Abstract
Fruit trees need to overcome harsh winter climates to ensure perennially; therefore, they are strongly influenced by environmental stress. In the present study, we focused on the pear homolog PcLEA14 belonging to the unique 5C late embryogenesis abundant (LEA) protein group for which information is limited on fruit trees. PcLEA14 was confirmed to belong to this protein group using phylogenetic tree analysis, and its expression was induced by low-temperature stress. The seasonal fluctuation in its expression was considered to be related to its role in enduring overwinter temperatures, which is particularly important in perennially. Moreover, the function of PcLEA14 in low-temperature stress tolerance was revealed in transgenic Arabidopsis. Subsequently, the pear homolog of dehydration-responsive element-binding protein/C-repeat binding factor1 (DREB1), which is an important transcription factor in low-temperature stress tolerance and is uncharacterized in pear, was analyzed after bioinformatics analysis revealed the presence of DREB cis-regulatory elements in PcLEA14 and the dormancy-related gene, both of which are also expressed during low temperatures. Among the five PcDREBs, PcDREB1A and PcDREB1C exhibited similar expression patterns to PcLEA14 whereas the other PcDREBs were not expressed in winter, suggesting their different physiological roles. Our findings suggest that the low-temperature tolerance mechanism in overwintering trees is associated with group 5C LEA proteins and DREB1.
Keywords: DREB, LEA14, low-temperature stress, pear, Pyrus communis
1. Introduction
Environmental stresses such as extreme temperatures and drought have a significant influence on the growth and yield of crops. Recently, due to concern about the extreme environmental changes arising from global warming [1], an increasing number of studies have focused on the mechanisms underlying herbaceous and woody crops’ tolerance to environmental stresses such as low temperature, drought, and high salinity and how these mechanisms can be utilized to improve crop yields [2,3,4,5,6,7]. It is important that researchers accelerate current studies on crops in this post-genomic era. The expression of genes that enable plants to adapt to environmental stress has been modeled using an herbaceous plant, Arabidopsis thaliana. However, particular attention must be paid to trees, especially fruit trees, as they are exposed to high environmental stress in harsh winter climates that need to be overcome to ensure perennially. Therefore, the present study focuses on the molecular mechanisms underlying low-temperature stress tolerance in fruit trees.
Late embryogenesis abundant (LEA) proteins accumulate in large quantities during the late stages of seed embryogenesis and, as predominantly hydrophilic proteins, are believed to be involved in environmental stress response [8]. LEA proteins are classified into seven groups according to their amino acid sequences; however, group 5 proteins are considered atypical LEA proteins because of their comparatively large number of hydrophobic residues and their lack of common sequences and important motifs found in other LEA proteins [8]. Group 5C proteins have a Water Stress and Hypersensitive (WHy) response domain, and their function is expected to differ from that of other LEA proteins [9,10,11]. Although some studies have investigated the stress response of group 5C LEA proteins in herbaceous plants [2,12,13,14], there is limited information available on trees in which a unique functionality of the gene is expected to ensure perennially. This study employs LEA14 to investigate the functionality of these stress proteins in trees.
The dehydration-responsive element-binding protein/C-repeat binding factor (DREB) is the most well-known transcription factor involved in abiotic stress responses. Studies have shown that DREB expression is induced by low temperatures, salt stress, as well as drought and that it interacts with NAC transcription factors and activates the expression of downstream genes [15,16]. Compared to herbaceous plants such as Arabidopsis, information regarding fruit trees is limited, and to our knowledge, no detailed analysis has been performed on pear trees (Pyrus communis). Although omics studies, including ones investigating of stress- and dormancy-related genes, have been conducted on pears [17], they have primarily involved the fruit itself and lack a detailed analysis of individual genes.
DREB1 and DREB2 are subgroups in the DREB family. DREB1 is categorized as a regulator of low-temperature stress responses, whereas DREB2 regulates dehydration and heat-shock responses [15,18]. Given DREB1’s role as a master regulator of cold stress responses [19], it is likely to be involved in inducing the expression of other stress response genes such as LEA14. Based on the above, phylogenetic and expression analyses were conducted on PcDREB1, the pear homolog of DREB1.
In order to understand the molecular mechanisms underlying low-temperature stress tolerance and to determine which genes are useful for enhancing stress tolerance for molecular breeding, we focused on a unique LEA belonging to the group 5C proteins from pears and characterized this protein with DREB1 homologs. The results in the present study contribute to the understanding of the role of these genes during the winter, which is of principal importance to ensure the perenniality of fruit trees.
