Abstract
Purpose
The coronavirus disease 2019 (COVID-19) pandemic severely challenges public health and necessitates the need for increasing our understanding of COVID-19 pathogenesis, especially host factors facilitating virus infection and propagation. The aim of this study was to investigate key factors for cellular susceptibility to severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) infection in the ocular surface cells.
Methods
We combined co-expression and SARS-CoV-2 interactome network to predict key genes at COVID-19 in ocular infection based on the premise that genes underlying a disease are often functionally related and functionally related genes are often co-expressed.
Results
The co-expression network was constructed by mapping the well-known angiotensin converting enzyme (ACE2), TMPRSS2, and host susceptibility genes implicated in COVID-19 genomewide association study (GWAS) onto a cornea, retinal pigment epithelium, and lung. We found a significant co-expression module of these genes in the cornea, revealing that cornea is potential extra-respiratory entry portal of SARS-CoV-2. Strikingly, both co-expression and interaction networks show a significant enrichment in mitochondrial function, which are the hub of cellular oxidative homeostasis, inflammation, and innate immune response. We identified a corneal mitochondrial susceptibility module (CMSM) of 14 mitochondrial genes by integrating ACE2 co-expression cluster and SARS-CoV-2 interactome. The gene ECSIT, as a cytosolic adaptor protein involved in inflammatory responses, exhibits the strongest correlation with ACE2 in CMSM, which has shown to be an important risk factor for SARS-CoV-2 infection and prognosis.
Conclusions
Our co-expression and protein interaction network analysis uncover that the mitochondrial function related genes in cornea contribute to the dissection of COVID-19 susceptibility and potential therapeutic interventions.
Keywords: COVID-19, SARS-CoV-2, ACE2, mitochondrial gene, cornea
A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) associated with severe human infected disease coronavirus disease 2019 (COVID-19) outbreak starting from December 2019, in China, and the disease is quickly spreading worldwide.1 Despite being primarily a respiratory virus, COVID-19 can also present with nonrespiratory signs, including ocular symptoms as conjunctival hyperemia, chemosis, epiphora, increased secretions, ocular pain, photophobia, and dry eye.2 The presence of virus in tears, conjunctival swab specimens, and animal models of infectious increasing clinical and scientific evidence that eyes may serve as a potential site of virus replication.3,4 Moreover, immunohistochemical studies and single-cell RNA-sequencing datasets revealed both extra- and intra-ocular localization of SARS-CoV-2 entry factors, ACE receptor, and TMPRSS2 protease in human eyes.5,6 Together, these results suggest that ocular surface cells are susceptible to infection by SARS-CoV-2. Moreover, a recent genomewide association study (GWAS) study identified 3p21.31 as a most significant genetic locus being associated with COVID-19 induced respiratory failure.7 This locus covers a cluster of six genes consisting of SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1, with the identified risk allele being associated with increased SLC6A20 and LZTFL1 expression. Of note, SLC6A20, LZTFL, and FYCO1 are known to associate with eye development, electroretinography abnormal, and anterior eye segment morphology. However, whether these key factors for cellular susceptibility to viral infection have correlation in ocular surface cell remains unclear. Herein, we constructed co-expression and interactome networks and mapped genes implicated by COVID-19 GWAS onto corneal co-expression network to infer the function of susceptibility gene.
