Abstract
Vascular endothelial growth factor A (VEGFA) stimulates angiogenesis in human endothelial cells, and increasing its expression is a potential treatment for heart failure. Here, we report the design of a small molecule (TGP-377) that specifically and potently enhances VEGFA expression by the targeting of a non-coding microRNA that regulates its expression. A selection-based screen, named two-dimensional combinatorial screening, revealed preferences in small-molecule chemotypes that bind RNA and preferences in the RNA motifs that bind small molecules. The screening program increased the dataset of known RNA motif–small molecule binding partners by 20-fold. Analysis of this dataset against the RNA-mediated pathways that regulate VEGFA defined that the microRNA-377 precursor, which represses Vegfa messenger RNA translation, is druggable in a selective manner. We designed TGP-377 to potently and specifically upregulate VEGFA in human umbilical vein endothelial cells. These studies illustrate the power of two-dimensional combinatorial screening to define molecular recognition events between ‘undruggable’ biomolecules and small molecules, and the ability of sequence-based design to deliver efficacious structure-specific compounds.
Main
Small molecules interacting with RNA (SMIRNAs) have broad potential to enable the study of RNA’s biological functions and to develop novel therapeutics1,2. The most common way to target RNA is through its sequence-based recognition by antisense oligonucleotides3,4. RNA, however, folds into three-dimensional (3D) structures that control its biology, including the causation of disease5,6. Thus, SMIRNAs are an alternative approach to targeting RNA at distinct target sites with potential advantages.
The design of SMIRNAs has been enabled by correlating RNA 3D folds with empirically derived interactions with small molecules7. These interactions are identified through a library-versus-library selection strategy dubbed two-dimensional combinatorial screening (2DCS)8. Our lead identification strategy, Inforna9, searches for overlap between binding partners defined by 2DCS and 3D folds in cellular RNAs, informing the design of bioactive ligands. This approach has the potential to be both general and scalable.
We have investigated the scalability of 2DCS to study the RNA-binding capacity of compound collections found within the pharmaceutical industry and whether the resulting data could be applied to a predetermined target. We deployed various technologies to probe quickly 63 million interactions between small molecules from the AstraZeneca compound library (n = 2,947) and RNA folds (n = 21,504). These studies defined not only the SMIRNAs themselves, but also features within them and the RNAs they bind. Our approach was then validated by identifying RNA folds of varying affinity in multiple cardiac-related microRNAs (miRNAs), and we show that the predictive power of the approach scales with both compound affinity in vitro and activity in cells. We then applied our privileged RNA motif–small molecule interactions to enhance vascular endothelial growth factor A (VEGFA) expression. Upregulation of VEGFA is a promising strategy for the management of coronary artery disease. Yet there are no known small molecules that increase VEGFA expression, as gene or messenger RNA therapies are the only approaches known to enhance its expression10,11,12,13. Collectively, these studies will probably inform the design of bioactive ligands targeting RNA broadly.
Results
Defining small molecule–RNA fold interactions
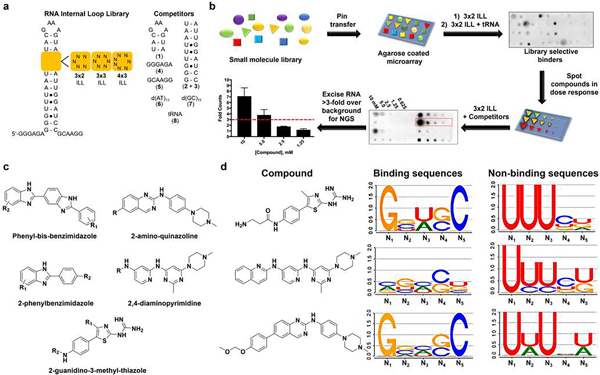
In silico analysis of the AstraZeneca corporate collection (2M compounds) was conducted using the physicochemical properties of 27 chemically diverse RNA binders from Inforna9 and the literature (Supplementary Table 1), generating a 1,967-member compound diversity set. The binding of these compounds to libraries of 3D RNA folds was then studied. The RNA motif libraries comprise a unimolecular hairpin that displays various structural elements, including 3 × 2 (3 × 2 internal loop library (ILL), n = 1,024 unique RNAs), 3 × 3 (3 × 3 ILL, n = 4,096 unique RNAs) and 4 × 3 (4 × 3 ILL, n = 16,384 unique RNAs) internal loops (Fig. 1a).
Fig. 1 |. Chemoinformatics and bioinformatics analysis of small molecules that bind RNA and the RNAs that bind small molecules.
a, RNA libraries used for screening the AstraZeneca collection. The 3 × 2 ILLs, 3 × 3 ILLs and 4 × 3 ILLs contain 1,024, 4,096 and 16,384 unique RNA sequences, respectively, totalling more than 63 million interactions probed by 2DCS. Competitor oligonucleotides used to restrict 2DCS selections to the randomized region are mimics of the hairpin (1), stem (2 + 3), 5′ tail (4), 3′ tail (5) as well as DNA (6, 7) and tRNA (8). b, Schematic of the AbsorbArray screening methodology14. NGS, next-generation sequencing. c, Tanimoto coefficient analysis identified five conserved classes of scaffolds, namely phenylbenzimidazoles, phenyl-bis-benzimidazoles, 2-aminoquinazolines, 4,6-diaminopyrimidines and 2-guanidinothiazoles, as RNA-biased binders. d, LOGOS analysis of the RNA sequences that bind compounds from the 3 × 2 ILLs; additional details can be found in the Supplementary Information. Examples are shown for the 2-guanidinothiazoles (C6), 4,6-diaminopyrimidine (C11) and 2-aminoquinazoline (C2), with compounds C6 and C2 being novel RNA-binding scaffolds. Nx denotes the position of the nucleotide in the randomized region, numbered in order from 5′ to 3′.
We applied our previously validated method14, AbsorbArray, to identify selective RNA binders from the AstraZeneca collection. The small-molecule library was absorbed onto an agarose coating and allowed to dry. After washing, radiolabelled RNA motif libraries were incubated on the surface in the presence of 100-fold excess of unlabelled competitor oligonucleotides that mimic the constant regions in the library (1–5), DNA (6, 7) and transfer RNA (8; Fig. 1a). The RNAs bound to each small molecule were excised, subjected to PCR with reverse transcription (RT–PCR) and analysed by RNA sequencing (RNA-seq; Fig. 1b). An aliquot of the RNA library that had not been incubated with the arrays was also subjected to RT–PCR and analysed by RNA-seq to remove potential biases in the downstream analysis of selected RNAs. That is, the frequency of an RNA in the RNA-seq analysis of selected RNAs was compared with the frequency of the same RNA in the RNA-seq analysis of the unhybridized library by a pooled population comparison. This rigorous statistical method is dubbed high-throughput structure–activity relationships through sequencing (HiT-StARTS). It identifies privileged RNA motif–small molecule interactions, defines the RNA-binding landscapes for a given small molecule and can predict small-molecule selectivity15. Due to the combinatorial nature of a library-versus-library screen, over 42 million RNA fold–small molecule binding interactions were probed simultaneously in this experiment. Of the 1,967 small molecules studied, 27 selectively bound the unique 3D folds displayed by the RNA libraries (1.4% hit rate).
Features in small molecules that bind RNA
By analysing the structural differences between binders and non-binders, features that confer RNA binding were defined. RNA binders were enriched in aromatic rings, such as benzimidazoles, phenylquinazolines, pyrrolopyridine, pyrimidine, pyridines and guanine-like moieties. Other enriched functionalities included piperazines, spirocycles (C3), guanidines (C5–C7), alkoxy groups (C11), exocyclic primary amines (C15–C25), tertiary amines (C22, C26) and nitrogen-rich heterocycles (Supplementary Table 2).
An analysis of the calculated molecular properties highlighted differences between the starting library and bona fide RNA binders (Supplementary Table 3). The RNA binders were on average more lipophilic (change in calculated partition coefficient ΔClogP = 1.3, where P is defined as the distribution of the compound in octanol versus water) and less flexible (that is, fewer rotational bonds, 4.3 vs 5.7). The RNA-binding small molecules also had on average two additional hydrogen-bond donors and a greater polar surface area. Further, many of the descriptors suggest a preference for a planar-like shape. That is, the hits include an additional ring, an increased occurrence of aromatic atoms at the expense of fewer aliphatic atoms, and fewer chiral centres. The average fraction of sp3 carbon atoms (a measure of carbon bond saturation) was 0.11, compared with 0.24 for the starting small-molecule library. Finally, a large number of RNA binders, ~50%, included a basic functionality.
We next compared our small molecules with those housed in databases of bioactive RNA-targeting ligands, namely SMMRNA, published in 2014, and R-BIND, published in 2019 (refs. 16,17). Interestingly, the comparison with the small molecules in R-BIND revealed that only three of our hits have a Tanimoto score >0.7, leaving 24 hits as highly dissimilar, novel RNA binders, as determined by fingerprint analysis (Supplementary Table 3)18,19. Despite their structural dissimilarity, the physicochemical properties of the RNA-binding small molecules reported herein are similar to those in the R-BIND database16 and those already housed in Inforna9,20 (Supplementary Table 4). Collectively, these data indicate that we have indeed identified novel chemical matter and suggest that a set of physicochemical properties might characterize RNA-binding ligands.
