Abstract
Meat diet plays a pivotal role in colorectal cancer (CRC). Hemin, a metabolite of myoglobin, produced after meat intake, has been involved in CRC initiation. The compound, 3,4-dihydroxyphenylacetic acid (3,4HPAA) is a scarcely studied microbiota-derived metabolite of the flavonoid quercetin (QUE), which exert antioxidant properties. The aim of this study was to determine the protective effect of 3,4HPAA against malignant transformation (increased cell proliferation, decreased apoptosis, DNA oxidative damage and augmented reactive oxidative species (ROS) levels) and mitochondrial dysfunction induced by hemin in normal colon epithelial cells and colon cancer cells. The effect of 3,4HPAA was assessed in comparison to its precursor, QUE and to a known CRC protective agent, sulforaphane (SFN). The results showed that both, tumor and normal cells, exposed to hemin, presented increased cell proliferation, decreased caspase 3 activity and cytochrome c release, as well as augmented production of intracellular and mitochondrial ROS. In addition, hemin decreased the mitochondrial membrane potential (MMP) and the activity of complexes I and II of the electron transport chain. These effects of hemin were prevented by the action of 3,4HPAA. The metabolite showed to be more active than QUE and slightly less active than SFN. In conclusion, 3,4HPAA administration could represent a promising strategy for preventing malignant transformation and mitochondrial dysfunction in colon epithelia induced by hemin.
Keywords: 3,4-dihydroxyphenylacetic acid; hemin; quercetin; sulforaphane; colorectal cancer; malignant transformation and mitochondrial dysfunction
1. Introduction
CRC is the third most commonly occurring cancer in men and the second most commonly occurring cancer in women [1]. The incidence has increased by 2% in the population aged <50 years. In fact, It has been estimated that the incidence rates for colon and rectal cancers will increase by 90.0%, and 124.2%, respectively by 2030 in people aged between 20 to 34 years [2]. In 2015, the Agency for Research on Cancer (IARC), an affiliated organization of the World Health Organization classified red meat as “probably carcinogenic to humans” and processed meat as “carcinogenic to humans”. The daily intake of 100 g of red meat increased the risk of CRC by 17% [3]. The excessive intake of heme iron should be considered, as it is an intrinsic factor of carcinogenicity, exhibited by dietary red meat. Hemin, a metabolite of heme produced in the gut after meat intake, has been involved in CRC initiation, as it promotes colon carcinogenesis through mechanisms involving oxidative damage [4,5,6,7].
There is a strong association with diets containing abundant vegetables, fruits, and grains, and a significant decline in the risk of colon cancer. The flavonol QUE (and QUE-glycosylated) is one of the most abundant polyphenols present in fruits and vegetables [8]. The anticancer effect of QUE in CRC is related to mechanisms involving cell cycle arrest, apoptosis, antioxidant response and regulation of signaling pathways [9,10]. QUE is barely absorbable in the gastrointestinal tract, and as a result, it accumulates in the colon lumen [11]. Colonic microbiota can degrade flavonoids, generating a large number of metabolites that can be absorbed and exert biological effects in the body, such as antioxidant, antimicrobial and anti-inflammatory activities, among others [12]. The major microbial metabolite of QUE and its glycosylated derivatives is 3,4-dyhydroxyphenylacetic acid (3,4HPAA) [13,14,15,16,17,18,19,20], which also has potent antioxidant properties. Although 3,4HPAA is abundantly produced in the colon, its role in CRC prevention remains unknown.
The isothiocyanate SFN is an isothiocyanate created from glucoraphanin (abundant in broccoli) by the action of myrosinase enzymes during crushing of the plant. Microbiota also has myrosinase activity, indicating that SFN can be formed from its precursor glucoraphanin in the colon [21]. SFN has extensively been shown to possess anticancer properties and to exert protection against CRC development in in vitro and in vivo models [22,23].
The present study aimed to determine the effect of 3,4HPAA against malignant transformation (increased cell proliferation, DNA oxidative damage and augmented reactive oxidative species (ROS) levels) and mitochondrial dysfunction induced by hemin in normal colon epithelial cells and colon cancer cells. We postulate that the beneficial effects of 3,4HPAA are associated with its antioxidant property and its ability to protect the mitochondrial function. This study also contributes to explaining the “health-promoting” effects of QUE, particularly against the development of CRC, and taking into account the metabolism that this flavonoid undergoes in the colon.
2. Results
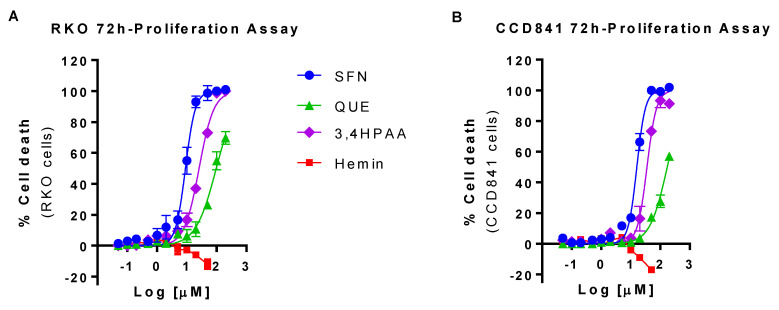
2.1. SFN, QUE and 3,4HPAA Reduced, and Hemin Increased RKO and CCD841 Cell Viability
SFN, QUE and 3,4HPAA more efficiently reduced the cell viability in RKO than in CCD841 cells. In comparison to RKO cells, 20 and 37% higher concentration of SFN, and 3,4HPAA, respectively, was required to reduce by 50% the viability of CCD841 cells (SFN = IC50 RKO cells 8.8 ± 0.21 μM and IC50 CCD841 cells 16.12 ± 0.90 μM, p = 0.0014 t-test; 3,4HPAA = IC50 RKO cells 25.8 ± 0.687 μM and IC50 CCD841 cells 35.4 ± 3.33 μM, p = 0.0460 t-test) (Figure 1A,B). QUE IC50 values were only estimated as it didn’t reach the 100% of inhibition and it didn’t plateau at the tested concentrations. Based on this estimation, 72% higher concentration of QUE is needed to reduce the cell viability of CCD841 cells by 50%, in comparison to the RKO cells (IC50 RKO cells 99.8 ± 13.1 μM and IC50 CCD841 cells 172 ± 13.3 μM, p = 0.018, t-test) (Figure 1A,B). Hemin at 50 μM increased the cell viability of RKO and CCD841 by 12 and 17% respectively (RKO: p = 0.0139; CCD841 cells: p < 0.0001) (Figure 1A,B), which was prevented by SFN, QUE and 3,4HPAA (Figure S1).
Figure 1.
