Abstract
Parasitic plants rely on neighboring host plants to complete their life cycle, forming vascular connections through which they withdraw needed nutritive resources. In natural ecosystems, parasitic plants form one component of the plant community and parasitism contributes to overall community balance. In contrast, when parasitic plants become established in low biodiversified agroecosystems, their persistence causes tremendous yield losses rendering agricultural lands uncultivable. The control of parasitic weeds is challenging because there are few sources of crop resistance and it is difficult to apply controlling methods selective enough to kill the weeds without damaging the crop to which they are physically and biochemically attached. The management of parasitic weeds is also hindered by their high fecundity, dispersal efficiency, persistent seedbank, and rapid responses to changes in agricultural practices, which allow them to adapt to new hosts and manifest increased aggressiveness against new resistant cultivars. New understanding of the physiological and molecular mechanisms behind the processes of germination and haustorium development, and behind the crop resistant response, in addition to the discovery of new targets for herbicides and bioherbicides will guide researchers on the design of modern agricultural strategies for more effective, durable, and health compatible parasitic weed control.
Keywords: Orobanche, Phelipanche, Striga, Cuscuta, germination, haustorium, crop resistance, bioherbicides, virulence, sustainable control
1. Introduction
Approximately 1% of all angiosperms are parasitic on other plants and these plants are distributed among 28 dicotyledonous families having evolved the parasitic lifestyle independently at least 12 times [1,2,3,4,5]. Some are facultative parasites, capable of living autotrophically until reproduction, but shifting to a parasitic life form when a host is available to obtain nutrients with less investment in assimilation system. In comparison, obligated parasitic plants require the infection of another plant to survive shortly after germination. Parasitic plants can be also grouped by their photosynthesis competency, being either photosynthetically active hemiparasites or achlorophyllous holoparasites; or separated based upon the type of vascular connections they form with their host, being either xylem feeders or phloem feeders. They can also be grouped by the host plant organ to which they attach, either root feeders or shoot feeders [4,6]. While many parasitic plants have remained part of larger ecological communities, a small number of species have evolved to weediness, becoming troublesome pests in the agricultural field and an important constraint to crop productivity. Besides colonizing agroecosystems, parasitic plants are also present in urban ecosystems (Figure 1).
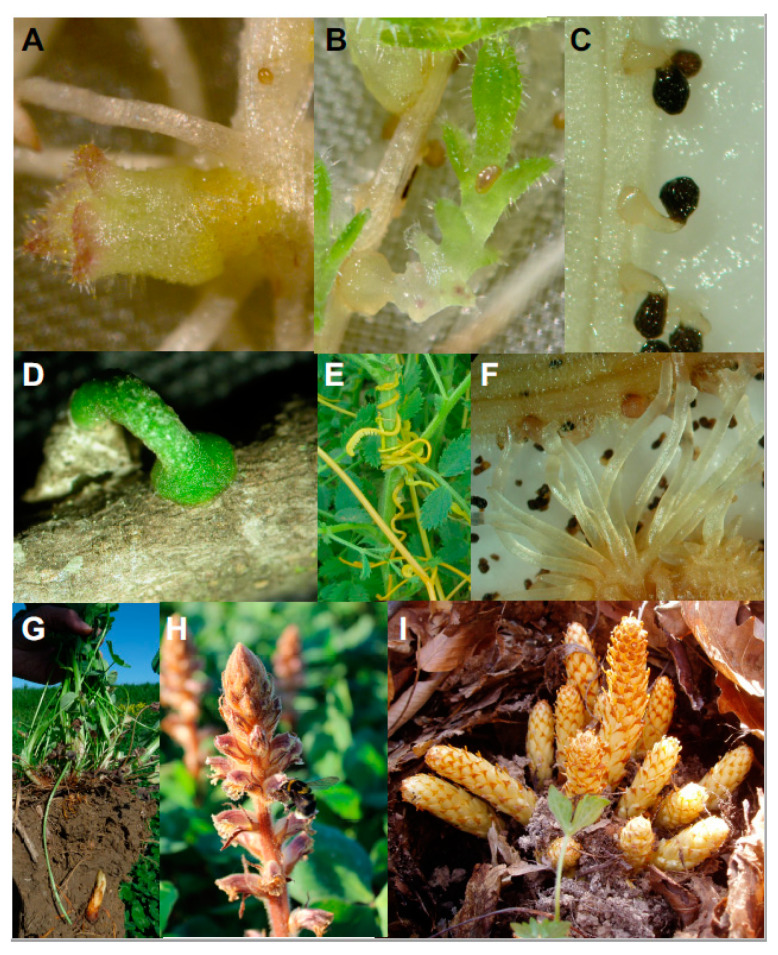
Figure 1.
Adaptation of parasitic plants to agricultural and urban ecosystems. Shown are representative photographs of parasitic plants in agricultural (A,B) and urban (C,D) locations. (A) Hemiparasitic shoot parasite Viscum sp. feeding on olive tree; (B) holoparasitic root parasite Orobanche sp. feeding on clover commercial field; (C) Viscum sp., in the city of Dijon, France; (D) Orobanche sp. parasitizing clover in a park in the city of Dijon, France.
While all agricultural weeds compete with crops for the space to obtain water, nutrients, and light, parasitic weeds are particularly noxious since they also directly extract valuable water and nutrients from the host plant. To extract nutrients from the host plants, parasitic weeds have evolved a unique multicellular structure termed the haustorium that invades the host, forms connections with the host vascular system, and withdraws its needed water and nutrients [7,8]. Successful haustorial connection to the host results in permanent damage during a large part of the crop life cycle, decreasing the crop value by reducing the harvested yield and contaminating it with parasitic seeds. Worldwide expansion of these noxious plant pests including the widely recognized genera Striga, Orobanche/Phelipanche, and Cuscuta has become a threat to food security [9].
There are few sources of crop resistance to parasitic plant infection [10,11]. The identified forms of resistance are classified as pre-attachment or post-attachment resistance according to whether the resistance occurs before or after the haustorium attaches the host surface [10]. Once the crop is infected, it is difficult to fend off the exhaustion of nutrients created by the attached parasite. Then, chemical control is the most frequent commercial method, albeit its application needs to observe specific recommendations for each parasitic life form [12].
In this review, we describe the current state of knowledge on the mechanisms controlling the infection process of parasitic weeds, and the defense systems and protection strategies employed by their hosts.
2. Infection by Parasitic Weeds
All forms of parasitic weeds, facultative or obligate parasites, hemiparasites or holoparasites, root parasites, or stem parasites interact with the host crop by means of the haustorium [7]. Two main types of haustoria exist in root parasitic plants [1,8]. The terminal haustorium is typically developed by obligated parasitic weeds at the tip of their embryonic radicle. The lateral haustorium is developed as an extension in hemiparasitic radicle and mature roots of hemiparasitic and holoparasitic weeds. In aboveground parasitic weeds that infect crop stems and leaves, lateral haustoria develop as lateral extensions of their parasitic stems (Figure 2). The formation of a mature haustorium involves four developmental stages: Haustorium initiation, host invasion, establishment of host-parasite vascular continuity, and the creation of a parasitic sink that will be accepted by the crop as one of its own. Several mechanisms of crop location adapt the timing of haustorium development to the resource availability of its hosts [13].
Figure 2.
Haustorial penetration of host plants and maturation of parasite. Shown are representative photographs of terminal (A–D,G–I) and lateral (E,F) haustorium penetration and maturation stages. (A) Young hemiparasitic weed Alectra vogelii infecting the root of cowpea; (B) young hemiparasitic weed Striga hermonthica infecting the root of sorghum; (C) young holoparasitic weed Phelipanche aegyptiaca infecting the root of Medicago; (D) young hemiparasitic weed Viscum cruciatum infecting an olive tree stem; (E) hemiparasitic weed Cuscuta campestris infecting a chickpea stem; (F) mature anchorage roots of Phelipanche aegyptiaca infecting the root of vetch; (G) underground shoot of Orobanche minor growing on clover roots towards soil surface; (H) emerged shoot of Orobanche minor; (I) emerged shoots of Conopholis americana feeding on oak roots.
2.1. Crop Location for Germination, Host Trophic Growth and Haustorium Initiation
In contrast to parasitic plants growing in wild ecosystems, where host plant communities provide predictable nutrition to the parasite [14], the supply of resources in agricultural lands for the persistence of parasitic weed communities is affected by changes of cropping systems attending market demands for specific crops, or novel market availability of resistant varieties. Sudden changes in cropping systems impact parasitic weed communities differently, depending on their degree of autotrophy and feeding strategy, being obligated root parasites of annual crops submitted to the higher pressure. To adapt their parasitic life with the timing of crop resource availability, parasitic weeds use mechanisms of host location, which, depending on the degree of autotrophy and feeding strategy, involve up to 3 steps: Host-induced germination, host-tropic growth, and host-induced haustorium initiation [13]. The high pressure to timely detect the host in resource-unpredictable agroecosystems promotes genetic and epigenetic changes, rapidly leading to new weed biotypes with adaptation capabilities to new agricultural practices, climatic areas, new hosts species, and resistant cultivars [15,16].
2.1.1. Germination
The root parasites in the genera Orobanche/Phelipanche and Striga are obligated parasitic weeds with a high degree in host dependency and host specificity [17]. In absence of host contact, the seedling has a limited growth capacity, allowing it to elongate only a few millimeters before its resources are exhausted, a few days after germination. In agricultural fields, the vulnerability of their seedlings is much higher than in any other weed. To survive, their germination strategy is built on the combination of four seed traits: Seed size, seed number, embryo longevity, and seed dormancy. Orobanche/Phelipanche and Striga seedlings do not compete for resources in their pre-attached young life stages and therefore the parent plants produce a tremendous number of tiny seeds with high colonization capacity [18]. These seeds penetrate easily into the soil, even in untilled soils, and are easily dispersed by wind, increasing the chances to encounter susceptible hosts in adjacent fields, allowing the population to persist. The seed viability is preserved over time by long embryo longevity and two cooperative processes of dormancy that delay germination until detection of a nearby host. The first dormancy process is a temperature-dependent cyclic dormancy, being non-dormant in the host-growing season and dormant in the next. During the non-dormant season, seed receptors are activated to detect germination stimulants exuded by host roots [19,20,21]. If the chemical signal is not detected, germination will remain inhibited during the non-dormant season and will re-enter the dormant phase the next season. The amount and nature of exudation of germination stimulants by crop roots varies depending on the crop species, the phenological stage, crop nutritional status, and the growing season [19,22,23,24,25,26,27,28,29,30]. For each parasitic weed species, there are multiple seed receptors within a family of α/β-hydrolases that specifically detect the stimulants differentially exuded by their host species [31].
Different feeding styles in other parasitic weeds, like in the host-generalist Cuscuta and the facultative parasite Ramphicarpa, make the likelihood of their finding nutrients less dependent on finding a specific host right after germination. Consequently, their seeds tend to be larger and are not dependent upon chemical signals for germination, although their seedlings will detect the host through different signals at later growth stages [13]. Their seeds have typical dormancy mechanisms in autotrophic weeds such as physical dormancy induced by a thick seed coat in Cuscuta, that preserves seedbank viability against seed predators and allows a staggered germination over time or a physiological dormancy in Ramphicarpa that inhibits germination in the dark, allowing the buried part of their seedbanks to delay germination. Once those regular mechanisms of dormancy are broken, their seeds germinate when temperature indicates the growing season for favorable establishment and reproduction [32,33,34].
2.1.2. Host-Tropic Growth
After Striga and Orobanche/Phelipanche germination, a radicle emerges from the seed and rapidly explores the nearby underground environment to reach the host surface. Striga radicle grows by cell growth and apical cell division while Orobanche/Phelipanche radicle grows by cell elongation [18]. Although chemotropism has not been demonstrated yet, in vitro observations of the seedling radicle redirecting its growth directly toward the host root have made different authors suggest that host chemi-localization by the radicle could be part of the Striga and Orobanche/Phelipanche infection process (Figure 2B,C) [13,18,35,36,37,38]. Seeds that germinate a distance longer than 4 mm lose their polarity and exhaust their viability before they can reach the host and therefore such chemotropism would be an advantageous mechanism [38,39]. Although the seedling viability rapidly expires in the soil or in rhizotron experiments, they can be grown on media culture to produce root cultures and regenerate flowering plants [40,41,42,43] (Figure 3). Cuscuta seedling viability expires without a host in 3 to 7 weeks depending on the photosynthetic activity of each Cuscuta species [44]. Cuscuta explores the nearby aerial environment using a rotative movement (Figure 4). Volatile chemicals and far-red light, indicative of proximity of vegetation, guides the Cuscuta movement towards the host [45,46,47].
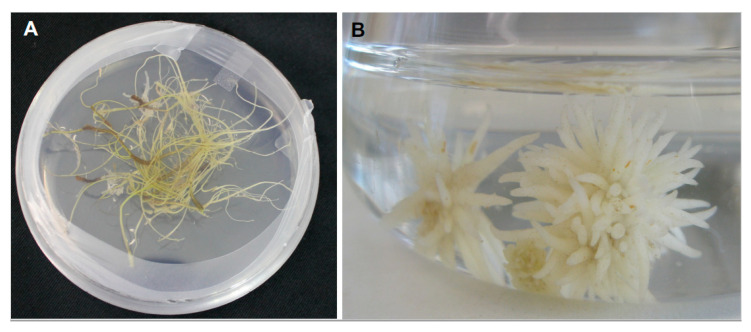
Figure 3.
