New Zealand: rapid emergence of sequence type 5 (ST5)-SCCmec-IV as the dominant community-associated MRSA clone. PLoS One. 2013;8(4):e62020. doi: 10.1371/journal.pone.0062020. doi: 10.1371/journal.pone.0062020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coombs GW, Daley DA, Lee YT, Pang S. Australian Group on Antimicrobial Resistance (AGAR) Australian Staphylococcus aureus Sepsis Outcome Programme (ASSOP) Annual Report 2017. Commun Dis Intell (2018) 2019:43. doi: 10.33321/cdi.2019.43.43. doi: 10.33321/cdi.2019.43.43. [DOI] [PubMed] [Google Scholar]
- 63.Coombs GW, Pearson JC, Nimmo GR, et al. Antimicrobial susceptibility of Staphylococcus aureus and molecular epidemiology of meticillin-resistant S. aureus isolated from Australian hospital inpatients: Report from the Australian Group on Antimicrobial Resistance 2011 Staphylococcus aureus Surveillance Programme. J Glob Antimicrob Resist. 2013;1(3):149–56. doi: 10.1016/j.jgar.2013.04.005. doi: 10.1016/j.jgar.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Hewagama S, Spelman T, Woolley M, McLeod J, Gordon D, Einsiedel L. The Epidemiology of Staphylococcus aureus and Panton-Valentine Leucocidin (pvl) in Central Australia, 2006-2010. BMC Infect Dis. 2016;16:382. doi: 10.1186/s12879-016-1698-5. doi: 10.1186/s12879-016-1698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouiller K, Gbaguidi-Haore H, Hocquet D, Cholley P, Bertrand X, Chirouze C. Clonal complex 398 methicillin-susceptible Staphylococcus aureus bloodstream infections are associated with high mortality. Clin Microbiol Infect. 2016;22(5):451–5. doi: 10.1016/j.cmi.2016.01.018. doi: 10.1016/j.cmi.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 66.Egyir B, Oteng AA, Owusu E, Newman MJ, Addo KK, Rhod Larsen A. Characterization of Staphylococcus aureus from human immunodeficiency virus (HIV) patients in Accra, Ghana. J Infect Dev Ctries. 2016;10:453–6. doi: 10.3855/jidc.7428. [DOI] [PubMed] [Google Scholar]
- 67.BenDarif E, Khalil A, Rayes A, et al. Characterization of methicillin-resistant Staphylococcus aureus isolated at Tripoli Medical Center, Libya, between 2008 and 2014. J Med Microbiol. 2016;65(12):1472–5. doi: 10.1099/jmm.0.000384. doi: 10.1099/jmm.0.000384. Epub 2016 Nov 1. [DOI] [PubMed] [Google Scholar]
- 68.Perovic O, Singh-Moodley A, Govender NP, et al. A small proportion of community-associated methicillin-resistant Staphylococcus aureus bacteraemia, compared to healthcare-associated cases, in two South African provinces. Eur J Clin Microbiol Infect Dis. 2017;36(12):2519–32. doi: 10.1007/s10096-017-3096-3. doi: 10.1007/s10096-017-3096-3. [DOI] [PubMed] [Google Scholar]
- 69.Conceição T, Coelho C, de Lencastre H, Aires-de-Sousa M. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J Antimicrob Chemother. 2015;70(12):3200–4. doi: 10.1093/jac/dkv261. doi: 10.1093/jac/dkv261. [DOI] [PubMed] [Google Scholar]
- 70.Boswihi SS, Udo EE, Al-Sweih N. Shifts in the Clonal Distribution of Methicillin-Resistant Staphylococcus aureus in Kuwait Hospitals: 1992-2010. PLoS One. 2016;11(9):e0162744. doi: 10.1371/journal.pone.0162744. doi: 10.1371/journal.pone.0162744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Omuse G, Van Zyl KN, Hoek K. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: a cross sectional study. Ann Clin Microbiol Antimicrob. 2016;15(1):51. doi: 10.1186/s12941-016-0171-z. doi: 10.1186/s12941-016-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alioua MA, Labid A, Amoura K, Bertine M, Gacemi-Kirane D, Dekhil M. Emergence of the European ST80 clone of community-associated methicillin-resistant Staphylococcus aureus as a cause of healthcare-associated infections in Eastern Algeria. Med Mal Infect. 2014;44(4):180–3. doi: 10.1016/j.medmal.2014.01.006. doi: 10.1016/j.medmal.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 73.Udo EE, Sarkhoo E. The dissemination of ST80-SCCmec-IV community-associated methicillin resistant Staphylococcus aureus clone in Kuwait hospitals. Ann Clin Microbiol Antimicrob. 2010;9:31. doi: 10.1186/1476-0711-9-31. doi: 10.1186/1476-0711-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Conceição T, Coelho C, Santos-Silva I, de Lencastre H, Aires-de-Sousa M. Epidemiology of methicillin-resistant and -susceptible Staphylococcus aureus in Luanda, Angola: first description of the spread of the MRSA ST5-IVa clone in the African continent. Microb Drug Resist. 2014;20(5):441–9. doi: 10.1089/mdr.2014.0007. doi: 10.1089/mdr.2014.0007. [DOI] [PubMed] [Google Scholar]
- 75.El-Mahdy TS, El-Ahmady M, Goering RV. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated over a 2-year period in a Qatari hospital from multinational patients. Clin Microbiol Infect. 2014;20(2):169–73. doi: 10.1111/1469-0691.12240. doi: 10.1111/1469-0691.12240. [DOI] [PubMed] [Google Scholar]
