Abstract
In December 2019, a novel coronavirus was isolated from the respiratory epithelium of patients with unexplained pneumonia in Wuhan, China. This pathogen, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causes a pathogenic condition that has been termed coronavirus disease 2019 (COVID-19) and has reached pandemic proportions. As of 17 September 2020, more than 30 million confirmed SARS-CoV-2 infections have been reported in 204 different countries, claiming more than 1 million lives worldwide. Accumulating evidence suggests that SARS-CoV-2 infection can lead to a variety of clinical conditions, ranging from asymptomatic to life-threatening cases. In the early stages of the disease, most patients experience mild clinical symptoms, including a high fever and dry cough. However, 20% of patients rapidly progress to severe illness characterized by atypical interstitial bilateral pneumonia, acute respiratory distress syndrome and multiorgan dysfunction. Almost 10% of these critically ill patients subsequently die. Insights into the pathogenic mechanisms underlying SARS-CoV-2 infection and COVID-19 progression are emerging and highlight the critical role of the immunological hyper-response — characterized by widespread endothelial damage, complement-induced blood clotting and systemic microangiopathy — in disease exacerbation. These insights may aid the identification of new or existing therapeutic interventions to limit the progression of early disease and treat severe cases.
Subject terms: Kidney diseases, Infectious diseases, SARS-CoV-2, Kidney
This Review describes our current understanding of the pathogenic mechanisms involved in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and the progression of coronavirus disease 2019 (COVID-19), focusing on the immunological hyper-response and the induction of widespread endothelial damage, complement-associated blood clotting and systemic microangiopathy, as well as the effects of these processes on the kidney. The authors also discuss therapeutic interventions that currently hold most promise.
Key points
Although most patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) experience mild to moderate symptoms that resolve after 6–10 days, almost 20% of patients develop severe illness characterized by atypical interstitial bilateral pneumonia and acute respiratory distress syndrome, with a high fatality rate.
Accumulating evidence suggests that an unbalanced and unrestrained innate immune response, which comes at the expense of effective adaptive immunity, underpins the progression of coronavirus disease 2019 (COVID-19).
In severe cases of COVID-19, massive endothelial dysfunction, widespread coagulopathy and complement-induced thrombosis can lead to the development of systemic microangiopathy and thromboembolism; these complications can be life-threatening and ultimately lead to multi-organ failure.
The kidney is one of the main targets of COVID-19 complications, and abnormal kidney function is associated with a significantly increased risk of death in severely ill patients.
Most repurposed anti-viral drugs have failed to improve clinical outcomes in COVID-19; by contrast, therapeutic interventions that target the host response, including the hyper-immune response, complement activation and systemic thrombosis, seem to be more promising approaches to the treatment of severe COVID-19.
Introduction
Coronaviruses are enveloped, single-stranded RNA viruses (Fig. 1a). The Orthocoronavirinae family encompasses several coronaviruses that can infect mammals and birds. Although such infections usually cause mild respiratory disease, in the past two decades, coronaviruses have caused two epidemic diseases in humans: severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) in 2003 and 2012, respectively. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus that was isolated from the respiratory epithelium of patients with unexplained pneumonia in Wuhan, China, in December 2019. As of 17 September 2020, the disease caused by SARS-CoV-2, named coronavirus disease 2019 (COVID-19), has reached pandemic proportions, affecting more than 30 million individuals and claiming more than 1 million lives worldwide.
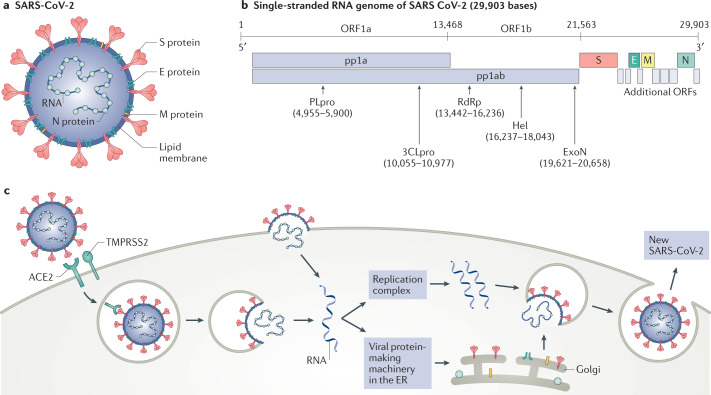
Fig. 1. SARS-CoV-2 structure, genome composition and life cycle.
a | Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a typical betacoronavirus belonging the Orthocoronavirinae family. Each SARS-CoV-2 virion has a diameter of approximately 100–200 nm. Like other coronaviruses, the SARS-CoV-2 envelope comprises a lipid membrane and three structural components: the spike (S) glycoprotein, the envelope (E) protein and the membrane (M) protein. Within the viral envelope, the nucleocapsid (N) protein holds the positive-sense single-stranded RNA, which is 29,903 bases in length. b | The SARS-CoV-2 genome is composed of ten open reading frames (ORFs). At least two-thirds of the viral genome are contained in ORF1a and ORF1b, which together encode a polyprotein, pp1ab, which is further cleaved into 16 non-structural proteins that are involved in genome transcription and replication. Of these proteins, papain-like protease (PLpro) and 3C-like protease (3CLpro) are encoded by ORF1a, whereas RNA-dependent RNA polymerase (RdRp), helicase (Hel) and exonuclease (ExoN) are encoded by ORF1b. The remaining ORFs encode the structural S glycoproteins, and the E protein, M protein and N protein, as well as several accessory proteins with unknown functions. c | SARS-CoV-2 enters the host cell using the endosomal pathway and the cell surface non-endosomal pathway. In the setting of endosomal entry, the SARS-CoV-2 virion attaches to its target cells by direct binding of the S glycoprotein to the host receptor angiotensin-converting enzyme 2 (ACE2). Upon binding, the transmembrane protease serine 2 (TMPRSS2) cleaves and primes S glycoprotein, leading to the fusion of the viral and cell membranes. In addition to canonical viral entry via the endosomal pathway, non-endosomal entry at the plasma membrane may be an additional infection route for SARS-CoV-2. Within the target cells, SARS-CoV-2 is disassembled to release nucleocapsid and viral RNA into the cytoplasm for translation and replication. Translated viral proteins are then assembled in the endoplasmic reticulum (ER) to form the new virions, which are then released from the Golgi membrane system by exocytosis into the extracellular compartment.
Bats are considered to be the most likely natural SARS-CoV-2 reservoir1, although phylogenetic studies have suggested that intermediate animal hosts may have mediated zoonotic transmission to humans2–5. The genome of SARS-CoV-2 is similar to that of a typical betacoronavirus and contains at least ten open reading frames (ORFs; Fig. 1b). The ORF1a and ORF1b encode non-structural proteins (NSPs) that are generated by proteolytic processing1,6. The NSP12 catalytic subunit, along with its cofactors NSP7 and NSP8, constitutes the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) complex, which is essential for viral replication7. The near-atomic resolution of the NSP12–NSP7–NSP8 complex revealed a molecular assembly similar to the equivalent complex in SARS-CoV, the virus responsible for SARS8, although biochemical analyses showed that the polymerase activity and thermostability of the SARS-CoV-2 RdRp complex was lower than that of the SARS-CoV RdRp complex, suggesting adaptation of SARS-CoV-2 towards humans, which have lower body temperatures than bats7. The other ORFs encode the four main structural proteins: the spike, envelope, nucleocapsid and membrane proteins, as well as several accessory proteins with unknown functions1,6.
SARS-CoV-2 infection (Fig. 1c) begins when the viral spike protein attaches to its complementary host cell receptor9. The host protease angiotensin-converting enzyme 2 (ACE2) has been identified as the main surface protein responsible for SARS-CoV-2 entry10. In contrast to SARS-CoV, the spike protein of SARS-CoV-2 has a molecular structure that eludes the host immune response11; moreover, its receptor binding domain harbours a single mutation that significantly enhances its ACE2 binding affinity12,13, suggesting that this novel coronavirus has evolved with an increased ability to infect and spread among humans14. In addition to ACE2, the MERS-CoV receptor dipeptidyl peptidase 4 (DPP4) may also act as a receptor for the SARS-CoV-2 spike protein15; however, this interaction remains to be confirmed. After attachment to ACE2, the transmembrane protease serine 2 (TMPRSS2) cleaves and primes the receptor-bound spike protein to mediate fusion of the viral envelope with the membrane of the target cell in the host10. Other proteases, expressed at different levels in various tissues, may also cleave the spike protein and facilitate virus entry11. Depending on the availability of protease on the host cell surface, this priming of the spike protein allows the virus to enter the host cell through endocytosis or via direct fusion of the viral envelope with the host cell membrane. Once inside its target cell, the viral particles are uncoated and the released viral genome is translated by the host ribosome in the endoplasmic reticulum9. The viral structural proteins move along the secretory pathway into the Golgi intermediate compartment for progeny assembly, and the progeny are then released from the host cell via secretory vesicles by exocytosis9.
Here, we review the current understanding of the pathogenic mechanisms involved in SARS-CoV-2 infection and progression of COVID-19, focusing on the critical role of the immunological hyper-response in disease exacerbation. We also examine emerging data in support of a role for SARS-CoV-2 infection in inducing widespread endothelial damage, complement-associated blood clotting and systemic microangiopathy, and describe how these consequences may contribute to the development of kidney complications in severely ill patients with COVID-19. Finally, we discuss the therapeutic interventions that currently hold most promise to limit progression of disease during the early phases and discuss currently available drugs that may be repurposed to treat severe cases of COVID-19.
SARS-CoV-2 infection and COVID-19
The human-to-human transmission of SARS-CoV-2 is primarily mediated by respiratory droplets or aerosols generated during coughing and sneezing. In the early phase of the infection, SARS-CoV-2 entry by inhalation is facilitated by the high expression of ACE2 in the mouth and on the tongue16,17. In the nasal mucosa, secretory goblet cells express high levels of ACE2 and TMPRSS2, suggesting a potential tropism of SARS-CoV-2 for these cells17,18. Virus replication can also occur in the throat19. SARS-CoV-2 generally has an incubation period of 5 to 6 days20. Once symptoms appear, peak viraemia occurs within 2–5 days of symptom onset20. For about 80% of infected patients, COVID-19 is a mild disease that is mostly restricted to the upper and conducting airways. These individuals normally exhibit modest symptoms, similar to the common flu, including fever and a dry cough, which resolve spontaneously within 6–10 days. However, in almost 20% of patients viral infection progresses down the trachea to the lungs21. Within the alveoli, the epithelial cells lining the lower airways are the main viral targets17,18, owing to their substantial expression of ACE2 and TMPRSS2. SARS-CoV-2 infection induces these epithelial cells to undergo apoptotic cell death as part of the viral replication cycle. This apoptotic response is associated with vascular leakage within alveoli that induces a first wave of local inflammation and recruits immune cells from the blood into the lungs to eliminate extra-cellular virus and destroy virus-infected cells22,23. The increase in pro-inflammatory cytokines within the lung leads to the recruitment of leukocytes, further propagating the local inflammatory response that underlies the pathology of atypical interstitial bilateral pneumonia observed in patients with COVID-19. Histology studies have revealed that the central part of the lung is the most affected region, with pneumocyte injury and diffuse alveolar damage, oedema and alveolar haemorrhage, and the presence of interstitial inflammatory infiltrates24–26. During this inflammatory phase in the lungs, the disease can rapidly progress to severe illness characterized by acute respiratory distress syndrome (ARDS), a hyperinflammatory state and multiorgan dysfunction with a fatality rate of nearly 10% among the most severe cases27,28. In this setting, lung-protective mechanical ventilation is the most effective strategy to reduce multiorgan failure and mortality29,30.
Although the lung is the primary site of pathology, an accumulating body of evidence shows that the virus can disseminate to other organs and induce additional pathological conditions. Kidneys, brain, heart, eye and gut can be affected by SARS-CoV-2 infection, potentially leading to specific disease manifestations such as anosmia and ageusia31,32, ocular conjunctivitis33, abdominal pain and diarrhoea34,35, or cardiac injury36. Many cell types present in these organs express ACE2 and proteases such as TMPRSS2 and have been shown to be infected by SARS-CoV-2 in in vitro and in vivo studies37–39. Greater insight into the molecular mechanisms underlying disease progression may aid the identification of therapeutic approaches to target different stages of COVID-19.
The SARS-CoV-2 immune response
Both the innate and the adaptive immune response have an essential role in recognizing and eliminating foreign pathogens. Accumulating evidence suggests that a suboptimal or unrestrained immune response during SARS-CoV-2 infection drives the clinical patterns, disease severity and progression of COVID-19. Identification of specific immune signatures may provide novel insights into the divergent disease trajectories observed in individual patients and aid the design of more targeted therapeutic approaches40–42.
