Abstract
The urgent need for a cure for early phase COVID-19 infected patients critically underlines drug repositioning strategies able to efficiently identify new and reliable treatments by merging computational, experimental, and pharmacokinetic expertise. Here we report new potential therapeutics for COVID-19 identified with a combined virtual and experimental screening strategy and selected among already approved drugs. We used hydroxychloroquine (HCQ), one of the most studied drugs in current clinical trials, as a reference template to screen for structural similarity against a library of almost 4000 approved drugs. The top-ranked drugs, based on structural similarity to HCQ, were selected for in vitro antiviral assessment. Among the selected drugs, both zuclopenthixol and nebivolol efficiently block SARS-CoV-2 infection with EC50 values in the low micromolar range, as confirmed by independent experiments. The anti-SARS-CoV-2 potential of ambroxol, amodiaquine, and its active metabolite (N-monodesethyl amodiaquine) is also discussed. In trying to understand the “hydroxychloroquine” mechanism of action, both pKa and the HCQ aromatic core may play a role. Further, we show that the amodiaquine metabolite and, to a lesser extent, zuclopenthixol and nebivolol are active in a SARS-CoV-2 titer reduction assay. Given the need for improved efficacy and safety, we propose zuclopenthixol, nebivolol, and amodiaquine as potential candidates for clinical trials against the early phase of the SARS-CoV-2 infection and discuss their potential use as adjuvant to the current (i.e., remdesivir and favipiravir) COVID-19 therapeutics.
Keywords: COVID-19, drug repositioning, drug repurposing, virtual screening, zuclopenthixol, nebivolol, amodiaquine
Officially declared a pandemic by the World Health Organization on 11 March 2020,1 the disease caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) spread across all continents and all countries2 by 30 June 2020, with over 35.6 million infected and over 1.044 million deaths (10/07/2020).3 Radical safety measures such as “work-from-home” and “safe social distancing”, implemented in many countries for 8–12 weeks or more, have de facto slowed down animal and clinical research worldwide. Research with “live” SARS-CoV-2 viruses requires Biosafety Level 3 (BSL-3) biocontainment conditions, which places additional burdens on our ability to identify new therapeutic interventions. To date, except for the emergency use authorization for remdesivir granted by the US FDA,4 no approved therapeutic approaches for COVID-19 exist. Given its catastrophic burden and associated deaths, there is a stringent need to rapidly find therapeutic interventions.
We developed and applied a virtual screening strategy with the goal of identifying currently approved drugs that could serve as treatment for the early phases of the COVID-19 disease.5 Since this is a fast-moving pandemic, drug repositioning is one of the highly active fields of biomedical research that parallels vaccine research and development in the fight to stop COVID-19.6 A number of drugs have been already proposed as new therapies for COVID-19 and are subject to clinical trials.7,8
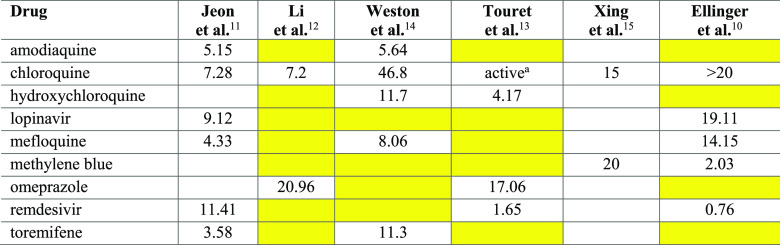
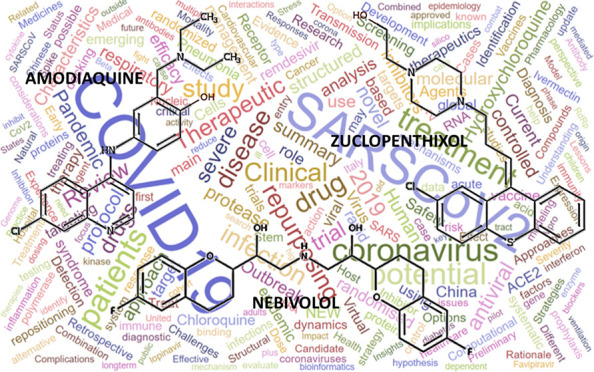
The scientific community continues to search for other approved drugs that may show an anti-SARS-CoV-2 activity, typically by combining high throughput screening (HTS) and in silico strategies. Kuleshov and co-workers recently compiled a database of such studies.9 While HTS studies have proven useful for the identification of active drugs, the reliability and the accuracy of HTS results are worthy of discussion. Indeed, there appears to be limited agreement between HTS measurements made across different laboratories, with a discouraging small overlap among the identified hits.9 Such inconsistencies may be due to differences in the experimental setup. In Table 1, nine hits extracted from six HTS publications are listed.10−15 For consistency, we list only drugs that actively block SARS-CoV-2 virus-induced host cell death (“cytopathic effect”, CPE) in at least two independent studies. All hits, except remdesivir, are FDA-approved and are typically included in drug libraries. The exact composition of the compound libraries screened in two studies was not disclosed.11,15 Thus, it is not possible to ascertain if these nine drugs were tested. The inconsistencies between studies summarized in Table 1 are highlighted using yellow background. For example, remdesivir, one of the promising anti-SARS-CoV-2 antivirals (still in clinical trials), was negative in two out of five studies, not counting the one where we could not ascertain whether it had been tested (see Table 1). This lack of consistency between different in vitro studies highlights the need for independent confirmation. Furthermore, Ellinger et al.10 used Caco-2 cells in their assay, whereas the other five studies used Vero E6 cells when evaluating anti-SARS-CoV-2 activity. Using a different cell line may account for the discrepancies between the activities of chloroquine and methylene blue (Table 1). Another reason might be differences in the initial viral load, or multiplicity of infection (MOI), when running the assays. Moreover, clinical relevance, such as pharmacokinetics (PK) and toxicity properties of these HTS hits are rarely mentioned. At the time of this writing, 488 peer-reviewed publications published between February and July 2020 on the topic of COVID-19 drug repositioning, were available through the Clarivate Analytics Web of Science.16 A word cloud summarizing the titles of these papers is shown in Figure 1, where the size of each word reflects the frequency of its occurrence in paper titles. The figure provides a snapshot of the topics related to current COVID-19 research and how frequently they are addressed. Therefore, it represents a visual summary of the state of the art of COVID-19 research.
Table 1. Activity Values (IC50 or EC50 in μM units) for the Nine HTS Hits That Were Detected in More than One Publication*.
Yellow cells indicate a negative result of the experiment (i.e., the drug resulted to be inactive). White cells indicate data not disclosed from the authors.
Chloroquine was confirmed to be active in the single concentration experiment, but the IC50 was not measured.
Figure 1.
A word cloud summarizing the titles of 488 peer-reviewed publications published between February and July 2020 on the topic of COVID19 drug repositioning listed on the Clarivate Analytics Web of Science.16
Given the complexity of disease–chemical biology interactions,17 this paper focuses on potential SARS-CoV-2 antiviral candidates by means of an experimentally validated ligand-based virtual screening (LBVS) protocol, with emphasis on clinical relevance. Our goal was to identify approved drugs that might have similar activity compared to the small molecule drugs that are currently in COVID-19 clinical trials. Consequently, we used LBVS to query a list of approved drugs, evaluating their structural similarity against hydroxychloroquine (HCQ), a drug with in vitro anti-SARS-CoV-2 activity.18 Starting with HCQ as a template, we virtually screened the online drug compendium DrugCentral,19 a database of about 4600 drugs approved worldwide.20 Candidates prioritized by in silico methods were then tested in vitro using two independent Vero E6 cell viability assays followed by a confirmatory titer reduction assay. We discuss the PK properties of our active hits, possible molecular determinants of their activity, and their potential therapeutic applications.
Materials and Methods
The main steps of the computational procedure applied in this study are summarized in the flowchart depicted in Figure 2.
Figure 2.
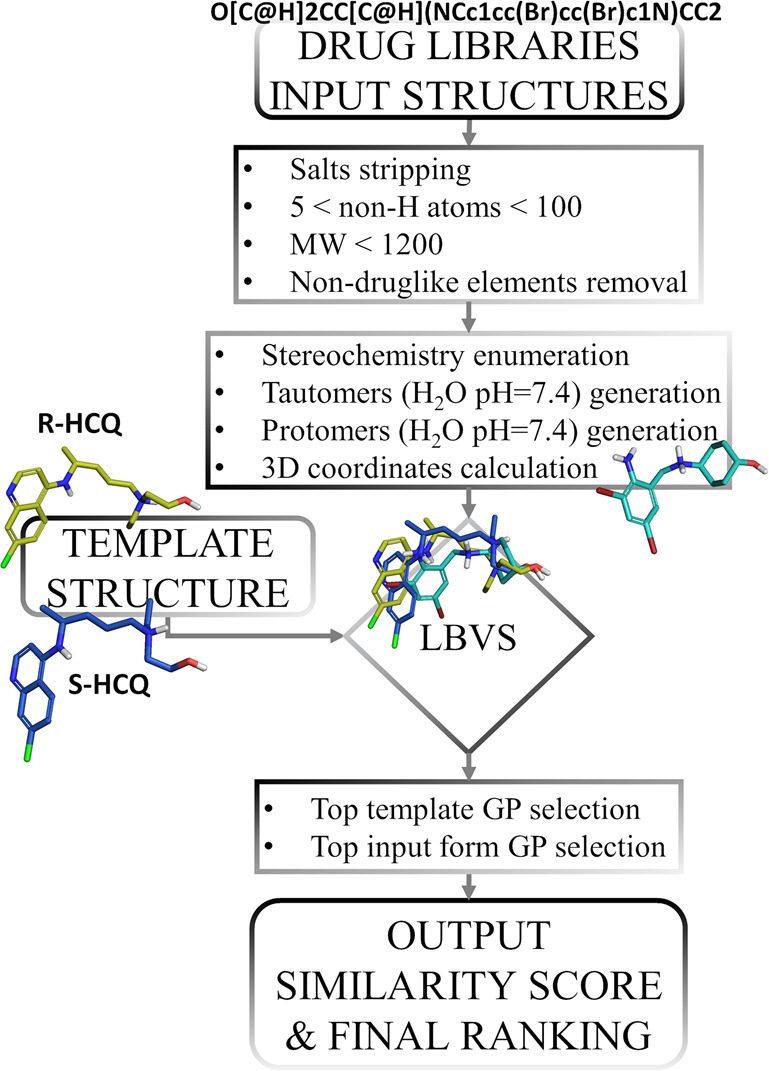
Flowchart showing the computational procedure designed and applied in this work.