- 76.Miragaia M, Carriço JA, Thomas JC, Couto I, Enright MC, de Lencastre H. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J Clin Microbiol. 2008;46(1):118–29. doi: 10.1128/JCM.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Willemse-Erix HF, Jachtenberg J, Barutçi H, et al. Proof of principle for successful characterization of methicillin-resistant coagulasenegative staphylococci isolated from skin by use of Raman spectroscopy and pulsed-field gel electrophoresis. J Clin Microbiol. 2010;48(3):736–40. doi: 10.1128/JCM.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saffari F, Widerström M, Gurram BK, Edebro H, Hojabri Z, Monsen T. Molecular and Phenotypic Characterization of Multidrug-Resistant Clones of Staphylococcus epidermidis in Iranian Hospitals: Clonal Relatedness to Healthcare-Associated Methicillin-Resistant Isolates in Northern Europe. Microb Drug Resist. 2016;22(7):570–577. doi: 10.1089/mdr.2015.0283. [DOI] [PubMed] [Google Scholar]
- 79.Du X, Zhu Y, Song Y, et al. Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS One. 2013;8(5):e62742. doi: 10.1371/journal.pone.0062742. doi: 10.1371/journal.pone.0062742. Print. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corredor NC, López C, Aguilera PA, et al. An epidemiological and molecular study regarding the spread of vancomycin-resistant Enterococcus faecium in a teaching hospital in Bogotá, Colombia 2016. BMC Infect Dis. 2019;19(1):258. doi: 10.1186/s12879-019-3877-7. doi: 10.1186/s12879-019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raven KE, Reuter S, Reynolds R, et al. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res. 2016;26(10):1388–96. doi: 10.1101/gr.204024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sundermann AJ, Babiker A, Marsh JW, et al. Outbreak of Vancomycin-resistant Enterococcus faecium in Interventional Radiology: Detection Through Whole Genome Sequencing-Based Surveillance. Clin Infect Dis. 2019 doi: 10.1093/cid/ciz666. pii: ciz666. doi: 10.1093/cid/ciz666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abbo L, Shukla BS, Giles A, et al. Linezolid- and Vancomycin-resistant Enterococcus faecium in Solid Organ Transplant Recipients: Infection Control and Antimicrobial Stewardship Using Whole Genome Sequencing. Clin Infect Dis. 2019;69(2):259–65. doi: 10.1093/cid/ciy903. doi: 10.1093/cid/ciy903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leal HF, Azevedo J, Silva GEO, et al. Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis. 2019;19(1):609. doi: 10.1186/s12879-019-4265-z. doi: 10.1186/s12879-019-4265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Can F, Kurt-Azap Ö, İspir P, et al. The clinical impact of ST131 H30-Rx subclone in urinary tract infections due to multidrug-resistant Escherichia coli. J Glob Antimicrob Resist. 2016;4:49–52. doi: 10.1016/j.jgar.2015.10.006. doi: 10.1016/j.jgar.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Merino I, Shaw E, Horcajada JP, et al. CTX-M-15-H30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J Antimicrob Chemother. 2016;71(8):2125–30. doi: 10.1093/jac/dkw133. doi: 10.1093/jac/dkw133. [DOI] [PubMed] [Google Scholar]
- 87.Ciesielczuk H, Jenkins C, Chattaway M, et al. Trends in ExPEC serogroups in the UK and their significance. Eur J Clin Microbiol Infect Dis. 2016;35(10):1661–6. doi: 10.1007/s10096-016-2707-8. doi: 10.1007/s10096-016-2707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burke L, Humphreys H, Fitzgerald-Hughes D. The Molecular Epidemiology of Resistance in Cefotaximase-Producing Escherichia coli Clinical Isolates from Dublin, Ireland. Microb Drug Resist. 2016;22(7):552–8. doi: 10.1089/mdr.2015.0154. Epub 2016 Mar 22. [DOI] [PubMed] [Google Scholar]
- 89.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect. 2014;20(5):380–90. doi: 10.1111/1469-0691.12646. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 90.Samuelsen Ø, Overballe-Petersen S, Bjørnholt JV, et al. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS One. 2017;12(11):e0187832. doi: 10.1371/journal.pone.0187832. doi: 10.1371/journal.pone.0187832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pitout JD. Infections with extended-spectrum beta-lactamase-producing enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs. 2010;70(3):313–33. doi: 10.2165/11533040-000000000-00000. doi: 10.2165/11533040-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 92.Kim YA, Kim JJ, Kim H, Lee K. Community-onset extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 at two Korean community hospitals: The spread of multidrug-resistant E. coli to the community via healthcare facilities. Int J Infect Dis. 2017;54:39–42. doi: 10.1016/j.ijid.2016.11.010. doi: 10.1016/j.ijid.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Croxall G, Hale J, Weston V, et al. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J Antimicrob Chemother. 2011;66(11):2501–8. doi: 10.1093/jac/dkr349. doi: 10.1093/jac/dkr349. [DOI] [PubMed] [Google Scholar]