Effect of hemin, SFN, QUE and 3,4HPAA in cell viability. (A) RKO cells and (B) CCD841 cells were incubated with increasing concentrations of hemin (0.05–50 μM), SFN, QUE or 3,4HPAA (0.05–200 μM). After 72 h, the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) reduction was detected by absorbance (MTS assay). The results were expressed as percentage of inhibition in relation to the positive control (30 µM puromycin) and calculated as: ((OD0.4% DMSO − ODsample) × 100)/(OD0.4% DMSO − ODpuromycin). Values are expressed as mean ± SEM, from three independent culture preparations. 3,4HPAA, 3,4-dihydroxyphenylacetic acid; QUE, quercetin; SFN, sulforaphane.
In order to determine the effect of the compounds in apoptosis, oxidative stress and mitochondrial dysfunction under conditions in which their effect on cell proliferation is marginal, the compounds were further studied at the concentrations that caused 10% of inhibition (IC10) of RKO cells proliferation (SFN = 1 μM; QUE = 10 μM and 3,4HPAA = 2.6 μM). The IC10 values were determined (interpolated using Prism) from the 72-hr proliferation assay with RKO cells, Figure 1A). Hemin was further evaluated at 10 μM, maximal concentration that did not increase cell proliferation either in RKO cell or CCD841 cells after 72 h incubation (Figure 1A,B).
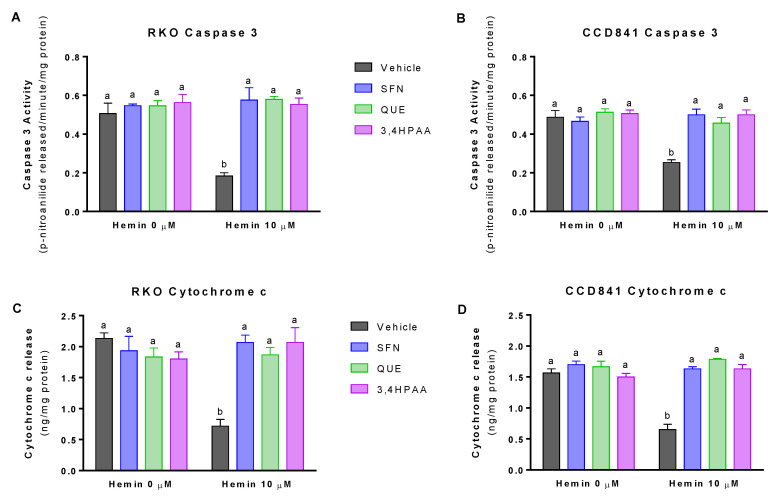
2.2. SFN, QUE and 3,4HPAA Prevented the Decrease of Apoptosis Induced by Hemin
At 10 μM, hemin decreased caspase 3 activity by 68% in RKO cells and 48% in CCD841 cells (Figure 2A,B). Hemin (10 μM) also decreased cytochrome c release by 66% in RKO cells and 59% in CCD841 cells (Figure 2C,D). SFN, QUE and 3,4HPAA, at the concentrations of 1, 10 and 2.6 μM, respectively prevented the decrease of caspase 3 and cytochrome c induced by hemin, in both cell lines (Figure 2A–D). Unlike 3,4HPAA, QUE at a concentration of 2,6 μM (Figure S2) did not prevent the decrease in apoptosis induced by hemin. In the same line, 3,4HPAA at 1 μM failed to protect antiapoptotic changes promoted by hemin (data not shown).
Figure 2.
Effect of hemin, SFN, QUE and 3,4HPAA in apoptosis. Cells were incubated with vehicle (0.4% DMSO), 1 μM SFN, 10 μM QUE or 2.6 μM 3,4HPAA in the absence or presence of 10 μM hemin. After 72 h, caspase 3 activity was measured in (A) RKO cells and (B) CCD841 cells; as well as cytochrome c levels in the media of (C) RKO cells; and (D) CCD841 cells. Caspase activity and cytochrome c release were expressed as p-nitroanilide/min/mg of protein, and ng (cytochrome c)/mg of protein, respectively. Values are expressed as mean ± SEM, from three independent culture preparations. For all bars with the same letter, the difference between the means is not statistically significant. Values with different letters indicate significant differences (p < 0.05) between bars (two-way ANOVA, Bonferroni post-test). 3,4HPAA, 3,4-dihydroxyphenylacetic acid; QUE, quercetin; SFN, sulforaphane.
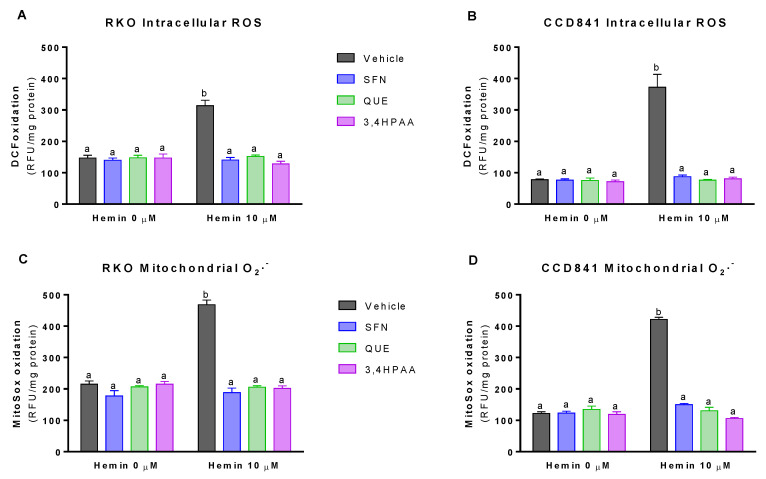
2.3. SFN, QUE and 3,4HPAA Inhibited the Hemin-Induced Increase in ROS Levels and DNA/RNA Oxidation
Basal (vehicle in the absence of hemin) intracellular ROS levels and mitochondrial O2− levels were 76 and 88% greater in RKO cells than in CCD841 cells, respectively (p = 0.0016 t-test, Figure 3A,B; p = 0.0013 t-test, Figure 3C,D). At 10 μM, hemin increased intracellular ROS levels by 113% in RKO cells and 383% in CCD841 cells (Figure 3A,B), which were completely inhibited by 1 μM SFN, 10 μM QUE and 2.6 μM 3,4HPAA. Same situation was observed for the mitochondrial O2− levels, in which hemin increased this free radical by 118 and 245% in mitochondria from RKO and CCD841 cells, respectively (Figure 3C,D).
Figure 3.