Innate immune response
The innate immune system is the first line of defence against viral infections43. When viruses colonize their target cells, viral single-stranded RNA, like that of SARS-CoV-2, activates innate immune cells by engaging any one of a number of intracellular pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors and melanoma differentiation-associated gene 5 (MDA5), which sense aberrant RNA structures that often form during viral replication44. Following PRR activation, molecular signalling cascades culminate in the activation of downstream transcription factors, such as interferon regulatory factors (IRFs) and nuclear factor-κB (NF-κB)45. These transcription factors trigger the initial cellular antiviral defences by inducing the transcriptional activation of type I and III interferons (IFN-I and IFN-III) and interferon-stimulated genes (ISGs), as well as cytokines and chemokines (Fig. 2a).
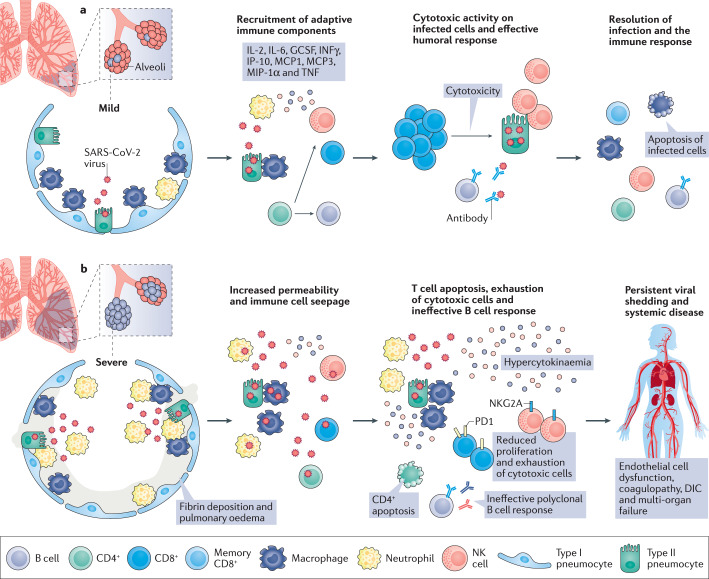
Fig. 2. Pathogenesis and outcomes of COVID-19.
a | Following infection of the lungs, (severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces the death of epithelial cells, in particular, type II pneumocytes, as part of the viral replication cycle. Macrophages and neutrophils elicit a specific innate immune response to eradicate the pathogen and kill virus-infected cells. The increase in pro-inflammatory cytokines within the lung leads to the recruitment of leukocytes, further propagating the local inflammatory response. Among these cytokines, interleukin-2 (IL-2), IL-6, granulocyte-colony-stimulating factor (GCSF), interferon-γ (IFNγ), IFNγ inducible protein 10 (IP-10), monocyte chemoattractant protein 1 and 3 (MCP1 and 3), macrophage inflammatory protein 1α (MIP-1α) and tumour necrosis factor (TNF) stimulate the adaptive immune response. At this stage, infiltration of lymphocytes (CD4+ and CD8+ T cells) and natural killer (NK) cells is required to ensure an optimal defence response against SARS-CoV-2. CD4+ T cells mediate antibody production by B cells and also enhance effector CD8+ T cell and NK responses during viral infections. This orchestrated immune response leads to viral eradication and resolution of the disease. In these patients, coronavirus disease 2019 (COVID-19) seems to manifest as a mild disease with symptoms similar to the common flu that resolve spontaneously. b | For unknown reasons, but possibly individual predisposition or differences in viral loads during primary SARS-CoV-2 infection, some patients experience more severe disease. A maladaptive immune response, characterized by lymphopenia and suppression of CD4+ T cells, is likely accountable for the poor prognosis of these patients. In the absence of robust CD4+ T cell activation, B cells generate a polyclonal antibody response that may be ineffective in neutralizing SARS-CoV-2. Increased numbers of exhausted T cells that express high levels of programmed cell death protein 1 (PD1), suggest decreased proliferation and activity of CD8+ T cells. Similarly, NK cells exhibit increased levels of the inhibitory CD94–NK group 2 member A (NKG2A). Impaired cytotoxic activity results in persistent viral shedding that amplifies macrophage and neutrophil activation, leading to the massive production of cytokines (a process referred to as hypercytokinaemia). In these patients, COVID-19 manifests as a severe disease, consisting of advanced pneumonia and acute respiratory distress syndrome. The generation of excess cytokines and persistent viral infection leads to systemic vascular damage, disseminated intravascular coagulation (DIC) and the failure of vital organs, including the kidney and the heart.
This broad antiviral response has placed selective pressure on viruses to develop countless countermeasures to avoid host immune responses, most of which are poorly understood46. Mechanistically, NSP1 of SARS-CoV-2 suppresses RIG-I-dependent innate immune responses by binding to the 40 S ribosomal subunit, resulting in shutdown of host mRNA translation47. SARS-CoV-2 may also evade immunological defence mechanisms through the indirect binding of ORF9b to the mitochondrial antiviral-signalling protein (MAVS) via the translocase of the outer mitochondrial membrane 70 (TOM70)48. The localization of MAVS on the outer mitochondrial membrane is essential for the activation of IFN-I after infection of RNA viruses49, and it is possible that SARS-CoV-2 binding to MAVS might prevent its translocation and IFN-I activation50. In support of this hypothesis, SARS-CoV-2 infection is associated with lower IFN-I and IFN-III levels and a moderate ISG response compared with other respiratory viruses, despite being characterized by vigorous production of inflammatory cytokines45,51.
In line with the above-described findings, the most frequent laboratory abnormalities in patients with unfavourable COVID-19 progression include a cytokine profile characterized by increased levels of IL-2, IL-6, granulocyte colony-stimulating factor (G-CSF), IFNγ inducible protein 10 (IP-10, also known as CXCL10), monocyte chemoattractant protein 1 and 3 (MCP1 and MCP3, also known as CCL2 and CCL7, respectively), macrophage inflammatory protein 1α (MIP-1α, also known as CCL3) and tumour necrosis factor (TNF)52–54. Of these dysregulated cytokines and chemokines, the combined evaluation of IP-10 and MCP3 had the highest predictive value of poor outcomes among patients with COVID-19 (ref.54). These findings are reminiscent of the hyper-inflammatory syndrome, secondary haemophagocytic lymphohistiocytosis, and consequently, the immune response to SARS-CoV-2 infection was initially considered to involve a form of cytokine storm, similar to that found in patients with other viral infections or sepsis, as well as in patients with autoimmune conditions such as arthritis55,56. However, this concept has since been revisited and the contribution of cytokines to COVID-19 questioned57. In COVID-19, macrophage activation seems to be integral to the initiation and propagation of this hyper-inflammatory reaction58 (Fig. 2b). One preliminary study detected ACE2 on the surface of macrophages and showed that SARS-CoV-2 spike proteins interact with CD68+ macrophages, suggesting direct viral infection of these cells59. Single-cell RNA sequencing of immune cells from bronchoalveolar lavage fluid from patients with COVID-19 suggested the expansion of pro-inflammatory monocytes and Ficolin-positive monocyte-derived macrophages, paralleled by a decrease in tissue-resident reparative alveolar macrophages in patients with severe disease compared with that seen in moderate cases60. A further elegant single-cell RNA sequencing study confirmed an enrichment of SARS-CoV-2 RNA in the lung macrophage population of infected patients, and differential gene expression analysis revealed that infected macrophages had higher chemokine expression than non-infected macrophages61. Moreover, use of untargeted metabolomics has shown that SARS-CoV-2 infection triggers profound alterations in macrophage phenotype and function by downregulating the expression of apolipoproteins that are important for proper macrophage polarization towards anti-inflammatory phenotype62,63. Mechanistically, virus–host interactome and proteomic analysis of peripheral blood cells from patients with varying symptoms of COVID-19 suggests that NSP9 and NSP10 of SARS-CoV-2 interact with the NF-kB repressor NKRF, which induces an over-exuberant innate immune response to SARS-CoV-2 infection64.
A transcriptome analysis of bronchoalveolar lavage fluid cells from eight patients with COVID-19 suggested that SARS-CoV-2 infection elicits more pronounced neutrophil infiltration than other forms of pneumonia65. In line with these findings, neutrophilia is predictive of poor outcomes in patients with COVID-19 (ref.66); moreover, studies published in preprint form identified a high neutrophil-to-lymphocyte ratio as an independent risk factor for severe disease67,68. A new study has also indicated that neutrophil activation and the formation of neutrophil extracellular traps (NETs) may have a prominent role in propagating the severe cytokine release observed in severe cases of COVID-19, representing a previously unrecognized, but powerful innate defence response that may exacerbate lung damage in affected patients69. These findings suggest that unbalanced innate immunity may exacerbate the hyper-inflammatory response in severe cases of the disease (Fig. 2b).
Adaptive immune response
Lymphopenia is a common feature of severe COVID-19, characterized by drastically reduced absolute numbers of CD4+ and particularly CD8+ T cells27,70,71; this depletion correlates with COVID-19 severity and associated mortality72,73. A proteomic analysis of peripheral blood cells from patients with COVID-19 reported decreased T cell counts, associated with reduced indices of T cell activation and function, including lower expression of T cell receptor (TCR) subunits (CD3e, CD3g, CD247, TRAC and TRBC1) in patients with severe disease than in patients with mild disease64. Levels of T cell surface molecules (CD4, CD8a, CD8b and CD2), T cell migration stimulators (DDP4), TCR signalling kinases (ZAP70, LCK and FYN) and MHC class II molecules (HLA-DRA, HLA-DRB1, HLA-DRB4 and HLA-DRB5) were also significantly lower in patients with severe disease, indicating a global impairment in the adaptive immune response64. Decreased numbers of peripheral T cells might result from increased migration to the lungs or other infected tissues. However, although CD4+ T cells can be detected in lung tissue of deceased patients with COVID-19, CD8+ T cell are infrequently observed31. Peripheral CD8+ T cells from patients with COVID-19 express high levels of exhaustion markers, including programmed cell death protein 1 (PD1) and T cell immunoglobulin mucin-3 (TIM3); of note, this expression pattern was more pronounced among patients who required intensive care than in patients with mild disease74. In addition to confirming the exhausted phenotype of CD8+ T cells in patients with COVID-19, another study demonstrated that natural killer (NK) cells were also exhausted with increased expression of inhibitory CD94–NK group 2 member A (NKG2A) receptor heterodimer. Of interest, numbers of CD8+ T cells and NK cells were restored in convalescent patients, with reduced expression of CD94–NKG2A73. These findings suggest that the rise in inflammatory cytokine levels induced by SARS-CoV-2 infection might drive the depletion of T and NK cells, which subsequently fail to destroy infected cells.
However, T cell lymphopenia may also result from SARS-CoV-2-induced cell death. Findings from a post-mortem examination of the spleen and lymph nodes of patients with COVID-19, published in preprint form, identified high levels of T cell apoptosis and increased expression of the death receptor FAS75, possibly suggesting that activation-induced cell death (AICD) is likely accountable for T cell depletion in patients with severe COVID-19. Flow cytometry analysis also revealed significantly higher FAS expression in circulating T cells from patients with COVID‐19 than in controls76. Of note, an earlier report indicated that ACE2 is not expressed by T cells, ruling out the possibility that SARS-CoV-2 has a direct toxic effect on these cells59 and further supporting the notion that T cell depletion in COVID-19 results from AICD. Regardless of the underlying cause, however, the absence of an effective anti-viral T cell and NK cell response could play a critical part in the maintenance of a high viral load and hyperactivation of the innate response77 (Fig. 2b). Severe infection caused by SARS-CoV-2 has been likened to lymphopenic community-acquired pneumonia78 — a specific immunological phenotype associated with a high risk of mortality79. Thus, T cell defects in the T cell response might contribute to sustained viral shedding and pathogenesis of multi-organ dysfunction in patients with COVID-19 with clinically severe disease80.
Despite the impaired T cell response associated with severe COVID-19, a study from the USA documented the presence of circulating SARS-CoV-2-specific CD4+ and CD8+ T cells in 100% and 70%, respectively, of patients who had recovered from the disease81. The functional CD4+ T cell response against SARS-CoV-2 was identified as a classical T helper (TH) type 1 response, which was robust against the spike protein, the membrane protein and the nucleocapsid protein. Additional CD4+ T cell responses were found to target NSP3, NSP4, ORF3a and ORF8 in convalescent patients with COVID-19 (ref.81). The SARS-CoV-2-specific CD8+ cells were IFNγ+ and recognized the spike and membrane proteins, and at least eight different SARS-CoV-2 ORFs81,82. Ex vivo stimulation with viral peptides induced SARS-CoV-2-specific CD4+ and CD8+ T cells; in the blood of patients with severe COVID-19, these SARS-CoV-2-specific lymphocytes were already detectable at ICU admission82,83. Notably, these studies have consistently identified the presence of SARS-CoV-2-specific T cells in a large subset of seronegative convalescent individuals, suggesting that T cells could represent a more sensitive indicator of SARS-CoV-2 exposure than antibodies81,84. An analysis of samples collected from 2015 to 2018 before the emergence of the COVID-19 pandemic detected SARS-CoV-2-specific T cells, including T cells that recognize NSP7 and NSP13, in uninfected individuals, suggesting possible cross-reactivity with common cold coronaviruses81,83,85.