- 94.Kim H, Kim YA, Park YS, Choi MH, Lee GI, Lee K. Risk Factors and Molecular Features of Sequence Type (ST) 131 Extended-spectrum β-Lactamase-producing Escherichia coli in Community-onset Bacteremia. Sci Rep. 2017;7(1):14640. doi: 10.1038/s41598-017-14621-4. doi: 10.1038/s41598-017-14621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weissman SJ, Hansen NI, Zaterka-Baxter K, Higgins RD, Stoll BJ. Emergence of Antibiotic Resistance-Associated Clones Among Escherichia coli Recovered From Newborns With Early-Onset Sepsis and Meningitis in the United States, 2008-2009. J Pediatric Infect Dis Soc. 2016;5(3):269–76. doi: 10.1093/jpids/piv013. doi: 10.1093/jpids/piv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother. 2009;63(1):72–9. doi: 10.1093/jac/dkn463. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 97.Salipante SJ, Roach DJ, Kitzman JO, et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 2015;25(1):119–28. doi: 10.1101/gr.180190.114. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153–63. doi: 10.1016/S1473-3099(16)30257-2. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 99.Roer L, Overballe-Petersen S, Hansen F, et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere. 2018;3(4) doi: 10.1128/mSphere.00337-18. pii: e00337-18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clancy CJ, Chen L, Shields RK, et al. Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients. Am J Transplant. 2013;13(10):2619–33. doi: 10.1111/ajt.12424. doi: 10.1111/ajt.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Endimiani A, Depasquale JM, Forero S, et al. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J Antimicrob Chemother. 2009;64(5):1102–10. doi: 10.1093/jac/dkp327. doi: 10.1093/jac/dkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rojas LJ, Weinstock GM, De La Cadena E, et al. An Analysis of the Epidemic of Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae: Convergence of Two Evolutionary Mechanisms Creates the “Perfect Storm”. J Infect Dis. 2017;217(1):82–92. doi: 10.1093/infdis/jix524. doi: 10.1093/infdis/jix524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zautner AE, Bunk B, Pfeifer Y, et al. Monitoring microevolution of OXA-48-producing Klebsiella pneumoniae ST147 in a hospital setting by SMRT sequencing. Antimicrob Chemother. 2017;72(10):2737–2744. doi: 10.1093/jac/dkx216. doi: 10.1093/jac/dkx216. [DOI] [PubMed] [Google Scholar]
- 104.Bowers JR, Kitchel B, Driebe EM, et al. Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic. PLoS One. 2015;10(7):e0133727. doi: 10.1371/journal.pone.0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013;19(1):E23–E30. doi: 10.1111/1469-0691.12070. doi: 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- 106.Palacios-Baena ZR, Oteo J, Conejo C, et al. Comprehensive clinical and epidemiological assessment of colonisation and infection due to carbapenemase-producing Enterobacteriaceae in Spain. J Infect. 2016;72(2):152–60. doi: 10.1016/j.jinf.2015.10.008. doi: 10.1016/j.jinf.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 107.Kim JO, Song SA, Yoon EJ, et al. Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn Microbiol Infect Dis. 2017;87(4):343–8. doi: 10.1016/j.diagmicrobio.2016.12.012. doi: 10.1016/j.diagmicrobio.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 108.Aqel AA, Giakkoupi P, Alzoubi H, Masalha I, Ellington MJ, Vatopoulos A. Detection of OXA-48-like and NDM carbapenemases producing Klebsiella pneumoniae in Jordan: A pilot study. J Infect Public Health. 2017;10(2):150–5. doi: 10.1016/j.jiph.2016.02.002. doi: 10.1016/j.jiph.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 109.Clarivet B, Grau D, Jumas-Bilak E, et al. Persisting transmission of carbapenemase-producing Klebsiella pneumoniae due to an environmental reservoir in a university hospital, France, 2012 to 2014. Euro Surveill. 2016;21(17) doi: 10.2807/1560-7917.ES.2016.21.17.30213. doi: 10.2807/1560-7917.ES.2016.21.17.30213. [DOI] [PubMed] [Google Scholar]
- 110.Cubero M, Grau I, Tubau F, et al. Molecular Epidemiology of Klebsiella pneumoniae Strains Causing Bloodstream Infections in Adults. Microb Drug Resist. 2018;24(7):949–57. doi: 10.1089/mdr.2017.0107. doi: 10.1089/mdr.2017.0107. [DOI] [PubMed] [Google Scholar]
