Abstract
Background
We evaluated the mutation status of c-Met in small cell lung cancer (SCLC) and neuroendocrine tumors (NET), for which relatively limited therapeutic targets have been explored.
Materials and Methods
c-Met was re-sequenced using cell lines and clinical samples. For in vitro studies, DNA constructs containing a juxtamembrane domain (JMD) and tyrosine kinase domain (TKD) were generated. Detected mutations were introduced into the construct and effects on c-Met phosphorylation and interaction with tyrosine kinase inhibitor drugs BMS777607 and SU11274 were assessed.
Results
97 specimens were analyzed: 13 SCLC and 2 pulmonary carcinoid cell lines, 46 SCLC and 36 NET clinical specimens. Mutations were only detected in the JMD. No mutations were detected in the TKD. Found mutations consisted of the previously reported R988C and T1010I mutations. One novel JMD mutation, P996S, was detected in a SCLC specimen. The mutation rate in SCLC cell lines was 25% (31% including a derivative cell line), and 6.5% in clinical specimens. The mutation rate in NET was 8.3%. In vitro, there were no differences between wild type, R988C or T1010I mutants regarding c-Met phosphorylation at Y1003, located in the JMD, and at Y1234/1235, located in the TKD. BMS777607 and SU11274 were shown to inhibit phosphorylation of c-Met in wild type and R988C and T1010I mutants in a similar fashion.
Conclusions
In SCLC and neuroendocrine tumors MET mutations are relatively rare. Detected mutations were located in the juxtamembrane domain and were of no functional relevance as they did not influence c-Met phosphorylation, regardless of TKI treatment.
Keywords: c-Met, mutation, neuroendocrine tumor, tyrosine kinase inhibitor
INTRODUCTION
Receptor tyrosine kinases (RTKs) play a key role in carcinogenesis and have been shown to be a good target for therapeutic intervention, e.g. Epidermal Growth Factor Receptor (EGFR) targeted therapy in non-small cell lung cancer using EGFR tyrosine kinase inhibitor (TKI) drugs such as erlotinib (Tarceva®) [1]. Overexpression and mutations of RTKs have been reported, leading to constitutive activation. The c-Met family of RTKs consists of two members, RON and c-Met [2]. The MET proto-oncogene encodes the hepatocyte growth factor/scatter factor (HGF/SF) receptor, c-Met (or HGFR), which was originally identified as a fusion oncogene, TPR-MET, resulting from chromosomal translocation [3]. c-Met/ HGFR signaling is triggered by binding to its ligand HGF (or “scatter factor”) and is involved in numerous important biological processes, including cell growth, transformation, invasion and notably “cell scattering”, and epithelial to mesenchymal transition (EMT) resulting in increased mobility [4].
Overexpression of c-Met and its activation by autocrine HGF expression is found in a variety of human tumors indicating coexpression of HGF and c-Met may be involved in tumor metastasis [5, 6]. Cross talk between c-Met and other RTKs has been reported and c-MET signaling is activated in some non-small cell lung cancer (NSCLC) tumors harboring EGFR activating mutations [7, 8]. Moreover, acquired resistance to EGFR-tyrosine kinase inhibitors in NSCLC has been associated with focal amplification of the MET proto-oncogene [9, 10].
Accumulating evidence suggests that targeting c-Met is a potentially effective cancer therapy and currently numerous compounds targeting c-Met signaling are in clinical development [11, 12]. Activating mutations in the c-Met tyrosine kinase domain have been reported. Hereditary papillary renal cell carcinoma was the first cancer for which germ line mutations were identified [13]. These mutations have been shown to result in constitutive activation of the tyrosine kinase without ligand stimulation and are involved in malignant transformation [14, 15]. In contrast, in small cell lung cancer, mutations have been predominantly found in the juxtamembrane domain of the c-Met protein [16, 17]. The juxtamembrane domain is of importance in c-Met signaling as it contains a c-Cbl (“Casitas B-lineage Lymphoma”)-binding domain which includes tyrosine 1003 (“Y1003”). Recruitment of c-Cbl, an E3 ubiquitin-protein ligase, induces polyubiquitination and subsequent degradation of the receptor [18]. Accordingly, the juxtamembrane domain is thought to negatively regulate c-Met activity and deletion of the juxtamembrane domain was reported in lung cancer [19, 20]. Missense juxtamembrane mutations, R988C and T1010I, have been reported in small cell lung cancer (SCLC) and non-small cell cancer (NSCLC) as oncogenic mutations [16]. However, others reported recently that the mutations were rather irrelevant “passenger” mutations without evidence of increased phosphorylation or transformative capacity [21]. Other mutations in the extracellular semaphorin domain and the immunoglobulin plexin transcription (IPT) domain have also been reported [13].