Methods
Dataset Summary and Quality Control
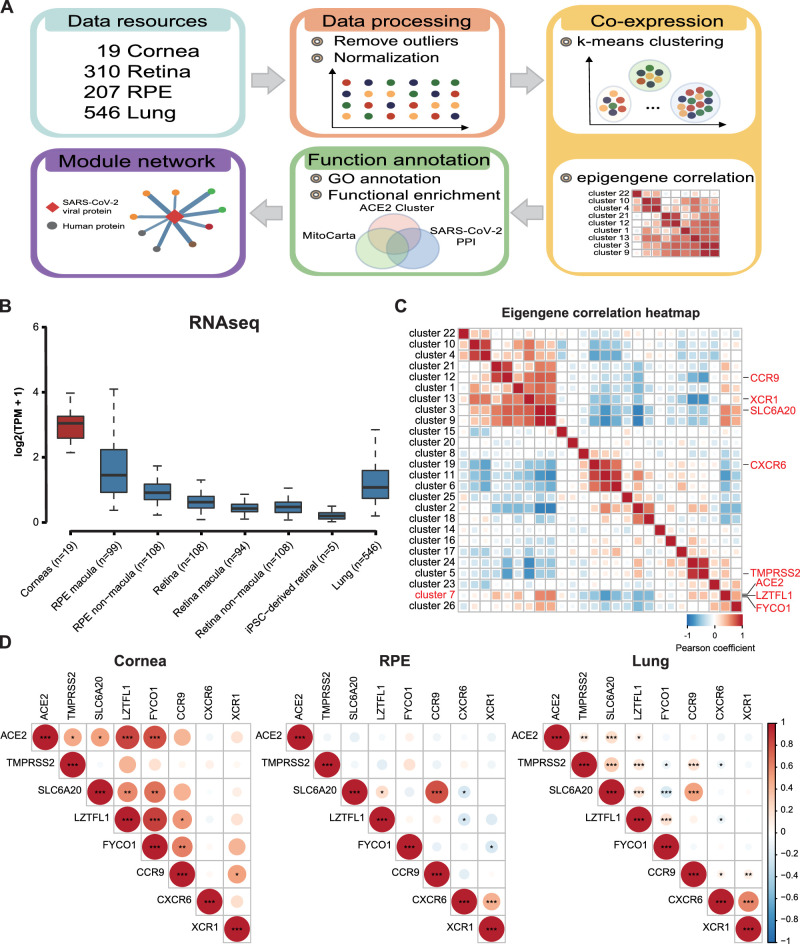
An overview of our strategy to inform corneal mitochondrial susceptibility module of SARS-CoV-2 infection in cornea is shown in Figure 1A. We began by collecting the several RNA-Seq datasets of the normal tissue of the cornea (n = 19 samples),8 retina (n = 310 samples),9,10 retinal pigment epithelium (RPE; n = 207 samples),10 induced pluripotent stem cell (iPSC)-derived retinal (n = 5 samples),11 and lungs (n = 546 samples)12 from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GSE77938, GSE115828, and GSE141531) and GTEx release version 8, respectively. Individual data sets underwent stringent quality control, outliers were removed as samples with standardized sample network connectivity Z scores < -2, as described,13 and were removed. Quantile normalization was then used to transform the statistical distributions across samples to be the same.
Figure 1.
Expression of ACE2 across different eye tissues and its co-expression pattern in the cornea. (A) Overview of the approach and pipeline used to predict susceptibility module responsible for SARS-CoV-2 infection. (B) Expression of ACE2 in the cornea, retina, RPE, and lungs. (C) Pearson's correlation analysis were estimated between ACE2 and TMPRSS2 and six susceptibility genes. The size of the circle scales with the correlation magnitude. The darker the color, the larger the magnitude of correlation coefficient. Star sign (*) indicates statistical significance. (D) Eigengene correlation heatmap representing the strength and significance of correlations between cluster eigengenes. Pearson's correlation coefficient is used as the correlation descriptor (red and blue for positive and negative correlations, respectively).
Co-Expression Analysis
In order to investigate the expression correlation of SARS-CoV-2 entry factors and GWAS susceptibility genes, we calculated the Pearson correlation coefficient to evaluate the linear correlation between any two genes. To specifically characterize the biological pathways involved, we performed k-means analysis,14 a popular unsupervised machine learning algorithm, to identify several co-expression clusters. The cluster eigengene was defined as the first principal component summarizing the expression patterns of all genes into a single expression profile within a given cluster. The cluster membership for each virus entry factors and disease susceptibility genes was determined by the correlation between the expression profile of a gene and the cluster eigengene of a module.