- 111.Fursova NK, Astashkin EI, Knyazeva AI, et al. The spread of bla OXA-48 and bla OXA-244 carbapenemase genes among Klebsiella pneumoniae, Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Ann Clin Microbiol Antimicrob. 2015;14:46. doi: 10.1186/s12941-015-0108-y. doi: 10.1186/s12941-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vanegas JM, Parra OL, Jiménez JN. Molecular epidemiology of carbapenem resistant gram-negative bacilli from infected pediatric population in tertiary care hospitals in Medellín, Colombia: an increasing problem. BMC Infect Dis. 2016;16:463. doi: 10.1186/s12879-016-1805-7. doi: 10.1186/s12879-016-1805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Apisarnthanarak A, Kiratisin P, Mundy LM. Clinical and molecular epidemiology of healthcare-associated infections due to extended-spectrum beta-lactamase (ESBL)-producing strains of Escherichia coli and Klebsiella pneumoniae that harbor multiple ESBL genes. Infect Control Hosp Epidemiol. 2008;29(11):1026–34. doi: 10.1086/591864. doi: 10.1086/591864. [DOI] [PubMed] [Google Scholar]
- 114.Abdul Momin MHF, Liakopoulos A, Phee LM, Wareham DW. Emergence and nosocomial spread of carbapenem-resistant OXA-232-producing Klebsiella pneumoniae in Brunei Darussalam. J Glob Antimicrob Resist. 2017;9:96–9. doi: 10.1016/j.jgar.2017.02.008. doi: 10.1016/j.jgar.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 115.O’Connor C, Cormican M, Boo TW, et al. An Irish outbreak of New Delhi metallo-β-lactamase (NDM)-1 carbapenemase-producing Enterobacteriaceae: increasing but unrecognized prevalence. J Hosp Infect. 2016;94(4):351–7. doi: 10.1016/j.jhin.2016.08.005. doi: 10.1016/j.jhin.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 116.Villa L, Feudi C, Fortini D, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3(4):e000110. doi: 10.1099/mgen.0.000110. doi: 10.1099/mgen.0.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Espenhain L, Jørgensen SB, Leegaard TM, et al. Travel to Asia is a strong predictor for carriage of cephalosporin resistant E. coli and Klebsiella spp. but does not explain everything; prevalence study at a Norwegian hospital 2014-2016. Antimicrob Resist Infect Control. 2018;7:146. doi: 10.1186/s13756-018-0429-7. doi: 10.1186/s13756-018-0429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18(1):6. doi: 10.1186/s12866-017-1148-6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meunier D, Findlay J, Doumith M, et al. FRI-2 carbapenemase-producing Enterobacter cloacae complex in the UK. J Antimicrob Chemother. 2017;72(9):2478–82. doi: 10.1093/jac/dkx173. doi: 10.1093/jac/dkx173. [DOI] [PubMed] [Google Scholar]
- 120.Noël A, Vastrade C, Dupont S, et al. Nosocomial outbreak of extended-spectrum β-lactamase-producing Enterobacter cloacae among cardiothoracic surgical patients: causes and consequences. J Hosp Infect. 2019;102(1):54–60. doi: 10.1016/j.jhin.2019.01.001. doi: 10.1016/j.jhin.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 121.Madueño A, González García J, Fernández-Romero S, Oteo J, Lecuona M. Dissemination and clinical implications of multidrug-resistant Klebsiella pneumoniae isolates producing OXA-48 in a Spanish hospital. J Hosp Infect. 2017;96(2):116–22. doi: 10.1016/j.jhin.2017.02.024. doi: 10.1016/j.jhin.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 122.Forde C, Stierman B, Ramon-Pardo P, Dos Santos T, Singh N. Carbapenem-resistant Klebsiella pneumoniae in Barbados: Driving change in practice at the national level. PLoS One. 2017;12(5):e0176779. doi: 10.1371/journal.pone.0176779. doi: 10.1371/journal.pone.0176779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zerr DM, Weissman SJ, Zhou C, et al. The Molecular and Clinical Epidemiology of Extended-Spectrum Cephalosporin- and Carbapenem-Resistant Enterobacteriaceae at 4 US Pediatric Hospitals. J Pediatric Infect Dis Soc. 2017;6(4):366–75. doi: 10.1093/jpids/piw076. doi: 10.1093/jpids/piw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Villa J, Arana DM, Viedma E, Perez-Montarelo D, Chaves F. Characterization of mobile genetic elements carrying VIM-1 and KPC-2 carbapenemases in Citrobacter freundii isolates in Madrid. Int J Med Microbiol. 2017;307(6):340–5. doi: 10.1016/j.ijmm.2017.07.001. doi: 10.1016/j.ijmm.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 125.Parcell BJ, Oravcova K, Pinheiro M, et al. Pseudomonas aeruginosa intensive care unit outbreak: winnowing of transmissions. with molecular and genomic typing. J Hosp Infect. 2018;98(3):282–8. doi: 10.1016/j.jhin.2017.12.005. doi: 10.1016/j.jhin.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martin K, Baddal B, Mustafa N, et al. Clusters of genetically similar isolates of Pseudomonas aeruginosa from multiple hospitals in the UK. J Med Microbiol. 2013;62(Pt 7):988–1000. doi: 10.1099/jmm.0.054841-0. doi: 10.1099/jmm.0.054841-0. Epub 2013 Apr 4. [DOI] [PubMed] [Google Scholar]
