Abstract
In response to the ongoing COVID-19 pandemic, there is a worldwide effort being made to identify potential anti-SARS-CoV-2 therapeutics. Here, we contribute to these efforts by building machine-learning predictive models to identify novel drug candidates for the viral targets 3 chymotrypsin-like protease (3CLpro) and RNA-dependent RNA polymerase (RdRp). Chemist-curated training sets of substances were assembled from CAS data collections and integrated with curated bioassay data. The best-performing classification models were applied to screen a set of FDA-approved drugs and CAS REGISTRY substances that are similar to, or associated with, antiviral agents. Numerous substances with potential activity against 3CLpro or RdRp were found, and some were validated by published bioassay studies and/or by their inclusion in upcoming or ongoing COVID-19 clinical trials. This study further supports that machine learning-based predictive models may be used to assist the drug discovery process for COVID-19 and other diseases.
1. Introduction
The ongoing COVID-19 pandemic has challenged the health system of many countries with its high rate of morbidity and mortality. Developing effective treatments against the disease is critical in the effort to save human lives and help society return to normal. In order to respond quickly, scientists have been exploring various ways to accelerate the drug development process, including drug repurposing and computational approaches in drug discovery. One promising way to achieve this goal is to build machine learning models by applying the quantitative structure–activity relationship (QSAR) methodology to suitable protein targets of the SARS-CoV-2 virus, which could predict possible drug candidates for treating COVID-19.
Among all the proteins in SARS-CoV-2, the 3 chymotrypsin-like protease (3CLpro) and RNA-dependent RNA polymerase (RdRp) are two ideal protein targets for QSAR modeling. 3CLpro is a protease that is required in order for the coronavirus to cleave the polyprotein peptides into individual functional nonstructural proteins.1 In addition, by comparing the amino acid sequences and protein structures, the 3CLpro was found to be highly conserved among SARS-CoV-2 and other human coronaviruses, with sequence identities of 96% with SARS-CoV-1, 87% with MERS-CoV, and 90% with Human-CoV.2−5 Therefore, the 3CLpro inhibitors identified in previous coronavirus-related research are promising inhibitors for SARS-CoV-2 3CLpro and their associated SAR study data are valuable information as training material for machine learning models in searching for new inhibitors of SARS-CoV-2 3CLpro. Consequently, it is reasonable to propose that broad-spectrum inhibitors against human coronaviruses are promising drug candidates that can be developed into suitable drugs for targeting SARS-CoV-2 and many other human coronaviruses.
RdRp is the major enzyme that is responsible for replicating viral genomic RNA in host cells. The amino acid residues of active sites in RdRp are highly conserved among single-stranded, positive-sense RNA [(+)ssRNA] viruses, including SARS-CoV-1 and hepatitis C virus (HCV).6 In addition, RdRp of SARS-CoV-2 shares an almost identical protein sequence with that of SARS-CoV-1, indicating that it also shares the highly conserved amino acid residues in the active sites among the (+)ssRNA viruses.1 Indeed, the EM-cryo structure of SARS-CoV-2 RdRp reveals that this enzyme contains the classic divalent-cation-binding residue D618, which is conserved in most viral polymerases including HCV (residue D220), and the catalytic residues [759 to 761 (SDD)], which are also conserved in most viral RdRps, such as HCV.7 Fortunately, various viral RdRps have been widely studied for inhibitors in the (+)ssRNA viruses, especially in HCV-related research.8 Therefore, these existing RdRp inhibitors for the (+)ssRNA viruses, such as HCV, may provide valuable insights for drug development for SARS-CoV-2 RdRp inhibition.8
QSAR and machine learning9a modeling have increasingly been used to facilitate drug discovery in recent years. QSAR is usually one of the first steps in the drug discovery process, in which large databases of chemical structures are screened through a variety of predictive mathematical models in order to narrow down the number of potential drug candidates. Simply put, QSARs are mathematical models approximating rather complicated biological or physicochemical properties of chemicals based on quantitative measures of the corresponding molecular structures. The underlying assumption is that the activity is directly related to the structure of the chemicals. Thus, molecules with similar structural features will exhibit similar physical properties or similar biological effects. One major challenge in QSAR is the selection of appropriate structural features to be used as molecular descriptors. To a large degree, the development of an accurate machine learning model depends on the results of the analysis of the factors that may be meaningful in the mechanism of action (MOA). Relevant molecular descriptors can be defined and calculated only after a solid understanding of the MOA has been achieved.
Many techniques have been developed to describe molecular structures in QSAR. Hansch et al.13−15 used 1-octanol/water partition coefficients and Hammett substituent constants to model hydrophobic and electronic effects. Methods that utilize topological indexes,16−20 3D geometry descriptors,21−24 electronic structural descriptors,25−27 molecular shape descriptors,28 fragment-based approaches,29−31 and 3D QSAR32,33 have been extensively used to model a wide range of biological end points.
Since the outbreak of COVID-19, different QSAR studies have been employed to predict the possibilities of FDA-approved drugs or investigational drugs to be repurposed for COVID-19, as well as the likelihood of repurposing inhibitors for other viruses related to SARS-CoV-2. QSAR modeling has been proven to be useful in virtual screening and lead optimization for drug discovery. Monte Carlo optimization-based and classification QSAR models of SARS-CoV main protease (Mpro) were developed and used for screening of some natural products.9b Multiple linear regression analysis combined with 2D-QSAR modeling was successfully employed to identify potential SARS-CoV-3CLpro enzyme inhibitors.10−12a Classification QSAR data mining of diverse SARS-CoV papain-like protease (PLpro) inhibitors were utilized to generate predictive QSAR models and later used for virtual screening to identify molecules that could be effective against SARS-CoV PLpro.12b
Development of a QSAR model also requires thorough collaboration between computational and experimental scientists who have deep knowledge of their respective fields and have made effort to understand and develop working knowledge of the complementary field. In this study, computational scientists and chemists with related background closely collaborated to curate training data and build highly predictive QSAR models for 3CLpro and RdRp. The two QSAR models presented here were validated with high sensitivity, specificity, and accuracy. After modeling protease inhibitor activity as a function of the substance structure, we identified some of the most promising candidates among substances predicted to be active inhibitors against coronavirus 3CLpro.
2. Methods
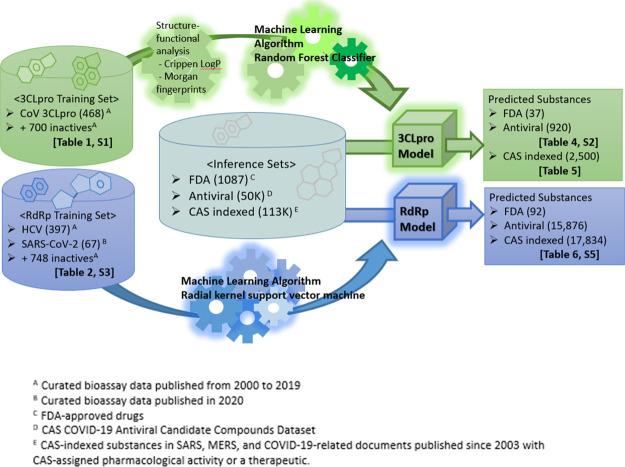
In this study, as illustrated in Figure 1, scientists curated over 1000 inhibitors with structure–bioactivity data as training molecules for 3CLpro and RdRp protein targets. We collected these data from the most current SARS-CoV-2 bioassay studies as well as existing studies with SARS-CoV-1, MERS-CoV, and other related viruses in the CAS data collection. Using this data, we applied a variety of machine learning algorithms to build several dozen QSAR models—selecting from among these, the strongest performing models—one targeting 3CLpro and one targeting RdRp.
Figure 1.
Overall process flow.
We used the resulting models to screen 1087 FDA-approved drugs,34 nearly 50,000 substances from the CAS COVID-19 Antiviral Candidate Compounds Dataset,35a and a list of 113,000 substances with CAS-assigned pharmacological activity or a therapeutic role indexed in SARS, MERS, and COVID-19-related documents published since 2003. Some predicted molecules of these models were validated by published bioassay studies and clinical trials as a positive indication of the predictive models.
2.1. 3CLpro & RdRp Training Data set Preparation
To develop training sets for predicting active substances to target SARS-CoV-2 3CLpro and RdRp, we examined bioassay data published from 2000 to 2020 in the CAS data collections. This included substance information, targets, activity measures [half maximal inhibitory concentration (IC50), half maximal effective concentration (EC50), inhibition constant (Ki), and dissociation constant (Kd)], source organisms, and assay details. We selected substances with assay data of IC50 ≤ 10 μM, EC50 ≤ 10 μM, Ki ≤ 10 μM, and/or Kd ≤ 10 μM toward the targets as active substances from these bioassay data—a threshold suggested by Deng et al.35b and by Zhou et al.35c Substances with IC50, EC50, Ki, and Kd values above 100 μM from these bioassay data were considered to be inactive.
