Abstract
Laboratory medicine provides an almost irreplaceable contribution to the diagnostic reasoning and managed care of most human pathologies. The novel coronavirus disease 2019 (COVID-19) is not an exception to this paradigm. Although the relatively recent emergence does not allow to draw definitive conclusions on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostics, some standpoints can be conveyed. First and foremost, it seems now clear that we will be living together with this virus for quite a long time, so that our vigilance and responsiveness against the emergence of new local outbreaks shall be maintained at the highest possible levels. The etiological diagnosis of COVID-19 is, and will remain for the foreseeable future, deeply based on direct identification of viral RNA by means of molecular biology techniques in biological materials, especially upper and lower respiratory tract specimens. Whether other materials, such as blood, urine, stools, saliva and throat washing, will become valid alternatives has not been unequivocally defined so far. As concerns serological testing, promising information can be garnered from preliminary investigations, showing that the vast majority of COVID-19 patients seem to develop a sustained immune response against the virus, characterized especially by emergence of anti-SARS-CoV-2 IgG and IgA, 1 to 2 weeks after the onset of fever and/or respiratory symptoms. Whether these antibodies will have persistent neutralizing activity against the virus is still to be elucidated on individual and general basis. The availability of rapid tests for detecting either viral antigens or anti-SARS-CoV-2 antibodies are a potentially viable opportunity for purposes of epidemiologic surveillance, though more information is needed on accuracy and reliability of these portable immunoassays. (www.actabiomedica.it)
Keywords: Coronavirus, COVID-19, laboratory medicine, laboratory tests
Introduction
A new viral outbreak, sustained by a member of the coronaviridae family that has been finally defined severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2) has recently emerged in Wuhan, China at the end of 2019 (1). The virus has since then spread all around the world, persuading the World Health Organization (WHO) to declare this infectious disease as the very last pandemic, 10 years after the H1N1 Swine Flu outbreak, in 2009-2010 (2). COVID-19 has already affected millions of people worldwide, causing such a high mortality that it may be responsible of over 50 million deaths if timely and appropriate measures, such as nationwide lockdown and social distancing (3), will not be undertaken by national health agencies and governments (1).
As other coronaviruses, SARS-CoV-2 is an enveloped virus with positive-sense, single-stranded RNA genome, containing four main structural proteins known as Spike (S, which contains the receptor-binding domain, known as RBD), Envelope (E), Membrane (M), and Nucleocapsid (N), along with additional genes such as ORF1a/b, ORF3a, ORF6, ORF7a/b, ORF8, and ORF10, which encode accessory proteins, including the RNA-dependent RNA polymerase (Figure 1 and Table 1) (4,5). This microorganism has likely emerged due to bats spillover, probably through another intermediate animal (pangolin, perhaps) (6). Human transition has been largely fostered by emergence of mutations in the S protein, which has amplified the affinity of this protein moiety (within a furin-cleavage site) for angiotensin converting enzyme 2 (ACE2) (7), its natural receptor at the surface of cells of a vast array of organs and tissues, especially alveolar type 2 cells in the lung (AT2), but also lymphocytes and cells of the heart, kidney and gastrointestinal system (8,9). Binding of SARS-CoV-2 to ACE2 is fostered by S protein priming catalyzed by transmembrane serine protease 2 (TMPRSS2) (10).
Figure 1.
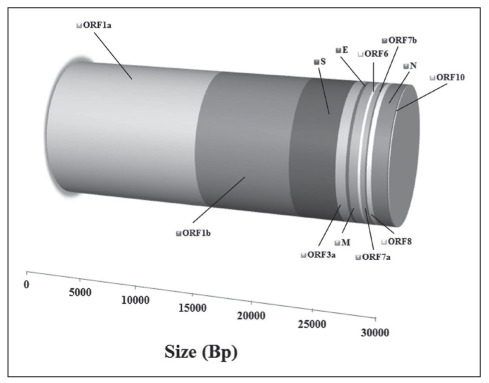
Structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gene.
Table 1.