- 127.Breathnach AS, Cubbon MD, Karunaharan RN, Pope CF, Planche TD. Multidrug-resistant Pseudomonas aeruginosa outbreaks in two hospitals: association with contaminated hospital waste-water systems. J Hosp Infect. 2012;82(1):19–24. doi: 10.1016/j.jhin.2012.06.007. doi: 10.1016/j.jhin.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 128.Giani T, Arena F, Pollini S, et al. Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J Antimicrob Chemother. 2018;73(3):664–71. doi: 10.1093/jac/dkx453. doi: 10.1093/jac/dkx453. [DOI] [PubMed] [Google Scholar]
- 129.Tissot F, Blanc DS, Basset P, et al. New genotyping method discovers sustained nosocomial Pseudomonas aeruginosa outbreak in an intensive care burn unit. J Hosp Infect. 2016;94(1):2–7. doi: 10.1016/j.jhin.2016.05.011. doi: 10.1016/j.jhin.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 130.Wright LL, Turton JF, Livermore DM, Hopkins KL, Woodford N. Dominance of international ‘high-risk clones’ among metallo-β-lactamase-producing Pseudomonas aeruginosa in the UK. J Antimicrob Chemother. 2015;70(1):103–10. doi: 10.1093/jac/dku339. doi: 10.1093/jac/dku339. [DOI] [PubMed] [Google Scholar]
- 131.Telling K, Laht M, Brauer A, et al. Multidrug resistant Pseudomonas aeruginosa in Estonian hospitals. BMC Infect Dis. 2018;18(1):513. doi: 10.1186/s12879-018-3421-1. doi: 10.1186/s12879-018-3421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tuan Anh N, Nga TV, Tuan HM, et al. Molecular epidemiology and antimicrobial resistance phenotypes of Acinetobacter baumannii isolated from patients in three hospitals in southern Vietnam. J Med Microbiol. 2017;66(1):46–53. doi: 10.1099/jmm.0.000418. doi: 10.1099/jmm.0.000418. [DOI] [PubMed] [Google Scholar]
- 133.Karaaslan A, Soysal A, Altinkanat Gelmez G, Kepenekli Kadayifci E, Söyletir G, Bakir M. Molecular characterization and risk factors for carbapenem-resistant Gram-negative bacilli colonization in children: emergejnce of NDM-producing Acinetobacter baumannii in a newborn intensive care unit in Turkey. J Hosp Infect. 2016;92(1):67–72. doi: 10.1016/j.jhin.2015.09.011. doi: 10.1016/j.jhin.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 134.Nawfal Dagher T, Al-Bayssari C, Chabou S, et al. Investigation of multidrug-resistant ST2 Acinetobacter baumannii isolated from Saint George hospital in Lebanon. BMC Microbiol. 2019;19(1):29. doi: 10.1186/s12866-019-1401-2. doi: 10.1186/s12866-019-1401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gómez RF, Castillo A, Chávez-Vivas M. Characterization of multidrug-resistant Acinetobacter ssp. strains isolated from medical intensive care units in Cali - Colombia. Colomb Med (Cali) 2017;48(4):183–90. doi: 10.25100/cm.v48i4.2858. doi: 10.25100/cm.v48i4.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.da Silva KE, Maciel WG, Croda J, et al. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PLoS One. 2018;13(12):e0209367. doi: 10.1371/journal.pone.0209367. doi: 10.1371/journal.pone.0209367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen Y, Yang Y, Liu L, et al. High prevalence and clonal dissemination of OXA-72-producing Acinetobacter baumannii in a Chinese hospital: a cross sectional study. BMC Infect Dis. 2018;18(1):491. doi: 10.1186/s12879-018-3359-3. doi: 10.1186/s12879-018-3359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saffari F, Monsen T, Karmostaji A, Azimabad FB, Widerström M. Significant spread of extensively drug-resistant Acinetobacter baumannii genotypes of clonal complex 92 among intensive care unit patients in a university hospital in southern Iran. J Med Microbiol. 2017;66(11):1656–62. doi: 10.1099/jmm.0.000619. doi: 10.1099/jmm.0.000619. [DOI] [PubMed] [Google Scholar]
- 139.El Bannah AMS, Nawar NN, Hassan RMM, Salem STB. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii in a Tertiary Care Hospital in Egypt: Clonal Spread of blaOXA-23. Microb Drug Resist. 2018;24(3):269–77. doi: 10.1089/mdr.2017.0057. doi: 10.1089/mdr.2017.0057. [DOI] [PubMed] [Google Scholar]
- 140.Ho PL, Ho AY, Chow KH, Lai EL, Ching P, Seto WH. Epidemiology and clonality of multidrug-resistant Acinetobacter baumannii from a healthcare region in Hong Kong. J Hosp Infect. 2010;74(4):358–64. doi: 10.1016/j.jhin.2009.10.015. doi: 10.1016/j.jhin.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 141.de Azevedo FKSF, Dutra V, Nakazato L, et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii infection in two hospitals in Central Brazil: the role of ST730 and ST162 in clinical outcomes. J Med Microbiol. 2019;68(1):31–40. doi: 10.1099/jmm.0.000853. doi: 10.1099/jmm.0.000853. Epub 2018 Dec. [DOI] [PubMed] [Google Scholar]