It was our aim to perform c-Met mutation analysis in an additional, larger cohort of small cell lung cancer specimens. As a potential therapeutic target, with specific TKI drugs available or in development, we wanted to achieve a more accurate estimation of the type and incidence of c-Met mutations in SCLC. Recently we have shown by array comparative genomic hybridization (aCGH)-based characterization the existence of shared copy number variations in small cell lung cancer as well as bronchial carcinoids and carcinoids of gastrointestinal origin, suggesting common oncogenic events in the carcinogenesis of neuroendocrine tumors [22]. Therefore, we also assessed the occurrence of c-Met mutations in our cohort of NETs. Few molecular targets for these less common tumors have been explored so far.
Functional implications of found mutations in terms of c-Met activation and interaction with the effects of c-Met tyrosine kinase inhibitor drugs BMS777607 and SU11274 were assessed in in vitro studies.
MATERIAL AND METHODS
Sample Acquisition
Thirteen SCLC cell lines were selected for this study: GLC4 and GLC4-CDDP (kindly provided by Dr. S. de Jong, Groningen, Netherlands), NCI-H69, NCI-H82, NCI-H128, NCI-H146, NCI-H187, NCI-H526, NCI-H592, NCI-H620, NCI-H678, NCI-H792, NCI-H1173 and 2 pulmonary carcinoid cell lines: H720 and H727 (all from National Cancer Institute, NIH, Bethesda, MD). Eightytwo formalin fixed paraffin-embedded (FFPE) and cytospin SCLC or neuroendocrine tumor samples were included. Samples originated from the Suburban Hospital and the National Cancer Institute, NIH, Bethesda, MD, the University of Pisa, Pisa, Italy, and the VU University Medical Center, Amsterdam, Netherlands. Use of human samples was approved by Institutional Review Boards according to the legal regulations of the participating countries.
DNA Isolation and Re-sequencing of the MET Gene
Total genomic DNA was isolated from cell lines, cytospins or formalin fixed paraffin embedded (FFPE) tissues using reagents of the DNeasy Blood and Tissue Kit, QIAquick PCR Purification Kit (Qiagen, Gaithersburg, MD) as well as Dako Target Retrieval Solution (Dako, Carpinteria, CA) in protocols optimized for maximum yield from the various types of sample preparations [23]. For cell lines, the codon regions of all 21 exons of the MET proto-oncogene (GenBank accession no. NM_000245) were re-sequenced, divided over 32 amplicons.
For clinical samples, re-sequencing was limited to either exons 11–19 (juxtamembrane domain and tyrosine kinase domain) or exon 14 only (juxtamembrane domain). A customized 96-well plate based protocol was used, allowing re-sequencing according to the VariantSEQr™ method (Applied Biosystems, Foster City, CA). PCR primers, based on sequences from the Oncogenomics Primer Database (http://ntddb.abcc.ncifcrf.gov/cgi-bin/Primers.pl) were further optimized and contained priming sites for M13 forward or reverse primers for subsequent sequencing reactions. The PCR cycling conditions were as follows: one cycle of 5 min at 96 °C/40 cycles of 30 s at 94 °C, 45 s at 60 °C and 45 s at 72 °C/one cycle at 72 °C for 10 min. Samples were sequenced using BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and analyzed on an ABI 3700 sequencer (Applied Biosystems), according to manufacturer’s guidelines. ExoSap-IT (USB Corp., Cleveland, OH) and the DyeEx 96-kit (Qiagen) were used for PCR-product and sequencing reaction clean-up, respectively, both according to the manufacturer’s guidelines. Dual directional sequence traces were analyzed for potential variants using Mutation Surveyor software (Softgenetics, State College, PA).
Antibodies and Compounds
The following antibodies were used: rabbit polyclonal anti-Met (c12) (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit monoclonal anti-phospho-Met (Tyr1234/1235) (D26) (Cell Signaling Technology, Danvers, MA); rabbit polyclonal anti-phospho-Met (Tyr1003) (Invitrogen, Carlsbad, CA); mouse monoclonal anti-beta-actin (AC-74) (Sigma, St. Louis, MO). Met inhibitor BMS777607 was obtained from Bristol-Myers Squibb (New York, NY). SU11274 was purchased from Sigma.
Plasmid Construction and Site-directed Mutagenesis
For functional in vitro studies, DNA constructs containing a YFP-tagged juxtamembrane (JM) domain and tyrosine kinase domain (TKD) were generated according to our previously described method [24, 25]. In order to generate the MET juxtamembrane-tyrosine kinase domain containing construct, a DNA fragment encoding MET residues 956–1408 was amplified by PCR using primers 5’-CCGGAATTCTGTTGTCTCAATATCAACAGC-3’ and 5’-CGCGGATCCCTATGA
TGTCTCCCAGAAGGAG-3’ and full-length human MET cDNA (kindly provided by Dr. D. Bottaro, National Cancer Institute) as template. The amplified product was digested with EcoRI and BamHI and cloned into the pEYFP-C1 mammalian expression vector (Clontech, Palo Alto, CA). SCLC- associated mutations were subsequently introduced into the YFP- MET construct using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following manufacturer’s protocol. In all cases, the sequence of the inserts was verified by DNA sequencing. The primers for R988C mutation were 5’- CTGGGCAGTGAATTAGTTTGC TACGATGCA AGAGTAC-3’and 5’-GTACTCTTGCATCGTA GCAAACTAATTCACTGCCCAG-3’.