Gene Enrichment Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed by the Metascape gene enrichment analysis tool.15 A Fisher's exact test was used to identify ACE2 cluster with significantly enriched in SARS-Cov-2 interactome16 and mitochondrial genes.17 The interactions between SARS-Cov-2 viral proteins and drug targets were visualized using Cytoscape version 3.6.1.18
Differential Expression Analysis
The transcriptome profiling of human keratoconus corneas was also downloaded from the GEO database (GSE77938).8 Only genes with Transcripts Per Million (TPM) > 1 were preserved in the down-stream analysis. The DESeq219 package was used to normalize expression levels and detect differential expressed genes (q value cutoff is 0.05). Statistical analysis was done using the R project for statistical computing (http://www.r-project.org).
Results
To understand the expression patterns of ACE2, TMPRSS2, and susceptibility genes in the cornea, we first compared the expression level of ACE2 in the cornea, retina, RPE, and lung tissues based on bulk RNA sequencing. As expected from prior literature,20 the cornea showed a higher ACE2 expression than the lungs both in terms of their TPM values (see Fig. 1A). ACE2 exhibits the highest co-expression correlation with TMPRSS2, SLC6A20C, and LZTFL1 in the cornea compared to the lungs and RPE (Fig. 1B). To gain more insight into the biological network of genes associated with SARS-CoV-2 entry factors and susceptibility gene, we performed k-means clustering algorithm to identify genes associated with ACE2 on cornea datasets (Fig. 1C). We identified 26 co-expression modules ranging in size from 111 to 1798 genes. One cluster contained ACE2, LZTFL1, and FYCO1 (cluster 7; 1434 genes). The strongest correlation with the eigengene (the principal component) of this ACE2 cluster was found for hub genes STK16 (r = 0.95, P = 1.07 × 10−9), a member of NAK family that activate the AP-2 scaffolding protein vital to viral entry and propagation.21 TMPRSS2 belonged to a separate cluster 5 (603 genes) with hub gene SLC25A1 (r = 0.91, P = 4.45 × 10−8), which involved in TNF-α and IFN- α triggered inflammation.22 Together these data suggest that the cornea may provide a susceptibility and entry portal for the SARS-CoV-2 entry.
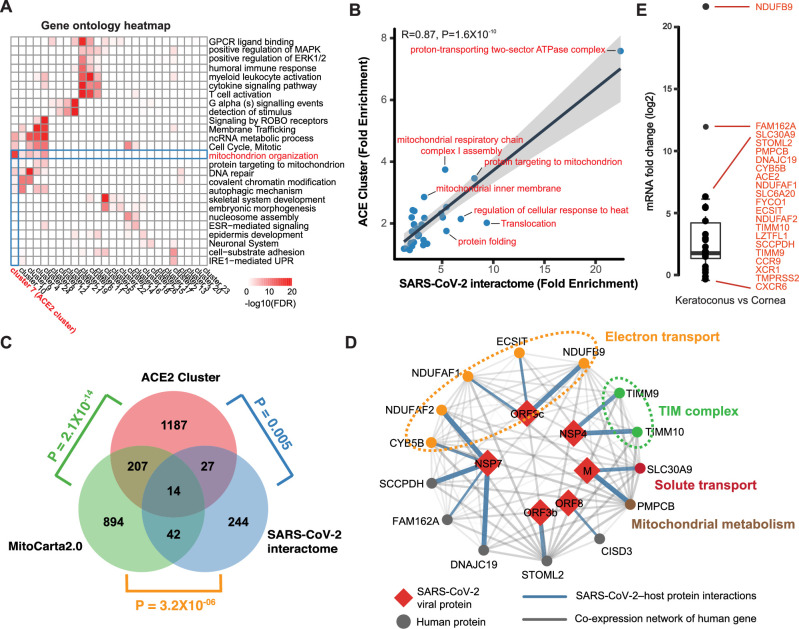
To gain insights into the molecular functionality related to ACE2 cluster, we integrated the ACE2 cluster and SARS-CoV-2 interactome and observed their shared a number of similarities. For instance, the ACE2 cluster and SARS-CoV-2 interactome eigengenes were highly correlated (r = 0.56, P = 0.014). Genes within the ACE2 cluster were mainly related to mitochondrion functions, such as the mitochondrial inner membrane and mitochondrial electron transport (P = 7.47 × 10−29; Fig. 2A). We further evaluated shared GO enrichments to determine whether the similarity in the behavior meant that both clusters contained functionally related genes. The ACE2 cluster and SARS-CoV-2 interactome