Effect of hemin, SFN, QUE and 3,4HPAA in ROS levels. Cells were incubated with vehicle (0.4% DMSO), 1 μM SFN, 10 μM QUE or 2.6 μM 3,4HPAA in the absence or presence of 10 μM hemin. After 72 h, DCF oxidation was measured by fluorescence in (A) RKO cells; and (B) CCD841 cells; as well as MitoSoxTM Red oxidation in (C) RKO cells; and (D) CCD841 cells. The results were expressed as RFU/mg of protein. Values are expressed as mean ± SEM, from three independent culture preparations. For all bars with the same letter, the difference between the means is not statistically significant. Values with different letters indicate significant differences (p < 0.05) between bars (two-way ANOVA, Bonferroni post-test). 3,4HPAA, 3,4-dihydroxyphenylacetic acid; DCF, dichlorofluorescein; QUE, quercetin; SFN, sulforaphane; RFU, relative fluorescence unit.
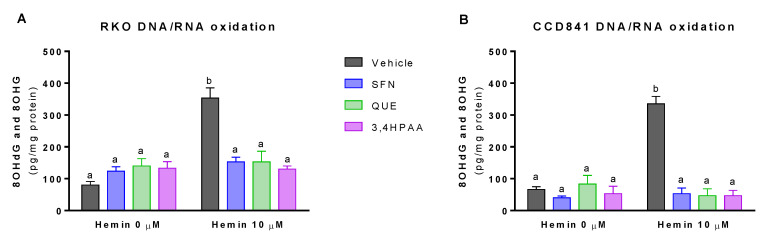
Basal levels of 8OHdG and 8OHG were 20% greater in RKO than in CCD841 cell supernatant (p = 0.0277 t-test, Figure 4A,B). Hemin, at a concentration of 10 μM, increased DNA/RNA oxidation by 4.4-fold and 5-fold in RKO cells and CCD841 cells, respectively (Figure 4A,B). This effect of hemin was fully inhibited by 1 μM SFN, 10 μM QUE and 2.6 μM 3,4HPAA in both cell lines (Figure 4A,B).
Figure 4.
Effect of hemin, SFN, QUE and 3,4HPAA in DNA/RNA damage. Cells were incubated with vehicle (0.4% DMSO), 1 μM SFN, 10 μM QUE or 2.6 μM 3,4HPAA in the absence or presence of 10 μM hemin. After 72 h, 8OHdG and 8OHG levels were assessed in (A) RKO; and (B) CCD841 cell supernatants. The results were expressed as pg (8OHdG and 8OHG)/mg of protein. Values are expressed as mean ± SEM, from three independent culture preparations. For all bars with the same letter, the difference between the means is not statistically significant. Values with different letters indicate significant differences (p < 0.05) between bars (two-way ANOVA, Bonferroni post-test). 3,4HPAA, 3,4-dihydroxyphenylacetic acid; 8OHdG, 8-hydroxy-2′-deoxyguanosine; 8OHG, 8-hydroxyguanosine; QUE, quercetin; SFN, sulforaphane.
SFN, QUE and 3,4HPAA had no effect on the ROS or 8OHdG and 8OHG levels in the absence of hemin (Figure 3A–D and Figure 4A,B). Unlike 3,4HPAA, QUE tested at 2.6 μM didn’t protect against the ROS levels and DNA/RNA oxidation induced by hemin in RKO or CCD841 cells (Figure S3). Same lack of effect was found with 1 μM 3,4HPAA (data not shown).
2.4. SFN, QUE and 3,4HPAA Inhibited the Hemin-Induced Decrease in MMP and Complex I and Complex II Activities
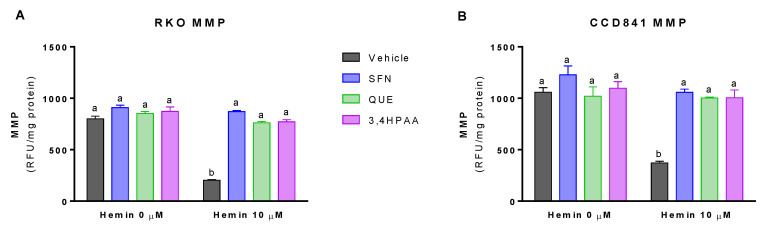
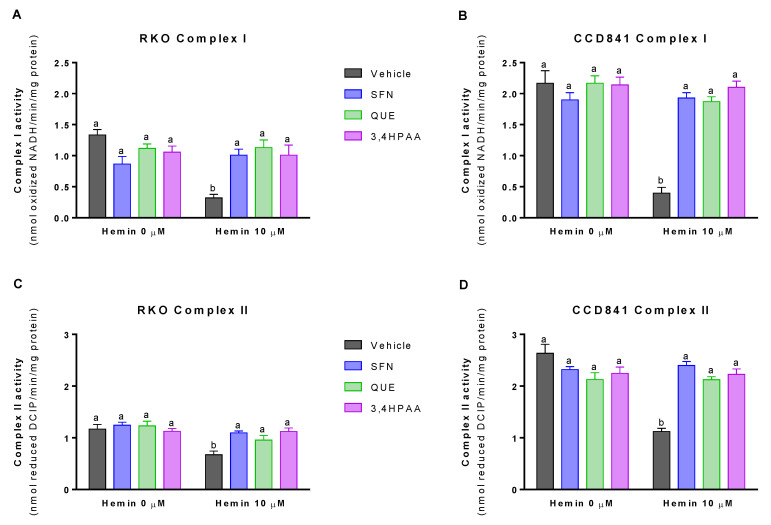
Basal (vehicle in the absence of hemin) levels of MMP was 32% greater in CCD841 than in RKO cells (p = 0.0072 t-test, Figure 5A,B). In addition, basal activities of the complex I and II were 63 and 125% higher in CCD841 than in RKO cells (Complex I: p = 0.0196 t-test, Figure 6A,B; Complex II: p = 0.018 t-test, Figure 6C,D). Hemin reduced MMP (Figure 5A) and the activities of complex I (Figure 6A) and complex II (Figure 6C) by 74, 76, and 43% in RKO cells, respectively; whilst in CCD841 cells, hemin reduced by 65% MMP (Figure 5B), 82% complex I activity (Figure 6B) and 57% complex II activity (Figure 6D). These effects of hemin were fully inhibited by 1 μM SFN, 10 μM QUE and 2.6 μM 3,4HPAA (Figure 5A,B and Figure 6A–D).
Figure 5.
Effect of hemin, SFN, QUE and 3,4HPAA in MMP. Cells were incubated with vehicle (0.4% DMSO), 1 μM SFN, 10 μM QUE or 2.6 μM 3,4HPAA in the absence or presence of 10 μM hemin. After 72 h, MMP was measured by fluorescence in (A) RKO; and (B) CCD841 cell supernatants. Results were expressed as RFU/mg of protein. Values are expressed as mean ± SEM, from three independent culture preparations. For all bars with the same letter, the difference between the means is not statistically significant. The values with different letters indicate significant differences (p < 0.05) between bars (two-way ANOVA, Bonferroni post-test3,4HPAA, 3,4-dihydroxyphenylacetic acid; MMP, mitochondrial membrane potential; QUE, quercetin; SFN, sulforaphane; RFU, relative fluorescence unit.