Haustorial competent root cultures of obligated root parasitic weeds. Shown are representative photographs of (A) Striga hermonthica; (B) Phelipanche aegyptiaca.
Figure 4.
Directional growth of five-day-old Cuscuta toward its host. Shown are representative photographs taken at 12-h intervals of the growth of Cuscuta and its host lentil. (A) Cuscuta seedlings rotate in a counterclockwise rotation; (B) Cuscuta seedling bending guided toward host stem; (C) Cuscuta coiling and adhesion; (D) formation of adhesive discs.
2.1.3. Initiation of Haustorium
The first stage in haustorium development is initiated upon host detection through chemical and physical signals, which develops an adhesive structure that cements the parasite to the host surface from which the invasive organ subsequently develops [7,48,49,50,51]. Perception of haustorium-inducing signals promotes a cessation of parasite root growth with a rapid swelling. Auxin biosynthesis genes are upregulated at the epidermal cells near the contact site [52]. These cells divide and elongate, becoming, depending on the parasitic species, either haustorial hairs or papillae covered by adhesive secretions to serve as the anchoring device (Figure 5) [53,54,55]. The chemical nature of the adhesive glues is not completely clear but literature indicates that it differs for different parasitic species. For example, the adhesive compounds have been described as hemicelluloses in Agalinis [56], pectinaceous mucilaginous material in Triphysaria [57], compounds that stain positive for carbohydrate material in Orobanche/Phelipanche [53], while in Striga these compounds stain negative for carbohydrate material but give a positive result with safranine [58]. In the stem parasite Cuscuta, epidermal cells at the parasite-host contact sites dedifferentiate into disk-like meristems forming the pre-haustorium (Figure 4D). Some of these cells become elongated and secrete pectinaceous substances to seal the parasite haustorium to the host surface. Tactile signals, light spectrum, and phytohormones play a pivotal role in the development of the Cuscuta pre-haustorium [13,50,59,60,61].
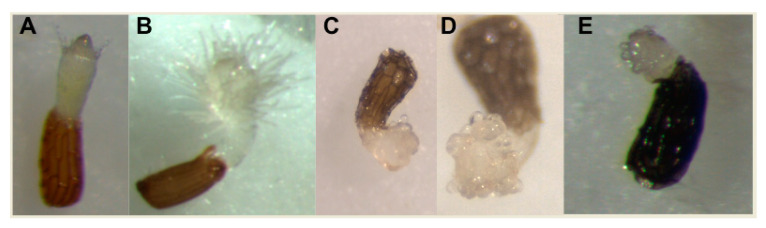
Figure 5.
Haustorial hairs and papillae on root parasitic weeds. Shown are representative photographs of haustorial hairs (A,B) and papillae (C–E) formed in seedlings of root parasitic weeds. (A) Ramphicarpa fistulosa; (B) Striga hermonthica; (C,D) Phelipanche ramose; (E) Orobanche cumana.
2.2. Host Invasion and Establishment of Vascular Connection
Infections by root and shoot parasites modify various cellular processes both in crop and parasitic tissues at the site of crop-parasite contact. Haustorium penetration upregulates genes such as expansins, arabinogalactan proteins, or xyloglucan-endotransglucosylases with a role of rearranging the cell wall [62,63,64]. Parasitic weed gene expression in the haustorium is similar to gene expression in roots of non-parasitic plants [65]. In some root parasites the haustorium initiates from root pericycle as it occurs for lateral roots of no-parasitic plants [66,67]. In other parasitic species the sequence of events initiates in cortical cells [7,68,69] Subsequently, the epidermal cells at the sealed parasite-host root interface become densely protoplasmic and enlarge rapidly, creating a mechanical force to enter the host. The radicle secretes cell wall-modifying enzymes such as pectinolytic enzymes, polygalacturonase, rhamnogalacturonase, and peroxidase [39,70,71]. The combination of mechanical force and enzymatic digestion of host cell walls pushes aside the cells of the host epidermis, cortex, and endodermis from the pathway of weed invasion [53]. Differentiation of vascular elements can occur concomitantly with penetration of endodermis [54] or when the intrusive cells reach the host xylem [72]. Comparative transcriptomics has shown that during haustorium evolution, parasitic plants may have recruited genes from unrelated plant structures with invasive functions (e.g., pollen tubes) and repurposed these genes in the haustorium [65]. Auxin flow is critical for xylem continuity and determines the polarity of the haustorium to allow its function as a root, absorbing water and solutes [73]. In parallel with the formation of xylem-xylem connections, depending on the parasite species, there is the development of sieve elements in haustoria [74]. Some holoparasites such as Orobanche/Phelipanche species develop direct symplastic connections through plasmodesmata [75]. The invasive haustorium of Cuscuta grows through the cells of the host stem, forming searching hyphae that elongate and reach the host xylem to become xylem cells that establish the crop-parasite bridge [32]. Cuscuta haustorium has been compared with a modified adventitious root; however, it develops in the pre-haustorium from dedifferentiation of cortical parenchyma in a different manner that the adventitious roots develop within the cambium [76,77,78]. Additionally, the SHOOT MERISTEMLESS-like factor plays a crucial role in Cuscuta haustorium but it is not involved in root development [78].
2.3. Maturation of Absorptive Sink
As in lateral roots, the vasculature of a mature haustorium extends from the corresponding host tissues oriented perpendicular to the main axis of host vascular system (Figure 6A). In addition, in the infestation site of some root parasites the host cambium is oriented in line with the cambium of the parasite, therefore perpendicular to the main axis of the host root. The same does not occur for endodermis, cortex, or epidermis, consistent with the fact that the haustorium assimilates nutrients and water from the crop vasculature rather than from the soil [8]. While hemiparasites are by definition photosynthetically active and usually considered parasites mainly for water and minerals, their photosynthesis is not sufficient to fulfill the growth requirements and their haustorium also extracts carbon from the host [79,80]. Holoparasitic weeds like Orobanche/Phelipanche and some species of Cuscuta do not photosynthesize and have low levels of transpiration and obtain all their carbon, nitrogen, and other minerals from the host mostly from the host phloem [81,82,83].
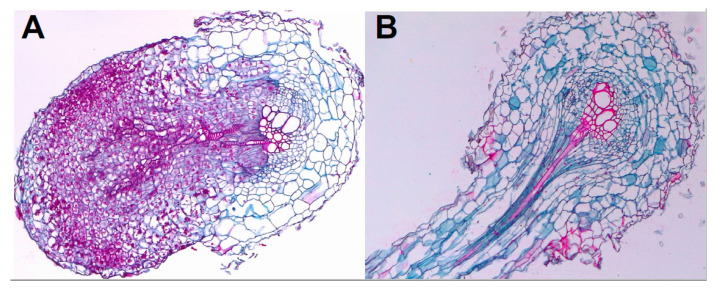
Figure 6.
Xylem to xylem junctures in sunflower root. (A) Longitudinal section of Orobanche cumana seedling connected to sunflower vascular system (shown in transversal section); (B) Longitudinal section of sunflower lateral root developed from main root sunflower root (shown in transversal section).
Despite several anatomical and biochemical studies of types of vascular connections and transferred nutrients between host and different parasitic weed species the physiology of parasitic phloem loading in parasitic weeds is not clear [6]. Direct symplastic connections through plasmodesmata [75] and raffinose series oligosaccharides [84,85] seem to indicate the symplastic loading style in holoparasites. In contrast, several indications support the hypothesis of an apoplastic pumping of extracellular sucrose into hemiparasite phloem: (i) The apoplastic loading is most common in herbaceous plants and in many hosts of hemiparasites [86]; (ii) symplastic connections have not been found in hemiparasitic weeds except for Striga gesnerioides [87]; and (iii) the apoplastic separation between hemiparasite and the host would reinforce a concentration gradient of solutes [6].
The relative sink strength among different plant organs is determined by factors such as the activity of invertase enzymes and sugar transporters, vascular pressure, the density of unloading sites, and the developmental coordination among competing sinks. The nutrient sink created by parasitic weeds gains competitive strength against authentic crop sinks in the host also by means of these same mechanisms. The haustorium cells metabolize the host nutrients into a parasite-specific metabolic profile, creating an osmotic potential favorable for the nutrient flow towards the parasite [84,88,89,90,91]. High levels of potassium, cytokinin, and abscisic acid in the parasite may keep the parasitic stomata open and decrease hydraulic conductivity to promote the host-to-parasite flow relative to other sinks [6,79,92,93,94]. The dense parasitic seedbanks in agricultural lands with germination synchronized with the specific phenology of the crop creates a coordinated attack of several parasites, creating multiple and simultaneous parasitic unloading sites before the crop has the opportunity to redirect the resource allocation towards host reproductive organs [28,95,96]. Breeding crops with coordinated and precocious pod filling relative to the timing of parasitic attachment suppresses the strength of the parasitic sink in susceptible crops [97].
3. Effect of Parasitic Weed Infection on the Crop
Crop development from germination to reproduction occurs according to a predicted alternation of priorities in sink organs ultimately destined to allocate crop resources toward crop reproduction. Parasitic weed infection strongly reduces crop harvest by either (i) disrupting the crop orchestration of resource allocation, altering dry matter partitioning between crop organs prioritizing those adjacent to the parasite (i.e., host roots in the case of root parasites and host shoots in the case of shoot parasites [83,98]) and subsequently diverting the destination of crop resources into the building of parasite biomass to fulfill the parasitic reproduction, or (ii) inducing pathogenic effects on the crop photosynthetic and nutrient uptake machineries [83,98,99,100]. The extent to which parasitic weeds affect the crop growth, biomass partitioning, and nutrient status differs depending on the differing feeding styles of the parasites [101]. The infection by some hemiparasitic weeds, such as Striga, induces crop biomass depression greater than the biomass accumulated by the parasite. Striga, like other plant herbivores and pathogens, decreases host productivity by lowering rates of crop photosynthesis [99,102,103]. In contrast, nutrient sinks created by the achlorophyllous Orobanche cernua or Cuscuta reflexa affect the crop in a fashion similar to mycorrhizal symbionts, increasing rates of net photosynthesis [98,100,104]. Orobanche-infected crops are smaller but have an increased rate of carbon acquisition with the end result that infected plants maintain an equivalent resource budget for the combined Orobanche-crop-infected biomass complex compared to that of an uninfected plant [100].
The parasitic sink strength increases with the density of unloading parasitic attachments up to maximum feeding capacity from which the parasites compete among them and a negative relation between number of parasites attached per plant and weight of individual parasites is observed [96,100,105,106]. For each parasitic species the maximum feeding capacity varies with the crop species they infect and may suggest a trade agreement by which the parasite allows the crop to grow and produce the resources the parasite needs to ensure parasitic reproduction [50,96]. Conserved symbiotic pathways that mediate mechanisms that regulate the extent of colonization by Rhizobium and arbuscular mycorrhizal fungi could also be in place in parasitic weeds [107]. The parasitic weed sink strongly competes with other host sinks at the greatest expense of host reproductive organs [95,96].
4. Phenotypic Expression of Resistance
Parasitic weeds differ in host range and crop species differ in their susceptibility to parasitic weeds, being resistant to some parasitic species but highly susceptible to the attack of others. Additionally, within host species, some genotypes will be capable of supporting growth and reproduction of a specific parasitic weed species, whereas others will have contrasting levels of resistance and susceptibility [97,108]. Defense-related genes are induced in the parasitized plant during parasitic weed penetration both in susceptible and resistant interactions, indicating that susceptible hosts detect at early stages the penetrating parasitic weed as an alien and not as coordinated growth of an attractive sink [109,110,111]. During the susceptible response, the parasite must circumvent the first response to later become a compatible sink. However, some wild species, landraces, and a few cultivars have innate or inducible resistance mechanisms to attack by Striga, Orobanche/Phelipanche, or Cuscuta (Figure 7) [10,38,112]. Post-attachment resistance operates at several levels. These processes initiate when the parasite haustorium attaches to the host surface and attempts to penetrate host tissues to connect with the vascular system. Among the processes described are: (i) Abiosis, the synthesis and release of cytotoxic compounds such as phenolic acids and phytoalexins; (ii) the rapid formation of physical barriers to prevent possible pathogen ingress and growth (e.g., lignification and other forms of cell wall modification at the host-parasite interface); (iii) the release of reactive oxygen species and activation of programmed cell death in the form of a hypersensitive response at the point of parasite attachment to limit parasite development and retard its penetration; and (iv) prevention of the parasite establishing the essential functional vascular continuity (i.e., xylem-to-xylem and/or phloem-to-phloem connections) with the host, delaying parasite growth followed by parasite developmental arrest and eventual death [113].
Figure 7.
Hypersensitive-like response developed on host in response to parasite attack. Shown are representative photographs of host root responses to attempted penetration by parasite haustorium. (A) Phelipanche aegyptiaca on Vicia arthropurpurea and (B) Cuscuta campestris on Vicia sativa. Red arrows point at hypersensitive-like response at host-parasite interface both in Phelipanche and Cuscuta. White arrows point at V. sativa sites where Cuscuta haustorium was manually removed to make the resistance response more visible for explanatory reasons.
5. Evolution of Host Specificities and Races
Some Orobanche/Phelipanche species are major phytopathogens on several important crops in throughout North Africa, the Middle East, and Southern and Eastern Europe, and their footprint continues to expand on these crops as well as their host range continues to evolve. Indeed, recent reports reveal that non-weedy Orobanche/Phelipanche species can also infest crop species as an ecological opportunity arises. For instance, the infestation of celery (Apium graveolens L.) by Orobanche nana, known as a non-weedy parasite, was reported for the first time in Italy in 2014 [114]. Several other reports indicated the infestation of new compatible targets by weedy Orobanche/Phelipanche species when the density of this new but unfavored hosts increase [115,116,117,118,119,120].