DrugCentral Preprocessing
The chemical structures of small molecule drugs were prepared as follows. The 2D structures for 3981 small molecule organic compounds, having received regulatory drug approval worldwide, were directly downloaded from the DrugCentral portal.19,20 For drugs lacking explicit chirality (e.g., ibuprofen is a racemic molecule which includes both the R and S enantiomers in equal amounts in all drug formulations), all enantiomers were generated with the software, Openeye-flipper.21 To better describe each structure in terms of polar and hydrogen-bond interactions, we computed tautomeric and charged (protomeric) forms of each chemical structure at pH 7.4 in water that are present with a minimum abundance of 25%. All tautomers and protomers were computed with MoKa.22 Finally, 3D structures were generated with the Openeye Omega software.21 Drugs having a number of non-hydrogen atoms below 5 or above 100, drugs having MW > 1200, and drugs that incorporate elements not associated with organic molecules (e.g., Hg, Pt, Fe, etc.) were not considered. Where present, salt forms were stripped as well, by saving only the largest fragment. A total of 6057 chemical structures underwent the virtual screening procedure. Note that this number is higher than the original due to the enumeration of drugs into enantiomer, tautomer, and protomer alternatives.
Ligand-Based Virtual Screening
To perform the ligand-based virtual screening, we used the FLAP23 (Fingerprints for Ligands and Proteins) software. FLAP uses the GRID molecular interaction fields (MIFs)24 for estimating the similarity between a template molecule, usually an active ligand, in this case hydroxychloroquine, and a collection of molecules to be screened, that is, the enumerated DrugCentral molecules. FLAP similarity should not be viewed as chemical or structural similarity, but rather as similarity of the way the two entities interact with their surrounding environment. The steps of the FLAP procedure were summarized in Figure 2. For each virtually screened drug, a certain number (up to 100) of conformers were generated; MIFs were computed for each of them. Four GRID probes were used with default settings for computing MIFs: The H probe maps the size and shape of the molecule, the N1 probe maps the hydrogen-bond acceptor areas around the molecule, the O probe maps the hydrogen-bond donor areas around the molecule and the DRY probe maps the hydrophobic interaction areas around the molecule. MIFs were then saved as three-dimensional geometrical entities called quadruplets, which are formed by four relevant points extracted from the molecule MIFs that are connected among each other. Depending on the molecule, a different number of quadruplets was extracted. Successively, the alignment to the template molecule was performed by overlapping the quadruplets of each conformer with the quadruples extracted from the template molecule. Once the alignment was done, similarity scores were computed from the original MIFs of the screened molecule and template. Among various similarity scores computed by FLAP, the Glob-Prod (GP) score measures the overall GRID fields similarity between the template and the screened molecule. A GP value of 0 indicates total dissimilarity, whereas a GP value of 1 indicates perfect similarity. HCQ is formulated as the racemate of its R and S enantiomers, and both were found to be active in vitro by blocking SARS-CoV-2 infection in the low micromolar range, with the S stereoisomer being slightly more active that the R stereoisomer.12 Therefore, both the S and R enantiomers served as templates for virtual screening, and only the top GP generated from R-HCQ and S-HCQ forms for the final ranking of the single screened drug were selected. When more than one form of the screened drug (e.g., more than one enantiomer, more than one protomer, etc.) was screened, only the form having the highest GP value was considered in the final ranking. Finally, drugs were ranked according to descending GP values. To perform the final selection of drugs, we evaluated the GP values of the active drugs reported in Table 1. Remdesivir, which is also listed in Table 1, was excluded from this operation because it was not an approved drug at the time. We identified a GP value of 0.34 (which corresponds to the HCQ-toremifene similarity) as the smallest GP value among the active drugs listed in Table 1. Therefore, we set this value as the cutoff for the minimal acceptable similarity above which we expected enrichment in anti-SARS-CoV-2 actives. Consequently, drugs having a GP value equal or above 0.34 were selected for in vitro testing against SARS-CoV-2.
Chemicals and Cells
Vero E6 cells were obtained from American Type Culture Collection and were grown in minimal essential media supplemented with 5% fetal bovine serum and 100 U/mL penicillin and 100 μg/mL streptomycin. Chemicals were obtained from compound libraries and stock sourced from Prestwick Chemical Library (PCL; Illkirch, France); MedChem Express Library (MCE; Monmouth Junction, NJ, USA); SelleckChem (SLK; Houston, TX, USA); Spectrum (Microsource Discovery systems; Gaylordsville, CT, USA); AdooQ Biosciences (Irvine, CA, USA); LC Laboratories (Woburn, MA, USA); Tocris Bioscience (Bristol, United Kingdom); Toronto Research Chemicals (North York, ON, Canada). Chemical purities ranged from 96% to 99%. Dilutions and dose response concentrations of the chemicals were prepared by hand. The following compounds were independently procured for confirmatory experiments: ambroxol, N-monodesethyl amodiaquine, amodiaquine, primaquine, and nebivolol (Sigma-Aldrich; St. Louis, MO, USA); zuclopenthixol and remdesivir (MedChem Express Library (MCE; Monmouth Junction, NJ, USA).
Vero E6 Cell Assay
Vero E6 cells were grown to ∼80% confluency in 96-well plates and treated with the indicated compounds (10 μM) in triplicate for 1 h prior to infection with a low multiplicity of infection (MOI = 0.05) of SARS-CoV-2 Isolate USA-WA1/2020 (deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH, NR-52281). At 48 h later, the SARS-CoV-2 mediated cytopathic effect (CPE) was assessed by XTT Cell Viability (ThermoFischer) using the manufacturer’s protocol. Negative controls were DMSO-treated infected cells. Positive controls were untreated infected cells and cells treated with 10 μM chloroquine. Hits from the Vero E6 cell assay at 10 μM of compounds were further assessed using a dose response assay. Compounds were added to cells at concentrations of 100, 10, 1, 0.1, and 0.01 μM and inhibition of virus-induced cell death was measured at 48 h as above.
Independent Confirmatory CPE Experiments
Compounds selected from the earlier assays were screened for their inhibition of the SARS-CoV-2 mediated CPE following infection in Vero E6 cells, using a dose response format adapted based on previously published SARS-CoV HTS methods.25−27 Briefly, Vero E6 cells were plated in a 384-well plate at 5000 cells/well. After 24 h, 2-fold serial dilutions of compounds were added to generate a 7-point dose response dilution series with DMSO = 0.5%. Cells were infected with SARS-CoV-2 with a higher MOI (0.1) and incubated for 2 days. The percent protection from SARS-CoV-2 CPE was assessed using Cell Titer Glo (measures cellular ATP), and the EC50 was calculated using a nonlinear regression using GraphPad. Remdesivir (positive reference control), cells alone (positive control) and cells plus virus (negative control) were included in each plate. This luminescence-based assay, validated in 384-well plates in the UTHSC RBL BSL-3 containment facility, is sensitive and robust, with Z values > 0.5, signal to background (S/B) > 19, and signal-to-noise (S/N) > 3.3. Cell viability was simultaneously measured using the Cell Titer Glo, with CC50 (cytotoxicity) calculated. From the EC50 and CC50 values, the selective index at 50% (SI50) was derived.
Titer Reduction Assay
A titer reduction assay was conducted to confirm antiviral potency of compounds at concentrations selected from dose–response curves. Each test compound was assessed in three independent biological replicates. Vero E6 cells were grown in 12 well plates, and were preincubated with an equal amount of infection media (Minimum Essential Media with Earle’s salts (MEM) + 2% FBS + 1% penicillin-streptomycin) and compound (2X concentration) for 2 h at 37 °C, 5% CO2. Cells were washed and infected with 0.2 mL of SARS-CoV-2 at 0.1 MOI for 1 h. At the end of infection, cells were washed and replenished with a 1:1 mixture of infection media and compound (2X). After 48 h incubation, the supernatants were collected and were quantified for residual virus titers by Tissue Culture Infectious Dose (TCID50) assay following the protocol described by Lee et al.28 Briefly, 10-fold dilutions of supernatants (10–1 to 10–8) were made, and 0.1 mL of each dilution was added to 96 well plates seeded with Vero E6 cells. Following 72 h incubation at 37 °C, 5% CO2, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich) was added to each well. Plates were read at a wavelength of 570 nm. Wells were scored as positive or negative, and TCID50 was calculated based on the Reed Muench method.29
Results and Discussion
During the preparation of this study, the FDA approved (3/28/20) and revoked (6/15/20) the emergency use authorization (EUA) that allowed for chloroquine and HCQ to be used for the treatment of certain hospitalized patients with COVID-19 when a clinical trial was unavailable, or when participation in a clinical trial was not feasible.30 On the basis of its ongoing analysis of the EUA and emerging scientific data, the FDA had determined that chloroquine and HCQ are unlikely to be effective and likely to be toxic in treating COVID-19 for the authorized uses in the EUA. Since the beginning of this pandemic, studies have been carried out to identify drugs that can be repurposed as treatment for COVID-19 either by employing in vitro high throughput screening (HTS) or by performing in silico structure-based virtual screening/docking on both human host targets and viral proteins.9 Yet, the clinical trials landscape is dominated by the initial list of therapeutics (e.g., HCQ, chloroquine, remdesivir, and azithromycin, respectively). The initial rush to publish results, often in the preprint (not peer-reviewed) format, inevitably resulted in the accumulation of comparatively weak evidence and the disclosure of suboptimally documented candidates for drug repositioning. Particularly in the context of this pandemic there is a stringent need for high-quality studies that can provide critical knowledge concerning the COVID-19 disease and reliable treatment proposals.31 With these caveats in mind, we conceived a computational workflow that included independent in vitro validation, followed by assessing emerging candidates in the context of available clinical pharmacology data, with the aim of proposing suitable candidates for clinical studies for early stage (incubation and symptomatic phases) patients infected by SARS-CoV-2.