Figure 6.
Effect of hemin, SFN, QUE and 3,4HPAA in complex I and II activities. Cells were incubated for 72 h with vehicle (0.4% DMSO), 1 μM SFN, 10 μM QUE or 2.6 μM 3,4HPAA in the absence or presence of 10 μM hemin. Complex I activity was measured in (A) RKO cells; and (B) CCD841 cells and results were expressed as nmol of oxidized NADH/minute/mg of protein. Complex II activity was measured in (C) RKO cells; and (D) CCD841 cells and results were expressed as nmol of reduced DCIP/minute/mg of protein. Values are expressed as mean ± SEM, from three independent culture preparations. For all bars with the same letter, the difference between the means is not statistically significant. Values with different letters indicate significant differences (p < 0.05) between bars (two-way ANOVA, Bonferroni post-test). 3,4HPAA, 3,4-dihydroxyphenylacetic acid; DCIP, 2,6-dichlorophenolin-dophenol; QUE, quercetin; SFN, sulforaphane.
SFN, QUE and 3,4HPAA had no effect on the cells in the absence of hemin (Figure 5A,B and Figure 6A–D). Unlike 3,4HPAA, QUE at a concentration of 2.6 μM (Figure S4) didn’t protect against the reduction of MMP and complex activities induced by hemin in RKO or CCD841 cells. 3,4HPAA at 1 μM did’t prevent these mitochondrial alterations promoted by hemin (data not shown)
3. Discussion
CRC is the third most commonly occurring cancer in men and the second most commonly occurring cancer in women [1]. Interestingly, more than 50% of all CRC cases and deaths are attributable to modifiable risk factors, such as unhealthy nutritional lifestyles [4]. In terms of bowel cancers, in 2015, the World Health Organization classified processed meats as a Group 1 carcinogen (known to cause cancer); while red meat has been classified as a Group 2A carcinogen (probable to cause cancer). Hemin, a metabolite of heme produced in the gut after meat intake, has been involved in CRC initiation, as it promotes colon carcinogenesis through mechanisms involving oxidative damage [4,5,6,7]. It has been proposed that iron present in heme or hemin is a key promotor of CRC through its ability to initiate lipid peroxidation and to produce nitroso compounds [24]. In the present study, we postulate that 3,4HPAA, a microbiota-derived metabolite of QUE, may protect colon cells in vitro, against malignant transformation and mitochondrial dysfunction induced by hemin. This protective effect could be associated with the antioxidant property of this metabolite and its ability to prevent mitochondrial dysfunction.
3.1. 3,4HPAA Prevented the Hemin-Induced Increase Proliferation of Normal Colon Epithelial Cells and Colon Cancer Cells
It has been widely demonstrated that heme induces colon epithelial hyperproliferation and decreases apoptosis [25,26,27]. Also it has been proposed that heme induces compensatory hyperproliferation by damaging surface epithelial cells and the signals to crypt cells to promote aberrant growth [25,26]. Interestingly, Sesink et al. showed that dietary heme enhances cytotoxicity in rat colons through the involvement of a heme-induced cytotoxic metabolite [7]. This metabolite, identified as hemin, is a covalently modified porphyrin formed from heme in the gastrointestinal tract [6,7]. Therefore, it has been suggested that heme iron, in the form of hemin may explain the link between the risk of CRC and red meat intake, and the lack of a link with white meat intake [5,7]. Hemin supplementation to non-initiated rats increases fat peroxidation and cytotoxic activity of fecal water, and epithelial proliferation by 70% [7]. Therefore, in terms of evaluating the role of meat diet in CRC development, studying hemin in in vitro models is more physiologically relevant than studying heme. Consistently, we found that hemin increases the proliferation of normal colon epithelia and colon cancer cells (Figure 1), by reducing apoptosis (Figure 2) and that it was prevented in the presence of 3,4HPAA.
Our results showed that 3,4HPAA reduces the proliferation of colon cancer RKO cells after 72 h incubation (Figure 1), which is consistent with previous studies. While, 3,4HPAA has been shown to exhibit a potent antiproliferative effect on colon cancer HCT116 cells (IC50 90 µM) after 24 h of incubation [28]. Skrbe et al. showed that the growth of HT29 colon cells decreased by 60% after 72 h of exposure to 100 µM 3,4HPAA [29]. Our results showed that 3,4HPAA reduces proliferation of colon cancer RKO cells more efficiently than of normal colon epithelia CCD841 cells (Figure 1). The clinical response of DNA-targeted chemotherapy is positively correlated to the proliferation rate of the tumour cells [30]. Although, more studies are needed to determine the mechanism of action underlying the anti-proliferative effect of 3,4HPAA, its effect could be DNA targeted, thus affecting cell division, as doubling time of RKO is 24 h and CCD841 is 72 h. Consistently, the anti-proliferative effect of 3,4HPAA is evidenced after allowing enough time for cell replication, as this effect is not observed in shorter incubation times. 3,4HPAA incubated for 6 h at 200 or 250 µM had no effect in the viability of mouse hepatoma Hepa1c1c7 cells [31] and Min-6 pancreatic β-cells [32], respectively. In addition, when incubated for only 2 h, 3,4HPAA (up to 1 mM concentration) did not affect the viability of Caco-2 cells [33].
3.2. 3,4HPAA Prevented the Intracellular and Mitochondrial ROS Increase and DNA Oxidative Damage Induced by Hemin in Normal Colon Epithelial Cells and Colon Cancer Cells
Our results showed that hemin increased ROS production in the cell and mitochondria of colon epithelial cells and colon cancer cells (Figure 3) [34,35,36,37]. Consistent with our finding, Gonzalez-Reyes et al. reported that 30 µM hemin increases ROS production in primary cultures of cerebellar granule neurons of rats [38]. This effect of hemin was completely abolished by the presence of the antioxidant molecules, curcumin [38] or 3,4HPAA (as we shown in Figure 3). 3,4HPAA has been shown to have the highest free radical scavenging activity among several flavonoid metabolites tested in vitro [39]. In addition, we have previously described that 3,4HPAA can enhance the cellular antioxidant defense, by increasing Nrf2 translocation to the nucleus in pancreatic β-cells [32]. Nrf2 regulates the component transcriptions of the glutathione and thioredoxin antioxidant systems, as well as the phase I and phase II detoxification systems [40]. Although, more studies are needed addressing the mechanism underlying the antioxidant effect of 3,4HPAA in this model, it is suggested that 3,4HPAA could reduce ROS levels induced by hemin through direct (ROS scavenging effect) and indirect antioxidant effects (Nrf2 antioxidant system).