Gene and protein structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
| Gene | Genomic Size (bp) | Protein size (aa) |
| ORF1a | 13542 | 4405 |
| ORF1b | 8021 | 2691 |
| S | 3821 | 1273 |
| ORF3a | 836 | 275 |
| E | 252 | 75 |
| M | 719 | 222 |
| ORF6 | 196 | 61 |
| ORF7a | 372 | 121 |
| ORF7b | 128 | 34 |
| ORF8 | 372 | 121 |
| N | 1274 | 419 |
| ORF10 | 141 | 38 |
The large and widespread diffusion of ACE2 at cell surface clearly explains the frequent lung involvement with interstitial pneumonia, occasionally evolving into acute respiratory distress syndrome (ARDS), along with possible injury of many other organs and tissues, thus justifying the risk of developing multiple organ failure (MOF), which is then associated with an extremely high death rate (11), especially in certain susceptible populations (12). The histological examination of lung tissue frequently shows diffuse alveolar damage, characterized by the presence of cellular fibromyxoid exudates, desquamation of pneumocytes and hyaline membrane formation, which is consistent with ARDS (13).
Although it has now been convincingly established that COVID-19 has an almost favorable clinical course in as many as 80-85% of infected patients, who can be totally asymptomatic or may only display mild respiratory symptoms, in 10-15% of SARS-CoV-2 positive patients the disease evolves into severe or even critical forms, needing mechanical ventilation, sub-intensive or even intensive care (14,15). This is probably dependent on some demographic (advanced age, male sex) and clinical risk factors (hypertension, diabetes, cardiovascular disease, chronic respiratory disorders, cancer, obesity) (12), but also on the presence of polymorphisms in the sequence of the ACE2 gene, which may variably influence virulence and pathogenecity of SARS-CoV-2 by influencing receptor binding (16).
Despite many biological aspects of this severe infectious disease remain largely obscure, it has now been clearly acknowledged that early management is associated with much better outcome, with lower progression towards systemic complications, including immunosuppression, development of a “cytokine storm” and severe inflammatory response syndrome (SIRS) (17,18). In this perspective, it is now almost unquestionable that laboratory diagnostics plays an essential, almost vital, role in COVID-19 as in many other human disorders (19), as will be further discussed in the following parts of this article.
Etiological diagnosis of COVID-19
Before specifically discussing the current armamentarium for etiological diagnosis, it is worthwhile mentioning here that the WHO currently defines a “confirmed case” of COVID-19 as patient who has received laboratory confirmation of SARS-CoV-2 infection, regardless of the presence of clinical signs and symptoms (20). The almost logical consequence of this straightforward connotation is that the etiological diagnosis of COVID-19 is only possible by detecting nucleic acid material (i.e., RNA) of SARS-CoV-2 in biological samples.
According to the WHO and the US Centers for Disease Control and Prevention (CDC), the material to be collected for initial COVID-19 testing include upper respiratory specimens (nasopharyngeal AND oropharyngeal swab, or wash in ambulatory patients) and/or lower respiratory specimens (sputum and/or endotracheal aspirate or bronchoalveolar lavage) (21-23). Additional biological samples that may be tested include blood, stool, urine, saliva and throat washing, though the significance of identifying the virus in these matrices remains undetermined (24,25) (Table 2). Once appropriately and accurately collected, the biological specimens (especially nasopharyngeal and oropharyngeal swabs) shall be placed into separate sterile tubes, containing 2-3 mL of viral transport media, and must be kept refrigerated at 2-4°C for less than 4 days, or frozen at -70°C (or below) until testing is carried out (26). Processing specimens not fulfilling these stringent pre-analytical requirements may be associated with generation of “false negative” tests results, and shall hence be avoided.
Table 2.
Biological sources where severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be detected in coronavirus disease 2019 (COVID-19) patients.