2. Results
2.1. Phylogenetic Analysis and Low-Temperature Induced Expression of the Pear Homolog of LEA14: PcLEA14
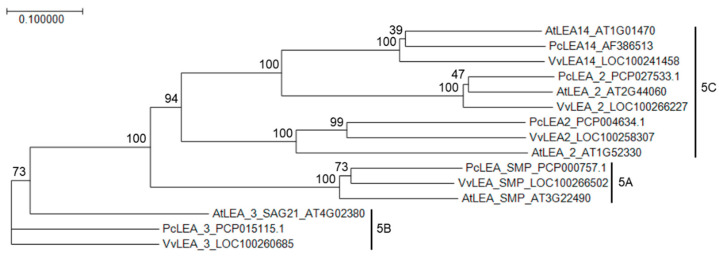
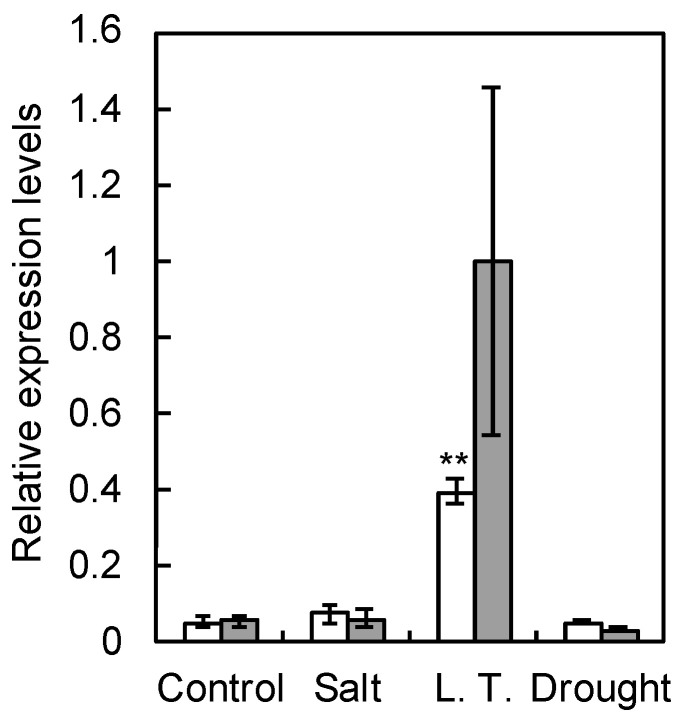
A phylogenetic tree based on the amino acid sequences of LEA proteins revealed that the pear homolog of LEA14 is included in the group 5C LEA proteins among groups 5A, 5B, and 5C (Figure 1). Therefore, the homolog was designated as PcLEA14. Subsequently, the stress response of PcLEA14 expression was investigated in pear leaves. The results showed that PcLEA14 mRNA expression levels increased to approximately eight times that of the control due to low-temperature stress in a period of 24 h, whereas no effects of salt and drought stress on expression levels were observed (Figure 2).
Figure 1.
A phylogenetic tree based on the amino acid sequences of PcLEA14 (AF386513) and group 5 late embryogenesis abundant (LEA) proteins from Arabidopsis, P. communis and V. vinifera: The tree was constructed using the neighbor-joining method after sequence alignment using ClustalW2, with Genetyx version 14 (GENETYX CO., Tokyo, Japan). Branch numbers refer to the percentage of replicates that support the branch using the bootstrap method (1000 replicates). The scale bar corresponds to 0.1 amino acid substitutions per residue.
Figure 2.
mRNA expression levels of PcLEA14 in pear leaves under environmental stress conditions for 24 h (white) and 48 h (grey): data shows the relative expression levels normalized against PcrRNA. Values indicate the mean ± standard error (n = 6). ** Significantly different from control conditions at p < 0.01 according to the Dunnett’s test.
2.2. Seasonal Changes in PcLEA14 Expression
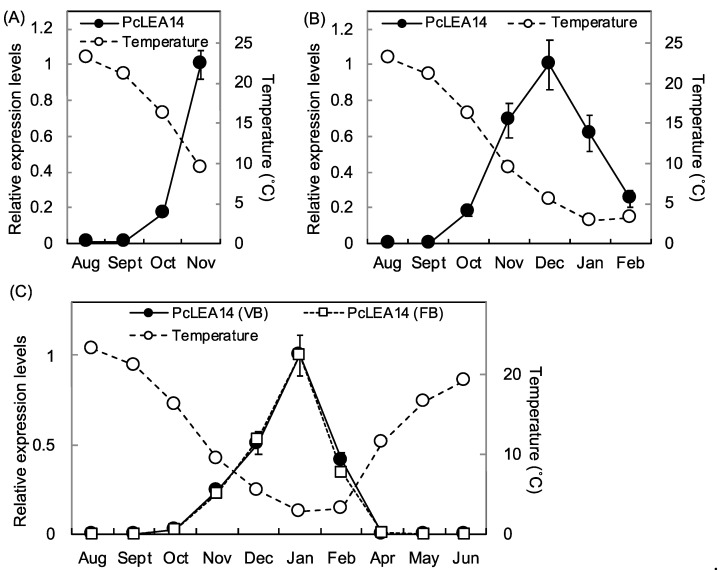
The expression of PcLEA14 was evaluated in mature leaves, current stems, flower buds, and vegetative buds using the pear trees in the orchard. The level of PcLEA14 mRNA expression in mature leaves was very low in August and September and then increased in October with decreasing temperatures (Figure 3A). No data were obtained for December and later months because of defoliation. The level of PcLEA14 mRNA expression in current stems also increased from October to December but then decreased after December (Figure 3B). The level of PcLEA14 mRNA expression in the flower buds and vegetative buds was very low in August and September, but it increased with decreasing temperatures (Figure 3C). These levels peaked in January, then remarkably decreased, and became very low from April onward.
Figure 3.
Seasonal changes in mRNA levels of PcLEA14 in wild-type pear leaves (A), stems (B), and buds (C) at air temperature: each value was determined from three independent biological replicates. Data show the relative expression levels normalized against PcActin for leaves and stems and PcGAPDH for buds. Values indicate the means ± standard error (n = 3). VB and FB indicate vegetative buds and flower buds, respectively (C).
