Abstract
T-cell prolymphocytic leukemia (T-PLL) is an aggressive malignancy characterized by chemotherapy resistance and a median survival of less than 2 years. Here, we investigated the pharmacological effects of the novel highly specific cyclin-dependent kinase 9 (CDK9) inhibitor LDC526 and its clinically used derivate atuveciclib employing primary T-PLL cells in an ex vivo drug sensitivity testing platform. Importantly, all T-PLL samples were sensitive to CDK9 inhibition at submicromolar concentrations, while conventional cytotoxic drugs were found to be largely ineffective. At the cellular level LDC526 inhibited the phosphorylation at serine 2 of the RNA polymerase II C-terminal domain resulting in decreased de novo RNA transcription. LDC526 induced apoptotic leukemic cell death through down-regulating MYC and MCL1 both at the mRNA and protein level. Microarray-based transcriptomic profiling revealed that genes down-modulated in response to CDK9 inhibition were enriched for MYC and JAK-STAT targets. By contrast, CDK9 inhibition increased the expression of the tumor suppressor FBXW7, which may contribute to decreased MYC and MCL1 protein levels. Finally, the combination of atuvecliclib and the BCL2 inhibitor venetoclax exhibited synergistic anti-leukemic activity, providing the rationale for a novel targeted-agent-based treatment of T-PLL.
Keywords: atuveciclib, CDK9 inhibitor, T-cell prolymphocytic leukemia, venetoclax
Introduction
T-cell prolymphocytic leukemia (T-PLL) is a rare aggressive T-cell neoplasia characterized by complex cytogenetic aberrations involving dysregulation of TCL1A/MTCP1, inactivation of ATM on chromosome 11q22.3, activating mutations affecting the JAK-STAT pathway (JAK3, STAT5B and PTPRC), and chromosomal gains at the MYC locus on 8q24.21.1–5 As the leukemic cells are largely resistant to conventional chemotherapy, intravenous treatment with the anti-CD52 monoclonal antibody alemtuzumab is currently considered the standard of care, yielding an overall response rate of 80–90% but an overall survival below 2 years.6,7 Allogeneic stem cell transplantation may be used to consolidate alemtuzumab-induced remissions, however, this option is available to only a minority of mostly elderly patients and those who are burdened with a high treatment related morbidity and mortality.8
Therefore, novel targeted therapeutic options that are capable of overcoming treatment resistance are urgently needed. In recently published drug screening efforts by our group9 and others10 targeting the transcriptomic machinery through inhibition of CDK9 emerged as a potentially promising novel therapeutic option. In these studies, suspension cultures of primary T-PLL cells were used to investigate the anti-leukemic effects of different CDK inhibitors including SNS-032, dinaciclib and alvocidib (flavopiridol). While all of these compounds exhibit anti-leukemic activity, their clinical use is limited by low target specificity and toxic side effects.11,12 To further increase CDK9 inhibitor specificity and to enable oral administration we recently developed the novel CDK9 inhibitor LDC526,12 starting from LDC06713 and characterized its therapeutic effects in different mouse models of chronic lymphocytic leukemia.12,14 In contrast to other tested CDK inhibitors, LDC526 shows high selectivity for its molecular target CDK9 as tested by cell culture experiments and in vitro selectivity kinase profiling.14 A further medicinal chemistry-based optimization of LDC526 resulted in atuveciclib,12 which is currently being studied in phase I trials in patients with acute leukemia and solid tumors/lymphomas [ClinicalTrials.gov identifiers: NCT02345382 and NCT01938638]. Following up on this work, we investigated the potential clinical value of LDC526 and atuveciclib in a preclinical model of T-PLL.
Materials and methods
Patients and samples
Patient samples were obtained from archived cryopreserved material from our institution. Patients were diagnosed in accordance with the WHO 2008 classification.5 The study was approved by ethical review committee of the University of Duisburg-Essen (14-6080-BO). All patients provided written informed consents according to the declaration of Helsinki.
The detection of an inv(14)/t(14;14) or t(X;14) by cytogenetic analysis and/or detection of TCL1 or MTCP1 breakpoints by fluorescence in situ hybridization (FISH) was required for inclusion into the study. Clinical data of 16 study patients are summarized in Table 1.
Table 1.
Clinical patient data.
| Patient ID | Sex | Age at diagnosis (years) | Genetic group | T-PLL immuno-phenotype | Absolute WBC count at diagnosis (109/L) | Absolute lymph. count at diagnosis (109/L) | Other sites of involvement (except blood and BM) | Type of treatment* | Allogenic transplant | CR at any time | Overall survival (months) | Death | Comorbidity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 74 | inv(14)/ t(14;14) |
CD4+ | 719 | 62.7 | none | 1; 2; 3; 4 | no | no | 34 | yes | Prior T-PLL: Cervical carcinoma (Radiotherapy – CR); Immunoblastic lymphoma (CTX – CR) |
| 2 | F | 64 | t(X;14) | CD4+ | 17.3 | 8.5 | skin | 1; 2; 4 | no | no | 18 | yes | Rheumatoid Arthritis; Myocardial Inf.; chronic renal disease, Std. III |
| 3 | M | 62 | inv(14)/ t(14;14) |
CD4+ | 148 | 121 | spleen, liver, lymph nodes, skin | 1; 2; 3; 4 | no | no | 7 | yes | none |
| 4 | M | 69 | inv(14) | CD4+ | n.a. | n.a. | n.a. | n.a. | no | no | 0 | yes | n.a. |
| 5 | M | 75 | inv(14) | CD8+ | 830 | 730 | spleen, liver, lymph nodes | 1; 2; 3; 4 | no | yes | 33 | yes | Ponsinfarction right; Vascular leukencephalopathie; Hypertension; Kidney-surgery right; Polyneuropathy |
| 7 | F | 51 | t(14;14) | CD8+ | 11.8 | 8 | spleen, liver, lymph nodes | 1; 2; 3; 4 | yes | yes | 16 | yes | Diabetes mellitus; Hypertension; Adiposity; Bronchial asthma |
| 8 | M | n.a. | t(X;14) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 33 | yes | n.a. | |
| 10 | M | 82 | inv(14) | CD4+ | 32 | 28 | spleen | 1; 2; 4 | no | no | 5 | yes | Hypertension; chronic renal disease; Depression; Prior to T-PLL: Prostate cancer |
| 11 | F | 62 | inv(14) | CD4+/CD8+ | 34 | 30 | spleen | 1; 2; 3; 4 | yes | no | 21 | yes | Hypertension; Diabetes mellitus; ulcerative colitis |
| 25 | F | 76 | inv(14) | CD4+ | 118 | 106 | spleen, lymph nodes | 3 | no | yes | 30 | no | CLL |
| 33 | F | 64 | inv(14) | CD4+ | 30.2 | 28.4 | spleen, liver | 3 | no | no | 17 | no | Chronic HepC infection, liver cirrhosis; Diabetes mellitus; chronic renal disease; Hypertension; Heart failure.; Pulmonary hypertension; Atrial fibrillation |
| 34 | M | 71 | n.a. | CD4+ | 76 | 68 | skin | 3 | yes | yes | 13 | yes | Hypertension |
| 35 | M | 76 | inv(14) | CD4+ | 407 | 387 | spleen | 3 | no | no | 19 | no | Hypertension; Hyperuricemia |
| 36 | M | 70 | t(X;14) | CD8+ | 185 | 180 | spleen, liver, lymph nodes, | 3; 4 | yes | yes | 8 | yes | Hypertension; Diabetes mellitus; chronic renal disease. |
| 37 | M | 74 | inv(14) | CD4+ | 53 | 44 | lymph nodes, skin | 4 | no | no | 51 | no | M. Bechterew, Psoriasis |
| 38 | F | 74 | inv(14) | CD4+ | 79 | 72 | skin | 1; 2; 3; 4 | no | yes | 29 | yes | Hypertension |
BM, bone marrow; CLL, chronic lymphocytic leukemia; CR, complete remission; CTX, chemotherapy; HepC, hepatitis C; Lymph., lymphocyte; n.a., not available; T-PLL, T-cell prolymphocytic leukemia; WBC, white blood cell; WT, wildtype.