The primers for T1010I mutation were 5’-GCCCGAAGTGT AAGCCCAATTACAGA AATGGTTTCAAATG-3’ and 5’-CATTTGAAACCATTTCTGTAATTGGGCTTAC ACTTCGGGC-3’. It must be noted that there are two isoforms of the MET gene. Isoform 2 is the longer variant with an additional 18 amino acids following S755. The amino acids are numbered according to isoform 2 in this report. For example, according to isoform 1, the R988C and T1010I mutations are numbered R970C and T992I, respectively (http://www.uniprot.org) [26].
Cell Culture, Transfection and drug Treatment
SCLC, pulmonary carcinoid cell lines and human breast cancer cells MCF-7 were grown in RPMI or Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen), supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco-Invitrogen). Cells were seeded onto twelve-well trays, and transfected with 0.3 μg of plasmid DNA using the Effectene reagent (Qiagen), according to the manufacturer’s protocol. The drugs were added at the indicated concentration 24 hours after transfection, and the cells were incubated for 1 hour before being processed for Western blot analysis. Drug treatment was always performed in standard culture medium containing 10% FCS.
RESULTS
Mutation analysis of the MET Gene in Small Cell Lung Cancer and Neuroendocrine Tumors
The MET gene was re-sequenced in 97 specimens consisting of 13 SCLC cell lines, 2 pulmonary carcinoid cell lines, 46 SCLC clinical specimens and 36 neuroendocrine tumor (NET)/ carcinoid tumor clinical specimens (Table 1). Excluding repeat samples 1164 amplicons were re-sequenced.
Table 1.
c-Met Re-sequencing Results in Cell Lines and Clinical Samples
| Sample | n | Exon | Mutation | Protein Domain | Mutation Rate |
|---|---|---|---|---|---|
| SCLC cell line | 12* | 1–21 | R988C (n=3) | JM | 25% |
| SCLC tumor | 26 | 11–19 | R988C (n=1) | JM | 6.5% |
| SCLC tumor | 20 | 14 | R988C (n=1) P996S (n=1) |
JM | |
| carcinoid cell line | 2 | 1–21 | none | n/a | n/a |
| NET# | 26 | 11–19 | T1010I (n=2) | JM | 8.3% |
| pulmonary carcinoid tumor | 10 | 14 | T1010I (n=1) | JM | |
| Total | 96* | P996S (n= 1) R988C (n= 5) T1010I (n = 3) |
JM | 9.4% |
Abbreviations: n: number; NET: neuroendocrine tumor; JM: juxtamembrane; n/a: not applicable.
number not including the GLC-4 CDDP cell line (see text)
including 6 pulmonary carcinoid tumors
In cell lines, all 21 exons were re-sequenced. The previously identified heterozygous missense mutation R988C was identified in the NCI-H69 cell line (“classic SCLC”), which was used as a positive control [16]. Additionally, the R988C mutation was detected in the NCI-H592 (“classic SCLC”), GLC-4 (“variant SCLC”) and GLC-4 CDDP cell lines (Fig. (1). GLC-4 CPPD is a cisplatin resistant cell line, derived from the parental GLC-4 cell line, which retained the R988C mutation. Mutation rate was 25% (3/12) in tested cell lines, 31% (4/12) when including the GLC-4 CDDP cell line. For the results listed in (Table 1) the GLC-4 CDDP cell line was excluded. Hence in (Table 1) the total number of cell lines is 12 and the total number of specimens 96.
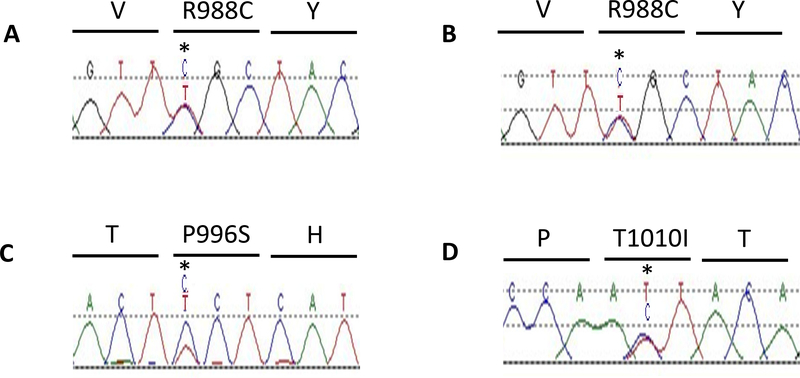
Fig. (1).
c-MET re-sequencing in SCLC and neuroendocrine tumors. A, JM heterozygous missense mutation of c-MET in SCLC cell line NCI-H69 and B, in a SCLC tumor. C, novel JM heterozygous missense mutation P996S in a SCLC tumor. D, JM heterozygous missense mutation T1010I in a carcinoid tumor.