The eco-evolutionary dynamics that cause parasitic plants to infest new hosts can be of two types: (i) An extension of the range of their natural hosts or (ii) a host specialization occurring as a result of genetic modifications impacting a major key step of the parasitic process (i.e., detection of germination stimulants or haustorium-inducing factors). In Phelipanche ramosa, it appears that host specialization is acting, rather than an extension of the host range [18]. This parasitic species is native to Europe and Western Asia and has been introduced to most continents. It is known for its broad host range as it can parasitize more than 50 different hosts including crops and vegetable [121]. However, at least three genetic groups of P. ramosa have been identified in France, differentiated in particular by their host preferences (oilseed rape, hemp, and tobacco, respectively) [122,123]. These different pathovars of P. ramosa are distinguished by their differential response to germination stimulants and by a higher fitness (i.e., greater reproductive success) on some crop hosts than on others [123,124,125]. Host specialization has also been observed in Orobanche cumana, which in contrast with P. ramosa exhibits a narrow host range. O. cumana is the most serious constraint for sunflower cultivation in Southern and Eastern Europe and in North Asia. The wild ancestor of O. cumana is found in Caucasus, Southern Russia, and on the Black Sea coast where isolated populations parasitized Artemisia spp. The first observation of O. cumana parasitizing sunflower was made in Russia in 1890. At the end of the 19th and beginning of the 20th century, the attacks of O. cumana in Russia were so severe that it threatened the production of sunflower oil. Breeders have therefore developed sunflower varieties or hybrids resistant to the forms of O. cumana, called races, which have proved to be increasingly aggressive because of their ability to bypass the successively introgressed resistant genes. To date, at least 7 races (A–G) have emerged along with seven resistant genes (Or1–Or7). The same type of “arms race” could be observed in both species of witchweeds, S. asiatica [126] and S. gesnerioides [127].
While significant progress has been made in understanding the biology behind the development of parasitic plants and their parasitic strategies, the biology underlying their adaptation to a host remains unknown. Host specialization probably involves genetic mutations. However, several lines of evidence, including rapid host adaptation and minimal or no genetic differentiation between pathovars and races of Orobanche/Phelipanche and Striga species, indicate that host changes could also involve epigenetic modifications. One major goal would be now to determine how host preferences arise and to evaluate the differential contributions of genetic change and epigenetic priming mechanisms for host specificity in parasitic weeds.
6. Resistance Genes to Parasitic Plants
The identification of differentially resistant genetic variants within host germplasms and construction of populations segregating for the resistance phenotype permitted researchers to identify individual genes or groups of genes conferring resistance [10,128,129,130]. In most cases, resistance to Striga spp. in the grasses (sorghum, millet, rice) appears to be polygenic with both major and minor genes with a large genotype by environment interaction [10]. Resistance often appears to involve several mechanisms, is often weak, and tends to break down in the presence of new geographic or physiologically specialized forms of the parasite. Resistance to S. gesnerioides, which primarily attacks dicots, appears to be mainly monogenic [131], and in cowpea where it has been extensively studied, resistance appears to be race-specific with multiple pathotypes of S. gesnerioides and multiple resistance genes in the cowpea genome [132]. The host differential response in cowpea cultivars allowed the construction of genetic populations segregating for the race-specific phenotype, and subsequently the use of a marker-assisted positional cloning strategy by [133] to isolate a dominant gene from cowpea termed RSG3-301, conferring resistance to S. gesnerioides race SG3. RSG3-301 encodes a typical nucleotide-binding domain and leucine-rich repeat containing (NLR) protein with a N-terminal coiled-coil domain (CC), followed by a central nucleotide binding site (NBS) and a C-terminal leucine-rich repeat (LRR) domain.
The characterization of RSG3-301 led Li and Timko [133] to suggest that race-specific Striga resistance in cowpea is an example of effector-triggered immunity (ETI) in which intracellular NLR proteins (such as RSG3-301) are activated either directly or indirectly upon recognition of pathogen/parasite effectors [134,135].
Membrane-bound receptors in tomato (CuRe) and sunflower (HaOr7) functioning as pattern recognition receptors (PRRs) were discovered [136,137] that appear to be involved in pathogen-associated molecular patterns (PAMP)-triggered immunity, the first level of plant defense responses [138,139]. Subsequently, Jhu et al. [140] identified a resistance gene in tomato dubbed CuRLR1 that appears to also be involved in ETI against Cuscuta. Most of the resistance genes to Orobanche have been discovered and used for sunflower breeding programs for O. cumana resistance [141]. Qualitative resistance to O. cumana is controlled by major genes and is race-specific. Thus, seven dominant resistance genes Or1 to Or7 have been described corresponding to 6 races of O. cumana A to F. A more virulent race G exists for which no resistance has been characterized to date. However, among these resistance genes only the Or5 and Or7 genes have been previously mapped. Or5 confers resistance to the race E and has been shown to interact with the AvrOr5 avirulence gene agreeing with the gene-for-gene hypothesis in the O. cumana-sunflower pathosystem [142,143]. Recently, Duriez et al. [137] deciphered the function of Or7 conferring resistance to O. cumana race F by preventing the connection of the parasite to the vascular system of the sunflower roots. Or7 encodes a membrane receptor-type LRR kinase, suggesting the existence of a parasitic AVROR7 avirulence protein. In P. ramosa, despite the identification of resistances in winter oilseed rape [144], nothing is known about the underlying molecular processes.
7. Parasite Effectors
The deployment of secreted effectors to block host resistance responses is observed in multiple host-pathogen interactions including pathogenic microbes, fungi, and nematodes [145,146,147]. Recently, Su et al. [148] used transcriptomic profiling and transgenic expression analysis to identify a novel secreted effector protein from S. gesnerioides termed SHR4z, which blocks host plant immunity in the multi-race resistance cowpea B301. SHR4z suppresses the hypersensitive response (HR) triggered by S. gesnerioides race SG4 by interfering with signal transduction pathway leading to HR activation upon parasite attack. The exact mechanism remains to be determined, but SHR4z is structurally similar within the N-terminal region to the SERK (somatic embryogenesis receptor-like kinase) subfamily of LRR-RLKs and is predicted to interact with the BTB/POZ interface of VuPOB1, a PUB E3 ligase family member that appears to be a positive regulator of HR in cowpea.
A set of five candidate effector proteins that suppress known plant defense pathways in Arabidopsis including two LRR SERK-like proteins similar to SHR4z have also been reported by Clarke et al. [149]. Additional characterization of the common aspects of signal transduction pathways involved in response to parasitic weeds is clearly needed.
8. Transcriptome and Proteome Analysis of Resistance
The immune response of host plants to parasitic plants is also likely multi-faceted. Similar suites of genes are observed to be upregulated during resistance responses to Orobanche and Striga and it is becoming increasingly clear that the SA signaling pathway and to a lesser extent the JA signaling pathway play important roles in the activation of resistance to parasitic plants. In other plant-plant pathogen interactions the SA and JA defense pathways can interact either antagonistically or synergistically [150]. The SA pathway is often activated in response to biotrophic fungal pathogens, leading to the expression of suites of PR genes, whereas the JA pathway is often important in resistance to necrotrophic pathogens and insect pests. A number of studies have evaluated the effectiveness of SA applications to hosts to prevent parasitism by Orobanche spp. SA consistently promoted resistance, in the case of clover roots infected with O. minor, by the activation of defense responses leading to lignification of the endodermis [151]. SA analogues have also been proved to induce resistance in oilseed rape against Phelipanche ramosa [152]. To date, SA-inducing chemicals have not been widely applied to field-grown crops to test their effectiveness as part of a control strategy. The induction of genes involved in JA biosynthesis has been observed in compatible interactions between Orobanche species and their hosts [153] but the involvement of JA in resistance is less clear [151]. A decade ago, in a study of resistance in tomato to the shoot parasite Cuscuta pentagona, a role for both JA and SA was also proposed [154]. The importance of several genes for A. thaliana resistance and susceptibility to P. aegyptiaca was examined by Clarke et al. [149] who found that functional crosstalk between SA and JA signaling is required to support the attachment and development of the parasite and that the parasite specifically manipulates these pathways to ensure success. Manipulation of the crosstalk can specifically limit the ability of P. aegyptiaca to parasitize A. thaliana. In particular, several host genes involved in JA and SA biosynthesis and signaling are required for full host susceptibility and the putative immunity hub protein PFD6 is a critical component of the host immune response that limits the progression of P. aegyptiaca parasitism.
9. Strategies for Effective Parasitic Weed Management
Due to the high fertility rate of parasitic weeds, their management must inhibit not only the loss of crop yield but also parasitic flowering. A given control method could not be considered completely successful if it does not inhibit parasitic weed seed bank replenishing at least by 95% [155,156,157]. Commercial strategies for parasitic weed are currently based in crop resistance and chemical control. The processes of host-induced parasitic germination and haustorium formation are the most frequent targets for the development of resistant cultivars [112,158,159,160]. Sources of resistance against these mechanisms are scarce among crop germplasm collections being more frequent in wild relatives to crop species [10,11]. The success of the slow and difficult process of traditional breeding methods that adds one resistance gene at a time is quickly overcome by parasitic weeds due to: (i) The existence of individuals among the dense and heterogeneous seed banks capable of overcoming the resistance of the new cultivar before its first cultivation and (ii) after the cultivation of the resistant cultivar, the capacity of stressed parasitic weeds to mutate, evolving virulence-enhanced parasitic biotypes much quicker than the breeders’ capacity to develop the resistant cultivar [141,161].
The use of herbicides as a strategy for parasitic weed control [12] is mainly determined by: (i) Whether the parasite and the crop are attached, the herbicide is translocatable, and if the herbicide is selective enough to kill the parasite without damaging the crop; (ii) if the parasite is holoparasitic and achlorophyllous since photosynthesis-inhibiting herbicides cannot be used for their control [157,162,163], and (iii) in the case of root parasitic weeds, the main crop damage is done during the underground parasitic life stages and therefore post-emergence herbicides do not prevent yield losses [164,165]. The close interaction of parasitic weeds with the host crop allows the use of systemic herbicides to control young parasites before they provoke the crop damage [166]. The systemic herbicide is applied to the crop foliage and delivered to the shoot or root parasites either via the haustorium or through exudation to the rhizosphere from the crop roots [167,168,169]. The systemic herbicides used for parasitic weeds include inhibitors of aromatic (glyphosate) or branched-chain amino acid synthesis (imidazolinones and sulfonylureas), inhibitors of vitamin folic acid (asulam), inhibitors of glutamine synthetase (glufosinate), or hormonal herbicides (2,4-D and dicamba) [169,170,171,172,173]. The efficacy of each herbicide varies with the parasitic weed species. For example, glyphosate and imazamox have herbicidal effects on root parasitic weeds Orobanche/Phelipanche, but do not kill Cuscuta seedlings; whereas Cuscuta are very sensitive to glufosinate [169]. In addition, the intrinsic killing effect of a given herbicide towards a specific weed genotype can be ameliorated by the quality of interaction between the host and parasite, through the quality of haustorium connection, or nutritious capacity of host genotypes [169]. Therefore, the use of herbicides as a strategy must be specifically crafted depending on which parasite-crop species combination is being targeted and on the availability of information on the specific herbicide and herbicidal doses that are sublethal for the crop but can be delivered in a lethal doses to the parasite, and the availability of crop varieties with herbicide resistance [157,166,174]. The success of herbicidal treatment for the control of parasitic weeds can be high, but its sustainability is compromised by the same general problems observed in the deployment of herbicides in general for weed control, namely (i) the predicted emergence of weed resistance to herbicides [175]; (ii) the lack of development of new herbicide mechanisms of action to counteract the emergence of resistance [176]; and (iii) unwanted non-target effects either disrupting biodiversity due to the low herbicide specificity [177,178] or compromising crop immunity against other pests [179,180].
Biotechnology has the potential to eliminate the bottleneck in parasitic weed management created by the scarcity in both herbicide mode of actions and resistance sources. As such, it could create opportunities to obtain control strategies based in biologically derived alternative chemistries, to protect the sustainability of existing resistant mechanisms by creating resistance-enhanced crops through gene pyramiding, and to expand the available number of resistant genes for breeding beyond the limited gene pool of a given crop and its wild interbreeding relatives [157,163,181]. Besides the above-mentioned control methods, other non-chemical strategies such as soil fertility amendments or solarization have proven effective in some cases of small-holder farmers or high-value specialty crops [2,12,164,182,183,184].
9.1. Bioherbicides
Given the low availability of herbicides against parasitic weeds and the prediction that parasitic weeds will evolve resistance to herbicides [175], the sustainability of chemical control depends on the discovery of new herbicides. The special biology of parasitic weeds limits the number of metabolic pathways that can be targeted by current commercial herbicides but at the same time generates opportunities for the discovery of parasitic weed-specific herbicide targets. For example, biologically-derived chemicals with activity of suicidal induction on essential life stages such as germination or haustorium formation away from the host have a specific killing effect in the parasitic weed while potentially minimizing unwanted health effects on non-target organisms and environment [185,186,187]. Biologically-based weed control methods will play a role on the future of weed management, because of their relatively low cost, long-term sustainability, and environmental friendliness [188]. According to the terminology of the U.S. Environmental Protection Agency, the following types of control strategies can be considered bioherbicides: (i) Microbial herbicides, (ii) biochemical herbicides, and (iii) allelopathic plants that secrete herbicidal compounds [188].