2.1.1. 3CLpro Inhibitors of SARS-CoV-1 and other Coronaviruses
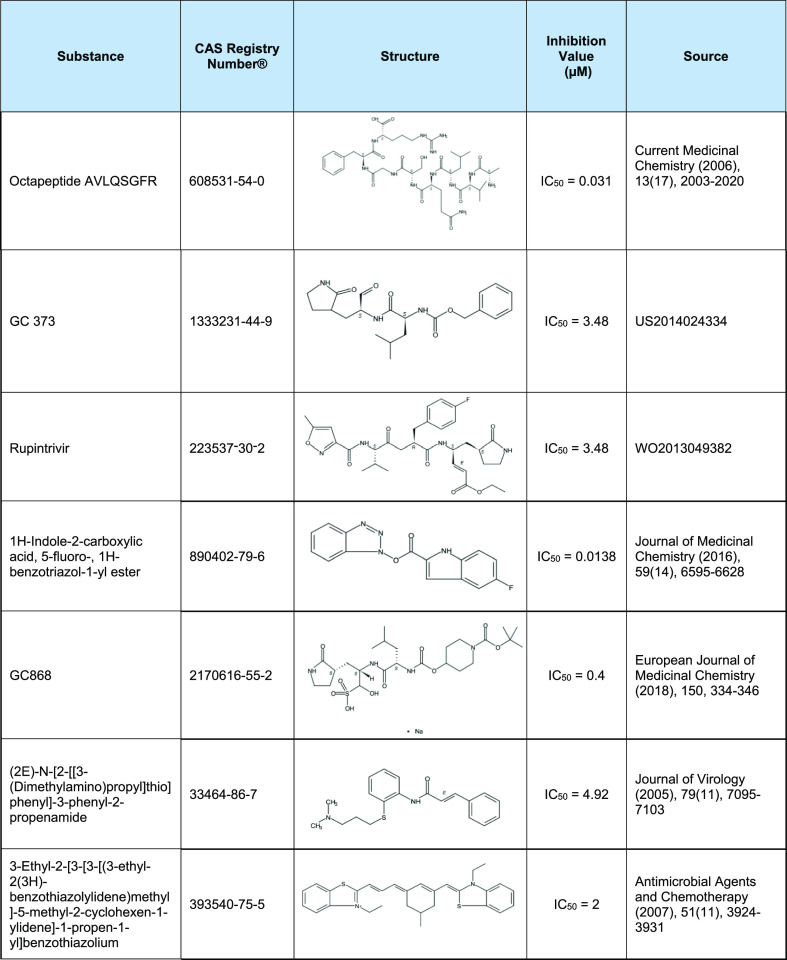
For the training set for the 3CLpro model, we collected 468 diverse active substances from 76 different documents published prior to 2020—and 700 inactive substances. Over 70% of active substances are from SARS-CoV-1 3CLpro studies. The rest of the active substances are from MERS-CoV, common human coronavirus, feline infectious peritonitis, transmissible gastroenteritis coronavirus, and infectious bronchitis virus 3CLpro studies. Seven unique small molecules representing the structural diversity of SARS-CoV-1 3CLpro inhibitors are shown in Table 1. See Supporting Information Table S1 for the complete training set.
Table 1. List of Example Training Substances with In Vitro Inhibitory Effect on SARS-CoV-1 3CLpro.
2.1.2. RdRp Inhibitors of SARS-CoV-1 and Other Viruses
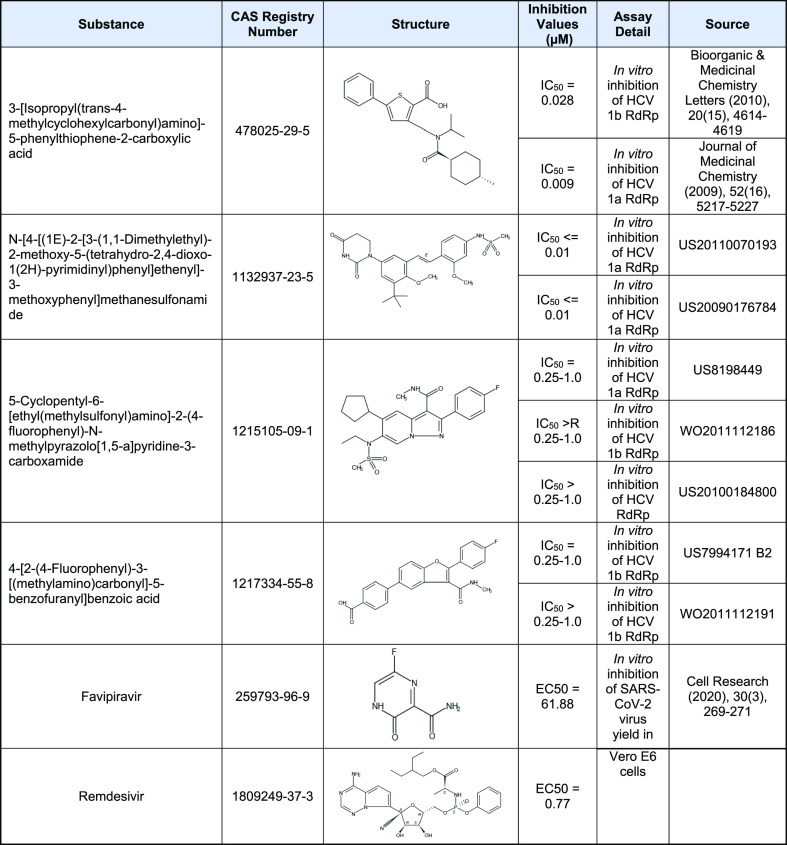
For the training set for the RdRp model, we collected 1212 diverse active substances from 51 documents published prior to 2020. We also collected 67 SARS-COV-2 RdRp inhibitors from journal articles published in 2020. Four unique small molecules representing the structural diversity of HCV RdRp inhibitors and two unique small molecules representing SARS-CoV-2 RdRp inhibitors are shown in Table 2. A full list of the training substances can be seen in Table S4.
Table 2. Examples of Training Substances with Inhibitory Effect on RdRp of HCV (a Positive-Stranded ssRNA Virus) in Various Biological Studies.
2.2. Machine Learning Methods
An automated machine learning platform, DataRobot v6.0.3 https://www.datarobot.com/, was used to train and evaluate performance of more than 40 different machine learning algorithms. DataRobot is a commercial tool that we used to build informative features selected from molecular descriptors.38 Model training and testing with fivefold cross-validation was utilized in the model selection process. Grid search was used as the default method for hyperparameter optimization.
Support Vector Classifier (Radial Kernel) is a robust algorithm that has been actively used to model biological activity of small molecules for various targets.39,40 DataRobot implementation is based on scikit-learn.41 This algorithm searches the optimal hypersurface in multidimensional feature space38 that separates the active and inactive classes of compounds. DataRobot tool identified SVM with Radial Kernel as the best model for our RdRp data set based on highest cross-validation area under curve (AUC) values among the other explored models. Hyperparameters Gamma and C were optimized using default DataRobot grid search.
XGBoost is a decision-tree-based ensemble ML algorithm that uses a gradient boosting framework.42,43 For building RdRp models, default DataRobot optimized parameters were used. Previously XGBoost demonstrated both high accuracy and robustness on bioactivity prediction tasks with highly imbalanced class distributions.44
Many ensemble models that are called “blenders” in DataRobot also demonstrated excellent performance that was comparable to top selected models. For example, the ENET Blender model combines the predictions generated by SVM and XGBoost models followed by Elastic-Net Classifier45—which is a linear model trained with L1 and L2 prior as regularizers. Very similar to that, average blender also combines the output of SVM and XGBoost predictions and returns mean that can improve performance of individual models.46 Ensemble-based models also were found to be an improvement for building QSAR models.47
2.3. Binary Classifiers
In our models, bioactivity prediction is a binary classification task that generates the probability of compounds, represented as a structural feature vector, being a member of the active or inactive class. An automated machine learning tool was used to train and evaluate performance of more than 40 different machine learning algorithms.
A number of common metrics were used to evaluate statistical performance of our models, including:
where: TP—true positives. TN—true negatives. FP—false positives. FN—false negatives.
To estimate the statistical performance of the models, a common fivefold cross-validation procedure has been used for all models in this study. In each of five iterations of this approach, 80% of the data is used to build a model, and 20% is held as a test set. Across the whole process, then, every record is held out as validation in one part of the process, yet all records are made available to the model.
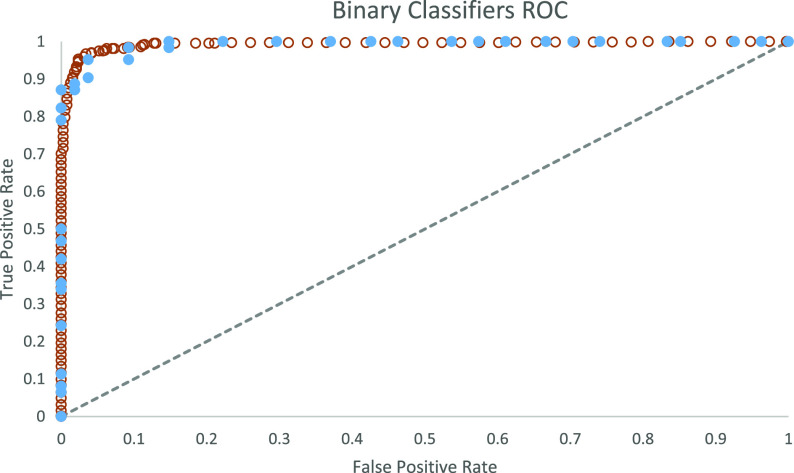
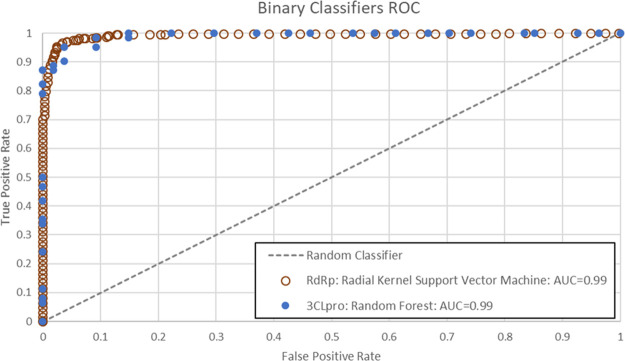
To measure the statistical quality of our models, we used the receiver–operator-characteristic (ROC)-AUC36 that is a measure of model fit ranging from 0 to 1 and created by plotting the true positive rate against the false positive rate at various threshold settings, with a perfect model having an ROC-AUC of 1.