Hemin-produced free iron reacts with hydrogen peroxide, through the Fenton reaction, to generate highly oxidizing species capable of initiating the oxidation of macromolecules, such as lipids, proteins and DNA/RNA [41]. In fact, we found that hemin induced DNA/RNA oxidation in colon epithelial cells and colon cancer cells (Figure 4). Consistently, it has been found that 25 µM hemin induces lipid peroxidation and protein oxidation in bovine aortic endothelial cells (BAEC), only after 4 h incubation [42]. This effect of hemin in lipid and protein oxidation was totally inhibited by the antioxidant α-tocopherol [42] In line with the protective effect exerted by antioxidants against oxidative stress induced by hemin, we found that 3,4HPAA fully prevented the DNA/RNA oxidation induced by hemin in colon epithelial cells and colon cancer cells (Figure 4).
In general, tumour cells exhibit higher levels of ROS and oxidative stress in comparison to normal cells [43], and the level of oxidative stress is directly correlated to the frequency of p53 mutation in CRC [44]. In our study, we found that colon cancer RKO cells, which is wild-type p53 [45,46], show similar ROS and oxidative stress levels than the normal CCD841 colon epithelia cells (Figure 3). Consistent with our finding, RKO cells have shown to exert the same ROS levels than the normal human umbilical vein endothelial cells (HUVEC) [47].
3.3. 3,4HPAA Prevented Mitochondrial Dysfunction Induced by Hemin in Normal Colon Epithelial Cells and Colon Cancer Cells
Our results showed that hemin caused mitochondrial dysfunction, by decreasing MMP (Figure 4) and complex I and II activities in normal colon epithelial cells and colon cancer cells (Figure 6). Hemin (50 µM for 1 h) has also been shown to reduce MMP in BAEC, which was associated to a decrease in the basal and maximal respiration [42]. This effect of hemin in mitochondrial respiration could be due to its ability to decrease complex I and II activities, as we reported here for the first time (Figure 6).
We showed that hemin induces the production of O2− in mitochondria, a fact consistent with the alteration of mitochondrial function induced by this molecule (Figure 3). The idea that hemin increases mitochondrial O2− production by inhibiting complex I and II is supported by the fact that rotenone and malonate (complex I, and II inhibitors, respectively) also promote the mitochondrial production of O2− [48,49,50]. Mitochondrial O2− is generated by the interaction between oxygen and reducing equivalents [51]. When the electron flow through the electron transport chain slows down, the proton efflux through the inner mitochondrial membrane diminishes (hemin decreased MMP), causing an increase of electron donors in mitochondria. In consequence, the concentration of oxygen increases (hemin decreased oxygen consumption [42]) and the formation of O2− by the respiratory chain also increases (Figure 3C,D).
We have shown that mitochondrial dysfunction induced by hemin is prevented by 3,4HPAA (Figure 5 and Figure 6). Notably, 3,4HPAA may protect against hemin-induced ROS increase (Figure 3A,B), oxidative damage (Figure 4) and mitochondrial dysfunction (Figure 5 and Figure 6) by preventing O2− production within mitochondria (Figure 3C,D), and protecting the activities of complex I and II (Figure 6).
3.4. Physiological Relevance of This Study and Limitations
The effect of 3,4HPAA reported here could be exerted in vivo, as the concentration, used in this study (2.6 μM), could be reached in the colon, considering that 6.98 µM of this metabolite has been detected in fecal water after the administration of a standard diet [52]. It was shown that 3,4HPAA is four times more effective than QUE in preventing the damage induced by hemin (Figures S2–S4). Therefore, it is possible that the metabolite is the main contributor to the protective effect against CRC observed with QUE in in vivo models [9,10]. SFN has extensively been shown to possess anticancer properties and to exert protection against CRC development in in vitro and in vivo models [22,23]. In our study, 3,4HPAA was shown to be only 2.6-fold less active than SFN, suggesting that the metabolite is of relevance for CRC protection and that the co-administration of SFN and QUE is highly beneficial to reduce CRC risk. However, further studies in vivo models are required to support the beneficial role of 3,4HPAA in CRC protection.
4. Materials and Methods
4.1. Materials
3,4-dihydroxyphenylacetic acid, quercetin, sulforaphane hemin, rhodamine123 (rhod), dichlorofluorescein (DCF) diacetate, triphenyltin,1,1,3,3-Tetramethoxypropane, 2,6-dichlorophenolin-dophenol (DCIP), rotenone, NADH, succinate, malonate and puromycin were purchased from Sigma-Aldrich (St. Louis, MO, USA). CellTiter96® AQueous Assay (Madison, WI, USA) was obtained from Promega (Madison, Wi, USA) and DNA/RNA oxidation ELISA kit from Cayman Chemicals (Ann Arbour, MI, USA). Pierce BCA Protein Assay Kit was purchased from Thermo Fisher Scientific (Waltham, MA, USA). MitoSoxTM Red mitochondrial was obtained from Molecular Probes (Eugene, OR, USA).
4.2. Cell Culture and Treatments
Cells were cultured in MEM supplemented with 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin in a humidified atmosphere of 95% air, and 5% CO2. Cells were obtained from ATCC. RKO cells were seeded in a 96-well plate at a density of 1 × 104 cells/well and the CCD841 at 4 × 104 cells/well. After 24 h, cells were incubated for 24 h, 48 h and 72 h with SFN, QUE and 3,4HPAA in the absence or presence of hemin. However, compounds incubated for 24 h and 48 h did not cause alterations in the parameters measured. Only the results of 72 h incubation are shown. Reagents were diluted in DMSO, giving a final concentration in the cell plate of 0.4% DMSO (vehicle).
4.3. Cell Viability
Cell viability was measured through the reduction of MTS to a colored formazan dye that is soluble in media. Cells were treated with increasing concentrations of SFN, QUE and 3,4HPAA (0.05–200 μM). Maximum concentration of hemin tested was 50 μM, due to solubility limitation and colour interference with the assay readouts. After washing, the ability of cells to reduce MTS was evaluated by measuring the optic density (OD) at λ 490 nm, using the CellTiter96® AQueous Assay according to the manufacturer’s instructions. The percentage of inhibition was calculated in relation to the positive control (30 µM puromycin) as: ((OD0.4% DMSO − ODsample) × 100)/(OD0.4% DMSO − ODpuromycin). OD values were normalized to the negative and positive control. Dose-response curves (log(inhibitor) versus response, four parameters variable slope) were obtained using Graphpad Prism 7.0 (GraphPad Inc., La Jolla, CA, USA).
4.4. Apoptosis
Apoptosis was quantitated by measuring the activities of caspase- 3 (Millipore Corporation, Bedford, MA, USA) and cytochrome c. The catalytic activity of caspase-3 was measured independently by using a commercial colorimetric assay according to the manufacturer’s instructions [53]. The determination is based on the detection of the chromophore p-nitroanilide at 405 nm after its enzymatic cleavage from the labeled substrate DEVD-p-nitroanilide. The specific caspase- 3 inhibitor,0.1 mM Ac-DEVD-CHO, were used as enzymatic activity controls, according to the manufacturer’s instructions. Enzyme activity was calculated as µmol of p-nitroanilide released/minute/mg of protein. Cytochrome c was measured using a Cytochromec Kit ELISA assay (Life Technologies, CA, USA), according to manufacturer’s instructions [53].