Microbial herbicides are plant-pathogenic or non-pathogenic microbes living or not, mixed in or not with their metabolites [188]. The numerous microbes studied for biological control of parasitic weeds have been reviewed by Watson [189]. There are many articles studying, under controlled conditions, the biological control of parasitic weeds by microbes, but few describing an adequate amount of control for farmers. Few biocontrol agents meet the conditions of sufficient virulence, host specificity, and ease of production and application for field use. These limitations could be overcome by biotechnological approaches of virulence enhancement. Mutants and transgenic variants of natural pathogenic isolates of parasitic weeds showed improved control levels through overproducing and secreting either toxic compounds or inductors of suicidal germination [181,190,191,192]. Biochemical herbicides are compounds of microbial or plant origin. Research with toxins from microbial and plant origin and certain naturally occurring chemicals has demonstrated that additional herbicides may be developed targeting the pre-attached parasitic seedlings and therefore inhibiting the haustorium formation on the host surface [49,193]. Recent developments of high-throughput screenings of metabolites on seeds and seedlings of parasitic weeds will speed up herbicide discovery efforts [194]. Allelopathic crops delivering in situ herbicidal action against parasitic weeds in agroecosystems should also be considered in future sustainable weed management [195]. The strategy of genetically engineering crops to overproduce and secrete either toxic chemicals or inductors of suicidal germination into the rhizosphere [157,196] is discussed in the section below. Finally, the use of RNA to silence essential weed genes through the process of RNA interference has great potential for weed management. Applied as a spray, RNAi can efficiently kill weeds with sufficient host specificity because sequences can be designed to selectively target a specific weed species [188]. For its application to parasitic weed control, genetically modified crops expressing RNAi (RNA interference) constructs or microRNAs (miRNAs) could also be considered bioherbicides (see below).
9.2. Gene Pyramiding
The search for new sources of resistance has always been a major process in breeding programs in order to generate new varieties resistant to pests. Vertical resistances are mainly governed by major genes that have a gene-for-gene relationship with a pathogen avirulence factor, thus generating a hypersensitive response (HR) to a specific race and most often preventing the development of the pathogen. Although their use is fairly simple, the history of O. cumana parasitizing sunflower specifically is remarkable [197]. Indeed, this example has shown that the resistances thus successively generated are unfortunately not sustainable. The other approach concerns the use of horizontal resistances which are regulated by several genes and which have the advantage of being non-specific (all races of parasitic plant weed are then affected) and durable. However, their use in breeding programs is too complex to implement. An alternative would therefore be to pyramid the genes involved in vertical resistance in order to mimic a kind of horizontal resistance. In O. cumana, it would therefore be interesting to accumulate resistance genes already identified such as HaDef1 encoding a defensin [198,199] or HaOr7 encoding a receptor-like LRR kinase protein and conferring resistance to race F [137]. Similarly, it would be relevant to revisit resistance alleles already characterized (Or1 to Or6) but whose underlying resistance gene remain to be identified [142]. In this way, the generated gene-pyramid sunflower lines could benefit from a potential synergistic effect among those genes, leading to activation of additional biological process. This could lead to increased resistance and prolonged durability of its effectiveness because the parasitic weed would need to overcome a larger number of sunflower genetic pathways to infect. This technology has proven effective to increase resistance to other plant pathogens [200,201,202] but has not been developed against parasitic weeds.
9.3. Genetic Engineering
There have been several attempts to use plant genetic engineering for improved parasitic weed control by introducing resistance to commercial herbicides into crop plants. The types of selectivity mode in herbicide-resistant crops can be metabolic resistance or target site resistance. In theory, crops with metabolic resistance that catabolize the herbicide in inactive forms could only be used in cases when the herbicide reaches the parasite directly, such as in the strategy of crop seed dressing where the herbicide reaches the parasite through the soil or during application of herbicides on crop canopy infected by stem parasitic weeds. However, Cuscuta survives the application of glyphosate and glufosinate to glyphosate-resistant and glufosinate-resistant crops [32,169,203,204]. In the study by Nadler-Hassar et al. [169] the authors determined that in the case of glufosinate applications on the canopy of Cuscuta-infected glufosinate-resistant oilseed rape carrying the bar gene, which encodes an enzyme that acetylates glufosinate to an inactive form, the herbicide had a reduced effect on Cuscuta even when unattached Cuscuta plants are very sensitive to glufosinate. These authors suggested that although attached Cuscuta plants were also sprayed with glufosinate, the amount of herbicide directly absorbed by Cuscuta could not have been sufficient to kill the parasite, indicating that in this case the herbicide primarily gets to the parasite through the host and not by direct application. Similarly, Cuscuta survives glufosinate treatment attached to LibertyLink® crops [203]. LibertyLink® crops resist glufosinate applications by transgenically expressing phosphinothricin acetyl transferase (PAT) that detoxifies the herbicide [205]. The work by Jiang et al. [206] determined that dodder acquires glufosinate tolerance from the LibertyLink® host through trafficking and inter-specific function of PAT. Engineered crops with target-site resistance are more appropriate for parasitic weeds because the herbicide is applied on the crop canopy and delivered in active form to the root- or stem-attached parasite through translocation. Transgenic tobacco engineered with a mutant form of acetolactate synthase (AS) conferring resistance to chlorsulfuron could be used as an effective means for controlling Orobanche/Phelipanche [207]. Similarly, transgenic carrots engineered for imazapyr resistance is effective in controlling P. aegyptiaca [208]. Glyphosate resistance conferred expression of mutant 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) appears to be readily translocated into developing Phelipanche and Orobanche spp. tubercles, thereby blocking their development [209,210].
Apart from herbicide resistance, among the earliest reports of successful control of parasitic weeds through genetic engineering was engineering the resistance against Phelipanche spp. with overexpression of the antibacterial peptide sarcotoxin IA [196,211]. The use of RNA interference was shown as a potential strategy for controlling plant parasitism by [212]. These investigators showed that target gene expression in Triphysaria versicolor could be controlled by expressing hpRNA against the gene in the host plant since the hpRNA could be effectively translocated across the haustorium junctions. They also showed that reciprocal movement of silencing RNA could move from parasite to host. In contrast, similar attempts to use RNA interference to block S. asiatica parasitism of maize engineered with 13 dsRNA constructs against five key parasite metabolic enzymes showed no significant effects on Striga [213]. Expression of multiple RNAi gene constructs (e.g., ACS, M6PR, and Prx1) targeting genes in P. aegyptiaca resulted in impairment of parasite growth on the engineered hosts [214]. As for herbicide efficacy, the reason for the difference among parasites in response RNAi expression in host plants could be due to the nature of the vascular connections formed in the different parasite interactions. Among those examined, Striga-host interactions do not exhibit inter-phloem connectivity that could limit the capacity to exchange small RNA molecules easily. Similar to root parasites, cross-species RNAi can be used to inhibit infection by stem parasites. SHOOT MERISTEMLESS-like RNA interference transgenic tobacco results in defects in establishment of Cuscuta haustorium [78]. To date, few reports have appeared describing gene editing as a means of controlling parasitic weeds. Those that have appeared have targeted pre-attachment resistance. For example, Butt et al. [215] used CRISPR/Cas9 to disrupt the CCD7 gene in rice (Oryza sativa). The CCD7 knockout plants showed a significant reduction in the stimulation of Striga germination, but also exhibited an increase in tillering and reduced height. Bari et al. [216] have recently reported that CRISPR/Cas9-mediated mutagenesis of the CCD8 gene in tomato resulted in less root stimulation activity of parasitic germination due to loss of root strigolactone, the main parasitic weed germination stimulant exuded by crops, and a significant reduction in parasite infestation compared to wild type tomato plants. Unfortunately, there were negative agronomic effects as a result of the gene editing, including dwarfing, excessive shoot branching, and adventitious root formation.
10. Conclusions
As their significance as a constraint to crop productivity worldwide has expanded, there has been an increased interest in the biology of parasitic weeds. The most notable advancements that have occurred in recent years is our better understanding the underlying chemical, molecular, and genetic factors that control pre- and post-attachment interactions between the parasite and its potential host plant; in particular, the exchange of small molecules, protein effectors, and small regulatory RNAs between parasite and host that define compatibility or incompatibility. This has certainly enlightened our appreciation of the complexity of these unique plant-plant interactions and the subtleties of what constitute success or failure during the process or parasitism. To date many similarities have emerged between the molecular mechanisms mediating parasitic plant-host interactions and other pathogen-plant interactions, including the elicitation of host innate immunity. While the intricacies of the interaction between parasitic weeds and their host plants remains in the early stages of discovery, relative to our understanding of other plant-pathogen interactions (e.g., bacterial, fungal, nematode-elicited pathologies), the availability and application of omics-scale data and related functional genomic tools have accelerated research progress in recent years, and pointed to new directions of research. While we have a much greater appreciation of the role of strigolactones and various haustorial initiation factors in the control of pre-attachment interactions, our knowledge of the factors and signaling processes governing post-attachment interactions is still in its infancy. There are still many open questions about exactly how avirulence and effector molecules move from parasite to host, how metabolic or other forms of feedback from the host conditions parasite development, and how input from the parasite modifies host processes and metabolism to make a favorable pairing to satisfy parasite growth and developmental needs. There is an urgent need for better methods and tools for culturing parasites free and in combinations with their host and for manipulating these associations to better dissect the fine points of successful and unsuccessful interactions. Given our current knowledge, the study of parasitic weeds offers exciting opportunities for young researchers, and will likely provide the plant biology community with new insights into the evolution of these plants. More importantly, the hope is that with increased knowledge about host-parasite interactions there will be greater opportunity and success in developing strategies to limit their spread and alleviate their damage to crops worldwide, thus promoting better food security for future generations.