Many binary classifiers from our training data showed high AUC, accuracy, sensitivity, and specificity in the cross-validation tests. AUCs are shown in Supporting Information Table S5, with a description of validation metrics. ROC plots for the final models are shown in Figure 2. Many models—including Support Vector Machine with Radial Kernel (SVM), Gradient Boosted Trees, ENET, and variations of AVG Blenders—showed AUC values of 0.99.
Figure 2.
ROC curves for 3CLpro and RdRp trained binary classifiers.
The high performance of these binary classifier models provides us with the ability to identify the most relevant active molecules in large data sets. This can be attributed to (i) the more separated distributions of actives and inactives (observations with a midrange of 10 μM < IC50 < 100 μM were excluded from the training data sets) and (ii) the high diversity of active and inactive examples in the training data that were prepared by CAS scientists.
2.4. Molecular Descriptors
In order to train the binary classification machine learning models that predict the probability of whether a new chemical will inhibit the human coronavirus 3CLpro or RdRp, the abovementioned training set molecules were used, and their molecular features were studied for the models. Molecular descriptors available in the software library RDKit,37 including Morgan48 (radius = 3; length = 2048), MACCS keys,49 Atom Pairs,50 Topological Torsion51 fingerprints, as well as Crippen LogP52 and MR,52 Molecular Quantum Numbers (MQN),38 PEOE_VSA and SMR_VSA,53 and FractionCSP3 (fraction of C atoms that are SP3 hybridized) were utilized to build the models.
2.5. Structure–Functional Analysis
In order to calculate and evaluate structural alerts, the following structure-functional analysis approach has been developed. The molecular graph of a given chemical structure is split into subfragments similar to Klopman.29 However, beyond just linear fragments, we further generated all possible subfragments with extended connectivity length from 1 to 9 atoms. The activity of the molecular structure is assigned to any of the generated subfragments. The above procedure is applied to every single molecule structure in the training set; thus, we end up with an extended list of fragments that may belong to several different molecules. In this study, we calculate the activity of a substructure as the mean value of the activities of the molecules that contain the fragment. Once the activity of the fragments is calculated, we order the list of fragments so that at the top of the list, are the most active structural alerts and at the bottom, the most inactive ones.
3. Results and Discussion
3.1. 3CLpro Model
After the calculation of the molecular descriptors, several machine learning algorithms, including Random Forest, Gradient Boosting, Neural Networks, and Support Vector Machine (SVM), to name a few (see Methods Section 2.2 above), were exploited in order to obtain robust machine learning models. The best model was obtained by using a Random Forest Classifier algorithm and utilizing Crippen LogP and Morgan fingerprints as molecular representations. This model achieved a ROC-AUC of 0.99, as previously shown in Figure 2.
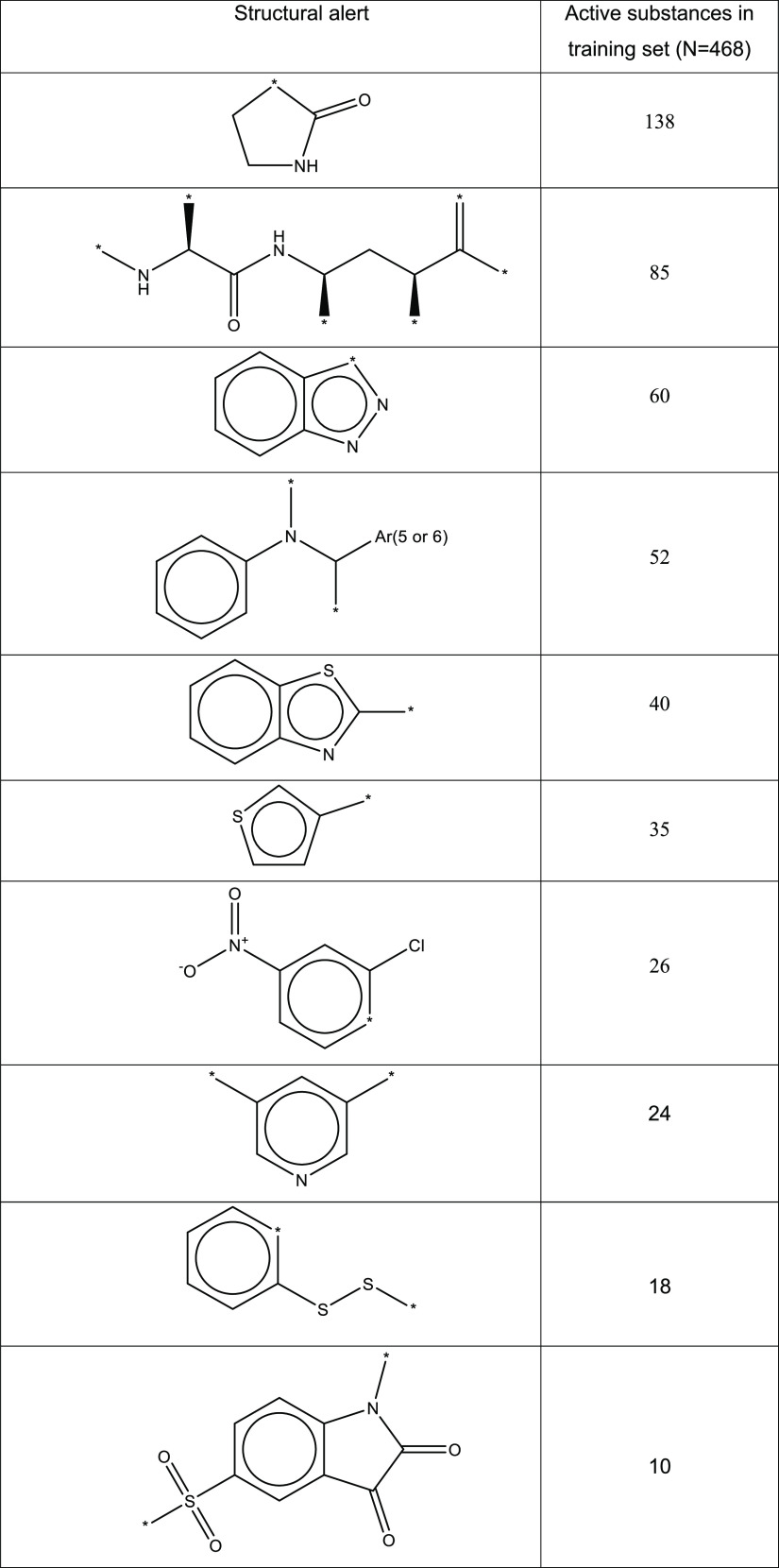
To assess the predictive ability of the 3CLpro model, we then performed a thorough structure–functional analysis of the substances in the training set. The following salient structural features, or “alerts”, were found to be prevalent among active substances and might be partly responsible for the 3CLpro inhibition activity (Table 3).
Table 3. Structural Alerts for the 3CLpro Model.
In order to predict the possibilities of FDA-approved drugs to be repurposed for COVID-19, this 3CLpro model was applied to predict inhibition activity in a data set of 1087 FDA-approved drugs. As approved drugs, these substances would be expected to have an acceptable ADME profile and side effects and might be likely to secure faster FDA approval as coronavirus or COVID-19 therapeutics. The model predicted that 37 of these drugs are likely to be active against 3CLpro of the coronavirus and, by extension, could be used as potential inhibitors of 3CLpro in SARS-CoV-2.