Typically, a robust T cell response is directed towards only a small subset of potentially immunogenic peptides in a foreign protein, and the major constraint that determines this immunodominance is dictated by antigen processing86. In SARS, the spike protein of SARS-CoV accounted for nearly two-thirds of CD4+ T cell reactivity whereas no reactivity was found against nucleocapsid and membrane proteins81. By contrast, the spike, nucleocapsid and membrane proteins of SARS-CoV-2 seem to be co-dominant immunogenic proteins, and marked CD4+ T cell responses directed against NSP and ORF proteins have also been reported81,87. The lack of a clear immunodominant CD4+ T cell response to SARS-CoV-2 could indicate aberrant antigen processing and consequent dysregulation of the T cell immune response81. In support of this hypothesis, the frequency of virus-specific memory T cells and the number of immunodominant virus epitopes they responded to were significantly higher in patients with severe disease than in patients with mild disease82,84, raising the question of why these virus-specific T cells are not able to control the disease.
In contrast to T cells, very few studies have demonstrated an effect of SARS-CoV-2 on B cell biology. To date, studies regarding B cell function in COVID-19 have mainly focused on the generation of specific antibodies against SARS-CoV-2. In a study of 285 patients with COVID-19, seroconversion for IgG and IgM against the nucleocapsid and the spike protein occurred simultaneously or sequentially and their titres plateaued within 6 days of seroconversion88. Within 19 days of symptom onset, 100% of patients were IgG-positive88. Based on the humoral response to SARS and MERS, the development of neutralizing antibodies against the SARS-CoV-2 spike protein is likely to be the most promising approach to ensure COVID-19 protective immunity89–91. Non-human primates reinfected with SARS-CoV-2 <1 month after their first infection displayed a reduced viral load and prevented disease recurrence relative to responses in primo-infected animals92,93; these beneficial effects were associated with potent neutralizing antibody responses. Moreover, treatment with two potent human monoclonal antibodies that recognize the spike protein protected rhesus macaques against a challenge with SARS-CoV-2 as reported by a preprint study94 — a finding that strongly suggests that a protective humoral response can be induced by vaccination.
However, the extent to which naturally induced antibodies against disease relapse may provide long-lasting protection is still unknown. The longevity of the humoral response against SARS-CoV-2 in humans may be transient, as demonstrated by the finding that 40% of asymptomatic patients and 13% of symptomatic patients became negative for anti-spike IgG in the early convalescent phase95. As shown in a preprint study, longitudinal evaluation of patients with SARS-CoV-2 infection reported a rapid decline in levels of neutralizing antibody during a 4-month follow-up period, with titres approaching the lower threshold of positivity in several individuals96. The decay of anti-SARS-CoV-2 antibodies was particularly rapid in patients with COVID-19 with mild disease97. However, these findings were challenged by a study in the Icelandic population showing that antibodies against SARS-CoV-2 remained stable for over 4 months98. More importantly, neutralizing antibodies from patients with COVID-19 are minimally mutated, suggesting that they bind SARS-CoV-2 with low affinity owing to limited affinity maturation99; indeed, these antibodies have modest neutralizing activity100,101, targeting epitopes outside the receptor binding domain100. This disparate antibody response may indicate failure to develop a robust, long-lasting protective humoral response against SARS-CoV-2.
In support of a dysregulated B cell response to SARS-CoV-2 infection, analyses of circulating B cells showed expansion of oligoclonal plasmablasts and reduced memory B cell frequencies in patients with severe COVID-19 compared with responses in patients with mild disease or healthy individuals42,102,103. The antibody sequences of the largest B cell clones detected in patients with severe COVID-19 were variable in terms of somatic hypermutation levels and VH gene usage, suggesting a polyclonal B cell response102, possibly arising via an extra-follicular pathway, as reported in a preprint study104. Plasmablast levels in patients with COVID-19 did not correlate with antibody levels against the spike protein, suggesting that these cells generated antibodies against antigens other than the spike protein or generated antibodies with low specificity and affinity.
Surprisingly, anti-SARS-CoV-2 antibodies are elevated in severely-ill patients with COVID-19 (refs95,105), whereas they are reportedly low in most recovered patients82. In a preprint study of individuals with mild COVID-19, titres of neutralizing antibodies correlated positively with age, and blood C-reactive protein levels, but correlated negatively with lymphocyte counts of patients at the time of admission106. Unfortunately, this study did not assess the potential correlation between viral load and disease symptoms with neutralizing antibody titres in individual patients to evaluate the actual pathogenic role of these antibodies106. In this context, a study that suggested that the humoral response against the SARS-CoV-2 spike protein may increase the severity of lung injury by skewing macrophage responses, abrogating wound-healing responses and promoting macrophage recruitment and accumulation in the lung parenchyma, is of particular interest107. Relative levels of IgA and IgG have also been demonstrated to be markedly higher in severely ill patients with COVID-19 than in patients with milder disease, suggesting that IgA may have synergistic effects with IgG in promoting antibody-dependent cellular cytotoxicity and COVID-19 exacerbation108.
Together, the available evidence indicates that a variety of dysregulated processes occur in innate and adaptive immunity, including the cellular and humoral response to SARS-CoV-2 infection (Fig. 2). These findings indicate the need for further studies to better understand COVID-19 immunopathology and to aid the design of the optimal vaccine and effective therapeutic interventions109.
Effects of SARS-CoV-2 on the endothelium
Endothelial cells contribute to the preservation of normal haemostasis by maintaining vessel wall integrity and balancing fibrinolysis through the expression of coagulation inhibitors and blood clot-lytic enzymes, and through maintenance of the glycocalyx110–112. Mounting evidence suggests that loss of vessel barrier integrity and the development of a pro-coagulative endothelium contributes to the initiation and propagation of ARDS in COVID-19, by inducing endotheliitis and mediating inflammatory cell infiltration in the lungs24,113,114. A substantial proportion of patients with severe COVID-19, including young individuals and children, develop venous and arterial complications. In New York City, over a 2-week period from 23 March to 7 April, 2020, researchers observed a sevenfold increase in large-vessel stroke among patients younger than 50 years of age compared with a 2-week time frame over the previous 12 months115,116. Although early evidence suggested that children may be at a lower risk of SARS-CoV-2 infection than adults owing to the lower expression of ACE2 in nasal epithelia117, an unprecedented cluster of hyper-inflammatory shock syndrome was observed in South East England, UK over a 10-day period in mid-April 2020 in eight children with likely family COVID-19 exposure118. This syndrome has been described as a pathological condition with features similar to Kawasaki disease — a rare condition that affects blood vessels, including those in the heart. All children tested negative for SARS-CoV-2 on bronchoalveolar lavage or nasopharyngeal aspirates, but laboratory tests were indicative of infection and inflammation, including high levels of C-reactive protein, ferritin and D-dimers118. More recent evidence suggested that this syndrome, now termed multisystem inflammatory syndrome in children (MIS-C), has unique haematological and immunological characteristics that are distinct from those of both typical COVID-19 in adults and Kawasaki disease119. Similar cases of MIS-C have been observed in 17 patients under 18 years of age in New York City120 and in Bergamo121, one of the hardest hit areas of northern Italy. A retrospective study of 35 children with MIS-C in France described acute heart failure as a result of the severe inflammatory state caused by SARS-CoV-2 infection; none of these children had underlying cardiovascular disease122.
SARS-CoV-2 may alter vascular homeostasis by directly infecting endothelial cells via ACE2 (Fig. 3a). In support of this proposal, SARS-CoV-2 can successfully infect engineered human blood vessel organoids, confirming SARS-CoV-2 tropism for endothelial cells123. This finding is further corroborated by electron microscopy analyses of post-mortem tissue, which have shown that SARS-CoV-2 can infect pulmonary endothelial cells and induce endotheliitis, at least in a subset of critically ill patients114. After binding by SARS-CoV-2, ACE2 is internalized and downregulated on endothelial cells, which favour the progression of inflammatory and pro-fibrotic processes in the lung triggered by local angiotensin II (Ang II) hyperactivity124. Given that ACE2 converts Ang II into Ang (1–7), which acts on the MAS receptor, it is conceivable that a reduction of ACE2 expression by endothelial cells in response to SARS-CoV-2 infection leads to reduced generation of Ang 1–7 and consequently, reduced activation of MAS, promoting a local pro-thrombotic endothelial cell phenotype (Fig. 3a). Furthermore, reduced expression of ACE2 might in turn indirectly activate the kallikrein–kinin system (KKS), which ultimately leads to increases in vascular permeability113,125. Indeed, several studies have documented that a tight balance between the KKS and renin–angiotensin systems is required to regulate the thromboresistance of endothelial cells126–128. The reduction in pulmonary ACE2 impairs the inactivation of des-Arg9 bradykinin (DABK), leaving it free to bind its cognate receptors129–131 (Fig. 3a). Moreover, high levels of bradykinin in patients with COVID-19 might explain the majority of severe symptoms, spanning from blood vessel injury to neurological complications132, suggesting that inhibition of the KKS, for example, with the bradykinin B2 receptor antagonist icatibant132, could be a possible therapeutic strategy133,134. Mechanistically, disruption of the endothelial barrier by SARS-CoV-2 spike protein is mediated by changes in the surface expression of intracellular adhesion molecule-1 (ICAM1), vascular cell adhesion protein-1 (VCAM1) and the tight junction scaffold protein zonula occludens-1 (ZO-1), as reported by a preprint study135.
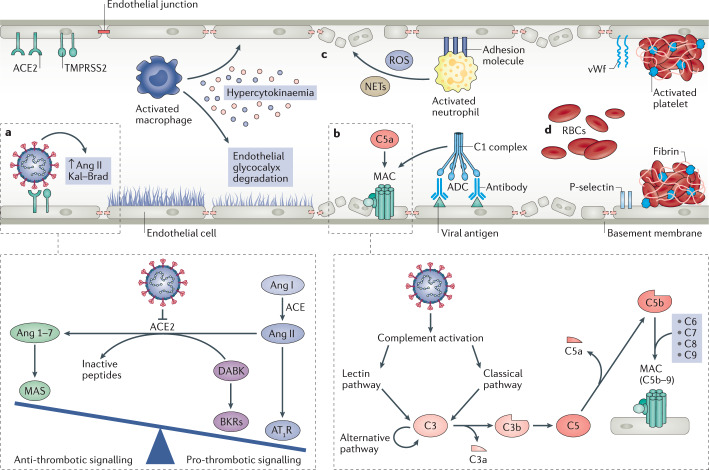
Fig. 3. Effect of SARS-CoV-2 infection on endothelial cell function, systemic coagulation and thrombosis.
a | Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly infects endothelial cells owing to their high expression levels of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2). After binding by SARS-CoV-2, ACE2 is internalized, and the lack of ACE2 on endothelial cells favours the progression of inflammatory and thrombotic processes triggered by local angiotensin II (Ang II) hyperactivity. Inhibition of ACE2 by binding of SARS-CoV-2 reduces the ACE2-mediated conversion of Ang II to Ang 1–7, the vasoactive ligand of the MAS receptor. The reduction of MAS receptor activation induces a pro-inflammatory phenotype through increased activation of type 1 angiotensin receptors (AT1R). Additionally, the reduction in levels of ACE2 limits the degradation of des-Arg9 bradykinin (DABK) into inactive peptides, ultimately leading to increased pro-thrombotic signalling via the activation of bradykinin receptors (BKRs). b | SARS-CoV-2 also activates the complement system — an integral component of the innate immune response. The complement cascade can be activated by three different pathways, the classical, the lectin and the alternative pathway, that resolve around the formation of the C3 convertases that cleave C3, generating the pro-inflammatory peptide C3a and large amount of C3b that opsonizes pathogens. C3b also forms the C5 convertase, which leads to release of the potent anaphylatoxin C5a, as well as the fragment C5b, responsible for the formation of the membrane attack complex (MAC) C5b–9 on target cells, which is considered to be the terminal event of complement activation. In addition, complexes of SARS-CoV-2-specific antibodies and viral antigens might induce endothelial cell injury through activation of the C1 complex of the classical pathway and induction of antibody-dependent cytotoxicity (ADC). c | Pro-inflammatory cytokines and chemokines released by activated macrophages amplify the vicious cycle of vascular integrity disruption, vessel coagulation and thrombosis by degrading the endothelial glycocalyx, activating the coagulation system and dampening anticoagulant mechanisms. The adhesive phenotype of endothelial cells induced by inflammatory cytokines and chemokines promotes infiltration of neutrophils, which produce large amounts of histotoxic mediators, including reactive oxygen species (ROS) and neutrophil extracellular traps (NETs), ultimately leading to injury of endothelial cells. d | Activated endothelial cells initiate coagulation by expressing P-selectin, von Willebrand factor (vWf) and fibrinogen, leading to massive platelet binding, fibrin formation and clotting of red blood cells (RBCs), ultimately resulting in systemic thrombosis and disseminated intravascular coagulation.