4.5. Cellular Redox Status
The redox status of RKO and CCD841 cells were evaluated by measuring the intracellular reactive oxygen species and the cytosolic and mitochondrial O2− production. The intracellular ROS was measured through the conversion of DCF into a fluorescent dye (λEx485 nm and λEm530 nm) [32,54]. Mitochondrial O2− production was evaluated through the oxidation of MitoSoxTM Red, a cationic lipophilic fluorochrome (λEx510 nm and λEm570 nm) which accumulates in the mitochondrial matrix [32]. After treatments, cells were incubated at 37 °C for 30 min with 50 mM DCFD or 5 μM MitoSox™. Cells were washed and the fluorescence was measured using a Multi-Mode Microplate Reader (SynergyHT,BioTek) and data were normalized to the protein content. In order to evidence the specificity of MitoSoxTM for O2−, a control incubating the samples with SOD 100 U/mL for 10 min was also carried out (data not shown) [55]. In the presence of SOD, the MitoSoxTM oxidation was only 10–15%, indicating that under this experimental condition the oxidation of this probe induced by H2O2 was marginal. The H2O2-independent MitoSoxTM oxidation was calculated as URFMitoSoxTM oxidation -URFMitoSoxTM oxidation in the presence of SOD.
4.6. DNA and RNA Oxidation
The levels of 8-hydroxy-2′-deoxyguanosine (8OHdG) and 8-hydroxyguanosine (8OHG) in cell media were assessed using DNA and RNA oxidation ELISA kit according to the manufacturer’s instructions. Values were normalized to the protein content using the Pierce BCA Protein Assay Kit.
4.7. Mitochondrial Membrane Potential
MMP was evaluated using Rhod, a cationic lipophilic fluorochrome (λEx500 nm and λEm530 nm) which accumulates in the mitochondrial matrix. After treatments, cells were incubated with 1 mg/mL Rhod for 10 min at 37 °C. The specific accumulation of Rhod in the mitochondria was evaluated using as control 3 mM triphenyltin, an inductor of mitochondrial membrane permeability transition pores [32]. The fluorescence was measured using a Multi-Mode Microplate Reader (Synergy HT, BioTek). Protein content was determined by using the Pierce BCA Protein Assay Kit.
4.8. Complexes I and II Activities
Activities of complexes I and II of the electron transport chain were measured in mitochondria isolated from RKO and CCD841 cells as previously described [32,49,56]. Briefly, cells were harvested, washed in a Ca2+/Mg2+-free PBS and centrifuged (10 min; 1000 g; 4 °C). The pellet was resuspended and homogenized in a buffer solution (pH 7.4) containing 250 mM sucrose, 1 mM EGTA, 10 mM HEPES and 1mg/mL BSA (fraction V). The homogenate was centrifuged (10 min; 12,000 g; 4 °C); the supernatant discarded; and the pellet was washed and centrifuged as above. Complex I activity was measured in isolated mitochondria by determining the changes of OD340 nm due to NADH oxidation (ε = 6.81 mM−1 cm−1). Rotenone (a complex I inhibitor) was used as a control; and the enzyme activity as rotenone-sensitive NADH–ubiquinone oxidoreductase was calculated. The activity of complex II was quantified by the increase in DCIP reduction measured at 600 nm (ε = 19.1 mM−1 cm−1); malonate (a complex II inhibitor, final concentration of 20 mM) was used as a control; and the enzyme activity as malonate-sensitive succinate-coenzyme Q reductase was calculated.
4.9. Statistical Analysis
Two-way ANOVA were performed followed by Bonferroni’s Multiple Comparison Test. The experiments were performed using three independent culture preparations; and in quadruplicate for RKO cell evaluations and duplicate for CCD841 cell assays. Values are expressed as mean ± SEM. Values with different superscript letters (a, b and c) indicate significant differences (p < 0.05) between groups. For all bars with the same letter, the difference between the means is not statistically significant.
5. Conclusions
In conclusion, this study shows that hemin induces cell proliferation, oxidative damage and mitochondrial dysfunction, as well as inhibits apoptosis in colon cancer and normal colon epithelia cell lines. 3,4HPAA, a microbial metabolite of QUE, due to its antioxidant and mitochondrial protective effects, prevented hemin-induced malignant transformation and mitochondrial impairment. Colonic metabolism of QUE seems to be critical for the CRC preventive effect of the flavonoid. We consider that 3,4HPAA administration could represent a promising strategy for preventing malignant transformation and mitochondrial dysfunction in colon epithelia induced by hemin. However, additional experiments are required to evaluate 3,4HPAA protective effects in in vivo models of CRC induced by meat diet.
Supplementary Materials
The following are available online, Figure S1: Effect of SFN, QUE and 3,4HPAA in cell viability, in the presence of hemin, Figure S2: Comparison of the effect of QUE and 3,4HPAA in apoptosis, Figure S3: Comparison of the effect of QUE and 3,4HPAA in ROS levels, Figure S4: Comparison of the effect of QUE and 3,4HPAA in mitochondrial parameters.
Author Contributions
M.C. and C.C.-P. designed and performed the experiments; collected and analyse the data; and wrote the manuscript. J.F. revised the manuscript and provided resources to perform the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to acknowledge the support of FONDECYT INICIACION 11160281 (M.C.) and FONDECYT Regular 1180296 (J.F.).