Type of treatment: 1 – Alkylators (Chlorambucile, etc.).
– Purine Analogs (Fludarabine, Pentostatin, etc.).
– Anti-CD52- Antibody (Alemtuzumab).
– Others.
Standard clinical criteria were applied for initiation of therapy. The patient cohort represents a subgroup of a previously published molecular profiling study investigating the mutational landscape in T-PLL, where the technical details of the molecular characterization of the samples are described.15 Overall, T-PLL peripheral blood samples enriched for CD3 positivity by magnetic cell separation (MACS, Miltenyi Biotech, Bergisch Gladbach, Germany) were characterized by RNA sequencing, targeted capture sequencing, and whole exome sequencing for somatic mutations, and by single nucleotide polymorphism arrays for detection of genomic imbalances in candidate regions. Data are available under gene expression omnibus (GEO) accession number GSE100882. The assignment of samples and experiments is given together with information on the karyotype of the T-PLL cases in Supplemental Table S1.
Tumor cell enrichment
We performed density gradient centrifugation of heparinized peripheral blood samples (Pancoll human, PAN-biotech, Aidenbach, Germany). Tumor cells were enriched from peripheral blood mononuclear cells by magnetic cell separation with an anti-CD3 antibody (MACS, Miltenyi Biotech), if the tumor cell content was <90% as detected by flow cytometry.
microRNA profiling
miRNAs were extracted from immunomagnetically purified peripheral blood CD3 positive T-cells from five healthy donors (HD) and ten patients with T-PLL using the miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. GeneChip miRNA 3.0 arrays (Affymetrix, Santa Clara, CA) were used for miRNA expression profiling. One μg of total RNA including miRNA from each sample was labeled with biotin using the FlashTag Biotin HSR Kit (Genisphere, Hatfield, PA).
Array hybridization, washing and scanning of the arrays were carried out according to the manufacturer’s recommendations. The data were extracted from the images, background corrected (RMA), quantile-normalized, summarized (median polish), and log2-transformed with Partek Genomic Suite (Partek Incorporated, St. Louis, MO, USA). Differentially expressed miRNAs were determined by Student’s t-test and p-values were corrected for multiple testing.
Western blotting
For analysis of MYC (n = 7), MCL1 (n = 7) and RNA Pol II (n = 4), proteins were extracted with the RIPA lysis buffer system including phenylmethylsulfonyl fluoride, protease inhibitor cocktail and sodium orthovanadate (Santa Cruz Biotechnology, Dallas, TX, USA). Protein concentrations were measured using Pierce bicinchoninic acid (BCA) protein assay (Thermo Scientific, Waltham, MA, USA). Equal amounts of protein lysates were separated by SDS-PAGE (Mini-Protean TGX gels, gradient 8–16%, Bio Rad Laboratories, Hercules, CA, USA) and transferred to a nitrocellulose membrane.
The following primary antibodies were used: rabbit monoclonal anti-cMYC (Y69, dilution 1:4000; ab32072, Abcam, Cambridge, UK), rabbit polyclonal anti-MCL-1 (S19, dilution 1:4000; sc-819, SantaCruz Biotechnology, Dallas, TX, USA), RNAPol II C-terminal domain (CTD) rat monoclonal anti-Ser2-P (3E10), rat monoclonal anti-Ser5-P (3E8) and rat monoclonal anti-Ser7-P (4E12, dilution 1:20 each, ChromoTek, Martinsried, Germany) and mouse monoclonal anti-β-actin-HRP (AC-15, dilution 1:500000; A3854, Sigma-Aldrich, St. Louis, MO, USA). As secondary antibodies the anti-rabbit-IgG-donkey-HRP (711-036-152, Jackson ImmunoResearch, Cambridgeshire, UK) and the anti-rat-IgG-goat-HRP (sc-2006. Santa Cruz Biotechnology) were used.
Quantitative reverse transcription PCR
RNA was transcribed into cDNA with the high capacity cDNA reverse transcription kit (Applied Biosystems, Life Technologies, Thermo-Fisher Scientific, Waltham, MA, USA). Each diluted reverse transcription reaction (1:20) was combined with TaqMan® Universal polymerase chain reaction (PCR) Master Mix, no AmpErase® UNG (Thermo Fisher) and TaqMan probes for MYC (Hs00153408, Thermo Fisher) and BCL2 (Hs00608023, Thermo Fisher) and GAPDH (Hs02758991, Thermo Fisher). Real-time PCR was performed using AriaMX (Agilent genomics, Santa Clara, CA, USA).
Drug sensitivity and cell viability assays
Compounds/dimethyl sulfoxide (DMSO) were prediluted at different concentrations in 25 μl cell culture medium in white 384 well-plates (Greiner Bio-One, Frickenhausen, Germany) on day 1. After incubation for 3 h at 37°C/5% CO2, 10 μl of cell suspension was added at cell numbers that assured assay linearity and optimal signal intensity. Cells were further incubated in humidified chambers for 48 h at 37°C and 5% CO2. Cells treated with the compound vehicle DMSO were used as positive controls and cells treated with 10 μM Staurosporine served as negative controls. At day 3, the CellTiter Glo reagent was prepared according to the instructions of the kit (Promega): Reagent was mixed 1:1 with cell culture medium. Thereon, mixture and assay plates were equilibrated at room temperature for 20 min. Equal volumes of the reagent medium mixture was added to the volume of culture medium present in each well. The plates were mixed at ~300 rpm for 2 min on an orbital shaker. The microplates were then incubated at room temperature for 10 min for stabilization of the luminescent signal. Following incubation, the luminescence was recorded on a Victor microplate reader (Perkin Elmer, Waltham, MA, USA) using a 200 ms integration time. The data were then analyzed with Excel using the XLFIT Plugin (dose response Fit 205) for IC50-determination. As quality control the Z'-factor was calculated from 16 positive and negative control values. Only assay results showing a Z'-factor ⩾ 0.5 were used for further analysis.