However, the finding that viral RNA is rarely detectable in blood19 indicates that additional host-dependent factors might contribute to systemic endothelial dysfunction and vasculopathy in COVID-19, rather than direct viral-dependent effects on endothelial cells. As reviewed elsewhere, the hypercytokinaemia and the massive pro-inflammatory response of the host may contribute to endothelial dysfunction in COVID-19, particularly via the actions of IL-6 and TNF — levels of which are strongly increased in critically ill patients113. Cytokines exert their pathogenic effects through various mechanisms. For example, IL-6 can increase vascular permeability and promote the secretion of pro-inflammatory cytokines by endothelial cells themselves, thereby amplifying the cytokine release113. TNF can activate glucuronidases that might degrade the endothelial glycocalyx but might also upregulate hyaluronic acid synthase 2, leading to increased deposition of hyaluronic acid in the extracellular matrix and promoting fluid retention113. Additional mechanisms, such as a reduction of endothelial nitric oxide synthase activity and nitric oxide levels, as well as the release of vascular endothelial growth factor (VEGF) as a consequence of the systemic hypoxia induced by ARDS, have also been proposed as key pathogenic processes underlying endothelial dysfunction following SARS-CoV-2 infection136. Below, we focus on the possible role of the complement system in inducing systemic endothelial dysfunction in COVID-19.
Excessive complement activation
The complement system is an integral component of the innate immune response and comprises over 30 different proteins137. This system can be activated by three different pathways: the classical, the lectin and the alternative pathway137. All of these pathways converge on the formation of C3 convertases that cleave C3 to generate the pro-inflammatory peptide C3a and a large amount of C3b, which opsonizes pathogens137. C3b also forms C5 convertase, which induces release of the potent anaphylatoxin C5a, as well as the fragment C5b that is responsible for the formation of the membrane attack complex C5b–9 on target cells — the terminal event of complement activation. Although complement, like other innate immune system components, helps to control bacterial and viral infections, its unrestrained activation — for example, in the context of prolonged SARS-CoV-2 infection — can be detrimental by causing direct damage of host tissues138 (Fig. 3b).
Most available data on complement activation in the context of coronavirus infection are derived from the SARS and MERS epidemics. A 2018 study of SARS-CoV demonstrated that C3-deficient mice experienced a less severe form of disease than wild-type mice, associated with reduced lung infiltration of inflammatory cells and lower hypercytokinaemia in both the lungs and sera139. SARS-CoV activates the lectin pathway by interacting with the complement component mannose-binding lectin (MBL) — a C-type lectin that activates mannose-binding protein-associated serine protease 2 (MASP-2) — enhancing the deposition of C4 activation fragments on virus-infected cells140. In mice infected with MERS-CoV, levels of circulating C5a increased post-infection and the expression of the C5a receptor (C5aR) was upregulated in pneumocytes and inflammatory leukocytes, associated with strong C5b–9 deposition on bronchiolar epithelial cells, pneumocytes and infiltrating leukocytes141. In line with these findings, immunohistochemistry analyses in post-mortem lung tissue from SARS-CoV-2-infected patients revealed high expression of the complement components MBL, C4, C3 and C5b–9, in alveolar epithelial cells and pneumocytes, as well as in inflammatory cells, as reported by a preprint study142. As mentioned above, the nucleocapsid protein of SARS-CoV-2 can interact indirectly with MASP-2 and initiate the lectin pathway of complement, resulting in aberrant complement activation and aggravated inflammatory lung injury142. Circulating levels of C5a were higher in patients with severe COVID-19 than in patients with mild disease who had levels of C5a similar to those of healthy controls142. Use of an untargeted metabolomic approach also demonstrated upregulation of complement activation proteins, including membrane attack complex proteins such as C5, C6 and C8, in sera from 46 patients with severe disease62.
In addition to the lectin pathway, the classical pathway may also contribute to tissue injury induced by coronavirus infection. Indeed, immune complexes formed by viral antigens and antibodies can activate C1 and initiate complement activation. In patients with SARS-CoV infection, the development of autoantibodies against human pulmonary epithelial cells and endothelial cells mediated complement-dependent cytotoxicity143; however, whether this process is also activated following SARS-CoV-2 infection is unknown. Use of an ultra-high-throughput clinical proteomic approach identified consistent activation of the classical complement pathway (specifically, complement components C1R, C1S and C8A) in the sera of patients with clinically severe COVID-19, along with increased activation of the alternative pathway component, complement factor B (CFB) and its modulators CFI and CFH144. Findings from another study suggest that enhanced IgA levels during SARS-CoV-2 infection might act synergistically with IgG to promote antibody-dependent cytotoxicity108. These researchers also proposed that COVID-19 might be at least in part an IgA-mediated disease108, possibly inducing a vasculitis phenotype via classical pathway-mediated cytotoxicity.
Although clinical insights into complement activation following SARS-CoV-2 infection are limited145, the above findings indicate that unrestrained complement activation — mainly at the downstream terminal phases of the complement cascades — may contribute to endothelial cell injury, thrombosis and intravascular coagulation, ultimately leading to multi-systemic organ failure in patients with COVID-19 (ref.137).
Microangiopathy and vessel thrombosis
Activation of the complement system leads to endothelial cell injury and death with subsequent vascular denudation and exposure of the thrombogenic basement membrane, which drives activation of clotting cascades. These events result in inflammation, microvascular thrombosis, vessel oedema and haemorrhagic sequelae — all of which are prominent features of the lung pathology of patients with COVID-19-associated pneumonia146. In an autopsy study of ten patients with COVID-19, small vessel thrombus formation in the lung periphery was associated with foci of alveolar haemorrhage147. In a larger series of post-mortem pulmonary analyses in Northern Italy, a predominant finding was the presence of platelet — fibrin thrombi in small arterial vessels, further suggesting that coagulopathy has an important role in outcomes following SARS-CoV-2 infection148. Indeed, patients with SARS-CoV-2 infection are at an increased risk of widespread coagulation of small and large vessels149, and venous thromboembolic complications — in particular acute pulmonary embolisms150–153 — are recognized as life-threatening complications in patients with COVID-19.
Pro-inflammatory cells, cytokines and chemokines amplify the vicious cycle of vessel coagulation and thrombosis, by acting to simultaneously activate the coagulation system and dampen anticoagulant mechanisms154. Infiltrating, activated neutrophils (Fig. 3c) produce large amounts of histotoxic mediators, including reactive oxygen species113, as well as NETs, which have been found in arterial microthrombi in the post-mortem lung specimens from four deceased patients with COVID-19 (ref.155). The expression of tissue factor (TF) by activated monocytes stimulates the coagulation cascade and the subsequent generation of thrombin; these processes are ineffectually counterbalanced by tissue factor pathway inhibitor (TFPI), which is present in plasma and platelets, and produced mainly by endothelial cells154. Concurrently, endothelial cell-associated anticoagulant pathways, in particular the protein C system, are impaired by pro-inflammatory cytokines154. In response to activation by IL-1β and TNF, endothelial cells promote coagulation by expressing P-selectin, von Willebrand factor and fibrinogen, leading to massive platelet binding113 (Fig. 3d). This process is further supported by the release of trophic cytokines by endothelial cells, which augment platelet production113. The recruited platelets release VEGF, which amplifies the expression of TF by endothelial cells, further stimulating the coagulation cascade113. Together, these events contribute to the increase in fibrin generation, inducing the deposition of blood clots in the microvasculature154 (Fig. 3).
Kidney involvement in COVID-19
The lungs are the main site for SARS-CoV-2 infection and the first affected organ in COVID-19. However, sustained viral shedding and hyperactivation of the immune response in severe cases greatly increases the risk of systemic multi-organ failure. Of the organs affected by COVID-19, the kidney is one of the main sites of COVID-19 complications (Fig. 4), and abnormal renal function is a significant risk factor for death in severely ill patients with COVID-19 in the ICU setting. In a prospective cohort study of 701 patients with COVID-19 in Wuhan, China, almost 17% (113) died in hospital156. On admission, 43.9% of patients had proteinuria and 26.7% haematuria, whereas 5.1% of patients experienced acute kidney injury (AKI) during the study156. Reports published later in the pandemic described a higher incidence of AKI (8–17%), with the highest ranges in the critically ill (14–35%)157,158. In most patients with COVID-19, abnormal kidney function in the presence of low-level proteinuria possibly reflects tubular injury. In some cases, however, proteinuria is abundant or contains albumin, suggesting glomerular impairment158. Patients with kidney disease are at a significantly higher risk of death than those without kidney disease, and elevated levels of baseline serum creatinine or blood urea nitrogen, AKI stage, proteinuria or haematuria were independent risk factors for in-hospital mortality156. A systematic review and meta-analysis suggested that, in patients with COVID-19, severe AKI is an ominous clinical predictor and is associated with high mortality159,160. As a result, dialysis is an important tool for the management of severely ill patients with AKI161,162.
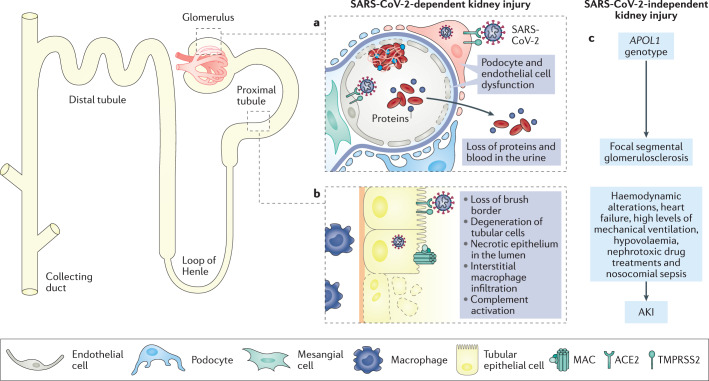
Fig. 4. The effects of COVID-19 on the kidney.
The kidney expresses high levels of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) and has been identified as a target organ for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection by several studies. a | Within the glomerulus, podocytes and endothelial cells have been identified as specific sites for viral infection. SARS-CoV-2-induced podocyte dysfunction is likely to induce impairment of glomerular filtration, leading to proteinuria and haematuria, which are often observed in patients with coronavirus disease 2019 (COVID-19). Infection of endothelial cells alters glomerular capillary haemostasis and induces the formation of fibrin thrombi. b | SARS-CoV-2 has also been found in proximal tubular cells, which exhibit high levels of ACE2 in the apical brush border. SARS-CoV-2 infection leads to loss of the brush border and vacuolar degeneration in tubular epithelial cells, with luminal debris composed of necrotic epithelium with evidence of complement activation and membrane attack complex (MAC) (C5b–9) deposition on tubular cells and massive macrophage infiltration in the kidney interstitium. c | In addition, a non-viral-dependent mechanism might also be accountable for kidney dysfunction in the context of COVID-19, including a possible contribution of the APOL1 ‘risk’ genotype in inducing focal segmental glomerulosclerosis following SARS-CoV-2 infection, as well as the role of haemodynamic factors, cardiac dysfunction, high levels of mechanical ventilation, hypovolaemia, nephrotoxic drug treatments and nosocomial sepsis resulting in COVID-19-associated acute kidney injury (AKI).
In autopsy specimens, the spectrum of renal pathological abnormalities includes loss of the brush border and vacuolar degeneration in tubular epithelial cells, with debris composed of necrotic epithelium in tubular lumens163. As seen in the lung164, post-mortem examination of kidney specimens in a preprint study also identified deposition of C5b–9 on tubule cells and massive interstitial infiltration of macrophages as hallmarks of kidney complications in patients with severe disease165. The relevance of microvascular thrombosis in the kidney is still a matter of investigation. One autopsy study identified limited fibrin thrombi in glomerular capillaries of kidney specimens from patients with COVID-19 with severe disease163. However, renal vasculopathy has not been consistently found in the kidney and no significant glomerular disease has been described, with the exception of collapsing focal segmental glomerulosclerosis, which has been reported in a few patients and seems to be associated with the presence of genetic ‘risk’ variants of APOL1166,167. Together, these findings suggest that the kidney is severely affected in some patients with COVID-19 and imply that further insight into disease pathogenesis may aid the identification of novel therapeutic targets to prevent kidney dysfunction and/or hasten renal recovery during SARS-CoV-2 infection.
Several non-specific mechanisms may contribute to the development of AKI in the setting of COVID-19, including haemodynamic alterations, cardiac dysfunction, hypovolaemia due to poor oral intake or high-grade fever, use of nephrotoxic drugs and nosocomial sepsis138. Additionally, high levels of mechanical ventilation can potentially induce AKI by inducing neuro-humoral-mediated alterations in intra-renal blood flow and other haemodynamic changes that impair renal perfusion, and through the induction of systemic inflammatory mediators as a consequence of ventilator-induced lung injury168.