2.3. Bioinformatic Analysis of Promoter Sequences
The DRE/CRT motifs, cis-elements targeted by DREB, were searched for in PcLEA14 to determine candidate genes that may play a role during dormancy and which are expected to be necessary for overwintering. To distinguish the motifs from their promoters, we analyzed the 1500-bp upstream sequences of those genes (Table 1). There were gap regions (NNN…) upstream of PCP000690.1 and PCP022935.1, and hence, we used the sequences between the start codon and the gap region. We identified two DRE/CRT motifs in the PcLEA14 promoter and either one or two DRE/CRT motifs in the promoters of the candidate genes, except PCP032234.1, PCP029251.1, PCP007373.1, and PCP002349.1. Some of the genes with the DRE/CRT motifs were homologous to genes with functions of interest. MADS-box protein AGL24-like contains the homolog of DAM/MADS13, which is known to regulate endodormancy in trees [20,21]. 12-Oxophytodienoate reductase 2-like gene may function in the synthesis of jasmonic acid, involved in dormancy and stress response [22]. Furthermore, protein phosphatase 2C 56-like is an ABI1 homologous gene and could be associated with ABA signaling [23]. On the other hand, the FLOWERING LOCUS T (FT)-interacting protein 1-like is a factor involved in the transport of flowering-inducing factor FT, and the presence of the DRE/CRT motifs in this gene suggests a novel research development regarding the role of DREB.
Table 1.
CRT/DRE motif and LTRE motif on the promoter of PcLEA14 and candidate genes playing a role during dormancy.
| Gene Name | Gene ID P. bretschneideri |
Gene ID P. communis |
CBFHV a | DRECRT- COREAT b |
LTRECORE-ATCOR15 c | Total Counts | Sequence Length (bp) |
|---|---|---|---|---|---|---|---|
| Motif: RYCGAC | Motif: RCCGAC | Motif: CCGAC | |||||
| PcLEA14 | - | PCP029268.1 | 2 | 1 | 1 | 2 | 1500 |
| FT-interacting protein 1-like | LOC103967842 | PCP000690.1 | 1 | - | - | 1 | 507 |
| MADS-box protein AGL24-like | LOC103964948 | PCP022935.1 | - | - | 1 | 1 | 866 |
| MADS-box protein AGL24-like | LOC103964950 | PCP022936.1 | 1 | 1 | 1 | 1 | 1500 |
| MADS-box protein AGL24-like | LOC103964952 | PCP005825.1 | 2 | 2 | 2 | 2 | 1500 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ-like | LOC103967963 | PCP000700.1 | - | - | 1 | 1 | 1500 |
| chlorophyll a-b binding protein CP24 10A, chloroplastic | LOC103967973 | PCP000701.1 | 2 | - | 2 | 4 | 1500 |
| 12-oxophytodienoate reductase 2-like | LOC103967564 | PCP032234.1 | - | - | - | - | 1500 |
| palmitoyl-monogalactosyldiacylglycerol delta-7 desaturase, chloroplastic-like | LOC103954983 | PCP032515.1 | 1 | 1 | 1 | 1 | 1500 |
| thymidine kinase a | LOC103955051 | PCP029243.1 | - | - | 1 | 1 | 1500 |
| chlorophyll a-b binding protein 151, chloroplastic-like | LOC103955064 | PCP029251.1 | - | - | - | - | 1500 |
| cytochrome b6-f complex iron-sulfur subunit, chloroplastic-like | LOC103944475 | PCP007373.1 | - | - | - | - | 1500 |
| protein phosphatase 2C 56-like | LOC103943902 | PCP028125.1 | 1 | - | 1 | 2 | 1500 |
| uncharacterized | LOC103964940 | PCP005820.1 | 1 | - | 1 | 2 | 1500 |
| uncharacterized | LOC103944526 | PCP040740.1 | 1 | - | - | 1 | 1500 |