These preferences were used to select compound nearest neighbours from the AstraZeneca collection, providing an additional 980 RNA-focused small molecules to be studied by AbsorbArray and 2DCS. In this second screen, 21 million interactions were probed, affording ten selective RNA-binding small molecules (1.1% hit rate). Tanimoto analysis revealed four additional conserved scaffolds that avidly bind RNA, namely 2-guanidinothiazoles, 2-aminoquinazolines, 4,6-diaminopyrimidines and phenylbenzimidazoles, which also includes phenyl-bis-benzimidazoles (Fig. 1c and Supplementary Fig. 1).
Features in RNA folds that bind small molecules
The RNA 3D folds that bound each small molecule were analysed by HiT-StARTS, which calculates the enrichment of a particular SMIRNA-bound RNA fold in 2DCS sequencing data as compared to the sequencing data derived for the starting RNA motif library. The statistical significance of that enrichment is reported as the Z-score (Zobs)16. HiT-StARTS precisely defines (1) the number of RNA folds that each small molecule binds selectively, (2) the relative affinity of each interaction and (3) a catalogue of RNA fold–small molecule binding partners that can inform drug/probe design7. Previous studies showed that avid SMIRNAs have a Zobs > 8 and that the relative affinity of the selected interactions directly correlates with Zobs (ref. 15). Small molecules with higher Zobs are more fit for binding an RNA than those with lower scores. In summary, the two screens defined 41,000 RNA fold–small molecule interactions that were deposited into Inforna, an increase of ~20-fold over the current Inforna server (Supplementary Table 5)9.
Some small molecules only bound members of certain RNA libraries, and some bound members from all three. For example, 4 × 3 ILLs bound 18 compounds (n = 3 unique) whereas 3 × 2 ILLs bound 16 (n = 1 unique) and 3 × 3 ILLs bound 15 small molecules (n = 1 unique). Perhaps 4 × 3 ILLs bound a larger subset of the small molecules because of the greater diversity in the RNA motif library. A detailed analysis of the types of RNA folds that bind small molecules, including a LOGOS analysis21, is provided in Supplementary Figs. 2–6. Some interesting observations include the following: (1) single nucleotide bulges bind a variety of small molecules (Fig. 1d and Supplementary Table 5), (2) randomized regions with additional base pairs and higher GC pairing appear more frequently in RNAs that bind small molecules than those that do not and (3) U-rich internal loops have lower small-molecule binding capacity.
Dynamics are a key determinant in RNA function, and local dynamics could affect ligand binding capacity22,23. If the structure of a target RNA is too dynamic, ligand binding potential could be diminished as a defined small-molecule binding pocket may not be present or the affinity for an ill-defined pocket could be low due to the energetic and entropic penalties associated with locking out multiple conformations. Previous studies have provided some insight into RNA structural dynamics; for example, smaller loops are not typically as conformationally dynamic as their larger counterparts and U-rich motifs can have dynamic character22,24. We were interested in whether the SMIRNAs studied herein had a preference for dynamic or structurally well-defined loops. Fortuitously, a set of structurally related small molecules showed overlap in the RNAs that they preferred and did not prefer to bind. We studied the structural dynamics of these loops by nuclear magnetic resonance (NMR) spectroscopy. As shown in Supplementary Fig. 7, RNAs that bind small molecules have defined structure, as evidenced by the presence of all possible imino proton peaks. By contrast, various peaks from imino protons were absent in the spectra of RNAs that generally do not bind small molecules (Supplementary Fig. 7). Our data suggest that a defined RNA structure is a contributing factor for SMIRNA binding, at least for the scaffolds studied herein.
Inforna identifies small molecules that selectively target functional sites
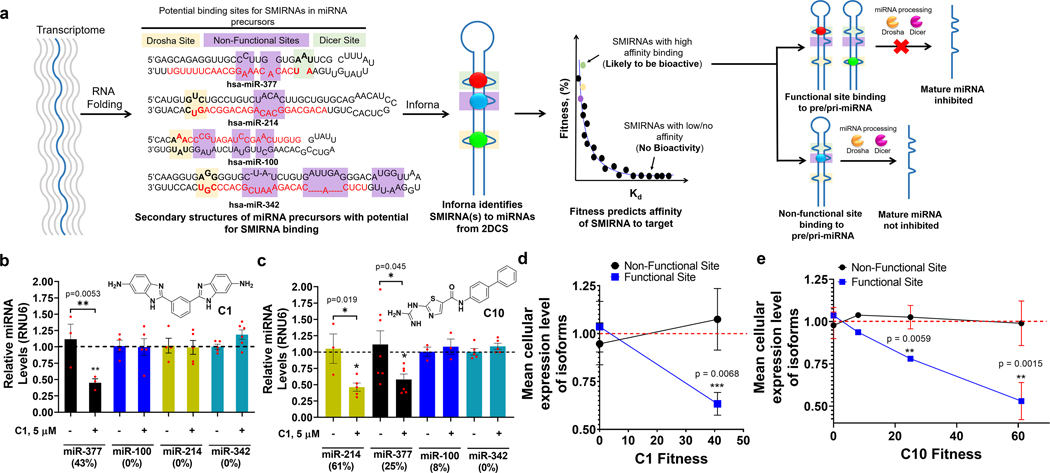
The Inforna pipeline9 compares structural elements within RNAs of potential therapeutic interest with its database of RNA motif–small molecule interactions. Lead interactions will have varying affinities15 and hence potential for bioactivity, as determined by Zobs. We have shown that selective, avid interactions have Zobs > 815,25, and that binding in a functional site is required for bioactivity (Fig. 2a)7,26,27.
Fig. 2 |. SMIRNA strategy and method validation in vitro and in HUVECs.
a, Schematic of how SMIRNAs are identified from sequencing using Inforna. Features with high fitness scores are likely to yield bioactive interactions. For miRNAs, SMIRNAs must have sufficient fitness and occupy a functional site (Drosha site, tan boxes; Dicer site, green boxes; non-functional sites, purple boxes). pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA. b,c, Effect of C1 (b) and C10 (c) on miRNAs with different fitness scores in HUVECs, as determined by measuring mature miRNA levels by RT–qPCR. Data are reported as mean ± s.e.m. (n= 4 biologically independent samples). d,e, Activity of C1 (d) and C10 (e) as a function of fitness score against all isoforms of miR-377, miR-214, miR-100 and miR-342. Data are reported as mean ± s.e.m. In d, n= 15 miRNAs where the SMIRNA binds to non-functional sites and n= 2 miRs where the SMIRNA binds to functional sites; in e, n= 15 miRNAs where the SMIRNA binds to non-functional sites and n= 4 miRNAs where the SMIRNA binds to functional sites. All P values were determined from a two-sided Student’s t-test; *, P<0.05; **, P< 0.01; ***, P< 0.001.
To validate that these observations hold for the 2DCS selections reported herein, we studied two representative compounds, C1 and C10, with phenylbenzimidazole and 2-guanidinothiazole scaffolds, respectively. We focused solely on motifs in Drosha or Dicer processing sites. C1 is predicted to bind the Dicer processing (functional) sites of miR-377 (5′-AAU/3′-U_A; fitness = 43%) and to have no avidity (fitness = 0%) for Drosha processing sites in miR-214 (5′-GUC/3′-CUG), miR-100 (5′-AAA/3′-UAU) and miR-342 (5′-AGG/3′-UGC) where underscored nucleotides and positions indicate single-stranded or non-canonically paired nucleotides in bulges or loops. These miRNAs were chosen due to their collective expression in human umbilical vein endothelial cells (HUVECs) and their modulation of cardiac-related phenotypes: (1) miR-377 inhibits VEGFA in heart failure blocking angiogenesis28, (2) miR-214 inhibits PPARα and XBP1 blocking cardiomyocyte metabolism and angiogenesis, respectively29,30, (3) miR-100 is upregulated in atherosclerotic tissue31 and (4) miR-342 regulates angiogenesis by modulation of TGFβ signalling in endothelial cells32.
Binding analysis by microscale thermophoresis showed that C1’s affinity for miR-377’s Dicer site is 29 ± 0.3 μM, with no saturable binding observed to a control base-paired RNA, miR-214, miR-100 or miR-342 (Supplementary Fig. 8). By contrast, C10 is predicted to bind the processing site of the four miRNAs with varying affinities based on their fitness scores: miR-214, fitness = 61%; miR-377, fitness = 25%; miR-100, fitness = 8%; miR-342, fitness = 0%. Binding analyses showed that fitness correlates with affinity (29 to >100 μM; Supplementary Fig. 8), in agreement with previous studies15.