However, emerging evidence suggests that SARS-CoV-2 may have a direct effect on kidney cells169–172. Tubule epithelial cells and podocytes exhibit high levels of ACE2 and TMPRSS2, suggesting that they are potential SARS-CoV-2 targets173,174. ACE2 internalization following SARS-CoV-2 infection could induce an imbalance in the renin–angiotensin–aldosterone system, with increased Ang II signalling leading to pro-inflammatory and profibrotic processes in the kidney14,175. The critical role of ACE2 in SARS-CoV-2 kidney infection is emphasized by findings from in vitro studies that show that SARS-CoV-2 can infect human kidney organoids and that this infection can be limited by expression of human recombinant soluble ACE2 — a decoy receptor that is undergoing evaluation in clinical trials123. Also, in support of the hypothesis that SARS-CoV-2 exerts tropism in the kidney, electron microscopy examination of autopsy samples from 26 patients who had died with COVID-19 demonstrated clusters of viral particles in the tubular epithelium and podocytes163. Another study that involved microdissection of autoptic tissue demonstrated detectable SARS-CoV-2 viral load in three of six deceased patients; within the three SARS-CoV-2-positive samples, virus was detected in all kidney compartments examined, with preferential targeting of glomerular cells176. However, most analyses of kidney tissue have used electron microscopy, light microscopy and/or immunohistochemistry, which cannot conclusively ascertain whether the identified particles are actually SARS-CoV-2 or merely viral-like structures. In particular, the identification of viral particles in kidney tissue by electron microscopy has been questioned given the resemblance of these particles to other cellular structures, such as clathrin-coated vesicles177, and the fact that several studies have been unable to confirm the presence of virus in the kidney178–181. Moreover, studies of SARS-CoV-2 in biological fluids have rarely shown viral shedding in the urine158, raising the possibility that a reservoir of SARS-CoV-2 might not exist in the kidney. Despite these discrepancies and unknowns, one elegant study has documented enrichment of SARS-CoV-2 RNA in glomerular epithelial and endothelial cells by in situ hybridization176. In line with this finding, active viral replication in different tissues, including kidney tissue, was found in a subset of patients with COVID-19 (ref.182), although it is still not clear whether this renal active replication contributes to the viral burden in the body.
As described earlier, innate and adaptive immune responses contribute to the pathogenesis of COVID-19. Some evidence exists to suggest that modulation of these processes in patients with kidney failure may have consequences for disease outcomes. Despite some early small, uncontrolled studies showing the opposite183,184, a report from Japan showed substantially higher mortality among patients on maintenance haemodialysis with COVID-19 than among non-dialysis patients, although this difference in mortality could be due to the older age of patients on haemodialysis185. Two further studies from the UK and Italy also reported significantly higher mortality among patients on haemodialysis than among the non-dialysis population162,186. In the Italian study, ARDS was probably the cause of death among hospitalized patients on haemodialysis162.
Therapeutic considerations
Huge effort has gone into developing an effective vaccine against SARS-CoV-2. However, despite the existence of more than 130 vaccine candidates currently under investigation187, our current understanding of SARS-CoV-2 biology and knowledge gained during the SARS and MERS epidemics suggests that the successful development of a vaccine will be challenging188. Until a safe and effective vaccine is available, it is imperative that alternative therapeutic strategies are identified that can be used during the current phase of the pandemic. From this point of view, understanding of the pathogenic mechanisms of COVID-19 offers important opportunities to identify and repurpose therapeutics that are already available to attenuate viral attachment and replication, hyperinflammation and the mechanisms underlying endothelial damage, blood clotting and systemic microangiopathy.
Antiviral therapies
The first week following SARS-CoV-2 infection — during which the virus enters alveolar cells and starts replicating in the lungs — represents a critical time for blocking the infection using antiviral agents. Of the repurposed antivirals investigated to date, the adenosine analogue remdesivir was initially identified as a promising therapy to induce premature termination of SARS-CoV-2 RNA replication by competing with endogenous nucleotides for the viral RdRp189. Despite initial, encouraging results190, the first placebo-controlled trial in COVID-19 failed to demonstrate a beneficial effect of remdesivir191. However, the US FDA granted emergency use authorization for remdesivir when review of the topline data from an ongoing randomized, double-blind, placebo-controlled trial192 showed that patients with COVID-19 who received remdesivir recovered 30% more quickly than those who had received placebo, although no statistical difference in mortality was found between the two study arms193. One major obstacle to the development of effective nucleotide-based drugs is our lack of understanding of the molecular mechanisms underlying substrate recognition during SARS-CoV-2 replication. New studies that have provided insights into the structural basis of RNA replication by the RdRp complex194 and the residues that are critical for nucleotide recognition and excision189 may aid the identification of more effective and targeted nucleotide-based therapies. The combination of HIV protease inhibitors lopinavir–ritonavir has also been investigated for their ability to treat COVID-19. However, these drugs specifically inhibit the HIV aspartyl protease (retropepsin), an enzyme that is essential HIV virion maturation195. Consistent with this high selectivity, a randomized, controlled, open-label trial reported no differences between treatment with lopinavir–ritonavir and standard care in time to clinical improvement196. Given the lack of specific SARS-CoV-2 protease inhibitors, preclinical studies are underway to develop more effective antiviral treatments for COVID-19, including a specific inhibitor of the SARS-CoV-2 protease Mpro (also called 3CLpro)197, although it may be some time before these novel targeted drugs are available for use in humans.
The finding that most repurposed antivirals are ineffective in COVID-19 is of utmost importance for clinical practice. Owing to their strong drug interactions, most antivirals cannot be administered in combination with life-saving drugs for other medical conditions. For example, lopinavir–ritonavir cannot be given in combination with immunosuppressive drugs198,199, which has consequences for specific patient groups such as kidney transplant recipients.
The anti-malaria drug hydroxychloroquine has also been investigated for its ability to inhibit viral infections200. However, available evidence does not support a beneficial effect of hydroxychloroquine over standard therapy in treating mild-to-moderate disease201, nor as a post-exposure prophylaxis strategy202. Moreover, two clinical trials have shown that hydroxychloroquine treatment poses serious safety concerns, increasing the risk of cardiovascular death by more than twofold203,204. Given the lack of definitive proof of efficacy for any of these drugs in patients with COVID-19, caution is warranted, particularly when the use of these compounds prevents the use of other, life-saving treatments for concurrent conditions.
Targeting hyperinflammation
In patients with adult-onset secondary haemophagocytic lymphohistiocytosis, early intervention with corticosteroids is of utmost importance for patient survival205. Corticosteroids exert anti-inflammatory effects by repressing a plethora of pro-inflammatory factors206, and prompt intervention with these drugs could be crucial for controlling the early stages of immune-mediated lung injury in COVID-19. Based on experience gained with previous coronavirus outbreaks, the use of high-dose corticosteroids in COVID-19 was initially discouraged because of the potential risk of secondary infections, long-term complications and prolonged virus shedding207. However, early reports from China indicated that treatment with methylprednisolone decreased the risk of death of patients with COVID-19 (ref.208) — a finding that is supported by other reports, indicating that a low to moderate dose of corticosteroids for a short period could be effective during the early stages of the disease209,210. In randomized case–control studies in adult patients with severe community-acquired pneumonia, early and short-term (from 5 to 7 days) corticosteroid therapy and intravenous administration of immunoglobulin after hospital admission reduced morbidity and rates of treatment failure211–213. A preliminary study found that an early, short course of methylprednisolone therapy improved outcomes of adult patients with COVID-19 with moderate or severe disease214. In a prospective meta-analysis of clinical trials of critically ill patients with COVID-19, use of systemic corticosteroids was associated with lower 28-day all-cause mortality compared with usual care or placebo215. A further study also documented an association between use of adjunctive steroid therapy plus intravenous immunoglobulin and recovery of cardiac function in children with MIS-C associated with SARS-CoV-2 infection122. A large randomized controlled trial showed that dexamethasone reduced 28-day mortality in severely ill patients who were receiving either invasive mechanical ventilation or oxygen alone at randomization, but not among those receiving no respiratory support216. These findings are encouraging since dexamethasone treatment is also highly beneficial in patients with pre-existing chronic kidney disease217,218.
Given the role of immune system hyperactivation and cytokines in the pathogenesis of COVID-19, the potential benefits of more targeted therapies should also be considered. For example, the observed suppression of interferon responses following SARS-CoV-2 infection suggests that interferon supplementation may be a potential therapeutic approach for COVID-19, as it is for other infectious diseases such as hepatitis. However, caution must be taken with this approach given the important immunomodulatory properties of interferon therapy, since interferon supplementation could induce or aggravate pre-existing autoimmune diseases or induce life-threatening complications, such as kidney and/or cardiac impairment or pulmonary interstitial fibrosis219. In addition, the finding that interferon stimulates expression of ACE2 in epithelial tissues indicates that interferon therapy could potentially promote virus entry into host cells17,18.
The first drug used to target specific components of the hyperactivated immune response in COVID-19 was the IL-6 inhibitor tocilizumab. Two retrospective reviews found that tocilizumab treatment was associated with a significant reduction in markers of inflammation and in the need for ventilatory support, and with improvements in radiological findings in patients with severe disease220,221. However, phase III trials of IL-6 inhibitors192,222, including tocilizumab and sarilumab, were inconclusive and their use is currently not recommended223. A small case series reported the beneficial effects of another targeted therapy — the IL-1 receptor antagonist anakinra — in severe patients with COVID-19 (ref.224). However, adequately powered, randomized controlled trials are still needed, and at least 16 different clinical trials are currently underway to assess the effects of other anti-interleukin drugs. Importantly, anti-interleukin agents will only limit propagation of the hypercytokinaemia but are unlikely to resolve the underlying cause of immune system dysregulation in COVID-19.
The clinical characteristics of lymphocytopenia and exhausted circulating T cells indicate that immunotherapies could represent another potential therapeutic option for COVID-19. As in cancer, PD1-mediated inhibitory signals contribute to T cell exhaustion and inhibit T cell effector functions in the early stages of viral infection225,226. Thus, PD1 blockade with the use of immune checkpoint inhibitors may enhance the effector function of T cells and stimulate viral clearance225,226. In patients with lung cancer, PD1-targeted immunotherapy increased proliferation of PD1+ CD8+ T cells and was associated with positive clinical outcomes227. Whether such PD1-targeted immunotherapies will also be effective in redirecting the cytotoxic activity of CD8+ T cells in COVID-19 is unknown, but will be assessed in three ongoing clinical trials underway to assess the efficacy of anti-PD1 antibodies in severely ill patients with COVID-19 (refs228–230). A more intriguing possibility is whether combining anti-PD1 antibodies with anti-cytotoxic T lymphocyte antigen 4 (CTLA4) blockade might increase the efficacy of the respective monotherapies. Indeed, combination therapy can promote switching of phenotypically exhausted CD8+ T cells towards an activated effector CD8+ T cell phenotype231, suggesting that this approach may have the potential to promote recovery of the antiviral activity of T cells against SARS-CoV-2. In line with this hypothesis, treatment of COVID-19 patients with a synthetic biological immune response modifier, thymosin alpha 1 (ref.232), effectively restored CD8+ and CD4+ T cell counts, as well as reversed T cell exhaustion by reducing expression of the T cell exhaustion markers PD1 and TIM-3 on CD8+ T cells233. Compared with untreated patients, supplementation of thymosin alpha 1 in COVID-19 patients with severe lymphocytopenia was associated with a significantly lower death rate and reduced need for invasive mechanical ventilation233.
Targeting endothelial dysfunction
In light of the major contribution of widespread coagulopathy and thrombosis to poor outcomes in COVID-19, the use of prophylactic doses of low-molecular-weight heparin (LMWH) has emerged as a valuable strategy for preventing the development of venous thromboembolisms. In a report from China published in preprint form, administration of LMWH to patients with COVID-19 improved indices of coagulation dysfunction and reduced levels of IL-6 (ref.234). As recommended by the International Society on Thrombosis and Haemostasis, the management of COVID-19- associated coagulopathy with LMWH should be accompanied by close monitoring of platelet counts, fibrinogen and the prothrombin time for the early diagnosis of bleeding235, although bleeding seems to be rare in patients with COVID-19 (ref.236). A more promising approach than LMWH may be the serine protease inhibitor nafamostat mesylate, which is under investigation in four different clinical trials237–240. This agent has short-acting anticoagulant effects and can limit viral infection and replication by inhibiting TMPRSS2 (ref.241). That fact TMPPRSS2 is a valuable therapeutic target in COVID-19 has been shown by an open-label randomized clinical trial documenting that early administration of oral bromhexine, a TMPPRSS2 inhibitor, reduces ICU admission, intubation and mortality242. Alternatively, inhibition of platelet aggregation may represent another therapeutic avenue for patients with COVID-19. In support of this proposal, a small clinical trial conducted in China and published in preprint form showed that prophylactic anti-coagulation therapy supplemented by the anti-platelet drug dipyridamole prevented the progression of COVID-19 to severe stages of the disease by significantly increasing platelet and lymphocyte counts and decreasing D-dimer levels243. Notably, through virtual screening of an approved drug library, dipyridamole was also found to suppress SARS-CoV-2 replication in vitro243. Despite these promising findings, available clinical data suggest that anti-coagulation therapies might have only a partial effect on coagulopathy and thromboembolism in severe forms of the disease244. For this reason, additional therapeutic interventions are needed to attenuate endothelial cell dysfunction and thromboembolic complications145.