Author Contributions
Conceptualized the manuscript, M.F.-A.; equally contributed to the writing of the manuscript, M.F.-A., P.D. and M.P.T. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support is acknowledged to M.F.-A. from the Spanish Ministry of Science, Innovation and Universities (AGL2017-87693-R and RYC-2015-18961), to P.D. from the French National Research Agency (ANR-16-CE20-0004), and to M.P.T. from the NSF (IOS-1213059 and IOS-1238057) and Kirkhouse Trust SCIO.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kuijt J. The Biology of Parasitic Plants. University of California Press; Berkeley, CA, USA: 1969. [Google Scholar]
- 2.Parker C., Riches C.R. Parasitic Weeds of the World: Biology and Control. CAB International; Wallingford, UK: 1993. [Google Scholar]
- 3.Nickrent D.L., Malécot V., Vidal-Russell R., Der J.P. A revised classification of Santalales. Taxon. 2010;59:538–558. doi: 10.1002/tax.592019. [DOI] [Google Scholar]
- 4.Westwood J.H., Yoder J.I., Timko M.P., de Pamphilis C.W. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Heide-Jørgensen H.S. Introduction: The parasitic syndrome in Higher Plants. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 1–18. [Google Scholar]
- 6.Westwood J.H. The physiology of the established parasite-host association. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 87–114. [Google Scholar]
- 7.Riopel J.L., Timko M.P. Haustorial initiation and differentiation. In: Press M.C., Graves J.D., editors. Parasitic Plants. Chapman & Hall; London, UK: 1995. pp. 39–79. [Google Scholar]
- 8.Joel D.M. Functional structure of the mature haustorium. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 25–60. [Google Scholar]
- 9.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. 2009;65:453–459. doi: 10.1002/ps.1713. [DOI] [PubMed] [Google Scholar]
- 10.Scholes J.D., Press M.C. Striga infestation of cereal crops—An unsolved problem in resource limited agriculture. Curr. Opin. Plant Biol. 2008;11:180–186. doi: 10.1016/j.pbi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Delavault P. Are root parasitic plants like any other plant pathogens? New Phytol. 2020;226:641–643. doi: 10.1111/nph.16504. [DOI] [PubMed] [Google Scholar]
- 12.Foy C.L., Jain R., Jacobsohn R. Recent approaches for chemical control of broomrape (Orobanche spp.) In: Foy C.L., editor. Reviews of Weed Science. Volume 4. Weed Science Society of America; Champaign, IL, USA: 1989. pp. 123–152. [Google Scholar]
- 13.Clarke C.R., Timko M.P., Yoder J.I., Axtell M.J., Westwood J.H. Molecular Dialog Between Parasitic Plants and Their Hosts. Ann. Rev. Phytopathol. 2019;57:279–299. doi: 10.1146/annurev-phyto-082718-100043. [DOI] [PubMed] [Google Scholar]
- 14.Schneeweiss G.M. Correlated evolution of life history and host range in the nonphotosynthetic parasitic flowering plants Orobanche and Phelipanche (Orobanchaceae) J. Evol. Biol. 2007;20:471–478. doi: 10.1111/j.1420-9101.2006.01273.x. [DOI] [PubMed] [Google Scholar]
- 15.Estabrook E.M., Yoder J.I. Plant-plant communications: Rhizosphere signalling between parasitic angiosperms and their hosts. Plant Physiol. 1998;116:1–7. doi: 10.1104/pp.116.1.1. [DOI] [Google Scholar]
- 16.Bruce T.B.A., Gressel J. Changing host specificities: By mutational changes or epigenetic reprogramming? In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 231–242. [Google Scholar]
- 17.Parker C. The parasitic weeds of the Orobanchaceae. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 313–344. [Google Scholar]
- 18.Joel D.M. The seed and the seedling. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 143–146. [Google Scholar]
- 19.Xie X., Yoneyama K., Yoneyama K. The strigolactone story. Ann. Rev. Phytopathol. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 20.Lechat M.M., Pouvreau J.B., Péron T., Gauthier M., Montiel G., Veronesi C., Todoroki Y., Le Bizec B., Monteau F., Macherel D., et al. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J. Exp. Bot. 2012;63:5311–5322. doi: 10.1093/jxb/ers189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun G., Thoiron S., Braem L., Pouvreau J.B., Montiel G., Lechat M.M., Simier P., Gevaert K., Goormachtig S., Delavault P. CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant Cell Environ. 2019;42:2612–2626. doi: 10.1111/pce.13594. [DOI] [PubMed] [Google Scholar]
- 22.Musselman L.J. The biology of Striga, Orobanche and other root parasitic weeds. Ann. Rev. Phytopathol. 1980;18:463–489. doi: 10.1146/annurev.py.18.090180.002335. [DOI] [Google Scholar]
- 23.López-Granados F., García-Torres L. Effects of environmental factors on dormancy and germination of crenate broomrape (Orobanche crenata) Weed Sci. 1996;44:284–289. [Google Scholar]
- 24.Kebreab E., Murdoch A.J. A quantitative model for loss of primary dormancy and induction of secondary dormancy in imbibed seeds of Orobanche spp. J. Exp. Bot. 1999;50:211–219. doi: 10.1093/jxb/50.331.211. [DOI] [Google Scholar]
- 25.Bouwmeester H.J., Roux C., Lopez-Raez J.A., Bécard G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 2007;12:224–230. doi: 10.1016/j.tplants.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Aparicio M., Flores F., Rubiales D. Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann. Bot. 2009;103:423–431. doi: 10.1093/aob/mcn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Aparicio M., Yoneyama K., Rubiales D. The role of strigolactones in host specificity of Orobanche and Phelipanche seed germination. Seed Sci. Res. 2011;21:55–61. doi: 10.1017/S0960258510000371. [DOI] [Google Scholar]
- 28.Fernández-Aparicio M., Kisugi T., Xie X., Rubiales D., Yoneyama K. Low strigolactone root exudation: A novel mechanism of broomrape (Orobanche and Phelipanche spp.) resistance available for faba bean breeding. J. Agric. Food Chem. 2014;62:7063–7071. doi: 10.1021/jf5027235. [DOI] [PubMed] [Google Scholar]
- 29.Yoneyama K., Xie X., Kim H.I., Kisugi T., Nomura T., Sekimoto H., Yokota T., Yoneyama K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235:1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auger B., Pouvreau J.B., Pouponneau K., Yoneyama K., Montiel G., Le Bizec B., Yoneyama K., Delavault P., Delourme R., Simier P. Germination Stimulants of Phelipanche ramosa in the Rhizosphere of Brassica napus are Derived from the Glucosinolate Pathway. Mol. Plant Microbe Interact. 2012;7:993–1004. doi: 10.1094/MPMI-01-12-0006-R. [DOI] [PubMed] [Google Scholar]
- 31.Conn C.E., Bythell-Douglas R., Neumann D., Yoshida S., Whittington B., Westwood J.H., Shirasu K., Bond C.S., Dyer K.A., Nelson D.C. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349:540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 32.Dawson J.H., Musselman L.J., Wolswinkel P., Dörr I. Biology and control of Cuscuta. Rev. Weed Sci. 1994;6:265–317. [Google Scholar]
- 33.Kaiser B., Vogg G., Fürst U.B., Albert M. Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Front. Plant Sci. 2015;6:45. doi: 10.3389/fpls.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabiri S., van Ast A., Rodenburg J., Bastiaans L. Host influence on germination and reproduction of the facultative hemi-parasitic weed Rhamphicarpa fistulosa. Ann. Appl. Biol. 2016;169:144–154. doi: 10.1111/aab.12288. [DOI] [Google Scholar]
- 35.Saunders A.R. Studies in phanerogamic parasitism, with particular reference to Striga lutea Lour. S. Afr. Dept. Agric. Sci. Bull. 1933;128:56. [Google Scholar]
- 36.Williams C.N. Tropism and morphogenesis of Striga seedlings in the host rhizosphere. Ann. Bot. 1961;25:406–415. doi: 10.1093/oxfordjournals.aob.a083761. [DOI] [Google Scholar]
- 37.Riopel J.L., Baird W.V. Morphogenesis of the early development of primary haustoria in Striga asiatica. In: Musselman L.J., editor. Parasitic Weeds in Agriculture. CRC; Boca Raton, FL, USA: 1987. pp. 107–125. [Google Scholar]
- 38.Yoshida S., Shirasu K. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 2009;183:180–189. doi: 10.1111/j.1469-8137.2009.02840.x. [DOI] [PubMed] [Google Scholar]
- 39.Veronesi C., Bonnin E., Calvez S., Thalouarn P., Simier P. Activity of secreted cell wall-modifying enzymes and expression of peroxidase-encoding gene following germination of Orobanche ramosa. Biol. Plant. 2007;51:391–394. doi: 10.1007/s10535-007-0084-y. [DOI] [Google Scholar]
- 40.Zhou W.J., Yoneyama K., Takeuchi Y., Iso S., Rungmekarat S., Chae S.H., Sato D., Joel D.M. In vitro infection of host roots by differentiated calli of the parasitic plant Orobanche. J. Exp. Bot. 2004;55:899–907. doi: 10.1093/jxb/erh098. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Aparicio M., Rubiales D., Bandaranayake P.C.G., Yoder J.I., Westwood J.H. Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods. 2011;7:36. doi: 10.1186/1746-4811-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Q., Zhang Y., Matusova R., Charnikhova T., Jamil M., Fernandez-Aparicio M., Huang K., Westwood J.H., Timko M., Ruyter-Spira C., et al. Striga hermonthica MAX2 restores branching but not the very low fluence response in the Arabidopsis thaliana max2 mutant. New Phytol. 2014;202:531–541. doi: 10.1111/nph.12692. [DOI] [PubMed] [Google Scholar]
- 43.Billard E., Goyet V., Delavault P., Simier S., Montiel G. Cytokinin treated microcalli of Phelipanche ramosa: An efficient model for studying haustorium formation in holoparasitic plants. Plant Cell Tissue Organ Cult. 2020;141:543–553. doi: 10.1007/s11240-020-01813-6. [DOI] [Google Scholar]
- 44.Heide-Jørgensen H.S. Parasitic Flowering Plants. Brill Leiden; Leiden, The Netherlands: 2008. [Google Scholar]
- 45.Runyon J.B., Mescher M.C., De Moraes C.M. Volatile chemical cues guide host location and host selection by parasitic plants. Science. 2006;313:1964–1967. doi: 10.1126/science.1131371. [DOI] [PubMed] [Google Scholar]
- 46.Holmes M.G., Smith H. The function of phytochrome in plants growing in the natural environment. Nature. 1975;254:512. doi: 10.1038/254512a0. [DOI] [Google Scholar]
- 47.Orr G.L., Haidar M.A., Orr D.A. Small seed dodder (Cuscuta planiflora) phototropism toward far-red when in white light. Weed Sci. 1996;44:233–240. doi: 10.1017/S0043174500093838. [DOI] [Google Scholar]
- 48.Bandaranayake P.C.G., Yoder J.I. Haustorium initiation and early development. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 61–74. [Google Scholar]
- 49.Fernández-Aparicio M., Masi M., Maddau L., Cimmino A., Evidente M., Rubiales D., Evidente A. Induction of haustorium development by sphaeropsidones in radicles of the parasitic weeds Striga and Orobanche. A structure-activity relationship study. J. Agric. Food Chem. 2016;64:5188–5196. doi: 10.1021/acs.jafc.6b01910. [DOI] [PubMed] [Google Scholar]
- 50.Hegenauer V., Körner M., Albert M. Plants under stress by parasitic plants. Curr. Opin. Plant Biol. 2017;38:34–41. doi: 10.1016/j.pbi.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Goyet V., Wada S., Cui S., Wakatake T., Shirasu K., Montiel G., Simier P., Yoshida S. Haustorium Inducing Factors for Parasitic Orobanchaceae. Front. Plant Sci. 2019;10:1056. doi: 10.3389/fpls.2019.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishida J.K., Wakatake T., Yoshida S., Takebayashi Y., Kasahara H., Wafula E., de Pamphilis C.W., Namba S., Shirasu K. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell. 2016;28:1795–1814. doi: 10.1105/tpc.16.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joel D.M., Losner-Goshen D. The attachment organ of the parasitic angiosperms Orobanche cumana and O. aegyptiaca and its development. Can. J. Bot. 1994;72:564–574. doi: 10.1139/b94-075. [DOI] [Google Scholar]
- 54.Hood M.E., Condom J.M., Timko M.P., Riopel J.L. Primary haustorial development of Striga asiatica on host and nonhost species. Phytopathology. 1998;88:70–75. doi: 10.1094/PHYTO.1998.88.1.70. [DOI] [PubMed] [Google Scholar]
- 55.Reiss G.C., Bailey J.A. Striga gesnerioides parasitizing cowpea: Development of infection structures and mechanisms of penetration. Ann. Bot. 1998;81:431–440. doi: 10.1006/anbo.1997.0577. [DOI] [Google Scholar]
- 56.Baird V.W., Riopel J. Experimental studies of the attachment of the parasitic angiosperm Agalinis purpurea to a host. Protoplasma. 1983;118:206–218. doi: 10.1007/BF01281804. [DOI] [Google Scholar]
- 57.Heide-Jørgensen H.S., Kuijt J. The haustorium of the root parasite Triphysaria (Scrophulariaceae), with special reference to xylem bridge ultrastructure. Am. J. Bot. 1995;82:782–797. doi: 10.1002/j.1537-2197.1995.tb15691.x. [DOI] [Google Scholar]
- 58.Musselman L.J., Dickinson W.C. The structure and development of the haustorium in parasitic Scrophulariaceae. Bot. J. Linn. Soc. 1975;70:183–212. doi: 10.1111/j.1095-8339.1975.tb01645.x. [DOI] [Google Scholar]