To assess whether fitness for a functional site correlates with bioactivity, the inhibitory effect of both compounds on all four miRNAs was studied in HUVECs. HUVECs were treated with 5 μM of C1 or 2.5 μM of C10, the maximum non-toxic doses as determined by cell viability assays, and the levels of each mature miRNA were measured by quantitative RT–PCR (RT–qPCR). HUVECs treated with C1 showed selective downregulation of miR-377 (~50% reduction), but no change in the levels of miR-214, miR-100 and miR-342 was observed because C1 does not bind these RNAs (Fig. 2b). In cells treated with C10, miR-214 was inhibited by ~60%, miR-377 by ~50% with miR-100 and miR-342 left unaffected, mirroring the pattern of C10’s fitness and avidity for each processing site (Fig. 2c).
We next studied the effects of C1 and C10 on miRNAs that contain the same motifs present in the Dicer site of miR-377 (n = 5) and the Drosha sites of miR-100 (n = 1), miR-214 (n = 8) and miR-342 (n = 4), or RNA isoforms (Supplementary Table 6), and whose expression levels in HUVECs are measurable. The motifs in the RNA isoforms are located in both functional processing sites and non-functional sites (Supplementary Table 6). For C1, no effect was observed on 11 of the 13 isoforms of miR-100, miR-214 and miR-342, regardless of the motif’s location in a functional or non-functional site, as the compound does not bind (Supplementary Table 6 and Supplementary Fig. 8). For the five miRNAs that have the same motif as miR-377’s Dicer site, four are located in non-functional sites and one is located in pre-miR-421’s Dicer processing site (Supplementary Table 6). In agreement with previous studies7,26,27, only the levels of miR-421 were affected (by ~40%; Supplementary Fig. 8).
For C10, no effect of compound treatment was observed for any miR-342 isoform, because the fitness for the 5′-AGG/3′-UGC motif is 0% (Supplementary Fig. 8). The best-fit interaction for C10 is with the 5′-GUC/3′-CUG motif in miR-214’s Drosha site (61%). Of its eight RNA isoforms, the motif is found in the processing site of only one miRNA, miR-2116, the levels of which were reduced significantly upon C10 treatment (P = 0.0006); the levels of the other seven isoforms were unaffected (Supplementary Fig. 8). This trend continued for isoforms of miR-377 and miR-100, that is, mature miRNA levels were unaffected for all isoforms in which the SMIRNA binding motif is in a non-functional site (Supplementary Fig. 8). The miR-377 motif is also present in the Dicer processing site of miR-421, however, its levels were unaffected by C10 treatment (Supplementary Fig. 8). These results contrast those observed for C1. This observation can be attributed to two factors, namely the difference in fitness (43 vs 25% for C1 and C10, respectively) and the high expression level of miR-421, which is 31-fold higher than that of miR-377.
Collectively, these studies show that SMIRNA binding to a functional site is required for bioactivity, as evidenced by the study of RNA isoforms, and that potency is correlated with the fitness of the SMIRNA’s interaction with the functional site (Fig. 2d,e).
Given these favourable results, we turned to a different target type, the r(G4C2) repeat expansion [r(G4C2)exp] found in the C9orf72 gene, the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia (c9ALS/FTD)33,34. This RNA folds into two structures that are in equilibrium in vitro, a G-quadruplex35 and hairpin displaying a periodic array of 1 × 1 G/G internal loops in its stem36,37. The hairpin structure causes toxicity by an RNA gain-of-function mechanism, in particular undergoing repeat-associated non-ATG (RAN) translation37 to form toxic dipeptide repeats that cause neuronal death (Supplementary Fig. 9)38. We identified a potential lead small molecule for the 1 × 1 G/G internal loops, C20, with a fitness score of 50%. C20 binds to r(G4C2)8 repeats with a dissociation constant (Kd) of 620 ± 110 nM, with no saturable binding to base-paired RNA (Supplementary Fig. 9). Using an r(G4C2)66–nanoluciferase and an SV40(ATG)-firefly luciferase fusion reporter, we studied C20’s effect on RAN translation of r(G4C2)exp and canonical translation, respectively (Supplementary Fig. 9). C20 selectively inhibited RAN translation with no effect on canonical translation (Supplementary Fig. 9). Together, these data highlight the broad applicability of Inforna.
Compound C1 targets pre-miR-377 and inhibits its processing
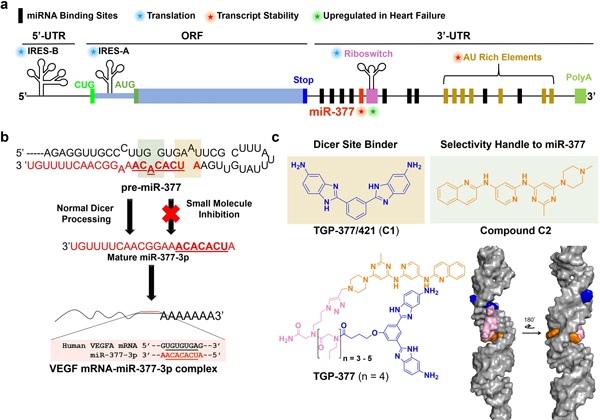
MiR-377 is a particularly interesting therapeutic target because one of its downstream targets is VEGFA, which stimulates vascular tissue repair. Bioinformatics analysis of patient tissue from failing hearts revealed miR-377 is upregulated, thus repressing VEGFA28. An oligonucleotide targeting miR-377 de-repressed VEGFA and stimulated angiogenesis in HUVECs28. Although there are multiple drugs that inhibit VEGFA and its receptor’s activity39,40,41, there are no known compounds that increase its levels. VEGFA expression could be regulated in various ways, including by an internal ribosome entry site (IRES) in its 5′ untranslated region (UTR) and multiple miRNA binding sites in its 3′ UTR, among others (Fig. 3a)42. Small-molecule binding to the IRES or 5′ UTR would most likely repress VEGFA expression; however, inhibition of an miRNA targeting the transcript, such as miR-377, would likely increase expression.
Fig. 3 |. Overview of Vegfa mRNA regulation, miR-377 pathway and targeting strategy.
a, Schematic of pathways that regulate VEGFA expression42. Of the known miRNAs that affect VEGFA, only miR-377 is upregulated post-ischemia. VegfA mRNA contains various regulatory elements, including (1) two IRESs that regulate translation, (2) a stress-responsive atypical riboswitch element that reacts to the binding of hnRNPL protein (activates translation) or interferon gamma-activated inhibitor of translation protein, (3) AU-rich elements (AREs) that control mRNA stability and (4) binding sites for miRNAs, including miR-377, miR-125a and miR-16, among others. b, Post-ischemic upregulation of miR-377 downregulates VEGFA, slowing growth and angiogenesis. A small molecule that binds pre-miR-377’s Dicer site (tan box, binding site for C1) and an adjacent bulge (green box, binding site for C2) can inhibit enzymatic processing and de-repress VEGFA, stimulating angiogenesis and proliferation. TargetScan-predicted binding site of miR-377’s seed sequence to Vegfa mRNA (aligned sequences are boxed); miR-377–3p indicates that the mature miRNA emanates from the 3′ end of the pre-miRNA. c, Design of a selective pre-miR-377 inhibitor by linking C1 (tan box in b indicates the binding site in pre-miR-377) and C2 (green box in b indicates the binding site in pre-miR-377) via compounds C1-COOH and C2-Ak, yielding TGP-377. MD simulations of the binding of TGP-377 to pre-miR-377 suggest that the compound intercalates into the two bulges with the linker threading through the major groove to support binding (bottom right). Both non-canonically paired adenines contribute to the stability of the complex by π−π stacking with the RNA-binding modules.
C1 bound miR-377’s Dicer processing site with a Kd of 29 ± 3 μM and reduced mature miR-377 levels in HUVECs by ~50% when the cells were dosed with 5 μM of the compound. To investigate whether the reduction of mature levels was due to the inhibition of biogenesis, we first studied whether C1 inhibited Dicer processing in vitro, affording a half-maximum inhibitory concentration (IC50) of 50 ± 10 μM, similar to its Kd (Supplementary Fig. 10). To assess the selectivity of this inhibition, we introduced point mutations into pre-miR-377. Pre-miR-377 has two A bulges, one at the Dicer site closed by AU pairs and another upstream closed by GC pairs. The first mutant was created by inserting a U nucleotide opposite the A bulge in the Dicer site to form an AU pair. The other is a double mutant in which both A bulges are replaced by AU pairs. Although properly processed by Dicer, C1 was is unable to inhibit Dicer processing of either mutant (Supplementary Fig. 10). These data support that C1 targets the native Dicer site miR-377 inhibit processing.
To further study whether C1 can inhibit the cellular biogenesis of miR-377, its effect was studied in HUVECs, as they differentiate into tubules in a miR-377- and VEGFA-dependent manner, which can be used to measure angiogenic capacity directly43. C1 reduced mature miR-377 levels in a dose-dependent fashion, with an IC50 of ~5 μM (Supplementary Fig. 11), tenfold more potently than in in vitro studies. The difference between the in vitro and cellular IC50 values can be traced to the active uptake of C1 into HUVECs, as liquid chromatography–tandem mass spectrometry showed an approximately threefold higher intracellular concentration in HUVECs than in the medium (Supplementary Fig. 12).