Of the available drugs, inhibitors of the complement system may have the most potential to preserve endothelial cell function and limit thromboembolic complications. A case report of a patient with COVID-19-associated ARDS and pneumonia showed that treatment with a C3 inhibitor was safe and associated with a favourable course245, suggesting that C3 blockade may have broad anti-inflammatory potential owing to the upstream positioning of C3 in innate immune cascade signalling. However, C3 inhibition prevents activation of the entire complement cascade, halting the opsonization of viral particles by C3b. Indeed, studies in knockout mice have demonstrated that C3 is required for protection from the highly pathogenic H5N1 influenza infection and proper viral clearance246, suggesting that C3 inhibition may have undesirable consequences.
Among other available inhibitors of the complement cascade247, the anti-C5 monoclonal antibody eculizumab could be the most valuable tool for preventing complement activation in COVID-19. C5 inhibitors block the formation of the terminal products that are common to the three complement pathways, while still allowing C3 activation and viral opsonization137. A 2015 study in an experimental model of sepsis found that eculizumab significantly lowered consumptive coagulopathy by inhibiting the formation of C5b–9, and by preventing inflammation and widespread intravascular coagulation248. Notably, C5 inhibition did not interfere with phagocytosis or the intracellular digestion of pathogens, ensuring the efficient removal of bacteria248. A clinical trial has now been initiated to evaluate the therapeutic effect of eculizumab in patients with COVID-19 (ref.249). C5a may represent an alternative therapeutic target. C5a stimulates cytokine release, increases vascular permeability and induces the upregulation of adhesion molecules that favour leukocyte extravasation and transmigration into lung parenchyma250 — pathological events that are all observed in severe cases of COVID-19. Interestingly, a preliminary study published in preprint form reported a promising immunosuppressive effect of the anti-C5a antibody BDB-001 in two severely ill patients with COVID-19 (ref.142). Of note, C5 blockade has demonstrated benefit in various kidney diseases, including C3 glomerulopathy and atypical haemolytic uraemic syndrome251,252, suggesting that targeting complement might protect both the lung and kidney in COVID-19.
Cautious oxygen support
Oxygen therapy has been used as first-line supportive treatment for breathing dysfunction and dyspnoea in patients with COVID-19 with pneumonia and ARDS. However, despite fulfilling most of the criteria of classic ARDS, COVID-19-associated pneumonia may be a discrete disease characterized by two distinct phenotypes253,254. In the early phase of the disease, the severe hypoxaemia induced by ARDS is often associated with near normal respiratory system compliance253,254. In these patients, gentle oxygen therapy — such as high flow nasal cannula, continuous positive airway pressure or non-invasive ventilation — is recommended253,254. During this phase, a patient’s condition may remain unchanged for a period of time and then either improve or suddenly deteriorate to a more severe disease stage. In the latter situation, the resulting ARDS phenotype is similar to that of acute fibrinous and organizing pneumonia — a rare form of acute lung injury that differs from the diffuse alveolar damage found in classic forms of ARDS255. The obliterative pattern of lung injury in these severe cases of COVID-19 raises concerns about the potential deleterious effect of invasive mechanical ventilation at a high level of positive end-expiratory pressure (PEEP)255,256. Indeed, if vascular alterations, such as pulmonary vasoconstriction, pulmonary emboli or thrombi, are the major driver of hypoxaemia in COVID-19, then increasing PEEP levels might not improve oxygenation but, conversely, might contribute to lung damage29,30,257. For this reason, oxygen therapy should be personalized in each patient because the response to PEEP differs on the basis of individual respiratory mechanics29,30,257. This issue is of particular relevance for nephrologists, given that mechanical ventilation is also associated with a threefold increase in the risk of AKI, and that patients with combined respiratory failure and AKI have a mortality of 60–80% in the ICU setting168. On the other hand, acute fibrinous and organizing pneumonia is a cortico-sensitive pathology and, as described above, appropriate corticosteroid therapy should be established for the clinical management of severely ill patients to avoid unnecessary ventilation.
Convalescent plasma
One virus-targeted approach under investigation is passive antibody therapy. This approach involves the infusion of sera from convalescent patients into infected patients with the aim of transferring neutralizing SARS-CoV-2 antibodies to patients with active disease258. To date, little is known about the efficacy of this treatment as most data are derived from uncontrolled case studies of patients with COVID-19. All case reports of patients with severe COVID-19 have described a rapid improvement in clinical symptoms over the course of 3 days, with resorption of lung lesions within a week, as revealed by radiological examination259–261. A small clinical trial of six critically ill patients with COVID-19 treated with convalescent plasma reported elimination of SARS-CoV-2 RNA within 3 days of therapeutic plasma infusion, although the mortality of these patients did not differ from that of 15 patients who received placebo262. Moreover, a larger randomized clinical trial of 103 patients with COVID-19 (ref.263), as well as a preprint report from India264, showed no significant clinical improvements after convalescent plasma therapy. Future studies should focus on developing approaches to isolating SARS-CoV-2 neutralizing antibodies from convalescent plasma, which may increase the efficacy of this treatment. In addition, two important factors mentioned earlier need to be taken into consideration when performing plasma collection. First, if protective neutralizing antibodies are short-lived, plasma should be collected early after recovery from COVID-19. Second, patients with more severe disease have higher antibody levels than patients with asymptomatic or mild disease; however, the extent of protection provided by antibodies in patients with severe disease is unclear, and it might therefore be counterproductive to select plasma donors based only on antibody levels. As an alternative approach, a number of neutralizing human monoclonal antibodies have been developed, some of which are entering phase I clinical trials94,265–270. If successful, such an approach would avoid the limitations and the time-consuming procedures required to obtain plasma or to purify polyclonal antibodies from convalescent patients.
Conclusions
Available evidence demonstrates that SARS-CoV-2 infection induces immune dysfunction, widespread endothelial injury, complement-associated coagulopathy and systemic microangiopathy. The clinical outcome of COVID-19 seems to be profoundly dependent on the individual response of the host. Although the contribution of differences in viral loads to clinical outcome cannot be excluded271, it is possible that predisposing genetic or biological factors may modulate the degree of disease severity. Additionally, socioeconomic conditions and structural racism may contribute to poor disease outcomes in some regions, as demonstrated by disproportionately poor outcomes among minority ethnic groups, including Black, Asian and Pakistani populations272. Improved insights into the mechanisms underlying predisposition to adverse disease outcomes might aid the identification of new strategies and targets for therapy, as well as identify ways to improve outcomes for susceptible individuals and those from minority ethnic backgrounds. However, the development of new drugs is a long process and may not be useful for dealing with the immediate challenge posed by the current COVID-19 pandemic.
Currently, no vaccines or effective antiviral drugs are available for COVID-19, although many are in development and some may become available soon. The paucity of available specific therapies has stimulated the search for existing drugs that can be repurposed for COVID-19. So far, these drugs have included steroids, several anti-interleukin drugs, complement inhibitors and agents that target coagulation and endothelial dysfunction. However, some of these next-generation targeted therapies are not devoid of adverse effects, which can be serious and sometimes life-threatening. The identification of inexpensive and effective drugs — including low-dose steroids and LWMH — is of utmost importance to prevent exacerbation of inflammatory and thrombotic processes and halt disease progression. Given the spectrum of pathogenic mechanisms involved in the development of severe COVID-19, ranging from immune hyperactivation to thromboembolic complications, it is unlikely that a single individual treatment will be effective. Although corticosteroids may be able to target most of these pathogenic pathways, the multifactorial pathogenic nature of the disease indicates that multiple avenues of treatment might be required and major effort should therefore be invested to determine the optimal timing and combinations in which these drugs should be administered to maximize their efficacy in severely ill patients with COVID-19.
Acknowledgements
The authors thank Kerstin Mierke, Istituto di Ricerche Mario Negri IRCCS, Italy, for help with English language editing of the manuscript before submission and Antonella Piccinelli, Istituto di Ricerche Mario Negri IRCCS, Italy, for preparing the figures before submission. L.P. is a recipient of the Career Development Program from Fondazione Aiuti per la Ricerca sulle Malattie Rare (ARMR), Bergamo, Italy. We are also grateful to Fondazione Aiuti per la Ricerca sulle Malattie Rare (ARMR) for research support for our studies on COVID-19 pathogenesis. L.F.P.N. and L.R. are supported by core funds at the Singapore Immunology Network (SIgN) through the Biomedical Medical Research Council (BMRC), A*STAR.
Glossary
- Betacoronavirus
One of four genera (α, β, γ and δ) of enveloped, positive-strand RNA viruses that infects humans and mammals, causing respiratory diseases that can be mild or severe.
- RNA-dependent RNA polymerase
(RdRp). An enzyme encoded by viral RNA that catalyses the synthesis of new RNA strands complementary to the initial RNA template included in the viral capsid.
- Incubation period
The number of days between the initial exposure to a pathogen and the day in which the infected individual experiences the first symptoms of the disease.
- Anosmia
Complete loss of the ability to detect one or more smells; anosmia can be temporary, as in the case of COVID-19, or permanent such as occurs in certain neurological conditions.
- Ageusia
Complete loss of taste on the tongue, particularly the inability to perceive sweetness, sourness, bitterness and saltiness; ageusia can be temporary, as in the case of COVID-19, or permanent such as occurs in certain neurological conditions.
- Pattern recognition receptors
(PRRs). Host sensors that detect pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) generated during infections or injury.
- Cytokine storm
Also called hypercytokinaemia. An excessive and uncontrolled release of pro-inflammatory molecules by the cellular components of the innate immune system in response to a number of infectious and non-infectious aetiologies that can lead to multi-organ damage and failure.
- VH gene
The heavy-chain-variable region (VH) gene gene is located along with diversity (D), joining (J) and constant (C) genes at three primary loci in the human genome and undergoes somatic rearrangements for the biosynthesis of the heavy (H) or light (L) chain of IgG.
- Glucuronidases
A class of enzyme that includes beta-glucuronidases — a glycosidases that catalyse the breakdown of complex carbohydrates by hydrolysing β-d-glucuronic acid residues from the non-reducing end of glycosaminoglycans, such as heparan sulfate.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Nephrology thanks C. Garlanda, C. Ince and A. Salama for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;6:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H, et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Q, et al. Structural and biochemical characterization of nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan, Y., Shang, J., Graham, R., Baric, R. S. & Li, F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 10.1128/JVI.00127-20 (2020). [DOI] [PMC free article] [PubMed]
- 13.Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23:101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perico L, Benigni A, Remuzzi G. Should COVID-19 concern nephrologists? Why and to what extent? The emerging impasse of angiotensin blockade. Nephron. 2020;144:213–221. doi: 10.1159/000507305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience. 2020;23:101160. doi: 10.1016/j.isci.2020.101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegler CGK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou YJ, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao X. COVID-19: immunopathology and its implications for therapy. Nat.Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguiar D, Lobrinus JA, Schibler M, Fracasso T, Lardi C. Inside the lungs of COVID-19 disease. Int. J. Legal Med. 2020;34:1271–1274. doi: 10.1007/s00414-020-02318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernazza A, et al. Early histologic findings of pulmonary SARS-CoV-2 infection detected in a surgical specimen. Virchows Arch. 2020 doi: 10.1007/s00428-020-02829-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan E, et al. COVID-19-associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir. Med. 2020;8:816–821. doi: 10.1016/S2213-2600(20)30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marini JJ, Gattinoni L. Management of COVID-19 respiratory distress. JAMA. 2020;323:2329–2330. doi: 10.1001/jama.2020.6825. [DOI] [PubMed] [Google Scholar]
- 31.Patel A, et al. New onset anosmia and ageusia in adult patients diagnosed with SARS-CoV-2. Clin. Microbiol. Infect. 2020;26:1236–1241. doi: 10.1016/j.cmi.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero-Sánchez CM, et al. Neurologic manifestations in hospitalized patients with COVID-19: The ALBACOVID registry. Neurology. 2020;95:e1060–e1070. doi: 10.1212/WNL.0000000000009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dockery DM, Rowe SG, Murphy MA, Krzystolik MG. The ocular manifestations and transmission of COVID-19: recommendations for prevention. J. Emerg. Med. 2020;59:137–140. doi: 10.1016/j.jemermed.2020.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suresh Kumar VC, et al. Novelty in the gut: a systematic review and meta-analysis of the gastrointestinal manifestations of COVID-19. BMJ Open Gastroenterol. 2020;7:e000417. doi: 10.1136/bmjgast-2020-000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J J Jin X, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi S, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020;5:802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang R, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grajewski RS, et al. Letter to the editor: a missing link between SARS-CoV-2 and the eye?: ACE2 expression on the ocular surface. J. Med. Virol. 2020 doi: 10.1002/jmv.26136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathew D, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27:863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vabret N, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park A, Iwasaki A. Type I. and type III interferons — induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoms M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang H, et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez C, Horner SM. MAVS coordination of antiviral innate immunity. J. Virol. 2015;89:6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trouillet-Assant S, et al. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020;146:206–208.e2. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Usmani GN, Woda BA, Newburger PE. Advances in understanding the pathogenesis of HLH. Br. J. Haematol. 2013;161:609–622. doi: 10.1111/bjh.12293. [DOI] [PubMed] [Google Scholar]
- 56.McGonagle D, Sharif K, O’Regan A, Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun. Rev. 2020;19:102537. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern. Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen SF, Ho Y-C. SARS-CoV-2: a storm is raging. J. Clin. Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, et al. Aveolar macrophage activation and cytokine storm in the pathogenesis of severe COVID-19. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 61.B Bost P, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giordano, S., Kramer, P., Darley-Usmar, V. M., & White C. R. in Apolipoprotein Mimetics in the Management of Human Disease (eds Anantharamaiah, G., Goldberg, D.)(Adis, Springer International Publishing, 2015).