Note: mutations confirmed by bi-directional sequencing, only forward sequences displayed.
After re-sequencing the first batch of 52 clinical specimens (26 SCLC and 26 NET) for the JM and TKD domains (exons 11–19), mutations were again detected exclusively in the JM domain. Therefore, in remaining clinical samples, only the JM domain (exon 14) was re-sequenced (20 SCLC and 10 carcinoid). In clinical SCLC specimens the R988C heterozygous missense mutation was detected in 2 samples, one from a cerebellar SCLC metastasis and one from a mediastinal tumor mass of small cell anaplastic/ undifferentiated lung cancer histology. One novel heterozygous missense juxtamembrane domain mutation, P996S, giving rise to an amino acid change of a proline to a serine, was also detected in a left lower lobe localization of small cell lung cancer. The total mutation rate in SCLC clinical samples [n = 46] was 6.5%. The R988C mutation rate in SCLC clinical samples was 4.3%. No mutations were detected in the two pulmonary carcinoid cell lines. In NET [n = 36] the T1010I juxtamembrane mutation was identified in 3 out of 36 samples (8.3%), consisting of a bladder paraganglioma, a pulmonary typical carcinoid specimen and another pulmonary carcinoid specimen. In pulmonary carcinoid tumors [n = 16] the mutation rate was 12.5%. Prior studies have reported mutation rates of 12.5% and 2.3% in clinical SCLC samples as well as higher mutation rates in SCLC cell lines [16, 17]. No prior reports exist on the mutation rate in carcinoids or other neuroendocrine tumors. See (Table 2) for an overview of reports on MET mutation analysis in SCLC (including current report). The mutation rate in clinical specimens is much lower compared to cell lines. In 32 matched tumor/ normal tissue SCLC specimens all found mutations were tumor specific (somatic mutation) [16]. Mutations were predominantly located in the JM domain.
Table 2.
c-Met Mutation Rate in SCLC
| Sample | n | Exon | Mutation Rate | Mutation, Type± | Mutated Protein Domain | Ref. |
|---|---|---|---|---|---|---|
| SCLC cell lines* | 17 | 1–21 | 5/17 (29.4%) | R988C (GLC-4, H69, H249, H592): missense Splice variant exon 10 (H128): deletion |
JM (23.5%) IPT(5.9%) |
[16] |
| SCLC tumors T/N# | 32 | 1–21 | 4/32 (12.5%) | E168D: missense (somatic) IVS13- (52–53)insCT**: insertion (somatic) T1010I: missense± (somatic) |
Sema (3.1%) Pre-JM (6.3%) JM (3.1%) |
[16] |
| SCLC tumors§ | 90 | 11–19 (n = 26) 2, 14 (n = 44) 14 (n = 20) |
1/26 (3.8%) 1/44 (2.3%) 2/20 (10%) |
T995I, P996S, R988C: missense© | JM (4.4%) | [17] |
| Total tumors | 122 | Variable | 8/122 (6.6%) | E168D, IVS13- (52–53)insCT, T995I, P996S, R988C | Sema (n = 76)¶ (1.3%) Pre-JM (n =32)¶ (6.3%) JM (n = 122)¶ (4.1%) |
Notes:
Cell lines: NCI-H69, H82, H128, H146, H187, H209, H249, H345, H446, H510, H526, (genomic DNA and cDNA), GLC-4, NCI-H592, H620, H678, H792, H1173 (genomic DNA)
genomic DNA and cDNA
genomic DNA
two-base-pair insertional mutation
two additional silent mutations/ polymorphisms were detected: S178S (ACT→ATT)
all missense mutations heterozygous
indicates number of samples analyzed for specific domain. Publication reference number indicated between brackets (first column).
Abbreviations: sema: semaphorin; IPT: immunoglobulin plexin transcription; JM: juxtamembrane T/N: tumor/ normal (paired).