In combinatorial experiments, atuveciclib and venetoclax were administered simultaneously in constant ratios and variable ratios and the effect on cell viability was determined as before. In addition, viable cells in all controls and treated cells were also determined by trypan blue dye exclusion. The best atuveciclib combinatorial ratio for the maximum killing effect was determined by a response-surface analysis matrix of the combination of 12 doses of each drug in the combination. Dose reduction index was determined using the Combenefit software.
Microarray-based gene expression profiling
Gene expression profiling (GEP) was performed using high-density oligonucleotide arrays (Clariom S, Affymetrix, Santa Clara, CA, USA). Total RNA was isolated from three different T-PLL samples cultured in serum containing medium (RPMI 1640, 10% fetal calf serum) for 90 min in the presence (10 µM) or absence (DMSO control) of LDC526 using the RNeasy mini purification kit (Qiagen, Hilden, Germany). Sample amplification and labeling with the Clariom S pico kit, hybridization, scanning, raw data extraction, and robust multi-array analysis of background-adjusted, normalized, and log-transformed probe-set values were conducted. Threshold settings for significantly regulated genes were ⩾2-fold difference from DMSO-treated control cells, with p < 0.05.
The datasets generated during the current study are available in the GEO repository, GSE155746 for miRNA data; GSE155747 for mRNA data.
Statistical and bioinformatics analysis
Gene ontology enrichment analysis
Gene ontology enrichment analysis was performed on the differentially expressed genes as indicated in the figure legends with the default Homo sapiens reference list using a Fisher’s exact test corrected by false discovery rate.
Hierarchical clustering of miRNA expression data
The 3% most variable miRNAs were defined from miRNAs with an expression median >6 over all samples and a coefficient of variation (CV = MAD/median, with MAD being median absolute deviation) larger than its median. Hierarchical clustering was performed using the complete agglomeration method and computing the Euclidian distance.
Ingenuity pathway analysis
A core analysis with default settings was conducted on the differentially expressed genes (fold change, FC ⩾ 2 or FC ⩽ –2, corrected p < 0.05) between T-PLL and HD determined by RNA-seq. The microRNA target filter was used to find potential targets of the n = 17 significantly differentially expressed miRNAs between T-PLL and HD.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed with the GSEA2-2.2.4 software using default settings and gene set permutation.16 Further analysis beyond GEP included a GeneTrail analysis.17
Results
Characterization of T-PLL cases
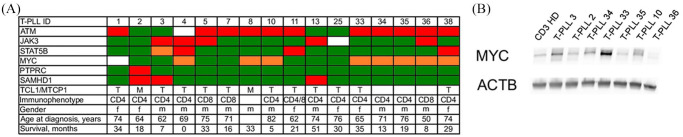
Figure 1A shows a selection of recurrent cytogenetic aberrations in n = 16 T-PLL combined with clinical characteristics. The T-PLL cases exhibited the typical molecular hallmarks of the disease including dysregulation of TCL1A/MTCP1, inactivation of ATM on chromosome 11q22.3, activating mutations affecting the JAK-STAT pathway (JAK3, STAT5B and PTPRC) and chromosomal gains at the MYC locus on 8q24. Activation of MYC was confirmed at the protein level by western blot analysis in a subset of patients (Figure 1B).
Figure 1.
Characterization of T-PLL samples. (A) genetic and clinical characteristics of 15 T-PLL samples subjected to molecular profiling. Color code: green – wildtype, red – mutated, orange – gain, blank – not determined. (B) Western blot analysis of MYC protein expression as compared with HD CD3+ T-cells. Blots were probed with antibodies MYC and Actin B used as a loading control.
ACTB, actin B; f, female; HD, healthy donor; M, mutated; m, male; n.a., not assessed; n.d., not determined; WT, wild type.
Transcriptomic profiling
RNA sequencing expression data from purified tumor cells of ten T-PLL patients were compared with CD3+ cells from 5 healthy blood donors yielding a panel of 674 differentially expressed genes (FC ⩾ 2 or FC ⩽ –2, corrected p ⩽ 0.05), the majority of which (603/674; 90.1%) were found to be down-regulated in the T-PLL cells (Supplemental Table S2).
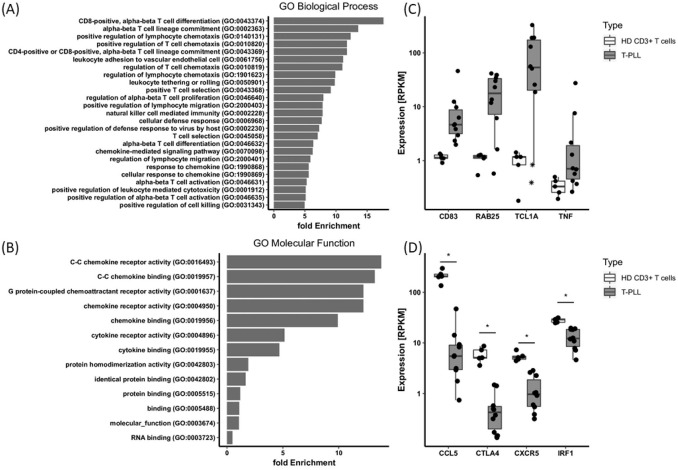
The panel of differentially expressed genes (Supplemental Table S2) was functionally analyzed for enrichment in the gene ontology categories biological process and molecular function (geneontology.org) revealing significant enrichment of T-cell activation, immune response, and chemokine-mediated signaling associated genes (Figure 2A, B). Deregulated molecular pathways identified by ingenuity pathway analysis (IPA) are shown in Supplemental Figure S1 and included STAT3, PTEN, TCR activation, WNT, and 4-1BB signaling. In all, the genomic and transcriptomic features of our T-PLL cohort were consistent with recently published data.2–4,10 At the gene-specific level, overexpression of TCL1A, RAB25, CD83, and TNF, as well as down-modulation of CTLA4, CXCR5, CCL5, and IRF1 in the T-PLL samples, as compared with healthy donor derived T-cells confirmed our own previous results and that of others (Figure 2C, D).2–4
Figure 2.
Comparative transcriptomic profiling of T-cell prolymphocytic leukemia (T-PLL) and healthy donor (HD) T-cells. RNA sequencing expression data from ten T-PLL samples were compared with CD3+ T-cells from five HD yielding a panel of 674 differentially expressed genes (DEG, FC ⩾ 2 or FC ⩽ –2, p ⩽ 0.05, Supplemental Table S2). (A), (B), Fold enrichment of differential expressed genes in gene ontology (GO) categories biological process and molecular function as identified by GO enrichment analysis (false discovery rate < 0.5). For biological process, only the top 25 GO terms are displayed. (C), (D), mRNA expression levels of individual DEG, bars indicate the median and lower and upper hinges mark the 25th and 75th percentile. Whiskers range to the smallest or highest value, respectively, but not further than 1.5 × interquartile range. *p < 0.05 (Student’s t-test, corrected). Note, outlier in the TCL1A plot marked by an asterisk represent T-PLL cases 2 and 8 with an t(X;14) resulting in overexpression of MTCP1 (not shown) but lacking cytogenetic evidence of TCL1A rearrangement.