- 142.Nhu NTK, Lan NPH, Campbell JI, et al. Emergence of carbapenem-resistant Acinetobacter baumannii as the major cause of ventilator-associated pneumonia in intensive care unit patients at an infectious disease hospital in southern Vietnam. J Med Microbiol. 2014;63(Pt 10):1386–94. doi: 10.1099/jmm.0.076646-0. doi: 10.1099/jmm.0.076646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fatima R, Aziz M. The Hypervirulent Strain of Clostridium Difficile: NAP1/B1/027 - A Brief Overview. Cureus. 2019;11(1):e3977. doi: 10.7759/cureus.3977. doi: 10.7759/cureus.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luo Y, Cheong E, Bian Q, et al. Different molecular characteristics and antimicrobial resistance profiles of Clostridium difficile in the Asia-Pacific region. Emerg Microbes Infect. 2019;8(1):1553–62. doi: 10.1080/22221751.2019.1682472. doi: 10.1080/22221751.2019.1682472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cartman ST, Heap JT, Kuehne SA, Cockayne A, Minton NP. The emergence of ‘hypervirulence’ in Clostridium difficile. Int J Med Microbiol. 2010;300(6):387–95. doi: 10.1016/j.ijmm.2010.04.008. doi: 10.1016/j.ijmm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 146.Freeman J1, Vernon J, Pilling S, et al. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014. Clin Microbiol Infect. 2018;24(7):724–31. doi: 10.1016/j.cmi.2017.10.008. doi: 10.1016/j.cmi.2017.10.008. Epub 2017 Oct 21. [DOI] [PubMed] [Google Scholar]
- 147.Rodriguez C, Fernandez J, Van Broeck J, et al. Clostridium difficile presence in Spanish and Belgian hospitals. Microb Pathog. 2016;100:141–8. doi: 10.1016/j.micpath.2016.09.006. doi: 10.1016/j.micpath.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 148.Del Prete R, Ronga L, Addati G, Magrone R, Miragliotta G. Prevalence of Clostridium difficile and ribotype 027 infection in patients with nosocomial diarrhoea in Southern Italy. New Microbiol. 2017;40(4):264–8. [PubMed] [Google Scholar]
- 149.Costa CL, Mano de Carvalho CB, González RH, et al. Molecular epidemiology of Clostridium difficile infection in a Brazilian cancer hospital. Anaerobe. 2017;48:232–6. doi: 10.1016/j.anaerobe.2017.10.001. doi: 10.1016/j.anaerobe.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 150.Jia H, Du P, Yang H, et al. Nosocomial transmission of Clostridium difficile ribotype 027 in a Chinese hospital, 2012-2014, traced by whole genome sequencing. BMC Genomics. 2016;17:405. doi: 10.1186/s12864-016-2708-0. doi: 10.1186/s12864-016-2708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Razavi B, Apisarnthanarak A, Mundy LM. Clostridium difficile: emergence of hypervirulence and fluoroquinolone resistance. Infection. 2007;35(5):300–7. doi: 10.1007/s15010-007-6113-0. [DOI] [PubMed] [Google Scholar]
- 152.Black SR, Weaver KN, Jones RC, et al. Clostridium difficile outbreak strain BI is highly endemic in Chicago area hospitals. Infect Control Hosp Epidemiol. 2011;32(9):897–902. doi: 10.1086/661283. doi: 10.1086/661283. [DOI] [PubMed] [Google Scholar]
- 153.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med. 2012;9(2):e1001172. doi: 10.1371/journal.pmed.1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dingle KE, Griffiths D, Didelot X, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One. 2011;6(5):e19993. doi: 10.1371/journal.pone.0019993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Foster NF, Collins DA, Ditchburn SL, et al. Epidemiology of Clostridium difficile infection in two tertiary-care hospitals in Perth, Western Australia: a cross-sectional study. New Microbes New Infect. 2014;2(3):64–71. doi: 10.1002/nmi2.43. doi: 10.1002/nmi2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Richardson C, Kim P, Lee C, Bersenas A, Weese JS. Comparison of Clostridium difficile isolates from individuals with recurrent and single episode of infection. Anaerobe. 2015;33:105–8. doi: 10.1016/j.anaerobe.2015.03.003. doi: 10.1016/j.anaerobe.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 157.Freeman J, Vernon J, Morris K, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21(3):248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 158.Santos A, Isidro J, Silva C, et al. Molecular and epidemiologic study of Clostridium difficile reveals unusual heterogeneity in clinical strains circulating in different regions in Portugal. Clin Microbiol Infect. 2016;22(8):695–700. doi: 10.1016/j.cmi.2016.04.002. doi: 10.1016/j.cmi.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 159.Zainul NH, Ma ZF, Besari A, et al. Prevalence of Clostridium difficile infection and colonization in a tertiary hospital and elderly community of North-Eastern Peninsular Malaysia. Epidemiol Infect. 2017;145(14):3012–9. doi: 10.1017/S0950268817002011. doi: 10.1017/S0950268817002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arvand M, Ruscher C, Bettge-Weller G, Goltz M, Pfeifer Y. Prevalence and risk factors for colonization by Clostridium difficile and extended-spectrum β-lactamase-producing Enterobacteriaceae in rehabilitation clinics in Germany. J Hosp Infect. 2018;98(1):14–20. doi: 10.1016/j.jhin.2017.07.004. doi: 10.1016/j.jhin.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 161.Sathyendran V, McAuliffe GN, Swager T, Freeman JT, Taylor SL, Roberts SA. Clostridium difficile as a cause of healthcare-associated diarrhoea among children in Auckland, New Zealand: clinical and molecular epidemiology. Eur J Clin Microbiol Infect Dis. 2014;33(10):1741–7. doi: 10.1007/s10096-014-2139-2. doi: 10.1007/s10096-014-2139-2. [DOI] [PubMed] [Google Scholar]