were nominally enriched (P < 0.001) for 41 and 35 GO terms, respectively. Of these, 17 terms were enriched in both modules. Furthermore, we observed a positive correlation in fold enrichments for shared terms (Fig. 2B). Most of the ontologies shared between the modules described cellular components, biological processes, and molecular functions pertinent to mitochondrion (see Fig. 2B). In both the ACE2 cluster and SARS-CoV-2 interactome, the observed numbers of mitochondrial genes were significantly higher than the number what would be expected by chance (P = 2.1 × 10−14 and P = 3.2 × 10−6, respectively; Fig. 2C). In addition, we found the observed number of ACE2 clusters that was significantly higher (P = 0.005) associated with interaction protein of SARS-CoV-2 compared to randomness (see Fig. 2C). Together, these results indicated that the ACE2 cluster possess the key factors required for cellular susceptibility to SARS-CoV-2 infection in the cornea. Therefore, the core 14 genes were co-occurred in all three datasets as the corneal mitochondrial susceptibility module (CMSM) of SARS-CoV-2 infection in cornea (Fig. 2D). Of the 14 CMSM genes, 5 genes have been shown to directly implicate in the function of electron transport. They include genes, such as NDUFB9, a member of mitochondrial respiratory-chain complex 1, and NDUFAF1, NDUFAF2, and ECSIT, which are involved in mitochondrial respiratory-chain complex 1 assembly. Next, we studied the expression alteration of these genes in the cornea from 23 patients with keratoconus and 19 healthy controls. We found that ACE2 expression was significantly increased in keratoconus compared to control cornea (log2FC = 2.8, P = 4.4 × 10−7; Fig. 2E). Other than the upregulation of ACE2 in keratoconus, there were 10 of 21 genes related to SARS-CoV-2 infection was significantly upregulated in keratoconus patients (see Fig. 2E, t-test P < 0.05). Based on the elevated expression of ACE2 and other susceptibility genes in keratoconus, we speculated that patients with keratoconus are more likely to be infected by the SARS-CoV-2.
Figure 2.
Identification of the cornea functional module and mitochondrial susceptibility genes . (A) Clustered heat map of gene ontology (GO) terms among cluster genes. Color coding according to legend at the bottom, only gene ontology terms with FDR < 0.05 were considered. (B) Gene ontology fold enrichments are correlated for GO terms shared between ACE2 cluster and SARS-CoV-2 interactome. (C) Venn diagram depicting the overlap between ACE2 cluster, SARS-CoV-2, interactome, and mitochondrial genes. P values computed using Fisher's exact test. (D) SARS-CoV-2 protein-protein interaction between 14 mitochondrial susceptibility genes (circles) and 6 SARS-CoV-2 proteins (red diamonds). Blue edge thickness proportional to interaction MiST score; Grey edge thickness proportional to correlation of gene expression. (E) mRNA abundance change of ACE2, TMPRSS2, and susceptibility genes in keratoconus.