RPKM, reads per kilobase per million mapped reads.
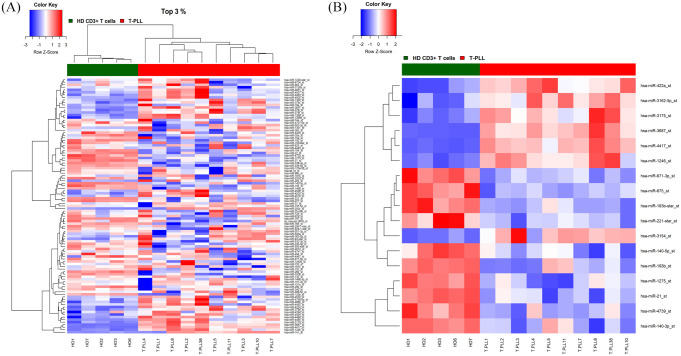
In another set of experiments, we compared the microRNA expression profiles of peripheral blood T-cells collected from 5 HD with 10 individual T-PLL samples employing the Affymetrix GeneChip miRNA 3.0 array platform. First, unsupervised hierarchical clustering considering the 3% most variable miRNAs across all samples clearly distinguished T-PLL from healthy donor derived T-cell controls (Figure 3A). Second, supervised comparison of the two groups (two-tailed unpaired Student’s t-test, corrected p-value <0.05, FC ⩾ 2 or FC ⩽ –2) identified a panel of n = 17 differentially expressed miRNAs (Figure 3B). Of note, two of the most prominently down-modulated miRNAs in the T-PLL cohort included mir-193b and mir-140-3p, which have been ascribed important functions as tumor suppressors in hematologic and solid malignancies.18,19 Conversely, mir-1246 and mir-3687, which were found to be over-expressed in the T-PLL samples, have been associated with tumor growth and metastasis in non-small cell lung20 and esophageal cancers,21 respectively.
Figure 3.
Comparative miRNA expression profiling of T-PLL and HD T-cells. (A) Unsupervised analysis comparing T-PLL versus CD3+ HD derived T-cells. The heat map diagram shows the result of the two-way unsupervised hierarchical clustering of miRNAs and samples based on the expression of the 3% most variably expressed miRNAs (n = 101). (B) Supervised analysis identified 17 differentially expressed miRNAs comparing T-PLL HD derived T-cells (corrected p < 0.05, FC ⩾ 2 or FC ⩽ –2). The color scale illustrates the relative expression level of a miRNA across all samples as a z-score: red represents an expression level above the mean and blue represents expression lower than the mean.
HD, healthy donors; T-PLL, T-cell prolymphocytic leukemia.
Ex vivo drug sensitivity testing validates CDK9 as a therapeutic target in T-PLL
To evaluate the therapeutic potential of CDK9 inhibition in native T-PLL cells we compared the cytotoxic effects of the specific CDK9 inhibitors LDC526 and atuveciclib (BAY 1143572) with those of a panel of chemotherapeutic compounds commonly used for the treatment of T-PLL patients in vivo. To this end, leukemic cells collected from 11 patients were exposed to 9 drugs at different concentrations for 48 h and analyzed for inhibition of cell viability using a metabolic-based assay. Consistent with the clinical situation in T-PLL we observed substantial heterogeneity in the drug responses between individual patient samples (Table 2). As a general phenomenon T-PLL cells tended to exhibit high IC50 values across most of the compounds indicating chemotherapy resistance and thereby mimicking the therapeutic situation in vivo. Interestingly, we observed a clear-cut difference in the response to the purine nucleoside analogues fludarabine and clofarabine, where four patient samples were found to be highly sensitive, whereas drug resistance was observed in the remaining seven cases (Table 2).
Table 2.
Drug sensitivity assays.
| IC50 (µM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound name | T-PLL_2 | T-PLL_3 | T-PLL_10 | T-PLL_11 | T-PLL_7 | T-PLL_36 | T-PLL_5 | T-PLL_25 | T-PLL_34 | T-PLL_37 | T-PLL_35 | Mean# |
| LDC526 | 0.937 | 1.605 | 2.275 | 2.398 | 1.548 | 1.378 | 3.526 | 1.505 | 1.064 | 1.39 | 1.74 | 1.65 |
| dinaciclib | 0.006 | 0.006 | 0.006 | 0.013 | 0.009 | 0.011 | 0.015 | 0.009 | 0.009 | 0.009 | 0.018 | 0.01 |
| atuveciclib | 0.537 | 0.946 | 1.034 | 0.983 | 0.848 | 0.651 | 1.653 | 0.896 | 0.849 | 0.966 | 0.836 | 0.89 |
| cytarabine | 16.09 | 1.875 | 3.212 | >30 | 14.72 | 3.897 | >30 | 7.761 | 2.034 | 24.826 | 6.233 | 8.3 |
| fludarabine | >30 | >30 | 0.468 | >30 | >30 | 0.76 | 25.828 | 39.91 | 0.241 | >30 | 0.363 | 6.4 |
| clofarabine | >30 | >30 | 0.046 | >30 | >30 | 0.225 | >30 | >30 | 0.372 | >30 | NA | 6.2 |
| cyclophosphamide | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | 29.9 |
| prednisone | ND | ND | ND | >30 | >30 | >30 | >30 | >30 | >30 | >30 | >30 | 29.9 |
| doxorubicine | 0.226 | 2.523 | 3.535 | 4.011 | 4.208 | 4.532 | 5.645 | 1.935 | 2.061 | 0.553 | 5.439 | 2.3 |
| TCL1A/MTCP1 | MTCP1 | TCL1A | TCL1A | TCL1A | TCL1A | n.d. | TCL1A | TCL1A | n.d. | TCL1A | n.d. | |
| ATM | WT | WT | M/loss | M | M | M/gain | M | WT* | M | WT* | M/loss | |
| MYC | WT | WT | gain | WT | WT | gain | WT | WT | gain | WT | gain | |
| JAK3 | WT | M | WT | WT | WT | M | M | n.d. | WT | M | WT | |
| STAT5B | WT | Gain | WT | M | WT | WT | WT | WT | WT | WT | WT | |
| SAMHD1 | mutated | mutated | WT | loss | WT | loss (UPD) | WT | WT | WT | mutated | WT | |
No copy number status available.
calculated as geometric mean.
M, mutated; NA, not available; ND, not determined; UPD, uniparental disomy; WT, wildtype.
25,000 cells/ well highly purified T-PLL cells with a purity >97% were used for each experiment.