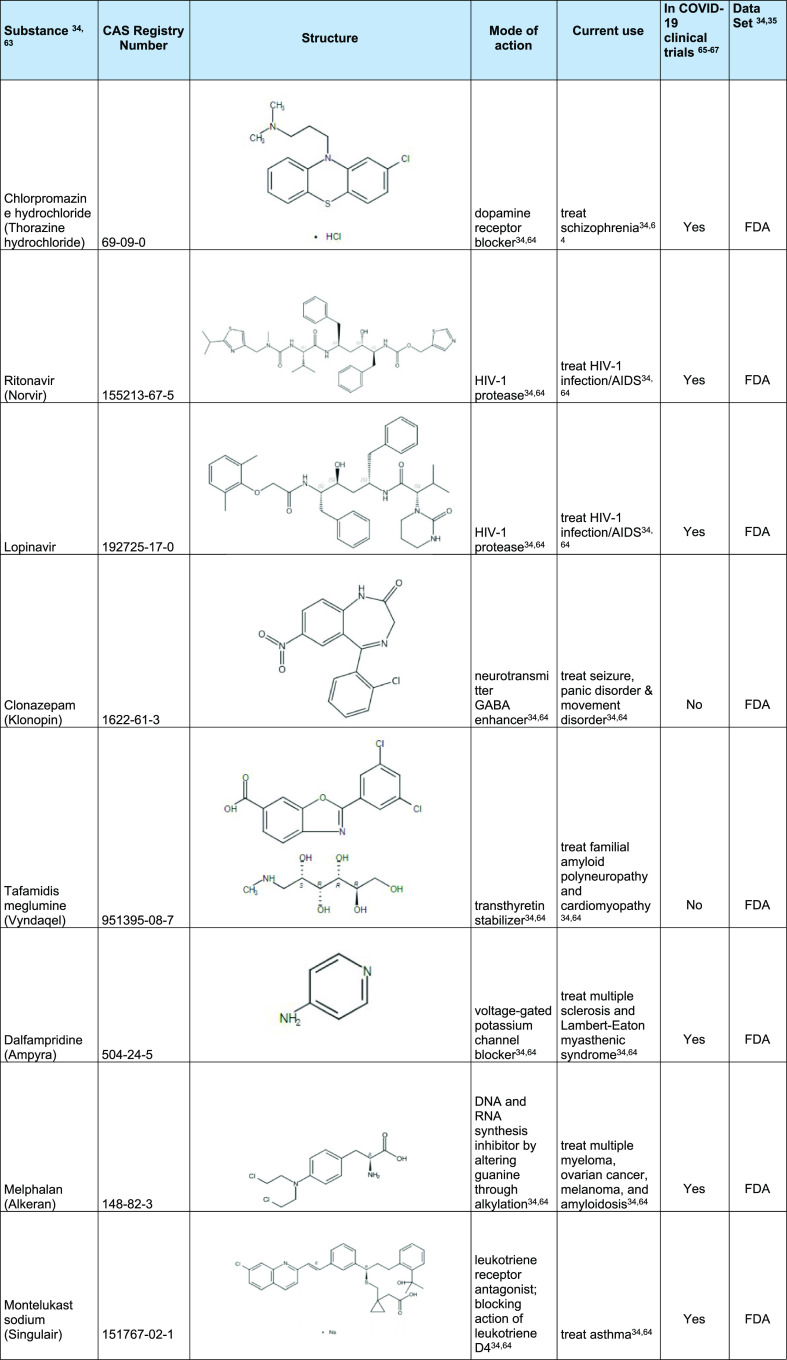
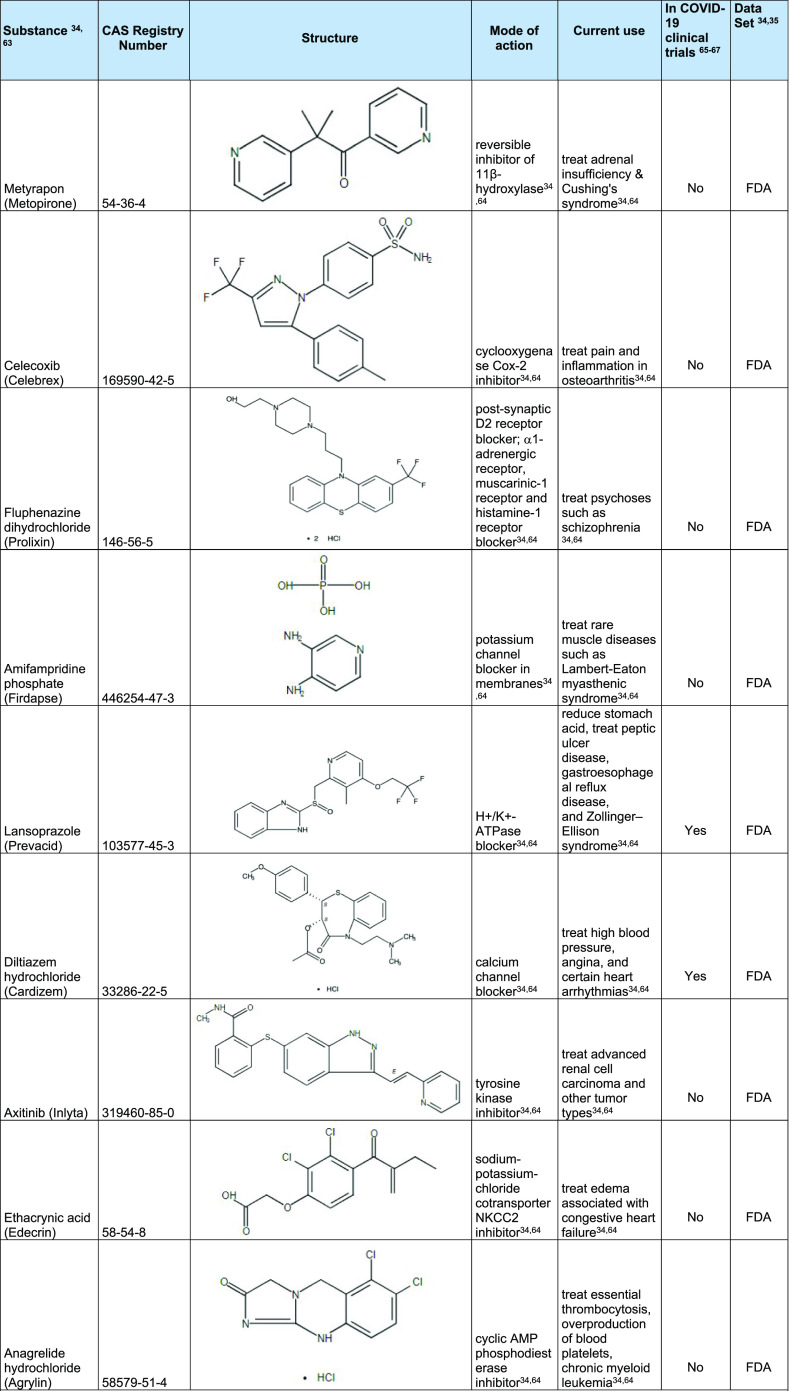
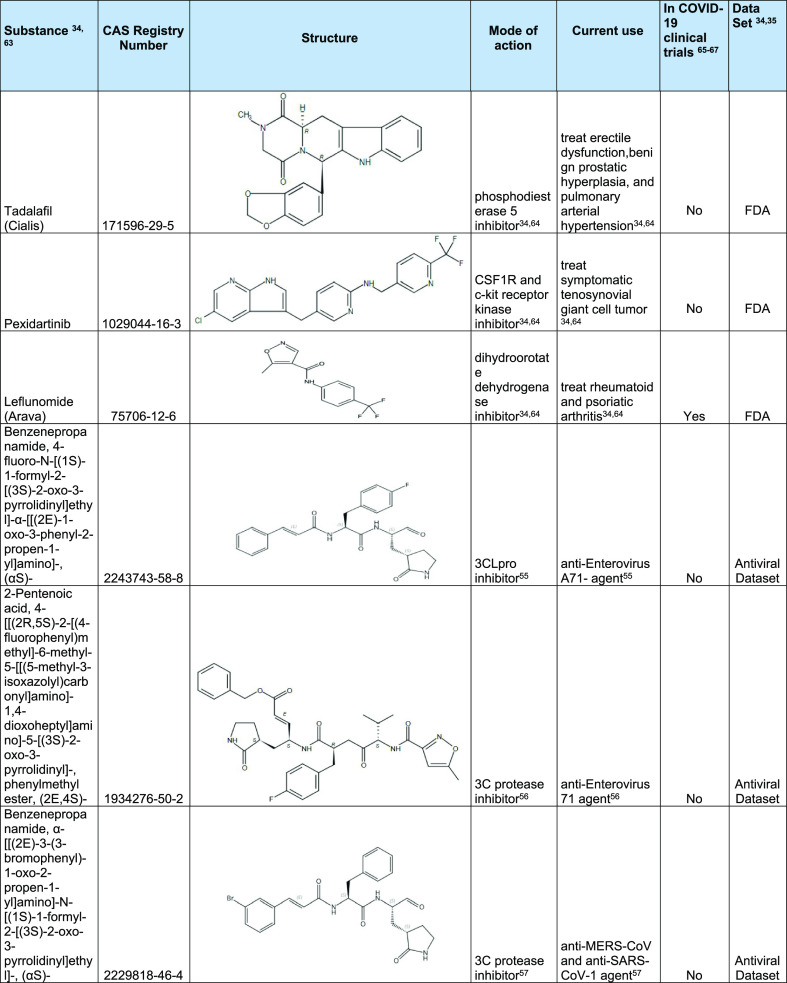
The model was then also applied to the CAS COVID-19 Antiviral Candidate Compounds Dataset, which contains 49,437 compounds with potential antiviral activity identified by CAS scientists. The model predicted that 970 of these chemical compounds are likely to be active against 3CLpro of the coronavirus. From each of these applications, a few selected molecules with the highest inhibition probability are shown in Table 4. A full list of chemical structures predicted to be active is shown in Table S3 of Supporting Information. As expected, the model identified several well-known HIV-1 protease inhibitors (ritonavir and lopinavir)54 and identified substances (RNs 2243743-58-8, 1934276-50-2, and 2229818-46-4) that target 3C protease/3CLpro and was shown to inhibit Enterovirus, MERS-CoV, and SARS-CoV-1 when tested in bioassays.55−57 These could represent new lead candidates as therapeutic agents for COVID-19 or other viral infections. The model further identified substances against host proteins involved in cellular processes, including diltiazem hydrochloride and leflunomide. Leflunomide is a dihydroorotate dehydrogenase inhibitor and involved in nucleotide synthesis.58 In a recent multiomics study, dihydroorotate dehydrogenase was identified as a possible target for SARS-CoV-2 infection.59 In addition, leflunomide has been shown to inhibit SARS-CoV-2 activity.60 Diltiazem hydrochloride is a calcium channel blocker that helps to increase blood flow and variably decrease the heart rate via strong depression of the A-V node. It has also been reported to be an inhibitor of the NF-κB signaling pathway. As hypertension is one of the common comorbidities seen in COVID-19 infected patients, and as diltiazem has dual effects (lowering high blood pressure and inhibiting NF-κB signaling), it may be an attractive substance to use in place of ACE inhibitors.61,62
Table 4. Examples of Substances Predicted to be Active Using 3CLpro Model.