To confirm the compound mode of action (MOA), the effect of C1 on pre-miR-377 levels was measured. An accumulation of pre-miR-377 was observed, suggesting that the Dicer site is unable to process the RNA, in agreement with in vitro data (Supplementary Fig. 11). We next investigated whether repression of miR-377 de-repressed Vegfa mRNA. Indeed, C1 caused an approximate 30% increase in Vegfa levels with a concomitant increase in VEGFA protein (Supplementary Fig. 11).
Silencing VEGFA inhibits the formation of tubules in HUVECs when grown on a Matrigel matrix12. To investigate the pro-angiogenic potential of compound C1, HUVECs treated with C1 were differentiated on a Matrigel support. Treatment with C1 stimulated angiogenesis by increasing tubule branching density (Supplementary Fig. 13). Furthermore, a stable HUVEC cell line was generated to ablate Vegfa mRNA by expression of a short hairpin RNA (shRNA) targeting this transcript. The shRNA-expressing cell line had reduced ability to differentiate into tubules as the number of tubules was reduced by >60% as compared with wild-type cells not expressing the shRNA (Supplementary Fig. 13). Addition of C1 to these cells showed ablated stimulation of tubule branching, indicating that the activity of C1 was VEGFA-dependent (Supplementary Fig. 13).
Inforna designs a compound with enhanced affinity and specificity
Inforna defined that C1 can target pre-miR-377, however, it also identified eight other miRNA precursors with the A bulge that C1 binds, of which only six are expressed in HUVECs (Supplementary Table 6)44. As shown in Supplementary Fig. 9, treatment with C1 inhibited miR-377 and miR-421, as this compound binds a functional site in both miRNAs. Because of the dual targeting nature of C1, it is henceforth referred to as Targapre-miR-377/421 (TGP-377/421).
To overcome this off-targeting, the structures of pre-miR-377 and pre-miR-421 were analysed, identifying a 5′-G_G/3′-CAC bulge adjacent to pre-miR-377’s Dicer site that is not found in pre-miR-421 (Supplementary Fig. 14). Mining Inforna identified C2 (Fig. 3c), which bound selectively to the bulge over a mutated base-paired control with a Kd of 3 ± 0.3 μM (Supplementary Fig. 14). Thus, linking TGP-377/421 and C2 appropriately could provide a ligand selective for pre-miR-377 (Supplementary Figs. 2 and 14). To do so, derivatives of each module were synthesized: a carboxylic acid was added to TGP-377/421 (C1-COOH) and an alkyne moiety was added to C2 (C2-Ak). C1-COOEt and C2-Ak bound selectively to their respective RNA targets with Kd values of 0.8 ± 0.1 and 1.6 ± 0.1 μM, respectively, with no measurable affinity for the corresponding mutant base-paired RNA control (Supplementary Fig. 14).
The two compounds were tethered to peptoid-based linkers of varying lengths displaying amine and azide handles to couple the compounds C1-COOH and C2-Ak, respectively. The binding of these compounds to a mimic of pre-miR-377 was studied and showed that the heterodimer with four N-n-propylglycine spacers bound with the highest affinity (Kd = 190 ± 50 nM), with no saturable binding to a base-paired mutant RNA (Supplementary Fig. 14). Thus, this optimal compound is dubbed Targapre-miR-377 (TGP-377).
To bolster our binding studies, we employed two methods developed by our laboratory named chemical cross-linking and isolation by pull-down (Chem-CLIP) and its competitive variant competitive Chem-CLIP (C-Chem-CLIP). To enable these studies, we synthesized a Chem-CLIP probe in which TGP-377/421 was conjugated to the cross-linking module chlorambucil (CA) and a biotin purification module (TGP-377/421-CA-Biotin; Supplementary Fig. 15). Incubation of pre-miR-377 with TGP-377-CA-Biotin afforded dose-dependent pull-down of pre-miR-377 (Supplementary Fig. 15). To study the binding of TGP-377 itself, pre-miR-377 was incubated with the heterodimer followed by addition of the Chem-CLIP probe. At a tenfold lower concentration than the Chem-CLIP probe (50 vs 500 nM), TGP-377 reduced the amount of pre-miR-377 pulled down by ~25% (Supplementary Fig. 1). An equimolar amount of TGP-377/421, 500 nM, was required to reduce pull-down to the same extent, indicating that TGP-377 binds more tightly (Supplementary Fig. 15).
Molecular dynamics (MD) simulations were performed to gain an understanding of the atomic level interaction of TGP-377 with the pre-miR-377 hairpin, revealing that the C1 RNA-binding module intercalates into the helix at the Dicer site, displacing the unpaired adenine residue and stacking with the AU closing base pairs (Supplementary Fig. 16). The model shows potential π–π interactions between the central benzene core and one of the benzimidazoles of C1 and the closing pairs of the A bulge. (The other benzimidazole protrudes from the minor groove of the helix; Supplementary Fig. 16.) The stacked benzimidazole also forms two hydrogen-bonding interactions: one between its heterocyclic imine and the U of the 3′ closing pair, and another between the exocyclic amine and the backbone phosphate (Supplementary Fig. 16). The C2 RNA-binding module intercalates into the helix at the adjacent A bulge (non-Dicer site), forming stacking interactions with the closing GC base pairs and causing partial unstacking of the adenine; that is, the interaction is primarily stabilized by stacking interactions (Supplementary Fig. 16). The tenfold higher affinity of TGP-377 compared with C1 can be traced to the combined effects of these interactions.
We next studied whether TGP-377 inhibits Dicer processing of pre-miR-377 in vitro. TGP-377 inhibited Dicer processing of pre-miR-377 with an IC50 of ~500 nM, a 100-fold improvement over TGP-377/421 (IC50 = ~50 μM). Furthermore, mutation of the two A bulges, the binding sites for the two RNA-binding modules, to base pairs ablated TGP-377’s inhibitory effect, demonstrating binding site dependence on activity (Supplementary Fig. 17).
TGP-377 potently and selectively inhibits pre-miR-377 biogenesis, stimulating VEGFA
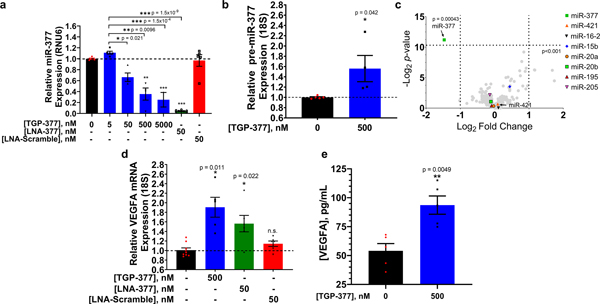
The delivery of TGP-377 into HUVECs decreased mature miR-377 levels tenfold more potently than TGP-377/421 (Fig. 4a). As expected based on its MOA, TGP-377 increased pre-miR-377 levels by 1.6-fold when HUVECs were treated with 500 nM of the compound (Fig. 4b).
Fig. 4 |. Activity of heterodimer TGP-377 in HUVECs.
a, Treatment of HUVECs with TGP-377 for 48 h reduced levels of mature miR-377 dose-dependently, as normalized to RNU6 (housekeeping gene). Data are reported as mean ± s.e.m. (n= 3 biologically independent replicates). b, Effect of TGP-377 treatment (500 nM) on pre-miR-377 levels, indicating the blockade of Dicer processing, as normalized to 18S rRNA. Data are reported as mean ± s.e.m. (n= 6 biologically independent replicates). c, Selectivity of TGP-377, as assessed by miRNA profiling. RNAs that share the structure of miR-377’s Dicer site, or RNA isoforms, are indicated by coloured shapes. TGP-377 is more selective than the substrate from which it was derived (C1), with exemplar cases of miR-421 and miR-16–2 shown, which are expressed at tenfold higher levels than miR-377. Data are reported as mean ± s.e.m. (n= 3 biologically independent replicates). d, Vegfa mRNA levels increased 1.8-fold upon TGP-377-treatment (500 nM). Data are reported as mean ± s.e.m. (n= 6 biologically independent replicates). n.s., not significant. e, Amount of VEGFA secreted from medium supernatant, as determined by the enzyme-linked immunosorbent assay (ELISA). Data are reported as mean ± s.e.m. (n= 3 biologically independent replicates). P values were determined by a two-sided Student’s t-test; *, P<0.05; **, P< 0.01; ***, P< 0.001. In c, the P values were not adjusted for multiple comparisons.
To demonstrate that TGP-377 binds pre-miR-377 in cellulis, we completed Chem-CLIP and C-Chem-CLIP studies in HUVECs. A dose-dependent enrichment of pre-miR-377 in the pulled-down fraction was observed: 500 nM TGP-377/421-CA-Biotin enriched pre-miR-377 levels by around threefold, whereas 2.5 μM of the Chem-CLIP probe enriched levels by around eightfold (Supplementary Fig. 15). To study target engagement of the parent compound TGP-377, HUVEC cells were pretreated with TGP-377 (500 nM) followed by the addition of an equimolar amount of TGP-377/421-CA-Biotin. TGP-377 reduced the enrichment of pre-miR-377 in the pulled-down fraction by ~50% (Supplementary Fig. 15), indicating that it competed for the same binding site as TGP-377/421-CA-Biotin. Collectively, these studies demonstrate that TGP-377 directly engages pre-miR-377 in HUVECs, thereby inhibiting its biogenesis.