| uncharacterized | LOC103944497 | PCP007115.1 | 1 | - | - | 1 | 1500 |
| uncharacterized | LOC103954139 | PCP002349.1 | - | - | - | - | 1500 |
| uncharacterized | LOC103943904 | PCP028129.1 | 2 | - | 2 | 4 | 1500 |
| uncharacterized | LOC103943918 | PCP033851.1 | 1 | - | - | 1 | 1500 |
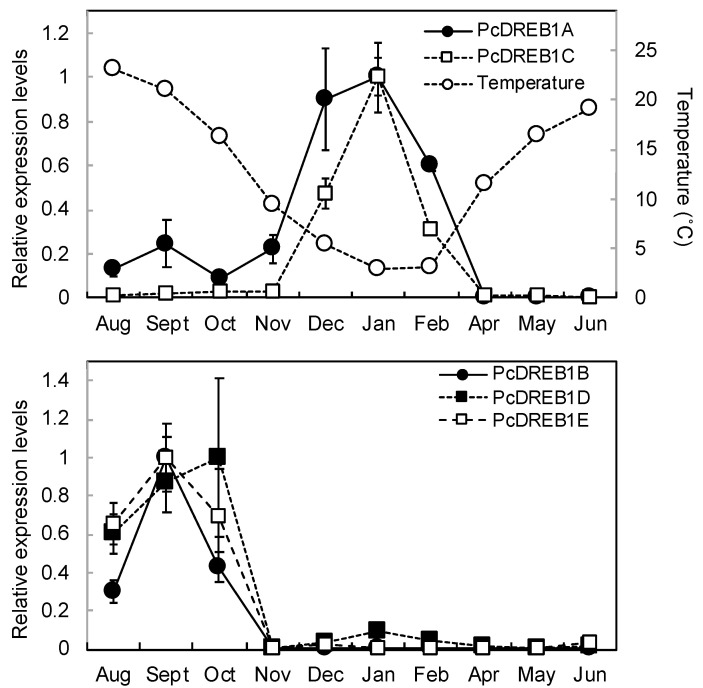
2.4. Seasonal Changes in the Expression of the Pear Homologs of DREB1: PcDREB1s
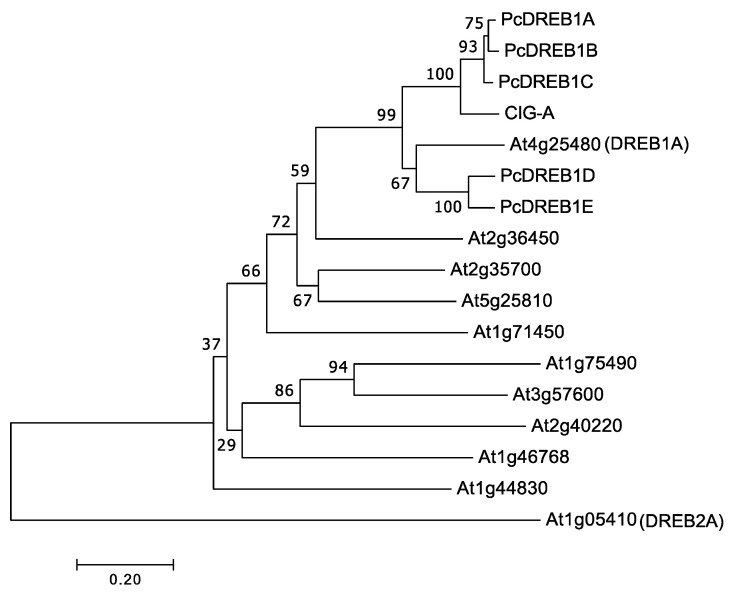
As indicated by the phylogenetic trees containing the Arabidopsis DREB proteins, these PcDREBs belonged to the DREB1 group together with CIG-A (Figure 4). CIG-A is a DREB1 homolog in sweet cherry, which is a rosaceous fruit tree like pear, and its low-temperature tolerance functional analysis has been performed in Arabidopsis [32]. Seasonal changes in the expression of these five genes were analyzed in the buds to compare them with PcLEA14 expression (Figure 5). The level of PcDREB1A mRNA expression began to increase in approximately November, peaked from December to January, and markedly decreased from February to April. The level of PcDREB1C mRNA expression increased in December, peaked in January, and then markedly decreased from February to April. PcDREB1B, PcDREB1D, and PcDREB1E exhibited similar expression patterns (i.e., their expression was observed from August to October but was very low thereafter).
Figure 4.
Phylogenetic tree based on the amino acid sequences of PcDREBs and dehydration-responsive element-binding protein/C-repeat binding factor (DREB) proteins from Arabidopsis: the tree was constructed using the neighbor-joining method, with MEGA version 7.0. Branch numbers refer to the percentage of replicates that support the branch using the bootstrap method (1000 replicates). The scale bar corresponds to 0.2 amino acid substitutions per residue. The sequences of Arabidopsis DREB proteins were obtained from Mizoi et al. [15]. CIG-A accession number: Q8H9A2. Gene names of PcDREBs in the Genome Database for Rosaceae are PCP009299.1 (PcDREB1A), PCP014363.1 (PcDREB1B), PCP011972.1 (PcDREB1C), PCP019717.1 (PcDREB1D), and PCP012284.1 (PcDREB1E).
Figure 5.
Seasonal changes in the mRNA levels of PcDREBs in wildtype pear vegetative buds at air temperature: each value was determined from three independent biological replicates. Data show the relative expression levels normalized against PcGAPDH. Values indicate the means ± standard error (n = 3).
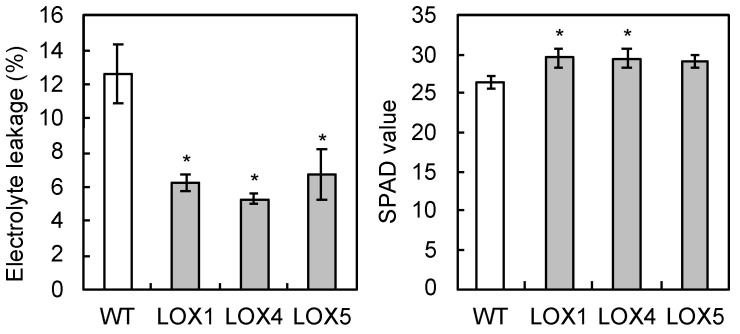
2.5. Stress Tolerance in Arabidopsis Expressing PcLEA14
Under the stress condition at −2 °C, electrolyte leakage was lower in Arabidopsis expressing PcLEA14, LOX lines, than that in the wildtype, and the chlorophyll content was higher in LOX1 and LOX4 than that in the wildtype (Figure 6). Although the expression of PcLEA14 was confirmed in LOX lines, the mRNA levels of AtLEA14, which is the endogenous LEA14 gene in Arabidopsis, were not higher in LOX lines than in the wildtype (Supplementary Figure S1). These results suggest that PcLEA14 may contribute to cell survival and maintenance of photosynthetic pigments under low-temperature stress.
Figure 6.