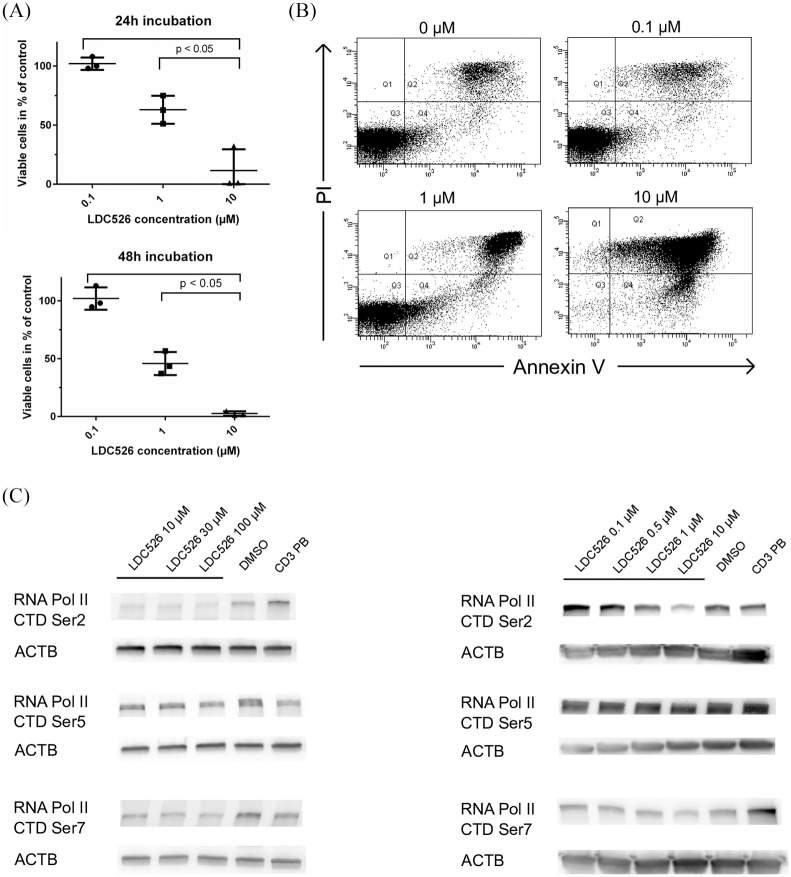
By contrast, targeting the general transcriptional machinery through inhibition of CDK9 (LDC526, atuveciclib and dinaciclib) proved to be highly effective showing consistent and potent anti-leukemic effects in all the patient samples investigated. Of note, atuveciclib exerted a 1.9-fold stronger anti-leukemic activity as compared with LDC526 (p = 0.0007, Table 2). To study the effects of LDC526 in more detail, leukemic cells from four T-PLL patients were exposed to LDC526 at three concentrations in suspension cultures. Employing flow cytometric Annexin V/ PI assays, we showed that LDC526 induced apoptosis in a dose- and time-dependent manner (Figure 4).
Figure 4.
LDC526 induces apoptosis in T-cell prolymphocytic leukemia (T-PLL) cells. T-PLL cells from three individuals were exposed to the indicated concentrations of LDC526 or dimethyl sulfoxide (DMSO) (control) in suspension cultures for 24 h and 48 h. Apoptosis was then analyzed by Annexin V and propidium iodide (PI; nuclear) staining using flow cytometry. The percentage of cells in each quadrant (Q) is indicated in the dot plot panels. Q3 defines the fraction of viable Annexin V -/ PI- cells. (A) representative analysis of T-PLL3 after 24 h incubation. (B) Summary of three independent experiments showing that LDC526 induces apoptosis in a concentration and time dependent manner. Bars represent medians, circles indicate results from individual measurements. (C) T-PLL cells were treated with the indicated concentrations of LDC526 for 90 min, and western blotting was performed. Blots were probed with antibodies to phospho-RNAPII Ser2 of the C-terminal domain (Ser2), phospho-RNAPII (Ser5), and phospho-RNAPII (Ser7). Actin was used as a loading control. One representative example of four independent experiments is shown. HD CD3+ T-cells were blotted as an additional control.
Effects of LDC526 on CDK9 function in T-PLL cells
Next, the effects of CDK9 inhibition by LDC526 in T-PLL cells were investigated. Based on previous experiments, cells were treated with LDC526 over a concentration range of 0.1–100 µM for 90 min and subsequently analyzed by western blot. LDC526 reduced the phosphorylation of Ser2 which is an established cellular target of CDK9 within the RNA polymerase CTD, while Ser5-P and Ser7-P exhibited this effect to a lesser extent (Figure 4C).
Effects of LDC526 on RNA synthesis in T-PLL cells
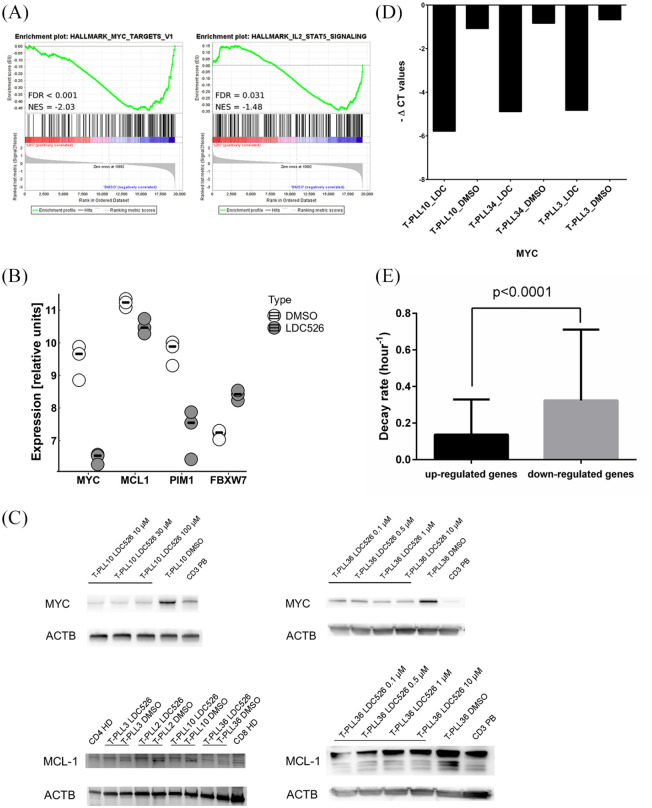
Following a previously published strategy22 microarray-based GEP was performed on RNA isolated from leukemia cells treated with LDC526 for 90 min. Comparing the transcriptomic profiles of three individual samples cultured in the presence or absence of LDC526 yielded a panel of 403 differentially expressed genes (FC ⩾ 2 or FC ⩽ –2, p < 0.05), the majority of which (n = 352; 87.3%) were found to be down-regulated in the T-PLL cells (Supplemental Table S3). GSEA revealed enrichment of 14 gene sets (p < 0.01, Supplemental Table S4) in the DMSO-treated samples including MYC targets and IL2-STAT5 signaling (Figure 5A).
Figure 5.