HCQ is an antimalarial drug that has been claimed as treatment for COVID-19.32,33 Several randomized clinical trials proved its lack of efficacy.34−36 The utility of HCQ in combination with azithromycin is also controversial,37−40 with some studies having been retracted.38 The overall consensus is that the risk of HCQ-induced toxicity, even death, outweighs its therapeutic benefit.36 However, HCQ appears to be effective in SARS-CoV-2 CPE assays (VeroE6 cells) and was thus deemed as suitable candidate for a LBVS study. Indeed, HCQ is often used as positive control in antiviral cell-based assays,9 despite lack of clarity regarding its antiviral mechanism of action. Whether its activity is due to inhibition of endocytic pathways through elevation of endosomal pH40 or to the protective binding to host cells receptors,41 it is not certain if the function of proteins involved in these processes can be subject to therapeutic manipulation with drugs other than HCQ or chloroquine. While finding alternatives to HCQ could be accomplished by elucidating these cellular and molecular processes, we focused on identifying alternatives to HCQ by means of molecular similarity.42
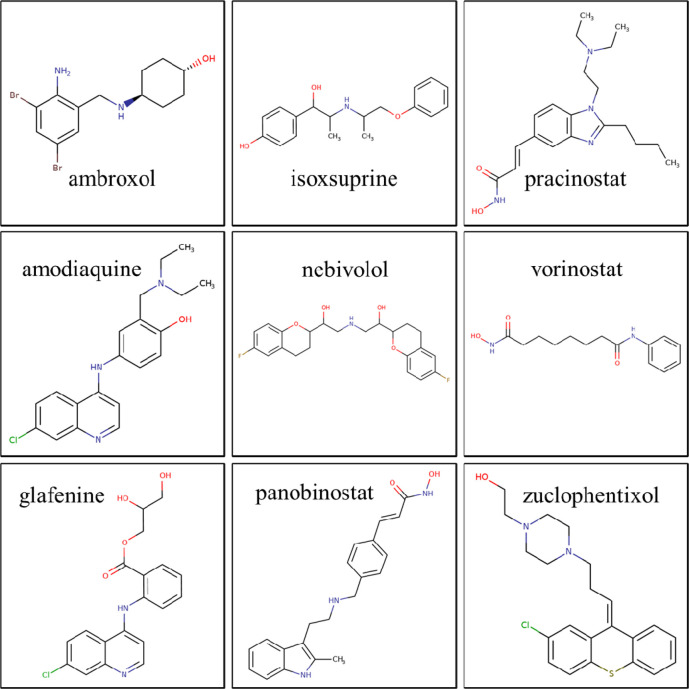
Therefore, we used HCQ as a template for LBVS and screened about 4000 approved drugs (Figure 2). Drugs were then ranked by similarity to HCQ and a minimum GP similarity cutoff of 0.34. Nine drugs having GP above 0.34 were identified: glafenine, AQ (amodiaquine), vorinostat, zuclopenthixol, isoxsuprine, nebivolol, ambroxol, panobinostat, and pracinostat (see Figure 3).
Figure 3.
Chemical structures of the nine LBVS hit drugs.
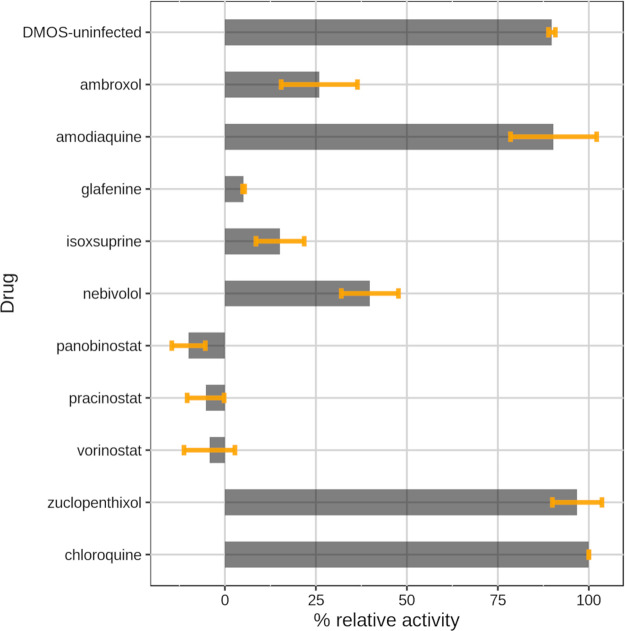
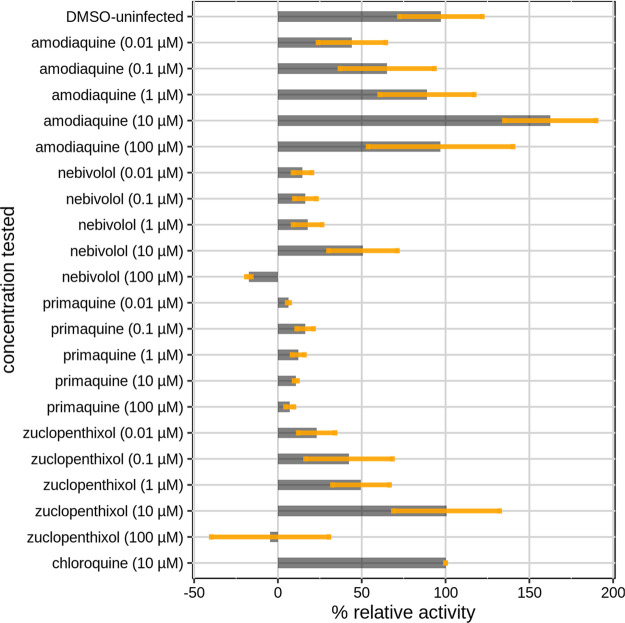
Their ability to block SARS-CoV-2 infection was subsequently evaluated in vitro on Vero E6 cells at a concentration of 10 μM. Uninfected cells and chloroquine were used as positive controls. Here, the choice of chloroquine (and not of HCQ) as an active reference drug was intended to provide an even more robust validation of our methodology. The results of the assay are displayed in Figure 4. Activity values were normalized to chloroquine (100% response) and infected cells (0% response) for a better comparison of potencies between the tested drugs and the well-known active drug chloroquine. Two LBVS hits, zuclopenthixol and AQ, show anti-SARS-CoV-2 activity comparable to chloroquine. Nebivolol, tested at 10 μM, displayed a moderate antiviral effect (∼40% of chloroquine activity) whereas ambroxol shows a mild/low signal of activity (∼25% of chloroquine activity). The other compounds, isoxsuprine, glafenine, vorinostat, panobinostat, and pracinostat did not seem to have any beneficial effect in blocking SARS-CoV-2 infection. GP values did not correlate in any way with the drug activities measured at 10 μM.
Figure 4.
Bar plot showing the antiviral activity of the selected drugs from the virtual screening tested at 10 μM. Data are the means ± SD of three replicates. The values shown are normalized to chloroquine (100% antiviral activity) and to infected cells with DMSO (0% antiviral activity).
The antiviral activity of zuclopenthixol (ZPX), nebivolol and AQ was further evaluated in dose–response experiments at five different concentrations (see Figure 5). At this stage we also chose to include primaquine in this experiment. Primaquine is an antimalarial drug that is structurally related to chloroquine, HCQ, and AQ. It was excluded by accident from the LBVS hit selection. The dose–response based EC50 values were estimated as 0.13 μM for AQ, 1.35 μM for ZPX, and 2.72 μM for nebivolol, respectively. We note that all these drugs, when given at a concentration of 100 μM, show decreased antiviral activity. We hypothesize that, at 100 μM concentrations, these drugs might exhibit a certain degree of cytotoxicity. AQ is similar to HCQ and chloroquine, both in terms of structural features (4-aminoquinoline derivatives) and drug profile (e.g., antimalarial). Its anti-SARS-CoV-2 activity, confirmed here, has first been discussed elsewhere.11,14 On the other hand, primaquine showed no appreciable activity in comparison with chloroquine (see Figure 5) despite their high structural similarity.
Figure 5.
Bar plot showing dose–response experiment results for amodiaquine, nebivolol, primaquine, and zuclopenthixol. Data are the means ± SD of three replicates. The values shown are normalized to chloroquine (100% antiviral activity) and to infected cells with DMSO (0% antiviral activity).
Comparison with other HTS Experiments
The National Center for Advancing Translational Sciences (NCATS) recently conducted an HTS study for detecting SARS-CoV-2 antiviral candidates.43 They performed SARS-CoV-2 cytopathic effect (CPE) experiments to screen a large library of chemicals and drugs. The results and the assay description can be accessed from their dedicated COVID-19 portal.44 When we examined the results of our hits in both assays (this study vs NCATS), seven out of nine were found to have the same outcome; thus, AQ displayed high activity and vorinostat, ambroxol, glafenine, isoxsuprine, pracinostat, and panobinostat displayed low activity, in CPE experiments. However, both ZPX and nebivolol were also reported with poor activity in the NCATS assay. At this point, we cannot explain these inconsistencies. Perhaps, differences in ZPX formulation (dihydrochloride in our experiments vs the decanoate prodrug in the NCATS assay) may be a possible explanation. However, we conducted additional experiments for both ZPX and nebivolol, as further discussed below.