Electrolyte leakage and chlorophyll content (SPAD value) under low-temperature stress conditions at −2 °C in LOX lines of transgenic Arabidopsis expressing PcLEA14: values indicate the means ± standard error (n = 3). * Significantly different from wildtype (WT) at p < 0.05 according to Dunnett’s test.
3. Discussion
The LEA14 pear homolog was confirmed to belong to the group 5C proteins of the LEA family, as shown in the phylogenetic tree analysis, and it was designated as PcLEA14. Group 5C proteins have a unique structure [9,10,11], and although their role is unclear, they have been the subject of a small number of studies. An earlier study reported that overexpression of the same gene group in maize, ZmLEA5C, improved low-temperature stress tolerance in tobacco plant [2]. Furthermore, in the present study, low temperatures induced the expression of PcLEA14 and low-temperature stress tolerance in Arabidopsis was improved after its overexpression. These results show that one of the roles of the group 5C LEA proteins is a response to low-temperature stress. Conversely, previous studies have reported that, in Arabidopsis and sweet potato, AtLEA14 and IbLEA14, which belong to the same group, are involved in both salt and drought stress responses [12,14]. As demonstrated in the expression analysis using pear leaves, the expression of PcLEA14 was induced only by low temperature. These different reactions suggest a variety of roles for LEA14. Based on these previous reports and the present study, the LEA14 protein itself is considered multifunctional, being effective at both low temperature and drought conditions, and in fact, PcLEA14 may also contribute to drought tolerance in transformed Arabidopsis (data not shown). Therefore, it is possible that the diversity of roles may be derived from the fact that the expression pattern differs depending on the species. The PcLEA14 promoter was found to have CRT/DRE motifs similar to dormancy-related genes in pear, suggesting that PcLEA14 could be co-expressed with such genes during overwintering.
DREB1 is known as a master regulator of the low-temperature stress response [19], and its homolog plays a role in the induction of the LEA14 homolog in strawberry, belonging to the same rosaceous family as pear [33]. Unlike herbaceous plants, the low-temperature stress response is more important in trees because they are perennial and must endure overwinter conditions; however, there is a lack of information regarding the seasonal expression patterns of LEA and DREB homologs. In the present study, the expression of PcLEA14 increased in leaves as temperature decreased in autumn, suggesting a contribution to the low-temperature stress response. Moreover, the expression of PcLEA14 increased in overwintering stems and buds as the temperature decreased, suggesting its contribution to low-temperature stress tolerance in winter. Among the five PcDREBs, the expression patterns of PcDREB1A and PcDREB1C were similar to that of PcLEA14. In particular, the expression pattern of PcDREB1C was most similar to that of PcLEA14 and PcDREB1C belonged to the same cluster on the phylogenetic tree as CIG-A, which is the DREB1 homolog of sweet cherry of the same rosaceous fruit tree and induced by low temperature [34,35]. PcLEA14, in fact, contained DREB cis-regulatory elements in its promoter region, suggesting that PcDREB1A and PcDREB1C are expected to regulate the expression of PcLEA14. Moreover, during our studies of sweet cherry DREB1, the introduction of CIG-A into pear was confirmed to induce PcLEA14 expression (data not shown). Conversely, DREB1B, DREB1D, and DREB1E were not expressed in winter but were expressed in summer; therefore, they were not involved in low-temperature stress response and their other possible roles were considered. The distinct difference in the expression patterns found in the DREB1 subfamily could be associated with their physiological roles. A recent study in P. pyrifolia conducted expression analysis of the DREB family gene [36]. Although data was not collected year-round, the expression levels of all genes peaked between October and February and no members with unique expression such as DREB1B, DREB1D, and DREB1E were reported. Because the response to high temperature, drought, and high salinity is related to the wide signal network involving GTPase and rhizosphere bacteria together with DREB and LEA [6,37], it is necessary to analyze DREB1B, DREB1D, and DREB1E from the same broad aspect.
The level of PcLEA14 mRNA expression increased with low temperature in the leaves, stems, and buds; however, the expression was not consistently high during low-temperature months. The level of PcLEA14 mRNA expression decreased in January and February, with the lowest temperature in the stems, and expression also decreased markedly in February, with the lowest temperature in the buds. The mRNA expression levels of PcDREB1A and DREB1C with PcLEA14 also decreased in February. These results indicate that the expression levels of PcDREB1A, PcDREB1C, and PcLEA14 are regulated by factors other than temperature. Based on data released by the Japan Meteorological Agency, the low-temperature requirement for breaking endodormancy in pear is approximately 1600 h at ≤7.2 °C [38]. During the year in which the expression analysis was performed, the low-temperature requirement reached 1600 h in late January. In fact, the relationship between LEA homolog expression and endodormancy has been reported in another rosaceous fruit tree, Japanese apricot [39], although the LEA homolog tested is not involved in the group 5C LEA proteins. These results suggest that the expression of PcLEA14 is regulated by both endodormancy and temperature.