Microarray-based transcriptomic analysis of T-cell prolymphocytic leukemia (T-PLL) cells treated with LDC526. (A) Two examples of gene set enrichment analysis plots with potential functional relevance for the pharmacological effects of LDC526 in T-PLL cells. Genes found to be down-regulated in response to LDC526 were enriched for MYC targets and the IL6-JAK-STAT3- pathway. (B) Selected differentially regulated genes as indicated below the x axis. Bars represent the median; circles indicate results from individual patients/controls. (C) Down-regulation of MYC and MCL1 after incubation of T-PLL cells with LDC526 was validated at the protein level using western blot in four different patients. Left: MCL1 - patient samples comparing LDC526 with dimethyl sulfoxide (DMSO) in a concentration of 10 µM, right: MYC and MCL-1 sample T-PLL36 treated with different concentrations of LDC526. (D) Down-regulation of MYC mRNA levels after incubation of T-PLL cells with LDC526 is confirmed by quantitative reverse transcription polymerase chain reaction. (E) Genes down-regulated by LDC526 exhibit higher RNA decay rates as compared with genes with stable transcript levels.
Importantly, the JAK-STAT targets PIM1, MYC and MCL1 were among the most strongly down-modulated genes (Figure 5B). LDC526 induced suppression of MYC and MCL1 were confirmed at the protein level using western blotting (Figure 5C). Down-regulation of MYC mRNA transcripts was further validated by quantitative reverse transcription-PCR (Figure 5D). The PIM1 oncogene encodes a short-lived serine/threonine kinase (PIM1), which has been found to closely cooperate with MYC in oncogenesis.23 Recently, high PIM1 expression was demonstrated to be a pro-survival factor and biomarker of JAK-STAT activation in T-cell acute lymphoblastic leukemia.24
Of note, the mRNA transcripts encoding MYC, PIM1, and MCL1 are all relatively short-lived22,25 which may render them particularly susceptible to small-molecule transcription inhibitors. In support of this observation, gene ontology analysis indicated an enrichment of DNA transcription factors with fast-decaying mRNAs in the down-regulated genes (Supplemental Figure S2). To further test the hypothesis that short-term CDK9 inhibition by LDC526 predominantly affects genes with intrinsically instable mRNA we investigated whether the set of genes down-regulated in T-PLL cells was enriched for transcripts with high decay rates. To this end we utilized publicly available mRNA degradation data, which were generated in human cell lines exposed to the RNA polymerase inhibitor actinomycin D.25 Since these experiments were performed in the year 2003 using Affymetrix U95Av2 arrays, we had to restrict our analysis to 5245 manually curated gene accessions (4835 genes). Considering these limitations, we compared the mRNA decay rates of 263 genes significantly down-regulated (p < 0.05, FC ⩾ 2) by LDC526 with the remaining genes with stable gene expression (Supplemental Table S5). As expected, we observed a highly significant difference between the two groups, where mRNA transcripts down-modulated by LDC526 exhibited ~ 2-fold higher decay rates, underpinning the proposed working mechanism as an inhibitor of de novo mRNA synthesis (Figure 5E).
Of note, LDC526 also induced the up-regulation of 51 genes including FBXW7 which has been shown to be an important tumor suppressor gene in several malignancies including T-cell neoplasias.26,27
Synergistic antileukemic activity of atuveciclib and venetoclax in primary T-PLL cells
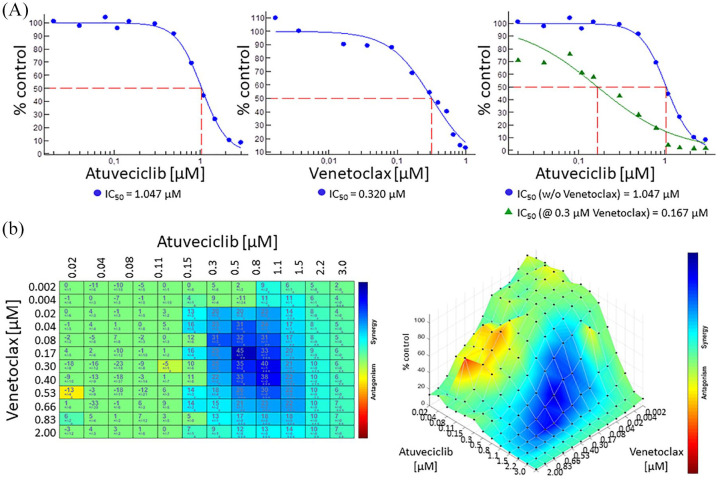
LDC526 potently and rapidly down-regulated MCL1 both at the mRNA and protein level, whereas BCL2 is characterized by a substantially lower mRNA decay rate and therefore remained unaffected (Supplemental Figure S3). Therefore, it may be tempting to target the apoptotic machinery by dual inhibition of CDK9 and the BCL2 protein. Here, we combined 12 concentrations of atuveciclib and 12 concentrations of venetoclax in our T-PLL cell culture model in order to define the optimal combined dose regimen for a maximum leukemic cell killing effect. Employing a response-surface analysis matrix we could indeed show a higher anti-leukemic effect in the combination as compared with each drug alone (Figure 6).
Figure 6.
Synergistic anti-leukemic effects of atuveciclib and venetoclax. (A) Separate and combined incubations of atuveciclib and venetoclax in dose-response format; combined incubation exemplified by atuveciclib dose-response treatment at 0.3 µM venetoclax concentration; y axis: relative viability (5 of vehicle control) of T-cell prolymphocytic leukemia cell sample 25 incubated for 48 h with respective compound determined by CellTiterGlo™ detection methods; half-maximal inhibition (IC50) was determined using Combenefit software and is depicted below each graph; x axis: respective compound concentration in µM. (B) Left: two-dimensional representations upon highest single agent (HSA) analysis using Combenefit software (coloration: red/orange pointed antagonism, green additive effects, and blue indicated synergy); right: relative viabilities (y axis) after incubation with different concentrations of atuveciclib (x axis; 20 nM to 3 µM) and venetoclax (z axis; 2 nM to 1 µM). Similar combinatorial effects were obtained with three different donors.
Discussion
In the first part of the study we aimed to characterize the pharmacological effects of LDC526 on primary leukemic cells isolated from the peripheral blood of patients with T-PLL and correlating their efficacy with the pattern of common genetic aberrations in this disease. As a novel unreported aspect, miRNA profiling of T-PLL in comparison with healthy donor derived T-cells led to the identification of 17 differentially expressed miRNAs, of which some candidates may play a role in the molecular pathogenesis of the disease. For example, miR-3687 found to be 56 fold up-regulated in the T-PLL samples has been demonstrated to increase the migratory and invasive ability of esophageal squamous cell carcinoma (ESCC) cells in vitro and correlate with the prognosis of patients with ESCC.21 Along the same line, mir-1246, which was also over-expressed in the T-PLL cases, has been associated with tumor initiation and progression in non-small cell lung cancer20 and other solid tumors including pancreatic and colorectal cancer.28,29 Probably more important, the most prominently down-modulated mir-193b in T-PLL has been recently characterized as a novel tumor suppressor gene in different hematological cancers including T-cell acute lymphoblastic leukemia (T-ALL)30 and also solid tumors.31 Bhayadia et al.18 showed that miR-193b is down-regulated in several cytogenetically defined subgroups of pediatric acute myeloid leukemia (AML), and low expression served as an independent adverse prognostic factor in this patient cohort. Conversely, experimental over-expression of miR-193b induced apoptosis in AML blasts by targeting the MAPK signaling pathway and the downstream cell cycle regulator CCND1.18 Based on the above, although requiring confirmation in an independent patient cohort, our results strongly suggest that these deregulated miRNAs could be relevant for the molecular pathogenesis of T-PLL. In light of recent data from the literature associating expression of mir-1246 and mir-193b with the clinical course of malignancy, it may be also interesting to study the potential prognostic value of miRNA signatures in larger homogenously treated cohorts of patients with T-PLL.