- 59.Dörr I. Feinstruktur intrazellular wachsender Cuscuta-Hyphen. Protoplasma. 1969;67:123–137. doi: 10.1007/BF01248735. [DOI] [Google Scholar]
- 60.Haidar M.A., Orr G.L., Westra P. Effects of light and mechanical stimulation on coiling and pre-haustoria formation in Cuscuta spp. Weed Res. 1997;37:219–228. doi: 10.1046/j.1365-3180.1997.d01-36.x. [DOI] [Google Scholar]
- 61.Vaughn K.C. Attachment of the parasitic weed dodder to the host. Protoplasma. 2002;219:227–237. doi: 10.1007/s007090200024. [DOI] [PubMed] [Google Scholar]
- 62.Honaas L., Wafula E., Yang Z., Der J., Wickett N., Altman N.S., Taylor C.G., Yoder J.I., Timko M.P., Westwood J.H., et al. Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 2013;13:9. doi: 10.1186/1471-2229-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranjan A., Ichihashi Y., Farhi M., Zumstein K., Townsley B., David-Schwartz R., Sinha N.R. De novo assembly and characterization of the transcriptome of the parasitic weed Cuscuta pentagona identifies genes associated with plant parasitism. Plant Physiol. 2014;166:1186–1199. doi: 10.1104/pp.113.234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ichihashi Y., Mutuku J.M., Yoshida S., Shirasu K. Transcriptomics exposes the uniqueness of parasitic plants. Brief Funct. Genom. 2015;14:275–282. doi: 10.1093/bfgp/elv001. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z., Wafula E.K., Honaas L.A., Zhang H., Das M., Fernández-Aparicio M., Huang K., Bandaranayake P.C.G., Wu B., Der J.P., et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 2014;32:767–790. doi: 10.1093/molbev/msu343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber H.C. Anatomische Studien an den Haustorien einiger parasitischer Scrophulariaceen Mitteleuropas. Ber. Dtsch. Bot. Ges. 1976;89:57–84. [Google Scholar]
- 67.Alexander T., Weber H.C. Zur parasitischen Lebensweise von Parentucellia latifolia (L.) Caruel (Scrophulariaceae) Beitr. Biol. Pflanz. 1985;60:23–34. [Google Scholar]
- 68.Riopel J., Musselman L. Experimental initiation of haustoria in Agalinis purpurea. Am. J. Bot. 1979;66:570–575. doi: 10.1002/j.1537-2197.1979.tb06259.x. [DOI] [Google Scholar]
- 69.Krause D. Ph.D. Dissertation. Philipps-University; Marburg, Germany: 1990. Vergleichende Morphologisch/Anatomische Untersuchungen an Striga-Arten (Scrophulariaceae) [Google Scholar]
- 70.Kim D., Kocz R., Boone L., Keyes W.J., Lynn D.G. On becoming a parasite: Evaluating the role of wall oxidases in parasitic plant development. Chem. Biol. 1998;5:103–117. doi: 10.1016/S1074-5521(98)90144-2. [DOI] [PubMed] [Google Scholar]
- 71.Losner-Goshen D., Portnoy V.H., Mayer A.M., Joel D.M. Pectolytic activity by the haustorium of the parasitic plant Orobanche L. (Orobanchaceae) in host roots. Ann. Bot. 1998;81:319–326. doi: 10.1006/anbo.1997.0563. [DOI] [Google Scholar]
- 72.Dörr I. How Striga parasitizes its host: A TEM and SEM study. Ann. Bot. 1997;79:463–472. doi: 10.1006/anbo.1996.0385. [DOI] [Google Scholar]
- 73.Bar-Nun N., Sachs T., Mayer A.M. A role for IAA in the infection of Arabidopsis thaliana by Orobanche aegyptiaca. Ann. Bot. 2008;101:261–265. doi: 10.1093/aob/mcm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ekawa M., Aoki K. Phloem-Conducting Cells in Haustoria of the Root-Parasitic Plant Phelipanche aegyptiaca Retain Nuclei and Are Not Mature Sieve Elements. Plants. 2017;6:60. doi: 10.3390/plants6040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dörr I., Kollmann R. Symplasmic sieve element continuity between Orobanche and its host. Bot. Acta. 1995;108:47–55. doi: 10.1111/j.1438-8677.1995.tb00830.x. [DOI] [Google Scholar]
- 76.Lee K.B. Structure and development of the upper haustorium in the parasitic flowering plant Cuscuta japonica (Convolvulaceae) Am. J. Bot. 2007;94:737–745. doi: 10.3732/ajb.94.5.737. [DOI] [PubMed] [Google Scholar]
- 77.Ahkami A.H., Lischewski S., Haensch K.T., Porfirova S., Hofmann J., Rolletschek H., Melzer M., Franken P., Hause B., Druege U., et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009;181:613–625. doi: 10.1111/j.1469-8137.2008.02704.x. [DOI] [PubMed] [Google Scholar]
- 78.Alakonya A., Kumar R., Koenig D., Kimura S., Townsley B., Runo S., Garces H.M., Kang J., Yanez A., David-Schwartz R., et al. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell. 2012;24:3153–3166. doi: 10.1105/tpc.112.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Press M.C., Smith S., Stewart G.R. Carbon acquisition and assimilation in parasitic plants. Funct. Ecol. 1991;5:278–283. doi: 10.2307/2389265. [DOI] [Google Scholar]
- 80.Tennakoon K.U., Pate J.S., Fineran B.A. Growth and partitioning of carbon and fixed nitrogen in the shrub legume Acacia littorea in the presence or absence of the root hemiparasite Olax phyllanthi. J. Exp. Bot. 1997;48:1047–1060. doi: 10.1093/jxb/48.5.1047. [DOI] [Google Scholar]
- 81.Seel W.E., Cechin I., Vincent C.A., Press M.C. Carbon partitioning in parasitic angiosperms and their hosts. In: Pollock C.J., Farrar J.F., Gordon A.J., editors. Carbon Partitioning Within and Between Organisms. BIOS Scientific Publishers Ltd.; Oxford, UK: 1992. [Google Scholar]
- 82.Jeschke W.D., Räth N., Bäumel P., Czygan F.C., Proksch P. Modelling the flow and partitioning of carbon and nitrogen in the holoparasite Cuscuta reflexa Roxb. & its host Lupinus albus L. I. Methods for estimating net flows. J. Exp. Bot. 1994;45:791–800. [Google Scholar]
- 83.Hibberd J.M., Quick W.P., Press M.C., Scholes J.D., Jeschke W.D. Solute fluxes from tobacco to the parasitic angiosperm Orobanche cernua and the influence of infection on host carbon and nitrogen relations. Plant Cell Environ. 1999;22:937–947. doi: 10.1046/j.1365-3040.1999.00462.x. [DOI] [Google Scholar]
- 84.Abbes Z., Kharrat M., Delavault P., Chaïbi W., Simier P. Nitrogen and carbon relationships between the parasitic weed Orobanche foetida and susceptible and tolerant faba bean lines. Plant Physiol. Biochem. 2009;47:153–159. doi: 10.1016/j.plaphy.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 85.Fu Q., Cheng L., Guo Y., Turgeon R. Phloem loading strategies and water relations in trees and herbaceous plants. Plant Physiol. 2011;157:1518–1527. doi: 10.1104/pp.111.184820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007;581:2309–2317. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 87.Dörr I. New results on interspecific bridges between parasites and their hosts. In: Moreno M.T., Cubero J.I., Berner D., Joel D., Musselman L.J., editors. Advances in Parasitic Plant Research. Junta de Andalucia; Cordoba, Spain: 1996. pp. 195–201. [Google Scholar]
- 88.Draie R., Péron T., Pouvreau J.-B., Véronési C., Jégou S., Delavault P., Thoiron S., Simier P. Invertases involved in the development of the parasitic plant Phelipanche ramosa: Characterization of the dominant soluble acid isoform, PrSAI1. Mol. Plant Pathol. 2011;12:638–652. doi: 10.1111/j.1364-3703.2010.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harloff H.J., Wegmann D. Evidence for a mannitol cycle in Orobanche ramosa and Orobanche crenata. J. Plant Physiol. 1993;141:513–520. doi: 10.1016/S0176-1617(11)80449-9. [DOI] [Google Scholar]
- 90.Delavault P., Simier P., Thoiron S., Véronési C., Fer A., Thalouarn P. Isolation of mannose 6- phosphate reductase cDNA, changes in enzyme activity and mannitol content in broomrape (Orobanche ramosa) parasitic on tomato roots. Physiol. Plant. 2002;115:48–55. doi: 10.1034/j.1399-3054.2002.1150105.x. [DOI] [PubMed] [Google Scholar]
- 91.Péron T., Candat A., Montiel G., Veronesi C., Macherel D., Delavault P., Simier P. New insights into phloem unloading and expression of sucrose transporters in vegetative sinks of the parasitic plant Phelipanche ramosa L. (Pomel) Front. Plant Sci. 2016;7:2048. doi: 10.3389/fpls.2016.02048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith S., Stewart G.R. Effect of potassium levels on the stomatal behavior of the hemiparasite Striga hermonthica. Plant Physiol. 1990;94:1472–1476. doi: 10.1104/pp.94.3.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lechowski Z. Stomatal response to exogenous cytokinin treatment of the hemiparasite Melampyrum arvense L. before and after attachment to the host. Biol. Plant. 1997;39:13–21. doi: 10.1023/A:1000392502943. [DOI] [Google Scholar]
- 94.Jiang F., Jeschke W.D., Hartung W. Abscisic acid (ABA) flows from Hordeum vulgare to the hemiparasite Rhinanthus minor and the influence of infection on host and parasite abscisic acid relations. J. Exp. Bot. 2004;55:2323–2329. doi: 10.1093/jxb/erh240. [DOI] [PubMed] [Google Scholar]
- 95.Lins R.D., Colquhoun J.B., Mallory-Smith C.A. Effect of small broomrape (Orobanche minor) on red clover growth and dry matter partitioning. Weed Sci. 2007;55:517–520. doi: 10.1614/WS-07-049.1. [DOI] [Google Scholar]
- 96.Fernández-Aparicio M., Flores F., Rubiales D. The effect of Orobanche crenata infection severity in faba bean, field pea and grass pea productivity. Front. Plant Sci. 2016;7:1049. doi: 10.3389/fpls.2016.01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernández-Aparicio M., Flores F., Rubiales D. Field response of Lathyrus cicera germplasm to crenate broomrape (Orobanche crenata) Field Crop. Res. 2009;113:321–327. doi: 10.1016/j.fcr.2009.06.009. [DOI] [Google Scholar]
- 98.Jeschke W.D., Hilpert A. Sink-stimulated photosynthesis and sink dependent increase in nitrate uptake: Nitrogen and carbon relations of the parasitic association Cuscuta reflexa–Ricinus communis. Plant Cell Environ. 1997;20:47–56. doi: 10.1046/j.1365-3040.1997.d01-2.x. [DOI] [Google Scholar]
- 99.Hibberd J.M., Quick W.P., Press M.C., Scholes J.D. The influence of the parasitic angiosperm Striga gesnerioides on the growth and photosynthesis of its host, Vigna unguiculata. J. Exp. Bot. 1996;47:507–512. doi: 10.1093/jxb/47.4.507. [DOI] [Google Scholar]
- 100.Hibberd J.M., Quick W.P., Press M.C., Scholes J.D. Can source–sink relations explain responses of tobacco to infection by the root holoparasitic angiosperm Orobanche cernua? Plant Cell Environ. 1998;21:333–340. doi: 10.1046/j.1365-3040.1998.00272.x. [DOI] [Google Scholar]
- 101.Press M.C., Scholes J.D., Watling J.R. Parasitic plants: Physiological and ecological interactions with their hosts. In: Press M.C., Scholes J.D., Barker M.G., editors. Physiological Plant Ecology: The 39th Symposium of the British Ecological Society. University of York; New York, NY, USA: 1999. pp. 175–197. [Google Scholar]
- 102.Press M.C. How do the parasitic weeds Striga and Orobanche influence host carbon relations? Asp. Appl. Biol. 1995;42:63–70. [Google Scholar]
- 103.Frost D.L., Gurney A.L., Press M.C., Scholes J.D. Striga hermonthica reduces photosynthesis in sorghum: The importance of stomatal limitations and a potential role for ABA? Plant Cell Environ. 1997;20:483–492. doi: 10.1046/j.1365-3040.1997.d01-87.x. [DOI] [Google Scholar]
- 104.Allen M.F., Smith W.K., Moore T.S., Christensen M. Comparative water relations and photosynthesis of mychorrhizal Bouteloua gracilis H.B.K. Lab Steud. New Phytol. 1981;88:683–693. doi: 10.1111/j.1469-8137.1981.tb01745.x. [DOI] [Google Scholar]
- 105.Manschadi A.M., Kroschel J., Sauerborn J. Dry matter production and partitioning in the host-parasite association Vicia faba–Orobanche crenata. J. Appl. Bot. 1996;70:224–229. [Google Scholar]
- 106.Barker E.R., Press M.C., Scholes J.D., Quick W.P. Interactions between the parasitic angiosperm Orobanche aegyptiaca and its tomato host: Growth and biomass allocation. New Phytol. 1996;133:637–642. doi: 10.1111/j.1469-8137.1996.tb01932.x. [DOI] [Google Scholar]
- 107.Fernández-Aparicio M., Rispail N., Prats E., Morandi D., García-Garrido J.M., Dumas-Gaudot E., Duc G., Rubiales D. Parasitic plant infection is partially controlled through symbiotic pathways. Weed Res. 2009;50:76–82. doi: 10.1111/j.1365-3180.2009.00749.x. [DOI] [Google Scholar]
- 108.Fernández-Aparicio M., Pérez-de-Luque A., Prats E., Rubiales D. Variability of interactions between barrel medic (Medicago truncatula) genotypes and Orobanche species. Ann. Appl. Biol. 2008;153:117–126. doi: 10.1111/j.1744-7348.2008.00241.x. [DOI] [Google Scholar]
- 109.Joel D.M., Portnoy V.H. The angiospermous root parasite Orobanche L. (Orobanchaceae) induces expression of a pathogenesis related (PR) gene in susceptible tobacco roots. Ann. Bot. 1998;81:779–781. doi: 10.1006/anbo.1998.0629. [DOI] [Google Scholar]
- 110.Westwood J.H., Yu X., Foy C.L., Cramer C.L. Expression of a defense-related 3-hydroxy-3-methylglutaryl CoA reductase gene in response to parasitism by Orobanche spp. Mol. Plant Microbe Interact. 1998;11:530–536. doi: 10.1094/MPMI.1998.11.6.530. [DOI] [PubMed] [Google Scholar]