Discussion
To control and mitigate the impact of the COVID-19 pandemic, it is vital to gain greater understanding of the routes and modes of transmission, including the role of the ocular surface. Using a cornea-relevant co-expression network to inform virus entry factors and GWAS interpretation, we were able to identify putative susceptibility genes for highly correlated with ACE2. Interestingly, we found that the ACE2 co-expression cluster and SARS-CoV-2 interactome were both enriched for mitochondrial functions. As previously described, the ACE2 not only serves as a critical determinant of CoV-2 transmissibility but also regulates mitochondrial functions.23 ACE2 overexpression regulates mitochondria-localized nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 4, which is known to produce reactive oxygen species (ROS) in the mitochondria.24 Oxidative stress caused by ROS excessiveness was indicated as a major player in COVID-19 pathogenesis and severity.25 Additionally, several lines of evidence have established a link between inflammation and oxidative stress,26–28 for example, TNF-α induces calcium-dependent increase in mitochondrial ROS.29 Once SARS-CoV-2 enters the host cell, its RNAs, such as ORF-9b, also can directly manipulate mitochondrial function to release mitochondrial DNA (mtDNA) in the cytoplasm and activate mtDNA-induced inflammasome and suppress innate and adaptive immunity.30,31 Together, the proinflammatory cytokines, such as TNF-α, IL-1β, IL-6, IL-10, and CXCL-8, affect diverse physiological processes by driving cellular oxidative stress ROS generation. In turn, increased ROS production stimulates proinflammatory mediator release that contributes to mitochondrial dysfunction. As we know, the corneal cell, especially corneal endothelial cells, is a mitochondria-rich cell. Given the highly exposed position, the cornea receives a significant amount of high-tension atmospheric oxygen and the ultraviolet range, which result in the generation of ROS and subsequent oxidative stress. Moreover, ROS are a by-product of oxidative phosphorylation in mitochondria, which can subsequently result in further mitochondrial damage and a further increase in ROS. Overall, a vicious oxidation / inflammatory cycle is more likely to have a potential impact of SARS-CoV-2 invasion and immune responses in corneal cells.
In our study, we predicted that the mitochondrial related CMSM gene set was vital susceptibility genes of COVID-19, including five genes involved in respiratory electron transport which are being targeted by metformin.32 A favorable effect of metformin in patients with COVID-19 has been hypothesized as the drug might prevent virus entry into target cells via adenosine monophosphate-activated protein kinase activation and the phosphatidylinositol-3-kinase-protein kinase B-mammalian target of rapamycin signaling pathway.33 Because metformin is found to have the properties of anti-inflammation and anti-oxidation,34 it has also been used in the treatment of eye diseases, including age-related macular degeneration, glaucoma, and diabetic retinopathy.35–37 These may give insight into metformin that may lower the COVID-19 risk in eye infection. Notably, ECSIT, one of the CMSM genes, is a cytosolic adaptor protein involved in inflammatory responses and plays a regulatory role as part of the TAK1-ECSIT-TRAF6 complex that is involved in the activation of NF-κB by the TLR4 signal. In the previous study, treatment with drugs that inhibited NF-κB activation led to a reduction in inflammation and significantly increased mouse survival after SARS-CoV infection.38 Additionally, ECSIT is also essential for the association of RIG-I-like receptors (RIG-I or MDA5) to VISA in innate antiviral responses.39 Therefore, ECSIT may be used as a new drug target to protect against the development of severe forms of COVID-19 infection. Based on our results, we believe that significant insight into COVID-19 in the cornea can be gained using co-expression and interaction networks.
Acknowledgments
Supported by the Key Program of National Natural Science Foundation of China (81830027) to J. Qu; the National Natural Science Foundation of China (61871294), Zhejiang Provincial Natural Science Foundation of China (LR19C060001) to J. Su.