4. Materials and Methods
4.1. Plant Materials
“La France” pear trees at age 12 years in 2008 were the primary models used for analyzing the expression of PcLEA14 and its nucleotide sequences. The trees were grown in the Tohoku University (Sendai, Japan) experimental field at 38°16′ N and 140°52′ E. For exploring the seasonal fluctuation in its expression, mature leaves, current stems, and buds were sampled from August to November 2008, from August 2008 to February 2009, and from August 2012 to July 2013, respectively. The samples were randomly obtained from four trees and stored at −80 °C until use. Mature leaves were used for investigating the effect of environmental stress on PcLEA14 expression. For the transformation experiment in Arabidopsis, PcLEA14 cDNA was cloned into a pBI-OX-GW vector containing a constitutive promoter for plants (Inplanta Innovations Inc., Yokohama, Japan) and the constructed vector was introduced into A. thaliana ecotype Columbia using the Agrobacterium-mediated transformation method (Inplanta Innovations Inc.). The transgenic plants were selected on media containing kanamycin, and the transgenes were confirmed by PCR. The transgenic Arabidopsis lines with PcLEA14, LOX1, LOX4, and LOX5 in the T2 generation were used with the wildtype Colombia plants as a control. The plants were grown in vermiculite/pearlite (1:1) at 22 °C under a 16 h photoperiod using a growth chamber [40] and used 4 weeks after sowing for experiments.
4.2. Sequences Used for Phylogenetic Analysis
The gene LEA14 of P. communis (accession number AF386513) was searched for in the National Center for Biotechnology Information (NCBI). The protein sequences of Arabidopsis were used from representative sequences of groups 5A, 5B, and 5C according to the corresponding PFAM number by Battaglia et al. [8]. The pear homologs of group 5 LEA genes were searched using BLAST in Genome Database for Rosaceae (GDR). The Vitis vinifera homologs of group 5 LEA genes were searched by BLAST in NCBI. The pear homologs of DREB1 were searched using BLAST+ in GDR, and the five genes found in this search were designated as PcDREB1A, 1B, 1C, 1D, and 1E.
4.3. Expression Analyses
Total RNA was extracted from pear using the cetyltrimethylammonium bromide method according to Hatsuda et al. [41] and from Arabidopsis using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The removal of genomic DNA and reverse transcription were performed using the Quantiscript Reverse Transcription Kit (QIAGEN) and Rever Tra Ace qPCR RT Master Mix (TOYOBO, Osaka, Japan). Real-time PCR was performed using the QuantiTect SYBR Green PCR Kit (QIAGEN) and THUNDERBIRD SYBR qPCR Mix (TOYOBO) according to Ikeda et al. [42]. The nucleotide sequences of all primers used for the expression analysis are shown in Supplementary Table S1.
4.4. Environmental Stress Treatment
The effect of environmental stress on the expression of PcLEA14 was investigated using detached mature leaves of the wildtype pear “La France” under the conditions of high salinity at 150 mM NaCl, low temperature at 4 °C, and drought. In the control, leaves cut in half horizontally were placed with the cut end facing down in water in tubes and left at 25 °C under dark conditions. The leaves were taken out 24 and 48 h after the start of the treatment and stored at −80 °C; 150 mM NaCl was used instead of water under conditions of high salinity. In the low-temperature stress treatment, the leaves were left at 4 °C instead of 25 °C. No water was added in the drought stress treatment.
Low-temperature stress tolerance was evaluated at −2 °C for LOX lines of transgenic Arabidopsis plants. Stress tolerance was evaluated by measuring electrolyte leakage after stress treatment as described by Kanayama et al. [43] and Feng et al. [44]. After the plants were left at 21 °C for 2 h, the temperature was lowered to −2 °C in 5 h and left for 30 h. Then, the temperature was raised to 21 °C in 5 h followed by cultivation at 21 °C for 2 h. Control plants were cultivated at 21 °C during the treatment. After stress treatment, the plants were grown as described in Section 4.1. for 5 days and used for stress tolerance evaluation.
4.5. Bioinformatic Analysis of Promoter Sequences
The upstream 1500-bp PcLEA14 promoter sequences of the start codon were obtained from P. communis Genome v1.0 using BLAST+ on GDR. The homologs of candidate genes associated with dormancy [22] were analyzed using BLAST+ on GDR, and the promoter sequences were obtained as in PcLEA14. The promoter sequences were analyzed using the PLACE database [45] for detecting DRE/CRT motifs RCCGAC, RYCGAC, or CCGAC [24,25,26,27,28,29,30,31].
5. Conclusion
We identified potential roles of PcLEA14 and PcDREB1s in pear trees using expression analysis, transgenic experiments, and bioinformatic analysis. The DRE/CRT motifs were, in fact, found in PcLEA14 and in many candidate genes playing a role during dormancy, including genes associated with DAM/MADS13, jasmonic acid synthesis, and ABA signaling, which are involved in dormancy and stress response. PcLEA14 is likely expressed coordinately with these genes and PcDREB1A and C to allow plants to endure harsh winter climates.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/9/1138/s1, Table S1: List of primer sequences, Figure S1: mRNA levels of PcLEA14 and AtLEA14 in LOX lines of transgenic Arabidopsis expressing PcLEA14.