The selectivity of TGP-377 was assessed by miRNA profiling. First, we studied the effect of TGP-377 on miRNAs predicted to modulate VEGFA production using TargetScan (ref. 45). These miRNAs included miR-15b, miR-16, miR-20a, miR-20b, miR-195, miR-377 and miR-205 (ref. 42). Analysis of their precursor hairpins showed that only pre-miR-377 contains the 5′-AAC/3′-U_A target site. Among >200 miRNAs in HUVECs, only miR-377 was significantly inhibited, by >50% (P < 0.001), upon treatment with TGP-377 (Fig. 4c). Importantly, and in contrast to the observations with TGP-377/421, pre-miR-421 was not affected (orange triangle in the volcano plot, Fig. 4c), demonstrating that TGP-377 is selective for pre-miR-377. As Vefga mRNA is directly regulated by miR-377 (ref. 28), inhibition of its biogenesis by TGP-377 enhanced levels of Vegfa mRNA by >50% at 500 nM, as expected (Fig. 4d).
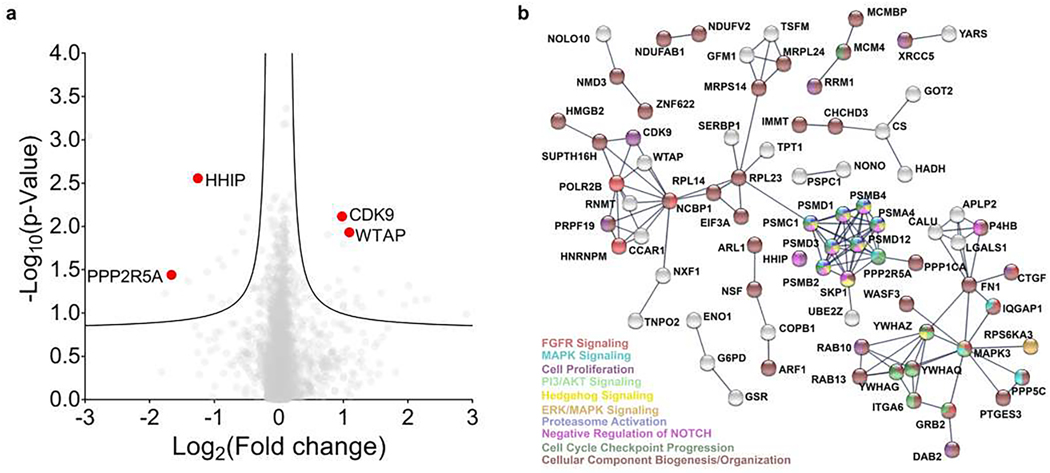
TGP-377 selectively alters the proteome in HUVECs
Treatment of HUVECs with 500 nM TGP-377 increased levels of excreted VEGFA in HUVEC media supernatants twofold (Fig. 4e), with a similar effect observed by the Western blot method (Supplementary Fig. 18). Global proteomics analysis of the HUVEC proteome (>4,000 unique proteins) revealed that only 160 proteins were affected by TGP-377 treatment (P < 0.05; Fig. 5a). A bioinformatic STRING analysis (string-db.org) was performed to study the protein association networks that are affected by TGP-377, which showed that proliferative pathways, including FGFR, Hedgehog, MAPK and ERK pathways, were upregulated (Fig. 5b). Previous studies have shown that upregulation of VEGFA causes a reduction of Hedgehog interacting protein (HHIP) levels, part of a larger feedback loop for angiogenesis33. Levels of HHIP were considerably reduced upon TGP-377 treatment (P < 0.01; Fig. 5a). Of interest, the levels of protein phosphatase 2 regulatory subunit B′ alpha (PPP2R5A) were also greatly reduced upon treatment (P < 0.05; Fig. 5a). PPP2R5A, which is implicated in a pathway that controls cell survival, has been previously shown to be downregulated upon upregulation of VEGFA46,47. Other affected proteins that are known to be modulated specifically within VEGFA-mediated pathways can be found in Supplementary Table 12. Collectively, these studies show that TGP-377 exerts selective effects on the proteome and indeed broadly affects signalling through the VEGFA pathway.
Fig. 5 |. Proteomics analysis of HUVECs treated with TGP-377.
a, Volcano plot showing the relative fold changes of >4,000 detected proteins. Data are reported as the mean of three biologically independent replicates (n= 3). The statistical significance was determined by a two-sided Student’s t-test and was not adjusted for multiple comparisons. b, STRING analysis of the 160 proteins affected by TGP-377. The analysis reported interaction scores of >0.9 (highest confidence) and is consistent with previous studies of VEGFA upregulation (Supplementary Table 7).
Effect of TGP-377 on VEGFA protein levels and stimulation of a pro-angiogenic phenotype
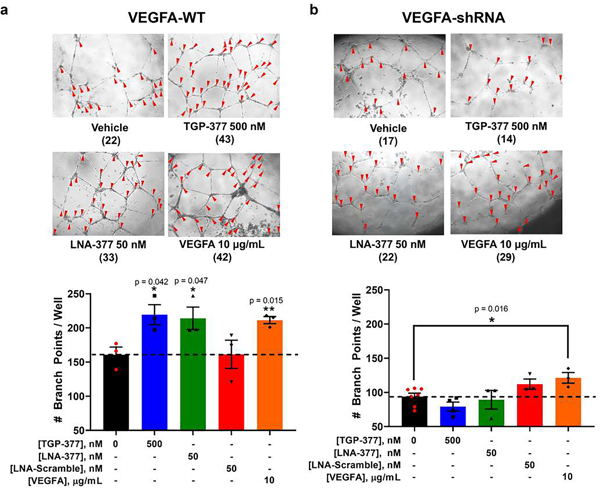
Because proteome- and miRNome-wide studies show that TGP-377 is selective at both the RNA and protein levels, we studied the effect of TGP-377 on phenotype, that is, tubule branching density. Treatment of HUVECs with TGP-377 (500 nM) increased tubule branching density by ~50% (Fig. 6a). Interestingly, the effect of TGP-377 was similar to that observed when recombinant VEGFA protein (10 μg ml–1) was added to the cell culture medium or an LNA antagomir against miR-377 (LNA-377; Fig. 6a). By contrast, a scrambled LNA control antagomir (50 nM) had no effect on tubule branching (Fig. 6a).
Fig. 6 |. TGP-377 stimulates angiogenesis.
a, Representative images of angiogenesis in wild-type HUVECs treated with vehicle (dimethyl sulfoxide < 0.1%), TGP-377 (500 nM), LNA-377 (50 nM) and recombinant VEGFA (10 μg ml–1) for 18 h. The number of branch points in each image is shown below the indicated treatment in parentheses. TGP-377 and LNA-377 stimulated angiogenesis to a similar extent as VEGFA-treated cells, whereas no effect was observed for cells treated with a scrambled oligonucleotide control. Data are reported as mean ± s.e.m. (n= 3 biologically independent replicates). b, Similarly, HUVECs stably expressing an anti-VEGF shRNA showed a 50% reduction in branching, with a loss of TGP-377 and LNA-377 efficacy, as expected. Exogenous VEGFA continued to stimulate branching, confirming VEGFA dependence. Data are reported as mean ± s.e.m. (n= 3 biologically independent replicates); *, P<0.05; **, P< 0.01, as determined by a two-sided Student’s t-test.
To investigate the dependence of TGP-377-mediated enhancement in tubule density on VEGFA expression, the compound’s effect was studied in a HUVEC Vegfa knockdown cell line as described above. TGP-377 was unable to stimulate tubule formation in this cell line, as was LNA-377, indicating miR-377-mediated effects (Fig. 6b). By contrast, exogenous addition of VEGFA (10 μg ml–1) to the cell culture medium stimulated tubule formation (Fig. 6b). Collectively, these studies support the hypothesis that TGP-377 affects phenotype by the miR-377-VEGFA circuit (Fig. 6).
To further validate TGP-377’s MOA, we treated HUVECs with bevacizumab (Avastin) alone or with Avastin and TGP-377. Avastin binds VEGFA and prevents its binding to the VEGF receptor (VEGFR). Because TGP-377 functions by increasing VEGFA levels, Avastin should inhibit TGP-377-mediated stimulation of angiogenesis. Indeed, co-treatment of HUVECs with Avastin (10 ng ml–1) and TGP-377 (500 nM) resulted in a reduction of ~25% in tubule branching and ablation of TGP-377 stimulation of tubule formation (Supplementary Fig. 19). Taken together, these data show that TGP-377’s MOA is VEGF-dependent.