Molecular Analysis of the LBVS Results
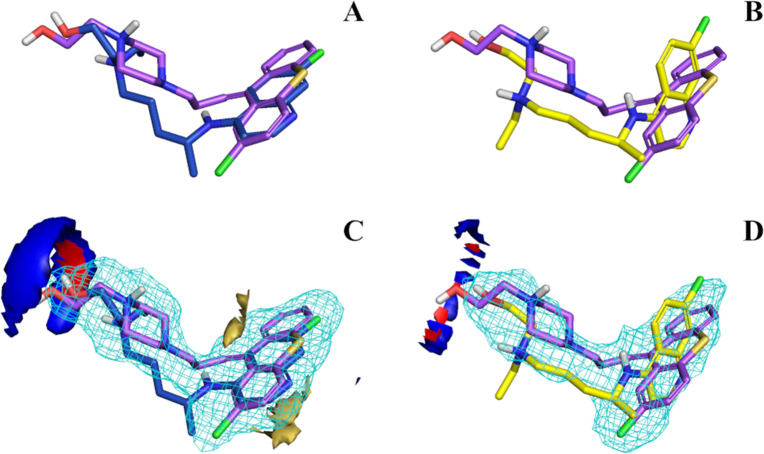
Details concerning the virtual screening hits and their ionization state are summarized in Table 2. As stated previously, we choose a GP cutoff of 0.34 below which drugs were not selected for in vitro testing. With GP = 0.411, ZPX meets this criterion due to the alignment of ZPX with the HCQ R isomer; The ZPX/S-HCQ alignment score was below the cutoff (GP = 0.32). ZPX could have been discarded, had we considered only the S isomer of HCQ as template for LBVS. Figure 6 shows the FLAP-generated alignments between ZPX and both HCQ enantiomers. Although the side chains are aligned, aromatic moieties are better matched with R-HCQ (Figure 6A,B). Matching the 4-aminoquinoline group of R-HCQ with the thioxanthene moiety of ZPX improves the similarity score. To further study this relationship, we examined the MIF overlap between ZPX and both HCQ enantiomers (see Figure 6C,D). The ZPX hydrogen bonding fields (donor and acceptor) are in better alignment with R-HCQ, while the hydrophobic field of ZPX does not align well with the S-HCQ equivalent. No evident differences were observed for the shape field. The different MIF configurations explain the higher GP score between R-HCQ and ZPX, vs ZPX/S-HCQ, and suggest that stereochemistry may play a role in the SARS-CoV-2 CPE activity.
Table 2. Molecular Details of the Virtual Screening Hits.
| drug | GPa | closest HCQ isomerb | top ranked drug isomerc | pKbd |
|---|---|---|---|---|
| ambroxol | 0.414 | R | N/A | 8.78 |
| amodiaquine | 0.429 | R | N/A | 9.48 |
| glafenine | 0.482 | R | S | N/A |
| isoxsuprine | 0.343 | R | RSS | 8.56 |
| nebivolol | 0.349 | S | RSSS | 7.88 |
| panobinostat | 0.349 | R | N/A | 9.33 |
| pracinostat | 0.345 | R | N/A | 8.59 |
| vorinostat | 0.417 | S | N/A | N/A |
| zuclopenthixol | 0.411 | R | N/A | 8.12 |
The similarity score GP (Glob-Prod) was computed with FLAP version 2.2.1.
The hydroxychloroquine isomer that is most similar (highest GP) to the screened drug.
If the screened drug is a racemate, we report the most similar (highest GP) drug isomer.
pKb values were computed with MoKa version 3.2.1.
Figure 6.
Aligned structures of zuclopenthixol with R-HCQ (A) and S-HCQ (B). GRID fields intersection of both R-HCQ (C) and S-HCQ (D) with zuclopenthixol. Structures can be identified by the different color of carbon atoms: violet for zuclopenthixol, blue for R-HCQ and yellow for S-HCQ. GRID fields are colored as follows: red for hydrogen-bond donor, blue for hydrogen-bond acceptor, yellow for hydrophobic interaction. The size/shape field is shown as a light blue wireframe. Energy levels of the fields were tuned similarly for a better comparison across the figures.
We further studied the lack of in vitro activity for the other drugs, by inspecting the intersection of glafenine and isoxsuprine MIFs with HCQ. According to FLAP, the most similar alignments are between R-HCQ, S-glafenine, and RSS-isoxsuprine isomers. Figure 7 shows their GRID fields intersections. Upon inspection, it is not obvious why glafenine is inactive, since MIFs overlap well compared to ZPX (glafenine GP > zuclopenthixol GP, see Table 2 and Figure 7A). The aromatic moiety of glafenine is also 4-amino-quinoline, the same as AQ, chloroquine, and HCQ, all of which are potent in vitro. However, glafenine does not share the positive charge featured in HCQ, ZPX, and AQ, lacking an aliphatic amine. Thus, we hypothesize that the ionization state of these drugs plays a critical role, and assume that the electrostatic potential generated by the protonated aliphatic amine group may be necessary for anti-SARS-CoV-2 CPE activity. The pKb values computed with MoKa are 9.48 for HCQ and 8.12 for ZPX, respectively. We hypothesize that a weak base (8 ≤ pKb ≤ 10), but not a quaternary amine may be required for drugs exhibiting CPE activity in this assay. As for isoxsuprine (Figure 7B), its inactivity does not seem to be due to the lack of a positive charge (pKb = 8.56), but rather to the mismatch of its hydrophobic and shape MIFs. Indeed, the MIF alignment is not optimal, placing the phenol moiety of isoxsuprine on the HCQ side-chain region. Moreover, the isoxsuprine methoxybenzene moiety does not appear sufficiently large or rigid for a good alignment with the HCQ 4-aminoquinoline. As the benzopyranyl group resembles the quinoline core of HCQ to a certain extent, this could explain the moderate activity of isoxsuprine (see also Figure 4). On the basis of these observations, we can suggest additional structural differences shared by the other poorly active drugs. For example, vorinostat lacks both a basic nitrogen and a quinolone-like aromatic core and pracinostat has bulky substituents attached to the aromatic core, whereas a bulky group is attached to the positive nitrogen in panobinostat.
Figure 7.
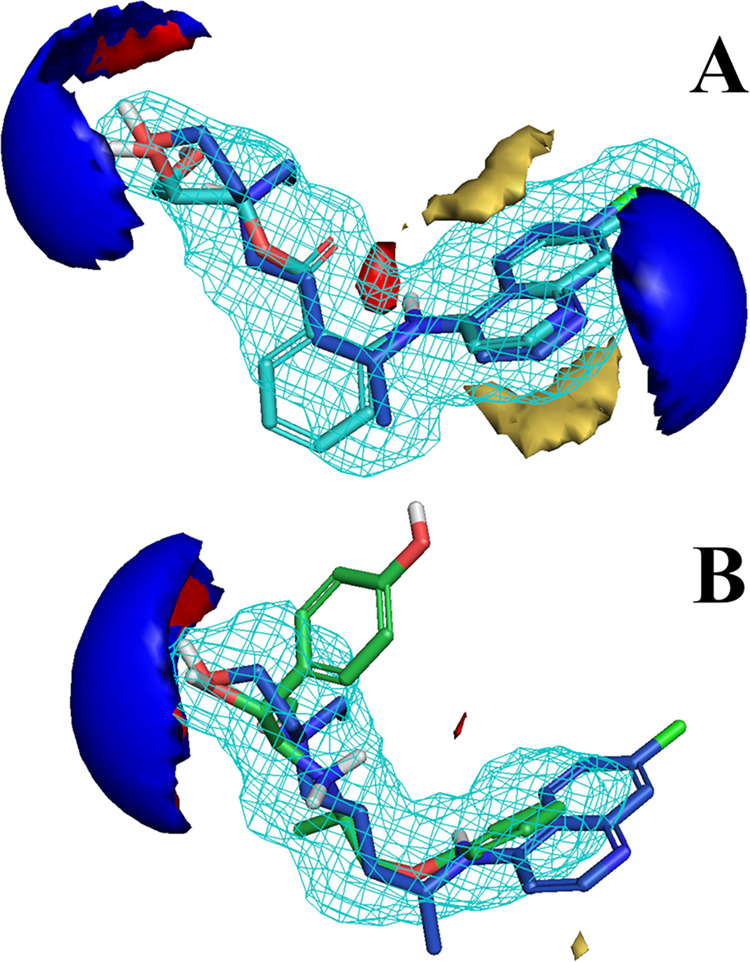
Aligned structures of R-HCQ with S-glafenine (A) and RSS-isoxsuprine (B). Structures can be identified by the different color of carbon atoms: blue for R-HCQ, cyan for S-glafenine, and green for RSS-isoxsuprine. GRID fields are colored as follows: red for hydrogen-bond donor, blue for hydrogen-bond acceptor, yellow for hydrophobic interaction. The size/shape field is shown as a light blue wireframe. Energy levels of the fields were tuned similarly for a better comparison across the figures.
With respect to ambroxol, the situation is less clear. Ambroxol is a drug and the active metabolite of bromhexine. Bromhexine ability to block SARS-CoV-2 infection has been assessed in a CPE assay, EC50 of 13.93 μM.44 The structural difference between the two drugs consists in the aliphatic 4-hydroxylation of the cyclohexane ring and the N-demethylation of the aliphatic amine. However, bromhexine and ambroxol remain chemically similar, and a reasonable hypothesis could be that both drugs would exhibit anti-SARS-CoV-2 activity. Finally, the structural features of nebivolol are borderline with respect to HCQ-like activity. Indeed, the 3,4-dihydro-6-fluoro-2H-chromene group is less aromatic (and more hydrophobic) than 4-aminoquinoline, yet similar to 4-aminoquinoline with regards to size and shape. The secondary aliphatic amine nitrogen pKb (7.88) is less basic, but close to our initial hypothesis for drugs that exhibit CPE activity in this assay (8 ≤ pKb ≤ 10). Nebivolol has an intrinsic symmetry (centered at the nitrogen) that may explain nebivolol’s HCQ-like activity, in contrast to the other drugs. However, nebivolol has four chiral centers. The drug formulation of nebivolol consists of d-nebivolol and l-nebivolol, stereochemically designated as (SRRR)-nebivolol and (RSSS)-nebivolol, respectively, which makes these enantiomers an almost mirror image.45
We conclude that simply evaluating the ionization state of a drug, prior to LBVS, might be a necessary but not sufficient requirement for anti-SARS-CoV-2 activity. Indeed, the presence of a hydrophobic core with no less than two fused aromatic rings, may also be required for HCQ-like antiviral activity.