| Biological source | Detection rate |
| Bronchoalveolar lavage fluid | >90% |
| Saliva | ~90% |
| Sputum | ~70% |
| Nasopharyngeal AND oropharyngeal swabs | ~70% |
| Nasal swabs | ~60% |
| Pharyngeal swabs | ~30% |
| Stool | ~30% |
| Throat washing | ~30% |
| Blood | 15-30% |
The definitive diagnosis of SARS-CoV-2 infection, as endorsed by both the WHO and CDC, shall then be performed using molecular biology techniques on upper and lower respiratory materials. Therefore, the diagnostic strategy encompasses the use of real-time reverse-transcription polymerase chain reaction (rRT-PCR) assays, targeting one or more genes in the SARS-CoV-2 genome. A typical RT-PCR procedure for detecting this coronavirus encompasses, in sequence, RNA isolation, its purification, reverse transcription to cDNA, cDNA amplification with RT-PCR instrumentation, followed by (fluorescent) signal detection (25). A validated diagnostic workflow, which has been endorsed by the WHO, and is hence now largely used in Europe, entails a first-line screening assay with amplification of E gene, followed by a confirmatory assay with amplification of RdRp (RNA-dependent RNA polymerase) gene, and then an additional potential confirmatory assay, entailing amplification of N gene (27). The CDC has also developed a molecular biology assay, that has been defined “Centers for Disease Control and Prevention (CDC) 2019-Novel Coronavirus (2019-nCoV) Real-Time Reverse Transcriptase (RT)-PCR Diagnostic Panel” (28). According to the CDC, the primers and probes for detecting SARS-CoV-2 have been identified from genetic regions belonging to N gene, encompassing the usage of two primer/probe sets. An additional primer/probe set can then be used for amplifying human RNase P gene (RP) in control specimens. Importantly, a recent study which has assessed the comparative performance of multiple primer/probe sets, revealed that the WHO and CDC protocols display exceptional sensitivity compared to other assays (29). Importantly, regardless of the technique that will be used, the identification of SARS-CoV-2 by molecular biology techniques in either upper or lower respiratory specimens enables the diagnosis of active infection from this coronavirus, but does not rule out any co-infection by other microorganisms (e.g., bacteria, fungi, viruses, and so forth) (27).
The accuracy and reliability of RT-PCR for diagnosing SARS-CoV-2 infection depends on many biological and technical variables (30). Beside the influence of procedures used for collecting, transporting and storing the specimens, as well as from concomitant antiviral therapy (26), virus detection is largely influenced by the biological source. Wang et al, for example, recently showed that the rate of RT-PCR detection of SARS-CoV-2 in patients diagnosed with COVID-19 is as high as 93% in bronchoalveolar lavage fluid, but then decreases to 72% in sputum and 63% in nasal swabs, respectively, whilst it is only 32% in pharyngeal swabs and 29% in stool (21). To et al also reported that the positive rate of RT-PCR for SARS-CoV-2 is 15-30% in blood and 14-38% in rectal swabs, respectively (31).
The suboptimal diagnostic accuracy of nasopharyngeal and oropharyngeal swabs has been confirmed in some other published studies. For example, Zhao et al (32) and Yang et al. (33), reported that the positive rate of RT-PCR for SARS-CoV-2 in these materials is only 70%, decreasing to approximately 60% in the study of Ai et al. (34). A major influence of the analytical techniques used for detecting viral RNA has also been recently highlighted by Wang et al, who showed that the limit of detection (i.e., the lowest detectable amount of virus) displayed by six commercial RT-PCR kits is extremely heterogeneous, so that the use of some of these tests may potentially generate false-negative results due to inadequate analytical sensitivity (35). This information is noteworthy, whereby would shed some light on the fact some symptomatic patients who were not originally diagnosed as having SARS-CoV-2 infection by RT-PCR (or who have then been diagnosed as re-infected after two consecutive negative RT-PCR tests) may have been misclassified due to the use of methods with inadequate analytical sensitivity. It is also important to mention here that some of these initially false-negative test results may then turn later positive, when swabs are re-collected some days after initial testing since the incubation of the virus is generally between 3-7 days (36). Interesting evidence has been published by Zhang et al (37), who showed that 14.1% patients who are later diagnosed with COVID-19 may have negative test results initially, but this rate would then decreases in parallel with the number of repeated tests on follow-up, from 6.9% to 0.3% from 2 up to 5 consecutive ensuing swab tests, respectively. Another interesting aspect that emerged from this study, is that the risk of progressing towards more severe disease stages was almost double in patients with initially positive swab test than in those with initially negative result (44.6% vs. 24.4%; p=0.015).
Recent studies have also been published on the possibility to use rapid reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays for SARS-CoV-2 detection, but additional evidence is needed at this point in time for validating their routine usage in COVID-19 diagnostics (38,39). Importantly, fully-automated commercial RT-PCR have also been recently introduced in the diagnostic market, which are characterized by high-throughput and fast turnaround time, thus enabling to reduce by nearly 90% the bench time per sample and allowing to analyze larger volume of patients in a shorter timeframe (40).