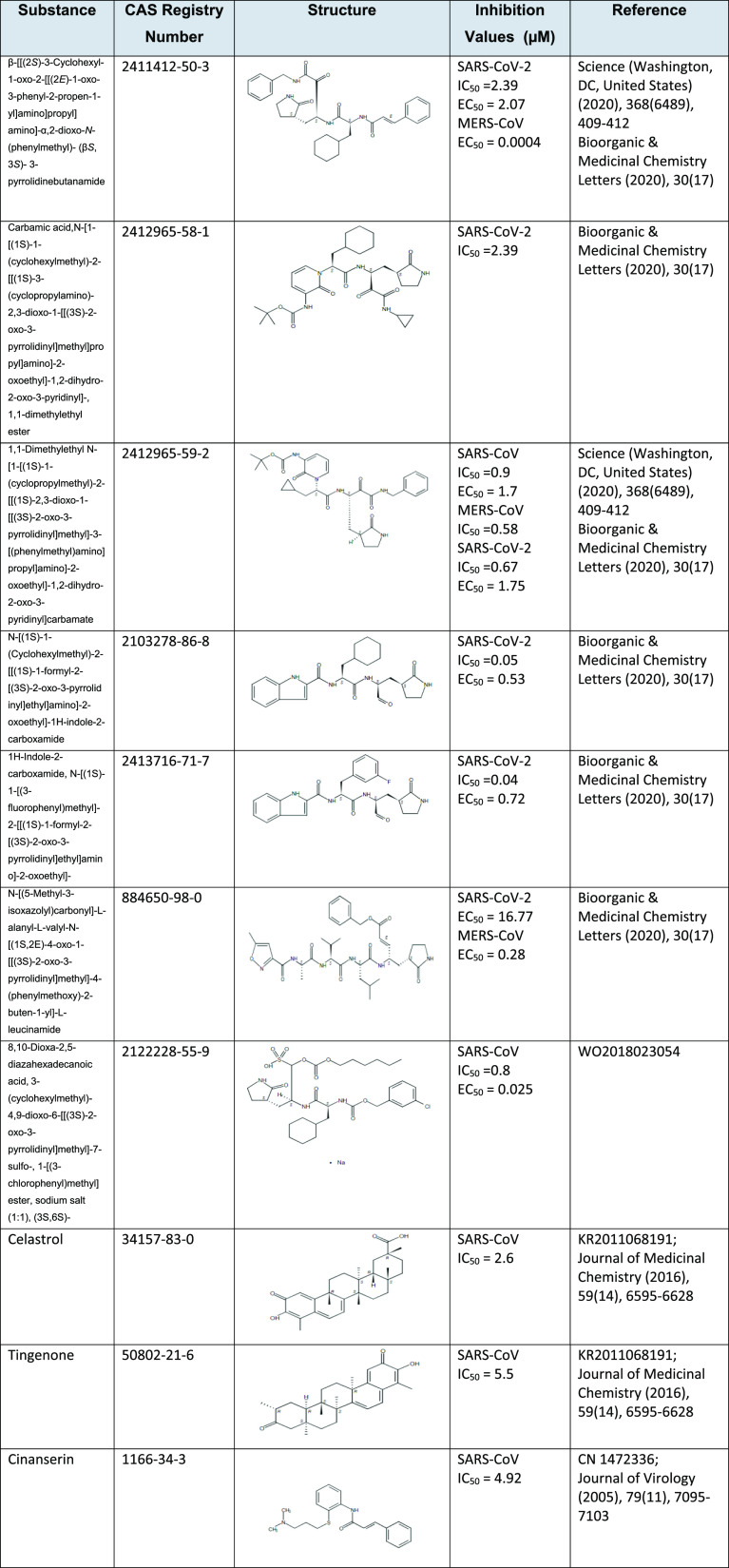
Furthermore, the 3CLpro model has been applied to screen-predicted active molecules from a data set that contains over 113 K substances with CAS-assigned pharmacological activity or a therapeutic role indexed in SARS-, MERS-, and COVID-19-related documents published since 2003. Beyond these substances known to be active from the training set, this process identified about 2500 additional substances predicted to be active against 3CLpro. Many of these compounds with predicted activities were previously shown as inhibitors of coronavirus 3CLpro, serving as a validating mechanism of the model that several substances predicted by the model are currently being pursued for repurposing against COVID-19. Table 5 shows several predicted substances with experimentally validated 3CLpro inhibition activity (IC50) and inhibition of RNA replication (EC50) for SARS-CoV-1, MERS-CoV, and/or SARS-CoV-2.
Table 5. Sample of Predicted Active Substances with Experimentally Validated Inhibitory Effect on Viral 3CLpro.
3.2. RdRp Target—Model and Results
In order to identify potential drug candidates of RdRp, we built machine learning models that can be used to predict the ability of a small molecule to inhibit the polymerase function of RdRp. The abovementioned training set molecules of RdRp were used as training materials for this model. Each molecule was represented as a 42-component vector using MQNs that were used previously for organic molecule classification tasks.38
A “Radial Kernel Support Vector Machine” model (SVM) was determined to be the most accurate and efficient model with a ROC-AUC value of 0.99 (Figure 2). Cumulative gain and lift analysis for this model is shown in Figures S2 and S3 and Tables S8 and S9 in Supporting Information. We applied this model for screening of RdRp inhibitors in the same three data sets screened above for 3CLpro: a data set of FDA-approved drugs, the CAS COVID-19 Antiviral data set, and a larger list of substances indexed by CAS from SARS, MERS. and COVID-19 published literature.
With a probability threshold of 0.50, we found more than 21,000 active candidates with diverse structural features when we applied the SVM model to the three data sets described above. Previously, CAS framework identifiers were used to evaluate the diversity of the CAS REGISTRY.68 We found more than 2000 unique framework IDs within the predicted active candidates, indicating a high structural diversity of the screened active compound data sets. The relative importance of several individual features can be seen in more detail in Figure S1 of Supporting Information.
An important class of the screened active compounds for RdRp that we found were nucleotide analogues with structural similarity to remdesivir. As it was recently demonstrated, cyano-substituted adenosines69 not only effectively compete with natural RdRp substrates but also inhibit RNA synthesis via steric effect. Several interesting nucleotide analogues including cyano-substituted adenosines that were found are listed in Table 6.
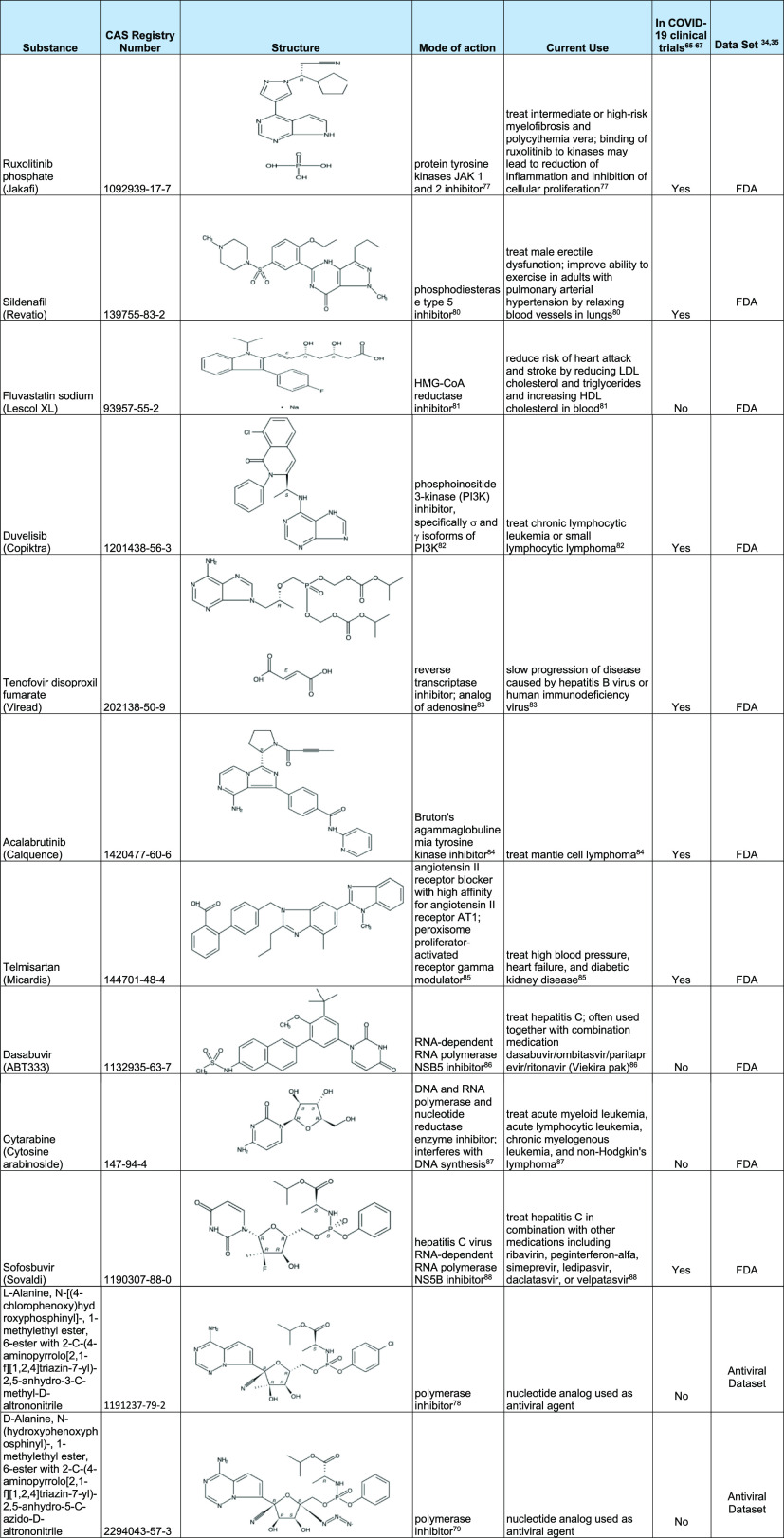
Table 6. Examples of Substances Predicted to be Active Using the RdRp Model64,77−88a.