Author Contributions
Conceptualization, H.K., K.I., and Y.K.; methodology, T.S. and K.K.; validation, T.S. and K.K.; formal analysis, T.S. and R.I.; investigation, R.I. and M.M.; resources, H.K., K.I., and Y.K.; data curation, T.S.; writing—original draft preparation, R.I. and M.M.; writing—review and editing, K.K. and Y.K.; visualization, R.I. and M.M.; supervision, H.K. and K.I.; project administration, Y.K.; funding acquisition, Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI grant number JP24248006.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kanayama Y., Kochetov A.V. Abiotic Stress Biology in Horticultural Plants. Springer; New York, NY, USA: 2015. [Google Scholar]
- 2.Liu Y., Wang L., Jiang S., Pan J., Cai G., Li D. Group 5 LEA protein, ZmLEA5C, enhance tolerance to osmotic and low temperature stresses in transgenic tobacco and yeast. Plant Physiol. Biochem. 2014;84:22–31. doi: 10.1016/j.plaphy.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Kanayama Y., Watanabe M., Moriguchi R., Deguchi M., Kanahama K., Yamaki S. Effects of low temperature and abscisic acid on the expression of the sorbitol-6-phosphate dehydrogenase gene in apple leaves. J. Jpn. Soc. Hortic. Sci. 2006;75:20–25. doi: 10.2503/jjshs.75.20. [DOI] [Google Scholar]
- 4.Kanayama Y., Mizutani R., Yaguchi S., Hojo A., Ikeda H., Nishiyama M., Kanahama K. Characterization of an uncharacterized aldo-keto reductase gene from peach and its role in abiotic stress tolerance. Phytochemistry. 2014;104:30–36. doi: 10.1016/j.phytochem.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 5.El-Esawi M.A., Alayafi A.A. Overexpression of StDREB2 transcription factor enhances drought stress tolerance in cotton (Gossypium barbadense L.) Genes. 2019;10:142. doi: 10.3390/genes10020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Esawi M.A., Al-Ghamdi A.A., Ali H.M., Alayafi A.A. Azospirillum lipoferum FK1 confers improved salt tolerance in chickpea (Cicer arietinum L.) by modulating osmolytes, antioxidant machinery and stress-related genes expression. Environ. Exp. Bot. 2019;159:55–65. doi: 10.1016/j.envexpbot.2018.12.001. [DOI] [Google Scholar]
- 7.Uozumi A., Ikeda H., Hiraga M., Kanno H., Nanzyo M., Nishiyama M., Kanahama K., Kanayama Y. Tolerance to salt stress and blossom-end rot in an introgression line, IL8-3, of tomato. Sci. Hortic. 2012;138:1–6. doi: 10.1016/j.scienta.2012.01.036. [DOI] [Google Scholar]
- 8.Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccarelli F.D., Bork P. The WHy domain mediates the response to desiccation in plants and bacteria. Bioinformatics. 2005;21:1304–1307. doi: 10.1093/bioinformatics/bti221. [DOI] [PubMed] [Google Scholar]
- 10.Hunault G., Jaspard E. LEAPdb: A database for the late embryogenesis abundant proteins. BMC Genom. 2010;11:221. doi: 10.1186/1471-2164-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaspard E., Hunault G. Comparison of amino acids physico-chemical properties and usage of late embryogenesis abundantproteins, hydrophilins and WHy domain. PLoS ONE. 2015;9:e109570. doi: 10.1371/journal.pone.0109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S.C., Kim Y.H., Jeong J.C., Kim C.Y., Lee H.S., Bang J.W., Kwak S.S. Sweet potato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta. 2011;233:621–634. doi: 10.1007/s00425-010-1326-3. [DOI] [PubMed] [Google Scholar]
- 13.He S., Tan L., Hu Z., Chen G., Wang G., Hu T. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol. Genet. Genom. 2012;287:39–54. doi: 10.1007/s00438-011-0660-x. [DOI] [PubMed] [Google Scholar]
- 14.Jia F., Qi S., Li H., Liu P., Wu C., Zheng C., Huang J. Overexpression of late embryogenesis abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014;454:505–511. doi: 10.1016/j.bbrc.2014.10.136. [DOI] [PubMed] [Google Scholar]
- 15.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Jin C., Li K.Q., Xu X.Y., Zhang H.P., Chen H.X., Chen Y.H., Hao J., Wang Y., Huang X.S., Zhang S.L. A novel NAC transcription factor, pbenac1, of Pyrus betulifolia confers cold and drought tolerance via interacting with pbeDREBs and activating the expression of stress-responsive genes. Front. Plant. Sci. 2017;8:1049. doi: 10.3389/fpls.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nashima K., Shimizu T., Nishitani C., Yamamoto T., Takahashi H., Nakazono M., Itai A., Isuzugawa K., Hanada T., Takashina T., et al. Microarray analysis of gene expression patterns during fruit development in European pear (Pyrus communis) Sci. Hortic. 2013;164:466–473. doi: 10.1016/j.scienta.2013.09.054. [DOI] [Google Scholar]
- 18.Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 19.Cao P.B., Azar S., SanClemente H., Mounet F., Dunand C., Marque G., Marque C., Teulieères C. Genome-wide analysis of the AP2/ERF family in eucalyptus grandis: An intriguing over-representation of stress-responsive. PLoS ONE. 2015;10:e0121041. doi: 10.1371/journal.pone.0121041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T., Bai S., Ito A., Sakamoto D., Saito T., Ubi B.E., Imai T., Moriguchi T. Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakai) that differ in their chilling requirement for endodormancy release. Tree Physiol. 2013;33:654–667. doi: 10.1093/treephys/tpt037. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Yan X., Yang Q., Ma Y., Yang B., Tian J., Teng Y., Bai S. PpCBFs selectively regulate PpDAMs and contribute to the pear bud endodormancy process. Plant Mol. Biol. 2019;99:575–586. doi: 10.1007/s11103-019-00837-7. [DOI] [PubMed] [Google Scholar]