Discussion
There are potential RNA drug targets for nearly every disease. Small molecules offer key potential advantages as chemical probes and lead drug molecules targeting RNA. Indeed, they typically target structured regions that often play critical roles in disease biology, and their properties can be more easily optimized compared with oligonucleotide-based therapeutic agents. One major challenge for SMIRNAs has been the perception that selectivity and potency are difficult to achieve. Evidently, selectivity and potency are indeed possible as now several SMIRNAs have been designed to affect RNA-mediated biology in cells and preclinical models of disease48. It is likely that as more data emerges defining the RNA folds that are bound by small molecules that the number of targets that can be affected by SMIRNAs will increase. Herein, we designed TGP-377 as a test case to develop approaches to drug a predefined RNA target rationally and predictably. A lead compound was quickly identified and then rapidly optimized. Translating lead compounds such as TGP-377 into medicines that reach patients is a more complicated endeavour.
The class of compounds developed herein could be suitable for the treatment of heart disease and myocardial infarctions by increasing VEGFA production. Both DNA and mRNA delivery are being tested in clinical trials for boosting VEGFA levels, with mRNA the most promising49,50. Interestingly, TGP-377 is a relatively small molecule compared with an mRNA. The nature of miRNA targeting may provide selectivity without directly delivering the therapeutic to affected tissues. That is, TGP-377 will only be effective in a limited number of tissues that express both miR-377 and Vegfa mRNA. Further, the increase in VEGFA production will have an upper limit based on the effect of miR-377 on Vegfa translation, not simply compound dose. Our proteomics studies have shown that TGP-377 exerts highly specific effects on the proteome. Although additional studies will be required to progress TGP-377 into animal models and later clinical studies, our data suggest that the compound shows promise for therapeutic development. There are many indications for which RNA-targeted small-molecule medicines will be applicable, and our studies suggest that molecular design can provide potent and selective small molecules to specifically recognize RNA targets in cells.
Methods
General
Reagents were obtained and used as received. DNA templates and primers were obtained from Integrated DNA Technologies. The RNA oligonucleotide competitors and precursor RNAs, obtained from Dharmacon, were 2′-ACE-protected (where ACE is 2′-bis(acetoxyethoxy)-methyl ether) and were deprotected according to the manufacturer’s protocol. All RNAs were desalted using PD10 Sephadex (GE Healthcare) columns by first pre-equilibrating the column with 25 ml water, then loading the RNA and eluting with 10 ml water, collecting 1 ml fractions. Autoradiography images were obtained using a Typhoon 9410 variable mode imager (GE Healthcare) and quantified using Quantity One (Bio-Rad) software. All UV-vis measurements for RNA quantification were obtained using a Beckman Coulter DU800 UV-vis spectrophotometer by heating the RNA to 90 °C and measuring the absorption at 260 nm. Complementary DNA (cDNA) samples were quantified using an Agilent Technologies 2100 Bioanalyzer (Model: G1939A) and an Invitrogen Qubit 2.0 Fluorimeter. Subsequent sequencing was carried out on a Life Technologies Ion Proton sequencer with at least 200-fold coverage. EGM-2 Bullet Kit tissue culture medium obtained from Lonza (CC-3162) was used to grow HUVECs directly. All cells were cultured at 37°C in 5% CO2 in 100-mm-diameter dishes unless stated otherwise.
Preparation of small-molecule libraries
To generate the first diverse compound set for initial screening, the AstraZeneca corporate collection (2M compounds) was clustered in an all-against-all process using the ECFI (Scitegic extended connectivity fingerprints) fingerprint method51. Clusters of molecules with three or more members were kept while making sure that any frequent hitters, controlled substances and impure compounds (<80%) were not present. Compounds with low permeability (Caco-2 assay < 1.0) and low solubility (<1.0 μM) were discarded due to the nature of the assay/target. The clustering process was set up in such a way that 20,000 clusters were generated. Three members were recorded in each cluster, affording a list of 60,000 possible compounds to choose from. Subsequently, a set of RNA binders were collected from the literature and used to define a ‘known RNA binder chemical space’, enabling a principal component analysis based on calculated molecular descriptors. Finally, a diverse selection of 1,967 compounds in the described principal component analysis space were selected for preliminary testing by 2DCS. The hits generated from 2DCS were used as reference compounds to further refine selections from the AstraZeneca corporate collection. An arsenal of similarity-based methods (maximum common substructure, 2D fingerprints, 3D shape) were employed, followed by visual inspection, to produce a second set of 980 near-neighbour compounds for follow-up testing. Again, the compounds were filtered for solubility, permeability, purity and restricted compounds.
Buffers for 2DCS
The following buffers were employed for 2DCS: 1× folding buffer (FB): 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM KCl and 1 mM MgCl2; 1× hybridization buffer (HB): 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2 and 40 μg ml−1 BSA; 10× PCR buffer: 100 mM Tris, pH 9.0, 500 mM KCl and 1% Triton X-100.
Preparation of RNA libraries
PCR amplification was carried out in 1× PCR buffer supplemented with 4.25 mM MgCl2, 0.33 μl of 5 mM 2′-deoxyribonucleoside 5′-triphosphates (dNTPs), 500 nM forward primer (5′- GGCCGGATCCTAATACGACTCACTATAGGGAGAGGGTTTAAT-3′), 500 nM reverse primer (5′-CCTTGCGGATCAAT-3′) and 20 nM DNA template with 2 μl Taq polymerase. Amplification was completed using three-stage cycling conditions as follows: 95 °C for 60 s, 50 °C for 30 s and 75 °C for 60 s. The PCR products were transcribed using an RNAMaxx High Yield transcription kit (Stratagene) according the manufacturers protocol, supplementing 50% of the cold ATP with [α-32P]ATP. Transcripts were purified on denaturing 15% polyacrylamide gels and the bands were excised, eluted with 300 mM NaCl, precipitated with ethanol, and quantified by UV-vis spectroscopy as described above. Extinction coefficients were estimated on the basis of 10,800 M−1 cm−1 nucleotide−1.
2DCS: primary screen
Microarrays were constructed by curing 25 ml of molten 1% (w/v) agarose onto glass plates followed by pinning 100 nl of the compound library onto the agarose using a Beckman Coulter Biomek NX workstation pin tool. After drying, the arrays were washed in 1× FB supplemented with 0.1% Tween-20 and Nano pure water and air-dried. Then, 5 μl labelled RNA was folded by heating at 95 °C for 45 s in 1× FB, cooled to room temperature and brought to a total volume of 2.5 ml with 1× FB. The arrays were pre-hybridized with 2.5 ml 1× HB for 5 min. Excess buffer was removed, and the arrays were incubated with labelled RNA libraries for 45 min. After hybridization, the slides were washed four times with 1× HB, air-dried and imaged.
2DCS: tRNA competition screen
Competition arrays were performed on microscope slides that were coated with 2 ml molten 1% agarose and allowed to cure for 2 h before manually spotting 200 nl of 10 mM compound directly into the agarose. The slides were air-dried overnight, washed with Nano pure water three times for 5 min each and then dried in a stream of air. Then, the slides were pre-hybridized with 1× HB for 5 min and then incubated with 100 pmol of radioactively labelled RNA and tRNA (equimolar relative to the total moles of compound spotted) in 400 μl 1× FB for 45 min followed by washing with 1× HB five times before air-drying and imaging by autoradiography.
2DCS: chase oligonucleotide competition screen
Microarrays were constructed as described above with compounds that still bound RNA motifs in the presence of excess tRNA. Compounds were spotted onto the agarose in a concentration gradient of 10, 5, 2.5, 1.25 and 0.625 mM, with a maximum of eight compounds per slide (~31 nmol total compound per slide). The chase oligomer mixture was prepared as follows. Competitor RNA oligonucleotides, bulk brewer’s yeast tRNA and d(GC)11 and d(AT)11 DNA oligonucleotides were folded separately in 1× HB without MgCl2, which was added to a final concentration of 1 mM after cooling. The folded nucleic acids were then combined with the labelled RNA libraries in a total volume of 400 μl. (All competitors were equimolar to the total moles of compound spotted.) Pre-hybridization and incubation with the library were carried out as described above. RNAs that bound to compounds were excised and sequenced as previously described15.
Statistical analysis of selected RNAs
Statistical analysis of sequencing data was carried out as previously described15. Briefly, the frequencies of the RNAs that were selected to bind a compound in RNA-seq data were compared with their frequencies in the entire starting library. A Z-score (Zobs) was then calculated according to equations (1) and (2):
| (1) |
| (2) |
where ɸ is the pooled sample proportion, n1 is the number of reads observed for all selected RNAs in RNA-seq data, n2 is the number of reads of the starting library, p1 is the observed proportion of reads for a selected RNA compared with the total number of reads for selected RNA and p2 is the observed proportion of the number of reads of a selected RNA compared with the total number of reads for the starting library. Zobs can then be used to rank RNA fold–small molecule interactions by affinity and selectivity.