Serological testing
Serological testing is conventionally defined as a diagnostic procedure used for identifying the presence of an immune response against an infectious agent (41). Inherent to this definition is the origin of many misunderstanding and misconceptions regarding the use of serological testing in COVID-19, whereby this type of testing is not meant to replace the identification of viral RNA for etiological diagnosis of COVID-19, but rather for establishing as to whether individuals have been infected by the virus and/or have developed an immune response. The CDC endorses a highly reasonable conception underlying serological testing in COVID-19, that is a strategy used mostly for epidemiological and surveillance purposes (28). To put this in the context of COVID-19, serology testing encompasses the identification (by qualitative assays) and/or measurement (using quantitative assays) of different classes of immunoglobulins (typically IgA, IgM, IgG) against SARS-CoV-2 for establishing whether a person has been infected by SARS-CoV-2, and has then developed antibodies which, if possessing neutralizing effects, may prevent future re-infection.
Although the emergence of COVID-19 is still too recent to enable us presenting definitive data on the individual response against this new coronavirus, some useful information has been published. Guo et al have first shown that the median time of antibodies appearance in serum or plasma of COVID-19 patients begins 3-6 days after the onset of symptoms for both IgM and IgA, whilst it is delayed to 10-18 days for IgG (42). The positive rate for the different classes of antibodies is 85.4% for IgM, 92.7% for IgA and 77.9% for IgG, respectively. In another recent study, Padoan et al studied the kinetics of anti-COVID-19 antibodies (43), concluding that IgM and IgG tend to appear 6-7 days after symptoms onset. Notably, although 100% of COVID-19 patients seem to develop anti-SARS-CoV-2 IgG antibodies 12 days after the onset of symptoms, IgM could only be found in <90% of this same group of patients. These important findings have been confirmed in a subsequent study, in which we showed that the rate of anti-SARS-CoV-2 antibody positivity up to two weeks after the onset of symptoms is as high as 100% for both IgA and IgM, whilst IgM could only be measured in 60% of COVID-19 patients after the same period (44). Similar data were published by Jin et al (45), who also showed that positivity for anti-SARS-CoV-2 IgM and IgG antibodies is 50% and 95%, respectively, and by Du et al, who reported that the rate of detectable anti-SARS-CoV-2 IgM and IgG antibodies in convalescent patients is 78% and 100%, respectively (46). In a more recent investigation, Pan et al also observed that the cumulative rate of positivity for anti-SARS-CoV-2 IgM and IgG antibodies 15 days from symptom onset is about 74% and 97%, respectively (47). An interesting aspect, recently highlighted, is that SARS-CoV-2 may trigger efficient generation of secretory IgA even in asymptomatic or mild infections, so that their assessment both in blood and saliva may complement and perhaps improve the diagnostic process (48).
One of the major unresolved issues, almost entirely attributable to the very recent emergence of this novel coronavirus disease, is establishing whether anti-SARS-CoV-2 antibodies shall be considered neutralizing (i.e., effective to neutralize virulence and/or pathogenicity), as well as their persistence in blood. Encouraging data on the former aspect have emerged from a recent publication, showing that human anti-SARS-CoV-2 antibodies seem to specifically target nucleocapsid and spike proteins, and thus possess neutralizing effect against the virus (49). In a separate investigation, Okba et al confirmed that serum collected from COVID-19 patients is capable to neutralize SARS-CoV-2 infection (50). As concerns the persistence of neutralizing antibodies in the circulation, some information can be translated from earlier findings on the former and relatively similar coronavirus disease SARS, whereby the titer of anti-SARS-CoV-1 neutralizing antibodies was found to be stably high for 16 months after infection, but progressively declined afterwards, falling to 50-75% after 4 years and ~10% after 6 years, respectively (51). A final issue that will need to be clarified is the possible cross-reaction of current anti-SARS-CoV-2 immunoassays with previous coronaviruses such as SARS-CoV-1, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-NL63, and HCoV-229E.
Rapid serological testing
The first serological strategy entails qualitative (or semi-quantitative) assessment by means of the so-called “rapid tests”, which are basically portable devices to be used singly, with non-automated procedures, for producing rapid test results (i.e., around 5-20 min). Since the leading advantages of these membrane-based immunoassays encompass low sample volume (a drop of blood may generally be sufficient), little operator training, low cost, easy performance and relatively simple interpretation, their usage is mostly reserved to bedside or near-to-patient rapid testing (52). These tests could conventionally entail two strategies, the former encompassing direct detection of SARS-CoV-2 antigens, the latter based instead on anti-SARS-CoV-2 antibodies identification. A comprehensive description of this technology has been provided, and is regularly updated, by the European Center for Disease Control and Prevention (ECDC) (53).