Functional Implications of R988C and T1010I Mutations on c-MET Activation Status with or without Exposure to c-Met Tyrosine Kinase Inhibitors BMS777607 and SU11274
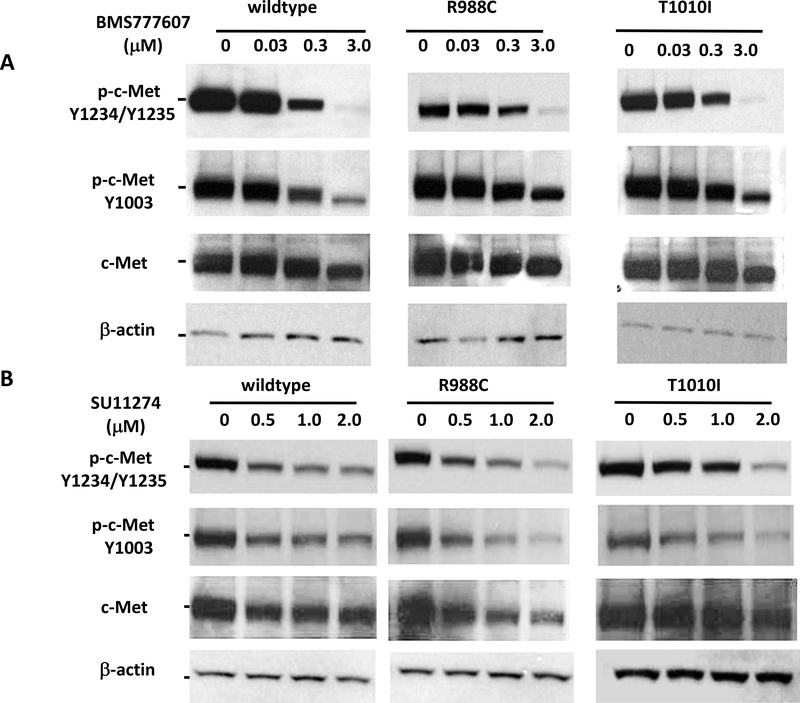
Juxtamembrane domain mutant forms, R988C and T1010I, were reported as oncogenic mutations [16]. To elucidate the mechanism of c-Met activation, we made truncated YFP-tagged c-Met constructs, containing only the juxtamembrane, the tyrosine kinase and C-terminal regulatory domains. Using these constructs and by introducing found mutations into the construct, we were able to observe the effects of mutations without the influence of ligand stimulation. Autophosphorylation of Y1234/1235 in the activation loop is an indicator of and required for c-Met activation [27]. c-Met phosphorylation status was determined by Western blot analysis. In untreated condition, there were no differences between wild type and mutant c-Met phosphorylation status at Y1003 and Y1234/1235 (Fig. 3).
Fig. (3).
MET JM-TK domain constructs, wild type or harboring either the R988C or T1010I mutation, which were detected in several clinical specimens, were transfected into MCF-7 cells. Transfected cells were treated with c-Met TKIs BMS777607 (A) or SU11274 (B). Met phosphorylation was determined by Western blot analysis. The drug response did not differ between wild type and juxtamembrane mutants.
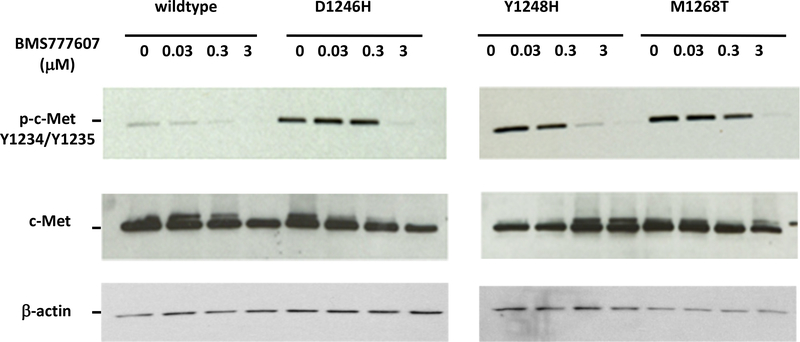
A great variety of agents targeting c-Met are currently in clinical development. BMS-777607, an ATP-competitive MET inhibitor binds to c-Met ATP-binding site, disrupting the MET signaling pathway, and potentially inducing cell death in tumor cells expressing c-Met. It is currently being investigated in a phase 1/2 study and has been shown to suppress the HGF-stimulated prostate cancer metastatic phenotype [28, 29]. SU11274 is an ATP-competitive small molecule inhibitor of the catalytic activity of the c-Met tyrosine kinase. Selective Met inhibitors, BMS777607 and SU11274, inhibited c-Met phosphorylation at indicated concentrations. First we wanted to demonstrate if the model was effective in showing a functional effect of mutations on phosphorylation status and drug sensitivity in terms of dephosphorylation. For this purpose YFP-tagged TK domain constructs were transfected transiently into MCF-7 cells. We used wild type as well as constructs harboring known activating TKD mutations, D1246, Y1248H and M1268T, all described to occur in hereditary papillary renal cell cancer. (Fig. (2) shows that c-Met phosphorylation is increased in mutants, e.g. D1246H, while the total c-Met levels are comparable to wild-type c-Met. BMS777607 inhibited c-Met phosphorylation in the wild type and Y1248H mutant at 0.3μM concentration with Y1248H being more sensitive to the drug than its wild type counterpart. At that concentration, phosphorylation of c-Met was not decreased in D1246H and M1268T mutants, which required a 3μM concentration of BMS777607. This suggests that the D1246H and M1268T mutants are less sensitive to BMS777607 than wild type or the Y1248 mutant c-Met.
Fig. (2).