Binding assays and determination of Kd
Using 5′-fluorescein-labelled RNA (see sequences in Supplementary Table 13), 250 nM RNA was folded by heating at 70 °C in 1× non-proprietary binding buffer (8 mM sodium phosphate, pH 7.4, 150 mM NaCl and 2 mM EDTA) with 40 μg ml–1 BSA. After cooling, the compounds were added at doses of 500 or 100 μM and serially diluted twofold. The samples were then incubated at room temperature in the dark and plated onto Greiner 384-well low-volume plates. Fluorescein was measured with a Biotek FLX800 plate reader at an excitation wavelength of 485 ± 20 nm and an emission wavelength of 520 ± 20 nm. The percentage change in fluorescence intensity was calculated according to equation (3), where F0 is the emission with no compound and FS is the emission with compound. All isotherms were fitted using equation (4):
| (3) |
| (4) |
where I and I0 are the observed fluorescence and initial fluorescence intensity in the presence and absence of RNA, respectively, Δε is the difference between the fluorescence intensity in the absence and presence of infinite RNA concentration, [FL]0 and [RNA]0 are the initial concentrations of the small molecule and RNA, respectively, and Kd is the dissociation constant.
Microscale thermophoresis binding
The sequences of the RNAs used for microscale thermophoresis binding (MST) binding assays can be found in Supplementary Table 13. Compounds C1, C10, C20 and TGP-377 (n = 3) were tested in dose response from 100 μM to 3 nM by 1:2 serial dilution. Premium capillaries were used for all binding assays conducted, and measurements were taken with a Monolith NT.115 (NanoTemper) capillary thermophoresis instrument. Briefly, the compounds were diluted twofold in Nanopure water, and 20 nM RNA was folded in 2× DNA buffer at 95 °C for 2 min with slow cooling to room temperature on the benchtop. After folding, 100× BSA (400 μg ml–1) was added to 1× and then 10 μl 2× RNA solution was mixed with 10 μl 2× compound. The samples were allowed to equilibrate for 1–2 h before loading into the capillaries. Note that binding assays for the cyanine 5-labelled (Cy5) RNAs Cy5-r(G4C2)8 and Cy5-r(GGCC)5 were performed in 1× binding buffer specific for G-rich RNAs (10 mM NaHPO4, 100 mM LiCl, pH 7.0) and folded for 5 min at 95 °C. The instrument settings were as follows: LED power 20–5% (the signal was kept below 14,000 relative fluorescence units (RFUs) for the capillary scan), MST power 80%, fluorescence before 5 s, MST at 30 s, fluorescence after 5 s, delay 25 s. The average of each measurement was fit according to equation (4).
miR-377 knockdown assay
HUVEC cells were plated in 12-well tissue culture plates (Corning) at 100,000 cells per well and allowed to adhere for 12 h. The medium was removed, and the cells were washed once with 1× PBS before treatment with the following: (1) DMSO (0.8%) in EGM-2 medium (Lonza), (2) Power LNA-anti-miR-377–3p (50 nM; Exiquon, now part of Qiagen), (3) Power-LNA-Scramble A (50 nM; Exiquon), (4) TGP-377/421 (1.25, 2.5, 5 and 10 μM) or (5) TGP-377 (5, 50, 500 and 5,000 nM) for 48 h. Total RNA was collected using a Zymo RNA Clean Mini-prep kit and assessed by RT–qPCR for expression of miR-377, pre-miR-377, VEGF mRNA and control genes (RNU6 for mature miRNA, and 18S or β-actin for mRNA, and pre-miR-377 levels).
Angiogenesis tubule formation assay
Tubule formation assays were performed using a Culturex In Vitro Angiogenesis Assay Kit (Trevigen) according to the manufacturer’s protocol. Briefly, approximately 3 × 105 HUVEC or HUVEC-VEGFA-shRNA cells were seeded onto a thick layer of basement membrane extract in a 96-well tissue culture plate as a single cell suspension in EGM-2 medium (Lonza). The cells were plated in medium supplemented with the following: (1) DMSO (0.8%), (2) Power LNA-anti-miR-377–3p (50 nM; Exiquon), (3) Power-LNA-Scramble A (50 nM; Exiquon), (4) TGP-377/421 (5 μM), (5) TGP-377 (500 nM) or (6) VEGFA (10 μg ml–1) for 18 h. Images of cells were taken on a Leica DM800 inverted microscope, nine fields of view per well. The number of branch points per well was counted as previously reported by Wen et al.28. Using the same setup as described above, we then treated HUVECs with Avastin (bevacizumab) at 100, 10 and 1 ng ml–1 to determine the concentration required to inhibit angiogenesis. To determine whether Avastin could inhibit TGP-377-mediated angiogenesis, we co-treated HUVECs with 10 ng ml–1 Avastin and 500 nM TGP-377 for 18 h. Branch points were counted as previously described. Please note the images taken for the Avastin study were taken with a BioTek Lionheart FX automated microscope with 20 fields of view and used to create stitched images of each well for counting. Image stitching was carried out directly on the Biotek imaging suite.
Immunoblotting of hVEGFA
HUVECs were seeded in six-well tissue culture plates (Corning) at 125,000 cells per well and allowed to adhere for 12 h. The cells were then treated with the following: (1) DMSO (0.8%) in EGM-2 medium (Lonza), (2) Power LNA-anti-miR-377–3p (50 nM; Exiquon), (3) Power-LNA-Scramble A (50 nM; Exiquon), (4) TGP-377/421 (5 μM) or (5) TGP-377 (500 nM) for 48 h. The cells were detached using 0.25% trypsin (Life Technologies) and washed with 1× PBS once. The cells were lysed using MPER lysis buffer (Life Technologies) supplemented to 1× with a protease inhibitor cocktail (Fisher Scientific) according to the manufacturer’s protocol. Total protein concentration was quantified using a Micro BCA Protein Assay Kit (Thermo Scientific) according to the manufacturer’s protocol. The proteins were resolved on 12.5% pH 8.3 SDS polyacrylamide gel with a 4% stacking layer (pH 6.8). SDS–PAGE gels were run in 1× running buffer according to the Abcam standard protocol for 2 h at 140 V. The protein size was assessed using a Spectra Multicolor Broad Range Protein Ladder (Thermo Scientific). Proteins were electroblotted onto a nitrocellulose membrane at 250 mA for 2.5 h.
All blots were blocked in Licor blocking buffer in 1× PBS. Rabbit anti-human VEGFA IgG (Abcam, ab46514) was used at 1:5,000 dilution in 3% (w/v) BSA in 1× Tris buffered saline (TBS; 20 mM Tris-HCl, pH 7.6, 150 mM NaCl) overnight at 4 °C. Blots were then washed five times with 1× TBS supplemented with 0.1% (v/v) Tween-20 (TBST) and then incubated with 1:5,000 goat-anti-rabbit-CW 800 IgG (Licor) or horseradish peroxidase (HRP) conjugate (Cell Signaling Technologies) for 2 h at room temperature. After washing three times in 1× TBST, α-tubulin was probed using 1:5,000 mouse anti-human α-tubulin IgG (Cell Signaling Technologies) for 2 h at room temperature and washed as described above. After washing, blots were incubated with donkey anti-mouse CW 680 IgG (Licor) or goat-anti-mouse HRP conjugate for 2 h at room temperature. All blots were imaged on a Licor Odyssey IR imager or by X-ray film exposure.
ELISA assay of secreted VEGFA
The assessment of hVEGFA protein levels was completed using the VEGF Human ELISA Kit (Thermo Fisher) according to the manufacturer’s protocol. Medium supernatants were concentrated in Amicon 3K MWCO microfuge protein concentrators (Millipore) to a volume of 50 μl. The protein concentration was then quantified by using a Micro BCA Assay as previously indicated. Briefly, strips with immobilized anti-human VEGFA were washed twice with 1× wash buffer (400 μl each, provided with the ELISA kit). Then, 170 μg total protein was loaded into each well in 50 μl H2O. Next, 50 μl sample diluent was added and the samples were incubated on a plate shaker for 2 h at 400 r.p.m. The wells were then aspirated and washed six times with 1× wash buffer followed by treatment with biotin-conjugated anti-hVEGFA in 1× assay buffer (provided in the ELISA kit) for 1 h on a plate shaker (400 r.p.m.). As before, the wells were aspirated and washed six times followed by treatment with streptavidin linked-HRP for 1 h while shaking at 400 r.p.m. The wells were washed six times and then incubated with substrate solution for 0.5 h in the dark. The absorbance was measured at 460 nm using a SpectraMax M5 plate reader (Molecular Devices). All samples were measured in duplicate and fitted to a standard curve of recombinant VEGFA.
RNA isoform analysis
To determine the selectivity of TGP-377/421, we identified all human miRNAs that contain the 5′-AAU/3′-U_A bulge. Using RT–qPCR, we assessed the expression of each miRNA in HUVECs and determined that hsa-miR-4272 and hsa-miR-568 were not expressed, whereas all other miRNAs were measurable. Similarly, we identified the isoforms for miR-214’s 5′-GUC/3′-CUG, miR-100’s 5′-AAA/3′-UAU and miR-342’s 5′-AGG/3′-UGC internal loops using the database of microRNA secondary structural elements created by Liu et al.44.
miRNA profiling by RT–qPCR for selectivity
Total RNA from HUVECs treated with TGP-377 (500 nM) was harvested as described above. The primers used for amplification and methods for data analysis were completed according to the work flow previously described by Costales et al.52.