Major concern has been recently raised on the analytical and diagnostic performance of these tests, especially after Spain and some other European countries complained that many rapid test kits are inaccurate and do not allow to obtain a reliable diagnosis and surveillance of COVID-19 (54). Additional emphasis has then been provided by the recent publication of a study by Cassaniti et al (55), who claimed that the sensitivity of one of these rapid tests was <20%, thus potentially leading to under-diagnosing COVID-19 in a large subset of patients. This would persuade us to conclude that the general paradigm that “one-size-fits-all” does not (and shall not) apply here, and that each single device must be adequately validated before entering routine clinical usage. The underlying problem is the fact that some of these tests underwent quick commercialization, without adequate analytical and clinical validation. Our straightforward suggestion, also endorsed by the ECDC, is that scientific publications shall be made urgently available for clarifying performance and limitations of each single rapid diagnostic test before its introduction into routine diagnostics, clinical management and public health or epidemiologic surveillance (53). It shall also be clear, that the most reasonable placement of these tests within the clinical decision making is for supporting decentralized testing capacity, but they shall not be considered a replacement of central laboratory diagnostics.
Centralized serological laboratory testing
The second serological option encompasses centralized testing within microbiological and clinical laboratories, by using fully-automated immunoassays (56). Although this alternative strategy is more expensive, requires the collection of whole blood samples by venipuncture rather than capillary blood, and is essentially dependent on availability of specific laboratory analyzers, it has some important advantages. These basically include better accuracy and reliability, the possibility to generate quantitative data (which are essential for longitudinal titer monitoring), performance by skilled laboratory personnel (thus inherently lowering the risk of errors and subjective interpretation), permanent storage of test results within the laboratory information system (LIS), along with more stringent quality monitoring as enabled by performance of internal quality control and, hopefully in a near future, external quality assessment (EQAs) schemes.
The modern generation of laboratory analyzers is characterized by exceptional throughput and very limited turnaround time (i.e., they can perform hundreds tests per hour). The use of centralized laboratory diagnostics shall hence be considered a robust and viable strategy for epidemiological surveillance purposes. Importantly, the University Hospitals of Padova and Verona (Italy) have been forerunners worldwide in conceiving and developing a project, which has been approved by the scientific committee of the Veneto Region and is now underway, entailing a vast epidemiological screening by means of validated fully-automated immunoassays of all healthcare personnel working in the Veneto region (i.e., between 50,000-70,000 people). Phase 2 of this project encompasses the possibility to broaden this epidemiological analysis to the nearly 5 million inhabitants of the entire Veneto region (57).
Laboratory monitoring and risk prediction
Since the current epidemiological figures contribute to raise several doubts that the pandemic will cease soon, it becomes imperative to identify reliable predictors of disease severity, which may enable earlier clinical interventions and more appropriate usage of healthcare resources within a system of care whose responsive capacity has been literally overwhelmed by this unprecedented and virtually unpredictable epidemiological crisis (58,59). Therefore, the possibility to identify a subset of subjects which will be more likely to progress towards severe/critical disease is an additional and almost essential contribution provided by laboratory medicine. This group of patients can be identified by discretional use of laboratory resources, whereby unfavorable clinical course has been associated with lymphopenia, thombocytopenia, neutrophilia, increased concentration of biomarkers of cardiac injury (i.e., cardiac troponins), C reactive protein and other inflammatory cytokines, liver and kidney function tests (60,61), as well as of D-dimer (62) and procalcitonin (63).
Conclusions
The fairly recent emergence of COVID-19, the third coronavirus outbreak after SARS in 2002-2003 and Middle East respiratory syndrome (MERS) in 2012, does not allow drawing definitive conclusions on SARS-CoV-2 diagnostics. Nevertheless, some standpoints can be conveyed (Figure 2).
Figure 2.
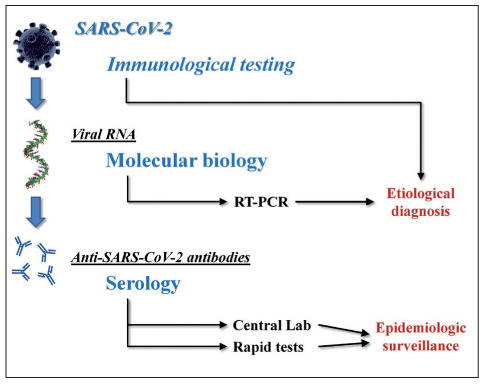
Laboratory diagnostics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gene infection and coronavirus disease 2019 (COVID-19).