Drug sensitivity testing of the T-PLL samples revealed high IC50 values across most of the compounds indicating chemotherapy resistance and thereby reflecting the clinical situation in the patient. Interestingly, we observed a clear-cut difference in the response to the purine nucleoside analogues fludarabine and clofarabine, where one third of the patient samples were found to be highly sensitive, while drug resistance was noted in the remaining cases. Unfortunately, due to the low number of samples tested we could not correlate the drug responses with the molecular features of the leukemic cells. All CDK9 inhibitors elicited a homogenous potent anti-leukemic effect in all patient samples investigated providing indirect evidence that their mechanism of action differs from that of conventional cytotoxic agents.
In order to better understand the effect of CDK9 inhibition on the transcriptional machinery in T-PLL cells we compared the mRNA gene expression profiles of leukemic cells cultured for 90 min in the absence or presence of LDC526. Consistent with its proposed mechanism of action and data reported by Albert et al.,22 we observed that LDC526 predominantly reduced mRNA transcripts characterized by high decay rates. This finding supports the concept that CDK9 inhibitors function through inhibition of de novo RNA synthesis and that their cytotoxic effects are mediated by down-modulation of genes like MYC or MCL1 that encode mRNAs and proteins with short half-lives.22 Indeed, we observed efficient down-modulation of MYC and MCL1 both at the mRNA and protein level, which may at least in part explain the onset of apoptosis in the leukemic cells. Furthermore, our microarray analysis uncovered additional novel CDK9 targets including the oncoprotein PIM1, which has been shown to cooperate with MYC in oncogenesis and exert pro-survival functions in T-cell acute lymphoblastic leukemia.23,24 GSEA of genes regulated in response to LDC526 in T-PLL cells revealed significant enrichment of MYC and JAK-STAT pathway target genes. In aggregate these results suggest that using a single inhibitor of CDK9 allows us to target multiple hubs of leukemia cell signaling including transcription factors simultaneously.
Unexpected from its function as a transcriptional inhibitor, our microarray analysis revealed that LDC526 induced the upregulation of multiple genes. Similar results have been obtained by GEP experiments investigating the effects of different CDK9 inhibitors in a variety of cellular models.22,32 The potential underlying mechanism remained unknown until recently Zhang et al.33 demonstrated that CDK9 contributes to gene silencing through binding to and phosphorylating SMARCA4. Consequently, CDK9 inhibition was found to de-repress epigenetically silenced genes including tumor suppressor genes. Along the same line, we observed that LDC526 up-regulates the expression of FBXW7 in T-PLL cells, which functions as a critical tumor suppressor through controlling proteasomal degradation of several oncoproteins including MYC, MCL-1, and NOTCH in a wide range of malignancies.26,27 As LDC526 significantly increased FBXW7 transcript levels in T-PLL cells compared with DMSO treated controls, it is tempting to speculate that this may have contributed to the down-regulation of MYC and MCL1 proteins as detected by western blot.
In contrast to MCL1, BCL2 is not affected by CDK9 inhibition, which may be explained by a higher stability of both the BCL2 transcript and protein.25 Given that T-PLL cells are efficiently killed by BCL2 inhibitors in vitro and the recent success of venetoclax in the treatment of patients with B-cells malignancies, we speculated that dual inhibition of CDK9 and BCL2 may be a promising strategy. Further support for this concept comes from an in vitro study demonstrating synergy between CDK7 inhibition by THZ1, which reduced STAT gene target gene transcription and sensitization to BCL2 inhibitors in T-cell leukemia/ lymphoma cell lines.34 The combination of atuvecliclib and venetoclax was tested in a cell culture system exposing primary T-PLL cells to each of the two compounds. We found a higher anti-leukemic effect in the combination as compared with each drug alone, thereby recapitulating the results obtained with THZ1 and venetoclax in the T-cell leukemia/ lymphoma cell line model.34 Preliminary data from a small first-in-human case series recently provided the first in vivo evidence of single-agent-activity and tolerability of venetoclax in patients with relapsed T-PLL.35
Conclusions
Results of preliminary data from a first in vivo study, which provides evidence for single-agent venetoclax, together with our data presented here provide a rationale for combining atuveciclib with venetoclax as a novel targeted-agent-based treatment of T-PLL.
Supplemental Material
Supplemental material, Figure-S1 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Figure-S2 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Figure-S3 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Supplement_to_CDK9_inhibition_in_T-PLL_2020-03-20 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S1 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S2 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S3 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S4 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S5 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Acknowledgments
We thank Sabine Senkel und Julia Jesdinsky-Elsenbruch for expert technical assistance. We thank Philip Abstoß for expert help performing the Western blots. We thank the Imaging Center Essen (IMCES).
Footnotes
Author contributions: PJ and LKH performed the sequencing and miRNA profiling experiments. AC, SM, and PH performed the cell viability assays. AC and BK supported the medicinal chemistry-based optimization strategy and selected the proper CDK9 inhibitors as chemical probes. AB performed genetic analyses. JD and LKH processed all sequencing data. LD performed bioinformatic analyses. PJ prepared cells from healthy donors and patients. PJ performed the protein analyses. JD and PJ designed the study. JD, PJ, LD, JS, UD, LKH, and AC interpreted data and wrote the manuscript. All authors read, edited, and approved the manuscript.
Conflict of interest statement: SM, PH, BK, and AC are employees of the Lead Discovery Center GmbH (LDC). PH, AC, and BK furthermore are co-inventors of the patent family for LDC526 and atuveciclib. These patents have been licensed to Bayer AG.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a research grant provided from the Dr. Werner Jackstädt-Stiftung (PJ) and the German Cancer Consortium (DKTK) for LD, JTS and JD. JTS. is supported by the German Cancer Consortium (DKTK), the German Cancer Aid (grant no. 70112505; PIPAC consortium) and the Deutsche Forschungsgemeinschaft (DFG) through grant SI1549/3-1 (Clinical Research Unit KFO337) and Collaborative Research Center SFB824 (project C4).
ORCID iDs: Patricia Johansson  https://orcid.org/0000-0002-3265-8689
https://orcid.org/0000-0002-3265-8689
Jens T. Siveke  https://orcid.org/0000-0002-8772-4778
https://orcid.org/0000-0002-8772-4778
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Patricia Johansson, Department of Hematology, University Hospital Essen, University of Duisburg-Essen, Hufelandstr. 55, Essen, D-45122, Germany.
Laura Dierichs, Institute for Developmental Cancer Therapeutics, West German Cancer Center, University Hospital Essen, Essen, Germany; Division of Solid Tumor Translational Oncology, German Cancer Consortium (DKTK, partner site Essen) and German Cancer Research Center, DKFZ, Heidelberg, Germany.