See Table S6 for additional substances predicted to be active.
We used CAS REGISTRY information such as framework and ring identifiers to highlight the structural similarity between candidates. In addition to cyano-substituted compounds, we found several azido-substituted nucleotide analogues. Azido-substituted nucleotide analogues are known for their ability to generate reactive intermediates that are often used to modify enzyme active sites.70
Beyond providing the molecular structures of these identified substances, Table 6 also provides information on their mode of action, current use, and inclusion in ongoing or upcoming clinical trials. As expected, the model identified several RdRp or polymerase inhibitors (dasabuvir, cytarabine, and sofosbuvir) and also identified several substances that inhibit enzymes or receptors involved in the host immune response (ruxolitinib phosphate, duvelisib, acalabrutinib, and telmisartan) and in cholesterol synthesis (fluvastatin sodium). In addition to the suggestion by the model that these substances could be useful in modulating RdRp, an overview of their current uses indicates they might have an ancillary benefit in treating COVID-19 patients. Of the substances inhibiting the host immune response, ruxolitinib phosphate and telmisartan are being studied in numerous current or upcoming COVID-19 clinical trials, including NCT04362137, NCT04348071, and NCT04360551.71−73
Ruxolitinib phosphate is an inhibitor of protein kinase JAK1 and JAK2 shown to be part of the severe immune overreaction called a cytokine storm that can lead to life-threatening respiratory complications in some patients with COVID-19.74 Telmisartan, an antagonist of angiotensin II receptor type 1 (AT1) and a modulator of peroxisome proliferator-activated receptor gamma, could be used to reduce the negative inflammatory response seen in COVID-19 patients.63 Duvelisib, a phosphoinositide 3-kinase (PI3K) inhibitor, is being studied in NCT04372602 to determine if it can reduce the aberrant hyperactivation of the innate immune system, preferentially polarize macrophages, reduce pulmonary inflammation, and limit viral persistence.75 Fluvastatin sodium, a HMG-CoA reductase inhibitor and cholesterol-lowing drug, is being tested in clinical trial NCT00664742 that includes patients with metabolic syndrome.76 As cardiovascular disease is a risk factor for COVID-19, this clinical study may provide some assistance for COVID-19 patients.
4. Summary and Perspectives
The COVID-19 pandemic caused by the new coronavirus SARS-CoV-2 represents a serious threat to the global health system. In order to aid development of efficient therapeutics against the disease, we use machine learning-based predictive modeling to identify novel drug candidates for the viral targets 3CLpro and RdRp. Chemist-curated training sets of substances were assembled from CAS data collections and integrated with curated bioassay data. Using suitable binary classifiers, we were able to screen a set of FDA-approved drugs, nearly 50,000 substances in the CAS COVID-19 Antiviral Candidate Compounds Dataset, and a list of 113,000 substances with CAS-assigned pharmacological activity or a therapeutic role indexed in SARS-, MERS-, and COVID-19-related documents. Through these screenings, we identified many potential inhibitors with potential activity against 3CLpro or RdRp. Many of these predicted active substances also have ancillary activities such as cardiovascular effects [diltiazem hydrochloride (cardizem)], cholesterol-reduction [fluvastatin sodium (lescol XL)], dihydroorotate dehydrogenase inhibition [leflunomide (arava)] and signal transduction inhibition [ruxolitinib phosphate (jakafi)], which may have other beneficial effects in treating COVID-19. For example, as noted above, heart disease is a known risk factor for COVID-19, so a candidate substance for treating COVID-19, which has known effectiveness treating heart disease, could potentially offer a dual benefit in certain cases. Dihydroorotate dehydrogenase inhibitors have been shown to have anti-SARS-CoV-2 activity, and Janus kinase inhibitors are being studied for inhibiting the cytokine storm process. Additionally, as mentioned above in this report, many of the predicted active substances have FDA approval for specific treatments, and several are included in current or future COVID-19 clinical trials, which could assist or accelerate finding therapeutic agents to use in COVID-19 treatment. In summary, it is hoped that the information provided in this study will be of value in the ongoing search for anticoronaviral therapeutic agents.
While this paper focused on identifying potential therapeutic compounds for use in the current COVID-19 crisis, it is not certain but possible that there will be additional pandemics in the years to come. Many of these will be of viral origin. Because viral transmission and spread can occur very fast, while the typical drug discovery process can take a decade or longer of costly development, it is urgent that preparation for future outbreaks begins now and that the current focus on antiviral agent research continues into the future. The ongoing development of computer-based drug discovery methods such as the machine learning procedures described here and elsewhere, molecular docking and virtual screening of potential therapeutics, and other artificial intelligence methods will be of central importance. Facilitating this is the ongoing increase in computer processing power, continued development of docking and structure prediction algorithms, and protein crystal structure determination. Additionally, the use of high-throughput screening, omics technologies, and the repurposing of already developed drugs will continue and increase in importance as these can shave years off the development time for antiviral drugs. These new research methods will not replace human-based laboratory research but will instead complement it. Finally, because different types of viruses can cause epidemics (e.g., coronavirus, influenza viruses, Ebola viruses, and retroviruses), the development of broad-spectrum antiviral agents or vaccines would be of great value. This would also be useful in that despite the use of faster drug discovery methods, safety and effectiveness testing in humans still takes time. Organ-on-a-chip methods will help but will not eliminate this time-consuming step. Therefore, research on broad-spectrum antivirals will be essential. We hope the current work combining human data curation and machine learning-based predictive models to identify potential small molecule drug candidates for COVID-19 will contribute in some small way to the ongoing efforts in antiviral research.
Acknowledgments
We sincerely appreciate Don Swartwout, Haitao Chen, and Chris Gessner for their technical assistance during preparation of this paper. We are also very grateful to Manuel Guzman, Gilles George, and Mark Grabau for their support. We thank Steven P. Watkins and Cristina Tomeo for their assistance in the preparation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03682.