Zuclopenthixol Therapeutics Analysis
ZPX is a typical antipsychotic of the thioxanthene class, with a piperazine side chain.46 Its antipsychotic effect has been linked to a dopamine receptors blockade. Thioxanthenes have a high affinity for both the dopamine D1 and D2 receptors. Currently, ZPX is neither FDA nor EMA approved, but it is approved to treat schizophrenia in several countries such as UK, Canada, Australia, Denmark, and India.47 According to our in vitro experiments, it blocks SARS-CoV-2 infection with a higher potency than either chloroquine or HCQ (see Table 3). To better understand the suitability of ZPX as a COVID-19 early treatment, here we discuss its PK profile.48 ZPX has a number of PK properties that make it suitable for clinical use (see Table 3). In addition to being available for intravenous and oral administration, ZPX has moderate to good oral bioavailability (about 50% of the dose reaches systemic circulation). Its half-life is approximately 20 h and its clearance is mainly metabolic by means of the cytochrome P450 isoenzymes 2D6 and 3A4.49 Maximum serum concentration (Cmax) is reached 4 h after oral administration; upon dosing of 20 mg/day to steady state, it is estimated to reach 0.03 μM (13 ng/mL).48 When administered intramuscularly, a direct correlation between dose given and plasma levels could be measured over a two-week period.50 Hence, we can expect that a higher dosing regimen (to reach the maximum approved dose of 150 mg/day)46 would yield proportionally increased plasma concentrations. Such a therapeutic regimen with the hydrochloride (not decanoate) formulation would be preferred, should ZPX be prescribed during the early incubation/symptomatic phases (first 5–12 days postinfection) of COVID-19. Overall, the PK profile of ZPX seems less promising when compared to chloroquine and HCQ (Table 3). Given the lack of comprehensive clinical PK studies, and because ZPX is a relatively old drug (first introduced in 1978), additional investigations are warranted. Several dose-dependent adverse reactions may occur upon ZPX administration.46,48 However, it is difficult to discuss ZPX drug safety in contrast to clinical trials candidates chloroquine and HCQ, given the adverse events recorded for both chloroquine51 and HCQ,52 and given that these drugs manifest toxic reactions in clinical trials.38
Table 3. Activity Values and PK Properties for Chloroquine, Hydroxychloroquine, and the Drugs Identified in This Studya.
| drug | EC50 (μM) | bioavailability (%) | t1/2 (hours) | Cmax (μM) |
|---|---|---|---|---|
| chloroquine | 4.575 | 80 | 570 | 2.62 |
| hydroxychloroquine | 2.775 | 79 | 1056 | 0.14 |
| zuclopenthixol | 1.35 | 50 | 20 | 0.0348 |
| nebivolol | 2.72 | 12 | 10 | 0.0254 |
| amodiaquine | 0.13 | 2976 | 7.960 | 0.177 |
EC50 values were measured in-house except where differently referenced. Concerning PK properties, values were retrieved from Goodman and Gillman,74 except where differently referenced.
Nebivolol Therapeutics Analysis
Nebivolol is a widely used β-blocker approved for treatment of high blood pressure and heart failure.53 As mentioned above, it is orally formulated as the racemic mixture of (SRRR)-nebivolol and (RSSS)-nebivolol.45 On the basis of our in vitro experiments, it blocks SARS-CoV-2 infection with a higher potency than chloroquine, and with a potency comparable to HCQ (Table 3). Regarding its PK properties, its absorption varies depending on the extent of gut metabolism, primarily via the 2D6 cytochrome P450 isoenzyme (CYP2D6).54 This further influences its half-life and Cmax (0.02 μM), which are both lower than chloroquine and HCQ. While its PK profile makes it less viable as a candidate, nebivolol is a relatively safe drug, with a relatively low number of adverse reactions having an incidence of at least 1% reported.53 The adverse events are generally mild, with an incidence similar to placebo, and with a lower incidence of side effects that are typical for other β-blockers.55
Amodiaquine Therapeutics Analysis
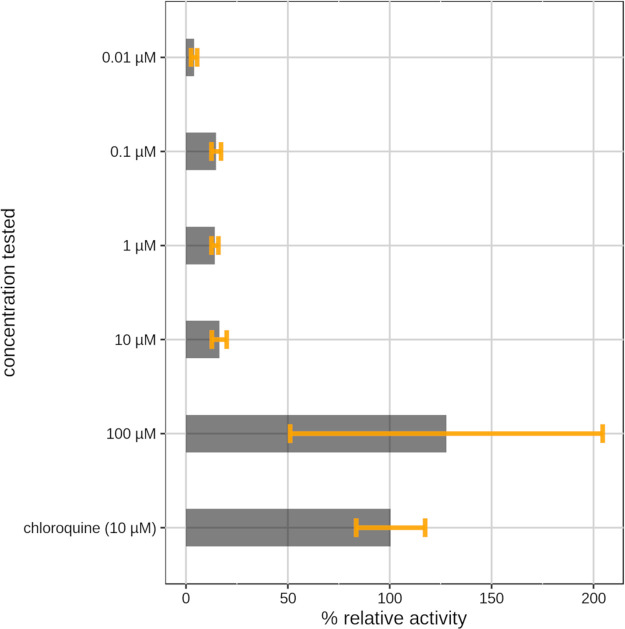
On the basis of efficacy, AQ displays better antiviral activity, blocking SARS-CoV-2 infection with an EC50 of 0.13 μM in the VeroE6 cell-based assay. In addition to our experiments, its activity has been recorded by other laboratories as well.11,14 AQ is on the WHO list of essential medicines for the treatment of malaria, in combination with artesunate.56 Can AQ be a valid antiviral candidate? AQ is rapidly converted by hepatic cytochrome P450s into DAQ (N-monodesethyl amodiaquine).57 The 2C8 cytochrome P450 isoform (CYP2C8) is the main route of metabolism for AQ.58 The major metabolite, DAQ, retains substantial antimalarial activity and has a much longer half-life compared to the parent drug (3 h vs ∼500 h for AQ and DAQ, respectively).59 Hence, the prolonged efficacy of the drug is ensured by its active metabolite, not by AQ. Our intention was then to verify if, similarly to antimalarial activity, DAQ retains antiviral activity as well. To test this hypothesis, we performed a dose–response experiment with DAQ (see Figure 8) in infected Vero E6 cells. The metabolite has anti-SARS-CoV-2 activity, with a lower potency compared to AQ (see Figures 3 and 4). In fact, an activity comparable to chloroquine is likely reached between 1 μM, where the metabolite seems almost inactive, and 100 μM, where the metabolite is as potent as 10 μM chloroquine in the cytopathic effect. The blood concentration of DAQ is much higher than that of AQ,60 and DAQ is equipotent with remdesivir61 in the titer reduction assay at 10 μM (vide infra). Given that the concentration of DAQ remains at high levels for over a week due to its prolonged (over 3 weeks) half-life, AQ may well be a viable therapeutic option for SARS-CoV-2 infections. A more detailed analysis is needed to explore the pros and cons of administering AQ (or DAQ) to COVID19 patients.
Figure 8.
Bar plot showing dose–response experiment results for N-monodesethyl amodiaquine. Data are the means ± SD of three replicates. The values shown are normalized to chloroquine (100% antiviral activity) and to infected cells with DMSO (0% antiviral activity).
The Ambiguity of Ambroxol
Ambroxol is a drug used to treat respiratory diseases involving abnormal mucus secretions and impaired mucus transport. It facilitates mucus clearance, thus, allowing easier breathing.62 Currently, ambroxol is under clinical trials investigation in China,63 but it is not clear whether it blocks viral infection or if it affects the course of the respiratory illness caused by the virus. When tested at 10 μM, ambroxol had relatively weak potency in blocking SARS-CoV-2, compared with other drugs (e.g., ZPX, AQ). Hence, one would suggest that there is not a relevant interaction between ambroxol and the virus, but rather that ambroxol could act in a complementary manner with the coadministered antivirals, or perhaps that it is the host response. We cannot exclude that hypothesis that ambroxol is active at higher concentrations. Bromhexine (another drug used in respiratory diseases and prodrug of ambroxol) was found active in the SARS-CoV-2 NCATS experiments.44 As the differences between the two drugs are minor, it is unlikely that bromhexine is active, while ambroxol is not, which highlights another inconsistency in experimental evaluations. Ambroxol had low activity in the NCATS single-dose experiments, supporting our hypothesis that ambroxol’s beneficial effect is not related to direct antiviral activity.64 Both bromhexine65 and ambroxol66 induce autophagy in vitro. Although the different cell systems in which these drugs were tested might influence the experimental results, ambroxol was measured to be at least twice as potent as bromhexine at clinically relevant concentrations. Moreover, ambroxol has been shown to induce autophagy in mice lung cells.66 Drugs that modulate autophagy could have a broad applicability on several human diseases, including the treatment of viral infections.65 However, autophagy modulation, as an alternative mechanism of action for COVID-19, has not been elucidated yet. Nevertheless, it should be noted that such modulation is induced by drugs (bromhexine and ambroxol) that have similar pharmacophoric features to chloroquine and HCQ, yet their antiviral mechanism could be different. To clarify the situation, confirmatory CPE experiments for both ambroxol and bromhexine are required, with ambroxol needing higher in vitro test concentrations to fully explore its effects in the Vero E6 assay. Since ambroxol (at 50 μM) is inactive in the titer reduction assay (vide infra), we speculate that the use of ambroxol or bromhexine in SARS-CoV-2 infected patients may be beneficial via another (not directly antiviral) mechanism.
Independent Confirmation
Running independent confirmatory experiments is a good practice that serves to strengthen the reliability of the experimental findings provided to the scientific community. Concerning COVID-19 drug repositioning, few, if any, papers claim to have confirmed their outcomes with a second, independent set of experiments, with different batches of compounds and a different lab. We previously discussed the high variability of in vitro SARS-CoV-2 experiments. The rush to find a cure may seem like a good reason, yet it did not yield significant advances on current COVID-19 drug therapies. Therefore, to confirm experimental findings from the University of New Mexico (UNM), we initiated a series of independent tests for the most promising drugs described above. These experiments were carried out at the University of Tennessee Health Science Center (UTHSC) where the antiviral activities of ZPX, nebivolol, ambroxol, primaquine, AQ, and DAQ were reassessed in terms of inhibitory activity toward the cytopathic effect (CPE) induced by SARS-CoV-2 and the titer reduction assay.