- 111.Vieira Dos Santos C., Letousey P., Delavault P., Thalouarn P. Defence gene expression analysis of Arabidopsis thaliana parasitized by Orobanche ramosa. Phytopathology. 2003;93:451–457. doi: 10.1094/PHYTO.2003.93.4.451. [DOI] [PubMed] [Google Scholar]
- 112.Yoder J.I., Scholes J.D. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr. Opin. Plant Biol. 2010;13:478–484. doi: 10.1016/j.pbi.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 113.Timko M., Scholes J. Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer; New York, NY, USA: 2013. Host reaction to attack by root parasitic plants; pp. 115–141. [Google Scholar]
- 114.Crescenzi A., Fanigliulo A., Fontana A., Fascetti S. First Report of Orobanche nana on Celery in Italy. Plant Dis. 2015;99:1188. doi: 10.1094/PDIS-11-14-1217-PDN. [DOI] [Google Scholar]
- 115.Musselman L.J., Bolin J.F. New Infestation of Branched Broomrape, Orobanche ramosa (Orobanchaceae), on Black Medic, (Medicago lupulina) (Fabaceae), in Virginia. Plant Dis. 2008;92:315. doi: 10.1094/PDIS-92-2-0315B. [DOI] [PubMed] [Google Scholar]
- 116.Rubiales D., Fernández-Aparicio M., Rodríguez M.J. First Report of Crenate Broomrape (Orobanche crenata) on Lentil (Lens culinaris) and Common Vetch (Vicia sativa) in Salamanca Province, Spain. Plant Dis. 2008;92:1368. doi: 10.1094/PDIS-92-9-1368B. [DOI] [PubMed] [Google Scholar]
- 117.Fernández-Aparicio M., Emeran A.A., Moral A., Rubiales D. First Report of Crenate Broomrape (Orobanche crenata) on White Lupine (Lupinus albus) Growing in Alkaline Soils in Spain and Egypt. Plant Dis. 2009;93:970. doi: 10.1094/PDIS-93-9-0970C. [DOI] [PubMed] [Google Scholar]
- 118.Tsialtas J.T., Eleftherohorinos I.G. First Report of Branched Broomrape (Orobanche ramosa) on Oilseed Rape (Brassica napus), Wild Mustard (Sinapis arvensis), and Wild Vetch (Vicia spp.) in Northern Greece. Plant Dis. 2011;95:1322. doi: 10.1094/PDIS-06-11-0462. [DOI] [PubMed] [Google Scholar]
- 119.Teimoury M., Karimmojeni H., Ehtemam M.H., Mehri H.R. First Report of Orobanche aegyptiaca Parasitism on Sesame in Iran. Plant Dis. 2012;96:1232. doi: 10.1094/PDIS-01-12-0068-PDN. [DOI] [PubMed] [Google Scholar]
- 120.Gibot-Leclerc S., Reibel C., Legros S. First Report of Branched Broomrape (Phelipanche ramosa) on Celeriac (Apium graveolens) in Eastern France. Plant Dis. 2014;98:1286. doi: 10.1094/PDIS-02-14-0148-PDN. [DOI] [PubMed] [Google Scholar]
- 121.Piwowarczyk R. A revision of distribution and historical analysis of preferred hosts of Orobanche ramose (Orobanchaceae) in Poland. Acta Agrobot. 2012;65:53–62. doi: 10.5586/aa.2012.043. [DOI] [Google Scholar]
- 122.Le Corre V., Reibel C., Gibot-Leclerc S. Development of Microsatellite Markers in the Branched Broomrape Phelipanche ramosa L. (Pomel) and Evidence for Host-Associated Genetic Divergence. Int. J. Mol. Sci. 2014;15:994–1002. doi: 10.3390/ijms15010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stojanova B., Delourme R., Duffé P., Delavault P., Simier P. Genetic differentiation and host preference reveal non-exclusive host races in the generalist parasitic weed Phelipanche ramosa. Weed Res. 2019;59:107–118. doi: 10.1111/wre.12353. [DOI] [Google Scholar]
- 124.Gibot-Leclerc S., Sallé G., Reboud X., Moreau D. What are the traits of Phelipanche ramosa (L.) Pomel that contribute to the success of its biological cycle on its host Brassica napus L? Flora–Morphol. Distrib. Funct. Ecol. Plants. 2012;207:512–521. doi: 10.1016/j.flora.2012.06.011. [DOI] [Google Scholar]
- 125.Gibot-Leclerc S., Dessaint F., Reibel C., Le Corre V. Phelipanche ramosa (L.) Pomel populations differ in life-history and infection response to hosts Flora-Morphol. Distrib. Funct. Ecol. Plants. 2013;208:247–252. doi: 10.1016/j.flora.2013.03.007. [DOI] [Google Scholar]
- 126.Botanga C.J., Timko M.P. Genetic structure and analysis of host and nonhost interactions of Striga gesnerioides (witchweed) from Central Florida. Phytopathology. 2005;95:1166–1173. doi: 10.1094/PHYTO-95-1166. [DOI] [PubMed] [Google Scholar]
- 127.Dube M.-P., Belzile F.J. Low genetic variability of Striga gesnerioides populations parasitic on cowpea might be explained by a recent origin. Weed Res. 2010;50:493–502. doi: 10.1111/j.1365-3180.2010.00804.x. [DOI] [Google Scholar]
- 128.Ramaiah K.V. Parasitic Weeds in Agriculture. Volume 1. Crc-Press; Boca Raton, FL, USA: 1987. Breeding cereal grains for resistance to witchweed; pp. 227–242. [Google Scholar]
- 129.Gebisa E. The Striga scourge in Africa: A growing pandemic. World Sci. 2007:3–15. doi: 10.1142/9789812771506_0001. [DOI] [Google Scholar]
- 130.Spallek T., Mutuku M., Shirasu K. The genus Striga: A witch profile. Mol. Plant Pathol. 2013;14:861–869. doi: 10.1111/mpp.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Timko M., Gowda B.S., Ouédraogo J., Ousmane B. Integrating New Technologies for Striga Control. World Scientific Publishing Co Pte Ltd.; Singapore: 2007. Molecular markers for analysis of resistance to Striga gesnerioides in cowpea; pp. 115–128. [Google Scholar]
- 132.Ohlson E.W., Timko M.P. Race structure of cowpea witchweed (Striga gesnerioides) in West Africa and its implications for Striga resistance breeding of cowpea. Weed Sci. 2020;68:125–133. doi: 10.1017/wsc.2020.3. [DOI] [Google Scholar]
- 133.Li J., Timko M.P. Gene-for-gene resistance in Striga-cowpea associations. Science. 2009;325:1094. doi: 10.1126/science.1174754. [DOI] [PubMed] [Google Scholar]
- 134.Oren M., Hudgell M.A.B., Golconda P., Lun C.M., Smith L.C. Evolution of the Immune System: Conservation and Diversification. Elsevier; Amsterdam, The Netherlands: 2016. pp. 295–310. [Google Scholar]
- 135.Jubic L.M., Saile S., Furzer O.J., El Kasmi F., Dangl J.L. Help wanted: Helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 2019;50:82–94. doi: 10.1016/j.pbi.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 136.Hegenauer V., Fürst U., Kaiser B., Smoker M., Zipfel C., Felix G., Stahl M., Albert M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science. 2016;353:478–481. doi: 10.1126/science.aaf3919. [DOI] [PubMed] [Google Scholar]
- 137.Duriez P., Vautrin S., Auriac M.C., Bazerque J., Boniface M.C., Callot C., Carrere S., Cauet S., Chabaud M., Gentou F., et al. A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat. Plants. 2019;5:1211–1215. doi: 10.1038/s41477-019-0556-z. [DOI] [PubMed] [Google Scholar]
- 138.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 139.Macho A.P., Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol. Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 140.Jhu M.-Y., Farhi M., Wang L., Philbrook R.N., Belcher M.S., Nakayama H., Zumstein K.S., Rowland S.D., Ron M., Shih P.M., et al. Lignin-based resistance to Cuscuta campestris in tomato. bioRxiv. 2019:706861. doi: 10.1101/706861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Molinero-Ruiz L., Delavault P., Pérez-Vich B., Pacureanu-Joita M., Bulos M., Altieri E., Domínguez J. History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed: A review. Span. J. Agric. Res. 2015;13:e10R01. doi: 10.5424/sjar/2015134-8080. [DOI] [Google Scholar]
- 142.Tang S.X., Heesacker A., Kishore V.K., Fernandez A., Sadik E.S., Cole G., Knapp S.J. Genetic mapping of the Or(5) gene for resistance to Orobanche Race E in sunflower. Crop Sci. 2003;43:1021–1028. doi: 10.2135/cropsci2003.1021. [DOI] [Google Scholar]
- 143.Rodriguez-Ojeda M.I., Pineda-Martos R., Alonso L.C., Fernandez-Escobar J., Fernandez-Martinez J.M., Perez-Vich B., Velasco L. A dominant avirulence gene in Orobanche cumana triggers Or5 resistance in sunflower. Weed Res. 2013;53:322–327. doi: 10.1111/wre.12034. [DOI] [Google Scholar]
- 144.Gauthier M., Véronési C., El-Halmouch Y., Leflon M., Jestin C., Labalette F., Simiera P., Delourmee R., Delavaulta P. Characterisation of resistance to branched broomrape, Phelipanche ramosa, in winter oilseed rape. Crop Prot. 2012;42:56–63. doi: 10.1016/j.cropro.2012.07.002. [DOI] [Google Scholar]
- 145.Hogenhout S.A., Van der Hoorn R.A.L., Terauchi R., Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009;22:115–122. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- 146.Kvitko B.H., Park D.H., Velásquez A.C., Wei C.-F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. Deletions in the Repertoire of Pseudomonas syringae pv. tomato DC3000 Type III Secretion Effector Genes Reveal Functional Overlap among Effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Eitas T.K., Dangl J.L. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Su C., Liu H., Wafula E.K., Honaas L., de Pamphilis C.W., Timko M.P. SHR4z, a novel decoy effector from the haustorium of the parasitic weed Striga gesnerioides, suppresses host plant immunity. New Phytol. 2020;226:891–908. doi: 10.1111/nph.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Clarke C.R., So-Yon P., Tuosto R., Jia X., Yoder A., Van Mullekom J., Westwood J. Multiple immunity-related genes control susceptibility of Arabidopsis thaliana to the parasitic weed Phelipanche Aegypt. PeerJ —J. Life Environ. Sci. PeerJ. 2020;8:e9268. doi: 10.7717/peerj.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mur L.A., Kenton P., Atzorn R., Miersch O., Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kusumoto D., Goldwasser Y., Xie X., Yoneyama K., Takeuchi Y., Yoneyama K. Resistance of red clover (Trifolium pratense) to the root parasitic plant Orobanche minor is activated by salicylate but not by jasmonate. Ann. Bot. 2007;100:537–544. doi: 10.1093/aob/mcm148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Véronési C., Delavault P., Simier P. Acibenzolar-S-methyl induces resistance in oilseed rape (Brassica napus L.) against branched broomrape (Orobanche ramosa L.) Crop Prot. 2009;28:104–108. [Google Scholar]
- 153.Hiraoka Y., Ueda H., Sugimoto Y. Molecular responses of Lotus japonicus to parasitism by the compatible species Orobanche aegyptiaca and the incompatible species Striga hermonthica. J. Exp. Bot. 2009;60:641–650. doi: 10.1093/jxb/ern316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Runyon J.B., Mescher M.C., Felton G.W., De Moraes C.M. Parasitism by Cuscuta pentagona sequentially induces JA and SA defence pathways in tomato. Plant Cell Environ. 2010;33:290–303. doi: 10.1111/j.1365-3040.2009.02082.x. [DOI] [PubMed] [Google Scholar]
- 155.Smith M.C., Webb M. Estimation of the seed bank of Striga spp. (Scrophulariaceae) in Malian fields and the implications for a model of biocontrol of S. hermonthica. Weed Res. 1996;36:85–92. doi: 10.1111/j.1365-3180.1996.tb01804.x. [DOI] [Google Scholar]
- 156.Kebreab E., Murdoch A.J. Simulation of integrated control strategies for Orobanche spp. based in a life cycle model. Exp. Agric. 2001;37:37–51. doi: 10.1017/S001447970100401X. [DOI] [Google Scholar]
- 157.Gressel J. Biotechnologies for directly generating crop resistant to parasites. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 433–458. [Google Scholar]
- 158.Fernández-Aparicio M., Westwood J.H., Rubiales D. Agronomic, breeding and biotechnological approaches for parasitic plant management by manipulating strigolactone levels in agricultural soils. Botany. 2011;89:813–826. doi: 10.1139/b11-075. [DOI] [Google Scholar]
- 159.Perez-De-Luque A., Moreno M.T., Rubiales D. Host plant resistance against broomrapes (Orobanche spp.): Defence reactions and mechanisms of resistance. Ann. Appl. Biol. 2008;152:131–141. doi: 10.1111/j.1744-7348.2007.00212.x. [DOI] [Google Scholar]
- 160.Pérez-de-Luque A., Fondevilla S., Pérez-Vich B., Aly R., Thoiron S., Simier P., Castillejo M.A., Fernández J.M., Jorrín J., Rubiales D., et al. Understanding Orobanche and Phelipanche-host plant interactions and developing resistance. Weed Res. 2009;49:8–22. doi: 10.1111/j.1365-3180.2009.00738.x. [DOI] [Google Scholar]
- 161.Li J.X., Lis K.E., Timko M.P. Molecular genetics of race-specific resistance of cowpea to Striga gesnerioides (Wilid.) Pest Manag. Sci. 2009;65:520–527. doi: 10.1002/ps.1722. [DOI] [PubMed] [Google Scholar]
- 162.Wickett N.J., Honaas L.A., Wafula E.K., Das M., Huang K., Wu B., Landherr L., Timko M.P., Yoder J., Westwood J.H., et al. Transcriptomes of the Parasitic Plant Family Orobanchaceae Reveal Surprising Conservation of Chlorophyll Synthesis. Curr. Biol. 2011;21:2098–2104. doi: 10.1016/j.cub.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 163.Westwood J.H., de Pamphilis C.W., Das M., Fernández-Aparicio M., Honaas L.A., Timko M.P., Wickett N.J., Yoder J.I. The Parasitic Plant Genome Project: New Tools for Understanding the Biology of Orobanche and Striga. Weed Sci. 2012;60:295–306. doi: 10.1614/WS-D-11-00113.1. [DOI] [Google Scholar]