3CLpro data set, AUCs of evaluated 3CLpro models, chemical structures predicted to be active by the 3CLpro model, RdRp data set, AUCs of evaluated RdRp models, examples of substances predicted active by the RdRp SVM model, RdRp validation tables and figures, RdRp cumulative gain analysis, RdRp cumulative lift analysis, MQN feature importance analysis for the RdRp model, RdRp cumulative gain chart, and RdRp cumulative lift chart (XLSX)
Author Contributions
† J.I. and D.P. contributed equally to this paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. V.; Guo J. Z.; Swaney D. L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X.-M.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; et al. Structure-Based Design, Synthesis and Biological Evaluation of Peptidomimetic Aldehydes as a Novel Series of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease. Science 2020, 368, 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Sun X.; Rox K.; Hilgenfeld R.. X-ray structure of main protease of the novel coronavirus SARS-CoV-2 enables design of α-ketoamide inhibitors, bioRxiv, doi: https://doi.org/ 10.1101/2020.02.17.952879. [DOI] [Google Scholar]
- Anand K.; Ziebuhr J.; Wadhwani P.; Mesters J. R.; Hilgenfeld R. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300, 1763–1767. 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Tahir ul Qamar M.; Alqahtani S. M.; Alamri M. A.; Chen L. L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313. 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A. J. W. Common and unique features of viral RNA-dependent Polymerases. Cell. Mol. Life Sci. 2014, 71, 4403–4420. 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARSCoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Vamathevan J.; Clark D.; Czodrowski P.; Dunham I.; Ferran E.; Lee G.; Li B.; Madabhushi A.; Shah P.; Spitzer M.; Zhao S. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ghosh K.; Amin S. A.; Gayen S.; Jha T. Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J. Mol. Struct. 2021, 1224, 129026. 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R.; Parves M. R.; Paul A. S.; Uddin N.; Rahman M. S.; Mamun A. A.; Hossain M. N.; Ali M. A.; Halim M. A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 1–12. 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.; Roy K. Development of a simple, interpretable and easily transferable QSAR model for quick screening antiviral databases in search of novel 3C-like protease (3CLpro) enzyme inhibitors against SARS-CoV diseases. SAR QSAR Environ. Res. 2020, 31, 511–526. 10.1080/1062936X.2020.1776388. [DOI] [PubMed] [Google Scholar]
- a Rahman M. M.; Saha T.; Islam K. J.; Suman R. H.; Biswas S.; Rahat E. U.; Hossen M. R.; Islam R.; Hossain M. N.; Mamun A. A.; et al. Virtual screening, molecular dynamics and structure–activity relationship studies to identify potent approved drugs for Covid-19 treatment. J. Biomol. Struct. Dyn. 2020, 1–11. 10.1080/07391102.2020.1794974. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Amin S. A.; Ghosh K.; Gayen S.; Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020, 1. 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansch C.; Maloney P. P.; Fujita T.; Muir R. M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. 10.1038/194178b0. [DOI] [Google Scholar]
- Hansch C.; Muir R. M.; Fujita T.; Maloney P. P.; Geiger F.; Streich M. The Correlation of Biological Activity of Plant Growth-Regulators and Chloromycetin Derivatives with Hammett Constants and Partition Coefficients. J. Am. Chem. Soc. 1963, 85, 2817–2824. 10.1021/ja00901a033. [DOI] [Google Scholar]
- Hansch C.; Fujita T. p-σ-π analysis—A method for the correlation of biological activity and chemical structure. J. Am. Chem. Soc. 1964, 86, 1616–1626. 10.1021/ja01062a035. [DOI] [Google Scholar]
- Gutman I.; Trinajstić N. Graph theory and molecular orbitals. Total φ-electron energy of alternant hydrocarbons. Chem. Phys. Lett. 1972, 17, 535–538. 10.1016/0009-2614(72)85099-1. [DOI] [Google Scholar]
- Hosoya H. Topological Index. A Newly Proposed Quantity Characterizing the Topological Nature of Structural Isomers of Saturated Hydrocarbons. Bull. Chem. Soc. Jpn. 1971, 44, 2332–2339. 10.1246/bcsj.44.2332. [DOI] [Google Scholar]
- Randic M. Characterization of molecular branching. J. Am. Chem. Soc. 1975, 97, 6609–6615. 10.1021/ja00856a001. [DOI] [Google Scholar]
- Balaban A. T. Highly discriminating distance-based topological index. Chem. Phys. Lett. 1982, 89, 399–404. 10.1016/0009-2614(82)80009-2. [DOI] [Google Scholar]
- Balaban A. T. Topological Indices Based on Topological Distances in Molecular Graph. Pure Appl. Chem. 1988, 55, 199–206. 10.1351/pac198855020199. [DOI] [Google Scholar]
- Gasteiger J.; Sadowski J.; Schuur J.; Selzer P.; Steinhauer L.; Steinhauer V. Chemical Information in 3D Space. J. Chem. Inf. Comput. Sci. 1996, 36, 1030–1037. 10.1021/ci960343+. [DOI] [Google Scholar]
- Yan A.; Gasteiger J. Prediction of Aqueous Solubility of Organic Compounds Based on a 3D Structure Representation. J. Chem. Inf. Comput. Sci. 2003, 43, 429–434. 10.1021/ci025590u. [DOI] [PubMed] [Google Scholar]
- Mekenyan O.; Peitchev D.; Bonchev D.; Trinajstic N.; Bangov I. P. Modeling the interaction of small organic-molecules with bio-macromolecules. Drug Res. 1986, 36, 176–183. [PubMed] [Google Scholar]
- Ivanov J.; Karabunarliev S.; Mekenyan O. 3DGEN: A system for exhaustive 3D molecular design proceeding from molecular topology. J. Chem. Inf. Comput. Sci. 1994, 34, 234–243. 10.1021/ci00018a001. [DOI] [Google Scholar]
- Debnath A. K.; Lopez de Compadre R. L.; Debnath G.; Shusterman A. J.; Hansch C. Corwin Hansch, Structure-activity relationship of mutagenic aromatic and heteroaromatic nitro compounds. Correlation with molecular orbital energies and hydrophobicity. J. Med. Chem. 1991, 34, 786–797. 10.1021/jm00106a046. [DOI] [PubMed] [Google Scholar]
- Clark T.Quantum Cheminformatics: An Oxymoron? (Part 1). In Chemical Data Analysis in the Large: The Challenge of the Automation Age; Hicks M. G., Ed.; Proceedings of the Beilstein-Institut Workshop May 22nd - 26th; Logos Verlag Berlin: Bozen, Italy, 2000. [Google Scholar]
- Clark T.Quantum Cheminformatics: An Oxymoron? (Part 2). In Rational Approaches to Drug Design; Höltje H.-D., Sippl W., Eds.; Prous Science: Barcelona, 2001. [Google Scholar]
- Hopfinger A. J. A QSAR investigation of dihydrofolate reductase inhibition by Baker triazines based upon molecular shape analysis. J. Am. Chem. Soc. 1980, 102, 7196–7206. 10.1021/ja00544a005. [DOI] [Google Scholar]
- Klopman G. Artificial Intelligence Approach to Structure–Activity Studies. Computer Automated Structure Evaluation of Biological Activity of Organic Molecules. J. Am. Chem. Soc. 1984, 106, 7315–7321. 10.1021/ja00336a004. [DOI] [Google Scholar]
- Klopman G.; Wang S. A Computer Automated Structure Evaluation (CASE) Approach to Calculation of Partition Coefficient. J. Comput. Chem. 1991, 12, 1025–1032. 10.1002/jcc.540120815. [DOI] [Google Scholar]
- Chakravarti S.; Klopman G.; Ivanov J.; Saiakhov R.. MC4PC—An Artificial Intelligence Approach to the Discovery of Quantitative Structure–Toxic Activity Relationships; Taylor & Francis Group, 2005. [Google Scholar]
- Cramer R. D. III; Patterson D. E.; Bunce J. D. Comparative Molecular Field Analysis (CoMFA). 1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- Cramer R. D. III; Bunce J. D.; Patterson D. E.; Frank I. E. Crossvalidation, Bootstrapping, and Partial Least Squares Compared with Multiple Regression in Conventional QSAR Studies. Quant. Struct.-Act. Relat. 1988, 7, 18. 10.1002/qsar.19880070105. [DOI] [Google Scholar]
- Drugs@FDA: FDA-Approved Drugs. https://www.accessdata.fda.gov/scripts/cder/daf/ (accessed May 26, 2020).
- a https://www.cas.org/covid-19-antiviral-compounds-dataset (accessed May 2020).; b Deng Y.-H.; Wang N.-N.; Zou Z.-X.; Zhang L.; Xu K.-P.; Chen A. F.; Cao D.-S.; Tan G.-S. Multi-Target Screening and Experimental Validation of Natural Products from Selaginella Plants against Alzheimer’s Disease. Front. Pharmacol. 2017, 8, 539. 10.3389/fphar.2017.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Zhou Y.; Hou Y.; Shen J.; Huang Y.; Martin W.; Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery 2020, 6, 14. 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- RDKit, Open-Source Cheminformatics. http://www.rdkit.org (accessed June 2020).
- Nguyen K. T.; Blum L. C.; van Deursen R.; Reymond J.-L. Classification of Organic Molecules by Molecular Quantum Numbers. ChemMedChem 2009, 4, 1803–1805. 10.1002/cmdc.200900317. [DOI] [PubMed] [Google Scholar]
- Ogura K.; Sato T.; Yuki H.; Honma T. Support Vector Machine model for hERG inhibitory activities based on the integrated hERG databaseusing descriptor selection by NSGA-II. Sci. Rep. 2019, 9, 12220. 10.1038/s41598-019-47536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. A.; Yangb Q.; BassyouniC A.; Butlerh C. R.; Houh X.; Jenkinson S.; Priceb D. A. Ligand biological activity predicted by cleaning positive and negative chemical correlations. Proc. Natl. Acad. Sci. 2019, 116, 3373. 10.1073/pnas.1810847116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://scikit-learn.org/stable/modules/svm.html#svm-kernels (accessed May 2020).
- Chen T.; Guestrin C.. XGBoost: A Scalable Tree Boosting System. In 22nd SIGKDD Conference on Knowledge Discovery and Data Mining, 2016.
- Chen T.; Tong H.. Higgs Boson Discovery with Boosted Trees. JMLR: Workshop and Conference Proceedings, 2015; Vol. 42, pp 69–80.
- Mustapha I. B.; Saeed F. Bioactive Molecule Prediction Using Extreme Gradient Boosting. Molecules 2016, 21, 983. 10.3390/molecules21080983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M.; Seki K.; Uehara K. Block coordinate descent algorithms for large-scale sparse multiclass classification. Mach. Learn. 2013, 93, 31–52. 10.1007/s10994-013-5367-2. [DOI] [Google Scholar]
- Bell R.; Koren Y. Lessons from the Netflix prize challenge. ACM SIGKDD Explor. Newsl. 2007, 9, 75–79. 10.1145/1345448.1345465. [DOI] [Google Scholar]
- Zhang Q.; Hughes-Oliver J. M.; Ng R. T. “A Model-Based Ensembling Approach for Developing QSARs”. J. Chem. Inf. Model. 2009, 49, 1857–1865. 10.1021/ci900080f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- RDKit SMARTS-based implementation of the 166 public MACCS keys.
- Carhart R. E.; Smith D. H.; Venkataraghavan R. Atom Pairs as Molecular Features in Structure-Activity Studies: Definition and Applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. 10.1021/ci00046a002. [DOI] [Google Scholar]
- Nilakantan R.; Bauman N.; Dixon J. S.; Venkataraghavan R. Topological Torsion: A New Molecular Descriptor for SAR Applications. Comparison with Other Descriptors. J. Chem. Inf. Comput. Sci. 1987, 27, 82–85. 10.1021/ci00054a008. [DOI] [Google Scholar]
- Wildman S. A.; Crippen G. M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. 10.1021/ci990307l. [DOI] [Google Scholar]
- MOE-type descriptors using partial charges and surface area contributions. http://www.chemcomp.com/journal/vsadesc.htm (accessed May 2020).