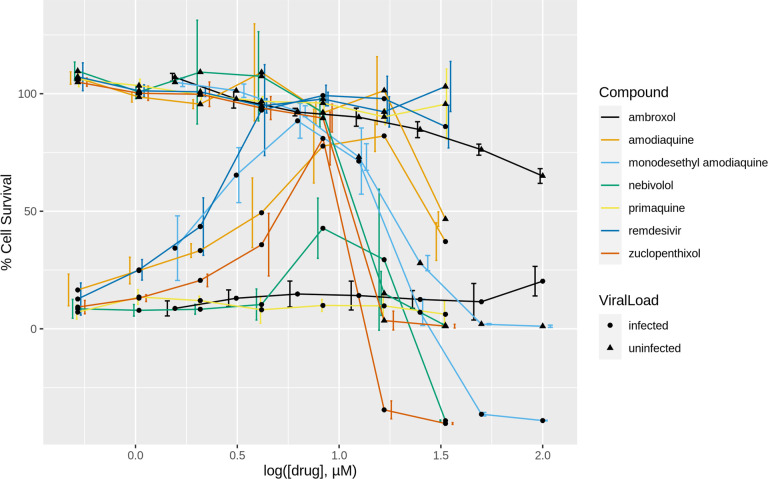
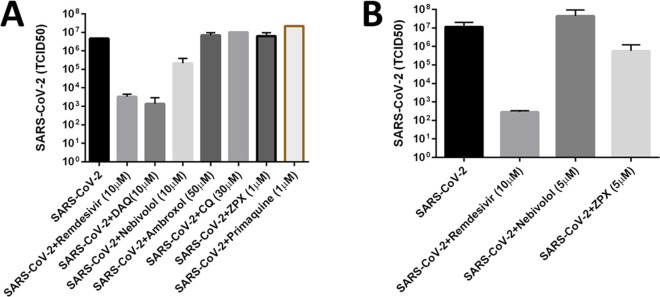
Dose–response profiles of the compounds tested both in the presence (dot lines) or absence (triangle lines) of the virus are shown in Figure 9. Antiviral potencies and CC50 and SI50 values are reported in Table 4. Primaquine did not exhibit appreciable activity at any of the tested concentrations. Since its activity in the former dose–response experiments was also low, we no longer plan to evaluate its anti-SARS-CoV-2 therapeutic potential. The potency of nebivolol is practically identical: 2.8 μM (UTHSC) vs 2.72 μM (UNM; Table 3), respectively. However, the EC50 values for ZPX were somehow different: 1.35 μM (UNM) vs 15 μM (UTHSC) experiments, despite a similar peak of activity: 100% at 10 μM in the UNM tests, and 80% at 8.33 μM in the UTHSC tests, respectively. This discrepancy may be due in part to the different MOI (virus load) used: 0.05 (UNM) vs 0.1 (UTHSC). Regardless of the exact value, independent experiments confirm ZPX as a potent anti-SARS-COV-2 drug. Although we did not conduct dose–response tests for ambroxol at UNM, we examined its antiviral activity at higher concentrations at UTHSC. Ambroxol exhibits antiviral behavior with an EC50 of 14.8 μM (Figure 9), and has the highest SI50 among the tested compounds, given its low cytotoxicity (Figure 9 and Table 4). Thus, higher concentrations of ambroxol may be achieved in vivo without incurring unwanted toxicity. These results confirm ambroxol as a valid antiviral candidate despite its apparently lower in vitro potency. ZPX also has the lowest SI50 (Table 4) among the tested drugs, which raises a warning about its potential toxicity in vivo when used at high concentrations. With respect to AQ and its metabolite DAQ, the former shows a higher EC50 value (5.4 μM) than the one reported in the UNM experiment (0.13 μM). However, this value is in agreement with other literature values (see Table 1).11,14 Here as well differences in the drug concentrations regimen and MOI may have influenced the results, but the anti-SARS-CoV-2 activity of AQ was nonetheless validated. The activity of DAQ was independently confirmed as well, with slightly different results. In Figure 9, DAQ peaks at 6.25 μM, with nearly 90% of the Vero E6 cells surviving, whereas earlier findings show activity peaking somewhere between 10 and 100 μM (Figure 8). In this case, UTHSC experiments provided a more accurate picture of the DAQ antiviral activity, with EC50 = 4 μM. Finally, we note that ZPX, AQ, DAQ, and nebivolol showed decreased antiviral activity at concentrations above 10 μM in both sets of experiments (see Figures 5 and 9). To summarize, independent experiments conducted at UTHSC with chemicals purchased from different vendors confirmed all the antiviral activities found at UNM, providing additional insights in the anti-SARS-CoV-2 activity of the drugs reported here.
Figure 9.
Confirmatory experiments, dose–response curves. Data are the means ± SD of four replicates. The values shown are normalized to uninfected cells (100% cell survival) and to infected cells (0% cell survival).
Table 4. Activity Values Measured from the Confirmatory Experiments for the Drugs Identified in This Study. Remdesivir Was Used as Positive Control.
| compound | CC50 (μM) | EC50 (μM) | SI |
|---|---|---|---|
| ambroxol | 202 | 14.8 | 13.6 |
| N-mono desethyl amodiaquine | 5.9 | 4 | 1.5 |
| amodiaquine | 32.9 | 5.4 | 6.1 |
| nebivolol | 12.5 | 2.8 | 4.5 |
| primaquine | ND | ND | ND |
| zuclopenthixol | 10.9 | 15 | 0.7 |
| remdesivir | ND | 1.8 | ND |
Titer Reduction Assay
We assessed the most promising compounds at UTHSC, using a SARS-CoV-2 titer reduction assay. The assay estimates the degree of reduction of infected cells at a fixed concentration of the drug and at various viral loads. The concentration of the tested drug is fixed, resembling the expected systemic drug concentration. Cells were infected with seven different viral loads, dose–response curves were drawn, and TCID50 values were determined. CQ, ambroxol, and primaquine were also tested in this assay. Results are summarized in Figure 10. In our first experiment (Figure 10A), we tested 10 μM of DAQ and nebivolol, 50 μM of ambroxol, 30 μM of CQ, and 1 μM of ZPX and primaquine, respectively. We used 10 μM remdesivir as positive control. Of these, DAQ reduced the viral titer by more than 3 log units, while nebivolol reduced the virus titer by more than 1 log unit. Neither primaquine nor ambroxol were found to reduce the virus titer. Surprisingly, CQ did not reduce the virus titer either, in disagreement with its known (potent) CPE effects. We repeated ZPX and nebivolol at 5 μM (see Figure 10B), and noted that the higher concentration of ZPX reduced the virus titer by more than 1 log unit. On the basis of these two experiments, nebivolol, ZPX, and DAQ showed promising in vitro antiviral efficacy. In both experiments, 10 μM remdesivir had a 3–4 log units reduction in infectious virus titer.
Figure 10.
Bar plots showing the results of the titer reduction experiments 1 (A) and 2 (B). Remdesivir is used as positive control.
Drug Combinations Therapy
One of the perils of drug repurposing is that the original drug target, for which the drug was approved in first place, can become an off target. This could potentially induce unwanted side effects that further complicate the previously understood side effects tableau for the drug in question. For example, were ZPX to be administered to COVID-19 patients, this drug would still interact with the dopamine D1 and D2 receptors (mode of action – MoA – targets for its antipsychotic activity). Thus, one cannot exclude undesired reactions due to the “re-targeting” of repurposed drugs. One way to address this is to lower the drug dosage with respect to the maximum recommended daily dosage approved for the original indication. This could however lead (in this case) to reduced antiviral efficacy. To compensate for the possible loss of efficacy due to lower dosage, combinations of repurposed drugs with antiviral activity could be used.
Drug combinations have been highly successful against the hepatitis C virus (e.g., Harvoni–ledipasvir and sofosbuvir) and against the human immunodeficiency virus (Atripla–efavirenz, tenofovir, and emtricitabine), for example. We anticipate that the anti-SARS-CoV-2 drugs remdesivir (currently approved in Japan) and favipiravir (currently approved in Russia and India) may prove more effective, particularly when combined with other drugs having anti-SARS-CoV-2 activity, preferably with another MoA. Thus, finding drugs that can be combined and administered at lower doses while maintaining a good overall antiviral efficacy can reduce the risk of side effects. The risk of drug–drug interactions needs to be addressed as well. Here we propose that drugs described here can be used as adjuvant anti-SARS-CoV-2 therapy for remdesivir and favipiravir.67,68 Both remdesivir69 and favipiravir70 block RNA-dependent RNA polymerase, but this MoA is most likely distinct from the drugs discussed here. Hence, we expect that combining two different MoAs may have a synergistic effect on the overall antiviral efficacy. Equally important, neither remdesivir71 nor favipiravir72 interact with the enzymes responsible for the drug metabolism of ZPX, nebivolol, and AQ, respectively. Thus, their combination is likely to be safe in terms of drug–drug interactions. Experiments to evaluate the anti-SARS-CoV-2 synergy of ZPX, nebivolol, and DAQ when combined with remdesivir and favipiravir are under way.
Conclusions
As of 2018, chloroquine was being considered for repurposing in no fewer than 392 diseases,73 and much uncertainty about its exact mechanism of action or that of its active metabolite, HCQ, remains. Absent a clear antiviral MoA target for HCQ, we resorted to ligand-based virtual screening using hydroxychloroquine as a reference drug, with the aim of repositioning approved drugs for treating SARS-CoV-2 infections. This protocol included independent experimental confirmation, which was carried out at UNM and UTHSC with compounds procured from independent sources. Our validated results identify zuclopenthixol and nebivolol as potentially novel, viable therapeutic options for the treatment of incubation and early stage COVID-19 infections. Both zuclopenthixol and nebivolol exhibit in vitro antiviral activities and potencies that are comparable or better than chloroquine and hydroxychloroquine. On the basis of the reported side effects, nebivolol appears safer than chloroquine/HCQ, while ZPX may require additional safety evaluation. In our opinion, both zuclopenthixol and nebivolol are viable candidates for COVID-19 clinical trials.
Furthermore, we show evidence that AQ, validated through its active metabolite DAQ, and ambroxol are promising repositioning candidates, with independently confirmed in vitro anti-SARS-CoV-2 activity. Ambroxol appears to have the safest PK profile, yet lacks direct antiviral activity. Indeed, SARS-CoV-2 titer reduction assay on Vero E6 cells confirms the antiviral efficacy of DAQ (equipotent to remdesivir), ZPX, and nebivolol, but not chloroquine or ambroxol. We are currently planning to evaluate the synergistic anti-SARS-CoV-2 effects of remdesivir and/or favipiravir with DAQ, ZPX and nebivolol, respectively. In parallel, we will explore other LBVS templates, such as AQ and ZPX, and prioritize weak bases, especially those incorporating a tertiary aliphatic amine and a fused aromatic core.