Conflicts of Interest
Authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Bailey C.E., Hu C.Y., You Y.N., Bednarski B.K., Rodriguez-Bigas M.A., Skibber J.M., Cantor S.B., Chang G.J. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan D.S., Lau R., Aune D., Vieira R., Greenwood D.C., Kampman E., Norat T. Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS ONE. 2011;6:e20456. doi: 10.1371/journal.pone.0020456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thanikachalam K., Khan G. Colorectal Cancer and Nutrition. Nutrients. 2019;11:164. doi: 10.3390/nu11010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastide N.M., Pierre F.H.F., Corpet D.E. Heme Iron from Meat and Risk of Colorectal Cancer: A Meta-analysis and a Review of the Mechanisms Involved. Cancer Prev. Res. 2011;4:177–184. doi: 10.1158/1940-6207.CAPR-10-0113. [DOI] [PubMed] [Google Scholar]
- 6.Sesink A.L. Ph.D. Thesis. Rijksuniversiteit Groningen; Groningen, The Netherlands: 2000. Groningen, Red Meat and Colon Cancer: A Possible Role for Heme. [Google Scholar]
- 7.Sesink A.L., Termont D.S., Kleibeuker J.H., Van der Meer R. Red meat and colon cancer: The cytotoxic and hyperproliferative effects of dietary heme. Cancer Res. 1999;59:5704–5709. [PubMed] [Google Scholar]
- 8.Kelly G.S. Quercetin. Monograph. Altern. Med. Rev. 2011;16:172–194. [PubMed] [Google Scholar]
- 9.Murakami A., Ashida H., Terao J. Multitargeted cancer prevention by quercetin. Cancer Lett. 2008;269:315–325. doi: 10.1016/j.canlet.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 10.Darband S.G., Kaviani M., Yousefi B., Sadighparvar S., Pakdel F.G., Attari J.A., Mohebbi I., Naderi S., Majidinia M. Quercetin: A functional dietary flavonoid with potential chemo-preventive properties in colorectal cancer. J. Cell. Physiol. 2018;233:6544–6560. doi: 10.1002/jcp.26595. [DOI] [PubMed] [Google Scholar]
- 11.Cai X., Fang Z., Dou J., Yu A., Zhai G. Bioavailability of quercetin: Problems and promises. Curr. Med. Chem. 2013;20:2572–2582. doi: 10.2174/09298673113209990120. [DOI] [PubMed] [Google Scholar]
- 12.Landete J.M. Updated knowledge about polyphenols: Functions, bioavailability, metabolism, and health. Crit. Rev. Food Sci. Nutr. 2012;52:936–948. doi: 10.1080/10408398.2010.513779. [DOI] [PubMed] [Google Scholar]
- 13.Aura A.M., O’Leary K.A., Williamson G., Ojala M., Bailey M., Puupponen-Pimia R., Nuutila A.M., Oksman-Caldentey K.M., Poutanen K. Quercetin derivatives are deconjugated and converted to hydroxyphenylacetic acids but not methylated by human fecal flora in vitro. J. Agric. Food Chem. 2002;50:1725–1730. doi: 10.1021/jf0108056. [DOI] [PubMed] [Google Scholar]
- 14.Benov L., Sztejnberg L., Fridovich I. Critical evaluation of the use of hydroethidine as a measure of superoxide anion radical. Free Radic. Biol. Med. 1998;25:826–831. doi: 10.1016/S0891-5849(98)00163-4. [DOI] [PubMed] [Google Scholar]
- 15.Blaut M., Schoefer L., Braune A. Transformation of flavonoids by intestinal microorganisms. Int. J. Vitam Nutr. Res. 2003;73:79–87. doi: 10.1024/0300-9831.73.2.79. [DOI] [PubMed] [Google Scholar]
- 16.Braune A., Gutschow M., Engst W., Blaut M. Degradation of quercetin and luteolin by Eubacterium ramulus. Appl. Environ. Microbiol. 2001;67:5558–5567. doi: 10.1128/AEM.67.12.5558-5567.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeBlanc L.M., Pare A.F., Jean-Francois J., Hebert M.J., Surette M.E., Touaibia M. Synthesis and antiradical/antioxidant activities of caffeic acid phenethyl ester and its related propionic, acetic, and benzoic acid analogues. Molecules. 2012;17:14637–14650. doi: 10.3390/molecules171214637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manach C., Williamson G., Morand C., Scalbert A., Remesy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005;81:230S–242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 19.Schneider H., Schwiertz A., Collins M.D., Blaut M. Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract. Arch. Microbiol. 1999;171:81–91. doi: 10.1007/s002030050682. [DOI] [PubMed] [Google Scholar]
- 20.Schoefer L., Mohan R., Schwiertz A., Braune A., Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl. Environ. Microbiol. 2003;69:5849–5854. doi: 10.1128/AEM.69.10.5849-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelino D., Dosz E.B., Sun J., Hoeflinger J.L., Van Tassell M.L., Chen P., Harnly J.M., Miller M.J., Jeffery E.H. Myrosinase-dependent and –independent formation and control of isothiocyanate products of glucosinolate hydrolysis. Front. Plant Sci. 2015;6:831. doi: 10.3389/fpls.2015.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke J.D., Dashwood R.H., Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269:291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullah M.F. Sulforaphane (SFN): An Isothiocyanate in a Cancer Chemoprevention Paradigm. Medicines. 2015;2:141–156. doi: 10.3390/medicines2030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg P. Red Meat-Derived Nitroso Compounds, Lipid Peroxidation Products and Colorectal Cancer. Foods. 2019;8:252. doi: 10.3390/foods8070252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.IJssennagger N., Rijnierse A., de Wit N., Jonker-Termont D., Dekker J., Müller M., van der Meer R. Dietary haem stimulates epithelial cell turnover by downregulating feedback inhibitors of proliferation in murine colon. Gut. 2012;61:1041–1049. doi: 10.1136/gutjnl-2011-300239. [DOI] [PubMed] [Google Scholar]
- 26.de Vogel J., van-Eck W.B., Sesink A.L.A., Jonker-Termont D.S.M.L., Kleibeuker J., van der Meer R. Dietary heme injures surface epithelium resulting in hyperproliferation, inhibition of apoptosis and crypt hyperplasia in rat colon. Carcinogenesis. 2008;29:398–403. doi: 10.1093/carcin/bgm278. [DOI] [PubMed] [Google Scholar]
- 27.de Vogel J., Jonker-Termont D.S.M.L., van Lieshout E.M.M., Katan M.B., van der Meer R. Green vegetables, red meat and colon cancer: Chlorophyll prevents the cytotoxic and hyperproliferative effects of haem in rat colon. Carcinogenesis. 2005;26:387–393. doi: 10.1093/carcin/bgh331. [DOI] [PubMed] [Google Scholar]
- 28.Gao K., Xu A., Krul C., Venema K., Liu Y., Niu Y., Lu J., Bensoussan L., Seeram N.P., Heber D., et al. Of the major phenolic acids formed during human microbial fermentation of tea, citrus, and soy flavonoid supplements, only 3,4-dihydroxyphenylacetic acid has antiproliferative activity. J. Nutr. 2006;136:52–57. doi: 10.1093/jn/136.1.52. [DOI] [PubMed] [Google Scholar]
- 29.Skrbek S., Rufer C.E., Marko D., Esselen M. Quercetin and its microbial degradation product 3,4-dihydroxyphenylacetic acid generate hydrogen peroxide modulating their stability under in vitro conditions. J. Food Nutr. Res. 2009;48:129–140. [Google Scholar]