- Cvetkovic R. S.; Goa K. L. Lopinavir/Ritonavir. Drugs 2003, 63, 769–802. 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- Nie Q.; Xu X.; Zhang Q.; Ma Y.; Yin Z.; Shang L. 3D-quantitative structure-activity relationship study for the design of novel enterovirus A71 3C protease inhibitors. Chem. Biol. Drug Des. 2018, 92, 1750–1762. 10.1111/cbdd.13344. [DOI] [PubMed] [Google Scholar]
- Li P.; Yang B.; Hao F.; Wang P.; He H.; Huang L.; Zhang X.; Zhang S.; Peng X.; Yin K.; et al. Design, synthesis, and biological evaluation of anti-EV71 agents. Bioorg. Med. Chem. Lett. 2016, 26, 3346–3350. 10.1016/j.bmcl.2016.05.036. [DOI] [PubMed] [Google Scholar]
- Kumar V.; Shin J. S.; Shie J.-J.; Ku K. B.; Kim C.; Go Y. Y.; Huang K.-F.; Kim M.; Liang P.-H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 2017, 141, 101–106. 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschner S.; Burst V. Leflunomide: a drug with a potential beyond rheumatology. Immunotherapy 2010, 2, 637–650. 10.2217/imt.10.52. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Zhang Y.; Liu Y.; Baird D.; Liu X.; Wang L.; Zhang H.; Davey Smith G.; Gaunt T.. Multi-omics study revealing tissue-dependent putative mechanisms of SARS-CoV-2 drug targets on viral infections and complex diseases, medRxiv, 2020, DOI: 10.1101/2020.05.07.20093286. [Google Scholar]
- Xiong R.; Zhang L.; Li S.; Sun Y.; Ding M.; Wang Y.; Zhao Y.; Wu Y.; Shang W.; Jiang X.; et al. Novel and potent inhibitors targeting DHODH, a rate-limiting enzyme in de novo pyrimidine biosynthesis, are broad-spectrum antiviral against RNA viruses including newly emerged coronavirus SARS-CoV-2, bioRxiv, 2020, DOI: 10.1101/2020.03.11.983056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor S. E.; Grosset A.; Janiak P. The pharmacological basis and pathophysiological significance of the heart rate-lowering property of diltiazem. Fundam. Clin. Pharmacol. 1999, 13, 145–153. 10.1111/j.1472-8206.1999.tb00333.x. [DOI] [PubMed] [Google Scholar]
- Gilmore T. D.; Herscovitch M. Inhibitors of NF-κB signaling: 785 and counting. Oncogene 2006, 25, 6887–6899. 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- Rothlin R. P.; Vetulli H. M.; Duarte M.; Pelorosso F. G. Telmisartan as tentative angiotensin receptor blocker therapeutic for COVID-19. Drug Dev. Res. 2020, 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH, U.S. National Library of Medicine, druginfo.nlm.nih.gov/drugportal/ (accessed on May 26, 2020).
- ClinicalTrials.gov, clinicaltrials.gov/ct2/home (accessed on July 29, 2020).
- The ISRCTN Registry, www.isrctn.com (accessed on July 29, 2020).
- EU Clinical Trials Register, www.clinicaltrialsregister.eu (accessed on July 29, 2020).
- Lipkus A. H.; Yuan Q.; Lucas K. A.; Funk S. A.; Bartelt W. F. III; Schenck R. J.; Trippe A. J. Structural Diversity of Organic Chemistry. A Scaffold Analysis of the CAS Registry. J. Org. Chem. 2008, 73, 4443–4451. 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- Gordon C. J.; Tchesnokov E. P.; Woolner E.; Perry J. K.; Feng J. Y.; Porter D. P.; Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. 10.1074/jbc.ra120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polshakov D.; Rai S.; Wilson R. M.; Mack E. T.; Vogel M.; Krause J. A.; Burdzinski G.; Platz M. S. Photoaffinity Labeling with 8-Azidoadenosine and Its Derivatives: Chemistry of Closed and Opened Adenosine Diazaquinodimethanes. Biochemistry 2005, 44, 11241–11253. 10.1021/bi050859z. [DOI] [PubMed] [Google Scholar]
- Phase 3 Randomized, Double-blind, Placebo-controlled Multi-center Study to Assess the Efficacy and Safety of Ruxolitinib in Patients With COVID-19 Associated Cytokine Storm (RUXCOVID), Clinical Trials.gov Identifier NCT04362137 clinicaltrials.gov/ct2/show/NCT0436213 (accessed on May 30, 2020).
- Safety and Efficacy of Ruxolitinib for COVID-19, Clinical Trials.gov Identifier NCT04348071 clinicaltrials.gov/ct2/show/NCT0434807 (accessed on May 30, 2020).
- Pilot Clinical Trial of the Safety and Efficacy of Telmisartan for the Mitigation of Pulmonary and Cardiac Complications in COVID-19 patients, Clinical Trials.gov Identifier NCT04360551 clinicaltrials.gov/ct2/show/NCT04360551 (accessed on May 30, 2020).
- Novartis announces plan to initiate clinical study of Jakavi in severe COVID-19 patients and establish international compassionate use program, Novartis, News release. April 2, 2020, novartis.com/news/media-releases/novartis-announces-plan-initiate-clinical-study-jakavi-severe-covid-19-patients-and-establish-international-compassionate-use-program.
- Duvelisib to Combat COVID-19, Clinical Trials.gov Identifier NCT04372602 clinicaltrials.gov/ct2/show/NCT04372602 (accessed on May 30, 2020).
- The Effect of Fluvastatin XL Treatment in Patients with Metabolic Syndrome, Clinical Trials.gov Identifier NCT00664742 clinicaltrials.gov/ct2/show/NCT00664742 (accessed on May 30, 2020).
- U.S. Food & Drug Administration, JAKAFI (ruxolitinib)-FDA, www.accessdata.fda.gov/drugsatfda_docs/label/2017/202192s015lbl.pdf (accessed on May 22, 2020).
- Butler T.; Cho A.; Kim C. U.; Saunders O. L.; Zhang L.. 1’-substituted carba-nucleoside analogs as antiviral agents. WO2009132135A1; World Intellectual Property Organization, 2009.
- Beigelman L.; Deval J.; Prhavc M.. Substituted nucleosides, nucleotides and analogs thereof. WO2019053696A1; World Intellectual Property Organization, 2019.
- Rust K. Y.; Wilkens H.; Kaiser R.; Bregel D.; Wilske J.; Kraemer T. Detection and Validated Quantification of the Phosphodiesterase Type 5 Inhibitors Sildenafil, Vardenafil, Tadalafil, and 2 of Their Metabolites in Human Blood Plasma by LC-MS/MS-Application to Forensic and Therapeutic Drug Monitoring Cases. Ther. Drug Monit. 2012, 34, 729–735. 10.1097/ftd.0b013e31827318b8. [DOI] [PubMed] [Google Scholar]
- Adams S. P.; Sekhon S. S.; Tsang M.; Wright J. M. Fluvastatin for lowering lipids. Cochrane Database Syst. Rev. 2018, 3, CD012282. 10.1002/14651858.CD012282.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn I. W.; Cherry M. A.; Maris M. B.; Matous J. V.; Berdeja J. G.; Patel M. Combination Trial of Duvelisib (IPI-145) With Rituximab or Bendamustine/Rituximab in Patients With non-Hodgkin Lymphoma or Chronic Lymphocytic Leukemia. Am. J. Hematol. 2019, 94, 1325–1334. 10.1002/ajh.25634. [DOI] [PubMed] [Google Scholar]
- Agarwal K.; Brunetto M.; Seto W. K.; Lim Y.-S.; Fung S.; Marcellin P.; Ahn S. H.; Izumi N.; Chuang W. L.; Bae H.; Sharma M.; et al. 96 weeks treatment of tenofovir alafenamide vs. tenofovir disoproxil fumarate for hepatitis B virus infection. J. Hepatol. 2018, 68, 672–681. 10.1016/j.jhep.2017.11.039. [DOI] [PubMed] [Google Scholar]
- Wu J.; Zhang M.; Liu D. Acalabrutinib (ACP-196): A Selective Second-Generation BTK Inhibitor. J. Hematol. Oncol. 2016, 9, 21. 10.1186/s13045-016-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S. C.; Pershadsingh H. A.; Ho C. I.; Chittiboyina A.; Desai P.; Pravenec M.; Qi N.; Wang J.; Avery M. A.; Kurtz T. W. Identification of telmisartan as a unique angiotensin II receptor antagonist with selective PPARgamma-modulating activity. Hypertension 2004, 43, 993–1002. 10.1161/01.hyp.0000123072.34629.57. [DOI] [PubMed] [Google Scholar]
- Ferenci P.; Bernstein D.; Lalezari J.; Cohen D.; Luo Y.; Cooper C.; Tam E.; Marinho R. T.; Tsai N.; Nyberg A.; Box T. D.; et al. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N. Engl. J. Med. 2014, 370, 1983–1992. 10.1056/nejmoa1402338. [DOI] [PubMed] [Google Scholar]
- Drug Information Panel, Summary for Cytarabine. druginfo.nlm.nih.gov/drugportal/name/cytarabine (accessed on May 28, 2020).
- Bourlière M.; Gordon S. C.; Flamm S. L.; Cooper C. L.; Ramji A.; Tong M.; Ravendhran N.; Vierling J. M.; Tran T. T.; Pianko S.; et al. Sofosbuvir, Velpatasvir, and Voxilaprevir for Previously Treated HCV Infection. N. Engl. J. Med. 2017, 376, 2134–2146. 10.1056/nejmoa1613512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.