In vitro Chem-CLIP and C-Chem-CLIP
Radiolabelled pre-miR-377 was folded in 1× DNA buffer and then treated with TGP-377/421-CA-Biotin in a dose response (0.5 μM, 5 μM, 25 μM and 50 μM) overnight. Reacted RNA was captured using magnetic streptavidin C1 dynabeads (Life Technologies), using 100 μl of slurry per sample, and the samples were shaken at room temperature for 2 h. The beads were captured on a magnetic rack and washed eight times with 1× high salt wash buffer (10 mM Tris-HCl, 1 mM EDTA, 4 M NaCl, 0.01% Tween-20, pH 7.4). All washes and supernatants were pooled, and bound and unbound RNA were quantified by liquid scintillation counting. Fold enrichment was calculated as the ratio of radioactivity associated with the beads to radioactivity in the washes and supernatant. To complete competitive Chem-CLIP in vitro, radioactively labelled pre-miR-377 was incubated with either TGP-377/421 or TGP-377 (50 nM and 500 nM) for 2 h before the addition of 500 nM TGP-377/421-CA-Biotin. Samples were incubated for 8 h before addition of magnetic streptavidin beads and shaking for 2 h. Bound RNA was collected by magnetic capture and the beads washed eight times before pooled washes and beads were counted by liquid scintillation counting.
In cellulis Chem-CLIP and C-Chem-CLIP
HUVECs were grown in 100-mm-diameter dishes until they reached ~25% confluency. For Chem-CLIP studies, the cells were treated with TGP-377/421-CA-Biotin for 48 h. For C-Chem-CLIP studies, HUVECs were pretreated with 500 nM TGP-377 for 15 min followed by the addition of 500 nM TGP-377/421-CA-Biotin. The cells were then incubated for an additional 48 h. Total RNA was collected using an miRNeasy RNA Extraction Kit (Qiagen) with DNase treatment according to the manufacturer’s protocol.
To pulldown, 60 μg total RNA was incubated with a slurry of 100 μl streptavidin C1 dynabeads (Life Technologies), and the samples were shaken at room temperature for 2 h. The beads were captured on a magnetic rack and washed eight times with 1× high salt wash buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 4 M NaCl and 0.01% (v/v) Tween-20). After washing, the pulled-down RNA was released from the beads by boiling with 95% formamide containing 1 mM EDTA for 20 min at 95 °C. The supernatant was then collected and the beads washed twice with 50 μl water. The RNA was concentrated in a vacuum concentrator and then cleaned using the Zymo Quick-RNA Miniprep Kit according to the manufacturer’s protocol. The RNA was reverse-transcribed using qScript cDNA Synthesis Kit (Quantabio) and the fold enrichment calculated for pre-miR-377 as previously described27.
Supplementary Material
Acknowledgements
This work was funded by the National Institutes of Health R01 GM097455 and R01 CA249180 (to M.D.D.) and AstraZeneca. We thank J. Childs-Disney and R. Rahaim, Jr. for editing the manuscript. We also thank U. Bauer for supporting the collaboration and K. Jennbacken for useful input on VEGFA biology. Correspondence and request for materials should be addressed to M.D.D.
Footnotes
Competing interests
M.D.D. is a founder of Expansion Therapeutics. M.L., J.B. and L.K. are employees of AstraZeneca.
Code availability
No unique code was used in the described data analyses.
Data availability
All data supporting this manuscript are contained within the main text, source data and Supplementary figures. Specific data are freely available upon reasonable request from the corresponding author. The Inforna database9 can be accessed via the following URL https://disney.florida.scripps.edu/software/. Users wishing to obtain access must complete a software license agreement with TSRI, upon which login credentials will be provided after approval.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
- 1.Angelbello AJ et al. Using genome sequence to enable the design of medicines and chemical probes. Chem. Rev 118, 1599–1663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disney MD & Angelbello AJ Rational design of small molecules targeting oncogenic noncoding RNAs from sequence. Acc. Chem. Res 49, 2698–2704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein CA & Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther 25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stojic L. et al. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 46, 5950–5966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watters KE, Abbott TR & Lucks JB Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 44, e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strobel EJ, Yu AM & Lucks JB High-throughput determination of RNA structures. Nat. Rev. Genet 19, 615–634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velagapudi SP, Gallo SM & Disney MD Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol 10, 291–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran T. & Disney MD Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun 3, 1125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Disney MD et al. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol 11, 1720–1728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrara N, Gerber H-P & LeCouter J. The biology of VEGF and its receptors. Nat. Med 9, 669–676 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Sun N. et al. Modified VEGF-A mRNA induces sustained multifaceted microvascular response and accelerates diabetic wound healing. Sci. Rep 8, 17509–17519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ylä-Herttuala S, Rissanen TT, Vajanto I. & Hartikainen J. Vascular endothelial growth factors: biology and current status of clinical applications in cardiovascular medicine. J. Am. Coll. Cardiol 49, 1015–1026 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Taimeh Z, Loughran J, Birks EJ & Bolli R. Vascular endothelial growth factor in heart failure. Nat. Rev. Cardiol 10, 519–530 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Velagapudi SP et al. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem. Biol 25, 1086–1094 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velagapudi SP et al. Defining RNA–small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Cent. Sci 3, 205–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan BS, Forte JE, Culver RN, Zhang Y. & Hargrove AE Discovery of key physicochemical, structural, and spatial properties of RNA-targeted bioactive ligands. Angew. Chem. Int. Ed 56, 13498–13502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta A. et al. SMMRNA: a database of small molecule modulators of RNA. Nucleic Acids Res. 42, D132–D141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordella LP, Foggia P, Sansone C. & Vento MA (sub)graph isomorphism algorithm for matching large graphs. IEEE Trans. Pattern Anal. Mac. Intell 26, 1367–1372 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich H-C & Rarey M. Systematic benchmark of substructure search in molecular graphs - from Ullmann to VF2. J. Cheminform 4, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haniff HS, Graves A. & Disney MD Selective small molecule recognition of RNA base pairs. ACS Comb. Sci 20, 482–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider TD & Stephens RM Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stelzer AC, Kratz JD, Zhang Q. & Al-Hashimi HM RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. Int. Ed 49, 5731–5733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelzer AC et al. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol 7, 553–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JL, VanEtten DM, Fountain MA, Yildirim I. & Disney MD Structure and dynamics of RNA repeat expansions that cause Huntington’s disease and myotonic dystrophy type 1. Biochemistry 56, 3463–3474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childs-Disney JL et al. A massively parallel selection of small molecule-RNA motif binding partners informs design of an antiviral from sequence. Chem 4, 2384–2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costales MG et al. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J. Am. Chem. Soc 139, 3446–3455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velagapudi SP et al. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl Acad. Sci. USA 113, 5898–5903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen Z. et al. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS ONE 9, e104666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.el Azzouzi H. et al. The hypoxia-inducible microRNA cluster miR-199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab. 18, 341–354 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Duan Q. et al. MicroRNA-214 is upregulated in heart failure patients and suppresses XBP1-mediated endothelial cells angiogenesis. J. Cell. Physiol 230, 1964–1973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Dong L-D, Meng X-B, Shi Q. & Sun W-Y Unique microRNA signatures associated with early coronary atherosclerotic plaques. Biochem. Biophys. Res. Commun 464, 574–579 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Yan XC et al. MiR-342–5p is a notch downstream molecule and regulates multiple angiogenic pathways including Notch, vascular endothelial growth factor and transforming growth factor β signaling. J. Am. Heart Assoc 5, e003042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renton AE et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brčić J. & Plavec J. NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism. Nucleic Acids Res. 46, 11605–11617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Z. et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 84, 1043–1050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang ZF et al. The hairpin form of r(G4C2)exp in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol 26, 179–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balendra R. & Isaacs AM C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol 14, 544–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrara N, Hillan KJ & Novotny W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun 333, 328–335 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Ivy SP, Wick JY & Kaufman BM An overview of small-molecule inhibitors of VEGFR signaling. Nat. Rev. Clin. Oncol 6, 569–579 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Ahmed SI, Thomas AL & Steward WP Vascular endothelial growth factor (VEGF) inhibition by small molecules. J. Chemother 16, 59–63 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Arcondéguy T, Lacazette E, Millevoi S, Prats H. & Touriol C. VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 41, 7997–8010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koransky ML, Robbins RC & Blau HM VEGF gene delivery for treatment of ischemic cardiovascular disease. Trends Cardiovasc. Med 12, 108–114 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Liu B. et al. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinf. 17, 112–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal V, Bell GW, Nam JW & Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes K, Roberts OL, Thomas AM & Cross MJ Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal 19, 2003–2012 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Mao Z, Liu C, Lin X, Sun B. & Su C. PPP2R5A: a multirole protein phosphatase subunit in regulating cancer development. Cancer Lett. 414, 222–229 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Angelbello AJ et al. Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model. Proc. Natl Acad. Sci. USA 116, 7799–7804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gan LM et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun 10, 871–879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zangi L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol 31, 898–907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassan M, Brown RD, Varma-O’Brien S. & Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Divers 10, 283–299 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Costales MG, Matsumoto Y, Velagapudi SP & Disney MD Small molecule targeted recruitment of a nuclease to RNA. J. Am. Chem. Soc 140, 6741–6744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.