- 64.Li J, et al. Virus-host interactome and proteomic survey of PMBCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Z Zhou Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu, J. et al. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage. Preprint at medRxiv10.1101/2020.02.10.20021584 (2020).
- 68.Carissimo, G. et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early prognostic marker for severe COVID-19. Preprint at bioRxiv10.1101/2020.06.11.147389 (2020). [DOI] [PMC free article] [PubMed]
- 69.Barnes BJ, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan L, et al. Validation of predictors of disease severity and outcomes in COVID-19 patients: a descriptive and retrospective study. Med. 2020 doi: 10.1016/j.medj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng M, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen, Y. et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. Preprint at medRxiv10.1101/2020.03.27.20045427 (2020).
- 76.Bellesi S, et al. Increased CD95 (Fas) and PD-1 expression in peripheral blood T lymphocytes in COVID-19 patients. Br. J.Haematol. 2020 doi: 10.1111/bjh.17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qin C, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bermejo-Martin JF, et al. Lymphopenic community acquired pneumonia as signature of severe COVID-19 infection. J. Infect. 2020;80:e23–e24. doi: 10.1016/j.jinf.2020.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bermejo-Martin JF, et al. Lymphopenic community acquired pneumonia (L-CAP), an immunological phenotype associated with higher risk of mortality. EBioMedicine. 2017;24:231–236. doi: 10.1016/j.ebiom.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Menéndez R, et al. simultaneous depression of immunological synapse and endothelial injury is associated with organ dysfunction in community-acquired pneumonia. J. Clin. Med. 2019;8:1404. doi: 10.3390/jcm8091404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng Y, et al. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020 doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 84.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;217:e20200206. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 86.Cassotta A, et al. Deciphering and predicting CD4+ T cell immunodominance of influenza virus hemagglutinin. J. Exp. Med. 2020;217:e20200206. doi: 10.1084/jem.20200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mateus J, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;26:845–848. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long QX, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 89.Wang Y-T, et al. Spiking pandemic potential: structural and immunological aspects of SARS-CoV-2. Trends Microbiol. 2020;28:605–618. doi: 10.1016/j.tim.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan M, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu Y, et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chandrashekar A, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deng W, et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zost, S. J. et al. Potently neutralizing human antibodies that block SARS-CoV-2 receptor binding and protect animals. Preprint at bioRxiv10.1101/2020.05.22.111005 (2020).
- 95.Long Q-X, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 96.Seow, J. et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. Preprint at medRxiv10.1101/2020.07.09.20148429 (2020).
- 97.Ibarrondo FJ, et al. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;83:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gudbjartsson DF, et al. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seydoux E, et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Juno JA, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 101.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wilk AJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Woodruff, M. et al. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. Preprint at medRxiv10.1101/2020.04.29.20083717 (2020).
- 105.Amrun SN, et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine. 2020;58:102911. doi: 10.1016/j.ebiom.2020.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu, F. et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. Preprint at medRxiv10.1101/2020.03.30.20047365 (2020).
- 107.Liu L, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu H, et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020;56:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hotez PJ, Corry DB, Bottazzi ME. COVID-19 vaccine design: the Janus face of immune enhancement. Nat. Rev. Immunol. 2020;20:347–348. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Olson JD. D-dimer: an overview of hemostasis and fibrinolysis, assays, and clinical applications. Adv. Clin. Chem. 2015;69:1–46. doi: 10.1016/bs.acc.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 111.Yau JW, Teoh H, Verma S. Endothelial cell control of thrombosis. BMC Cardiovasc. Disord. 2015;15:130. doi: 10.1186/s12872-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reitsma S, Slaaf DW, Vink H, van Zandvoort MAMJ, oude Egbrink MGA. The endothelial glycocalyx: composition, functions, and visualization. Pflugers Arch. 2007;454:345–359. doi: 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 2020;20:389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Varga Z, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oxley TJ, et al. Large-vessel stroke as a presenting feature of covid-19 in the Young. N. Engl. J. Med. 2020;382:e60. doi: 10.1056/NEJMc2009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng. M. European Doctors Warn Rare Kids’ Syndrome May Have Virus Tie. (Associated Press, 2020).
- 117.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting Enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Riphagen S, Gomez X, Gonzalez-Martinez C, Wilkinson N, Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Diorio C, et al. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 2020 doi: 10.1172/JCI140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cheung EW, et al. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA. 2020;324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Verdoni L, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zahra B, et al. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 123.Monteil V, et al. Inhibition of SARS-CoV-2 Infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garvin MR, et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. eLife. 2020;9:e59177. doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmaier AH. The contact activation and kallikrein/kinin systems: pathophysiologic and physiologic activities. J. Thromb. Haemost. 2016;14:28–39. doi: 10.1111/jth.13194. [DOI] [PubMed] [Google Scholar]
- 127.Schmaier AH. A novel antithrombotic mechanism mediated by the receptors of the kallikrein/kinin and renin–angiotensin systems. Front. Med. 2016;3:61. doi: 10.3389/fmed.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stavrou EX, et al. Reduced thrombosis in Klkb1−/− mice is mediated by increased Mas receptor, prostacyclin, Sirt1, and KLF4 and decreased tissue factor. Blood. 2015;125:710–719. doi: 10.1182/blood-2014-01-550285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sodhi CP, et al. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am. J. Physiol. Lung Cell Mol. Physiol. 2018;314:L17–L31. doi: 10.1152/ajplung.00498.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chung MK, et al. SARS-CoV-2 and ACE2: the biology and clinical data settling the ARB and ACEI controversy. EBioMedicine. 2020;58:102907. doi: 10.1016/j.ebiom.2020.102907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fang C, et al. Angiotensin 1-7 and Mas decrease thrombosis in Bdkrb2−/− mice by increasing NO and prostacyclin to reduce platelet spreading and glycoprotein VI activation. Blood. 2013;121:3023–3032. doi: 10.1182/blood-2012-09-459156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Veerdonk FLvande, et al. Outcomes associated with use of a kinin B2 receptor antagonist among patients with COVID-19. JAMA Netw. Open. 2020;3:e2017708–e2017708. doi: 10.1001/jamanetworkopen.2020.17708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van de Veerdonk FL, et al. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. eLife. 2020;9:e57555. doi: 10.7554/eLife.57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roche JA, Roche R. A hypothesized role for dysregulated bradykinin signaling in COVID-19 respiratory complications. FASEB J. 2020;34:7265–7269. doi: 10.1096/fj.202000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buzhdygan, T. P. et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in vitro models of the human blood–brain barrier. Preprint at bioRxiv10.1101/2020.06.15.150912 (2020). [DOI] [PMC free article] [PubMed]
- 136.Martini R. The compelling arguments for the need of microvascular investigation in COVID-19 critical patients. Clin. Hemorheol. Microcirc. 2020;75:27–34. doi: 10.3233/CH-200895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Noris M, Benigni A, Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98:314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song WC, FitzGerald GA. COVID-19, microangiopathy, hemostatic activation, and complement. J. Clin. Invest. 2020;130:3950–3953. doi: 10.1172/JCI140183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gralinski LE, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9:e01753–18. doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ip WKE, et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191:1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang Y, et al. Blockade of the C5a-C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg. Microbes Infect. 2018;7:77. doi: 10.1038/s41426-018-0063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao, T. et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. Preprint at medRxiv10.1101/2020.03.29.20041962 (2020).
- 143.Yang YH, et al. Autoantibodies against human epithelial cells and endothelial cells after severe acute respiratory syndrome (SARS)-associated coronavirus infection. J. Med. Virol. 2005;77:1–7. doi: 10.1002/jmv.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Messner CB, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11:11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Risitano AM, et al. Complement as a target in COVID-19? Nat. Rev. Immunol. 2020;20:343–344. doi: 10.1038/s41577-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McGonagle D, O’Donnell JS, Sharif K, Emery P, Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fox, S. E. et al. Pulmonary and cardiac pathology in Covid-19: the first autopsy series from New Orleans. Preprint at medRxiv10.1101/2020.04.06.20050575 (2020).
- 148.Carsana, L. et al. Pulmonary post-mortem findings in a large series of COVID-19 cases from Northern Italy. Preprint at medRxiv10.1101/2020.04.19.20054262 (2020). [DOI] [PMC free article] [PubMed]
- 149.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Danzi GB, Loffi M, Galeazzi G, Gherbesi E. Acute pulmonary embolism and COVID-19 pneumonia: a random association? Eur. Heart J. 2020;41:1858. doi: 10.1093/eurheartj/ehaa254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Llitjos JF, et al. High incidence of venous thromboembolic events in anticoagulated severe COVID-19 patients. J. Thromb. Haemost. 2020;18:1743–1746. doi: 10.1111/jth.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zuckier LS, Moadel RM, Haramati LB, Freeman L. Diagnostic evaluation of pulmonary embolism during the COVID-19 pandemic. J. Nucl. Med. 2020;61:630–631. doi: 10.2967/jnumed.120.245571. [DOI] [PubMed] [Google Scholar]
- 153.Beun R, Kusadasi N, Sikma M, Westerink J, Huisman A. Thromboembolic events and apparent heparin resistance in patients infected with SARS-CoV-2. Int. J. Lab. Hematol. 2020;42:19–20. doi: 10.1111/ijlh.13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Levi M, van der Poll T. Coagulation and sepsis. Thromb. Res. 2017;149:38–44. doi: 10.1016/j.thromres.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 155.Radermecker C, et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217:e20201012. doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cheng Y, et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97:829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gabarre P, et al. Acute kidney injury in critically ill patients with COVID-19. Intensive Care Med. 2020;46:1339–1348. doi: 10.1007/s00134-020-06153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chan L, et al. AKI in hospitalized patients with COVID-19. J. Am. Soc. Nephrol. 2020 doi: 10.1681/ASN.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ali H, et al. Survival rate in acute kidney injury superimposed COVID-19 patients: a systematic review and meta-analysis. Ren. Fail. 2020;42:393–397. doi: 10.1080/0886022X.2020.1756323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lim J-H, et al. Fatal outcomes of COVID-19 in patients with severe acute kidney injury. J. Clin. Med. 2020;9:1718. doi: 10.3390/jcm9061718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Division of Nephrology, Columbia University Vagelos College of Physicians Disaster Response to the COVID-19 Pandemic for Patients with Kidney Disease in New York City. J. Am. Soc. Nephrol. 2020;31:1371–1379. doi: 10.1681/ASN.2020040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alberici F, et al. Management of patients on dialysis and with kidney transplantation during the SARS-CoV-2 (COVID-19) pandemic in Brescia, Italy. Kidney Int. Rep. 2020;5:580–585. doi: 10.1016/j.ekir.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Su H, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Magro C, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Diao, B. et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Preprint at medRxiv10.1101/2020.03.04.20031120 (2020).