- 22.Gabay G., Faigenboim A., Dahan Y., Izhaki Y., Itkin M., Malitsky S., Elkind Y., Flaishman M.A. Transcriptome analysis and metabolic profiling reveal the key role of α-linolenic acid in dormancy regulation of European pear. J. Exp. Bot. 2019;70:1017–1031. doi: 10.1093/jxb/ery405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in anegative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 24.Xue G.P. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 2002;30:e77. doi: 10.1093/nar/gnf076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue G.P. The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J. 2003;33:373–383. doi: 10.1046/j.1365-313X.2003.01630.x. [DOI] [PubMed] [Google Scholar]
- 26.Svensson J.T., Crosatti C., Campoli C., Bassi R., Stanca A.M., Close T.J., Cattivelli L. Transcriptome analysis of cold acclimation in barley albina and xantha mutants. Plant Physiol. 2006;141:257–270. doi: 10.1104/pp.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- 28.Qin F., Sakuma Y., Li J., Liu Q., Li Y.Q., Shinozaki K., Yamaguchi-Shinozaki K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004;45:1042–1052. doi: 10.1093/pcp/pch118. [DOI] [PubMed] [Google Scholar]
- 29.Skinner J.S., von Zitzewitz J., Szucs P., Marquez-Cedillo L., Filichkin T., Amundsen K., Stockinger E.J., Thomashow M.F., Chen T.H., Hayes P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- 30.Baker S.S., Wilhelm K.S., Thomashow M.F. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 31.Jiang C., Iu B., Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- 32.Kitashiba H., Ishizaka T., Isuzugawa K., Nishimura K., Suzuki T. Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J. Plant Physiol. 2004;161:1171–1176. doi: 10.1016/j.jplph.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Gu X., Gao Z., Zhuang W., Qiao Y., Wang X., Mi L., Zhang Z., Lin Z. Comparative proteomic analysis of rd29A:RdreB1BI transgenic and non-transgenic strawberries exposed to low temperature. J. Plant Physiol. 2013;170:696–706. doi: 10.1016/j.jplph.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Kitashiba H., Matsuda N., Ishizaka T., Nakano H., Suzuki T. Isolation of genes similar to DREB1/CBF from sweet cherry (Prunus avium L.) J. Jpn. Soc. Hortic. Sci. 2002;71:651–657. doi: 10.2503/jjshs.71.651. [DOI] [Google Scholar]
- 35.Kitashiba H., Ban Y., Honda C., Moriguchi T. Nuclear localization of sweet cherry DREB1/CBF ortholog (CIG) and low temperature-inducible activity of CIG promoter. Plant Biotechnol. 2006;23:515–518. doi: 10.5511/plantbiotechnology.23.515. [DOI] [Google Scholar]
- 36.Ahmad M., Li J., Yang Q., Jamil W., Teng Y., Bai S. Phylogenetic, molecular, and functional characterization of PpyCBF proteins in Asian pears (Pyrus pyrifolia) Int. J. Mol. Sci. 2019;20:2074. doi: 10.3390/ijms20092074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Esawi M.A., Alayafi A.A. Overexpression of rice Rab7 gene improves drought and heat tolerance and increases grain yield in rice (Oryza sativa L.) Genes. 2019;10:56. doi: 10.3390/genes10010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura F., Tanabe K., Itai A., Morimoto M. Variation in the chilling requirements for breaking leaf bud endodormancy in wild pear species and pear cultivars. J. Jpn. Soc. Hortic. Sci. 2001;70:596–598. doi: 10.2503/jjshs.70.596. [DOI] [Google Scholar]
- 39.Yamane H., Kashiwa Y., Kakehi E., Yonemori K., Mori H., Hayashi K., Iwamoto K., Tao R., Kataoka I. Differential expression of dehydrin in flower buds of two Japanese apricot cultivars requiring different chilling requirements for bud break. Tree Physiol. 2006;26:1559–1563. doi: 10.1093/treephys/26.12.1559. [DOI] [PubMed] [Google Scholar]
- 40.Kagaya H., Ito N., Shibuya T., Komori S., Kato K., Kanayama Y. Characterization of FLOWERING LOCUS C homologs in apple as a model for fruit trees. Int. J. Mol. Sci. 2020;21:4562. doi: 10.3390/ijms21124562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatsuda Y., Nishio S., Komori S., Nishiyama M., Kanahama K., Kanayama Y. Relationship between MdMADS11 gene expression and juvenility in apple. J. Jpn. Soc. Hortic. Sci. 2011;80:396–403. doi: 10.2503/jjshs1.80.396. [DOI] [Google Scholar]
- 42.Ikeda H., Shibuya T., Imanishi S., Aso H., Nishiyama M., Kanayama Y. Dynamic metabolic regulation by a chromosome segment from a wild relative during fruit development in a tomato introgression line, IL8-3. Plant Cell Physiol. 2016;57:1257–1270. doi: 10.1093/pcp/pcw075. [DOI] [PubMed] [Google Scholar]
- 43.Kanayama Y., Sato K., Ikeda H., Tamura T., Nishiyama M., Kanahama K. Seasonal changes in abiotic stress tolerance and concentrations of tocopherol, sugar, and ascorbic acid in sea buckthorn leaves and stems. Sci. Hortic. 2013;164:232–237. doi: 10.1016/j.scienta.2013.09.039. [DOI] [Google Scholar]
- 44.Feng H.L., Ma N., Meng X., Zhang S., Wang J., Chai S., Meng Q. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant. Physiol. Biochem. 2013;73:309–320. doi: 10.1016/j.plaphy.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.