MET TK domain constructs, wildtype or harboring either the D1246H, Y1248H, or M1268T mutation (all previously detected in renal cancer), were transfected into MCF-7 cells. Transfected cells were treated with the TKI BMS777607 and Western blot analysis was performed using a c-Met or phoshorylated c-Met antibody. The Y1248H mutant was shown to be most sensitive.
Next, we assessed the effects of the juxtamembrane domain mutations R988C and T1010I in our model. There were no differences between R988C or T1010I mutants of c-Met phosphorylation at Y1003, located in the juxtamembrane domain, and at Y1234/1235, located in the tyrosine kinase domain (Figs. (2 and 3). In Western blots BMS777607 and SU11274 showed concentration-dependent inhibition of c-Met phosphorylation in both wild type and R988C and T1010I mutants.
We previously reported on array comparative genomic hybridization (aCGH)-based genome wide copy number analysis in the currently described cohort of SCLC and neuroendocrine tumors. A copy number gain of the MET gene was detected in 18.2% of SCLC tumors, 53.8% of SCLC cell lines and in 26.3% of bronchial carcinoids [22]. Of 97 samples re-sequenced for the critical domains of the MET gene, array CGH data was available in 80 cases. There was no association between c-Met mutation status and copy number gain of the MET gene.
DISCUSSION
c-Met is a potential therapeutic target and currently many therapeutic interventions are developed aimed at disrupting c-Met signaling. Specific tyrosine kinase inhibitor drugs and monoclonal antibodies have become available for clinical testing and development. MET mutations have been described in various tumor types, spanning all domains of the MET gene. Tyrosine kinase domain gain-of-function mutations are known oncogenic drivers in hereditary papillary renal cell cancer and mutations status may be an important factor influencing therapeutic outcome with inhibitor drugs.
We aimed to evaluate the mutation status of c-Met in neuroendocrine tumors, notably small cell lung cancer and carcinoid tumors, for which relatively limited therapeutic targets have been explored so far. In our study we have detected only mutations in the c-Met juxtamembrane domain. We observed an increased mutation rate in cell lines compared to clinical specimens. However, this difference may be due to the limited number of samples and therefore no definite conclusions can be drawn in this regard. We describe for the first time c-Met mutation analysis in a variety of neuroendocrine tumors including pulmonary and gastrointestinal carcinoid tumors.
As reviewed by Sattler et al., juxtamembrane domain mutations of c-Met have been described in an increasing number of malignancies, including small cell and non-small cell lung cancer, mesothelioma, melanoma, breast cancer, gastric cancer, renal cancer and acute myeloid leukemia [13]. Recently it has been shown as well that novel juxtamembrane mutations in RON (MST1R), one of the two members of the MET receptor tyrosine kinase family, were found in 11% of cases of gastrosoephageal cancer [30].
Prior reports have described somatic as well as germline juxtamembrane mutations of c-Met. In Rottweiler dogs the G966S juxtamembrane mutation was reported to predispose to the development of a variety of cancers [31]. Interestingly, in humans, there seem to be ethnic differences in the occurrence of c-Met mutations. In a study in NSCLC patients it was demonstrated that a germline R988C mutation occurs in Caucasians (1.3%, n = 76) as well as African Americans (1.5%, n = 66), but not in Asians (n = 144) [26]. In an association study in autistic vs. healthy individuals, the R988C and T1010I germline mutation was found in 1.8% of autistic individuals (5/277) and in 0.6% of healthy controls (2/319) (no significant difference) [32].
The exact molecular sequelae of juxtamembrane mutations are yet to be elucidated. In the current study we aimed to assess whether there was an effect of c-Met juxtamembrane mutations on c-Met phosphorylation following treatment with c-Met tyrosine kinase inhibitor drugs. We could not demonstrate such an effect.
The oncogenic effect of juxtamembrane mutations is controversial. The T1010I and R988C mutations have been associated with enhanced tumor formation and lung tumorigenesis in animal studies [33, 34]. Ma et al. showed in vitro that introduction of the R988C and T1010I mutation resulted in a more aggressive phenotype with a “small but significant” growth factor independence, enhanced tumorigenicity and increased phosphorylation of focal adhesion protein paxicillin on tyrosine residue Y31, resulting in increased motility [16]. Both the R988C and T1010I mutation have been associated with increased reactive oxygen species formation [35]. However, in another study no evidence of increased phosphorylation or transformative capacity of the R988C and T1010I mutation was found in a variety of tumor types and it was proposed that these mutations are merely passenger mutations [21]. Finally, a mutation of the juxtamembrane tyrosine residue 2 (Y1001) was shown to result in constitutively mobile, fibroblastoid cells [36]. See (Table 3) for an overview of literature including current study results.
Table 3.