Acknowledgments
We thank OpenEye Scientific Software for providing access to their software. Dr. Graham Timmins (UNM College of Pharmacy) provided valuable input regarding the mechanistic evaluation of chloroquine-like CPE activities. The DrugCentral component of this work is funded by NIH Common Fund U24 CA224370 (TIO). This work was supported in part by UTHSC funding made available to CBJ.
Glossary
Abbreviations Used
- AQ
amodiaquine
- DAQ
N-monodesethyl amodiaquine
- FLAP
fingerprints for ligands and proteins
- HCQ
hydroxychloroquine
- LBVS
ligand-based virtual screening
- MoA
mode of action
- MIF
molecular interaction fields
- MOI
multiplicity of infection
- PK
pharmacokinetics
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- UNM
University of New Mexico
- UTHSC
University of Tennessee Health Sciences Center
- TCID50
tissue culture infectious dose
- ZPX
zuclopenthixol
The authors declare the following competing financial interest(s): T.I.O. has received honoraria or consulted for Abbott, AstraZeneca, Chiron, Genentech, Infinity Pharmaceuticals, Merz Pharmaceuticals, Merck Darmstadt, Mitsubishi Tanabe, Novartis, Ono Pharmaceuticals, Pfizer, Roche, Sanofi and Wyeth. He is on the Scientific Advisory Board of ChemDiv Inc.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
References
- Coronavirus outbreak officially declared a pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (accessed 2020).
- COVID-19 World Map. https://www.cdc.gov/coronavirus/2019-ncov/global-covid-19/world-map.html.
- WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/.
- Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment.
- Sohrabi C.; Alsafi Z.; O’Neill N.; Khan M.; Kerwan A.; Al-Jabir A.; Iosifidis C.; Agha R. (2020) World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 76, 71–76. 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie N.; Saville M.; Hatchett R.; Halton J. (2020) Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 382, 1969–1973. 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- Liu C.; Zhou Q.; Li Y.; Garner L. V.; Watkins S. P.; Carter L. J.; Smoot J.; Gregg A. C.; Daniels A. D.; Jervey S.; Albaiu D. (2020) Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 6, 315–331. 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. M.; Monogue M. L.; Jodlowski T. Z.; Cutrell J. B. (2020) Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 323, 1824–1836. 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Kuleshov M. V.; Clarke D. J. B.; Kropiwnicki E.; Jagodnik K. M.; Bartal A.; Evangelista J. E.; Zhou A.; Ferguson L. B.; Lachmann A.; Ma’ayan A.; et al. (2020) The COVID-19 Gene and Drug Set Library. Patterns 1, 100090. 10.1016/j.patter.2020.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger B., Bojkova D., Zaliani A., Cinatl J., Claussen C., Westhaus S., Reinshagen J., Kuzikov M., Wolf M., Geisslinger G., Gribbon P., and Ciesek S. (2020) Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection, Res. Square, 10.21203/rs.3.rs-23951/v1. [DOI] [Google Scholar]
- Jeon S.; Ko M.; Lee J.; Choi I.; Byun S. Y.; Park S.; Shum D.; Kim S. (2020) Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 64, e00819 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Sun J., Huang Y.-Y., Li Y., Shi Y., Li Z., Li X., Yang F. H., Zhao J., Luo H.-B., Zhang T. Y., and Zhang X. (2020) Enantiomers of Chloroquine and Hydroxychloroquine Exhibit Different Activities Against SARS-CoV-2 in vitro, Evidencing S-Hydroxychloroquine as a Potentially Superior Drug for COVID-19, bioRxiv, 10.1101/2020.05.26.114033. [DOI] [Google Scholar]
- Touret F.; Gilles M.; Barral K.; Nougairède A.; van Helden J.; Decroly E.; de Lamballerie X.; Coutard B. (2020) In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 10, 13093. 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Haupt R., Logue J., Matthews K., and Frieman M. B. (2020) FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro, bioRxiv, 10.1101/2020.03.25.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Shankar R., Drelich A., Paithankar S., Chekalin E., Dexheimer T., Rajasekaran S., Tseng C.-T. K., and Chen B. (2020) Reversal of Infected Host Gene Expression Identifies Repurposed Drug Candidates for COVID-19, bioRxiv, 10.1101/2020.04.07.030734. [DOI] [Google Scholar]
- Clarivate Analytics.. Web of Science, https://clarivate.com/webofsciencegroup/solutions/web-of-science.
- Taboureau O.; Nielsen S. K.; Audouze K.; Weinhold N.; Edsgärd D.; Roque F. S.; Kouskoumvekaki I.; Bora A.; Curpan R.; Jensen T. S.; Brunak S.; Oprea T. I. (2011) ChemProt: a disease chemical biology database. Nucleic Acids Res. 39, D367–72. 10.1093/nar/gkq906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner R. E.; Aronson J. K. (2020) Chloroquine and hydroxychloroquine in covid-19. BMJ. 369, m1432 10.1136/bmj.m1432. [DOI] [PubMed] [Google Scholar]
- Ursu O.; Holmes J.; Knockel J.; Bologa C. G.; Yang J. J.; Mathias S. L.; Nelson S. J.; Oprea T. I. (2017) DrugCentral: online drug compendium. Nucleic Acids Res. 45, D932–D939. 10.1093/nar/gkw993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursu O.; Holmes J.; Bologa C. G.; Yang J. J.; Mathias S. L.; Stathias V.; Nguyen D.-T.; Schürer S.; Oprea T. (2019) DrugCentral 2018: an update. Nucleic Acids Res. 47, D963–D970. 10.1093/nar/gky963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. C. D.; Skillman A. G.; Warren G. L.; Ellingson B. A.; Stahl M. T. (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 50, 572–584. 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletti F.; Storchi L.; Sforna G.; Cruciani G. (2007) New and original pKa prediction method using grid molecular interaction fields. J. Chem. Inf. Model. 47, 2172–2181. 10.1021/ci700018y. [DOI] [PubMed] [Google Scholar]
- Baroni M.; Cruciani G.; Sciabola S.; Perruccio F.; Mason J. S. (2007) A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J. Chem. Inf. Model. 47, 279–294. 10.1021/ci600253e. [DOI] [PubMed] [Google Scholar]
- Goodford P. J. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28, 849–857. 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Goetz D. H.; Choe Y.; Hansell E.; Chen Y. T.; McDowell M.; Jonsson C. B.; Roush W. R.; McKerrow J.; Craik C. S. (2007) Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry 46, 8744–8752. 10.1021/bi0621415. [DOI] [PubMed] [Google Scholar]
- Severson W. E.; Shindo N.; Sosa M.; Fletcher T. 3rd; White E. L.; Ananthan S.; Jonsson C. B. (2007) Development and validation of a high-throughput screen for inhibitors of SARS CoV and its application in screening of a 100,000-compound library. J. Biomol. Screening 12, 33–40. 10.1177/1087057106296688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A. O.; Severson W.; Jonsson C.; Singh K.; Weiss S. R.; Sarafianos S. G. (2013) Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 87, 8017–8028. 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Parvathareddy J., Yang D., Bansal S., O’Connell K., Golden J. E., and Jonsson C. B. (2020) Emergence and Magnitude of ML336 Resistance in Venezuelan Equine Encephalitis Virus Depends on the Microenvironment, J. Virol. 10.1128/JVI.00317-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J.; Muench H. (1938) A simple method for estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Hinton D. M. (2020) Letter revoking EUA for chloroquine phosphate and hydroxychloroquine sulfate. https://www.fda.gov/media/138945/download.
- Levin J. M.; Oprea T. I.; Davidovich S.; Clozel T.; Overington J. P.; Vanhaelen Q.; Cantor C. R.; Bischof E.; Zhavoronkov A. (2020) Artificial intelligence, drug repurposing and peer review. Nat. Biotechnol. 38, 1127–1131. 10.1038/s41587-020-0686-x. [DOI] [PubMed] [Google Scholar]
- Colson P.; Rolain J.-M.; Lagier J.-C.; Brouqui P.; Raoult D. (2020) Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 55, 105932. 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M. D. S.; Rathod J.; Gernsheimer J. (2020) A Rapid Systematic Review of Clinical Trials Utilizing Chloroquine and Hydroxychloroquine as a Treatment for COVID-19. Acad. Emerg. Med. 27, 493–504. 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J.; Sun Y.; Platt J.; Zucker J.; Baldwin M.; Hripcsak G.; Labella A.; Manson D. K.; Kubin C.; Barr R. G.; Sobieszczyk M. E.; Schluger N. W. (2020) Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 382, 2411–2418. 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., and Zhang Z. (2020) Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial, MedRxiv, 10.1101/2020.03.22.20040758. [DOI] [Google Scholar]
- Cavalcanti A. B., Zampieri F. G., Rosa R. G., Azevedo L. C. P., Veiga V. C., Avezum A., Damiani L. P., Marcadenti A., Kawano-Dourado L., Lisboa T., Junqueira D. L. M., de Barros E Silva P. G. M., Tramujas L., Abreu-Silva E. O., Laranjeira L. N., Soares A. T., Echenique L. S., Pereira A. J., Freitas F. G. R., Gebara O. C. E., Dantas V. C. S., Furtado R. H. M., Milan E. P., Golin N. A., Cardoso F. F., Maia I. S., Hoffmann Filho C. R., Kormann A. P. M., Amazonas R. B., Bocchi de Oliveira M. F., Serpa-Neto A., Falavigna M., Lopes R. D., Machado F. R., and Berwanger O., Coalition Covid-19 Brazil I Investigators . (2020) Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate Covid-19, N. Engl. J. Med. 10.1056/NEJMoa2019014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.; Cao Z.; Han M.; Wang Z.; Chen J.; Sun W.; Wu Y.; Xiao W.; Liu S.; Chen E.; Chen W.; Wang X.; Yang J.; Lin J.; Zhao Q.; Yan Y.; Xie Z.; Li D.; Yang Y.; Liu L.; Qu J.; Ning G.; Shi G.; Xie Q. (2020) Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 369, m1849 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra M. R., Desai S. S., Ruschitzka F., and Patel A. N. (2020) RETRACTED: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis, Lancet, 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boulware D. R.; Pullen M. F.; Bangdiwala A. S.; Pastick K. A.; Lofgren S. M.; Okafor E. C.; Skipper C. P.; Nascene A. A.; Nicol M. R.; Abassi M.; Engen N. W.; Cheng M. P.; LaBar D.; Lother S. A.; MacKenzie L. J.; Drobot G.; Marten N.; Zarychanski R.; Kelly L. E.; Schwartz I. S.; McDonald E. G.; Rajasingham R.; Lee T. C.; Hullsiek K. H. (2020) A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 383, 517–525. 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M.; Orhon I.; Rocchi C.; Zhou X.; Luhr M.; Hijlkema K.-J.; Coppes R. P.; Engedal N.; Mari M.; Reggiori F. (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455. 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J.; Di Scala C.; Chahinian H.; Yahi N. (2020) Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 55, 105960. 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert H.; Bajorath J. (2007) Molecular similarity analysis in virtual screening: foundations, limitations and novel approaches. Drug Discovery Today 12, 225–233. 10.1016/j.drudis.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Brimacombe K. R., Zhao T., Eastman R. T., Hu X., Wang K., Backus M., Baljinnyam B., Chen C. Z., Chen L., Eicher T., Ferrer M., Fu Y., Gorshkov K., Guo H., Hanson Q. M., Itkin Z., Kales S. C., Klumpp-Thomas C., Lee E. M., Michael S., Mierzwa T., Patt A., Pradhan M., Renn A., Shinn P., Shrimp J. H., Viraktamath A., Wilson K. M., Xu M., Zakharov A. V., Zhu W., Zheng W., Simeonov A., Mathé E. A., Lo D. C., Hall M. D., and Shen M. (2020) An OpenData portal to share COVID-19 drug repurposing data in real time, bioRxiv, 10.1101/2020.06.04.135046. [DOI] [Google Scholar]
- OpenData COVID-19. (2020) https://opendata.ncats.nih.gov/covid19/.