- 166.Larsen CP, Bourne TD, Wilson JD, Saqqa O, Sharshir M. A. collapsing glomerulopathy in a patient with COVID-19. Kidney Int. Rep. 2020;5:935–939. doi: 10.1016/j.ekir.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Magoon S, et al. COVID-19–related glomerulopathy: a report of 2 cases of collapsing focal segmental glomerulosclerosis. Kidney Med. 2020;2:488–492. doi: 10.1016/j.xkme.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hepokoski ML, Malhotra A, Singh P, Crotty Alexander LE. Ventilator-induced kidney injury: are novel biomarkers the key to prevention? Nephron. 2018;140:90–93. doi: 10.1159/000491557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Durvasula R, Wellington T, McNamara E, Watnick S. COVID-19 and kidney failure in the acute care setting: our experience from Seattle. Am. J. Kidney Dis. 2020;76:4–6. doi: 10.1053/j.ajkd.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Naicker S, et al. The novel coronavirus 2019 epidemic and kidneys. Kidney Int. 2020;97:824–828. doi: 10.1016/j.kint.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Farkash EA, Wilson AM, Jentzen JM. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31:1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Abbate M, Rottoli D, Gianatti A. COVID-19 attacks the kidney: ultrastructural evidence for the presence of virus in the glomerular epithelium. Nephron. 2020;144:341–342. doi: 10.1159/000508430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu D, et al. Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: a study based on single-cell transcriptome analysis. Intensive Care Med. 2020;46:1114–1116. doi: 10.1007/s00134-020-06026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ye M, et al. Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: implications for albuminuria in diabetes. J. Am. Soc. Nephrol. 2006;17:3067–3075. doi: 10.1681/ASN.2006050423. [DOI] [PubMed] [Google Scholar]
- 175.Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020;10:317. doi: 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Puelles VG, et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miller SE, Goldsmith CS. Caution in identifying coronaviruses by electron microscopy. J. Am. Soc. Nephrol. 2020;31:2223–2224. doi: 10.1681/ASN.2020050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu H, et al. AKI and collapsing glomerulopathy associated with COVID-19 and APOL1 high-risk genotype. J. Am. Soc. Nephrol. 2020;31:1688–1695. doi: 10.1681/ASN.2020050558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kudose S, et al. Kidney biopsy findings in patients with COVID-19. J. Am. Soc. Nephrol. 2020;31:1959–1968. doi: 10.1681/ASN.2020060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Santoriello D, et al. Postmortem kidney pathology findings in patients with COVID-19. J. Am. Soc. Nephrol. 2020;31:2158–2167. doi: 10.1681/ASN.2020050744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sharma P, et al. COVID-19-associated kidney injury: a case series of kidney biopsy findings. J. Am. Soc. Nephrol. 2020;31:1948–1958. doi: 10.1681/ASN.2020050699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hanley B, et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ma, Y. et al. 2019 novel coronavirus disease in hemodialysis (HD) patients: Report from one HD center in Wuhan, China. Preprint at medRxiv10.1101/2020.02.24.20027201 (2020).
- 184.Wang R, et al. COVID-19 in hemodialysis patients: a report of 5 cases. Am. J. Kidney Dis. 2020;76:141–143. doi: 10.1053/j.ajkd.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kikuchi K, et al. COVID-19 in dialysis patients in Japan: current status and guidance on preventive measures. Ther. Apher. Dial. 2020;24:361–365. doi: 10.1111/1744-9987.13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Corbett RW, et al. Epidemiology of COVID-19 in an urban dialysis center. J. Am. Soc. Nephrol. 2020;31:1815–1823. doi: 10.1681/ASN.2020040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.World Health Organization. Draft landscape of COVID-19 candidate vaccines https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (2020).
- 188.Begum J, et al. Challenges and prospects of COVID-19 vaccine development based on the progress made in SARS and MERS vaccine development. Transbound Emerg. Dis. 2020 doi: 10.1111/tbed.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shannon A, et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antivir. Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu Z, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04280705 (2020).
- 193.Gilead. Gilead’s Investigational Antiviral Remdesivir Receives U.S. Food and Drug Administration Emergency Use Authorization for the Treatment of COVID-19. https://www.gilead.com/news-and-press/press-room/press-releases/2020/5/gileads-investigational-antiviral-remdesivir-receives-us-food-and-drug-administration-emergency-use-authorization-for-the-treatment-of-covid19 (2020).
- 194.Wang Q, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;82:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shen Y, Radhakrishnan ML, Tidor B. Molecular mechanisms and design principles for promiscuous inhibitors to avoid drug resistance: lessons learned from HIV-1 protease inhibition. Proteins. 2015;83:351–372. doi: 10.1002/prot.24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cao B, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gandolfini I, et al. COVID-19 in kidney transplant recipients. Am. J. Transpl. 2020;20:1941–1943. doi: 10.1111/ajt.15891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bartiromo M, et al. Threatening drug-drug interaction in a kidney transplant patient with Coronavirus Disease 2019 (COVID-19) Transpl. Infect. Dis. 2020;22:e13286. doi: 10.1111/tid.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vincent MJ, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cavalcanti AB, et al. Hydroxychloroquine with or without azithromycin in mild-to-moderate Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2019014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Boulware DR, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Magagnoli J, et al. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. Med. 2020 doi: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Borba MGS, et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw. Open. 2020;3:e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 205.Kleynberg RL, Schiller GJ. Secondary hemophagocytic lymphohistiocytosis in adults: an update on diagnosis and therapy. Clin. Adv. Hematol. Oncol. 2012;10:726–732. [PubMed] [Google Scholar]
- 206.Cruz-Topete D, Cidlowski JA. One hormone, two actions: anti- and pro-inflammatory effects of glucocorticoids. Neuroimmunomodulation. 2015;22:20–32. doi: 10.1159/000362724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wu C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:1–11. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Russell B, et al. Associations between immune-suppressive and stimulating drugs and novel COVID-19 — a systematic review of current evidence. Ecancermedicalscience. 2020;14:1022. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shang L, Zhao J, Hu Y, Du R, Cao B. On the use of corticosteroids for 2019-nCoV pneumonia. Lancet. 2020;395:683–684. doi: 10.1016/S0140-6736(20)30361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Blum CA, et al. Adjunct prednisone therapy for patients with community-acquired pneumonia: a multicentre, double-blind, randomised, placebo-controlled trial. Lancet. 2015;385:1511–1518. doi: 10.1016/S0140-6736(14)62447-8. [DOI] [PubMed] [Google Scholar]
- 212.Torres A, et al. Effect of corticosteroids on treatment failure among hospitalized patients with severe community-acquired pneumonia and high inflammatory response: a randomized clinical trial. JAMA. 2015;313:677–686. doi: 10.1001/jama.2015.88. [DOI] [PubMed] [Google Scholar]
- 213.Lee KY, Rhim J-W, Kang J-H. Early preemptive immunomodulators (corticosteroids) for severe pneumonia patients infected with SARS-CoV-2. Clin. Exp. Pediatr. 2020;63:117–118. doi: 10.3345/cep.2020.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fadel R, et al. Early short course corticosteroids in hospitalized patients with COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.The WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) working group Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA. 2020 doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.RECOVERY Collaborative Group. Horby P, et al. Dexamethasone in hospitalized patients with covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jacob KA, et al. Intraoperative high-dose dexamethasone and severe AKI after cardiac surgery. J. Am. Soc. Nephrol. 2015;26:2947–2951. doi: 10.1681/ASN.2014080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ponticelli C, Locatelli F. Glucocorticoids in the treatment of glomerular diseases: pitfalls and pearls. Clin. J. Am. Soc. Nephrol. 2018;13:815–822. doi: 10.2215/CJN.12991117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dusheiko G. Side effects of alpha interferon in chronic hepatitis C. Hepatology. 1997;26:112S–121S. doi: 10.1002/hep.510260720. [DOI] [PubMed] [Google Scholar]
- 220.Alattar R, et al. Tocilizumab for the treatment of severe coronavirus disease 2019. J. Med. Virol. 2020 doi: 10.1002/jmv.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Guaraldi G, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04341870 (2020).
- 223.Furlow B. COVACTA trial raises questions about tocilizumab’s benefit in COVID-19. Lancet Rheumatol. 2020;2:e592. doi: 10.1016/S2665-9913(20)30313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dimopoulos G, et al. Favorable anakinra responses in severe Covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell HostMicrobe. 2020;28:117–123.e1. doi: 10.1016/j.chom.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ahn E, et al. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl Acad. Sci. USA. 2018;115:4749–4754. doi: 10.1073/pnas.1718217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jin H-T, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl Acad. Sci. USA. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kamphorst AO, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl Acad. Sci. USA. 2017;114:4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04413838 (2020).
- 229.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04356508 (2020).
- 230.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04268537 (2020).
- 231.Wei SC, et al. Combination anti-CTLA-4 plus anti–PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc. Natl Acad. Sci. USA. 2019;116:22699–22709. doi: 10.1073/pnas.1821218116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Goldstein AL, Goldstein AL. From lab to bedside: emerging clinical applications of thymosin α1. Expert Opin. Biol. Ther. 2009;9:593–608. doi: 10.1517/14712590902911412. [DOI] [PubMed] [Google Scholar]
- 233.Liu Y, et al. Thymosin alpha 1 (Tα1) reduces the mortality of severe COVID-19 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Shi, C. et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: a retrospective clinical study. Clin. Transl Sci.10.1111/cts.12880 (2020). [DOI] [PMC free article] [PubMed]
- 235.Stein, B. L. Coagulopathy associated with COVID-19. (NEJM Journal Watch, 2020).
- 236.Thachil J, et al. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J. Thromb. Haemost. 2020;8:1023–1026. doi: 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04473053 (2020).
- 238.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04352400 (2020).
- 239.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04390594 (2020).
- 240.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04418128 (2020).
- 241.Yamamoto M, et al. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ansarin K, et al. Effect of bromhexine on clinical outcomes and mortality in COVID-19 patients: a randomized clinical trial. Bioimpacts. 2020;10:209–215. doi: 10.34172/bi.2020.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liu X, et al. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B. 2020;10:1205–1215. doi: 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Porfidia A, Pola R. Venous thromboembolism and heparin use in COVID-19 patients: juggling between pragmatic choices, suggestions of medical societies. J. Thromb. Thrombolysis. 2020;50:68–71. doi: 10.1007/s11239-020-02125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mastaglio S, et al. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;215:108450. doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.O’Brien KB, Morrison TE, Dundore DY, Heise MT, Schultz-Cherry S. A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza A virus infection. PLoS ONE. 2011;6:e17377. doi: 10.1371/journal.pone.0017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ricklin D, Mastellos DC, Reis ES, Lambris JD. The renaissance of complement therapeutics. Nat. Rev. Nephrol. 2018;14:26–47. doi: 10.1038/nrneph.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Keshari RS, et al. Complement C5 inhibition blocks the cytokine storm and consumptive coagulopathy by decreasing lipopolysaccharide (LPS) release in E. coli sepsis. Blood. 2015;126:765–765. [Google Scholar]
- 249.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04288713 (2020).
- 250.Woodruff TM, Nandakumar KS, Tedesco F. Inhibiting the C5–C5a receptor axis. Mol. Immunol. 2011;48:1631–1642. doi: 10.1016/j.molimm.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 251.Williams AL, et al. C5 inhibition prevents renal failure in a mouse model of lethal C3 glomerulopathy. Kidney Int. 2017;91:1386–1397. doi: 10.1016/j.kint.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Noris M, Remuzzi G. Terminal complement effectors in atypical hemolytic uremic syndrome: C5a, C5b-9, or a bit of both? Kidney Int. 2019;96:13–15. doi: 10.1016/j.kint.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 253.Gattinoni L, et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020;46:1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gattinoni L, Chiumello D, Rossi S. COVID-19 pneumonia: ARDS or not? Crit. Care. 2020;24:154. doi: 10.1186/s13054-020-02880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Copin M-C, et al. Time to consider histologic pattern of lung injury to treat critically ill patients with COVID-19 infection. Intensive Care Med. 2020;46:1124–1126. doi: 10.1007/s00134-020-06057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Aloisi, S., Beasley, D., Borter, G., Escritt, T. & Kelland K. Special report: as virus advances, doctors rethink rush to ventilate. Tildeshttps://www.reuters.com/article/us-health-coronavirus-ventilators-specia/special-report-as-virus-advances-doctors-rethink-rush-to-ventilate-idUSKCN2251PE (2020).
- 257.Robba C, et al. Distinct phenotypes require distinct respiratory management strategies in severe COVID-19. Respir. Physiol. Neurobiol. 2020;279:103455. doi: 10.1016/j.resp.2020.103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Casadevall A, Pirofski L. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Duan K, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl Acad. Sci. USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Shen C, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ye M, et al. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J. Med. Virol. 2020 doi: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zeng QL, et al. Effect of convalescent plasma therapy on viral shedding and survival in COVID-19 patients. J. Infect. Dis. 2020;222:38–43. doi: 10.1093/infdis/jiaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li L, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Agarwal, A. et al. Convalescent plasma in the management of moderate COVID-19 in India: An open-label parallel-arm phase II multicentre randomized controlled trial (PLACID Trial). Preprint at medRxiv10.1101/2020.09.03.20187252 (2020).
- 265.Chi X, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wec AZ, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hansen J, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ju B, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 269.Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 270.Wang C, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Liu Y, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kirby T. Evidence mounts on the disproportionate effect of COVID-19 on ethnic minorities. Lancet Respir. Med. 2020;8:547–548. doi: 10.1016/S2213-2600(20)30228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]