- 162.Novak A, Spigaglia P, Barbanti F, Goic-Barisic I, Tonkic M. First clinical and microbiological characterization of Clostridium difficile infection in a Croatian University Hospital. Anaerobe. 2014;30:18–23. doi: 10.1016/j.anaerobe.2014.07.007. doi: 10.1016/j.anaerobe.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 163.Collins DA, Putsathit P, Elliott B, Riley TV. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology. 2017;49(3):309–13. doi: 10.1016/j.pathol.2016.10.013. doi: 10.1016/j.pathol.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 164.Drabek J, Nyc O, Krutova M, Stovicek J, Matejkova J, Keil R. Clinical features and characteristics of Clostridium difficile PCR-ribotype 176 infection: results from a 1-year university hospital internal ward study. Ann Clin Microbiol Antimicrob. 2015;14:55. doi: 10.1186/s12941-015-0114-0. doi: 10.1186/s12941-015-0114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kuijper EJ, Coignard B, Tüll P. ESCMID Study Group for Clostridium difficile; EU Member States; European Centre for Disease Prevention and Control. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. Review. [DOI] [PubMed] [Google Scholar]
- 166.Mutters NT, Heeg K, Späth I, Henny N, Günther F. Improvement of infection control management by routine molecular evaluation of pathogen clusters. Diagn Microbiol Infect Dis. 2017 May;88(1):82–87. doi: 10.1016/j.diagmicrobio.2017.01.013. doi: 10.1016/j.diagmicrobio.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 167.La Fauci V, Costa GB, Arena A, et al. Trend of MDR-microorganisms isolated from the biological samples of patients with HAI and from the surfaces around that patient. New Microbiol. 2018;41:42–6. [PubMed] [Google Scholar]
- 168.Squeri R, Genovese C, Trimarchi G, et al. Nine years of microbiological air monitoring in the operating theatres of a university hospital in Southern Italy. Ann Ig. 2019;31(2 Suppl 1):1–12. doi: 10.7416/ai.2019.2272. doi: 10.7416/ai.2019.2272. [DOI] [PubMed] [Google Scholar]
- 169.La Fauci V, Genovese C, Facciolà A, et al. Five-year microbiological monitoring of wards and operating theatres in southern Italy. J Prev Med Hyg. 2017;58:E166–E172. [PMC free article] [PubMed] [Google Scholar]
- 170.Montagna MT, Mascipinto S, Pousis C, et al. Knowledge, experiences, and attitudes toward Mantoux test among medical and health professional students in Italy: a cross-sectional study. Ann Ig. 2018;30(5 Suppl 2):86–98. doi: 10.7416/ai.2018.2253. doi: 10.7416/ai.2018.2253. [DOI] [PubMed] [Google Scholar]
- 171.Squeri R, Genovese C, Palamara MA, Trimarchi G, La Fauci V. “Clean care is safer care”: correct handwashing in the prevention of healthcare associated infections. Ann Ig. 2016;28(6):409–15. doi: 10.7416/ai.2016.2123. doi: 10.7416/ai.2016.2123. [DOI] [PubMed] [Google Scholar]
- 172.La Fauci V, Costa GB, Genovese C, Palamara MAR, Alessi V, Squeri R. Drug-resistant bacteria on hands of healthcare workers and in the patient area: an environmental survey in Southern Italy’s hospital. Rev Esp Quimioter. 2019 Jun 28 pii: fauci28jun2019. [PMC free article] [PubMed] [Google Scholar]
- 173.Squeri R, Grillo OC, La Fauci V. Surveillance and evidence of contamination in hospital environment from meticillin and vancomycin-resistant microbial agents. J Prev Med Hyg. 2012;53(3):143–5. [PubMed] [Google Scholar]
- 174.La Fauci V, Riso R, Facciolà A, Merlina V, Squeri R. Surveillance of microbiological contamination and correct use of protective lead garments. Ann Ig. 2016;28(5):360–6. doi: 10.7416/ai.2016.2116. doi: 10.7416/ai.2016.2116. [DOI] [PubMed] [Google Scholar]
- 175.La Fauci V, Costa GB, Facciolà A, Conti A, Riso R, Squeri R. Humidifiers for oxygen therapy: what risk for reusable and disposable devices. J Prev Med Hyg. 2017 Jun;58(2):E161–E165. [PMC free article] [PubMed] [Google Scholar]