Implications of c-Met Juxtamembrane Domain Mutations
| Mutation | Tumor Type | Molecular Sequelae | Ref. |
|---|---|---|---|
| R988C & T1010I | SCLC | Transfection with mutant form resulted in increased paxicillin tyrosine Y31 phosphorylation, motility and tumorigenicity in vitro | [16] |
| G966S | n/a | Predisposition to cancer in Rottweiler dogs carrying this germline mutation; mutation rate 74% | [31] |
| T1010I | Murine | Increased tumorigenicity of mutant transfected NIH3T3 cells when injected in athymic nude mice | [32] |
| R968C | Murine | SWR/J mice, importance in lung tumorigenesis | [34] |
| R988C & T1010I | SCLC | Increased levels of reactive oxygen (ROS) formation | [35] |
| P991S | Gastric | Increased persistent response to HGF stimulation when expressed in fibroblasts | [33] |
| R988C & T1010I | MCF-7 cells | No effect on c-MET phosphorylation upon treatment with tyrosine kinase inhibitors BMS777067 and SU11274 (unstimulated condition) | current study |
| R988C & T1010I | Various* | No evidence of increased phosphorylation or transformative capacity | [21] |
| Splice mutation | NSCLC | Somatic intronic mutation causing a deletion of juxtamembrane domain resulting in loss of Cbl-E3-ligase binding resulting in decreased ubiquitination, delayed downregulation and enhanced c-Met activation | [19] |
| Y1001 | Epithelial cells (MDCK) | Y1001 mutation was shown to produce constitutively mobile, fibroblastoid cells | [36] |
| R988C & T1010I | Healthy individuals | Autism association study. Germline mutation found in 1.8% of autistic individuals and in 0.6% of healthy controls (not significant) | [32] |
Notes:
CLL, AML, CMML, colorectal, endometrial, thyroid, melanoma, healthy control.
The Y1003 residue is a binding site for ubiquitin E3 ligase c-Cbl. Ubiquitination of c-Met is important in the regulation of c-Met signaling. The effects of juxtamembrane domain mutations, e.g. by potential conformational changes, on c-Met ubiquitination are unknown and should be investigated in future studies as they might contribute to the more aggressive phenotype of the juxtamembrane mutants. It was shown by Peschard et al. that reduced c-Met ubiquitination in their Y1103F mutant led to oncogenic transformation [18]. Furthermore, it should be investigated whether juxtamembrane mutations lead to an enhanced or altered response to hepatocyte growth factor exposure which was described for the P991S mutation in gastric carcinoma [33]. In our model however we made use of a truncated c-Met construct lacking the extracellular domain. Therefore we could not assess the effects of HGF stimulation in this model.
CONCLUSION
In SCLC and neuroendocrine tumors mutations in the MET gene are relatively rare. Found mutations were all located in exon 14, which encodes the juxtamembrane domain. Detected mutations are of no functional relevance as they did not influence c-Met phosphorylation, regardless of TKI treatment, as determined by functional assays in vitro. Functional implications of juxtamembrane domain mutations need to be further elucidated in future studies.
ACKNOWLEDGEMENTS
Mutation studies were in part supported by the Clinical and Molecular Profiling Core (CMPC), National Cancer Institute, with special thanks to Marbin Pineda, Cancer Genetics Branch, National Cancer Institute.
ABBREVIATIONS
- HGFR
Hepatocyte growth factor receptor
- IPT
Immunoglobulin-like regions, plexins and transcriptional factors
- JMD
Juxtamembrane domain
- NET
Neuroendocrine tumor
- sema
Semaphorin
- RTKI
Receptor tyrosine kinase inhibitor
- SF
Scatter factor
- TKD
Tyrosine kinase domain
- TMD
Transmembrane domain
- Y
Tyrosine
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- [1].Giaccone G, Rodriguez JA. EGFR inhibitors: what have we learned from the treatment of lung cancer? Nat Clin Pract Oncol 2005; 2: 554–61. [DOI] [PubMed] [Google Scholar]
- [2].Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene 1993; 8: 1195–202. [PubMed] [Google Scholar]
- [3].Gonzatti-Haces M, Seth A, Park M, Copeland T, Oroszlan S, Vande Woude GF. Characterization of the TPR-MET oncogene p65 and the MET protooncogene p140 protein-tyrosine kinases. Proc Natl Acad Sci USA 1988; 85: 21–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 2010; 11: 834–48. [DOI] [PubMed] [Google Scholar]
- [5].Tsao MS, Zhu H, Giaid A, Viallet J, Nakamura T, Park M. Hepatocyte growth factor/scatter factor is an autocrine factor for human normal bronchial epithelial and lung carcinoma cells. Cell Growth Differ 1993; 4: 571–9. [PubMed] [Google Scholar]
- [6].Rong S, Segal S, Anver M, Resau JH, Vande Woude GF. Invasiveness and metastasis of NIH 3T3 cells induced by Methepatocyte growth factor/scatter factor autocrine stimulation. Proc Natl Acad Sci USA 1994; 91: 4731–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kubo T, Yamamoto H, Lockwood WW, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int J Cancer 2009; 124: 1778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem 2000; 275: 8806–11. [DOI] [PubMed] [Google Scholar]