Disclosure: J. Yuan, None; D. Fan, None; Z. Xue, None; J. Qu, None; J. Su, None
References
- 1. Zhu N, Zhang D, Wang W, et al.. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020; 382(8): 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu P, Duan F, Luo C, et al.. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020; 138: 575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang R, Liu M, Chen Y, et al.. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020; 182: 50–58.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang X, Chen X, Chen L, et al.. The evidence of SARS-CoV-2 infection on ocular surface. Ocul Surf. 2020; 18: 360–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collin J, Queen R, Zerti D, et al.. Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul Surf, 10.1016/j.jtos.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou L, Xu Z, Castiglione G, Soiberman U, Eberhart C, Duh E. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf. 2020; 18(4): 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellinghaus D, Degenhardt F, Bujanda L, et al.. Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med, 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kabza M, Karolak J, Rydzanicz M, et al.. Collagen synthesis disruption and downregulation of core elements of TGF-β, Hippo, and Wnt pathways in keratoconus corneas. Eur J Hum Genet. 2017; 25: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Orozco LD, Chen H-H, Cox C, et al.. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 2020; 30: 1246–1259.e1246. [DOI] [PubMed] [Google Scholar]
- 10. Ratnapriya R, Sosina O, Starostik M, et al.. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genetics. 2019; 51: 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao M, Lei X, Han F, et al.. Patient-specific retinal organoids recapitulate disease features of late-onset retinitis pigmentosa. Front Cell Dev Biol. 2020; 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oldham M, Langfelder P, Horvath S. Network methods for describing sample relationships in genomic datasets: application to Huntington's disease. BMC Syst Biol. 2012; 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit Lett. 2010; 31: 651–666. [Google Scholar]
- 15. Zhou Y, Zhou B, Pache L, et al.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019; 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon DE, Jang GM, Bouhaddou M, et al.. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020; 583(7816): 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calvo S, Clauser K, Mootha V. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016; 44: D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011; 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sungnak W, Huang N, Bécavin C, et al.. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020; 26: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stebbing J, Krishnan V, de Bono S, et al.. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Molec Med. 2020; 12: e12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Infantino V, Iacobazzi V, Menga A, Avantaggiati M, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFα- and IFNγ-triggered inflammation. Biochim Biophys Acta. 2014; 1839: 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi T, Yang F, Liu C, et al.. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic β-cells. Biochem Biophys Res Commun. 2018; 495: 860–866. [DOI] [PubMed] [Google Scholar]
- 24. Kim S, Kim Y, Jeong K, et al.. Angiotensin II-induced mitochondrial Nox4 is a major endogenous source of oxidative stress in kidney tubular cells. PloS One. 2012; 7: e39739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delgado-Roche L, Mesta F.. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch Med Res. 2020; 51: 384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Naik E, Dixit V. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J Exp Med. 2011; 208: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khomich O, Kochetkov S, Bartosch B, Ivanov A. Redox biology of respiratory viral infections. Viruses. 2018; 10(8): 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao H, Lan D, Duan Z, et al.. Upregulation of mitochondrial gene expression in PBMC from convalescent SARS patients. J Clin Immunol. 2006; 26: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dada LA, Sznajder JI. Mitochondrial Ca 2+ and ROS take center stage to orchestrate TNF-α–mediated inflammatory responses. J Clin Invest. 2011; 121: 1683–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi C, Qi H, Boularan C, et al.. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol (Baltimore, MD: 1950). 2014; 193: 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh K, Chaubey G, Chen J, Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am J Physiol Cell Physiol. 2020; 319: C258–C267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soberanes S, Misharin A, Jairaman A, et al.. Metformin targets mitochondrial electron transport to reduce air-pollution-induced thrombosis. Cell Metab. 2019; 29: 335–347.e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma S, Ray A, Sadasivam B. Metformin in COVID-19: a possible role beyond diabetes. Diabetes Res Clin Pract. 2020; 164: 108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cameron A, Morrison V, Levin D, et al.. Anti-inflammatory effects of metformin irrespective of diabetes status. Circ Res. 2016; 119: 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown EE, Ball JD, Chen Z, Khurshid GS, Prosperi M, Ash JD. The common antidiabetic drug metformin reduces odds of developing age-related macular degeneration. Invest Ophthalmol Vis Sci. 2019; 60: 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richards JE, Lin H-C, Nan B, et al.. Targeting aging: geroprotective drug metformin reduces risk of adult-onset open-angle glaucoma. Invest Ophthalmol Vis Sci. 2014; 55: 1668–1668. [Google Scholar]
- 37. Munie M, Noorulla S, Rana S, et al.. Effect of metformin on the development and severity of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014; 55: 1069–1069. [Google Scholar]
- 38. DeDiego M, Nieto-Torres J, Regla-Nava J, et al.. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014; 88: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lei C, Zhang Y, Li M, et al.. ECSIT bridges RIG-I-like receptors to VISA in signaling events of innate antiviral responses. J Innate Immun. 2015; 7: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]