RT-PCR, reverse transcription polymerase chain reaction
First and foremost, it seems now rather clear that we will be living together with this virus for a quite a long time, so that our vigilance and responsiveness against the emergence of new local outbreaks must be maintained at the highest possible levels. That said, the etiological diagnosis of COVID-19 is, and will remain for long, deeply based on direct identification - by means of molecular biology techniques - of viral RNA in biological materials, especially upper and lower respiratory specimens. Whether other biological matrices, such as blood, urine, stools and saliva, will represent valid alternatives has not been unequivocally defined, so far. As concerns serological testing, promising information can be garnered from preliminary investigations, showing that the vast majority of COVID-19 patients seem to develop a sustained immune response against the virus, characterized by emergence of anti-SARS-CoV-2 IgG and IgA, 1 to 2 weeks after the onset of fever and/or respiratory symptoms. Whether these antibodies will have persistent neutralizing activity against the virus is still to be elucidated on an individual and general basis. The availability of rapid tests for detecting either viral antigens or anti-SARS-CoV-2 antibodies shall then be seen as a potentially viable opportunity for purposes of epidemiologic surveillance, though more information is needed on accuracy and reliability of the many portable immunoassays that are now widely available in the market.
One final consideration shall be clearly highlighted. Laboratory medicine, along with many other clinical disciplines, has demonstrated an extraordinary resilience in managing the current crisis. All laboratory professionals have supplied to the lack of human and technical resources, caused by unreasonable cuts suffered during the past decades, with fearless work and spasmodic devotion (64). This crisis has hence once more demonstrated that laboratory diagnostics has always been, is still, and will ever remain at the very core of the clinical decision making. Policymakers and hospital administrators shall take the unfortunate example of COVID-19 pandemic as a firm paradigm for more reasonably planning the future of this discipline.
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1.Lippi G, Sanchis-Gomar F, Henry BM. Coronavirus disease 2019 (COVID-19): the portrait of a perfect storm. Ann Transl Med. 2020 doi: 10.21037/atm.2020.03.157. doi: 10.21037/atm.2020.03.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippi G, Henry BM, Bovo C, et al. Health risks and potential remedies during prolonged lockdowns for coronavirus disease 2019 (COVID-19) Diagnosis (Berl) 2020 doi: 10.1515/dx-2020-0041. Doi: 10.1515/dx-2020-0041. [DOI] [PubMed] [Google Scholar]
- 4.Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GenBank. Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. https://www.ncbi.nlm.nih.gov/nuccore/NC_045512 . Last accessed, April 15. [Google Scholar]
- 6.Cagliani R, Forni D, Clerici M, et al. Computational inference of selection underlying the evolution of the novel coronavirus, SARS-CoV-2. J Virol. 2020 Apr 1 doi: 10.1128/JVI.00411-20. pii: JVI.00411-20. doi: 10.1128/JVI.00411-20. (Epub ahead of print) 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Zhang Y, Wu L, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020 Apr 7 doi: 10.1016/j.cell.2020.03.045. doi: 10.1016/j.cell.2020.03.045. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 Mar 12 doi: 10.1007/s11684-020-0754-0. doi: 10.1007/s11684-020-0754-0. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 Mar 4 doi: 10.1016/j.cell.2020.02.052. doi: 10.1016/j.cell.2020.02.052. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020 Feb 17:105924. doi: 10.1016/j.ijantimicag.2020.105924. doi: 10.1016/j.ijantimicag.2020.105924. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippi G, Mattiuzzi C, Sanchis-Gomar F, et al. Clinical and demographic characteristics of patients dying from COVID-19 in Italy versus China. J Med Virol. 2020 Apr 10 doi: 10.1002/jmv.25860. doi: 10.1002/jmv.25860. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-9 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020 Feb 18:pii. doi: 10.1016/S2213-2600(20)30076-X. S2213-2600(20)30076-X. doi: 10.1016/S2213-2600(20)30076-X. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 Feb 24 doi: 10.1001/jama.2020.2648. doi: 10.1001/jama.2020.2648. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 15.Onder G, Rezza G, Brusaferro S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA. 2020 Mar 23 doi: 10.1001/jama.2020.4683. doi: 10.1001/jama.2020.4683. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 16.Hussain M, Jabeen N, Raza F, et al. Structural Variations in Human ACE2 may Influence its Binding with SARS-CoV-2 Spike Protein. J Med Virol. 2020 Apr 6 doi: 10.1002/jmv.25832. doi: 10.1002/jmv.25832. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta P, McAuley DF, Brown M, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham AC, Goh HP, Koh D. Treatment of COVID-19: old tricks for new challenges. Crit Care. 2020;24:91. doi: 10.1186/s13054-020-2818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippi G, Plebani M. A modern and pragmatic definition of Laboratory Medicine. Clin Chem Lab Med. 2020 Feb 18 doi: 10.1515/cclm-2020-0114. doi: 10.1515/cclm-2020-0114. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization Coronavirus disease 2019 (COVID-19) Situation Report. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ . Last accessed, April 15, 2020. [Google Scholar]
- 21.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 Mar 11 doi: 10.1001/jama.2020.3786. doi: 10.1001/jama.2020.3786. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo WL, Jiang Q, Ye F, et al. Effect of throat washings on detection of 2019 novel coronavirus. Clin Infect Dis. 2020 Apr 9 doi: 10.1093/cid/ciaa416. pii: ciaa416. doi: 10.1093/cid/ciaa416. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To KK, Tsang OT, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 Feb 12 doi: 10.1093/cid/ciaa149. doi: 10.1093/cid/ciaa149. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim guidance. 2 March 2020 [Google Scholar]
- 25.Centers for Disease Control and Prevention. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons Under Investigation (PUIs) for Coronavirus Disease 2019 (COVID-19) Available at: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html . Last accessed, April 15, 2020. [Google Scholar]
- 26.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020 Mar 16 doi: 10.1515/cclm-2020-0285. doi: 10.1515/cclm-2020-0285. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 27.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention CDC Tests for COVID-19. Available at: https://www.cdc.gov/coronavirus/2019-ncov/about/testing.html . Last accessed, April 15, 2020. [Google Scholar]
- 29.Nalla AK, Casto AM, Huang MW, et al. Comparative Performance of SARS-CoV-2 Detection Assays using Seven Different Primer/Probe Sets and One Assay Kit. J Clin Microbiol. 2020 Apr 8 doi: 10.1128/JCM.00557-20. doi: 10.1128/JCM.00557-20. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galli C, Plebani M. Clinical laboratory and SARS-CoV-2 infection: where do we stand? Clin Chem Lab Med. 2020 Apr 2 doi: 10.1515/cclm-2020-0372. doi: 10.1515/cclm-2020-0372. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 31.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 Mar 23 doi: 10.1016/S1473-3099(20)30196-1. doi: 10.1016/S1473-3099(20)30196-1. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease. Clin Infect Dis. 2019 doi: 10.1093/cid/ciaa344. 2020 Mar 28. pii: ciaa344. doi: 10.1093/cid/ciaa344. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. MedRxiv. 2020.02.11.20021493. [Google Scholar]
- 34.Ai T, Yang Z, Hou H, et al. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020 Feb 26:200642. doi: 10.1148/radiol.2020200642. doi: 10.1148/radiol.2020200642. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Yao H, Xu X, et al. Limits of Detection of Six Approved RT-PCR Kits for the Novel SARS-coronavirus-2 (SARS-CoV-2) Clin Chem. 2020 Apr 13 doi: 10.1093/clinchem/hvaa099. doi: 10.1093/clinchem/hvaa099. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JJ, Cao YY, Dong X, et al. Distinct characteristics of COVID-19 patients with initial rRT-PCR positive and negative results for SARS-CoV-2. Allergy. 2020 Apr 13 doi: 10.1111/all.14316. doi: 10.1111/all.14316. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020 Apr 7 doi: 10.1016/j.cmi.2020.04.001. doi: 10.1016/j.cmi.2020.04.001. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park GS, Ku K, Baek SH, et al. Development of Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) Assays Targeting SARS-CoV-2. J Mol Diagn. 2020 Apr 7 doi: 10.1016/j.jmoldx.2020.03.006. doi: 10.1016/j.jmoldx.2020.03.006. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poljak M, Korva M, Knap Gašper N, et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020 Apr 10:pii. doi: 10.1128/JCM.00599-20. JCM.00599-20. doi: 10.1128/JCM.00599-20. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fierz W. Basic problems of serological laboratory diagnosis. Methods Mol Med. 2004;94:393–427. doi: 10.1385/1-59259-679-7:393. [DOI] [PubMed] [Google Scholar]