- 164.Joel D.M. The long-term approach to parasitic weeds control: Manipulation of specific developmental mechanisms of the parasite. Crop Prot. 2000;19:753–758. doi: 10.1016/S0261-2194(00)00100-9. [DOI] [Google Scholar]
- 165.Eizenberg H., Colquhoun J.B., Mallory-Smith C.A. A predictive degree-days model for small broomrape (Orobanche minor) parasitism in red clover in Oregon. Weed Sci. 2005;53:37–40. doi: 10.1614/WS-04-018R1. [DOI] [Google Scholar]
- 166.Eizenberg H., Colquhoun J.B., Mallory-Smith C.A. Imazamox application timing for small broomrape (Orobanche minor) control in red clover. Weed Sci. 2006;54:923–927. doi: 10.1614/WS-05-151R.1. [DOI] [Google Scholar]
- 167.Kanampiu F.K., Ransom J.K., Friesen D., Gressel J. Imazapyr and pyrithiobac movement in soil and from maize seed coats controls Striga while allowing legume intercropping. Crop Prot. 2002;21:611–619. doi: 10.1016/S0261-2194(01)00151-X. [DOI] [Google Scholar]
- 168.Colquhoun J.B., Eizenberg H., Mallory-Smith C.A. Herbicide placement site affects small broomrape (Orobanche minor) control in red clover (Trifolium pratense) Weed Technol. 2006;20:356–360. doi: 10.1614/WT-04-327R2.1. [DOI] [Google Scholar]
- 169.Nadler-Hassar T., Shaner D.L., Nissen S., Westra P., Rubin B. Are herbicide-resistant crops the answer to controlling Cuscuta? Pest Manag. Sci. 2009;65:811–816. doi: 10.1002/ps.1760. [DOI] [PubMed] [Google Scholar]
- 170.Schloss J.V. Acetolactate synthase, mechanism of action and its herbicide binding site. Pestic. Sci. 1990;29:283–292. doi: 10.1002/ps.2780290305. [DOI] [Google Scholar]
- 171.Awad A.E., Worsham A.D., Corbin F.T., Eplee R.E. Absorption, translocation and metabolism of foliarly applied 14-C dicamba in sorghum (Sorghum bicolor) and corn (Zea mays) parasitized with witchweed (Striga asiatica) In: Ransom J.K., Musselman L.J., Worsham A.D., Parker C., editors. Proceedings of the 5th International Symposium of Parasitic Weeds, Nairobi, Kenya, 24–30 June 1991. CIMMYT; Mexico City, Mexico: 1991. pp. 535–536. [Google Scholar]
- 172.Odhiambo G.D., Ransom J.K. Effect of Dicamba on the control of Striga hermonthica in maize in western Kenya. Afr. Crop Sci. 1993;1:105–110. [Google Scholar]
- 173.Joel D.M., Kleifeld Y., Losner-Goshen D., Herzlinger G., Gressel J. Transgenic crops against parasites. Nature. 1995;374:220–221. doi: 10.1038/374220a0. [DOI] [Google Scholar]
- 174.Eizenberg H., Hershenhorn J., Ephrath J.H., Kanampiu F. Chemical control. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 415–432. [Google Scholar]
- 175.Gressel J., Segel L., Ransom J.K. Managing the delay of evolution of herbicide resistance in parasitic weeds. Int. J. Pest Manag. 1996;42:113–129. doi: 10.1080/09670879609371981. [DOI] [Google Scholar]
- 176.Duke S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag Sci. 2012;68:505–512. doi: 10.1002/ps.2333. [DOI] [PubMed] [Google Scholar]
- 177.Druille M., Omacini M., Golluscio R.A., Cabello M.N. Arbuscular mycorrhizal fungi are directly and indirectly affected by glyphosate application. Appl. Soil Ecol. 2013;72:143–149. doi: 10.1016/j.apsoil.2013.06.011. [DOI] [Google Scholar]
- 178.Barzman M., Barberi P., Birch A.N.E., Boonekamp P., Dachbrodt-Saaydeh S., Graf B., Hommel B., Jensen J.E., Kiss J., Kudsk P. Eight principles of integrated pest management. Agron. Sustain. Dev. 2015;35:1199–1215. doi: 10.1007/s13593-015-0327-9. [DOI] [Google Scholar]
- 179.Lévesque C.A., Rahe J.E. Herbicide interactions with fungal root pathogens, with special reference to glyphosate. Ann. Rev. Phytopathol. 1992;30:579–602. doi: 10.1146/annurev.py.30.090192.003051. [DOI] [PubMed] [Google Scholar]
- 180.Sharon A., Amsellem Z., Gressel J. Glyphosate suppression of an elicited defense response. Plant Physiol. 1992;98:654–659. doi: 10.1104/pp.98.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sands D.C., Pilgeram A.L. Methods for selecting hypervirulent biocontrol agents of weeds: Why and how? Pest Manag. Sci. 2009;65:581–587. doi: 10.1002/ps.1739. [DOI] [PubMed] [Google Scholar]
- 182.Eizenberg H., Plakhine D., Ziadne H., Tsechansky L., Graber E.R. Non-chemical Control of Root Parasitic Weeds with Biochar. Front. Plant Sci. 2017;8:939. doi: 10.3389/fpls.2017.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tippe D.E., Bastiaans L., van Ast A., Dieng I., Cissoko M., Kayeke J., Makokha D.W., Rodenburg J. Fertilisers differentially affect facultative and obligate parasitic weeds of rice and only occasionally improve yields in infested fields. Field Crop. Res. 2020;254:107845. doi: 10.1016/j.fcr.2020.107845. [DOI] [Google Scholar]
- 184.Scavo A., Mauromicale G. Integrated Weed Management in Herbaceous Field Crops. Agronomy. 2020;10:466. doi: 10.3390/agronomy10040466. [DOI] [Google Scholar]
- 185.Eplee R.E., Norris R.S. Chemical control of Striga. In: Musselman L.J., editor. Parasitic weeds in agriculture: Vol. I. Striga. CRC Press; Boca Raton, FI, USA: 1987. pp. 173–182. [Google Scholar]
- 186.Zwanenburg B., Mwakaboko A.S., Reizelman A., Anilkumar G., Sethumadhavan D. Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Manag. Sci. 2009;65:478–491. doi: 10.1002/ps.1706. [DOI] [PubMed] [Google Scholar]
- 187.Fernández-Aparicio M., Bernard A., Falchetto L., Marget P., Chauvel B., Steinberg C., Morris C.E., Gibot-Leclerc S., Boari A., Vurro M., et al. Investigation of Amino Acids As Herbicides for Control of Orobanche minor Parasitism in Red Clover. Front. Plant Sci. 2017;8:842. doi: 10.3389/fpls.2017.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Westwood J.H., Charudattan R., Duke S.O., Fennimore S.A., Marrone P., Slaughter D.C., Swanton C., Zollinger R. Weed Management in 2050: Perspectives on the Future of Weed Science. Weed Sci. 2018 doi: 10.1017/wsc.2017.78. [DOI] [Google Scholar]
- 189.Watson A.K. Biocontrol. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobanchaceae. Springer; Berlin/Heidelberg, Germany: 2013. pp. 469–497. [Google Scholar]
- 190.Cohen B., Amsellem Z., Maor R., Sharon A., Gressel J. Transgenically enhanced expression of indole-3-acetic acid (IAA) confers hypervirulence to plant pathogens. Phytopathology. 2002;92:590–596. doi: 10.1094/PHYTO.2002.92.6.590. [DOI] [PubMed] [Google Scholar]
- 191.Meir S., Amsellem Z., Al-Ahmad H., Safran E., Gressel J. Transforming a NEP1 toxin gene into two Fusarium spp. to enhance mycoherbicide activity on Orobanche–Failure and success. Pest Manag. Sci. 2009;65:588–595. doi: 10.1002/ps.1736. [DOI] [PubMed] [Google Scholar]
- 192.Nzioki H.S., Oyosi F., Morris C.E., Kaya E., Pilgeram A.L., Baker C.S., Sands D. Striga Biocontrol on a Toothpick: A Readily Deployable and Inexpensive Method for Smallholder Farmers. Front. Plant Sci. 2016;7:1121. doi: 10.3389/fpls.2016.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cimmino A., Fernández-Aparicio M., Andolfi A., Basso S., Rubiales D., Evidente A. Effect of fungal and plant metabolites on broomrapes (Orobanche and Phelipanche spp.) seed germination and radicle growth. J. Agric. Food Chem. 2014;62:10485–10492. doi: 10.1021/jf504609w. [DOI] [PubMed] [Google Scholar]
- 194.Pouvreau J., Gaudin Z., Auger B., Lechat M.M., Gauthier M., Delavault P., Simier P. A high-throughput seed germination assay for root parasitic plants. Plant Methods. 2013;9:32. doi: 10.1186/1746-4811-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Macías F.A., Mejías F.J.R., Molinillo J.M.G. Recent advances in allelopathy for weed control: From knowledge to appplications. Pest Manag. Sci. 2019;75:2413–2436. doi: 10.1002/ps.5355. [DOI] [PubMed] [Google Scholar]
- 196.Aly R. Conventional and biotechnological approaches for control of parasitic weeds. Vitr. Cell. Dev. Biol. Plant. 2007;43:304–317. doi: 10.1007/s11627-007-9054-5. [DOI] [Google Scholar]
- 197.Cvejić S., Radanović A., Dedić B., Jocković M., Jocić S., Miladinović D. Genetic and Genomic Tools in Sunflower Breeding for Broomrape Resistance. Genes. 2020;11:152. doi: 10.3390/genes11020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Letousey P., De Zélicourt A., Vieira Dos Santos C., Thoiron S., Monteau F., Simier P., Thalouarn P., Delavault P. Molecular analysis of resistance mechanisms to Orobanche cumana in sunflower. Plant Pathol. 2007;56:536–546. doi: 10.1111/j.1365-3059.2007.01575.x. [DOI] [Google Scholar]
- 199.De Zélicourt A., Letousey P., Thoiron S., Campion C., Simoneau P., Elmorjani K., Marion D., Simier P., Delavault P. Ha-DEF1, a sunflower defensin, induces cell death in Orobanche parasitic plants. Planta. 2007;226:591–600. doi: 10.1007/s00425-007-0507-1. [DOI] [PubMed] [Google Scholar]
- 200.McCarville M.T., O′Neal M.E., Potter B.D., Tilmon K.J., Cullen E.M., McCornack B.P., Tooker J.F., Prischmann-Voldseth D.A. One Gene Versus Two: A Regional Study on the Efficacy of Single Gene Versus Pyramided Resistance for Soybean Aphid Management. J. Econ. Entomol. 2014;107:1680–1687. doi: 10.1603/EC14047. [DOI] [PubMed] [Google Scholar]
- 201.Varenhorst A.J., Pritchard S.R., O′Neal M.E., Hodgson E.W., Singh A.K. Determining the Effectiveness of Three-Gene Pyramids Against Aphis glycines (Hemiptera: Aphididae) Biotypes. J. Econ. Entomol. 2017;110:2428–2435. doi: 10.1093/jee/tox230. [DOI] [PubMed] [Google Scholar]
- 202.Zhang S.C., Wen Z.X., DiFonzo C., Song Q.J., Wang D.C. Pyramiding different aphid-resistance genes in elite soybean germplasm to combat dynamic aphid populations. Mol. Breed. 2018;38:29. doi: 10.1007/s11032-018-0790-5. [DOI] [Google Scholar]
- 203.Guza C.J. Master’s Thesis. Oregon State University; Corvallis, OR, USA: 2000. Weed Control with Glyphosate and Glufosinate in Herbicide-Resistant Sugarbeets (Beta vulgaris L.) [Google Scholar]
- 204.Nadler-Hassar T., Rubin B. Natural tolerance of Cuscuta campestris to herbicides inhibiting amino acid biosynthesis. Weed Res. 2003;43:341–347. doi: 10.1046/j.1365-3180.2003.00350.x. [DOI] [Google Scholar]
- 205.De Blok M., Botterman J., Vanderwiele M., Dockx J., Thoen C., Gossele V., Rao Movva N., Thompson C., Van Montagu M., Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987;6:2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jiang L., Qu F., Li Z., Doohan D. Inter-species protein trafficking endows dodder (Cuscuta pentagona) with a host-specific herbicide-tolerant trait. New Phytol. 2013;198:1017–1022. doi: 10.1111/nph.12269. [DOI] [PubMed] [Google Scholar]
- 207.Slavov S., Valkov V., Batchvarova R., Atanassova S., Alexandrova M., Atanassov A. Chlorsulfuron resistant transgenic tobacco as a tool for broomrape control. Transgenic Res. 2005;14:273–278. doi: 10.1007/s11248-004-8081-9. [DOI] [PubMed] [Google Scholar]
- 208.Aviv D., Amsellem Z., Gressel J. Transformation of carrots with mutant acetolactate synthase for Orobanche (broomrape) control. Pest Manag. Sci. 2002;58:1187–1193. doi: 10.1002/ps.567. [DOI] [PubMed] [Google Scholar]
- 209.Nandula V.K., Foy C.L., Orcutt D.M. Glyphosate for Orobanche aegyptiaca control in Vicia sativa and Brassica napus. Weed Sci. 1999;47:486–491. doi: 10.1017/S0043174500092158. [DOI] [Google Scholar]
- 210.Senseman S.A. Herbicide Handbook. Weed Science Society of America; Champaign, IL, USA: 2007. pp. 155–157. [Google Scholar]
- 211.Hamamouch N., Westwood J.H., Banner I., Cramer C.L., Gepstein S., Aly R. A peptide from insects protects transgenic tobacco from a parasitic weed. Transgenic Res. 2005;14:227–236. doi: 10.1007/s11248-004-7546-1. [DOI] [PubMed] [Google Scholar]
- 212.Tomilov A.A., Tomilova N.B., Wroblewski T., Michelmore R., Yoder J.I. Trans-specific gene silencing between host and parasitic plants. Plant J. 2008;56:389–397. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- 213.De Framond A.R.P., Mcmillan J., Ejeta G. Effects on Striga parastitism of transgenic maize armed with RNAi constructs targeting essential S. asiatica genes. In: Ejeta G., Gressel J., editors. Integrating New Technologies for Striga Control-Towards Ending the Witch-Hunt. World Scientific; Singapore: 2007. pp. 185–196. [Google Scholar]
- 214.Dubey N.K., Eizenberg H., Leibman D., Wolf D., Edelstein M., Abu-Nassar J., Marzouk S., Gal-On A., Aly R. Enhanced host-parasite resistance based on down-regulation of Phelipanche aegyptiaca target genes is likely by mobile small RNA. Front. Plant Sci. 2017;8:1574. doi: 10.3389/fpls.2017.01574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Butt H., Jamil M., Wang J.Y., Al-Babili S., Mahfouz M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 2018;18:174. doi: 10.1186/s12870-018-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bari V.K., Nassar J.A., Kheredin S.M., Gal-On A., Ron M., Britt A., Steele D., Yoder J., Aly R. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019;9:e11438. doi: 10.1038/s41598-019-47893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]