Ludger Klein-Hitpass, Institute of Cell Biology (Cancer Research), University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Anke K. Bergmann, Department of Human Genetics, Hannover Medical School, Hannover, Germany
Michael Möllmann, Department of Hematology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Sascha Menninger, Lead Discovery Center GmbH, Dortmund, Germany.
Peter Habenberger, Lead Discovery Center GmbH, Dortmund, Germany.
Bert Klebl, Lead Discovery Center GmbH, Dortmund, Germany.
Jens T. Siveke, Institute for Developmental Cancer Therapeutics, West German Cancer Center, University Hospital Essen, Essen, Germany Division of Solid Tumor Translational Oncology, German Cancer Consortium (DKTK, partner site Essen) and German Cancer Research Center, DKFZ, Heidelberg, Germany.
Ulrich Dührsen, Department of Hematology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Axel Choidas, Lead Discovery Center GmbH, Dortmund, Germany.
Jan Dürig, Department of Hematology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany; German Cancer Consortium (DKTK, partner site Essen) and German Cancer Research Center (DKFZ), Heidelberg, Germany.
References
- 1. Bergmann AK, Schneppenheim S, Seifert M, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer 2014; 53: 309–316. [DOI] [PubMed] [Google Scholar]
- 2. Dürig J, Bug S, Klein-Hitpass L, et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32). Leukemia 2007; 21: 2153–2163. [DOI] [PubMed] [Google Scholar]
- 3. Kiel MJ, Velusamy T, Rolland D, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood 2014; 124: 1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schrader A, Crispatzu G, Oberbeck S, et al. Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat Commun 2018; 9: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: IARC Press, 2008. [Google Scholar]
- 6. Dearden C. Management of prolymphocytic leukemia. Hematology Am Soc Hematol Educ Program 2015; 2015: 361–367. [DOI] [PubMed] [Google Scholar]
- 7. Dearden CE, Khot A, Else M, et al. Alemtuzumab therapy in T-cell prolymphocytic leukemia: comparing efficacy in a series treated intravenously and a study piloting the subcutaneous route. Blood 2011; 118: 5799–5802. [DOI] [PubMed] [Google Scholar]
- 8. Wiktor-Jedrzejczak W, Drozd-Sokolowska J, Eikema DJ, et al. EBMT prospective observational study on allogeneic hematopoietic stem cell transplantation in T-prolymphocytic leukemia (T-PLL). Bone Marrow Transplant 2019; 54: 1391–1398. [DOI] [PubMed] [Google Scholar]
- 9. Dietrich S, Oles M, Lu J, et al. Drug-perturbation-based stratification of blood cancer. J Clin Invest 2018; 128: 427–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andersson EI, Putzer S, Yadav B, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia 2018; 32: 774–787. [DOI] [PubMed] [Google Scholar]
- 11. Lanasa MC, Andritsos L, Brown JR, et al. Final results of EFC6663: a multicenter, international, phase 2 study of alvocidib for patients with fludarabine-refractory chronic lymphocytic leukemia. Leuk Res 2015; 39: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lucking U, Scholz A, Lienau P, et al. Identification of atuveciclib (BAY 1143572), the first highly selective, clinical PTEFb/CDK9 inhibitor for the treatment of cancer. ChemMedChem 2017; 12: 1776–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bragelmann J, Dammert MA, Dietlein F, et al. Systematic kinase inhibitor profiling identifies CDK9 as a synthetic lethal target in NUT midline carcinoma. Cell Rep 2017; 20: 2833–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Göthert JR, Imsak R, Mollmann M, et al. Potent anti-leukemic activity of a specific cyclin-dependent kinase 9 inhibitor in mouse models of chronic lymphocytic leukemia. Oncotarget 2018; 9: 26353–26369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson P, Klein-Hitpass L, Choidas A, et al. SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J 2018; 8: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Backes C, Keller A, Kuentzer J, et al. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res 2007; 35: W186–W192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhayadia R, Krowiorz K, Haetscher N, et al. Endogenous tumor suppressor microRNA-193b: therapeutic and prognostic value in acute myeloid leukemia. J Clin Oncol 2018; 36: 1007–1016. [DOI] [PubMed] [Google Scholar]
- 19. Huang H, Wang Y, Li Q, et al. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett 2019; 446: 81–89. [DOI] [PubMed] [Google Scholar]
- 20. Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun 2016; 7: 11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagio K, Furuya S, Nakamura J, et al. High miR-3687 expression affects migratory and invasive ability of oesophageal carcinoma. Anticancer Res 2019; 39: 557–565. [DOI] [PubMed] [Google Scholar]
- 22. Albert TK, Rigault C, Eickhoff J, et al. Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. Br J Pharmacol 2014; 171: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Kim J, Roh M, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene 2010; 29: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. La Starza R, Messina M, Gianfelici V, et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia 2018; 32: 1807–1810. [DOI] [PubMed] [Google Scholar]
- 25. Yang E, van Nimwegen E, Zavolan M, et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 2003; 13: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu RB, Guo JG, Liu TZ, et al. Meta-analysis of the clinical characteristics and prognostic relevance of NOTCH1 and FBXW7 mutation in T-cell acute lymphoblastic leukemia. Oncotarget 2017; 8: 66360–66370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer 2018; 17: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hasegawa S, Eguchi H, Nagano H, et al. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer 2014; 111: 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang S, Zeng Y, Zhou JM, et al. MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol Med Rep 2016; 13: 273–280. [DOI] [PubMed] [Google Scholar]
- 30. Mets E, Van der Meulen J, Van Peer G, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia 2015; 29: 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mazzu YZ, Hu Y, Shen Y, et al. miR-193b regulates tumorigenesis in liposarcoma cells via PDGFR, TGFβ, and Wnt signaling. Sci Rep 2019; 9: 3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garriga J, Xie H, Obradovic Z, et al. Selective control of gene expression by CDK9 in human cells. J Cell Physiol 2010; 222: 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H, Pandey S, Travers M, et al. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell 2018; 175: 1244–1258e1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cayrol F, Praditsuktavorn P, Fernando TM, et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat Commun 2017; 8: 14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boidol B, Kornauth C, van der Kouwe E, et al. First-in-human response of BCL-2 inhibitor venetoclax in T-cell prolymphocytic leukemia. Blood 2017; 130: 2499–2503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Figure-S1 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Figure-S2 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Figure-S3 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Supplement_to_CDK9_inhibition_in_T-PLL_2020-03-20 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S1 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S2 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S3 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S4 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology
Supplemental material, Table_S5 for Anti-leukemic effect of CDK9 inhibition in T-cell prolymphocytic leukemia by Patricia Johansson, Laura Dierichs, Ludger Klein-Hitpass, Anke K. Bergmann, Michael Möllmann, Sascha Menninger, Peter Habenberger, Bert Klebl, Jens T. Siveke, Ulrich Dührsen, Axel Choidas and Jan Dürig in Therapeutic Advances in Hematology