- 42.Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis. 2020 Mar 21:pii. doi: 10.1093/cid/ciaa310. ciaa310. doi: 10.1093/cid/ciaa310. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padoan A, Cosma C, Sciacovelli L, et al. Analytical performances of a chemiluminescence immunoassay for 2019-nCov IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020 Apr 16 doi: 10.1515/cclm-2020-0443. doi: 10.1515/cclm-2020-0443. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44.Lippi G, Salvagno GL, Pegoraro M, et al. Assessment of immune response to SARS-CoV-2 with fully-automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med. 2020 Apr 16 doi: 10.1515/cclm-2020-0473. doi: 10.1515/cclm-2020-0473. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 45.Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020 Apr 3 doi: 10.1016/j.ijid.2020.03.065. pii: S1201-9712(20)30198-3. doi: 10.1016/j.ijid.2020.03.065. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Z, Zhu F, Guo F, et al. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J Med Virol. 2020 Apr 3 doi: 10.1002/jmv.25820. doi: 10.1002/jmv.25820. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020 Apr 10 doi: 10.1016/j.jinf.2020.03.051. pii: S0163-4453(20)30175-4. doi: 10.1016/j.jinf.2020.03.051. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Béné MC, de Carvalho M, Eveillard M, et al. Good IgA bad IgG in SARS-CoV-2 infection? Clin Infect Dis. 2020 Apr 11 doi: 10.1093/cid/ciaa426. pii: ciaa426. doi: 10.1093/cid/ciaa426. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020 Mar;25(11) doi: 10.2807/1560-7917.ES.2020.25.11.2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okba NMA, Müller MA, Li W, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg Infect Dis. 2020 Apr 8;26(7) doi: 10.3201/eid2607.200841. doi: 10.3201/eid2607.200841. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q, Zhu L, Ni Z, et al. Duration of serum neutralizing antibodies for SARS-CoV-2: Lessons from SARS-CoV infection. J Microbiol Immunol Infect. 2020 Mar 25 doi: 10.1016/j.jmii.2020.03.015. pii: S1684-1182(20)30075-X. doi: 10.1016/j.jmii.2020.03.015. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vashist SK. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics. 2020;10:202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.European Center for Disease Control and Prevention An overview of the rapid test situation for COVID-19 diagnosis in the EU/EEA. Last Update: 1 April 2020. [Google Scholar]
- 54. [No Author Listed]. Spain and Other Countries Return Defective COVID-19 Test Kits to China. Available at: https://www.sciencetimes.com/articles/25145/20200329/spain-and-other-countries-return-defective-covid-19-test-kits-to-china.htm . Last accessed, April 20, 2020. [Google Scholar]
- 55.Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiagTM COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020 Mar 30 doi: 10.1002/jmv.25800. doi: 10.1002/jmv.25800. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lippi G, Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. 2020 Mar 19 doi: 10.1515/cclm-2020-0240. doi: 10.1515/cclm-2020-0240. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 57. [No Author Listed]. Coronavirus, the “immunity license” in Veneto: carpet antibody test. Available at: https://www.news1.news/i/2020/04/coronavirus-the-immunity-license-in-veneto-carpet-antibody-test.html . Last accessed: Last Update 15 April, 2020. [Google Scholar]
- 58.Remuzzi A, Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020 Mar 13 doi: 10.1016/S0140-6736(20)30627-9. doi: 10.1016/S0140-6736(20)30627-9. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moghadas SM, Shoukat A, Fitzpatrick MC, et al. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc Natl Acad Sci U S A. 2020 Apr 3 doi: 10.1073/pnas.2004064117. pii: 202004064. doi: 10.1073/pnas.2004064117. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020 Mar 3 doi: 10.1515/cclm-2020-0198. doi: 10.1515/cclm-2020-0198. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 61.Henry BM, Santos de Oliveira MH, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0369. Doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 62.Lippi G, Favaloro EJ. D-dimer is Associated with Severity of Coronavirus Disease 2019: A Pooled Analysis. Thromb Haemost. 2020 Apr 3 doi: 10.1055/s-0040-1709650. doi: 10.1055/s-0040-1709650. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clin Chim Acta. 2020;505:190–1. doi: 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lippi G, Plebani M. Laboratory Medicine resilience during coronavirus disease 2019 (COVID-19) pandemic. Adv Lab Med. 2020 doi: 10.1515/almed-2020-0035. Doi: 10.1515/almed-2020-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]