- 30.Mitchison T.J. The proliferation rate paradox in antimitotic chemotherapy. Mol. Biol. Cell. 2012;23:1–6. doi: 10.1091/mbc.e10-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Kurita A., Nakashima S., Zhu B., Munemasa S., Nakamura T., Murata Y., Nakamura Y. 3,4-Dihydroxyphenylacetic acid is a potential aldehyde dehydrogenase inducer in murine hepatoma Hepa1c1c7 cells. Biosci. Biotechnol. Biochem. 2017;81:978–1983. doi: 10.1080/09168451.2017.1361809. [DOI] [PubMed] [Google Scholar]
- 32.Carrasco-Pozo C., Gotteland M., Castillo R.L., Chen C. 3,4-Dihydroxyphenylacetic acid, a microbiota-derived metabolite of quercetin, protects against pancreatic beta-cells dysfunction induced by high cholesterol. Exp. Cell Res. 2015;334:70–282. doi: 10.1016/j.yexcr.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Tralma K., Gotteland M., Carrasco-Pozo C. 3, 4-Dihydroxyphenyl Acetic Acid, A Microbial Metabolite of Quercetin, Protects Intestinal and Pancreatic Beta Cell Lines from the Cytotoxicity Induced by Rotenone. SL Nutr. Metab. 2017;1:112. [Google Scholar]
- 34.Martín P.L., Ceccatto P., Razori M.V., Francés D.E., Arriaga S.M., Pisani G.B., Martínez A.I., Sánchez Pozzi E.J., Roma M.G., Basiglio C.L. Heme oxygenase-1 induction by hemin prevents oxidative stress-induced acute cholestasis in the rat. Clin. Sci. 2019;133:117–134. doi: 10.1042/CS20180675. [DOI] [PubMed] [Google Scholar]
- 35.Kumar D., Jena G.R., Ram M., Lingaraju M.C., Singh V., Prasad R., Kumawat S., Kant V., Gupta P., Tandan S.K., et al. Hemin attenuated oxidative stress and inflammation to improve wound healing in diabetic rats. Naunyn-Schmiedebergs Arch. Pharmacol. 2019;392:1435–1445. doi: 10.1007/s00210-019-01682-7. [DOI] [PubMed] [Google Scholar]
- 36.Tronel C., Rochefort G.Y., Arlicot N., Bodard S., Chalon S., Antier D. Oxidative stress is related to the deleterious effects of heme oxygenase-1 in an in vivo neuroinflammatory rat model. Oxid. Med. Cell. Longev. 2013;2013:264935. doi: 10.1155/2013/264935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson S.R., Dang T.N., Dringen R., Bishop G.M. Hemin toxicity: A preventable source of brain damage following hemorrhagic stroke. Redox Rep. 2009;14:228–235. doi: 10.1179/135100009X12525712409931. [DOI] [PubMed] [Google Scholar]
- 38.González-Reyes S., Guzmán-Beltrán S., Medina-Campos O.N., Pedraza-Chaverri J. Curcumin Pretreatment Induces Nrf2 and an Antioxidant Response and Prevents Hemin-Induced Toxicity in Primary Cultures of Cerebellar Granule Neurons of Rats. Oxid. Med. Cell. Longev. 2013;2013:801418. doi: 10.1155/2013/801418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice-Evans C.A., Miller N.J., Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 40.Tonelli C., Chio L.I.C., Tuveson D. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKenzie E.L., Iwasaki K., Tsuji Y. Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:997–1030. doi: 10.1089/ars.2007.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higdon A.N., Benavides G.A., Chacko B.K., Ouyang X., Johnson M.S., Landar A., Zhang J., Darley-Usmar V.M. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: The protective role of autophagy. Am. J. Physiol. Heart Circ. Physiol. 2012;302:13. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sreevalsan S., Safe S. Reactive Oxygen Species and Colorectal Cancer. Curr. Colorectal Cancer Rep. 2013;9:350–357. doi: 10.1007/s11888-013-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartsch H., Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: Role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch. Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 45.Mal’tseva D.V., Makarova Y.A., Raigorodskaya M.P., Rodin S.A. Effects of Laminins 332 and 411 on the Epithelial—Mesenchymal Status of Colorectal Cancer Cells. Bull. Exp. Biol. Med. 2019;166:377–382. doi: 10.1007/s10517-019-04354-x. [DOI] [PubMed] [Google Scholar]
- 46.Murley J.S., Miller R.C., Weichselbaum R.R., Grdina D.J. TP53 Mutational Status and ROS Effect the Expression of the Survivin-Associated Radio-Adaptive Response. Radiat. Res. 2017;188:579–590. doi: 10.1667/RR14831.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu X., Yu H., Ma Q., Shen S., Das U.N. Linoleic acid suppresses colorectal cancer cell growth by inducing oxidant stress and mitochondrial dysfunction. Lipids Health Dis. 2010;9:9–106. doi: 10.1186/1476-511X-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrasco-Pozo C., Gotteland M., Speisky H. Apple peel polyphenol extract protects against indomethacin-induced damage in Caco-2 cells by preventing mitochondrial complex I inhibition. J. Agric. Food Chem. 2011;59:11501–11508. doi: 10.1021/jf202621d. [DOI] [PubMed] [Google Scholar]
- 50.Grivennikova V.G., Kozlovsky V.S., Vinogradov A.D. Respiratory complex II: ROS production and the kinetics of ubiquinone reduction. Biochim. Biophys. Acta BBA Bioenerg. 2017;1858:109–117. doi: 10.1016/j.bbabio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Turrens J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenner A.M., Rafter J., Halliwell B. Human fecal water content of phenolics: The extent of colonic exposure to aromatic compounds. Free Radic. Biol. Med. 2005;38:763–772. doi: 10.1016/j.freeradbiomed.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Carrasco-Pozo C., Tan K.N., Reyes-Farias M., De La Jara N., Ngo S.T., Garcia-Diaz D.F., Llanos P., Cires M.J., Borges K. The deleterious effect of cholesterol and protection by quercetin on mitochondrial bioenergetics of pancreatic beta-cells, glycemic control and inflammation: In vitro and in vivo studies. Redox. Biol. 2016;9:229–243. doi: 10.1016/j.redox.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrasco-Pozo C., Gotteland M., Speisky H. Protection by apple peel polyphenols against indometacin-induced oxidative stress, mitochondrial damage and cytotoxicity in Caco-2 cells. J. Pharm. Pharmacol. 2010;62:943–950. doi: 10.1211/jpp.62.07.0017. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez-Candia A., Veliz M., Carrasco-Pozo C., Castillo R.L., Cardenas J.C., Ebensperger G., Reyes R.V., Llanos A.J., Herrera E.A. Antenatal melatonin modulates an enhanced antioxidant/pro-oxidant ratio in pulmonary hypertensive newborn sheep. Redox. Biol. 2019;22:101128. doi: 10.1016/j.redox.2019.101128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carrasco-Pozo C., Tan K.N., Borges K. Sulforaphane is anticonvulsant and improves mitochondrial function. J. Neurochem. 2015;135:932–942. doi: 10.1111/jnc.13361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.