- 176.Spagnolo EV, Cannavò G, Mondello C, Cardia L, Bartoloni G, Cardia G. Unexpected death for Takayasu aortitis associated with coronary ostial stenosis: case report. Am J Forensic Med Pathol. 2015;36(2):88–90. doi: 10.1097/PAF.0000000000000154. doi: 10.1097/PAF.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 177.Rini MS, Argo A, Spagnolo EV, Zerbo S, Bucci MB, D’Urso D. When is necessary not to apply guidelines? Pitfalls in dentistry practice. Dental Cadmos. 2018;86(8):686–95. http://www.odontoiatria33.it/ doi:10.19256/d.cadmos.08.2018.07 . [Google Scholar]
- 178.Ventura Spagnolo E, Mondello C, Roccuzzo S, et al. A unique fatal case of Waterhouse-Friderichsen syndrome caused by Proteus mirabilis in an immunocompetent subject: Case report and literature analysis. Medicine (Baltimore) 2019;98(34):e16664. doi: 10.1097/MD.0000000000016664. doi: 10.1097/MD.0000000000016664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ventura Spagnolo E, Stassi C, Mondello C, Zerbo S, Milone L, Argo A. Forensic microbiology applications: A systematic review. Leg Med (Tokyo) 2019;36:73–80. doi: 10.1016/j.legalmed.2018.11.002. doi: 10.1016/j.legalmed.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 180.Genovese C, LA Fauci V, Squeri A, Trimarchi G, Squeri R. HPV vaccine and autoimmune diseases: systematic review and meta-analysis of the literature. J Prev Med Hyg. 2018;59(3):E194–E199. doi: 10.15167/2421-4248/jpmh2018.59.3.998. doi: 10.15167/2421-4248/jpmh2018.59.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Genovese C, Picerno IAM, Trimarchi G, et al. Vaccination coverage in healthcare workers: a multicenter cross-sectional study in Italy. J Prev Med Hyg. 2019;60(1):E12–E17. doi: 10.15167/2421-4248/jpmh2019.60.1.1097. doi: 10.15167/2421-4248/jpmh2019.60.1.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Squeri R, Genovese C, Trimarchi G, Palamara MAR, La Fauci V. An evaluation of attitude toward vaccines among healthcare workers of a University Hospital in Southern Italy. Ann Ig. 2017;29(6):595–606. doi: 10.7416/ai.2017.2188. doi: 10.7416/ai.2017.2188. [DOI] [PubMed] [Google Scholar]
- 183.La Fauci V, Sindoni D, Grillo OC, Calimeri S, Lo Giudice D, Squeri R. Hepatitis E virus (HEV) in sewage from treatment plants of Messina University Hospital and of Messina City Council. J Prev Med Hyg. 2010;51(1):28–30. [PubMed] [Google Scholar]
- 184.Genovese C, La Fauci V, Costa GB, et al. A potential outbreak of Measles and chickenpox among healthcare workers of a university Hospital. EMBJ. 2019;14(10):045–8. [Google Scholar]
- 185.Squeri R, Genovese C, Trimarchi G, Palamara MAR, La Fauci V. An evaluation of attitude toward vaccines among healthcare workers of a University Hospital in Southern Italy. Ann Ig. 2017;29(6):595–606. doi: 10.7416/ai.2017.2188. doi: 10.7416/ai.2017.2188. [DOI] [PubMed] [Google Scholar]
- 186.Squeri R, La Fauci V, Sindoni L, Cannavò G, Ventura Spagnolo E. Study on hepatitis B and C serologic status among municipal solid waste workers in Messina (Italy) J Prev Med Hyg. 2006 Sep;47(3):110–3. [PubMed] [Google Scholar]
- 187.Ferrera G, Squeri R, Genovese C. The evolution of vaccines for early childhood: the MMRV. Ann Ig. 2018;30(4 Suppl 1):33–7. doi: 10.7416/ai.2018.2232. doi: 10.7416/ai.2018.2232. [DOI] [PubMed] [Google Scholar]
- 188.Lo Giudice D, Capua A, La Fauci V, Squeri R, Grillo OC, Calimeri S. Congenital rubella syndrome and immunity status of immigrant women living in southern Italy: a cross-sectional, seroepidemiological investigation. Travel Med Infect Dis. 2014 May-Jun;12(3):253–7. doi: 10.1016/j.tmaid.2014.01.003. doi: 10.1016/j.tmaid.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 189.La Fauci V, Riso R, Facciolà A, et al. Response to anti-HBV vaccine and 10-year follow-up of antibody levels in healthcare workers. Public Health. 2016 Oct;139:198–202. doi: 10.1016/j.puhe.2016.08.007. doi: 10.1016/j.puhe.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 190.Facciolà A, Squeri R, Genovese C, Alessi V, La Fauci V. Perception of Rubella risk in pregnancy: an epidemiological survey on a sample of pregnant women. Ann Ig. 2019;31(2 Suppl 1):65–71. doi: 10.7416/ai.2019.227. doi: 10.7416/ai.2019.2278. [DOI] [PubMed] [Google Scholar]
- 191.Squeri R, La Fauci V, Picerno IAM, et al. Evaluation of Vaccination Coverages in the Health Care Workers of a University Hospital in Southern Italy. Ann Ig. 2019;31(2 Suppl 1):13–24. doi: 10.7416/ai.2019.2273. doi:10.7416/ai.2019.2273. [DOI] [PubMed] [Google Scholar]
- 192.Squeri R, Riso R, Facciolà A, et al. Management of two influenza vaccination campaign in health care workers of a university hospital in the south Italy. Ann Ig. 2017;29(3):223–31. doi: 10.7416/ai.2017.2150. doi: 10.7416/ai.2017.2150. [DOI] [PubMed] [Google Scholar]
- 193.De Luca F, Aversa T, Alessi L, et al. Thyroid nodules in childhood: indications for biopsy and surgery. Ital J Pediatr. 2014;40:48. doi: 10.1186/1824-7288-40-48. doi: 10.1186/1824-7288-40-48. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]