- [9].Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA 2007; 104: 20932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–43. [DOI] [PubMed] [Google Scholar]
- [11].Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov 2008; 7: 504–16. [DOI] [PubMed] [Google Scholar]
- [12].Gherardi E, Birchmeier W, Birchmeier C, Vande WG. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012; 12: 89–103. [DOI] [PubMed] [Google Scholar]
- [13].Sattler M, Reddy MM, Hasina R, Gangadhar T, Salgia R. The role of the c-Met pathway in lung cancer and the potential for targeted therapy. Ther Adv Med Oncol 2011; 3: 171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jeffers M, Schmidt L, Nakaigawa N, et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci USA 1997; 94: 11445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET protooncogene in papillary renal carcinomas. Nat Genet 1997; 16: 68–73. [DOI] [PubMed] [Google Scholar]
- [16].Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003; 63: 6272–81. [PubMed] [Google Scholar]
- [17].Aguirre de I, Salvatierra A, Font A, et al. c-Met mutational analysis in the sema and juxtamembrane domains in small-cell-lung-cancer. Translational Oncogenomics 2006; 1: 11–8. [PMC free article] [PubMed] [Google Scholar]
- [18].Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J Biol Chem 2004; 279: 29565–71. [DOI] [PubMed] [Google Scholar]
- [19].Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006; 66: 283–9. [DOI] [PubMed] [Google Scholar]
- [20].Lee CC, Yamada KM. Alternatively Spliced Juxtamembrane Domain of a Tyrosine Kinase Receptor Is a Multifunctional Regulatory Site. Journal of Biological Chemistry 1995; 270: 507–10. [DOI] [PubMed] [Google Scholar]
- [21].Tyner JW, Fletcher LB, Wang EQ, et al. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res 2010; 70: 6233–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Voortman J, Lee JH, Killian JK, et al. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci USA 2010; 107: 13040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Killian JK, Walker RL, Suuriniemi M, et al. Archival fine-needle aspiration cytopathology (FNAC) samples: untapped resource for clinical molecular profiling. J Mol Diagn 2010; 12: 739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harada T, Lopez-Chavez A, Xi L, Raffeld M, Wang Y, Giaccone G. Characterization of epidermal growth factor receptor mutations in non-small-cell lung cancer patients of African-American ancestry. Oncogene 2011; 30: 1744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].de Gunst MM, Gallegos-Ruiz MI, Giaccone G, Rodriguez JA. Functional analysis of cancer-associated EGFR mutants using a cellular assay with YFP-tagged EGFR intracellular domain. Mol Cancer 2007; 6: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 2009; 15: 5714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhen Z, Giordano S, Longati P, Medico E, Campiglio M, Comoglio PM. Structural and functional domains critical for constitutive activation of the HGF-receptor (Met). Oncogene 1994; 9: 1691–7. [PubMed] [Google Scholar]
- [28].Schroeder GM, An Y, Cai ZW, et al. Discovery of N-(4-(2-Amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a Selective and Orally Efficacious Inhibitor of the Met Kinase Superfamily. J Med Chem 2009; 52: 1251–4. [DOI] [PubMed] [Google Scholar]
- [29].Dai Y, Siemann DW. BMS-777607, a Small-Molecule Met Kinase Inhibitor, Suppresses Hepatocyte Growth FactorΓÇôStimulated Prostate Cancer Metastatic Phenotype In vitro. Molecular Cancer Therapeutics 2010; 9: 1554–61. [DOI] [PubMed] [Google Scholar]
- [30].Catenacci DV, Cervantes G, Yala S, et al. RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer Biol Ther 2011; 12: 9–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liao AT, McMahon M, London CA. Identification of a novel germline MET mutation in dogs. Anim Genet 2006; 37: 248–52. [DOI] [PubMed] [Google Scholar]
- [32].Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci USA 2006; 103: 16834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee JH, Han SU, Cho H, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene 2000; 19: 4947–53. [DOI] [PubMed] [Google Scholar]
- [34].Zaffaroni D, Spinola M, Galvan A, et al. Met proto-oncogene juxtamembrane rare variations in mouse and humans: differential effects of Arg and Cys alleles on mouse lung tumorigenesis. Oncogene 2005; 24: 1084–90. [DOI] [PubMed] [Google Scholar]
- [35].Jagadeeswaran R, Jagadeeswaran S, Bindokas VP, Salgia R. Activation of HGF/c-Met pathway contributes to the reactive oxygen species generation and motility of small cell lung cancer cells. Am J Physiol Lung Cell Mol Physiol 2007; 292: L1488–L1494. [DOI] [PubMed] [Google Scholar]
- [36].Weidner KM, Sachs M, Riethmacher D, Birchmeier W. Mutation of juxtamembrane tyrosine residue 1001 suppresses loss-of-function mutations of the met receptor in epithelial cells. Proc Natl Acad Sci USA 1995; 92: 2597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]