- Siebert C. D.; Hänsicke A.; Nagel T. (2008) Stereochemical comparison of nebivolol with other β-blockers. Chirality 20, 103–109. 10.1002/chir.20509. [DOI] [PubMed] [Google Scholar]
- Zuclopenthixol Drug Label. (2020) https://www.medicines.org.uk/emc/product/994/#PRODUCTINFO.
- Bryan E. J.; Purcell M. A.; Kumar A. (2017) Zuclopenthixol dihydrochloride for schizophrenia. Cochrane Database Syst. Rev. 11, CD005474. 10.1002/14651858.CD005474.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLOPIXOL Product Information. (2020) https://gp2u.com.au/static/pdf/C/CLOPIXOL-PI.pdf.
- Davies S. J. C.; Westin A. A.; Castberg I.; Lewis G.; Lennard M. S.; Taylor S.; Spigset O. (2010) Characterisation of zuclopenthixol metabolism by in vitro and therapeutic drug monitoring studies. Acta Psychiatr. Scand. 122, 444–453. 10.1111/j.1600-0447.2010.01619.x. [DOI] [PubMed] [Google Scholar]
- Poulsen J. H.; Olesen O. V.; Larsen N. E. (1994) Fluctuation of serum zuclopenthixol concentrations in patients treated with zuclopenthixol decanoate in viscoleo. Ther. Drug Monit. 16, 155–159. 10.1097/00007691-199404000-00008. [DOI] [PubMed] [Google Scholar]
- Aralen. https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/006002s043lbl.pdf.
- Plaquenil. (2020) https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/009768s037s045s047lbl.pdf.
- Bystolic. (2020) https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/021742s013lbl.pdf.
- Chen C. L.; Desai-Krieger D.; Ortiz S.; Kerolous M.; Wright H. M.; Ghahramani P. (2015) A Single-Center, Open-Label, 3-Way Crossover Trial to Determine the Pharmacokinetic and Pharmacodynamic Interaction Between Nebivolol and Valsartan in Healthy Volunteers at Steady State. Am. J. Ther. 22, e130 10.1097/MJT.0000000000000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft J. (2007) A review of the safety and efficacy of nebivolol in the mildly hypertensive patient. Vasc. Health Risk Manag. 3, 909–917. [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2019) World Health Organization Model List of Essential Medicines: 21st List, WHO, https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf.
- Zhang Y.; Vermeulen N. P. E.; Commandeur J. N. M. (2017) Characterization of human cytochrome P450 mediated bioactivation of amodiaquine and its major m etabolite N-desethylamodiaquine. Br. J. Clin. Pharmacol. 83, 572–583. 10.1111/bcp.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Q.; Björkman A.; Andersson T. B.; Ridderström M.; Masimirembwa C. M. (2002) Amodiaquine Clearance and Its Metabolism toN-Desethylamodiaquine Is Mediated by CYP2C8: A New High Affinity and Turnover Enzyme-Specific Probe Substrate. J. Pharmacol. Exp. Ther. 300, 399–407. 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- Krishna S.; White N. J. (1996) Pharmacokinetics of Quinine, Chloroquine and Amodiaquine. Clin. Pharmacokinet. 30, 263–299. 10.2165/00003088-199630040-00002. [DOI] [PubMed] [Google Scholar]
- Jullien V.; Ogutu B.; Juma E.; Carn G.; Obonyo C.; Kiechel J.-R. (2010) Population pharmacokinetics and pharmacodynamic considerations of amodiaquine and desethylamodiaquine in Kenyan adults with uncomplicated malaria receiving artesunate-amodiaquine combination therapy. Antimicrob. Agents Chemother. 54, 2611–2617. 10.1128/AAC.01496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. (2020) Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 6, 672–683. 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba M.; Ragnoli B. (2008) Ambroxol in the 21st century: pharmacological and clinical update. Expert Opin. Drug Metab. Toxicol. 4, 1119–1129. 10.1517/17425255.4.8.1119. [DOI] [PubMed] [Google Scholar]
- Zambon SpA . (2020) NCT03843541. A Clinical Trial to Compare the Efficacy and Safety of 1-week Treatment of Intravenous N-acetylcysteine (NAC) 600 mg Twice Daily, Ambroxol Hydrochloride 30 mg Twice Daily and Placebo as Expectorant Therapies in Adult Chinese Patients With Respiratory Tract Diseases and Abnormal Mucus Secretions. https://clinicaltrials.gov/ct2/show/NCT03843541.
- Depfenhart M.; de Villiers D.; Lemperle G.; Meyer M.; Di Somma S. (2020) Potential new treatment strategies for COVID-19: is there a role for bromhexine as add-on therapy?. Int. Emerg. Med. 15, 801–812. 10.1007/s11739-020-02383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S.; Ahmed Z.; Bradfute S. B.; Arko-Mensah J.; Mandell M. A.; Won Choi S.; Kimura T.; Blanchet F.; Waller A.; Mudd M. H.; Jiang S.; Sklar L.; Timmins G. S.; Maphis N.; Bhaskar K.; Piguet V.; Deretic V. (2015) Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat. Commun. 6, 8620. 10.1038/ncomms9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. W.; Gu Y.; Peters R. S.; Salgame P.; Ellner J. J.; Timmins G. S.; Deretic V. (2018) Ambroxol Induces Autophagy and Potentiates Rifampin Antimycobacterial Activity. Antimicrob. Agents Chemother. 62, e01019 10.1128/AAC.01019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frediansyah A., Nainu F., Dhama K., Mudatsir M., and Harapan H. (2020) Remdesivir and its antiviral activity against COVID-19: A systematic review, Clin. Epidemiol. Glob. Health, 10.1016/j.cegh.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H. (2020) Favipiravir: A Possible Pharmaceutical Treatment for COVID-19. J. Endocrinol. Metab. 10, 33–34. 10.14740/jem645. [DOI] [Google Scholar]
- Gordon C. J.; Tchesnokov E. P.; Feng J. Y.; Porter D. P.; Götte M. (2020) The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295, 4773–4779. 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y.; Komeno T.; Nakamura T. (2017) Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad., Ser. B 93, 449–463. 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. (2020) What do we know about remdesivir drug interactions?. Clin. Transl. Sci. 13, 842–844. 10.1111/cts.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madelain V.; Nguyen T. H. T.; Olivo A.; de Lamballerie X.; Guedj J.; Taburet A.-M.; Mentré F. (2016) Ebola Virus Infection: Review of the Pharmacokinetic and Pharmacodynamic Properties of Drugs Considered for Testing in Human Efficacy Trials. Clin. Pharmacokinet. 55, 907–923. 10.1007/s40262-015-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. C.; Ekins S.; Williams A. J.; Tropsha A. (2018) A bibliometric review of drug repurposing. Drug Discovery Today 23, 661–672. 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel K. E., Shen D. D., and Isoherranen N. (2017) Appendix II in Goodman & Gilman’s the Pharmacological Basis of Therapeutics (Brunton L. L., Hilal-Dandan R., and Knollman B. C., Eds.) pp 1325–1378, Mc Graw Hill, NewYork. [Google Scholar]
- Liu J.; Cao R.; Xu M.; Wang X.; Zhang H.; Hu H.; Li Y.; Hu Z.; Zhong W.; Wang M. (2020) Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 6, 16. 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) Artesunate/Amodiaquine 100/270 mg tablets. https://extranet.who.int/prequal/sites/default/files/documents/MA058part6v2.pdf.
- Quang N. N.; Chavchich M.; Anh C. X.; Birrell G. W.; van Breda K.; Travers T.; Rowcliffe K.; Edstein M. D. (2018) Comparison of the Pharmacokinetics and Ex Vivo Antimalarial Activities of Artesunate-Amodiaquine and Artemisinin-Piperaquine in Healthy Volunteers for Preselection Malaria Therapy. Am. J. Trop. Med. Hyg. 99, 65–72. 10.4269/ajtmh.17-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Clarivate Analytics.. Web of Science, https://clarivate.com/webofsciencegroup/solutions/web-of-science.